Abstract
The alpha C protein is a protective surface-associated antigen of group B streptococci (GBS). The prototype alpha C protein of GBS (strain A909) contains nine identical tandem repeats, each comprising 82 amino acids, flanked by N- and C-terminal domains. Clinical isolates of GBS show variable numbers of repeats with a normal distribution and a median of 9 to 10 repeats. Here, we show that escape mutants of GBS expressing one-repeat alpha C protein were 100-fold more pathogenic than GBS expressing wild-type nine-repeat alpha C protein in neonatal mice whose dams were immunized with antiserum elicited to nine-repeat alpha C protein (50% lethal doses of 1.6 × 103 and 1.8 × 105, respectively; P = 0.0073). There was no difference in pathogenicity in nonimmune mice. Enzyme-linked immunosorbent assay inhibition showed that nine-repeat but not one-repeat alpha C protein is readily available for antibody binding on the surface of intact GBS. Immune electron microscopy studies with antibodies to the capsular polysaccharide (CPS) and to the alpha C protein demonstrated localization of the nine-repeat alpha C protein and the CPS at similar distances from the cell wall. The one-repeat alpha C protein was visualized poorly and only in close proximity to the cell wall, thus suggesting that antibody binding to the protein was hindered by CPS or other cell surface components. We concluded that deletion in the repeat region of the alpha C protein enhanced the pathogenicity of GBS in immune mice by (i) loss of a protective (conformational) epitope(s) and (ii) loss of antibody binding to the alpha C protein due to a decrease in antigen size relative to cell wall components and/or CPS.
Group B streptococci (GBS) are the leading cause of meningitis, pneumonia, and sepsis in neonates (1). GBS also account for substantial morbidity in peripartum women and immunocompromised adults. The alpha C protein is a protective cell surface antigen present in many clinical GBS isolates (11), and it is the prototype for a family of repeat-containing proteins present in most GBS strains (13, 30). The nucleotide sequence of the bca gene encoding the alpha C protein contains nine identical tandem repeats, each composed of 246 nucleotides, flanked by N- and C-terminal regions (22). Clinical isolates of GBS show variable sizes of the alpha C protein (62.5 to 167 kDa) (16), and the size of the expressed protein was found to correspond to the number of tandem repeats within the gene (21). Recently, the occurrence of deletions of repeats in the alpha C protein during transmission of GBS from human mother to neonate was described (17), and an animal model demonstrated that deletions in the repeat region of bca enabled GBS to escape alpha C protein-specific host antibodies. GBS with fewer repeats in the alpha C protein escape antibody-mediated immunity because of a loss of protective epitopes (including conformationally determined epitopes) that results in diminished antibody binding (8, 17). While these findings implied a selective advantage for GBS expressing alpha C protein with few repeats, no conditions that select for increased repeat number have been found. It is possible that a larger repeat content is advantageous in colonization or in some undetermined niche. We have shown that higher repeat numbers impart lower immunogenicity to the alpha C protein, particularly to the N-terminal domain (9), which may in turn impart a selective advantage to the organisms displaying alpha C proteins with more repeats.
Several cell wall-associated proteins containing repeated amino acid sequences have been shown both to vary in repeat number and to play a role in virulence. Best described are the antiphagocytic M proteins of group A streptococci, which bind fibrinogen and inhibit complement activation (10, 12). PspA of Streptococcus pneumoniae, another repeat-containing surface protein, has also been shown to play a role in virulence (20, 29). Similarly, GBS lacking alpha C protein expression demonstrated attenuation in nonimmune mice (15). However, the relationship between antigenic variation (i.e., variability in repeat number) and pathogenicity of these bacteria has never been explored, particularly in immune hosts.
In this study, we directly explored the effect of antigenic variation on pathogenicity of GBS in immune and nonimmune mice. With the term pathogenicity, we refer here to the capacity of GBS to cause disease and/or death (31). We studied the lethality of GBS expressing one- or nine-repeat alpha C protein in neonatal mice immunized with rabbit antiserum elicited to one- or nine-repeat alpha C protein or with preimmune serum. Escape mutants were isolated from the spleens of neonatal mice and analyzed for their repeat contents by Western blotting. Finally, we examined the availability of epitopes for antibody binding in one- or nine-repeat alpha C protein at the cell surface of intact GBS by (i) enzyme-linked immunosorbent assay (ELISA) inhibition and (ii) electron microscopy (EM) of immunolabelled bacteria.
MATERIALS AND METHODS
Bacterial strains.
Wild-type GBS strain A909 (Ia/C alpha+, beta+), expressing 9 repeats, and mouse-passaged mutants of GBS strain A909 with 1 repeat (A909-1), 2 repeats (A909-2), and 18 repeats (A909-18) were used in ELISA inhibition and the neonatal mouse lethality study. Mutant A909-1 was obtained from spleens after immunization of mice with antiserum elicited to nine-repeat alpha C protein and challenge with wild-type GBS, as described previously (17), and mutant A909-2 was obtained in this study under similar conditions. Escape mutant A909-18 was isolated once after immunization of mice with antiserum elicited to two-repeat alpha C protein and challenged with wild-type GBS (unpublished results).
Purification of alpha C proteins and antibodies.
Recombinant 1-, 2-, 9-, and 16-repeat alpha C proteins were expressed and purified as described previously (8). Rabbit antisera were elicited to purified alpha C proteins with 1, 2, 9, or 16 repeats (8). 4G8, a monoclonal antibody directed to the repeat region of the alpha C protein, was used in Western blots (19).
SDS-PAGE and Western blotting.
Sodium dodecyl sulfate-polyacrylamide (10%) gel electrophoresis (SDS-PAGE) was performed by the method of Laemmli (14). Electrophoresis was performed with a Mighty Small apparatus (Hoeffer Scientific Instruments, San Francisco, Calif.) by standard methods. Proteins were transferred to nitrocellulose filters by electroblotting (Hoeffer Scientific model TE70), using 1.8 mA/cm2 for 1 h. After blocking with 5% skim milk in phosphate-buffered saline (PBS) containing 0.5% Tween 20, the blots were allowed to react with antiserum at a dilution of 1:500 in the same buffer. After reaction with goat anti-rabbit immunoglobulin G–alkaline phosphatase conjugate diluted 1:1,000, bands were visualized by the method of Blake et al. (2), utilizing the indoxyl phosphatase-Nitro Blue Tetrazolium reagent.
ELISA inhibition with intact bacteria.
Mouse-passed mutants of GBS strain A909, expressing 1, 2, 9, or 18 repeats on the cell surface, were grown in Todd-Hewitt broth (THB) until the A650 was 0.3. Cells were washed for 2 min in an Eppendorf centrifuge at 13,000 × g with 40 mM phosphate buffer twice, concentrated fivefold (final concentration, 5 × 108 CFU/ml), and used to inhibit antibody binding to plates coated with purified one- or nine-repeat alpha C protein, as described previously (8). The percent inhibition by ELISA was calculated as follows:{[A405(uninhibited control) − A405(sample)]/A405(uninhibited control)} × 100%
Quantitation of cell-associated CPS and alpha C protein.
The amount of capsular polysaccharide (CPS) in mutanolysin cell wall extracts (3, 4) was determined by ELISA inhibition (23), with minor modifications (18). In this assay, type Ia CPS–poly-l-lysine (1 μg/ml) was applied to the microtiter plate, and rabbit antiserum was raised to the type Ia CPS coupled to tetanus toxoid (1:100,000 final dilution); purified Ia CPS was used as the standard. The concentration of purified type Ia CPS in mutanolysin extracts required to inhibit 50% of antiserum binding (50% inhibitory concentration) was determined from the linear portion of the standard curve. Duplicate samples were processed for each growth rate, and results are reported as specific type Ia CPS (micrograms of CPS/milligrams of cells [dry weight]).
Quantitation of cell surface-associated alpha C protein was similarly determined by ELISA inhibition of mutanolysin extracts of GBS with methods adapted from earlier work (19, 28). GBS strains A909-1 and A909-9 were grown in THB. Overnight cultures were diluted until the A650 was 0.3, and a 5-ml suspension was washed with PBS in an Eppendorf centrifuge at 13,000 × g for 2 min. The pellets were resuspended in 300 μl of 40% (wt/vol) sucrose–0.03 M potassium phosphate (pH 7.0)–0.01 M MgCl2–1,500 Units of mutanolysin with protease inhibitors (benzamidine hydrochloride [0.2 M], iodoacetic acid [0.5 M and phenylmethylsulfonyl fluoride [0.2 M]; Gibco BRL, Gaithersburg, Md.). The solution was incubated at 37°C for 1 h and centrifuged at 13,000 × g for 20 min.
The supernatants were adjusted to 1 ml with PBS, and the concentration of alpha C protein was determined by ELISA inhibition (8) as follows. Rabbit antiserum elicited to one-repeat alpha (final dilution, 1:8,000) was used to measure the inhibition of antibody binding to one-repeat alpha (1 μg/ml) on microtiter plates by one- or nine-repeat alpha C proteins in the mutanolysin surface extracts (inhibiting antigens). Purified one- or nine-repeat alpha C proteins were used as inhibiting antigens to generate standard curves (with a 10-μg/ml starting concentration and 10 twofold serial dilutions). The concentration of alpha C protein in the mutanolysin-extracted supernatant was determined from the linear portion of the standard curve. The number of molecules of surface-associated one-repeat or nine-repeat alpha C protein per CFU was calculated with the following formula:
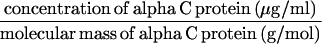 |
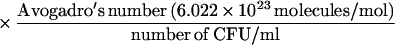 |
Determination of LD50 in the presence of immune antiserum.
A neonatal mouse model of GBS infection was adapted from that of Rodewald et al. (25). Pregnant dams were injected intraperitoneally (1 to 2 days before delivery) with 0.5 ml of rabbit antiserum elicited to one-repeat or nine-repeat alpha C protein or with preimmune rabbit serum. Pups were challenged intraperitoneally with 0.05 ml of serial 10-fold dilutions of GBS strain A909-1 or A909-9 (5 × 107 to 5 × 102 GBS/ml) within 48 h of birth. Survival was assessed 48 h after challenge. The 50% lethal dose (LD50) for each strain combined with each antiserum was calculated by logistic regression analysis (26). To this aim the following logistic regression model was fit: loge p/(1 − p) = α + β2x2 + β3x1x2, where x1 = 1 for strain A909-9 and 0 for strain A909-1, x2 = log10 GBS per milliliter, and p = probability of survival after 48 h. The LD50 for strain A909-1 was established by −α/β2; the LD50 for strain A909-9 was established by −α/(β2 + β3). The significance of the coefficient β3 was used to assess whether there were significant differences in the LD50s between strain A909-1 and strain A909-9.
Isolation of escape mutants of GBS.
Pregnant mice were immunized by intraperitoneal injection of 0.5 ml of rabbit antiserum elicited to one- or nine-repeat alpha C protein 1 to 2 days before delivery. Pups were challenged intraperitoneally with 0.05 ml containing 3 × 104 CFU of GBS strain A909 within 48 h of birth; 48 h after challenge, the pups were killed and each spleen was macerated between frosted glass slides and resuspended in 1 ml of THB. One hundred microliters of the spleen suspension was inoculated on a blood agar plate and cultured overnight at 37°C. Colonies were counted, and individual colonies were analyzed by Western blotting for their molecular mass. The repeat number was calculated from the molecular mass by the following formula and rounded to the nearest whole number: number of repeats = [M − (MN-terminal + MC-terminal)]/Msingle repeat, where M is the molecular mass of the largest band seen on the Western blot, MN-terminal is the predicted molecular mass of the N-terminal domain (20.4 kDa), MC-terminal is the predicted molecular mass of the C-terminal domain including the partial repeat (5.7 kDa), and Msingle repeat is the predicted molecular mass of a single repeating subunit (8.7 kDa).
EM.
Immunogold labelling was applied to intact GBS without fixation by the following method. GBS were grown in THB to mid-log phase (A650 = 0.3). Cells were washed three times in PBS for 1 min at 13,000 × g and resuspended in 5 ml of PBS (final concentration of GBS = 3 × 108 cells/ml). EM grids were placed upside down in a drop of the resuspended GBS solution for 1 min to bind the GBS to the grids, and the grids with the cells were placed in blocking buffer (0.5% fish skin gelatin in 0.1× PBS–0.1% Tween 20) for 10 min, followed by incubation with the primary antibody for 30 min at room temperature (final dilution, 1:2,000). The cells on the grids were washed 10 times by immersion in drops of 0.1× PBS, and the grids were placed in drops with protein A-labelled gold (dilution of 1:50) for 30 min. For alpha C protein antisera, gold particles 15 nm in diameter were used, and for antiserum to Ia CPS, 20-nm gold particles were used. Finally, the cells on the grids were washed 10 times by immersion in drops of distilled water. Air-dried grids were subjected to transmission EM (JEOL model 1200Ex), and representative fields were photographed.
Isolation of chromosomal DNA, PCR, and DNA sequencing.
Chromosomal DNA was prepared from the escape mutant of GBS with one-repeat alpha C protein as described previously (17). The bca gene was amplified from chromosomal DNA of the escape mutant with the same primers and PCR conditions used for amplification of wild-type (nine-repeat) bca (17). The PCR product of the one-repeat alpha C protein gene was sequenced with an automatic sequencer (model 373; Applied Biosystems, Foster City, Calif.) at the Beth Israel Deaconess Medical Center sequencing facility. The DNA sequence of one-repeat alpha C protein was aligned with one repeat in the DNA sequence of nine-repeat alpha C protein (22) with the software program from the Genetics Computer Group (Madison, Wis.) package.
RESULTS
LD50 study.
Since escape mutants expressing one-repeat alpha C protein occur in mice immunized with alpha C protein, we hypothesized that these mutants would be more pathogenic in such mice. We compared the difference in lethality of GBS with one- or nine-repeat alpha C protein in immunized and nonimmune mice. LD50s were determined by logistic regression analysis. In mice immunized with nine-repeat-elicited antibodies, one-repeat GBS was, in fact, approximately 100 times more pathogenic than nine-repeat GBS (Table 1). There was a trend toward greater pathogenicity of the one-repeat relative to the nine-repeat GBS when mice were immunized with one-repeat-elicited antiserum, but this was not statistically significant. No difference was observed when the mice were immunized with preimmune antiserum.
TABLE 1.
Pathogenicity of GBS with one- or nine-repeat alpha C protein in immune and nonimmune micea
| Challenge strain | LD50 after immunization with:
|
||
|---|---|---|---|
| One-repeat antiserum | Nine-repeat antiserum | Preimmune antiserum | |
| One-repeat A909 | 5.5 × 104 | 1.6 × 103 | 1.0 × 103 |
| Nine-repeat A909 | 2.4 × 105 | 1.8 × 105 | 2.4 × 103 |
| P value | 0.3842 | 0.0073b | 0.5813 |
LD50s were calculated by using the logistic regression procedure. Mice were immunized with rabbit antiserum elicited to one- or nine-repeat alpha C protein. Pups were challenged with 0.05 ml intraperitoneally within 48 h of birth. The range of the challenge dose was 5 × 102 to 5 × 107/mouse. Survival was determined 48 h after challenge.
Significant difference between LD50s of GBS with one- and nine-repeat alpha C proteins.
ELISA inhibition with purified alpha C protein and intact GBS.
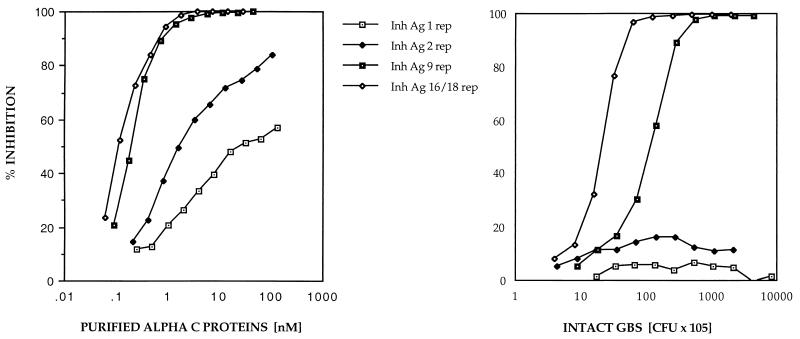
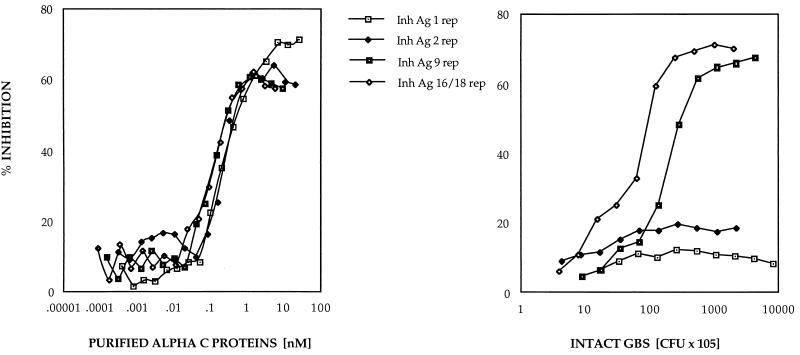
Previous studies have shown that purified alpha C protein constructs containing few repeats poorly inhibited antiserum raised to the wild-type, nine-repeat protein. In order to study this phenomenon in intact GBS, ELISA inhibition was performed to determine the availability of epitopes in the alpha C protein on the cell surface of intact GBS. ELISA inhibition with nine-repeat-elicited antiserum and with purified alpha C protein or intact GBS as the inhibiting antigen gave similar patterns of inhibition; i.e., nine-repeat-elicited antibodies recognized higher-repeat alpha C proteins both as purified antigens and on the cell surface of GBS better than alpha C proteins containing small numbers of repeats (Fig. 1). However, when one-repeat antiserum was used, there was a discrepancy in inhibition between purified alpha C proteins and the protein at the cell surface of intact GBS (Fig. 2): one- and two-repeat alpha C proteins on the cell surface of intact GBS failed to inhibit antibody binding, in contrast to the purified one- and two-repeat alpha C proteins.
FIG. 1.
ELISA inhibition with purified alpha C protein with different repeat numbers (1, 2, 9, and 16 repeats [rep]) as inhibiting antigen (Inh Ag) (left) and intact GBS with different repeat numbers (1, 2, 9, and 18 repeats) as inhibiting antigen (right). In both experiments rabbit antiserum to nine-repeat alpha C protein (final dilution, 1:8,000) was used. Microtiter plates were coated with purified nine-repeat alpha C protein at 0.125 μg/ml. Twofold dilutions were made from the inhibiting antigen (10-μg/ml starting dilution of purified alpha C protein; 109-cell/ml starting dilution of intact GBS). The left panel has been modified from reference 8 and is shown here for comparison.
FIG. 2.
ELISA inhibition with purified alpha C protein with different repeat numbers (1, 2, 9, and 16 repeats [rep]) as inhibiting antigen (Inh Ag) (left) and intact GBS with different repeat numbers (1, 2, 9, and 18 repeats) as inhibiting antigen (right). In both experiments rabbit antiserum to one-repeat alpha C protein was used (final dilution, 1:8,000). Microtiter plates were coated with purified one-repeat alpha C protein at 1 μg/ml. Twofold dilutions were made from the inhibiting antigen (10-μg/ml starting dilution of purified alpha C protein; 109-cell/ml starting dilution of intact GBS).
Availability of one- and nine-repeat alpha C proteins in GBS.
We hypothesized that the lower binding affinity of alpha C protein-specific antibodies to GBS expressing one-repeat alpha C protein relative to wild-type GBS expressing nine-repeat alpha C protein was due to structural differences in the alpha C protein that were related to its number of repeats. Alternatively, the lower binding affinity could have been due to decreased expression of the protein on the bacterial cell surface. In order to distinguish these possibilities, we quantified the surface-expressed alpha C protein in each strain by using ELISA inhibition with mutanolysin surface extracts.
The concentration of one-repeat alpha C protein in the mutanolysin extract was 3.2 μg/ml at a GBS concentration of 1.2 × 109 CFU/ml, the equivalent of 42,500 molecules of one-repeat alpha C protein/CFU. The concentration of nine-repeat alpha C protein on the wild-type A909 was 2 μg/ml at a bacterial density of 1.7 × 109 CFU/ml, or 6,000 molecules of nine-repeat alpha C protein/CFU. The number of bacteria per CFU was determined by counting individual bacteria in each of 98 bacterial clusters on EM. The average number of bacteria per CFU was between three and four. Because somewhat more one-repeat protein than nine-repeat protein was expressed at the cell surface, this difference in expression could not account for the lower antibody binding to the one-repeat-expressing GBS.
EM.
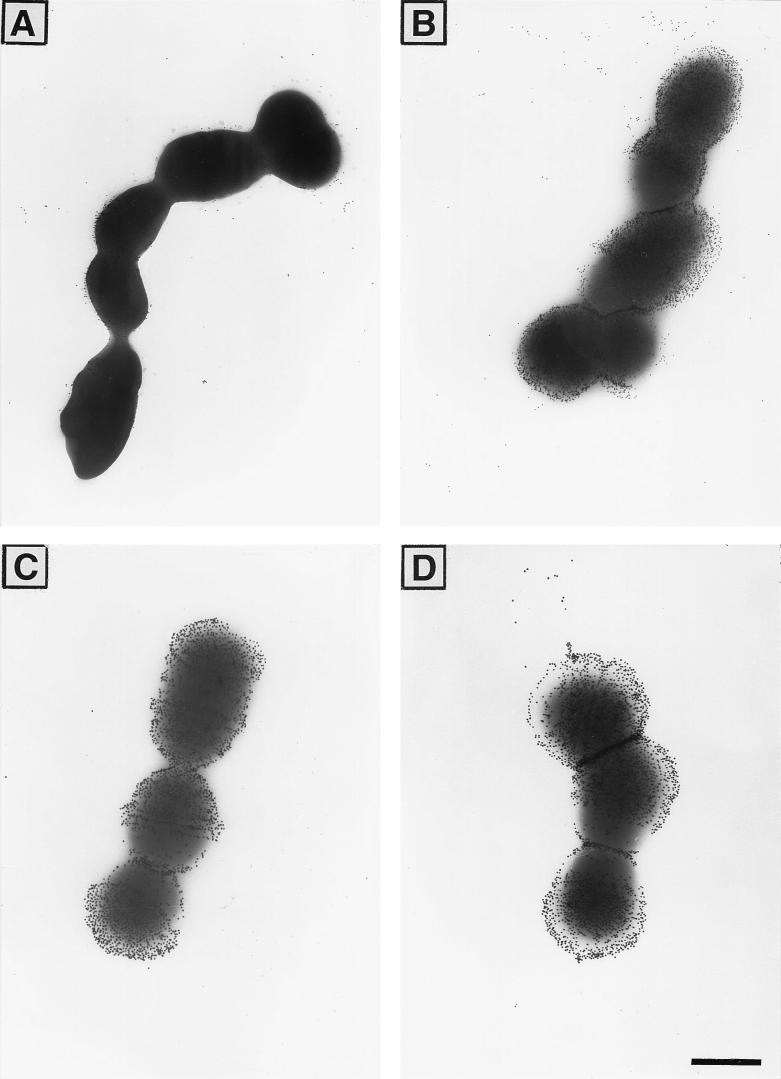
The discrepancy between the availabilities of one-repeat alpha C protein for binding to antibody in intact GBS and after mutanolysin extraction of the cell surface components suggested that the one-repeat alpha C protein at the cell surface of GBS could be obscured by other cell surface components, such as CPS. To test this hypothesis, we analyzed one- and nine-repeat alpha C protein-expressing GBS for binding to one-repeat-elicited antibodies and for binding to Ia CPS by EM with immunogold labelling (Fig. 3). Alpha C protein-specific antibodies bound readily to the wild-type GBS expressing nine-repeat alpha C protein but bound only poorly to the escape mutant expressing one-repeat alpha C protein. Both GBS strains readily bound antibodies to the capsule, as shown in Fig. 3. The few antibodies binding to the alpha C protein in the one-repeat mutant appeared (Fig. 3A) to bind close to the cell membrane, in contrast to the antibodies to the polysaccharide (Fig. 3C and D) and in contrast to the alpha C protein-specific antibodies binding to the nine-repeat strain (Fig. 3B). Equal amounts of Ia CPS (9.5 μg/mg [dry weight]) were measured in mutanolysin extracts from GBS with one- or nine-repeat alpha C protein. These findings, taken together, indicate that the one-repeat alpha C protein is present on the GBS surface but is poorly available for antibody binding, most likely as a result of steric hindrance by polysaccharides or other surface components.
FIG. 3.
EM photographs of GBS with one-repeat alpha C protein (A and C) and nine-repeat alpha C protein (B and D) at the cell surface, incubated with one-repeat alpha C protein-specific rabbit antiserum (A and B) or with rabbit antiserum to CPS type Ia (C and D). For protein staining, 15-nm-diameter-gold-labelled protein A was used, and for the CPS type Ia staining, 20-nm-diameter-gold-labelled protein A was used (final dilution, 1:50). Bar, 500 nm.
Sequence analysis of bca in A909-1.
The gene encoding the one-repeat alpha C protein was amplified from genomic DNA of the escape mutant A909-1 by PCR. Subsequently, 12 forward and reverse primers were developed to obtain the complete nucleotide sequence of the bca gene amplified from the escape mutant of GBS. The nucleotide sequence of the bca gene from the escape mutant A909-1 showed an open reading frame of 1,092 bp corresponding to 364 amino acids. The deduced amino acid sequence of the resultant protein included N-terminal and C-terminal regions identical to those of the wild-type A909. A single nucleotide substitution in the C-terminal region did not result in an amino acid change. The signal peptide amino acid sequence contained one conservative substitution (leucine was replaced by tryptophan at position 38). The mutant contained one repeat of 246 nucleotides (encoding 82 amino acids) and a partial repeat of 33 nucleotides (encoding 11 amino acids) with the exact sequence of the parent strain. Thus, the mutant contained a bca gene with the exact deletion of eight repeats from the parent strain. This finding is most compatible with deletion by homologous recombination among intragenic repeats.
Escape mutants.
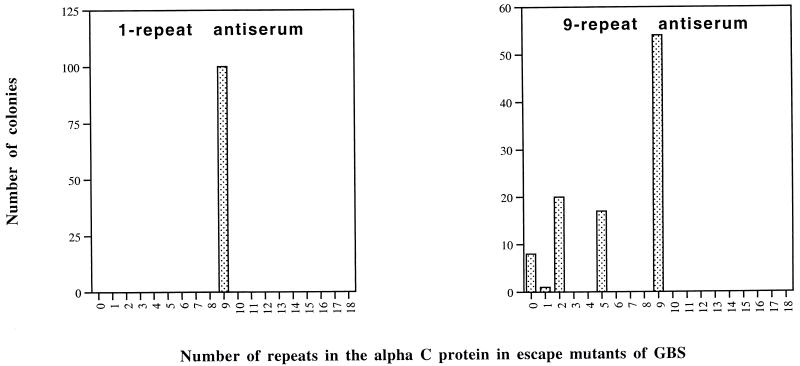
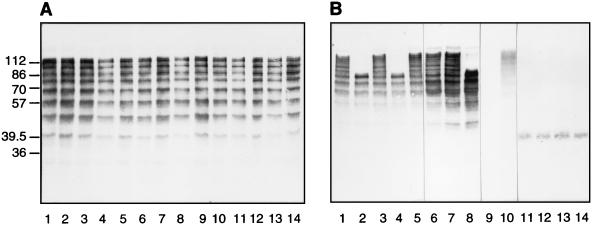
Pregnant mice were immunized with antiserum elicited to one- or nine-repeat alpha C protein, and their pups were challenged with wild-type GBS strain A909-9. Colonies were isolated from the spleens of the pups, individually cultured, and analyzed for their repeat contents by Western blotting. To obtain colonies from mice immunized with antiserum to one-repeat alpha C protein, the challenge dose needed to be 10-fold higher than for immunization with antiserum to nine-repeat alpha C protein. Eighty and 112 colonies were analyzed by Western blotting after immunization with antisera to one-repeat alpha and to nine-repeat alpha C protein, respectively (Fig. 4). Immunization with antiserum to nine-repeat alpha C protein resulted in escape mutants with one, two, five, or nine repeats and some mutants without any repeats. However, immunization with antiserum to one-repeat alpha C protein did not result in escape mutants with lower repeat numbers. In the latter case, all colonies showed nine repeats. Figure 5 shows an example of escape mutants of GBS isolated after immunization with antiserum to one- or nine-repeat alpha C protein.
FIG. 4.
Distribution of repeat number in the alpha C proteins from escape mutants of GBS isolated from the spleens of mice immunized with one-repeat alpha C protein antiserum (left) or nine-repeat alpha C protein antiserum (right). Repeat numbers were determined by Western blotting with monoclonal antibody 4G8 or rabbit antiserum to nine-repeat alpha C protein.
FIG. 5.
Western blots of escape mutants of GBS isolated from the spleens of pups immunized with antiserum to one-repeat alpha C protein (A) or to nine-repeat alpha C protein (B). These blots were incubated with antiserum to nine-repeat alpha C protein. (A) Wild-type GBS with nine-repeat alpha C protein (lane 1) and escape mutants with nine-repeat alpha C protein (lanes 2 to 14); (B) wild-type GBS with nine repeats (lane 1) and escape mutants with zero repeats (lane 9), with two repeats (lanes 11 to 14), with five repeats (lanes 2, 4, and 8), and with nine repeats (lanes 3, 5, 6, 7, and 10). Numbers on the left are molecular masses in kilodaltons.
Distribution of repeats in clinical isolates of GBS.
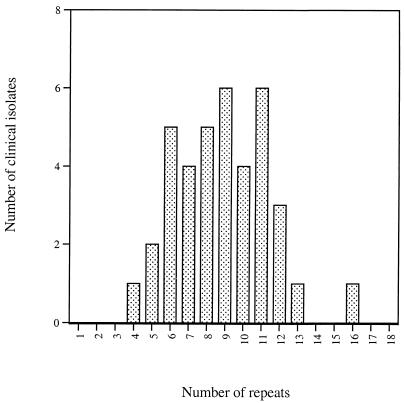
We analyzed 38 clinical isolates of GBS for their repeat content by Western blotting with monoclonal antibody 4G8. The repeat number was determined from the molecular mass obtained by SDS-PAGE and Western blotting, and a histogram was drawn (Fig. 6). Relatively few GBS with alpha C proteins of very high or very low repeat number were found, with the greatest number of isolates showing 9 or 10 repeats, which resembles a normal distribution with a mean and mode of 9 or 10 repeats.
FIG. 6.
Distribution of repeat number in the alpha C protein in clinical isolates of GBS. Repeat numbers were determined by Western blotting with 4G8, a repeat-specific monoclonal antibody. This figure was derived from data in a previous study (16).
DISCUSSION
Bacterial pathogens employ a number of genetic strategies that enhance their virulence and result in infection and/or disease in the host. Avoidance of the host immune defenses is key to the success of a pathogen. Most virulence factors are found on the bacterial surface or secreted in the immediate environment (5). Antigenic variation is a common microbial strategy used to avoid the host immune response. One of the best-studied examples of antigenic variation is the gonococcal pilus (27, 32). Continuous switching of pilus type occurs by removal of an expressed gene from the expression locus pilE and its replacement, by genetic rearrangement, with a copy of one silent gene generating a different pilus. Group A streptococci undergo variation in the repeat region of M proteins, thereby resulting in antigenic variation (6, 7, 12). GBS undergo deletions in the repeat region of the alpha C protein both during passage from human mothers to neonates and in an immune mouse model; these deletions result in altered antigenicity of the protein. Implicit in these findings is the hypothesis that such antigenic variation results in enhanced pathogenicity of the bacteria, at least in the immune host. However, this hypothesis has not previously been tested.
This study demonstrates that a deletion of eight of the nine tandem repeats in the alpha C protein of GBS enhances the pathogenicity of GBS (as defined by neonatal mouse lethality) about 100-fold in mice immunized with antiserum to the wild-type, nine-repeat alpha C protein. In nonimmune mice, pathogenicity was the same for the wild type and the mutant. We have previously shown that antigenic differences exist between the wild-type nine-repeat alpha C protein and variant proteins with different numbers of repeats. Specifically, when antiserum is raised to the wild-type, nine-repeat protein, it poorly recognizes constructs that contain one or two repeats. These differences appear to result from both the loss of epitopes and a change in conformation of the remaining epitopes. Similarly, GBS strains expressing one-repeat alpha C protein are poorly recognized by alpha C protein-specific antiserum, and it is likely that the enhanced pathogenicity in the immune mice is a result of this lack of recognition. Sequence analysis of the bca gene in the escape mutant showed no changes in the deduced amino acid sequence of the mature protein other than the loss of repeats, and the strain was shown to express the antigen on the bacterial surface. Capsule expression, a major virulence determinant, was identical in the mutant strain. No other phenotypic differences were observed in the mutant strain. Thus, modulation of repeat number is a mechanism for enhancement of pathogenicity in the immune host without diminution of pathogenicity in the naive host. Li et al. (15) showed a five- to sevenfold diminution in virulence of the strain with the alpha C protein deleted strain in nonimmune neonatal mice, which may suggest a specific physiological function for alpha C protein in nonimmune mice.
ELISA inhibition with intact GBS showed that antibodies elicited to one-repeat alpha C protein could bind well to the wild-type GBS strain expressing the nine-repeat protein but showed essentially no binding to the mutant strains expressing alpha C proteins with one or two repeats. These findings contrast with the results of ELISA inhibition with purified alpha C proteins and one-repeat-elicited antibodies, which show high affinity to alpha C proteins independent of their repeat number. This discrepancy between the in vivo and in vitro data prompted us to examine the presentation of the antigens on the surface of GBS by using EM. EM with immunogold labelling demonstrated that nine-repeat alpha C protein was readily available for antibody binding on the surface of GBS, at a location similar to that of the CPS. In contrast, the one-repeat alpha C protein was poorly visualized, and then only in close proximity to the cell wall. However, when protoplasts were formed (by disruption of the cell wall with mutanolysin), abundant one-repeat alpha C protein was present in the supernatant, in fact in greater molar quantity than the nine-repeat protein. Taken together, these findings suggest that deletion of repeats in the alpha C protein results not just in loss of epitopes but in a repositioning of the protein close to the GBS surface, sterically hidden from antibody binding by other cell surface components such as CPS.
In earlier work, we showed greater protection against GBS challenge when mice were immunized against one-repeat alpha C protein than when they were immunized with nine-repeat alpha C protein. We hypothesized that this result was due to high affinity of the one-repeat-elicited antiserum for alpha C proteins independent of repeat number, thereby reducing the ability of low-repeat-number mutants to escape antibody-mediated opsonization and phagocytosis. In the current study, we directly examined the types of mutants arising after immunization with antiserum to one- or nine-repeat alpha C protein and challenge with wild-type (nine-repeat alpha C protein) GBS. Lower-repeat-number mutants were not observed after immunization with antiserum to one-repeat alpha C protein, while about 50% of the escape mutants after immunization with antiserum to nine-repeat alpha C protein showed lower repeat numbers or expressed no alpha C protein. These findings show that escape of lower-repeat-number mutants after challenge with wild-type GBS was prevented by immunization with antiserum to one-repeat alpha C protein.
Interestingly, antiserum to one-repeat alpha C protein could prevent escape of one-repeat GBS mutants, while EM and ELISA inhibition showed poor binding of these antibodies to one-repeat protein at the surface of GBS. We hypothesized several possible mechanisms to explain this paradox. It is possible that at some phase(s) of growth of GBS, protein antigens may be more available for binding on the surface than at other growth phases. It has been shown in vitro that there is a direct correlation between density of type III CPS and the growth rate of GBS strain M781 (24). In other words, GBS cultured at a lower growth rate produce less CPS than GBS cultured under high-growth-rate conditions. It may be possible that the density of the CPS may vary in vivo and thus may affect the availability of the alpha C protein for antibody binding. As an alternative explanation, we showed that mutants with fewer than nine repeats but greater than one repeat were more frequently found than GBS mutants with one-repeat alpha C protein after immunization with antiserum to nine-repeat alpha C protein (Fig. 4). Such mutants might serve as intermediates to single-repeat mutants and would bind to antibodies with greater affinity than the one-repeat mutant in mice immunized with antiserum to one-repeat alpha C protein. Thus, this antiserum would inhibit the emergence of lower-repeat-number mutants and simultaneously prevent escape of single-repeat mutants even without binding to the latter mutants directly.
It is interesting that, with rare exceptions, only repeat deletion mutants (i.e., mutants with lower repeat numbers) have been observed under experimental conditions, yet clinical isolates show diverse repeat contents of up to 16 repeats. Our results showed that antiserum to nine-repeat alpha C protein is an efficient tool to select for mutants expressing fewer repeats. However, we have been unable to determine conditions that select for an increase in repeat number. It is possible that a larger repeat content is advantageous in a colonization setting or in some other undetermined niche. We have also shown that higher repeat numbers impart lower immunogenicity to the alpha C protein, particularly the N-terminal domain, and thus may allow GBS displaying alpha C proteins with larger numbers of repeats a selective advantage.
The existence of diversity in repeat number and the apparent presence of a normal distribution of repeat numbers in clinical isolates imply the existence in nature of selection for increased as well as decreased repeat number. If the ability to vary repeat number imparts a survival advantage to GBS, then the presence of multiple repeats would confer adaptability by providing mutable loci for homologous recombination. Thus, the one-repeat mutants would be at a disadvantage, since they would be unable to further undergo antigenic variation by homologous recombination. We isolated 88 individual colonies from the spleens of neonatal mice immunized with antiserum to one- or nine-repeat protein, or with preimmune serum, and then challenged with a GBS mutant expressing one-repeat alpha C protein. In all 88 colonies, the repeat number continued to be one (data not shown), which suggests that the one-repeat mutant cannot readily vary its repeat content.
The study presented here shows the role of the repeat number of the alpha C protein in pathogenicity of GBS. We have demonstrated enhanced pathogenicity in immune neonatal mice of an escape mutant that has undergone exact deletion of eight of nine tandem repeats in the bca gene. This finding is most compatible with homologous recombination of intragenic repeats as the mechanism for deletion of the repeats. The mechanisms accounting for the enhanced pathogenicity of this GBS strain after deletion in the repeat region of alpha C protein appeared to be (i) loss of protective conformational epitopes, as shown by ELISA inhibition, and (ii) steric hindrance of antibody binding to alpha C protein epitopes by cell wall components such as CPS after decrease in antigen size, as shown by immunolabelling and EM.
ACKNOWLEDGMENTS
We thank Johannes Heubner for generously providing the protocol for the immunogold staining procedure used in the EM studies, Larry C. Paoletti for providing rabbit antiserum to the GBS Ia CPS, Maria Ericsson for her excellent teaching of EM, and Dennis Kasper and Mike Wessels for fruitful discussions.
This research was supported by NIH grant AI38424 and PHS contracts N01 AI25152 and N01 AI732600.
REFERENCES
- 1.Baker C J, Edwards M S. Group B streptococcal infections. In: Remington J S, Klein J O, editors. Infectious diseases of the fetus and newborn infant. 3rd Edition ed. Philadelphia, Pa: W.B. Saunders; 1990. pp. 742–811. [Google Scholar]
- 2.Blake M S, Johnston H, Russell-Jones G J, Gotschlich E C. A rapid, sensitive method for detection of alkaline phosphatase-conjugated anti-antibody on western blots. Anal Biochem. 1984;136:175–179. doi: 10.1016/0003-2697(84)90320-8. [DOI] [PubMed] [Google Scholar]
- 3.Calandra G B, Cole R M. Lysis and protoplast formation of group B streptococci by mutanolysin. Infect Immun. 1980;28:1033–1037. doi: 10.1128/iai.28.3.1033-1037.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Cueninck B, Shockman G D, Swenson R M. Group B, type III streptococcal cell wall: composition and structural aspects revealed through endo-N-acetylmuramidase-catalyzed hydrolysis. Infect Immun. 1982;35:572–581. doi: 10.1128/iai.35.2.572-581.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Finlay B B, Falkow S. Common themes in microbial pathogenicity revisited. Microb Mol Biol Rev. 1997;61:136–169. doi: 10.1128/mmbr.61.2.136-169.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fischetti V A, Jarymowycz M, Jones K F, Scott J R. Streptococcal M protein size mutants occur at high frequency within a single strain. J Exp Med. 1986;164:971–980. doi: 10.1084/jem.164.4.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fischetti V A, Jones K F, Scott J R. Size variation of the M protein in group A streptococci. J Exp Med. 1985;161:1384–1401. doi: 10.1084/jem.161.6.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gravekamp C, Horensky D S, Michel J L, Madoff L C. Variation in repeat number within the alpha C protein of group B streptococci alters antigenicity and protective epitopes. Infect Immun. 1996;64:3576–3583. doi: 10.1128/iai.64.9.3576-3583.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gravekamp C, Kasper D L, Michel J L, Kling D E, Carey V, Madoff L C. Immunogenicity and protective efficacy of the alpha C protein of group B streptococci are inversely related to the number of repeats. Infect Immun. 1997;65:5216–5221. doi: 10.1128/iai.65.12.5216-5221.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hollingshead S K, Fischetti V A, Scott J R. Size variation in group A streptococcal M protein is generated by homologous recombination between intragenic repeats. Mol Gen Genet. 1987;207:196–203. doi: 10.1007/BF00331578. [DOI] [PubMed] [Google Scholar]
- 11.Johnson D R, Ferrieri P. Group B streptococcal Ibc protein antigen: distribution of two determinants in wild-type strains of common serotypes. J Clin Microbiol. 1984;19:506–510. doi: 10.1128/jcm.19.4.506-510.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones K F, Hollingshead S K, Scott J R, Fischetti V A. Spontaneous M6 protein size mutants of group A streptococci display variation in antigenic and opsonogenic epitopes. Proc Natl Acad Sci USA. 1988;85:8271–8275. doi: 10.1073/pnas.85.21.8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lachenauer C S, Madoff L C. A protective surface protein from type V group B streptococci shares N-terminal sequence homology with the alpha C protein. Infect Immun. 1996;64:4255–4260. doi: 10.1128/iai.64.10.4255-4260.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 15.Li J, Kasper D L, Ausubel F M, Rosner B, Michel J L. Inactivation of the alpha C protein antigen, bca, by a novel shuttle/suicide vector results in attenuation of virulence and immunity in group B Streptococcus. Proc Natl Acad Sci USA. 1997;94:13251–13256. doi: 10.1073/pnas.94.24.13251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Madoff L C, Hori S, Michel J L, Baker C J, Kasper D L. Phenotypic diversity in the alpha C protein of group B streptococcus. Infect Immun. 1991;59:2638–2644. doi: 10.1128/iai.59.8.2638-2644.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Madoff L C, Michel J L, Gong E W, Kling D E, Kasper D L. Group B streptococci escape host immunity by deletion of tandem repeat elements of the alpha C protein. Proc Natl Acad Sci USA. 1996;93:4131–4136. doi: 10.1073/pnas.93.9.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Madoff L C, Michel J L, Gong E W, Rodewald A K, Kasper D L. Protection of neonatal mice from group B streptococcal infection by maternal immunization with beta C protein. Infect Immun. 1992;60:4989–4994. doi: 10.1128/iai.60.12.4989-4994.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Madoff L C, Michel J L, Kasper D L. A monoclonal antibody identifies a protective C-protein alpha-antigen epitope in group B streptococci. Infect Immun. 1991;59:204–210. doi: 10.1128/iai.59.1.204-210.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McDaniel L S, Yother J, Vijayakumar M, McGarry L, Guild W R, Briles D E. Use of insertional inactivation to facilitate studies of biological properties of pneumococcal surface protein A (PspA) J Exp Med. 1987;154:1703–1708. doi: 10.1084/jem.165.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michel J L, Beseth B D, Madoff L C, Olken S K, Kasper D L, Ausubel F M. Genotypic diversity and evidence for two distinct classes of the C protein alpha antigen of group B Streptococcus. In: Totolian A, editor. Pathogenic streptococci: present and future. St. Petersburg, Russia: Lancer Publications; 1994. pp. 331–332. [Google Scholar]
- 22.Michel J L, Madoff L C, Olson K, Kling D E, Kasper D L, Ausubel F M. Large, identical, tandem repeating units in the C protein alpha gene, bca, of group B streptococci. Proc Natl Acad Sci USA. 1992;89:10060–10064. doi: 10.1073/pnas.89.21.10060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paoletti L C, Kasper D L, Michon F, DiFabio J, Jennings H J, Tosteson T D, Wessels M R. Effects of chain length on the immunogenicity in rabbits of group B Streptococcus type III oligosaccharide-tetanus toxoid conjugates. J Clin Invest. 1992;89:203–209. doi: 10.1172/JCI115564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paoletti L C, Ross R A, Johnson K D. Cell growth rate regulates expression of group B streptococcus type III capsular polysaccharide. Infect Immun. 1996;64:1220–1226. doi: 10.1128/iai.64.4.1220-1226.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodewald A K, Onderdonk A B, Warren H B, Kasper D L. Neonatal mouse model of group B streptococcal infection. J Infect Dis. 1992;166:635–639. doi: 10.1093/infdis/166.3.635. [DOI] [PubMed] [Google Scholar]
- 26.Rosner B. Fundamentals of biostatistics. 3rd ed. Boston, Mass: PWS-Kent; 1990. [Google Scholar]
- 27.Seifert H S, Wright C J, Jerse A E, Cohen M S, Cannon J G. Multiple gonococcal pilin antigenic variants are produced during experimental human infections. J Clin Invest. 1994;93:2744–2749. doi: 10.1172/JCI117290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stalhammar C M, Stenberg L, Lindahl G. Protein rib: a novel group B streptococcal cell surface protein that confers protective immunity and is expressed by most strains causing invasive infections. J Exp Med. 1993;177:1593–1603. doi: 10.1084/jem.177.6.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waltman W D, McDaniel L S, Gray B M, Briles D E. Variation in the molecular weight of PspA (pneumococcal surface protein A) among Streptococcus pneumoniae. Microb Pathog. 1990;8:61–69. doi: 10.1016/0882-4010(90)90008-e. [DOI] [PubMed] [Google Scholar]
- 30.Wastfelt M, Stalhammar-Carlemalm M, Delisse A M, Cabezon T, Lindahl G. Identification of a family of streptococcal surface proteins with extremely repetitive structure. J Biol Chem. 1996;271:18892–18897. doi: 10.1074/jbc.271.31.18892. [DOI] [PubMed] [Google Scholar]
- 31.Wood W B, Davis B D. Host-parasite relations in bacterial infections. In: Davis B D, Dulbecco R, Eisen H N, Ginsburg H S, editors. Microbiology. 3rd ed. Philadelphia, Pa: Harper & Row Publishers; 1980. pp. 552–553. [Google Scholar]
- 32.Zhang Q, DeRyckere D, Lauer P, Koomey M. Gene conversion in Neisseria gonorrhoeae: evidence for its role in pilus antigenic variation. Proc Natl Acad Sci USA. 1992;89:5366–5370. doi: 10.1073/pnas.89.12.5366. [DOI] [PMC free article] [PubMed] [Google Scholar]