Abstract
Current guidelines recommend single variant testing in relatives of patients with known pathogenic or likely pathogenic germline variants in cancer predisposition genes. This approach may preclude the use of risk-reducing strategies in family members who have pathogenic or likely pathogenic germline variants in other cancer predisposition genes. Cascade testing using multigene panels was performed in 3696 relatives of 7433 probands. Unexpected pathogenic or likely pathogenic germline variants were identified in 230 (6.2%) relatives, including 144 who were negative for the familial pathogenic or likely pathogenic variant but positive for a pathogenic or likely pathogenic variant in a different gene than the proband and 74 who tested positive for the familial pathogenic or likely pathogenic variant and had an additional pathogenic or likely pathogenic variant in a different gene than the proband. Of the relatives with unexpected pathogenic or likely pathogenic germline variants, 36.3% would have qualified for different or additional cancer screening recommendations. Limiting cascade testing to only the familial pathogenic or likely pathogenic variant would have resulted in missed, actionable findings for a subset of relatives.
After detection of a pathogenic or likely pathogenic variant in a cancer predisposition gene, cascade testing, which includes pretest education and germline genetic testing, should be offered to all blood relatives of the index patient. Cascade testing can be used to promote increased surveillance and risk-reducing strategies for relatives who test positive for the familial pathogenic or likely pathogenic variant and general population recommendations for those who test negative. Current guidelines recommend limiting cascade testing to the familial pathogenic or likely pathogenic variant (1), rather than using multigene panel testing in family members. However, a 2019 study of 1084 first-degree relatives undergoing cascade testing via multigene panel testing found that 4.9% of relatives had different pathogenic or likely pathogenic germline variants than those of the proband (2). The aim of this study was to determine the rate of unexpected pathogenic or likely pathogenic germline variants in a cohort of approximately 4000 individuals undergoing cascade testing, using multigene panel testing rather than a single gene approach for the familial pathogenic or likely pathogenic variant(s), and to characterize the individuals and genes with unexpected pathogenic or likely pathogenic germline variants.
Clinician-selected multigene panel testing for all probands was performed using the 47 gene Invitae Common Hereditary Cancers Panel. Sequencing and variant classification were performed as previously reported (3) and described in the Supplementary Materials (available online). Review and analysis of de-identified and aggregated data were approved for waiver of authorization by the WCG institutional review board (study number 1167406). Relatives with variants of uncertain significance in the absence of pathogenic or likely pathogenic variant were grouped with patients with negative results. Genes were classified as high risk (>50% absolute lifetime cancer risk), moderate risk (20%-50% lifetime risk), low risk (<20% lifetime cancer risk), rare cancer risk (elevated risk of uncommon cancers), or undefined cancer risk. To estimate how many relatives would have a change to clinical management based on unexpected pathogenic or likely pathogenic germline variants, guideline-directed screening modalities for relevant organs based on pathogenic or likely pathogenic germline variants and sex were assigned for each proband and relative. Demographics, relationship to the proband, and cancer history data were extracted from test requisition forms. When evaluating race and ethnicity, all race and ethnicities that represented less than 1% of the cohort were grouped as other. R Statistical Software (v.4.2.1; R Core Team 2021) was used for logistic regression analysis; a 2-sided P value less than .05 was considered statistically significant.
Of 15 362 individuals who underwent clinician-ordered cascade testing for a familial pathogenic or likely pathogenic variant between January 2017 and March 2021, a total of 3696 (24%) underwent multigene panel testing with the panel chosen at the discretion of the provider (Supplementary Figure 1, available online). The median number of genes evaluated was 47 (range = 10-156 genes). The average age at testing was 50.5 years, and the majority of relatives tested were female (76%), non-Hispanic White (74%), first-degree relatives (74%), and without a personal history of cancer (81%).
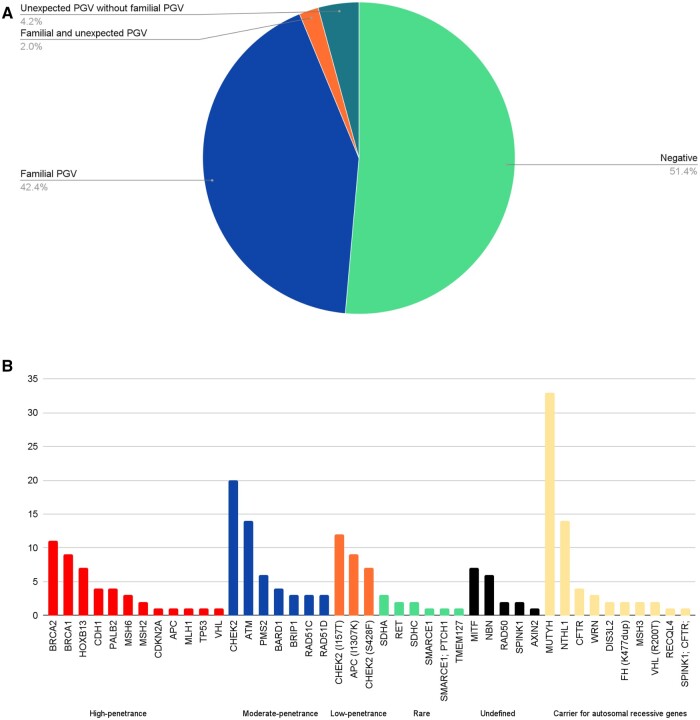
Most of the relatives had no (51%) or 1 (45%) pathogenic or likely pathogenic variant (Figure 1, A). Of relatives, 42% only had pathogenic or likely pathogenic germline variants shared with the proband, with 38.4% of relatives sharing all of the pathogenic or likely pathogenic germline variants detected in the proband and 4.0% sharing a subset of pathogenic or likely pathogenic germline variants with probands who had multiple pathogenic or likely pathogenic germline variants. A total of 144 (3.9%) relatives had unexpected pathogenic or likely pathogenic germline variants but did not share pathogenic or likely pathogenic germline variants with the proband, and 74 (2.0%) had at least 1 pathogenic or likely pathogenic variant present in the proband as well as unexpected pathogenic or likely pathogenic germline variants. For 11 (0.3%) relatives, the pathogenic or likely pathogenic variant within a given gene was different from that detected in the proband.
Figure 1.
Genetic results of 3696 relatives undergoing multigene panel testing. A) Overall frequency of pathogenic or likely pathogenic variant and B) genes with unexpected pathogenic or likely pathogenic variant. CHEK2 alleles were partitioned into those with moderate penetrance and those with low penetrance, in particular the I157T and S428F alleles. The 12 individuals who had unexpected pathogenic or likely pathogenic germline variants detected in the same gene as the proband were not included in this graph as the unexpected pathogenic or likely pathogenic variant would have been reported, per Invitae protocol. PGV = pathogenic or likely pathogenic variant.
Individuals of Ashkenazi Jewish ancestry were 2 times more likely to have unexpected pathogenic or likely pathogenic germline variants compared with non-Ashkenazi White individuals (odds ratio [OR] = 2.0, 95% confidence interval [CI] = 1.1 to 3.4; P = .017; Table 1). In addition, second-degree or more distant relatives and those with a personal history of cancer were statistically significantly more likely to have unexpected pathogenic or likely pathogenic germline variants than first-degree relatives (OR = 1.9, 95% CI = 1.4 to 2.5; P = 9.9 x 10−6) and unaffected relatives (OR = 1.6, 95% CI = 1.1 to 2.3; P = .012).
Table 1.
Multivariable logistic regression analysis to identify the association of demographic and clinical factors with unexpected pathogenic or likely pathogenic germline variants
| Variablea | Coefficient | Standard error | OR (95% CI) | P |
|---|---|---|---|---|
| Sex (referent, female) | ||||
| Male | 0.01 | 0.17 | 1.0 (0.7 to 1.4) | .956 |
| Race and ethnicity (referent, White) | ||||
| Ashkenazi Jewish | 0.67 | 0.28 | 2.0 (1.1 to 3.4) | .017 |
| Asian and Pacific Islanders | −15.17 | 508.76 | 0.0 (0.0 to ∞) | .976 |
| Black | −1.71 | 1.01 | 0.2 (0.0 to 1.3) | .090 |
| Hispanic | −0.24 | 0.38 | 0.8 (0.4 to 1.6) | .528 |
| Multiethnic | −0.57 | 0.30 | 0.6 (0.3 to 1.0) | .056 |
| Otherb | −14.83 | 582.75 | 0.0 (0.0 to ∞) | .980 |
| Unknown | −0.58 | 0.52 | 0.6 (0.2 to 1.5) | .260 |
| Degree of relationship (referent, first-degree relative) | ||||
| Second-degree relative or greater | 0.64 | 0.14 | 1.9 (1.4 to 2.5) | 9.9 × 10−6 |
| Cancer history (referent, no) | ||||
| Yes | 0.46 | 0.18 | 1.6 (1.1 to 2.3) | .012 |
| Age | −0.01 | 0.005 | 1.0 (1.0 to 1.0) | .014 |
| No. of genes analyzed | 0.02 | 0.004 | 1.0 (1.0 to 1.0) | 1.4 × 10−6 |
In the multivariable logistic regression analysis, n = 3695 after excluding 1 individual with missing data for sex. Sex, race and ethnicity, degree of relationship, cancer history, number of genes analyzed, and age were included in the model. CI = confidence interval; OR = odds ratio.
All race and ethnicities that represented less than 1% of the cohort were grouped in the other category.
The 218 unexpected pathogenic or likely pathogenic germline variants that were detected in genes other than those of the probands were distributed across 37 cancer predisposition genes (Figure 1, B). Among these 218 relatives, 98 (44.9%) had unexpected pathogenic or likely pathogenic germline variants in high-penetrance (n = 45, 20.6%) or moderate-penetrance (n = 53, 24.3%) genes. Most importantly, 79 of the 98 (81%) relatives with an unexpected high-penetrance or moderate-penetrance pathogenic or likely pathogenic variant would have a change to screening modalities or risk-reducing surgery compared with probands. Unexpected pathogenic or likely pathogenic germline variants were found in low-risk or recessive cancer predisposition genes in 120 (55.1%) patients, with the most common being in MUTYH (monoallelic).
One strength of this study is the sample size, which was approximately 4 times larger than that of an earlier study that investigated the rate of unexpected pathogenic or likely pathogenic germline variants in relatives undergoing multigene panel testing (2). The 4.9% rate of unexpected pathogenic or likely pathogenic germline variants (including 2.8% of pathogenic or likely pathogenic germline variants in high- or moderate-penetrance genes) in that study was not statistically significantly different (P = .12) from the 6.2% (2.7% in high- or moderate-penetrance genes) reported in our study, suggesting that 3% of relatives undergoing limited single-variant cascade testing would have an unexpected high- or moderate-risk pathogenic or likely pathogenic variant, with 80% of them missing the opportunity to employ preventive interventions. Of note, in the study by Caswell-Jin et al. (2), first-degree relatives of probands with pathogenic or likely pathogenic variant were invited to undergo cascade testing at a cost of $50 via an email sent by the testing laboratory. In contrast, in our study, multigene panel testing in relatives was initiated at the providers’ discretion, with the cost for multigene panel testing billed to the relative’s health insurance or paid for by the relative. This study is thus limited in its ability to determine rates of or reasons for test uptake, including how the cost of multigene panel testing, which is not currently covered as a form of familial variant testing, affects uptake. In addition, data regarding alterations to the clinical management of relatives were not collected. In a pilot study of 25 relatives with a pathogenic or likely pathogenic variant in a hereditary cancer gene identified by testing only for the proband’s variant, 72% completed at least 1 recommended screening recommendation, and 40% underwent risk-reducing surgeries within 2 years of genetic testing (4). Future studies are therefore needed to evaluate the impact of multigene panel testing in cascade testing on clinical management.
In conclusion, multigene panel testing identified 6.2% of relatives with unexpected pathogenic or likely pathogenic variant in cancer predisposition genes. Nearly half (2.7% of the relatives who were tested) of the unexpected pathogenic or likely pathogenic germline variants were in high- or moderate-penetrance genes, and 80% of them would be offered a change in clinical management. With the goal of providing all blood relatives with the genetic information necessary to optimize clinical management, current recommendations for single-variant testing in blood relatives may lead to missed opportunities compared with multigene panel testing for prevention in family members undergoing cascade testing.
Supplementary Material
Acknowledgements
We thank the patients and their relatives who have agreed to share their data for this research study. Employees of Invitae Corporation served as co-investigators in this study and participated in the design of the study; the collection, analysis, and interpretation of the data; the writing of the article and the decision to submit the article for publication. Preliminary results from this study were presented at American College of Medical Genetics and Genomics, National Society of Genetic Counselors and American Society of Human Genetics 2023 annual meetings.
Contributor Information
Brandie Heald, Medical Affairs, Invitae Corp, San Francisco, CA, USA.
Sara Pirzadeh-Miller, Cancer Genetics, University of Texas Southwestern/Simmons Comprehensive Cancer Center, Dallas, TX, USA.
Rachel E Ellsworth, Medical Affairs, Invitae Corp, San Francisco, CA, USA.
Sarah M Nielsen, Medical Affairs, Invitae Corp, San Francisco, CA, USA.
Emily M Russell, Medical Affairs, Invitae Corp, San Francisco, CA, USA.
Peter Beitsch, Medical Affairs, Invitae Corp, San Francisco, CA, USA.
Edward D Esplin, Medical Affairs, Invitae Corp, San Francisco, CA, USA.
Robert L Nussbaum, Medical Affairs, Invitae Corp, San Francisco, CA, USA.
Daniel E Pineda-Alvarez, Medical Affairs, Invitae Corp, San Francisco, CA, USA.
Allison W Kurian, Division of Oncology, Stanford University School of Medicine, Stanford, CA, USA.
Heather Hampel, Division of Clinical Cancer Genomics, Department of Medical Oncology, City of Hope, Duarte, CA, USA.
Data availability
When not prohibited by patient permissions or privacy laws, the de-identified individual data that underlie the results reported in this article will be made available to researchers. Researchers will be asked to submit a short proposal outlining objectives, research question, and analytical methods and submit institutional review board approval or determination of exempt status or nonhuman subjects research. For more information on how to access the Invitae data, please contact the corresponding author.
Author contributions
Brandie Heald, MS, CGC (Data curation; Formal analysis; Project administration; Writing—review & editing), Sara Pirzadeh-Miller, MS, CGC (Writing—review & editing), Rachel E Ellsworth, PhD (Writing—original draft), Sarah M Nielsen, MS, CGC (Formal analysis; Writing—review & editing), Emily R Russell, PhD (Data curation; Formal analysis; Writing—review & editing), Peter Beitsch, MD (Writing—review & editing), Edward D Esplin, MD, PhD (Writing—review & editing), Robert L Nussbaum, MD (Conceptualization; Writing—review & editing), Daniel E Pineda-Alvarez, MD (Writing—review & editing), Allison W Kurian, MD, MSc (Writing—review & editing), and Heather Hampel, MS, CGC (Writing—review & editing).
Funding
This work was supported by Invitae Corporation.
Conflicts of interest
Brandie Heald, MS, CGC; Rachel E. Ellsworth, PhD; Sarah M. Nielsen, MS, CGC; Emily M. Russell, PhD; Peter Beitsch, MD; Edward D. Esplin, MD, PhD; Robert L. Nussbaum, MD; Daniel E. Pineda-Alvarez, MD, are employees and shareholders of Invitae Corp. Edward D. Esplin is an advisor and stockholder of Taproot Health and Exir Bio. Heather Hampel is on the scientific advisory boards for Genome Medical and Natera. She does consulting for GI OnDemand, Carelon, and 23andMe. She has stock/stock options in Genome Medical and GI OnDemand.
References
- 1.National Comprehensive Cancer Network. Genetic/Familial High-Risk Assessment: Breast, Ovarian, and Pancreatic. National Comprehensive Cancer Network. 2023. https://www.nccn.org/professionals/physician_gls/pdf/genetics_bop.pdf. Accessed May 24, 2023.
- 2. Caswell-Jin JL, Zimmer AD, Stedden W, Kingham KE, Zhou AY, Kurian AW.. Cascade genetic testing of relatives for hereditary cancer risk: results of an online initiative. J Natl Cancer Inst. 2019;111(1):95-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schmidlen TJ, Bristow SL, Hatchell KE, Esplin ED, Nussbaum RL, Haverfield EV.. The impact of proband indication for genetic testing on the uptake of cascade testing among relatives. Front Genet. 2022;13:867226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Frey MK, Ahsan MD, Badiner N, et al. What happens in the long term: uptake of cancer surveillance and prevention strategies among at-risk relatives with pathogenic variants detected via cascade testing. Cancer. 2022;128(24):4241-4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
When not prohibited by patient permissions or privacy laws, the de-identified individual data that underlie the results reported in this article will be made available to researchers. Researchers will be asked to submit a short proposal outlining objectives, research question, and analytical methods and submit institutional review board approval or determination of exempt status or nonhuman subjects research. For more information on how to access the Invitae data, please contact the corresponding author.