Abstract
Background
The rate of esophagogastric cancer is rising among individuals under 50 years of age. It remains unknown whether early-onset esophagogastric cancer represents a unique entity. This study investigated the clinical and molecular characteristics of early-onset and average-onset esophagogastric cancer .
Methods
We reviewed the Memorial Sloan Kettering Cancer Center gastric, esophageal, and gastroesophageal junction cancer database. Associations between baseline characteristics and tumor and germline molecular alterations were compared between those with early-onset and average-onset esophagogastric cancer using Fisher exact tests and the Benjamini-Hochberg method for multiple-hypothesis correction.
Results
We included 1123 patients with early-onset esophagogastric cancer (n = 219; median age = 43 years [range = 18-49 years]) and average-onset esophagogastric cancer (n = 904; median age = 67 years [range = 50-94 years]) treated between 2005 and 2018. The early-onset group had more women (39% vs 28%, P = .002). Patients with early-onset esophagogastric cancer were more likely to have a gastric primary site (64% vs 44%, P < .0001). The signet ring cell and/or diffuse type was 3 times more common in the early-onset esophagogastric cancer group (31% vs 9%, P < .0001). Early-onsite tumors were more frequently genomically stable (31% vs 18%, P = .0002) and unlikely to be microsatellite instability high (2% vs 7%, P = .003). After restricting to adenocarcinoma and signet ring cell and/or diffuse type carcinomas, we observed no difference in stage (P = .40) or overall survival from stage IV diagnosis (median = 22.7 vs 22.1 months, P = .78).
Conclusions
Our study supported a preponderance of gastric primary disease sites, signet ring histology, and genomically stable molecular subtypes in early-onset esophagogastric cancer. Our findings highlight the need for further research to define the underlying pathogenesis and strategies for early detection and prevention.
The rates of gastric and esophageal cancers in patients younger than 50 years of age have increased by 30% and 50%, respectively, in the United States since the 1990s (1,2). Together, these cancers are responsible for more than 100 000 deaths per year globally among individuals younger than 50 years (3,4). Although the rate of esophageal adenocarcinoma is rising similarly in the population, the overall incidence of gastric adenocarcinoma has decreased by more than 30% during this time, suggesting an etiology specific to those under age 50. Clinical features of early-onset esophagogastric cancer that may ultimately enable earlier detection have been incompletely described in the literature.
Although several germline alterations have been associated with the risk of esophagogastric cancer, germline abnormalities account for less than one-third of early-onset esophagogastric cancer cases (5,6). This rise in incidence of esophagogastric cancer among young patients correlates with increased rates of obesity (7), changes in diet (8), and alterations in the environment, though varied evidence of the role of sex, obesity and other environmental factors has led to an incomplete understanding of this trend (9,10). Prior studies have been limited by small sample size (9,10), lack of access to patient-level data in those studies generated from large national registries (11), and varied patient access to care (12). Data are conflicting with regard to age as a prognostic indicator (13,14). It therefore remains unknown whether early-onset esophagogastric cancer is a distinct entity relative to average-onset esophagogastric cancer.
Understanding whether early-onset esophagogastric cancer is distinct from its average-onset counterpart is essential. This knowledge will affect whether standard diagnostic and treatment paradigms designed for older individuals apply to these young patients. The purpose of this study was to compare the clinical, pathologic, molecular, and germline characteristics of early-onset and average-onset esophagogastric cancer.
Methods
Definition of early-onset esophagogastric cancer
Current indications for upper gastrointestinal (GI) endoscopy based on American Society for Gastrointestinal Endoscopy guidelines include dysphagia or odynophagia; persistent or recurrent reflux, despite therapy; suspected chronic blood loss in the context of iron deficiency–related anemia; and upper abdominal symptoms that are unresponsive to empiric therapy or are new in onset in a patient older than 50 years of age (15). The guidelines use 50 years of age as the cutoff for urgent endoscopy referral based on the low incidence of esophageal adenocarcinoma in men younger than 55 years of age (<5 per 100 000 between 1973 and 2002) (10,16). Although the incidence remains less than 5 per 100 000 among individuals under 50 years of age based on 2018 data, the absolute incidence and mortality are increasing (3). For these reasons, we define early-onset esophagogastric cancer as an upper GI malignancy occurring before age 50.
Patients
We identified patients with a pathologic diagnosis of esophageal, gastroesophageal junction, or gastric cancer at Memorial Sloan Kettering Cancer Center. We included a sequential cohort of all patients diagnosed in 2018 in addition to the cohort of all patients who had undergone genomic profiling using the US Food and Drug Administration–authorized Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) assay of either their primary or metastatic tumor between 2005 and 2018. Adult patients (aged ≥18 years) were included. We abstracted clinical data from individual patient records, including demographics, pathology, sites of metastasis, stage at diagnosis, and surgical information. The study was approved by the Memorial Sloan Kettering Cancer Center Institutional Review Board. Written informed consent was obtained from patients for all genomic analyses.
Clinical data collection
To answer the question of whether patients with early-onset esophagogastric cancer present with distinct clinical characteristics and risk factors compared with those with average-onset esophagogastric cancer, we conducted an additional retrospective review of the patients’ presenting clinical symptoms (including time from symptom onset to diagnosis, body mass index [BMI] at diagnosis, presence or absence of weight loss [>5% body weight] at diagnosis, medical comorbidities, and smoking and alcohol use history).
Somatic mutation analysis
Patient tumors were sequenced using the MSK-IMPACT, a Clinical Laboratory Improvement Amendments–approved next-generation sequencing assay performed as part of standard management by physician decision for each patient. Genomic DNA was extracted from formalin-fixed, paraffin-embedded tumor samples, and matched normal blood samples were collected. After capture of exons and selected introns of the genes included in the sequencing panel, pooled libraries were sequenced on the Illumina HiSeq 2500 system, which included up to 505 genes (17,18). The resulting sequences were run through an optimized informatic pipeline to identify somatic mutations, copy number alterations, and select structural rearrangements. Genomic alterations were filtered for oncogenic variants using OncoKB (19), and genes were grouped into signaling pathways using curated pathway templates (20). The cases were subclassified into 4 groups by The Cancer Genome Atlas Network gastric adenocarcinoma criteria: Epstein-Barr virus, microsatellite instability (MSI), genomically stable, and chromosomal instability (21). Genomic data will be available online at the time of publication.
Germline analysis
Blood-derived DNA was sequenced for germline analysis using a 76- to 88-gene MSK-IMPACT panel (22). Germline analysis was offered prospectively at clinician discretion to patients who consented to tumor genetic analysis on an institutional review board–approved protocol (17,18).
Statistical analysis
Categorical variables were compared using a 2-sided Fisher exact test to identify significant differences between patient groups—specifically, between those with early-onset (age <50 years) and average-onset disease. Continuous variables were compared between groups using a Wilcoxon test. Overall survival and progression-free survival were calculated using the Kaplan-Meier method and compared using a log-rank test. Cox proportional hazards models were used to generate hazard ratios and confidence intervals. Subset analyses, including only patients with adenocarcinoma, were performed to determine whether the overall findings were influenced by inclusion of a small population of patients with squamous and other histologies. Clinical and genomic features statistically significant on univariable analysis were then included in a multivariable model. R statistical software (R Foundation for Statistical Computing, Vienna, Austria) was used for analyses.
Results
Clinicopathologic characteristics of early-onset esophagogastric cancer
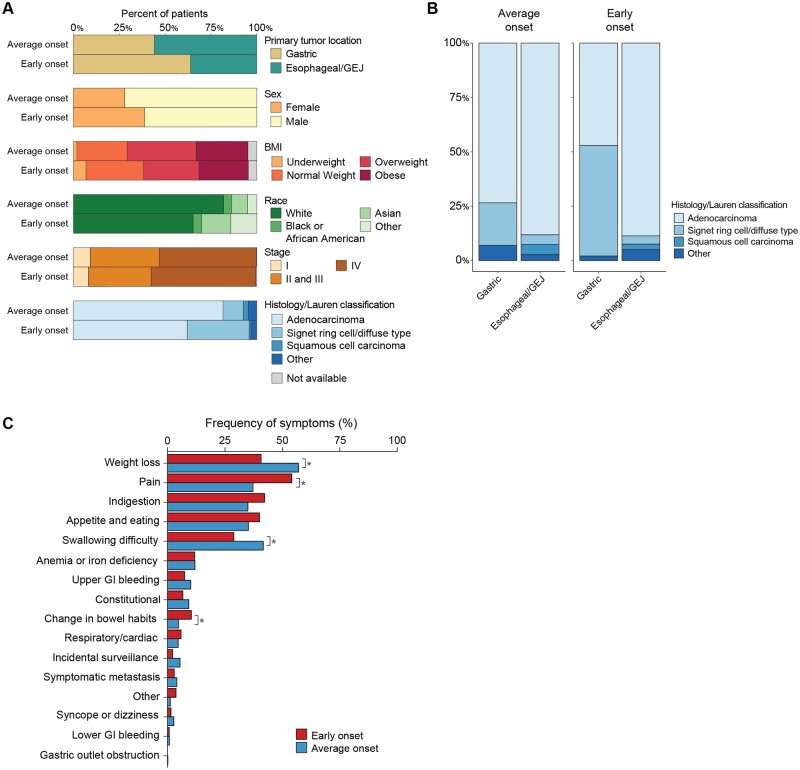
We compared the demographic, clinical, and pathologic features of a total of 1123 patients (median age = 64 years [range = 18-94 years]) with early-onset esophagogastric cancer (n = 219; median age = 43 years [range = 18-50 years]) and average-onset esophagogastric cancer (n = 904; median age = 67 years [range = 50-94 years]) (Figure 1, A, Table 1, Supplementary Figure 1, A, available online) (23). Compared with the average-onset esophagogastric cancer cohort, the early-onset group consisted of a higher proportion of women (39% vs 28%, P = .002). Asian race was more common in the early-onset group (16% vs 9%, P = .002), and White race was less common (65% vs 82%, P < .001); there was no difference in the proportion of Black or African American patients between the 2 groups (Figure 1, A, Table 1). Ethnicity also did not differ statistically significantly between the groups. Median BMI (early onset = 26.2 vs average onset = 27.1 kg/m2, P = .009), and the proportion of patients who were overweight or obese at the time of diagnosis was higher in the average-onset group than in the early-onset group (66% vs 57%, P = .02) (Figure 1, A, Table 1).
Figure 1.
Clinical characteristics of early-onset (age <50 years, n = 219) and average-onset (age ≥50 years, n = 904) esophagogastric cancer. A) Comparison of the clinical and pathologic characteristics demonstrates a higher percentage of gastric primary tumor site (64% vs 44%, P < .0001), proportion of women (39% vs 28%, P = .002), Asian race (16% vs 9%, P = .002), and signet ring histology or Lauren diffuse type cancer (31% vs 9%, P < .001) in the early-onset group. B) Histologic subtype distribution in the average-onset group compared with the early-onset group demonstrates a higher proportion of signet ring cell and/or diffuse type in the early-onset gastric cancer than in the average-onset gastric cancer group. C) Presenting symptoms that occurred statistically significantly more frequently in the early-onset group included pain and change in bowel habits, while weight loss and swallowing difficulty occurred more frequently in the average-onset group. BMI = body mass index; GEJ = gastroesophageal junction; GI = gastrointestinal.
Table 1.
Baseline characteristics of patients with early-onset and average-onset esophagogastric cancer
| Overall | Average onset | Early onset | P | |
|---|---|---|---|---|
| (N = 1123) | (n = 904) | (n = 219) | ||
| Age at diagnosis, y | ||||
| Mean (SD) | 61.6 (13.5) | 66.8 (8.7) | 40.4 (7.5) | <.0001 |
| Median (min, max) | 64.0 (18.3, 94.0) | 67.0 (50.0, 94.0) | 42.8 (18.3, 50.0) | |
| Sex, No. (%) | ||||
| Female | 338 (30.1) | 253 (28.0) | 85 (38.8) | .002325 |
| Male | 785(69.9) | 651 (72.0) | 134 (61.2) | |
| Race, No. (%) | ||||
| Asian | 113 (10.1) | 78 (8.6) | 35 (16.0) | .0014 |
| Black or African American | 50 (4.4) | 40 (4.4) | 10 (4.6) | .85 |
| White | 884 (78.7) | 741 (82.0) | 143 (65.3) | <.0001 |
| Other | 38 (3.4) | 20 (2.2) | 18 (8.2) | <.0001 |
| Unknown | 38 (3.4) | 25 (2.8) | 13 (5.9) | |
| Ethnicity, No. (%) | ||||
| Hispanic or Latino | 91 (8.1) | 55 (6.1) | 36 (16.4) | <.0001 |
| Not Hispanic or Latino | 985 (87.7) | 807 (89.3) | 178 (81.3) | |
| Unknown | 47 (4.2) | 42 (4.6) | 5 (2.3) | |
| Body mass index, kg/m2 | ||||
| Mean (SD) | 27.6 (5.63) | 27.8 (5.36) | 26.9 (6.62) | .0099 |
| Median (Min, Max) | 26.9 (11.6, 56.8) | 27.1 (15.2, 56.4) | 26.2 (11.6, 56.8) | |
| Missing, No. (%) | 33 (2.9) | 24 (2.7) | 9 (4.1) | |
| Smoking status, No. (%) | ||||
| Yes | 401 (35.7) | 352 (38.9) | 49 (22.4) | <.0001 |
| No | 334 (29.7) | 246 (27.2) | 88 (40.2) | |
| Missing | 388 (34.6) | 306 (33.9) | 82 (37.4) | |
| Alcohol use, No. (%) | ||||
| Yes | 340 (30.3) | 271 (30.0) | 69 (31.5) | .69 |
| No | 191 (17.0) | 158 (17.5) | 33 (15.1) | |
| Missing | 592 (52.7) | 475 (52.5) | 117 (53.4) | |
| Histology/Lauren classification, No. (%) | ||||
| Adenocarcinoma | 874 (77.8) | 738 (81.6) | 136 (62.1) | <.0001 |
| Signet ring and/or diffuse type | 174 (15.5) | 100 (11.1) | 74 (33.8) | <.0001 |
| Squamous cell carcinoma | 26 (2.3) | 24 (2.7) | 2 (0.9) | .21 |
| Other | 49 (4.4) | 42 (4.7) | 7 (0.9) | .46 |
| Primary site binary, No. (%) | ||||
| Proximal | 584 (52.0) | 505 (55.9) | 79 (36.1) | <.0001 |
| Distal | 539 (48.0) | 399 (44.1) | 140 (63.9) | |
| Primary site trinary, No. (%) | ||||
| Esophageal | 286 (25.5) | 252 (27.9) | 34 (15.6) | .0001 |
| Gastric | 539 (48.0) | 399 (44.1) | 140 (63.9) | <.0001 |
| Gastroesophageal junction (Siewert I-II) | 298 (26.5) | 253 (28.0) | 45 (20.5) | .03 |
| ERBB2 (formerly HER2 or HER2/neu) status, No. (%) | ||||
| Positive | 226 (20.1) | 195 (21.6) | 31 (14.2) | .02 |
| Negative | 722(64.3) | 570 (63.1) | 152 (69.4) | |
| Unknown | 175 (15.6) | 139 (16.9) | 36 (16.4) | |
| Primary resected, No. (%) | ||||
| Yes | 502 (44.7) | 406 (44.9) | 96 (43.8) | .82 |
| No | 619 (55.1) | 497 (55.0) | 122 (55.7) | |
| Unknown | 2 (0.2) | 1 (0.1) | 1 (0.5) | |
| Stage at diagnosis, No. (%) | ||||
| I | 102 (9.1) | 84 (9.3) | 18 (8.2) | .70 |
| II | 126 (11.2) | 103 (11.4) | 23 (10.5) | .81 |
| III | 289 (25.7) | 237 (26.2) | 52 (23.9) | .49 |
| IV | 603 (53.7) | 478 (52.9) | 125 (57.3) | .26 |
| Unknown | 3 (0.3) | 2 (0.2) | 1 (0) | |
| Metastatic or nonmetastatic at diagnosis, No. (%) | ||||
| Metastatic | 603 (53.7) | 478 (52.9) | 125 (57.1) | .26 |
| Nonmetastatic | 519 (46.2) | 426 (47.1) | 93 (42.5) | |
| Unknown | 1 (0.9) | 0 (0.0) | 1(0.4) |
Patients with early-onset esophagogastric cancer were more likely to have a gastric (distal) primary tumor site (64% vs 44%, P < .0001) and less likely to have esophageal (16% vs 28%, P < .001) or gastroesophageal junction (20% vs 28%, P = .03) primary sites. Although non–signet ring cell adenocarcinoma was the predominant histologic subtype in both groups (early onset = 65% vs average onset = 83%, P < .001), there was a higher proportion of patients with signet ring cell/diffuse–type adenocarcinoma in the early-onset group (34% vs 11%, P < .001). When comparing only patients with a gastric primary tumor site, the proportion with signet ring cell and/or diffuse type was higher in the early-onset group (Figure 1, B). Excluding the small population of patients (n = 26) with squamous cell carcinoma and nonadenocarcinoma or signet ring cell and/or diffuse type histologies (n = 49), the tendency for early-onset esophagogastric cancer to present with gastric primary site and signet ring cell and/or diffuse type adenocarcinoma did not change (Supplementary Figure 1, B, available online). When we restricted the analysis to patients of White race, comprising 79% of the overall cohort, gastric primary tumor site remained predominant in the early-onset group (55% vs 45%, P < .001), while esophageal/gastroesophageal junction cancers were more common in the average-onset group (38% genomically stable vs 62% esophageal/gastroesophageal junction).
Most patients in both groups had metastatic disease at diagnosis (early onset = 57% and average onset = 53%, P = .29), and a similar proportion of patients had stage I or locally advanced (stage II-III) disease in both groups (Supplementary Table 1, available online). Accordingly, there was no difference in the proportion of patients who underwent resection of the primary tumor (early onset = 44% vs average onset = 45%, P = .82).
Presenting symptoms and time from diagnosis to symptom onset were collected for a total of 737 patients (n = 138 early onset, n = 599 average onset). Pain (54% vs 37%, P = .0003), and change in bowel habits (10% vs 5%, P = .02) as presenting symptoms at diagnosis were statistically significantly more frequent among patients with early-onset than among average-onset esophagogastric cancer (Figure 1, C). In contrast, weight loss (40% vs 57%, P = .0003) and swallowing difficulty (29% vs 42%, P = .0067) occurred more frequently in patients with average-onset esophagogastric cancer, likely reflecting the difference in primary tumor location. The frequencies of other presenting symptoms, including constitutional symptoms, upper GI bleeding, lower GI bleeding, change in bowel habits, symptomatic metastases, incidental findings, and respiratory or cardiac symptoms, were not statistically significantly different between groups. When considering the subgroup of patients with gastric cancer, changes in bowel habits were more common in the early-onset group (13% vs 5%, P = .01), while constitutional symptoms (including fever, night sweats, weakness, and fatigue) occurred more frequently in the average-onset group (3% vs 14%, P = .004) (Supplementary Figure 2, A, available online). In the esophageal/gastroesophageal junction subgroup, weight loss was enriched in patients with average-onset vs early-onset disease (59.7% vs 38.9%, P = .009), whereas other symptoms were statistically significantly enriched in patients with early-onset disease (6.8% vs 1.3%, P = .04) (Supplementary Figure 2, A, available online). These findings remained consistent when restricting the analysis to include only patients with adenocarcinoma and signet ring cell/diffuse–type tumors (Supplementary Figure 1, C, available online).
An exact date of symptom onset was identified by retrospective chart review for 131 patients with early-onset esophagogastric cancer and 575 patients with average-onset esophagogastric cancer. Time from symptom onset to diagnosis was longer in the early-onset than in the average-onset group (median = 90 days [interquartile range = 28.5-189.0] vs 64 days [interquartile rage = 26.5-129], P = .03) (Supplementary Figure 2, B, available online), suggesting a delay in diagnosis. In the subset of patients with esophageal/gastroesophageal junction cancers, this difference in time from symptom onset to diagnosis remained statistically significant (median time to diagnosis of early-onset disease = 100 days vs median time to diagnosis of average-onset disease = 64 days, P = .02); however, the difference was not statistically significant in the gastric cancer subgroup (median time to diagnosis of early-onset disease = 76 days vs median time to diagnosis of average-onset disease = 64.5 days, P = .21) (Supplementary Figure 2, B, available online). To determine whether patients with early-onset esophagogastric cancer were more likely to be incorrectly diagnosed with or empirically treated for a non–esophagogastric cancer diagnosis upon presentation with symptoms, we reviewed patients’ initial diagnoses and management, as documented by their health-care professionals. We found that 35% (48/138) of patients with early-onset esophagogastric cancer were treated for an alternative diagnosis compared with 21% (125/599) of those with average-onset esophagogastric cancer. The most common alternative diagnosis overall was reflux, and patients with early-onset disease were more likely to be given diagnoses of Helicobacter pylori infection or genitourinary tract pathology than patients with average-onset disease (Supplementary Figure 2, C, available online).
Considering those patients whose social history was adequately documented, a higher proportion—59% (352/599)—of patients with average-onset esophagogastric cancer had a history of smoking than the 36% (49/137) of those with early-onset esophagogastric cancer (P < .0001) (Table 1). Among smokers, 71% (250/352) and 55% (27/49) in the average-onset and early-onset groups, respectively, confirmed having quit before or at the time of their cancer diagnosis (for patients with average-onset disease, median = 22.5 years before diagnosis [range = 0-60 years]; for patients with early-onset disease, median = 2 years before diagnosis [range = 0-30 years]). In contrast, the proportion of patients reporting a history of drinking alcohol was similar between the groups (63% [271/429] vs 68% [69/102], P = .69), though a higher proportion of patients with average-onset disease consuming alcohol had a history of heavy drinking (9% [22/316] vs 3% [2/77]), defined as greater than or equal to 2 drinks/day for men or 1 drink/day for women, or reported history of alcohol use disorder.
The most frequently observed comorbidity in the early-onset group was gastroesophageal reflux disease (19%), which was also common in the average-onset group (23%). Other comorbidities that were statistically significantly more common in the average-onset group included hypertension (47% vs 13.2%), hyperlipidemia (35% vs 10%), presence of a second cancer diagnosis (24% vs 7%), and diabetes (19% vs 5%) (Supplementary Figure 2, D, available online).
Somatic mutational profile of early-onset esophagogastric cancer
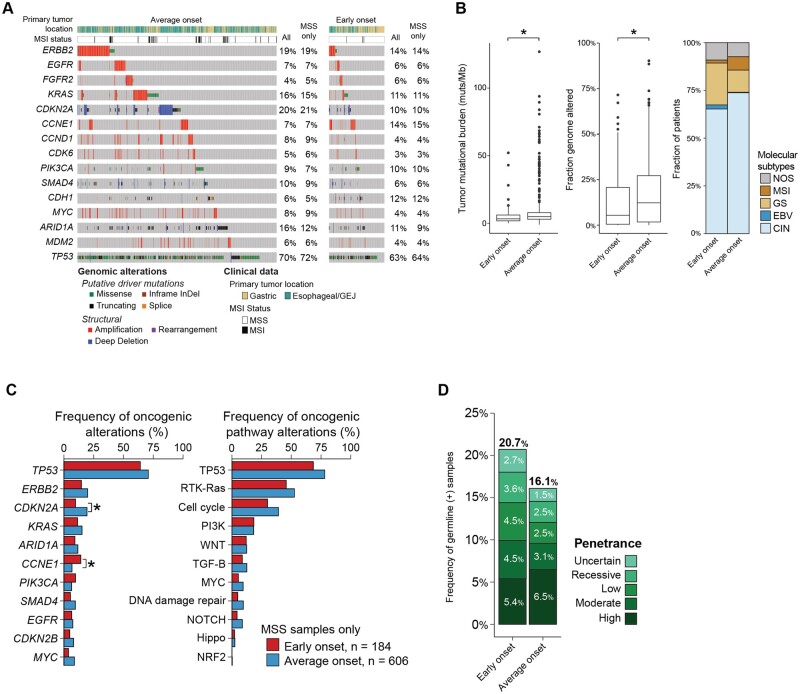
Somatic mutational profiling using MSK-IMPACT was available for 196 patients with early-onset esophagogastric cancer (90%, 23 not performed) and 706 patients with average-onset esophagogastric cancer (78%, 198 not performed) (P < .001) (Figure 2, A). Tumor mutational burden and fraction genome altered were statistically significantly higher in the average-onset group than in the early-onset group (tumor mutational burden median = 3.3 vs 4.9 alterations/megabase, P < .001; fraction genome altered median = 0.055 vs 0.132, P < .001) (Figure 2, B, Supplementary Figure 3, A, available online). We then examined molecular subtype and found that early-onset esophagogastric cancer was characterized by a higher proportion of the genomically stable and Epstein-Barr virus subtypes and a lower proportion of chromosomal instability and MSI-high subtypes than average-onset disease. Because MSI tumors are hypermutated (Supplementary Table 2, available online), we limited the remainder of the analysis to microsatellite stable tumors (early-onset group, 193/196 [98%]; average-onset group, 654/706 [93%]) (Figure 2, A).
Figure 2.
Molecular characteristics of early-onset (age <50 years) and average-onset (age ≥50 years) esophagogastric cancer. A) Oncoprint demonstrates that the most frequent oncogenic alterations were similar across the early-onset and average-onset groups (n = 196 early onset, n = 706 average onset). The genes that were altered at a statistically significantly higher frequency in the early-onset group compared with the average-onset group were CCNE1 (16% vs 7%, P = .001, Q = 0.011) and CDH1 (12% vs 6%, P = .004, Q = 0.03). B) Comparison of the tumor mutational burden (mutations/megabase), fraction genome altered, and The Cancer Genome Atlas molecular subtypes demonstrates statistically significantly lower tumor mutational burden and fraction genome altered in the early-onset group than in the average-onset group (tumor mutational burden median = 3.3 vs 4.9 mutations/megabase, P < .001; fraction genome altered median = 0.055 vs 0.132, P < .001) and a higher predominance of the genomically stable subtype. C) Comparison of the frequency of gene- and pathway-level alterations after restricting to adenocarcinoma and signet ring cell/diffuse–type tumors. CDKN2A was found to be statistically significantly enriched in average-onset tumors, whereas CCNE1 and CDH1 were enriched in early-onset tumors. D) Germline genomic analysis using a 76- to 88-gene panel showed that patients in both groups were similarly likely to have high or moderate penetrance alterations (9.9% vs 9.6%, P > .99). CIN = chromosomal instability; EBV = Epstein-Barr virus; GEJ = gastroesophageal junction; GS = gastric cancer; MSI = microsatellite instability; MSS = microsatellite stabile; NOS = not otherwise specified; ∗= P < .001.
The most frequent oncogenic alterations (>10% of patients) were similar across the early-onset and average-onset groups and included ERBB2, CDKN2A, KRAS, CCNE1, PIK3CA, CDH1, and ARID1A (Figure 2, A). The genes that were altered at a statistically significantly higher frequency in the early-onset group compared with the average-onset group were CCNE1 (16% vs 7%, P = .001, Q = 0.011) and CDH1 (12% vs 6%, P = .004, Q = 0.03) (Figure 2, A, Supplementary Figure 3, B, available online). The increased rate of CCNE1 gene alteration was driven by the chromosomal instability esophageal/gastroesophageal junction subgroup (Supplementary Figure 3, F, available online). In contrast, CDKN2A was altered at a higher frequency in the average-onset group (22% vs 11%, P < .001, Q = 0.011), and other genes with a trend toward a higher frequency of alterations in the average-onset group included ERBB2 (20% vs 15%, P = .16, Q = 0.228) and KRAS (15% vs 12%, P = .23, Q = 0.29) (Figure 2, A, Supplementary Figure 3, D, available online). After restricting the analysis to adenocarcinoma and signet ring cell/diffuse–type tumors only, CDKN2A was found to be statistically significantly enriched in average-onset tumors, whereas CCNE1 and CDH1 were enriched in early-onset tumors (Figure 2, C, Supplementary Figure 3, D, available online). No difference in rates of whole-genome duplication were observed (Supplementary Figure 3, E, available online).
Germline analysis of early-onset esophagogastric cancer
We performed germline genomic analysis on a total of 466 patients (n = 116 early onset; n = 350 average onset), of which 434 were adenocarcinoma and signet ring cell/diffuse–type tumors (n = 111 early onset, n = 323 average onset). Germline alterations were classified by penetrance as uncertain, recessive, low, moderate, or high penetrance. Of the 111 patients with early-onset esophagogastric cancer, 20.7% had a germline alteration compared with 16.1% of patients with average-onset disease (P = .3081). There was no difference in the frequency of moderate- or high-penetrance alterations between the 2 groups (9.9% vs 9.6%, P > .99) (Figure 2, D, Supplementary Figure 3, C and G, available online).
Clinical outcomes of metastatic early-onset esophagogastric cancer
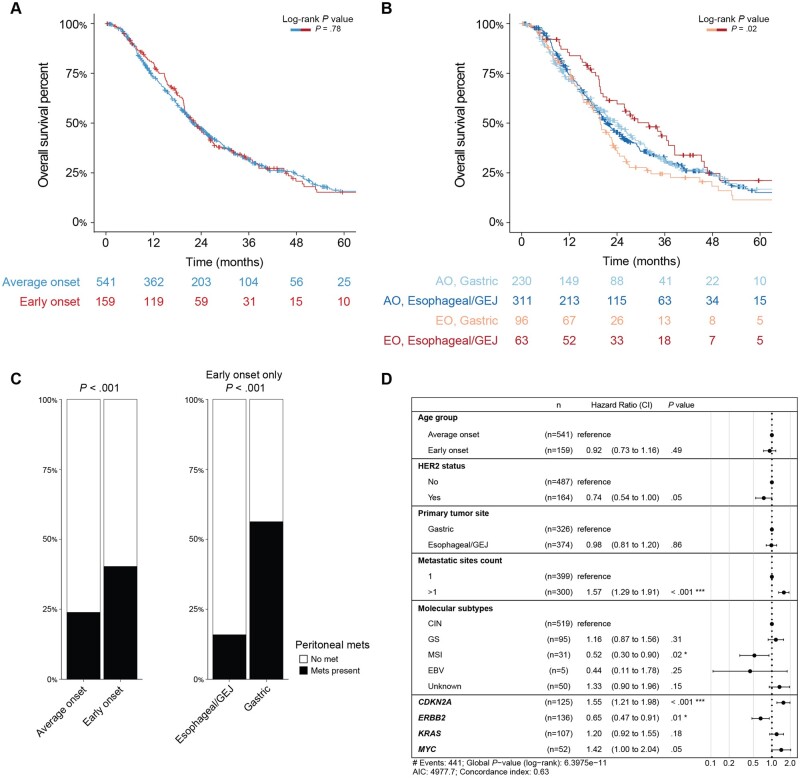
To assess whether clinical outcomes differ by age, we compared overall survival between the 2 groups. To reduce the bias introduced by longer survival for patients with earlier-stage disease, we compared overall survival from the time of metastasis to the time of death or last follow-up. In addition, patients included in the analysis were restricted to those with either adenocarcinoma or signet ring cell/diffuse–type tumors. Overall survival did not differ statistically significantly between patients with early-onset disease compared with average-onset disease (median overall survival = 22.7 vs 22.1 months; P = .78 by log-rank test) (Figure 3, A). When patients with early-onset and average-onset disease were stratified by disease site, survival time was statistically significantly longer in patients with early-onset esophageal/gastroesophageal junction cancers than in those with early-onset gastric cancer (median survival time = 32.0 vs 19.8 months, P = .024) (Figure 3, B). We hypothesized that the shorter survival time in the early-onset gastric cancer group was related to enrichment in signet ring cell/diffuse–type disease in the early-onset gastric cancer group and therefore more frequent peritoneal metastasis (40.3% vs 23.8%, P < .001) (Figure 3, C). These trends were maintained when the analysis was expanded to include squamous cell carcinoma and other tumor histologies that were previously excluded (Supplementary Figure 4, A-D, available online).
Figure 3.
Clinical outcomes of metastatic early-onset (age <50 years) and average-onset (age ≥50 years) esophagogastric cancer, restricted to adenocarcinoma and signet ring cell/diffuse–type tumors. A) Overall survival from the time of metastasis to the time of death or last follow-up did not differ statistically significantly between patients with early-onset compared with average-onset disease (median overall survival = 22.7 vs 22.1 months; P = .78 by log-rank test). B) Overall survival stratified by disease site showed that overall survival from time of metastasis to time of death or last follow-up was statistically significantly longer in patients with early-onset esophageal/gastroesophageal junction cancers and statistically significantly shorter in patients with early-onset gastric cancer (median overall survival = 32.0 vs 19.8 months, P = .02). C) Peritoneal metastasis was statistically significantly more common in the early-onset group than in the average-onset group (40.3% vs 23.8%, P < .001) and within the early-onset group occurred much more frequently in the gastric cancer group than in the esophageal/gastroesophageal junction cancer group (56.3% vs 15.9%, P < .001). D) Multivariate model generated based on variables that were statistically significant on univariate analysis demonstrated that more than 1 metastasis site and CDKN2A alterations were associated with worse outcomes. AO = average onset; CI = 95% confidence interval; CIN = chromosomal instability; EBV = Epstein-Barr virus; EO = early onset; GEJ = gastroesophageal junction; GS = gastric cancer; met = metastasis; MSI = microsatellite instability.
We examined the composite effects of clinical and genomic characteristics on survival using both univariable and multivariable analysis. Age and primary tumor site were not statistically significantly associated with survival. We created a multivariable model, including only adenocarcinoma and signet ring cell/diffuse–type tumors and selecting for those features that were statistically significant on univariable analysis as well as age and primary tumor site, which were clinically relevant. After adjustment for the statistically significant variables, age and primary tumor site were not statistically significantly associated with outcome (Figure 3, D). Genomic alterations associated with worse survival included CDKN2A (hazard ratio = 1.55, 95% confidence interval = 1.21 to 1.98; P = <.001), while ERBB2 was associated with improved survival (hazard ratio = 0.65, 95% confidence interval = 0.47 to 0.91; P = .01).
Discussion
In the largest single-institution study of early-onset esophagogastric cancer with comprehensive characterization of its clinical and genomic features, we found that differences in demographics, presenting symptoms, tumor location, and overall genomic subtype differed, while the molecular characteristics and outcomes of early-onset and average-onset esophagogastric cancer are similar.
Patients with early-onset esophagogastric cancer are more likely to have signet ring cell/diffuse–type gastric cancer, which correlates with a higher risk of occult peritoneal involvement and nondiagnostic endoscopic biopsies (24). Although the predominance of gastric cancer in the early-onset group was driven in part by the higher proportion of patients of Asian race, the gastric predominance in early-onset esophagogastric cancer persisted in the White cohort. This finding suggests that an additional factor other than race is contributing to the higher proportion of signet ring cell/diffuse–type gastric cancer in the early-onset group. As in prior reports, we found a higher proportion of women in the early-onset group. This finding is specifically concordant with that of Bautista et al. (12), a Surveillance, Epidemiology, and End Results registry study that included 1366 patients with newly diagnosed noncardia gastric adenocarcinoma and noted a distinct male to female ratio (0.84 in the <50 years age group compared with 1.52 in the >50 years age group). We found that early-onset esophagogastric cancer is more likely to be genomically stable, while average-onset esophagogastric cancer is more likely to be of the chromosomal instability or MSI subtype. These differences may in part be driven by tumor location rather than age at diagnosis, however, as the differences in molecular breakdown by age are less prominent compared with the gastric or esophageal/gastroesophageal junction subgroups.
Patients in both groups were most likely to present with stage IV disease, and survival from time of first metastasis was similar; as such, we do not find convincing evidence that esophagogastric cancer is more aggressive if it develops at a younger age, unlike other published data (12,25). The finding that 35% of patients with early-onset esophagogastric cancer were first treated for an alternative diagnosis, however, does suggest a low level of clinician awareness of the increasing incidence of esophagogastric cancer in this age group and potential delays in treatment initiation. Although some have hypothesized that early-onset esophagogastric cancer may be related to traditional risk factors, such as obesity (10), gastroesophageal reflux disease, and smoking or alcohol use, we found no difference in BMI, smoking and alcohol history, and preexisting gastroesophageal reflux disease in the early-onset group compared with the average-onset group. These risk factors may explain the overall rise in cancers of the gastric cardia or gastroesophageal junction, although we found in this study that early-onset esophagogastric cancer is characterized by a higher proportion of genomically stable and signet ring cell/diffuse–type gastric cancer, suggesting that it arises from an alternative pathway.
One potential explanation for the higher proportion of female patients and prevalent signet ring cell/diffuse–type gastric cancer in the early-onset group is the influence of endogenous or exogenous hormone exposure. Several reports have suggested that estrogen is associated with diffuse-type gastric cancer (26) and that differential exposure of estrogen receptors ɑ and β may affect tumor growth and prognosis (27-30). In organoid models of diffuse-type gastric cancer, estradiol enhances growth, particularly among cell lines with high expression of estrogen receptor α (31). Further, several bacterial species have been associated with expression of β-glucuronidases, which favor reabsorption of free estrogen; these bacteria are differentially enriched based on clinical characteristics in patients with breast cancer (32). This finding represents a potential diagnostic and therapeutic target specific to early-onset gastric cancer.
Although esophagogastric cancer confers a high mortality rate, it remains a rare disease for which screening with upper endoscopy for the general population in the United States has not been demonstrated to be cost-effective (33). Upper endoscopy may have a similarly low yield in the early-onset population in which signet ring cell/diffuse–type gastric cancer is most prevalent. Our findings raise the question of whether an alternative screening strategy is needed.
Several factors limit the generalizability of this study. First, it is a single-institution cohort with a relatively homogenous racial and ethnic distribution relative to that of the New York City area. In addition, the study population was enriched for those with ERBB2 amplification, which reflects the research expertise of the institution and may account for the relatively long median overall survival in this cohort. In addition, this study selected for patients with more advanced disease because of preferential inclusion of patients with available molecular analysis because those with more advanced disease are more likely to undergo molecular testing.
The emphasis of future research should be on identifying novel strategies to diagnose and manage early-onset esophagogastric cancer, with a focus first on young female patients with signet ring cell/diffuse–type gastric cancer. Notably, it is important to keep in mind that this young adult population presents with unique needs, similar to those with early-onset colorectal cancer (34,35). Given that overall survival from the time of first metastasis does not differ between the early-onset and average-onset groups, there is no evidence to suggest that intensifying treatment for young patients will be beneficial.
Early-onset esophagogastric cancer is characterized by a higher frequency of several presenting symptoms, a trend toward longer time from symptom onset to diagnosis, gastric primary site of disease, signet ring cell/diffuse type, and genomically stable molecular subtype compared with average-onset esophagogastric cancer. These distinctions are hypothesis generating and may provide insight into the underlying environmental mechanisms responsible for the rising incidence of early-onset esophagogastric cancer.
Supplementary Material
Acknowledgments
The funder had no role in the design of the study; the collection, analysis, or interpretation of the data; or the writing of the manuscript and decision to submit it for publication.
Contributor Information
Melissa A Lumish, Gastrointestinal Oncology Service, Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, NY, USA; Department of Medicine, Weill Cornell Medical College, New York, NY, USA.
Henry Walch, Computational Oncology, Department of Epidemiology and Biostatistics, Memorial Sloan Kettering Cancer Center, New York, NY, USA; Marie-Josée and Henry R. Kravis Center for Molecular Oncology, Memorial Sloan Kettering Cancer Center, New York, NY, USA.
Steven B Maron, Gastrointestinal Oncology Service, Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, NY, USA; Department of Medicine, Weill Cornell Medical College, New York, NY, USA.
Walid Chatila, Computational Oncology, Department of Epidemiology and Biostatistics, Memorial Sloan Kettering Cancer Center, New York, NY, USA; Marie-Josée and Henry R. Kravis Center for Molecular Oncology, Memorial Sloan Kettering Cancer Center, New York, NY, USA.
Yelena Kemel, Robert and Kate Niehaus Center for Inherited Cancer Genomics, Memorial Sloan Kettering Cancer Center, New York, NY, USA.
Anna Maio, Robert and Kate Niehaus Center for Inherited Cancer Genomics, Memorial Sloan Kettering Cancer Center, New York, NY, USA.
Geoffrey Y Ku, Gastrointestinal Oncology Service, Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, NY, USA; Department of Medicine, Weill Cornell Medical College, New York, NY, USA.
David H Ilson, Gastrointestinal Oncology Service, Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, NY, USA; Department of Medicine, Weill Cornell Medical College, New York, NY, USA.
Elizabeth Won, Gastrointestinal Oncology Service, Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, NY, USA; Department of Medicine, Weill Cornell Medical College, New York, NY, USA.
Jia Li, Gastrointestinal Oncology Service, Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, NY, USA; Department of Medicine, Weill Cornell Medical College, New York, NY, USA.
Smita S Joshi, Gastrointestinal Oncology Service, Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, NY, USA; Department of Medicine, Weill Cornell Medical College, New York, NY, USA.
Ping Gu, Gastrointestinal Oncology Service, Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, NY, USA; Department of Medicine, Weill Cornell Medical College, New York, NY, USA.
Mark A Schattner, Gastroenterology, Hepatology and Nutrition Service, Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, NY, USA.
Monika Laszkowska, Gastroenterology, Hepatology and Nutrition Service, Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, NY, USA.
Hans Gerdes, Gastroenterology, Hepatology and Nutrition Service, Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, NY, USA.
David R Jones, Department of Surgery Memorial, Sloan Kettering Cancer Center, New York, NY, USA.
Smita Sihag, Department of Surgery Memorial, Sloan Kettering Cancer Center, New York, NY, USA.
Daniel G Coit, Department of Surgery Memorial, Sloan Kettering Cancer Center, New York, NY, USA.
Laura H Tang, Department of Pathology, Memorial Sloan Kettering Cancer Center, New York, NY, USA.
Vivian E Strong, Department of Surgery Memorial, Sloan Kettering Cancer Center, New York, NY, USA.
Daniela Molena, Department of Surgery Memorial, Sloan Kettering Cancer Center, New York, NY, USA.
Zsofia K Stadler, Gastrointestinal Oncology Service, Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, NY, USA; Department of Medicine, Weill Cornell Medical College, New York, NY, USA; Robert and Kate Niehaus Center for Inherited Cancer Genomics, Memorial Sloan Kettering Cancer Center, New York, NY, USA.
Nikolaus Schultz, Computational Oncology, Department of Epidemiology and Biostatistics, Memorial Sloan Kettering Cancer Center, New York, NY, USA; Marie-Josée and Henry R. Kravis Center for Molecular Oncology, Memorial Sloan Kettering Cancer Center, New York, NY, USA.
Yelena Y Janjigian, Gastrointestinal Oncology Service, Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, NY, USA; Department of Medicine, Weill Cornell Medical College, New York, NY, USA.
Andrea Cercek, Gastrointestinal Oncology Service, Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, NY, USA; Department of Medicine, Weill Cornell Medical College, New York, NY, USA.
Data availability
The data underlying this article are available in the cBioportal repository at https://www.cbioportal.org/study/summary?id=egc_msk_2023.
Author contributions
Melissa Lumish, MD (Conceptualization; Data curation; Investigation; Project administration; Writing—original draft; Writing—review & editing), Nikolaus Schultz, PhD (Data curation; Formal analysis; Methodology; Resources), Zsofia K. Stadler, MD (Data curation; Investigation; Methodology; Writing—review & editing), Daniela Molena, MD (Investigation; Resources), Vivian E. Strong, MD (Investigation; Methodology; Resources), Laura H. Tang, MD, PhD (Data curation; Investigation; Methodology; Resources), Daniel G. Coit, MD (Investigation; Resources), Smita Sihag, MD (Investigation; Resources), David R. Jones, MD (Investigation; Resources), Hans Gerdes, MD (Investigation; Resources), Monika Laszkowska, MD, MS (Conceptualization; Investigation; Resources), Yelena Y. Janjigian, MD (Conceptualization; Formal analysis; Methodology; Project administration; Resources; Supervision; Writing—review & editing), Mark A. Schattner, MD (Investigation; Resources), Smita S. Joshi, MD (Investigation; Resources), Jia Li, MD (Investigation; Resources), Elizabeth Won, MD (Investigation; Resources), David H. Ilson, MD, PhD (Investigation; Writing—review & editing), Geoffrey Y. Ku, MD (Conceptualization; Writing—review & editing), Anna Maio, RN (Data curation), Yelena Kemel, MS (Data curation; Formal analysis; Writing—review & editing), Walid Chatila, PhD (Conceptualization; Formal analysis), Steven B. Maron, MD (Data curation; Methodology; Writing—review & editing), Henry Walch, MS (Formal analysis; Methodology; Writing—original draft; Writing—review & editing), Ping Gu, MD (Investigation; Resources), Andrea Cercek, MD (Conceptualization; Formal analysis; Methodology; Project administration; Resources; Supervision; Writing—review & editing).
Funding
Conquer Cancer, the ASCO Foundation, Young Investigator Award 2022 (M.A.L.), Tow Center for Developmental Oncology Career Development Award, 2021-2023 (M.A.L.), National Institutes of Health/National Cancer Institute Cancer Center Support Grant/Core grant (P30 CA008748) (Memorial Sloan Kettering Cancer Center).
Conflicts of interest
G.Y.K. provides services for AstraZeneca, Bristol-Myers Squibb, Merck & Co Inc, and Zymeworks Inc. D.H.I. provides services for Amgen, Astellas, Bayer, Bristol Myers Squibb, Daiichi Sankyo, Lilly Oncology, MacroGenics, Inc, Merck & Co Inc, Natera Inc, Roche, and Taiho. N.S. provides services for Cambridge Innovation Institute, Harvard T. H. Chan School of Public Health, Innovation in Cancer Informatics, and Seoul National University. Y.Y.J. has received research funding from Bayer, Bristol Myers Squibb, Cycle for Survival, the US Department of Defense, Eli Lilly, Fred’s Team, Genentech/Roche, Merck, the National Cancer Institute, and RGENIX and has served on advisory boards or in a consulting role for Amerisource Bergen, Arcus Biosciences, Astra Zeneca, Basilea Pharmaceutica, Bayer, Bristol Myers Squibb, Daiichi Sankyo, Eli Lilly, Geneos Therapeutics, GlaxoSmithKline, Imedex, Imugene, Lynx Health, Merck, Merck Serono, Michael J. Hennessy Associates, Paradigm Medical Communications, PeerView Institute, Pfizer, Research to Practice, RGENIX, Seagen, Silverback Therapeutics, and Zymeworks Inc. A.C. has consulting or advisory roles with Bayer, GlaxoSmithKline, Incyyte, Merck, Janssen, Seattle Genetics, and G1 Therapeutics and research funding from Seattle Genetics, Rgenix (Inst), and GlaxoSmithKline.
M.A.L., H.W., W.C., Y.K., A.M., E.W., J.L., S.S.J., P.G., M.A.S., M.L., H.G., D.R.J, S.S., D.G.C., L.H.T., V.E.S., D.M., and Z.K.S. have no relevant financial relationships to disclose. No other potential conflicts of interest relevant to this article exist.
References
- 1. Howlader N, Noone AM, Krapcho M, et al. Cancer Statistics Review, 1975-2017 - SEER Statistics. SEER Cancer Statistics Review, 1975-2017. Bethesda, MD: National Cancer Institute; 2019. https://seer.cancer.gov/csr/1975_2017/.
- 2. Islami F, DeSantis CE, Jemal A.. Incidence trends of esophageal and gastric cancer subtypes by race, ethnicity, and age in the United States, 1997–2014. Clin Gastroenterol Hepatol. 2019;17(3):429-439. [DOI] [PubMed] [Google Scholar]
- 3. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394-424. [DOI] [PubMed] [Google Scholar]
- 4. Ferlay J, Colombet M, Soerjomataram I, et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144(8):1941-1953. [DOI] [PubMed] [Google Scholar]
- 5. Ku GY, Kemel Y, Maron SB, et al. Prevalence of germline alterations on targeted tumor-normal sequencing of esophagogastric cancer. JAMA Netw Open. 2021;4(7):e2114753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pocurull A, Herrera-Pariente C, Carballal S, et al. Clinical, molecular and genetic characteristics of early onset gastric cancer: analysis of a large multicenter study. Cancers (Basel). 2021;13(13):3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Flegal KM, Carroll MD, Ogden CL, Curtin LR.. Prevalence and trends in obesity among US adults, 1999-2008. JAMA. 2010;303(3):235-241. [DOI] [PubMed] [Google Scholar]
- 8. Nielsen SJ, Popkin BM.. Patterns and trends in food portion sizes, 1977-1998. JAMA. 2003;289(4):450-453. [DOI] [PubMed] [Google Scholar]
- 9. Hashemi N, Loren D, Dimarino AJ, Cohen S.. Presentation and prognosis of esophageal adenocarcinoma in patients below age 50. Dig Dis Sci. 2009;54(8):1708-1712. [DOI] [PubMed] [Google Scholar]
- 10. Wu I-C, Zhao Y, Zhai R, et al. Association between polymorphisms in cancer-related genes and early onset of esophageal adenocarcinoma. Neoplasia. 2011;13(4):386-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bergquist JR, Leiting JL, Habermann EB, et al. Early-onset gastric cancer is a distinct disease with worrisome trends and oncogenic features. Surgery (United States). 2019;166(4):547-555. [DOI] [PubMed] [Google Scholar]
- 12. Bautista MC, Jiang SF, Armstrong MA, Postlethwaite D, Li D.. Impact of age on clinicopathological features and survival of patients with noncardia gastric adenocarcinoma. J. Gastric Cancer. 2014;14(4):238-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hamouda A, Forshaw M, Rohatgi A, et al. Presentation and survival of operable esophageal cancer in patients 55 years of age and below. World J Surg 2010;34(4):744-749. [DOI] [PubMed] [Google Scholar]
- 14. Brown LM, Devesa SS, Chow W-H.. Incidence of adenocarcinoma of the esophagus among white Americans by sex, stage, and age. J Natl Cancer Inst 2008;100(16):1184-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Early DS, Ben-Menachem T, Decker GA, et al. ; ASGE Standards of Practice Committee Appropriate use of GI endoscopy. Gastrointest Endosc. 2012;75(6):1127-1131. [DOI] [PubMed] [Google Scholar]
- 16. Holmes RS, Vaughan TL.. Epidemiology and pathogenesis of esophageal cancer. Semin Radiat Oncol 2007;17(1):2-9. [DOI] [PubMed] [Google Scholar]
- 17. Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor. Molecular Oncology. J Mol Diagn. 2015;17(3):251-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017;23(6):703-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chakravarty D, Gao J, Phillips S, et al. OncoKB: a precision oncology knowledge base. J Clin Oncol Precis. Oncol. 2017;1:1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sanchez-Vega F, Mina M, Armenia J, et al. ; Cancer Genome Atlas Research Network. Oncogenic signaling pathways in the cancer genome atlas. Cell. 2018;173(2):321-337.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bass A, Thorsson V, Shmulevich I, et al. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513(7517):202-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cheng D, Prasad M, Chekaluk Y, et al. Comprehensive detection of germline variants by MSK-IMPACT, a clinical diagnostic platform for solid tumor molecular oncology and concurrent cancer predisposition testing. BMC Med Genomics. 2017;10(33):1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lumish MA, Sabwa S, Maron SB, et al. Clinical and molecular characteristics of early-onset versus average-onset esophagogastric cancer. J Clin Oncol. 2021;39:250-250. doi: 10.1200/JCO.2021.39.3_suppl.250 [DOI] [Google Scholar]
- 24. Beck M, Bringeland EA, Qvigstad G, Fossmark R.. Gastric cancers missed at upper endoscopy in central Norway 2007 to 2016—a population-based study. Cancers (Basel). 2021;13(22):5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334(6052):105-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kameda C, Nakamura M, Tanaka H, et al. Oestrogen receptor-α contributes to the regulation of the hedgehog signalling pathway in ERα-positive gastric cancer. Br J Cancer. 2010;102(4):738-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Takano N, Iizuka N, Hazama S, et al. Expression of estrogen receptor-α and -β mRNAs in human gastric cancer. Cancer Lett. 2002;176(2):129-135. [DOI] [PubMed] [Google Scholar]
- 28. Xu CY, Guo JL, Jiang ZN, et al. Prognostic role of estrogen receptor alpha and estrogen receptor beta in gastric cancer. Ann Surg Oncol. 2010;17(9):2503-2509. [DOI] [PubMed] [Google Scholar]
- 29. Wang M, Pan J-Y, Song G-R, et al. Altered expression of estrogen receptor alpha and beta in advanced gastric adenocarcinoma: Correlation with prothymosin alpha and clinicopathological parameters. Eur J Surg Oncol. 2007;33(2):195-201. [DOI] [PubMed] [Google Scholar]
- 30. Ryu W-S, Kim J-H, Jang Y-J, et al. Expression of estrogen receptors in gastric cancer and their clinical significance. J Surg Oncol. 2012;106(4):456-461. [DOI] [PubMed] [Google Scholar]
- 31. Kang S, Park M, Cho JY, et al. Tumorigenic mechanisms of estrogen and Helicobacter pylori cytotoxin-associated gene A in estrogen receptor α-positive diffuse-type gastric adenocarcinoma. Gastric Cancer. 2022;25(4):678-696. [DOI] [PubMed] [Google Scholar]
- 32. Luu TH, Michel C, Bard J-M, et al. Intestinal proportion of Blautia sp. is associated with clinical stage and histoprognostic grade in patients with early-stage breast cancer. Nutr Cancer. 2017;69(2):267-275. [DOI] [PubMed] [Google Scholar]
- 33. Gupta N, Bansal A, Wani S, Gaddam S, Rastogi A, Sharma P. Endoscopy for upper GI cancer screening in the general population: a cost-utility analysis. Gastrointest. Endosc. 2011;74(3):610-624. [DOI] [PubMed] [Google Scholar]
- 34. Mendelsohn R, Palmaira RL, Lumish M, et al. A coordinated clinical center for young onset colorectal cancer. Oncologist. 2021;26(8):625-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lumish MA, Cercek A.. Practical considerations in diagnosing and managing early-onset GI cancers. J Clin Oncol. 2022;40(24):2662-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the cBioportal repository at https://www.cbioportal.org/study/summary?id=egc_msk_2023.