Abstract
Background:
This study aimed to explore the biomarkers of Alzheimer’s disease (AD).
Methods:
The microarray data of GSE16759 were from the expression profile samples of 4 parietal lobe tissues from patients with AD and 4 ones from age-matched control participants. The differentially expressed micro RNAs (miRNAs) and genes (DEGs) underwent hierarchical clustering and function analysis followed by target genes prediction. Finally, DEGs were mapped to the target genes to construct miRNA-regulated networks.
Results:
A total of 427 DEGs were obtained and clustered into 5 functions. After DEGs were mapped to the predicted target genes, 313 regulatory pairs were established. The target genes SEC22 vesicle trafficking protein homolog B (SEC22B) and SEC63 homolog (SEC63) regulated by miRNA-206, RAB10, member RAS oncogene family (RAB10) regulated by miRNA-655, and fms-related tyrosine kinase 1 (FLT1) regulated by miRNA-30e-3p and miRNA-369-3p were involved in the biological processes of protein transport and regulation of cell motion.
Conclusion:
The target genes SEC22B, RAB10, and FLT1 may be potential biomarkers of AD.
Keywords: Alzheimer’s disease, micro RNA, differentially expressed genes, functional cluster, target genes, regulatory network
Introduction
Alzheimer’s disease (AD) is a progressive and ultimately fatal neurodegenerative disease affecting around 35 million people worldwide. 1 Neuropathology results in premature neuronal loss. 2 Due to the nature of the primarily affected neuronal circuits, the clinical symptoms of AD are progressive impairment in memory, judgment, decision making, distorted language, and orientation to physical surroundings. 3 This illness is the leading cause of dementia in older people. At present, there is no definitive diagnostic test or therapy for this disease, and its pathogenesis is still debated and may be heterogeneous. 4,5
For the pathogenetic mechanisms of AD, 1 mechanism centers on the “amyloid cascade” hypothesis that the origins of AD involve an imbalance between production and clearance of β-amyloid (Aβ) initiating pathogenesis. 6 Other hypotheses for AD initiation include pathogen infection and epigenetic modifications caused by environmental insults. 5 Recently, an identified class of nonprotein-coding small RNAs, micro RNAs (miRNAs), may provide new insights into AD research. Micro RNAs are small regulatory RNAs that participate in posttranscriptional gene regulation in a sequence-specific manner. They bind complementary sequences in target messenger RNAs (mRNAs) and can thus lead to their selective degradation or selective inhibition of translation. 7 Meta-analysis of 100 public microarray data sets has suggested that AD progresses with changes in functional gene groups, miRNA, and epigenetic changes. 5 Genome-wide association study indicates that AD-specific miRNA expression patterns can be identified in the brain, cerebral spinal fluid, and blood. 8,9 Hébert et al showed that the decreased levels of miRNA-29a/b in AD caused the increased levels of Aβ cleaving enzyme 1 (BACE1) in vitro. The BACE1 is an essential protein in the generation of Aβ from amyloid precursor protein (APP). 10 The deposition of APP in plaques in brain tissue may lead to AD. 6 Wang et al found that decreased levels of miRNA-107 in AD also targeted BACE1. 11 In addition, studies have confirmed that some miRNAs, such as miRNA-29, miRNA-15, and miRNA-107, are specifically altered in the AD brain. 12,13 However, the present knowledge about the molecular mechanism of AD seems to be insufficient.
In the present study, we aimed to extend our understanding of the mechanisms of AD. We downloaded the microarray data of miRNA and mRNA in the parietal lobe cortex of patients with AD and age-matched controls from Gene Expression Omnibus (GEO) database and analyzed the differentially expressed miRNAs and genes (DEGs). Then, we analyzed the functional similarity of DEGs and miRNA target prediction. Besides, we constructed interaction networks to study and identify the potential target genes for diagnosis and treatment of AD. Figure 1 outlines the detailed workflow of the entire procedure.
Figure 1.

Analysis process of this study.
Materials and Methods
Affymetrix Microarray Data
Alzheimer’s disease miRNA and mRNA expression profile data were accessible at National Center for Biotechnology Information GEO (http://www.ncbi.nlm.nih.gov/geo/) database using the series accession number GSE16759. 8 The base data are based on the platform of GPL570 [HG-U133_Plus_2] Affymetrix Human Genome and U133 Plus 2.0 Array and of GPL8757 USC/XJZ Human 0.9 K miRNA-940-v1.0. Eight samples including 4 parietal lobe tissues from patients with AD and 4 ones from age-matched control participants were available for every platform. The average age of patients with AD was 85 years (range 75-92 years) and 91.8 years (90-96 years) for controls. The raw data and the annotation files were downloaded for further study.
Data Preprocessing and Analysis of Differentially Expressed miRNAs and DEGs
The original Simple Omnibus Format in Text-formatted expression data were first converted into identifiable expression form. The miRNAs were corresponded to genes to remove the empty probes. Then, the expression levels of the data were log2-transformed. 14 The limma package (Linear Models for Microarray Data) 15 in R language was used to identify differentially expressed miRNAs and DEGs between patients with AD and healthy controls. The multiple testing correction was performed using a Beniamini-Hochberg false discovery rate (HB FDR). 16 Only miRNAs and DEGs with FDR < 0.05 and |log fold change (FC)| > 1 were selected.
Comparison of Differential Expression Values
Hierarchical clustering 17,18 is a common analytical tool used to determine the closest associations among specimens under evaluation and gene profiles in multidimensional spaces. According to the corresponding probe information of DEGs and differentially expressed miRNAs, the expression values of genes with significant characteristics of expression differences in each samples were extracted from the expression values file. Two-way hierarchical clustering 18 based on the Euclidean distance 19 was performed using the Pretty Heatmaps package 20 in R language. Two separate heatmaps of differentially expressed miRNA and DEGs were generated.
Functional Similarity Analysis of DEGs
Ground-Operation Simulation (GOSim) 21 package is used to measure the functional similarity of genes based on a variety of information-theoretic similarity concepts for Gene Ontology (GO) terms. In this article, GOSim package in R language was used to cluster DEGs according to their biological process, with respect to the GO annotation. This method was based on calculating the distance between genes and ancestor nodes. Genes with similar function were located on the same nodes, and P value < 0.05 was chosen as the threshold.
Micro RNA Target Prediction
Identification of the predicted target mRNA genes of miRNAs provides the basis for understanding the miRNA functions. miRDB 22 is used for miRNA target prediction and functional annotation of animals. MicroRNA.org (http://www.microrna.org) 23 that is exploited by Memorial Sloan-Kettering Cancer Center is a comprehensive resource of miRNA target predictions and expression profiles. In this article, the intersection of target genes in both of the databases mentioned earlier was selected as predictive target genes. Then, the DEGs were mapped to the intersection of target genes to select the genes that were both predicted target genes and DEGs. Finally, the selected genes and their corresponding miRNAs were retained to perform functional clusters analysis.
Regulatory Interaction Analysis and Network Construction
Search Tool for the Retrieval of Interacting Genes (STRING) 24 is an online database that has been designed as a comprehensive perspective to evaluate interaction information of proteins. In this article, the STRING online tool was applied to analyze the interaction information of DEGs, and only the experimentally validated interactions with a confidence score > 0.6 were considered to be significant. The miRNA regulatory networks were then constructed using the Cytoscape 25 software based on the interactome.
Results
Identification of Differentially Expressed miRNAs and DEGs
A total of 52 differentially expressed miRNAs including 20 upregulated and 32 downregulated miRNAs and 427 DEGs including 158 upregulated and 269 downregulated DEGs were selected. This set of differentially expressed miRNAs and DEGs was used for hierarchical clustering analysis.
Comparison of Differential Expression Values
After hierarchical clustering analysis of differentially expressed miRNAs and DEGs, significant differences were found in both differentially expressed miRNAs and DEGs between the AD and the control groups (Figure 2). Simultaneously, hierarchical clustering distinguished AD and normal tissues according to the expressing values.
Figure 2.
The cluster heat map of differentially expressed microRNAs (miRNAs; A) and differentially expressed genes (DEGs; B). Red stands for high expression value and blue stands for low expression value. The changes of color from blue to red stands for the changes in expression value from low to high.
Functional Similarity Analysis of DEGs
According to functional similarity clustering, the 427 DEGs were clustered into 5 functions including protein transport, RNA splicing, intracellular protein transport, regulation of cell motion, and protein complex assembly with a P value of < 0.05 (Table 1).
Table 1.
Functional Classification of Differentially Expressed Genes.a
| Cluster Name | Count | Enrichment P value | Biological Function |
|---|---|---|---|
| C1 | 112 | 2.71 × 104 | GO:0015031 protein transport |
| GO:0045184 establishment of protein localization | |||
| GO:0008104 protein localization | |||
| C2 | 33 | 1.7 × 103 | GO:0000375 RNA splicing via transesterification reactions |
| GO:0000377 RNA splicing via transesterification reactions with bulged adenosine as nucleophile | |||
| GO:0000398 nuclear mRNA splicing via spliceosome | |||
| C3 | 54 | 6.19 × 103 | GO:0006886 intracellular protein transport |
| GO:0034613 cellular protein localization | |||
| GO:0070727 cellular macromolecule localization | |||
| C4 | 32 | 1.22 × 102 | GO:0051270 regulation of cell motion |
| GO:0030334 regulation of cell migration | |||
| GO:0040012 regulation of locomotion | |||
| C5 | 60 | 3.27 × 102 | GO:0070271 protein complex biogenesis |
| GO:0006461 protein complex assembly | |||
| GO:0065003 macromolecular complex assembly |
Abbreviation: mRNA, messenger RNA.
aCount stands for the number of enriched differentially expressed genes.
Micro RNA Target Prediction
A total of 8609 regulatory pairs between the differentially expressed miRNA and their predictive target genes were obtained. After DEGs were mapped to the predicted target genes, 313 regulatory pairs were established between 52 differentially expressed miRNAs and 427 DEGs. The miRNA target genes in 313 regulatory pairs were then clustered into 2 function classifications, C1 and C2 (Table 2) that were related to protein transport and regulation of cell motion, respectively.
Table 2.
Functional Classification of Differentially Expressed Target Genes.a
| Cluster Name | Count | Enrichment P value | Biological Function |
|---|---|---|---|
| C1 | 55 | 1.58 × 103 | GO:0015031 protein transport |
| GO:0045184 establishment of protein localization | |||
| GO:0008104 protein localization | |||
| C2 | 19 | 2.55 × 102 | GO:0051270 regulation of cell motion |
| GO:0030334 regulation of cell migration | |||
| GO:0040012 regulation of locomotion |
aCount stands for the number of enriched differentially expressed target genes.
Regulatory Interaction Analysis and Network Construction
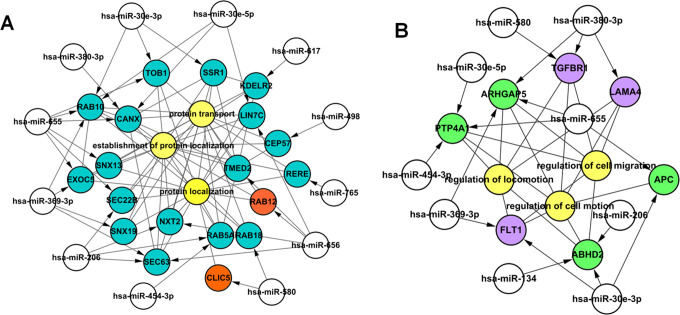
After the interaction association of DEGs in C1 and C2 was analyzed using STRING online tool, only 55 DEGs in C1 had interaction association. Combining the biological processes and interaction association of DEGs in C1 and C2, 2 regulatory networks were constructed using Cytoscape software. The network of C1 contained 34 nodes and 12 miRNAs, and the network of C2 contained 19 nodes and 9 miRNAs (Figure 3). The target genes SEC22 vesicle trafficking protein homolog B (SEC22B) and SEC63 homolog (SEC63) regulated by miRNA-206; RAB10, member RAS oncogene family (RAB10) regulated by miRNA-369-3p, miRNA-30e-5p, miRNA-30e-3p, and miRNA-655; and fms-related tyrosine kinase 1 (FLT1) regulated by miRNA-30e-3p and miRNA-369-3p were involved in the biological processes of protein transport and regulation of cell motion.
Figure 3.
The constructed regulatory networks of microRNAs (miRNAs). A, The regulatory network of miRNAs was constructed based on function of protein transport. B, The regulatory network of miRNAs was constructed based on function of regulation of cell motion. Yellow nodes: Gene Ontology biological process terms; white nodes: differentially expressed genes (DEGs); orange and purple nodes: upregulated target genes; blue and green nodes: downregulated target genes.
Discussion
This study aimed to explore the potential role of miRNAs in pathogenesis of AD. Combined with miRNA and mRNA expression profile information, we further screened the miRNA target genes. A total of 52 differentially expressed miRNAs and 427 DEGs were obtained, and 313 pairs of regulatory association were constructed. The miRNA-206 participated in the biological processes of both protein transport and regulation of cell motion. The target genes such as RAB10 and FLT1 regulated by miRNA-369-3p and miRNA-30e-3p were also involved in the biological processes of protein transport and regulation of cell motion.
Micro RNAs are believed to suppress target gene expression through the binding of miRNA “seed” regions to complementary sequences of 3’-untranslated regions of target genes. 26 In the central nervous system, balanced expression of miRNA plays an important role in preventing neurodegeneration. In this study, miRNA-206 was overexpressed in AD. Interestingly, miRNA-206 appears to be present at a very low level in normal brains but at an aberrantly high levels in diseased brains, such as in AD. 27 The miRNA-206 has also been found upregulated in cerebral ischemia and neurotoxicant exposure models. 28 Lee et al found that miRNA-206 participated in the pathogenesis of AD by suppressing the expression of brain-derived neurotrophic factor. 27 Additionally, the current study showed that target genes, SEC22B and SEC63, regulated by miRNA-206 participated in the biological processes of protein transport and regulation of cell motion. SEC22B and SEC63 have been found to be associated with protein trafficking and translocation and downregulation in the hippocampus of aging and AD brains. 29,30 In patients with AD, a disequilibrium of proteostasis in the brain leads to the accumulation of Aβ. 1,31 There’s a growing theme that defects in transport are closely associated with neurodegeneration. Krohn et al have determined that transport protein has a central role in clearing Aβ from the brain of mice. 32 Thus, miRNA-206 and its target genes SEC22B and SEC63 may be key regulators underlying the development of AD.
Our results also revealed that miRNA-655 was another significant miRNA enriched with DEGs such as RAB10. 33 RAB proteins have been found to be responsible for controlling the level of Aβ which are generated from APP by β- and γ-secretases. 34 The accumulation of Aβ in the brain is central to the pathogenesis of AD. 35 RAB10 has been confirmed to decrease Aβ that may be realized by affecting γ-secretase cleavage or the fate of Aβ. 34 RAB11, one of the RAB proteins, has high homology with RAB10. Udayar et al showed that RAB11 controlled β-secretase endosomal recycling to the plasma membrane and thus affected Aβ production. 34 Study suggests that miRNA-655 participates in the process of neuronal differentiation, 36 which may be related to the pathophysiology of AD. In addition, from the regulatory network, we found that RAB10 was also the target gene of miRNA-369-3p, -30e-3p, and -30e-5p. Satoh who analyzed molecular network of AD miRNA also extracted differentially expressed miRNAs of miRNA-655, -30e-3p, and -30e-5p in AD brains versus normal brains. 37 Although the present evidence of direct association between miRNA-655, -369-3p, -30e-3p, -30e-5p, and RAB10 progression are rare, miRNA-655, -369-3p, -30e-3p, -30e-5p, and its target gene RAB10 may be critical for progression of AD based on our results.
In addition, except for target gene RAB10, miRNA-369-3p and miRNA-30e-3p were also found to regulate FLT1. Ryu et al reported that the expression of FLT1 was significantly increased in the tissue of patients with AD. The enhanced FLT1 expression contributes to Aβ deposits and microglial autocrine or paracrine processes. 38 Chronic inflammation is integral to AD pathology. 39 A critical component of inflammatory response is an increased microglial mobility and chemotactic activity to Aβ deposition in the AD brain. FLT1 can mobilize microglia and cellular chemotactic to serve critical response functions in the AD brain. 38 The regulatory network suggested that FLT1 was regulated by miRNA-369-3p and miRNA-30e-3p, but there were few studies about the relationship between AD and miRNA of miRNA-369-3p and miRNA-30e-3p. Therefore, we speculate that these miRNAs and the target genes related to the key biological processes may be the therapeutic target for AD.
In summary, the target genes SEC22B and SEC63 regulated by miRNA-206; RAB10 regulated by miRNA-369-3p, miRNA-30e-5p, miRNA-30e-3p, and miRNA-655; and FLT1 regulated by miRNA-30e-3p and miRNA-369-3p may play key roles in the progression and development of AD. These significant miRNAs showed potential perspective in the treatment of AD. The miRNAs identified in our work may be potential therapeutic targets for AD.
However, there were a few limitations in our preliminary study. The sample size for microarray analysis was small which may cause a high rate of false-positive result. Besides, further studies are necessary for verifying the clinical applications of these miRNAs as biological targets for treatment of AD.
Footnotes
This article was accepted under the editorship of the former Editor-in-Chief, Carol F. Lippa.
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors received no financial support for the research, authorship, and/or publication of this article.
References
- 1. Querfurth HW, LaFerla FM. Alzheimer’s disease. N Engl J Med. 2010;362(4):329–344. [DOI] [PubMed] [Google Scholar]
- 2. Wenk GL. Neuropathologic changes in Alzheimer’s disease. J Clin Psychiatry. 2003;64(suppl 9):7–10. [PubMed] [Google Scholar]
- 3. Thies W, Bleiler L. 2011 Alzheimer’s disease facts and figures. Alzheimers Dement. 2011;7(2):208–244. [DOI] [PubMed] [Google Scholar]
- 4. Rosales-Corral SA, Acuña-Castroviejo D, Coto-Montes A, et al. Alzheimer’s disease: pathological mechanisms and the beneficial role of melatonin. J Pineal Res. 2012;52(2):167–202. [DOI] [PubMed] [Google Scholar]
- 5. Anderson DC. Alzheimer’s disease biomarkers: more than molecular diagnostics. Drug Dev Res. 2013;74(2):92–111. [Google Scholar]
- 6. Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297(5580):353–356. [DOI] [PubMed] [Google Scholar]
- 7. Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. [DOI] [PubMed] [Google Scholar]
- 8. Nunez-Iglesias J, Liu CC, Morgan TE, Finch CE, Zhou XJ. Joint genome-wide profiling of miRNA and mRNA expression in Alzheimer’s disease cortex reveals altered miRNA regulation. PloS One. 2010;5(2): e8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cogswell JP, Ward J, Taylor IA, et al. Identification of miRNA changes in Alzheimer’s disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis. 2008;14(1):27–41. [DOI] [PubMed] [Google Scholar]
- 10. Hébert SS, Horré K, Nicolaï L, et al. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/β-secretase expression. Proc Natl Acad Sci. 2008;105(17):6415–6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang WX, Rajeev BW, Stromberg AJ, et al. The expression of microRNA miR-107 decreases early in Alzheimer’s disease and may accelerate disease progression through regulation of β-site amyloid precursor protein-cleaving enzyme 1. J Neurosci. 2008;28(5):1213–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Delay C, Mandemakers W, Hébert SS. MicroRNAs in Alzheimer’s disease. Neurobiol Dis. 2012;46(2):285–290. [DOI] [PubMed] [Google Scholar]
- 13. Lukiw WJ, Andreeva TV, Grigorenko AP, Rogaev EI. Studying micro RNA function and dysfunction in Alzheimer’s disease. Front Genet. 2012;3:327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fujita A, Sato JR, Rodrigues LO, Ferreira CE, Sogayar MC. Evaluating different methods of microarray data normalization. BMC Bioinformatics. 2006;7:469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Smyth GK. Limma: Linear Models for Microarray Data. Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer; 2005. p. 397–420. [Google Scholar]
- 16. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B. 1995;57(1):289–300. [Google Scholar]
- 17. Yamaguchi U, Nakayama R, Honda K, et al. Distinct gene expression-defined classes of gastrointestinal stromal tumor. J Clin Oncol. 2008;26(25):4100–4108. [DOI] [PubMed] [Google Scholar]
- 18. Szekely GJ, Rizzo ML. Hierarchical clustering via joint between-within distances: extending Ward’s minimum variance method. J Classif. 2005;22(2):151–183. [Google Scholar]
- 19. Deza MM, Deza E. Encyclopedia of Distances. Berlin, Heidelberg: Springer; 2009. [Google Scholar]
- 20. Kolde R. pheatmap: Pretty Heatmaps. R package version 0.7. 7. 2012.
- 21. Fröhlich H, Speer N, Poustka A, Beißbarth T. GOSim–an R-package for computation of information theoretic GO similarities between terms and gene products. BMC Bioinformatics. 2007;8:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang X. miRDB: a microRNA target prediction and functional annotation database with a wiki interface. RNA. 2008;14(6):1012–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36(database issue):D149–D153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Szklarczyk D, Franceschini A, Kuhn M, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39( database issue ):D561–D568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Smoot ME, Ono K, Ruscheinski J, Wang P-L, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27(3):431–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cai Y, Yu X, Hu S, Yu J. A brief review on the mechanisms of miRNA regulation. Genomics Proteomics Bioinformatics. 2009;7(4):147–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee ST, Chu K, Jung KH, et al. miR-206 regulates brain-derived neurotrophic factor in Alzheimer disease model. Ann Neurol. 2012;72(2):269–277. [DOI] [PubMed] [Google Scholar]
- 28. Jeyaseelan K, Lim KY, Armugam A. MicroRNA expression in the blood and brain of rats subjected to transient focal ischemia by middle cerebral artery occlusion. Stroke. 2008;39(3):959–966. [DOI] [PubMed] [Google Scholar]
- 29. Chagoyen M, Carmona-Saez P, Shatkay H, Carazo JM, Pascual-Montano A. Discovering semantic features in the literature: a foundation for building functional associations. BMC Bioinformatics. 2006;7:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Berchtold NC, Coleman PD, Cribbs DH, Rogers J, Gillen DL, Cotman CW. Synaptic genes are extensively downregulated across multiple brain regions in normal human aging and Alzheimer’s disease. Neurobiol Aging. 2013;34(6):1653–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319(5865):916–919. [DOI] [PubMed] [Google Scholar]
- 32. Krohn M, Lange C, Hofrichter J, et al. Cerebral amyloid-β proteostasis is regulated by the membrane transport protein ABCC1 in mice. J Clin Invest. 2011;121(10):3924–3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bao S, Shen X, Shen K, Liu Y, Wang XF. The mammalian Rad24 homologous to yeast Saccharomyces cerevisiae Rad24 and Schizosaccharomyces pombe Rad17 is involved in DNA damage checkpoint. Cell Growth Differ. 1998;9(12):961–967. [PubMed] [Google Scholar]
- 34. Udayar V, Buggia-Prévot V, Guerreiro Rita L, et al. A Paired RNAi and RabGAP Overexpression Screen Identifies Rab11 as a Regulator of β-Amyloid Production. Cell Reports. 2013;5(6):1536–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Marzolo MP, Bu G. Lipoprotein receptors and cholesterol in APP trafficking and proteolytic processing, implications for Alzheimer’s disease. Semin Cell Dev Biol. 2009;20(2):191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pallocca G, Fabbri M, Nerini-Molteni S, et al. Changes in miRNA expression profiling during neuronal differentiation and methyl mercury-induced toxicity in human in vitro models. Toxics. 2014;2(3):443–463. [Google Scholar]
- 37. Satoh JI. Molecular network analysis of human microRNA targetome: from cancers to Alzheimer’s disease. BioData Min. 2012;5(1):1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ryu JK, Cho T, Choi HB, Wang YT, McLarnon JG. Microglial VEGF receptor response is an integral chemotactic component in Alzheimer’s disease pathology. J Neurosci. 2009;29(1):3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mrak RE, Griffin WST. Common inflammatory mechanisms in Lewy body disease and Alzheimer disease. J Neuropath Exp Neur. 2007;66(8):683–686. [DOI] [PubMed] [Google Scholar]