Abstract
The incidence of Alzheimer’s disease (AD) has been increasing in the recent years but the underlying mechanisms remain uncertain. This study aimed to analyze the differentially expressed genes (DEGs) in entorhinal cortex with AD and identify featured genes related to AD. Gene expression profile GSE5281 was downloaded from Gene Expression Omnibus, including 10 AD and 13 control samples. Differentially expressed genes were identified by Student t test including 119 upregulated and 591 downregulated DEGs. Then, we obtained 14 enrichment Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways among which 11 pathways were significantly enriched (adjusted P value < .05). The KEGG pathway network which was constructed by 14 KEGG pathways showed that 6-phosphofructokinase, muscle type, phosphoglucomutase 1, aldolase A, and adolase C had high degree. Glycometabolism pathways network which was constructed by 4 glycometabolism pathways showed that adenosine triphosphate (ATP) synthase, H+transporting, mitochondrial F1 complex ATP5B, ATP5C1, ATP5D, and ATP5G1 had high degree related to ATP metabolism. These findings suggested that these genes with high degree may be the underlying potential therapeutic targets for AD.
Keywords: Alzheimer’s disease, entorhinal cortex, glycometabolism pathway, differentially expressed genes
Introduction
Alzheimer’s disease (AD) is the most progressive neurodegenerative disorder 1 and a leading cause of dementia in the elderly patients. 2 It is estimated that 27 million people are affected worldwide. 3 The typical clinical presentation of patients with AD is progressive loss of memory and cognitive function, ultimately leading to a loss of independence and causing a heavy personal toll on the patient. 4 Although extensive investigations into this disease have taken place over the past few decades, the exact cause of this disease is yet to be elucidated.
To the best of our knowledge, the brain is a heavy user of metabolic energy requiring 25% of the body’s glucose supplies. It mainly relies on glucose for energy. The major energy metabolism systems of brain includes glycolytic pathway, tricarboxylic acid cycle, and oxidative phosphorylation. 5 Longitudinal studies show that metabolic insufficiency has been proposed to be involved in AD from the early stage of the disease, 6 and treatment of targeting metabolic insufficiency can increase cognitive performance. On a molecular-level analysis, the expression of some genes involved in energy metabolism has been shown to be significantly downregulated in the AD hippocampus. 5 Glyceraldehyde 3-phosphate dehydrogenase, downregulated in AD, 5,7 plays a role in glycolysis and phosphorylation. Glutamic–oxaloacetic transaminase 1, containing a susceptibility locus for late-onset AD, has also been shown to be downregulated in AD. 8 In addition, a number of downregulated genes in hippocampus were related to metabolism, including triosephosphate isomerase, adenosine triphosphate (ATP) citrate lyase, and malate dehydrogenase 2. The entorhinal cortex (EC), a part of the temporal cortex, is divided into superficial (I-III) and deep layers (IV-VI) that show differential anatomical and functional organization. 9 The superficial layers are the main recipient of intracortical information and the major output source to the hippocampus. Entorhinal dysfunction is involved in several brain diseases, including AD. 10 The AD-related neuronal loss and atrophy of the EC are well documented in patients. 11 –13 However, little is known about AD-associated gene expression changes related to metabolism in EC.
In order to further evaluate the role of energy metabolism in EC associated with AD, the present study analyzed differentially expressed genes (DEGs) between AD and controls (CTs). Then, we applied bioinformatics tools to identify featured DEGs involved in glycometabolism. We believed that molecular evaluation of the EC from metabolically affected brain could provide new information about the pathogenesis of AD and new therapeutic targets for AD.
Materials and Methods
Affymetrix Microarray Data
The microarray data were downloaded from Gene Expression Omnibus (GEO) database and the accession number was GSE5281 14,15 including 10 AD and 13 CT samples. All the analytical tissue samples were from EC. The microarray expression platform was Affymetrix Human Genome U133 plus 2.0.
Data Preprocessing
The data in CEL files were converted into expression profile and background correction and quartile data normalization were performed by the robust multiarray average 16 with affy package. For genes corresponding to multiple probe sets which have a plurality of expression values, the gene expression values of those probe sets were averaged. Eventually, expression profiles of 19 803 genes were obtained from 23 specimens.
Differentially Expressed Genes Analysis
Student t test was used to identify genes that were significantly differentially expressed between AD and CT samples. The P value less than .01 adjusted by Benjamin and Hochberg (BH) method 17 and fold change (FC) value larger than 2 was set as the threshold criteria for screening DEGs. 18
Gene Ontology Enrichment Analysis
Gene Ontology (GO) is a bioinformatics project developed by the GO Consortium that aims to introduce consistency in the description of functional information pertaining to gene products. 19 The GO consists of 3 ontologies used to describe the biological processes, molecular functions, and cellular component of a gene product. 20 The Database for Annotation, Visualization and Integrated Discovery (DAVID) consists of an integrated biological knowledgebase and functional annotation chart or table. It provides a comprehensive set functional annotation tools for investigators to integrate remarkable genes of specific function. 21 The EASE method 22 was used to calculate a significant value for GO categories after all DEGs being input into DAVID. The P value adjusted by BH less than .05 and EASE score of 0.01 were chosen as cutoff criteria.
Kyoto Encyclopedia of Genes and Genomes Pathway Analysis
The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database (http://www.kegg.jp/) consists of graphical diagrams of biochemical pathways including most of the known metabolic pathways and some known regulatory pathways. 23 We mapped these DEGs to this database and searched important pathways associated with AD. The P value adjusted by BH less than .05 and EASE score of 0.01 were chosen as cutoff criteria. Then, some featured pathways were screened to construct perturbing pathway network closely related to AD by Cytoscape. 24 To get a better understanding of selected DEGs in pathway network, the degree was calculated.
Results
Screening of DEGs
We obtained publicly available microarray data set GSE5281 from GEO database. Student t test was used to analyze DEGs between 10 AD and 13 CT samples. According to threshold criteria (adjusted P value < .01 and FC value > 2) for DEGs, in total, 710 DEGs were identified including 119 upregulated and 591 downregulated DEGs.
Gene Ontology Enrichment Analysis of DEGs
To investigate AD gene expression on a functional level, DEGs (adjusted P value < 0.01 and FC value > 2) between AD and CT samples were significantly enriched in 136 GO terms. The top 10 GO terms (P value < .01) are shown in Table 1, including ribonucleotide metabolic process and ATP metabolic process.
Table 1.
Gene Ontology Terms (Top 10) With Adjusted P Value < .05.a
| Terms | Biological Process | P Value |
|---|---|---|
| GO:0009259 | Ribonucleotide metabolic process | 1.77E-06 |
| GO:0009260 | Ribonucleotide biosynthetic process | 2.73E-06 |
| GO:0009150 | Purine ribonucleotide metabolic process | 2.90E-06 |
| GO:0009152 | Purine ribonucleotide biosynthetic process | 5.53E-06 |
| GO:0009199 | Ribonucleoside triphosphate metabolic process | 6.19E-06 |
| GO:0009201 | Ribonucleoside triphosphate biosynthetic process | 1.44E-05 |
| GO:0009142 | Nucleoside triphosphate biosynthetic process | 2.03E-05 |
| GO:0009141 | Nucleoside triphosphate metabolic process | 2.37E-05 |
| GO:0009205 | Purine ribonucleoside triphosphate metabolic process | 2.38E-05 |
| GO:0046034 | ATP metabolic process | 2.84E-05 |
Abbreviations: GO, Gene Ontology; ATP, adenosine triphosphate; BH, Benjamin and Hochberg.
a Adjusted P value means P value adjusted by BH.
Kyoto Encyclopedia of Genes and Genomes Pathway Analysis and Network Construction
We mapped 710 DEGs into KEGG pathway database to screen enrichment pathways and obtained 14 significant pathways, among which 11 pathways were significantly enriched (adjusted P value < .05, Table 2). There were 4 glycometabolism pathways (adjusted P value < .05, Table 2) including oxidative phosphorylation, glycolysis/gluconeogenesis, pentose phosphate pathway, and fructose and mannose metabolism. By integrating these 14 pathways and 4 glycometabolism pathways, respectively, 2 perturbing pathway networks were constructed using Cytoscape (Figures 1 and 2). Then, the degrees of DEGs in these 2 networks were calculated, respectively. Our results showed that 17 DEGs and 16 DEGs with the degree of no less than 5 were screened, respectively (Tables 3 and 4). Several featured DEGs with higher degree were identified including 6-phosphofructokinase, muscle type (PFKM), phosphoglucomutase 1 (PGM1), aldolase A (ALDOA), and adolase C (ALDOC), which were all downregulated in AD samples compared with CT samples (the adjusted P value was 6.82E-05, 9.85E-05, 9.01E-05, and .0002, respectively, and FC was 0.2405, 0.3382, 0.2340, and 0.3013, respectively). In addition, we found a network module of glycometabolism network closely related to ATP metabolism (Figure 3). Moreover, these DEGs, such as ATP synthase, H+transporting, mitochondrial F1 complex ATP5B, ATP5C1, ATP5D, and ATP5G1, related to ATP metabolism were downregulated (the adjusted P value was .0045, .0077, 2.37E-05, and 4.99E-05, respectively and FC was 0.3167, 0.3510, 0.2652, and 0.2932, respectively). They may play a crucial role in the occurrence of AD.
Table 2.
Kyoto Encyclopedia of Genes and Genomes Pathways of Differentially Expressed Genes.a,b
| Terms | Description | Count | Adjusted P Value |
|---|---|---|---|
| hsa00190 | Oxidative phosphorylation | 19 | 1.59E-06 |
| hsa05016 | Huntington’s disease | 21 | 1.32E-05 |
| hsa05010 | Alzheimer’s disease | 17 | 0.0004 |
| hsa05012 | Parkinson’s disease | 14 | 0.0011 |
| hsa04260 | Cardiac muscle contraction | 10 | 0.0027 |
| hsa00010 | Glycolysis/gluconeogenesis | 8 | 0.0073 |
| hsa04144 | Endocytosis | 15 | 0.0101 |
| hsa00030 | Pentose phosphate pathway | 5 | 0.0139 |
| hsa00970 | Aminoacyl-tRNA biosynthesis | 6 | 0.0189 |
| hsa00051 | Fructose and mannose metabolism | 5 | 0.0390 |
| hsa05120 | Epithelial cell signaling in Helicobacter pylori infection | 7 | 0.0440 |
| hsa04540 | Gap junction | 8 | 1.115760112 |
| hsa05110 | Vibrio cholerae infection | 6 | 0.836820084 |
| hsa05130 | Pathogenic Escherichia coli infection | 6 | 0.836820084 |
Abbreviation: BH, Benjamin and Hochberg.
a Count means the number of candidate genes with that annotation.
b Adjusted P value means P value adjusted by BH.
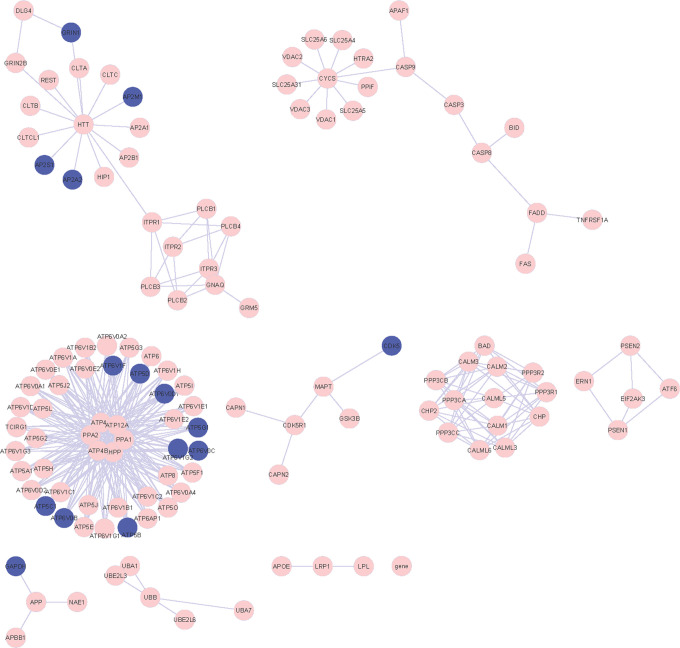
Figure 1.
Network of 10 KEGG pathways. The red nodes indicated annotated upregulated differentially expressed genes (DEGs) and the blue nodes indicated annotated downregulated DEGs. KEGG indicates Kyoto Encyclopedia of Genes and Genomes.
Figure 2.
Network of 4 glycometabolism pathways. The blue nodes indicated annotated downregulated DEGs and the pink nodes indicated genes in network. DEGs indicate differentially expressed genes.
Table 3.
Differentially Expressed Genes With Degree Larger Than 5 in KEGG Pathways.
| Gene | Degree | Gene | Degree |
|---|---|---|---|
| PFKM | 20 | ATP6V0C | 6 |
| PGM1 | 19 | ATP6V0B | 6 |
| SH3GL2 | 15 | ATP6V1G2 | 6 |
| ALDOA | 14 | ATP6V0D1 | 6 |
| ALDOC | 14 | ATP6V1F | 6 |
| GAPDH | 8 | LDHA | 5 |
| ATP5B | 6 | TPI1 | 5 |
| ATP5C1 | 6 | ATP5G1 | 6 |
| ATP5D | 6 |
Abbreviations: ATP, adenosine triphosphate; ALDOA, aldolase A; ALDOC, aldolase C; PFKM, 6-phosphofructokinase, muscle type; PGM1, phosphoglucomutase 1; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; LDHA, lactate dehydrogenase A; TPI1, triosephosphate isomerase.
Table 4.
Differentially Expressed Genes With Degree Larger Than 5 in Glycometabolism Pathways.
| Gene | Degree | Gene | Degree |
|---|---|---|---|
| PFKM | 20 | ATP5G1 | 6 |
| PGM1 | 19 | ATP6V0C | 6 |
| ALDOA | 14 | ATP6V0B | 6 |
| ALDOC | 14 | ATP6V1G2 | 6 |
| GAPDH | 7 | ATP6V0D1 | 6 |
| ATP5B | 6 | ATP6V1F | 6 |
| ATP5C1 | 6 | LDHA | 5 |
| ATP5D | 6 | TPI1 | 5 |
Abbreviations: ATP, adenosine triphosphate; ALDOA, aldolase A; ALDOC, aldolase C; PFKM, 6-phosphofructokinase, muscle type; PGM1, phosphoglucomutase 1; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; LDHA, lactate dehydrogenase A; TPI1, triosephosphate isomerase.
Figure 3.
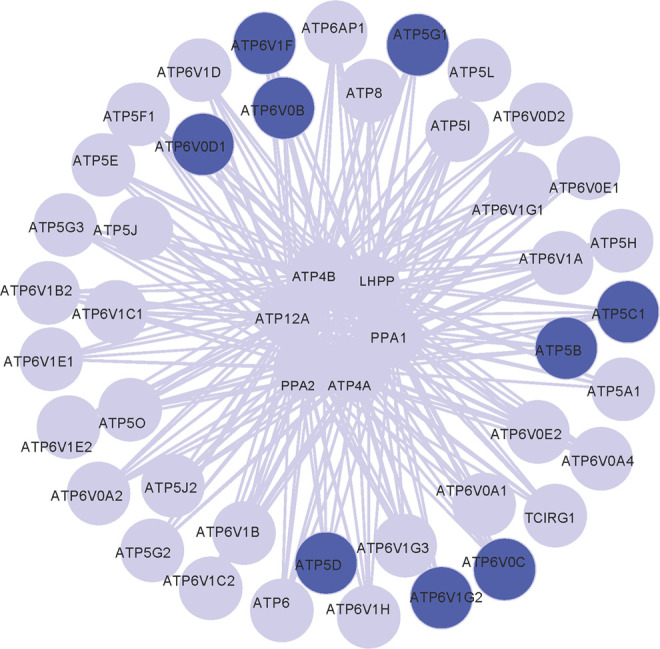
Network module related to ATP metabolism. The blue nodes indicated annotated downregulated DEGs. ATP indicates adenosine triphosphate; DEGs, differentially expressed genes.
Discussion
The EC is fundamental for cognitive functions. Thus, damage to this area appears as a key element in the progression of AD resulting in memory deficits arising from neuronal and synaptic alterations as well as glial malfunction. In this study, a total of 710 DEGs between AD and CT were identified. After GO analysis, we found these DEGs were involved in ribonucleotide metabolic process and biosynthetic process. The KEGG pathway analysis revealed most of DEGs participated in glycometabolism-related pathways including oxidative phosphorylation, glycolysis/gluconeogenesis, pentose phosphate pathway, and fructose and mannose metabolism. By constructing perturbing glycometabolism pathway network, several featured downregulated biomarkers associated with glycometabolism were identified including PFKM, PGM1, ALDOA, and ALDOC. In addition, we found an ATP metabolism network module with many downregulated genes such as ATP5B, ATP5C1, ATP5D, and ATP5G1.
It has been demonstrated that there was a close relation between energy metabolism and brain function. There are now several evidences suggesting that glucose metabolism is disrupted in AD brains. 25,26 Moreover, reduced regional cerebral glucose metabolism is correlated with the severity of dementia in AD. 27,28 These facts are consistent with our results of 4 downregulated DEGs associated with glycometabolism involved in the occurrence of AD. Related study has shown that the combination of glycolytic genes, including PFKM, ALDOA, ALDOC, and PGM1, participates in the glycolytic process. 29 6-Phosphofructokinase, muscle type is a regulatory protein coded by PFKM. It can transform fructose-6-phosphate into fructose-1,6-bisphosphate (F-1, 6-BP) in glycolysis, which is a key procedure in glycolysis. 30 Then F-1, 6-BP, as a substrate, can be broken down into glyceraldehyde 3-phosphate and dihydroxyacetone phosphate under catalysis of ALDOA or ALDOC enzymes. These 2 kinds of enzymes are coded by ALDOA and ALDOC genes. 29 Phosphoglucomutase 1 is also an enzyme involved in step 8 of glycolysis which can catalyze the conversion between 1,3-bisphosphoglycerate and 2,3-bisphosphoglycerate. 31 In line with our finding, Brooks et al reported that the messenger RNAs expression levels of ALDOA and ALDOC, as well as PFKM gene expression level, were decreased in AD. 5 The downregulation of these genes may suggest an interference of glucose utilization in AD brains. 32 Therefore, we conclude that the metabolism effect of these glycolytic genes plays a crucial role in the occurrence of AD.
To the best our knowledge, many cellular functions rely on ATP consumption and a high rate of ATP is fundamental to maintain signaling pathways, such as synapse. Moreover, there was a close relation between glycolysis and generation of ATP. According to the point of Kounelakis et al, glycolysis, a sugar splitting process, involves a series of biochemical reactions in which glucose is broken down into pyruvate with the release of usable energy in the form of ATP molecules. 29 Phosphoglucomutase 1, as an enzyme involved in ATP production, has been demonstrated to show a loss in enzymatic activity ultimately leading to decrease in ATP production. 33,34 Then, impairment of energy metabolism can selectively contribute to neurodegenerative processes. 35 Consistent with this notion, the report of Lin et al suggests that a reduction in ATP generation may contribute significantly to the cognitive impairment associated with AD. 36 According to these evidences, we can infer DEGs, such as ATP5B, ATP5C1, ATP5D, and ATP5G1, are downregulated in ATP metabolism network associated with the occurrence of AD.
Based on our analysis of metabolism-related DEGs associated with AD, we concluded that signaling pathways and genes modulated by metabolism may be potential therapeutic targets for AD. However, further experiments are needed to confirm our results in this study.
Acknowledgments
We wish to express our warm thanks to Fenghe (Shanghai) Information Technology Co, Ltd. Their ideas and help gave a valuable added dimension to our research.
Footnotes
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors received no financial support for the research, authorship, and/or publication of this article.
References
- 1. Alzheimer's Association, Thies W, Bleiler L. 2011. Alzheimer's disease facts and figures. Alzheimers Dement. 2011;7(2):208–244. [DOI] [PubMed] [Google Scholar]
- 2. Birks J. Cholinesterase inhibitors for Alzheimer’s disease Cochrane Database Syst Rev. 2006;1:CD005593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wimo A, Jonsson L, Winblad B. An estimate of the worldwide prevalence and direct costs of dementia in 2003. Dement Geriatr Cogn Disord. 2006;21(3):175–181. [DOI] [PubMed] [Google Scholar]
- 4. Bateman RJ, Xiong C, Benzinger TL, et al. Clinical and biomarker changes in dominantly inherited Alzheimer's disease. N Engl J Med. 2012;367(9):795–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brooks WM, Lynch PJ, Ingle CC, et al. Gene expression profiles of metabolic enzyme transcripts in Alzheimer's disease. Brain Res. 2007;1127(1):127–135. [DOI] [PubMed] [Google Scholar]
- 6. He X, Huang Y, Li B, Gong C-X, Schuchman EH. Deregulation of sphingolipid metabolism in Alzheimer's disease. Neurobiol Aging. 2010;31(3):398–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mazzola JL, Sirover MA. Reduction of glyceraldehyde - 3 - phosphate dehydrogenase activity in Alzheimer's disease and in Huntington's disease fibroblasts. J Neurochem. 2001;76(2):442–449. [DOI] [PubMed] [Google Scholar]
- 8. Augustin R, Lichtenthaler SF, Greeff M, Hansen J, Wurst W, Trümbach D. Bioinformatics identification of modules of transcription factor binding sites in Alzheimer's disease-related genes by in silico promoter analysis and microarrays. Int J Alzheimers Dis. 2011;2011:154325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Suzuki WA, Amaral DG. Topographic organization of the reciprocal connections between the monkey entorhinal cortex and the perirhinal and parahippocampal cortices. J Neurosci. 1994;14(3 pt 2):1856–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cunningham MO, Hunt J, Middleton S, et al. Region-specific reduction in entorhinal gamma oscillations and parvalbumin-immunoreactive neurons in animal models of psychiatric illness. J Neurosci. 2006;26(10):2767–2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. de Toledo-Morrell L, Goncharova I, Dickerson B, Wilson RS, Bennett DA. From healthy aging to early Alzheimer's disease: in vivo detection of entorhinal cortex atrophy. Ann N Y Acad Sci. 2000;911:240–253. [DOI] [PubMed] [Google Scholar]
- 12. Du A, Schuff N, Amend D, et al. Magnetic resonance imaging of the entorhinal cortex and hippocampus in mild cognitive impairment and Alzheimer's disease. J Neurol Neurosurg Psychiatry. 2001;71(4):441–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ribé EM, Pérez M, Puig B, et al. Accelerated amyloid deposition, neurofibrillary degeneration and neuronal loss in double mutant APP/tau transgenic mice. Neurobiol Dis. 2005;20(3):814–822. [DOI] [PubMed] [Google Scholar]
- 14. Liang WS, Dunckley T, Beach TG, et al. Gene expression profiles in anatomically and functionally distinct regions of the normal aged human brain. Physiol Genomics. 2007;28(3):311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liang WS, Reiman EM, Valla J, et al. Alzheimer's disease is associated with reduced expression of energy metabolism genes in posterior cingulate neurons. Proc Natl Acad Sci U S A. 2008;105(11):4441–4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. [DOI] [PubMed] [Google Scholar]
- 17. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc Series B (Methodological). 1995;57(1):289–300. [Google Scholar]
- 18. Mutch DM, Berger A, Mansourian R, Rytz A, Roberts MA. The limit fold change model: a practical approach for selecting differentially expressed genes from microarray data. BMC Bioinformatics. 2002;3:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25(1):25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mutowo-Meullenet P, Huntley RP, Dimmer EC, et al. Use of gene ontology annotation to understand the peroxisome proteome in humans. Database(Oxford). 2013;2013:bas062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Quentien MH, Delemer B, Papadimitriou DT, et al. Deficit in anterior pituitary function and variable immune deficiency (DAVID) in children presenting with adrenocorticotropin deficiency and severe infections. J Clin Endocrinol Metab. 2012;97(1):E121–E128. [DOI] [PubMed] [Google Scholar]
- 22. Hosack DA, Dennis Jr G, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4(10):R70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Altermann E, Klaenhammer TR. PathwayVoyager: pathway mapping using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. BMC Genomics. 2005;6:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Heiss W, Szelies B, Kessler J, Herholz K. Abnormalities of energy metabolism in Alzheimer's disease studied with PET. Ann N Y Acad Sci. 1990;640:65–71. [DOI] [PubMed] [Google Scholar]
- 26. Hoyer S, Nitsch R, Oesterreich K. Predominant abnormality in cerebral glucose utilization in late-onset dementia of the Alzheimer type: a cross-sectional comparison against advanced late-onset and incipient early-onset cases. J Neural Transm Park Dis Dement Sect. 1991;3(1):1–14. [DOI] [PubMed] [Google Scholar]
- 27. Xiao S, Cao Q, Xue H, et al. Measurement of regional cerebral metabolism rate of glucose in patients with Alzheimer's disease in different levels of severity]. Zhonghua Yi Xue Za Zhi. 2005;85(42):2975–2979. [PubMed] [Google Scholar]
- 28. Mielke R, Herholz K, Grond M, Kessler J, Heiss W. Clinical deterioration in probable Alzheimer's disease correlates with progressive metabolic impairment of association areas. Dement Geriatr Cogn Disord. 1994;5(1):36–41. [DOI] [PubMed] [Google Scholar]
- 29. Kounelakis M, Zervakis M, Giakos G, Postma G, Buydens L, Kotsiakis X. On the relevance of glycolysis process on brain gliomas. IEEE J Biomed Health Inform. 2013;17(1):128–135. [DOI] [PubMed] [Google Scholar]
- 30. Mutuku JM, Nose A. Changes in the contents of metabolites and enzyme activities in rice plants responding to Rhizoctonia solani Kuhn infection: activation of glycolysis and connection to phenylpropanoid pathway. Plant Cell Physiol. 2012;53(6):1017–1032. [DOI] [PubMed] [Google Scholar]
- 31. Sultana R, Butterfield DA. Role of oxidative stress in the progression of Alzheimer's disease. J Alzheimers Dis. 2010;19(1):341–353. [DOI] [PubMed] [Google Scholar]
- 32. Hashimoto M, Bogdanovic N, Nakagawa H, Volkmann I, Aoki M, Winblad B, et al. Analysis of microdissected neurons by 18O mass spectrometry reveals altered protein expression in Alzheimer's disease. J Cell Mol Med. 2012;16(8):1686–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Castegna A, Aksenov M, Aksenova M, et al. Proteomic identification of oxidatively modified proteins in Alzheimer’s disease brain. part I: creatine kinase BB, glutamine synthase, and ubiquitin carboxy-terminal hydrolase L-1. Free Rad Biol Med. 2002;33(4):562–571. [DOI] [PubMed] [Google Scholar]
- 34. Aksenova M, Butterfield DA, Zhang S-X, Underwood M, Geddes JW. Increased protein oxidation and decreased creatine kinase BB expression and activity after spinal cord contusion injury. J Neurotrauma. 2002;19(4):491–502. [DOI] [PubMed] [Google Scholar]
- 35. Sultana R, Perluigi M, Butterfield DA. Redox proteomics identification of oxidatively modified proteins in Alzheimer's disease brain and in vivo and in vitro models of AD centered around Aβ (1–42). J Chromatogr B Analyt Technol Biomed Life Sci. 2006;833(1):3–11. [DOI] [PubMed] [Google Scholar]
- 36. Lin AP, Shic F, Enriquez C, Ross BD. Reduced glutamate neurotransmission in patients with Alzheimer's disease–an in vivo 13C magnetic resonance spectroscopy study. MAGMA. 2003;16(1):29–42. [DOI] [PubMed] [Google Scholar]