Abstract
Background:
Alzheimer’s disease (AD) is a complex neurodegenerative disorder with largely unknown genetic mechanisms. Identifying altered neuronal gene expression in brain regions differentially affected by AD may provide the diagnostic or therapeutic targets of AD.
Methods:
The gene expression profile of AD was analyzed with bioinformatics. Function analysis was performed with Database for Annotation, Visualization and Integrated Discovery (DAVID), and TransFind was used to predict the possible transcriptional regulators in AD. Finally, connectivity map (cMap) database was used to explore small molecules targeted for AD.
Results:
The AD gene signatures associated with 6 different brain regions were identified. Functional analysis revealed that biological processes involved with metabolism, protein ubiquitination, and vasculature development were found dysregulated, and synaptic signaling pathways were found perturbated in AD. The WT1 was identified as an important transcriptional regulator in AD, and cMap database predicted that small molecules, such as histone deacetylase (HDAC) inhibitor, may be candidate drugs in the treatment of AD.
Conclusion:
According to our in silico analysis, Wilms' tumor suppressor may play regulatory roles in AD development and progress. The HDAC inhibitor could possibly be used to treat AD.
Keywords: Alzheimer’s disease, gene expression profile, connectivity map, functional analysis
Introduction
Alzheimer’s disease (AD) is the most common cause of dementia. In the past 20 years, deposition of the amyloid-β peptide in the brain was recognized as a central event in the pathogenesis of AD. 1 The clinical symptom of early AD is episodic memory impairment. Specific memory-functioning regions of brain, including the precuneus and posterior cingulate, are selectively vulnerable to early amyloid deposition in AD. 2 The most significant toxicity of amyloid-β peptide is toward the synapse, 3 and AD is increasingly recognized as a “synaptic disease.”
Studies suggested that there were significantly difference between early-onset and late-onset AD. For example, perfusion reduction was more prominent in patients with early-onset AD than patients with late-onset AD in the posterior cingulate gyrus precuneus and the parietal cortex. 4 Accumulating evidences also suggested that late-onset AD could result from epigenetic abnormalities such as histone modification or DNA methylation. 5
Although these are no cure for AD at present, efforts have been made to develop therapeutic strategies. The finding demonstrated that vaccination of antibody in PP-transgenic mice can prevent and reduce β amyloidosis deposited in cerebral and has stimulated the development of antibody-based immunotherapeutics for AD. 6 In recent clinical trials, continued treatment with donepezil was associated with cognitive benefits that exceeded the minimum clinically important difference and with significant functional benefits over the course of 12 months in patients with moderate or severe AD. 7
Gene expression profiling with microarray may provide opportunities for a better understanding of pathogenesis and progression of AD. Silva et al studied the gene expression profile of postmortem brain tissue from patients with AD and found that genes involved in energy metabolism, oxidative stress, DNA damage/repair, senescence, and transcriptional regulation were implicated with the neuropathology of AD. 8 Focusing on genes associated with energy metabolism, Liang et al found that patients with AD had significantly lower expression of 70% of the nuclear genes encoding subunits of the mitochondrial electron transport chain in posterior cingulate cortex. 9
In this present study, we reevaluated the gene expression profile referred by Liang et al 9 and performed bioinformatics analysis to reveal perturbated pathways and transcription factors in the pathogenesis and progression of AD. Potential drugs for AD treatment were also predicted.
Materials and Methods
Microarray Data Set
Laser-capture microdissection was performed on 6 brain regions with about 14 biological replicates per brain region. The brain regions are as follows: entorhinal cortex (EC), hippocampus (HIP), middle temporal gyrus (MTG), posterior cingulate cortex (PC), superior frontal gyrus (SFG), and visual cortex (VCX). Gene expression profiling was performed on Affymetrix Human Genome U133 Plus 2.0 Array (Shanghai, China). Microarray data set was previously generated and analyzed by Liang et al. 9 We downloaded the raw data GSE5281 from public database Gene Expression Omnibus and recalculated the gene expression intensities with robust multiarray average 10 method using EntrezGene-based center custom Chip Description File developed by University of Michigan (version 16). 11
Identification of Differentially Expressed Genes
Student’s t test was performed for identification of differentially expressed (DE) genes and the resulting P values were submitted to R package q value 12 to estimate false discovery rate. Genes with false discovery rate <1% were considered as significantly DE (also referred to as AD gene signature). The DE genes in each region were denoted by “region_up” or “region_dn.” For example, genes significantly upregulated in PC were denoted as PC_UP and those downregulated as PC_DN.
DAVID Functional Analysis of AD Gene Signature
The AD gene signature was submitted to Database for Annotation, Visualization and Integrated Discovery (DAVID) 13 to identify enriched Gene Ontology biological processes. Redundant/similar/hierarchical annotation terms were grouped into functional annotation clusters by fuzzy clustering. We considered annotation clusters with enrichment score >3 as significant annotation cluster.
Signaling Pathway Impact Analysis of AD Gene Signature
The AD gene signature was also submitted to R package 14,15 to identify perturbated signaling pathways. Most of the existing pathway analysis methods focus on either the number of DE genes observed in a given pathway (enrichment analysis methods) or on the correlation between the pathway genes and the class of the samples (functional class scoring methods). Both the approaches treat the pathways as simple sets of genes, disregarding the complex gene interactions that these pathways are built to describe. Signaling pathway impact analysis (SPIA) not only considers the overrepresentation of DE genes in a signaling pathway but also measures the pathway-level perturbation by considering the magnitude of each gene’s expression change, their position and interactions in the given pathways, and so on. A bootstrap procedure is used to assess the significance of the observed total pathway perturbation. Global pathway significance P value and false discovery rate were then calculated by combining the enrichment and perturbation P values. In our analysis, only pathways with false discovery rate <5% were considered as significantly perturbated pathways.
TransFind Analysis of AD Gene Signature
The AD gene signature was used to query TransFind 16 for finding transcription factors that may play regulatory roles in AD. TransFind uses Fisher’s exact test to quantify enrichment of putative high-affinity targets of a transcription factor in the query gene set and uses an analytical approach 17 to determine the false discovery rate. We submitted AD gene signature for each region to TransFind separately and used remaining genes as background. Only predictions with a false discovery rate (FDR) <0.05 were considered as significant in our analysis.
Connectivity Map Analysis of AD Gene Signature
The AD gene signature was used to query connectivity map (cMap) for finding potential drugs. The cMap 18 is an in silico method to predict potential drugs that could possibly reverse, or induce, the biological state encoded in particular gene expression signatures. The cMap provides a collection of more than 7000 genome-wide transcriptional expression data from cultured human cells treated with 1309 bioactive small molecules. Gene expression profiles were organized into instances that represent a treatment and control pair and the list of genes ordered by their extent of differential expression between this treatment and control pair. The query gene signature is then compared to each rank-ordered list to determine whether upregulated query genes tend to appear near the top of the list and downregulated query genes near the bottom (“positive connectivity”) or vice versa (“negative connectivity”), yielding a “connectivity score” ranging from −1 to 1. A high positive connectivity score indicates that the corresponding perturbagen induced the expression of the query signature. A high negative connectivity score indicates that the corresponding perturbagen reversed the expression of the query signature. All instances in the database are then ranked according to their connectivity scores; those at the top are most strongly correlated to the query signature, and those at the bottom are most strongly anticorrelated.
Entrez gene IDs for AD gene signature were converted into Affymetrix probeset IDs as cMap requires. Because a single gene could be represented by multiple probe sets and cMap could only take up to 1000 probe sets per input, we ranked DE genes by their fold changes and used the top 400 upregulated (or downregulated) genes for querying when necessary.
Results
Differentially Expressed Genes Between AD and Controls
At a false discovery rate of 1%, the largest number of DE genes between patients with AD and controls was observed in PC. In most regions, the number of downregulated genes exceeds upregulated genes except in SFG and VCX. The largest number of downregulated genes was found in MTG and PC.
Functional Analysis of AD Gene Signature
In most regions, downregulated genes were enriched with more functional annotation clusters except SFG. Downregulated genes were frequently associated with energy metabolism that is consistent with the original study. Besides, protein ubiquitination was also found to be downregulated in HIP and PC, while synaptic transmission was found to be downregulated in MTG. The SFG_UP genes were enriched with genes involved in embryonic development, transcription, and vasculature development.
Pathway Analysis of AD Gene Signature
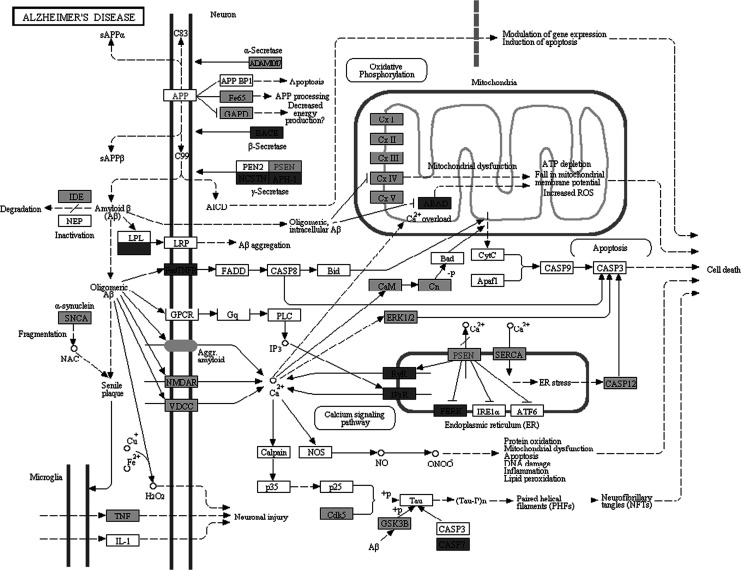
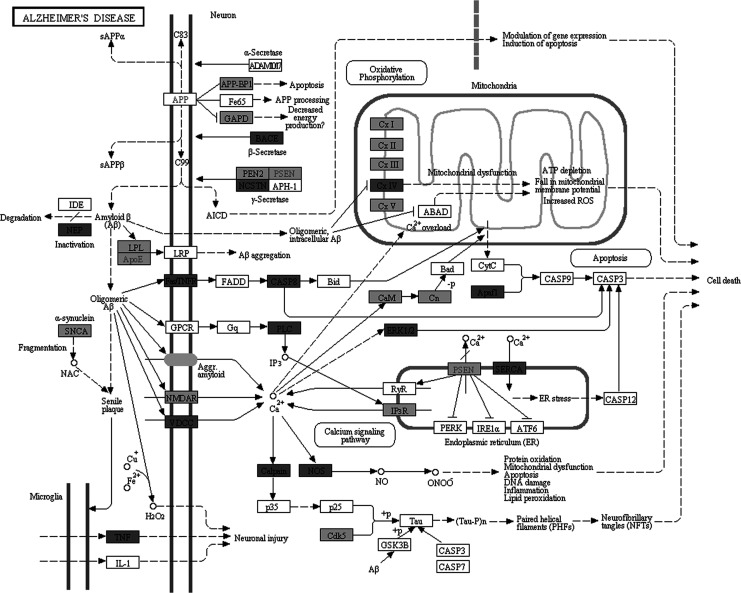
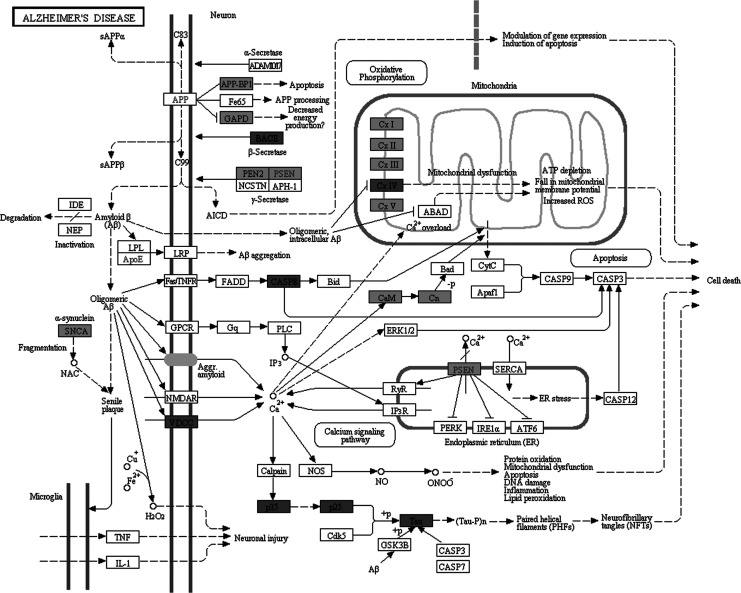
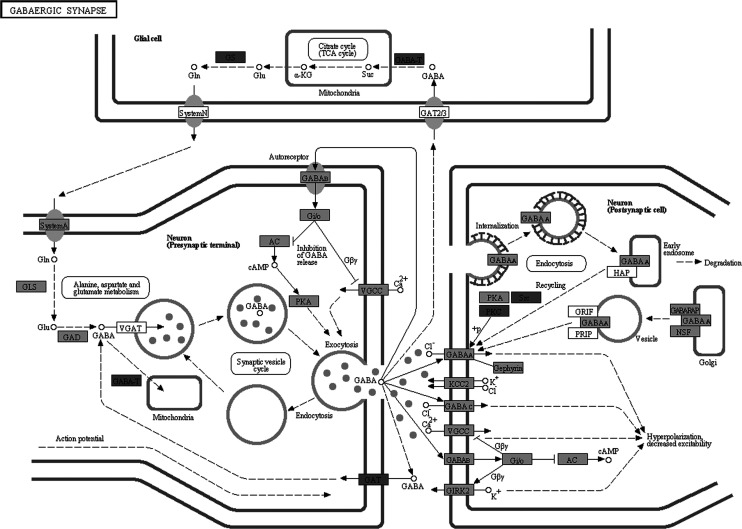
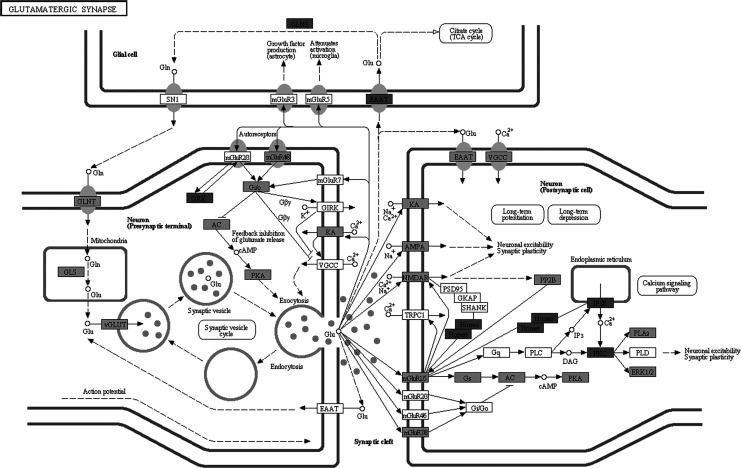
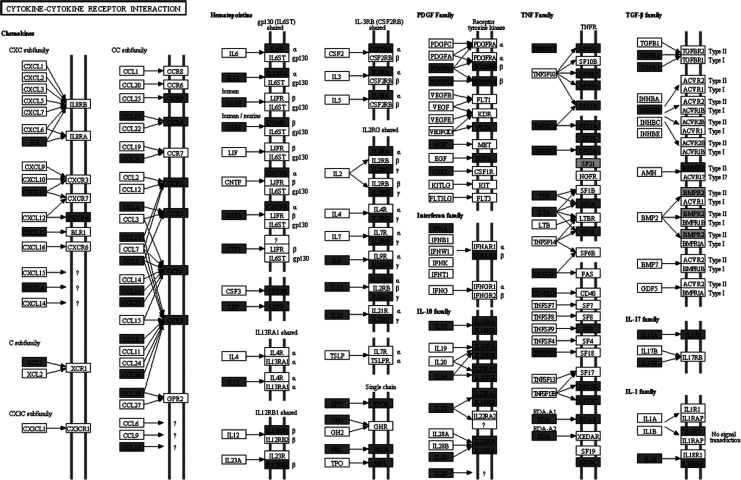
Molecular pathway associated with AD is already characterized by Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.jp/kegg/pathway/hsa/hsa05010.html), but SPIA analysis only found AD pathway perturbated in PC, MTG, and HIP significantly (Figures 1 –3). This may be due to the incomplete pathway of annotation. In addition, pathways associated with Huntington’s disease and Parkinson’s disease were also found perturbated in PC, MTG, and HIP. The MTG was also characterized by perturbation of GABAergic synaptic pathway (Figure 4) and glutamatergic synaptic pathway (Figure 5). Many dysregulated cytokine–cytokine receptor interaction was found in PC (Figure 6).
Figure 1.
Perturbation of the KEGG Alzheimer's disease pathway. Dark gray nodes are genes upregulated in middle temporal gyrus and light gray nodes are those downregulated in middle temporal gyrus.
Figure 2.
Perturbation of the KEGG Alzheimer's disease pathway. Dark Gray nodes are genes upregulated in posterior cingulate cortex and light gray nodes are those downregulated in posterior cingulate cortex.
Figure 3.
Perturbation of the KEGG Alzheimer's disease pathway. Dark gray nodes are genes upregulated in hippocampus and light gray nodes are those downregulated in hippocampus.
Figure 4.
Perturbated GABAergic synaptic signaling pathway in middle temporal gyrus. Dark gray nodes are genes upregulated in hippocampus and light gray nodes are those downregulated in middle temporal gyrus.
Figure 5.
Perturbated Glutamateergic synaptic signaling pathway in middle temporal gyrus. Dark gray nodes are genes upregulated in hippocampus and light gray nodes are those downregulated in middle temporal gyrus.
Figure 6.
Dysregulated cytokine-cytokine receptor interactions in posterior cingulate cortex. Dark gray nodes are genes upregulated in hippocampus and light gray nodes are those downregulated in posterior cingulate cortex.
TransFind Predicted Possible Transcriptional Regulators in AD
TransFind predicted transferrins (TFs) that could possibly perform transcriptional regulation of HIP_UP, MTG_UP, PC_UP, SFG_UP, and PC_DN genes. Unusually, high GC content was found in promoter region among SFG_UP genes. Wilms' tumor suppressor (WT1) was predicted to be a transcriptional regulator for HIP_UP, MTG_UP, and PC_UP genes.
Conectivity Map Identified AD-Related Bioactive Small Molecules
Functional connections between AD gene signatures and gene signatures induced by small molecules were explored using the cMap database. Gene signature induced by thioridazine was found significantly similar with AD gene signatures in EC, HIP, MTG, and SFG. On the contrary, gene signatures induced by 2 histone deacetylase (HDAC) inhibitors vorinostat and trichostatin A were significantly anticorrelated with AD gene signature in HIP and PC, which indicated that these drugs could possibly reverse the AD gene signature.
Discussion
Dysregulated Biological Processes Were Associated With Metabolism, Protein Ubiquitination, and Vasculature Development
We found that posterior cingulate cortex, MTG, hippocampal, and EC were all significantly enriched with downregulated mitochondria genes, in particular, oxidative phosphorylation genes. Actually, DAVID functional annotation found that downregulated genes in PC, MTG, HIP, and EC were significantly enriched with more biological processes than up-regulated genes. This is consistent with the fact that AD is a degenerative disease. Besides metabolic processes, we also found genes involved in protein ubiquitination was downregulated in HIP and PC, which was reminiscent of the increased proteolytic resistance and accumulation of potentially toxic proteins in neurodegenerative diseases, 19 in particular accumulation of the amyloid-β peptide in AD brains. Interestingly, genes participated in nerve impulse transmission, and vesicle-mediated transport was found to be downregulated in MTG. This is coincident with the fact that AD is associated with synaptic depression that could be caused by increased amyloid-β. 20
On the contrary, no significant annotation clusters was found in downregulated genes in SFG. We found that genes participated in embryonic development, transcription, and blood vessel development were upregulated in SFG. Disrupted perfusion is not only evident throughout disease manifestation but also demonstrated during the preclinical phase of AD (ie, mild cognitive impairment) as well as in cognitively healthy persons at high risk of developing AD due to family history or genetic factors. 21 Since hypoperfusion in AD is associated with both structural and functional changes in the brain, genes involved in vasculature development may be upregulated as a compensatory mechanism.
Signaling Pathway Impact Analysis Found Dysregulated Synaptic Pathways in AD
Alzheimer’s disease is increasingly recognized as a “synaptic disease.” We performed SPIA and found KEGG pathways of GABAergic synapse, glutamatergic synapse, calcium signaling, and long-term potential were dysregulated in MTGs. Although GABAergic neurotransmission was previously generally thought to be well preserved in AD, Limon et al 22 found an age-dependent reduction in GABA currents in the AD brain. Furthermore, GABA receptors from AD brains were slightly, but significantly, less sensitive to GABA than receptors from non-AD brains. They further proved that the reduction in GABA currents in AD was associated with reductions in mRNA and protein of the principal GABA receptor subunits. Glutamate is the most abundant neurotransmitter in the brain and glutamatergic system is impaired in AD. 23 Not only glutamate influences amyloid-β production, but amyloid-β can also alter the levels of glutamate at the synapse. 23 Glutamate is essential for memory formation through processes such as long-term potentiation (LTP). Reeves et al extracted pure populations of tau oligomers directly from the cerebral cortex of AD brain and found that these oligomers are potent inhibitors of LTP in hippocampal brain slices and disrupt memory in wild-type mice. 24
Several pathways associated with infection of pathogens or virus were found perturbated in AD. In fact, there is a significant association between AD and various types of spirochete and other pathogens such as Chlamydophila pneumoniae and herpes simplex virus type 1. 25 Miklossy 25 further suggested that dementia might be prevented by combined antibiotic, antiviral, and anti-inflammatory therapy.
TransFind Predicted Abnormal TFs in AD
Interestingly, predicted targets of transcription factor WT1 was found enriched in upregulation genes in HIP, MTG, and PC. The WT1 was a mediator of neuronal degeneration associated with the pathogenesis of AD. 26 We also noticed that predicted targets of LEF1 were enriched in genes upregulated in PC. The Wnt signaling function starts during the development of the nervous system and is crucial for synaptic plasticity in the adult brain. Inestrosa et al suggested that a sustained loss of Wnt signaling function may be a key relevant factor in the pathology of AD. 27 It was also shown that calcium/calmodulin-dependent protein kinase IV was regulated by the Wnt signaling pathway and that its expression could play a role in the neuroprotective function of the Wnt signaling against the Alzheimer’s amyloid peptide. 28
Histone Deacetylase Inhibitor May Be Candidate Drugs in the Treatment of AD
Thioridazine and trifluoperazine were predicted to play roles in inducing AD. Early clinical trial has demonstrated that for most patients with AD, withdrawal of neuroleptics, including thioridazine and trifluoperazine, had no overall detrimental effect on functional and cognitive status. 29 In fact, the manufacturer Novartis/Sandoz/Wander of the brands of thioridazine, Mellaril (USA and Canada) and Melleril (Europe), discontinued the drug worldwide in June 2005.
Venkataramani et al 30 found that treating pancreatic and colon cancer cells with valproic acid, a known HDAC inhibitor, leads to upregulation of GRP78, an endoplasmic reticulum chaperone immunoglobulin-binding protein. The GRP78 is involved in β-amyloid precursor protein (APP) maturation and inhibition of tumor cell growth by downregulation of APP and secreted soluble APPα. Trichostatin A, a pan-HDAC inhibitor, also lowered APP and increased GRP78 levels. In contrast, treating cells with valpromide, a VPA derivative lacking HDAC inhibitory properties, had no effect on APP levels. In our analysis, HDAC inhibitor trichostatin A was predicted to possibly reverse AD gene signature in HIP, PC, and SFG, while HDAC inhibitor vorinostat was predicted to possibly reverse AD gene signature in HIP and PC. Together, these studies propose a role for HDAC inhibitors in treating AD.
Footnotes
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship and/or publication of this article: Supported by the Fundamental Research Funds for the Central Universities (1508219048) and China Medical Foundation.
References
- 1. Karran E, Mercken M, De Strooper B. The amyloid cascade hypothesis for Alzheimer's disease: an appraisal for the development of therapeutics. Nat Rev Drug Discov. 2011;10(9):698–712. [DOI] [PubMed] [Google Scholar]
- 2. Sperling RA, Dickerson BC, Pihlajamaki M, et al. Functional alterations in memory networks in early Alzheimer's disease. Neuromolecular Med. 2010;12(1):27–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Eisenberg D, Jucker M. The amyloid state of proteins in human diseases. Cell. 2012;148(6):1188–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Matsuda H. Neuroimaging for patients with Alzheimer disease in routine practice [in Japanese]. Brain Nerve. 2010;62(7):743–755. [PubMed] [Google Scholar]
- 5. Bihaqi SW, Schumacher A, Maloney B, Lahiri DK, Zawia NH. Do epigenetic pathways initiate late onset Alzheimer disease (LOAD): towards a new paradigm. Curr Alzheimer Res. 2012;9(5):574–588. [DOI] [PubMed] [Google Scholar]
- 6. Brody DL, Holtzman DM. Active and passive immunotherapy for neurodegenerative disorders. Annu Rev Neurosci. 2008;31:175–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Howard R, McShane R, Lindesay J, et al. Donepezil and memantine for moderate-to-severe Alzheimer's disease. N Engl J Med. 2012;366(10):893–903. [DOI] [PubMed] [Google Scholar]
- 8. Silva AR, Grinberg LT, Farfel JM, et al. Transcriptional alterations related to neuropathology and clinical manifestation of Alzheimer's disease. PLoS One. 2012;7(11):e48751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liang WS, Reiman EM, Valla J, et al. Alzheimer's disease is associated with reduced expression of energy metabolism genes in posterior cingulate neurons. Proc Natl Acad Sci USA. 2008;105(11):4441–4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. [DOI] [PubMed] [Google Scholar]
- 11. Dai M, Wang P, Boyd AD, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33(20):e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100(16):9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. [DOI] [PubMed] [Google Scholar]
- 14. Tarca AL, Draghici S, Khatri P, et al. A novel signaling pathway impact analysis. Bioinformatics. 2009;25(1):75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Draghici S, Khatri P, Tarca AL, et al. A systems biology approach for pathway level analysis. Genome Res. 2007;17(10):1537–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kielbasa SM, Klein H, Roider HG, Vingron M, Bluthgen N. TransFind—predicting transcriptional regulators for gene sets. Nucleic Acids Res. 2010;38(Web Server issue):W275–W280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bluthgen N, Kielbasa SM, Herzel H. Inferring combinatorial regulation of transcription in silico. Nucleic Acids Res. 2005;33(1): 272–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lamb J, Crawford ED, Peck D, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313(5795):1929–1935. [DOI] [PubMed] [Google Scholar]
- 19. Ihara Y, Morishima-Kawashima M, Nixon R. The ubiquitin-proteasome system and the autophagic-lysosomal system in Alzheimer disease. Cold Spring Harb Perspect Med. 2012;2(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Origlia N, Bonadonna C, Rosellini A, et al. Microglial receptor for advanced glycation end product-dependent signal pathway drives beta-amyloid-induced synaptic depression and long-term depression impairment in entorhinal cortex. J Neurosci. 2010;30(34):11414–11425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Austin BP, Nair VA, Meier TB, et al. Effects of hypoperfusion in Alzheimer's disease. J Alzheimers Dis. 2011;26(suppl 3):123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Limon A, Reyes-Ruiz JM, Miledi R. Loss of functional GABA(A) receptors in the Alzheimer diseased brain. Proc Natl Acad Sci USA. 2012;109(25):10071–10076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Revett TJ, Baker GB, Jhamandas J, Kar S. Glutamate system, amyloid β peptides and tau protein: functional interrelationships and relevance to Alzheimer disease pathology. J Psychiatry Neurosci. 2013;38(1):6–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lasagna-Reeves CA, Castillo-Carranza DL, Sengupta U, et al. Alzheimer brain-derived tau oligomers propagate pathology from endogenous tau. Sci Rep. 2012;2:700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Miklossy J. Emerging roles of pathogens in Alzheimer disease. Expert Rev Mol Med. 2011;13:e30. [DOI] [PubMed] [Google Scholar]
- 26. Lovell MA, Xie C, Xiong S, Markesbery WR. Wilms' tumor suppressor (WT1) is a mediator of neuronal degeneration associated with the pathogenesis of Alzheimer's disease. Brain Res. 2003;983(1-2):84–96. [DOI] [PubMed] [Google Scholar]
- 27. Inestrosa NC, Montecinos-Oliva C, Fuenzalida M. Wnt signaling: role in Alzheimer disease and schizophrenia. J Neuroimmune Pharmacol. 2012;7(4):788–807. [DOI] [PubMed] [Google Scholar]
- 28. Arrazola MS, Varela-Nallar L, Colombres M, et al. Calcium/calmodulin-dependent protein kinase type IV is a target gene of the Wnt/beta-catenin signaling pathway. J Cell Physiol. 2009;221(3):658–667. [DOI] [PubMed] [Google Scholar]
- 29. Ballard C, Lana MM, Theodoulou M, et al. A randomised, blinded, placebo-controlled trial in dementia patients continuing or stopping neuroleptics (the DART-AD trial). PLoS Med. 2008;5(4):e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Venkataramani V, Rossner C, Iffland L, et al. Histone deacetylase inhibitor valproic acid inhibits cancer cell proliferation via down-regulation of the alzheimer amyloid precursor protein. J Biol Chem. 2010;285(14):10678–10689. [DOI] [PMC free article] [PubMed] [Google Scholar]