Abstract
The UspA1 and UspA2 proteins of Moraxella catarrhalis are potential vaccine candidates for preventing disease caused by this organism. We have characterized both proteins and evaluated their vaccine potential using both in vitro and in vivo assays. Both proteins were purified from the O35E isolate by Triton X-100 extraction, followed by ion-exchange and hydroxyapatite chromatography. Analysis of the sequences of internal peptides, prepared by enzymatic and chemical cleavage of the proteins, revealed that UspA1 and UspA2 exhibited distinct structural differences but shared a common sequence including an epitope recognized by the monoclonal antibody 17C7. By sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), purified UspA1 exhibited a molecular weight of approximately 350,000 when unheated and a molecular weight of 100,000 after being heated for 10 min at 100°C. In contrast, purified UspA2 exhibited an apparent molecular weight of 240,000 by SDS-PAGE that did not change with the length of time of heating. Their sizes as determined by gel filtration were 1,150,000 and 830,000 for UspA1 and UspA2, respectively. Preliminary results indicate the proteins have separate functions in bacterial pathogenesis. Purified UspA1 was found to bind HEp-2 cells, and sera against UspA1, but not against UspA2, blocked binding of the O35E isolate to the HEp-2 cells. UspA1 also bound fibronectin and appears to have a role in bacterial attachment. Purified UspA2, however, did not bind fibronectin but had an affinity for vitronectin. Both proteins elicited bactericidal antibodies in mice to homologous and heterologous disease isolates. Finally, mice immunized with each of the proteins, followed by pulmonary challenge with either the homologous or a heterologous isolate, cleared the bacteria more rapidly than mock-immunized mice. These results suggest that UspA1 and UspA2 serve different virulence functions and that both are promising vaccine candidates.
Moraxella catarrhalis is a human pathogen of the middle ear and the respiratory tract. It causes significant morbidity in the very young and the very old. In the very young, it is associated with approximately 15% of all cases of otitis media. It is the third leading bacterial cause of this disease after Streptococcus pneumoniae and Haemophilus influenzae (19). It is also a significant cause of sinusitis (26) and persistent cough (8) in children. In the elderly, it infects patients with predisposing conditions such as chronic obstructive pulmonary disease and other chronic cardiopulmonary conditions (3, 5, 10).
In previous reports (11, 14), including one from our laboratory (4), it was noted that a protein called UspA or high-molecular-weight outer membrane protein (HMWP-OMP) was a promising vaccine candidate. This protein was characterized as having a molecular weight of 100,000 or greater by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and reactivity with a monoclonal antibody (MAb) designated 17C7. This MAb exhibits complement-dependent bactericidal activity (4) and passive administration of it to mice promotes pulmonary clearance of an M. catarrhalis challenge (11). It was recently found that UspA was not a single protein but two distinct proteins, UspA1 and UspA2, encoded by separate genes (2). The molecular weight of the protein encoded by the uspA1 gene was predicted to be 88,271 while that of the uspA2 was 62,483. These values are much smaller than those determined by SDS-PAGE. The predicted amino acid sequences of the two proteins have 43% identity; however, there is 93% identity for a stretch of 140 amino acid residues. The epitope recognized by the 17C7 MAb has been mapped to this region (2).
We report here the purification of the two proteins, UspA1 and UspA2, their biochemical characteristics, some immunological properties, and properties that may be relevant for host colonization. Particular attention has been given to determining if the proteins might be suitable vaccine candidates.
MATERIALS AND METHODS
Bacteria.
Eric Hansen of the University of Texas Southwestern Medical Center provided the TTA24 and O35E isolates of M. catarrhalis. Additional isolates were obtained from Dwight Hardy of the University of Rochester and the American Type Culture Collection. The bacteria were passaged by incubation on Mueller-Hinton agar (Difco, Detroit, Mich.) incubated at 35°C with 5% carbon dioxide. The bacteria used for the purification of the protein were grown in sterile broth containing 10 g of Casamino Acids (Difco) per liter and 15 g of yeast extract (BBL, Cockeysville, Md.) per liter. The isolates were stored at −70°C in Mueller-Hinton broth containing 40% glycerol.
Purification of UspA2.
The bacterial cells (∼400 g [wet weight] of M. catarrhalis O35E) were washed twice with 2 liters of 0.03 M sodium phosphate buffer (pH 6.0) containing 1.0% Triton X-100 (TX-100) (J. T. Baker Inc., Philipsburg, N.J.) with stirring at room temperature for 60 min. The UspA2-containing cells were pelleted by centrifugation at 13,700 × g for 30 min at 4°C. Following centrifugation, the pellet was resuspended in 2 liters of 0.03 M Tris(hydroxymethyl)amino-methane HCl (Tris-HCl) (pH 8.) containing 1.0% TX-100 and stirred overnight at 4°C to extract the UspA2 protein. Cells were removed by centrifugation at 13,700 × g for 30 min at 4°C. The supernatant, containing UspA2 protein, was collected and further clarified by sequential microfiltration through a 0.8-μm-pore-size membrane (CN.8; Nalge, Rochester, N.Y.) and then through a 0.45-μm-pore-size membrane (cellulose acetate, low protein binding; Corning, Inc., Corning, N.Y.).
The entire filtered crude extract preparation was loaded onto a (∼200-ml) trimethylaminoethyl (TMAE) column [50 by 217 mm, model 650(S), (particle size) 0.025 to 0.4 mm; EM Separations, Gibbstown, N.J.] equilibrated with 0.03 M Tris-HCl buffer (pH 8.0) containing 0.1% TX-100 (THT). The column was washed with 400 ml of equilibration buffer followed by 600 ml of 0.25 M NaCl in 0.03 M THT. UspA2 was subsequently eluted with 800 ml of 1.0 M NaCl in 0.03 M THT. Fractions were screened for UspA2 by SDS-PAGE and pooled. Pooled fractions (∼750 ml) containing UspA2 were concentrated to approximately half their volume by ultrafiltration in stirred cell with a YM-100 membrane (Amicon Corp., Beverly, Mass.) under nitrogen pressure. The TMAE concentrate was split into two 175-ml aliquots, and each part buffer was exchanged by passage over a (∼550 ml) Sephadex G-25 (Coarse) column (50 by 280 mm; Pharmacia Biotech, Piscataway, N.J.) equilibrated with 10 mM sodium phosphate buffer (pH 7.0) containing 0.1% TX-100 (10 mM PT). The buffer-exchanged material was subsequently loaded onto a (∼425-ml) ceramic hydroxyapatite column (type I, 50 by 217 mm, 40 μm; Bio-Rad Laboratories, Hercules, Calif.) equilibrated with 10 mM PT. The column was washed with 450 ml of the equilibration buffer followed by 900 ml of 0.1 M NaPO4 (pH 7.0) containing 0.1% TX-100. UspA2 was then eluted with a linear gradient of 0.1 to 0.2 M sodium phosphate buffer containing 0.1% TX-100. An additional volume of 0.2 M sodium phosphate buffer (pH 7.0) containing 0.1% TX-100 was applied to the column and collected to maximize the recovery of UspA2. Fractions were screened for UspA2 by SDS-PAGE and pooled. The column was then washed with 900 ml of 0.5 M sodium phosphate buffer (pH 7.0) containing 0.1% TX-100. Removal of residual endotoxin was accomplished by passing the protein over either an ion-exchange or gel filtration column equilibrated with buffer containing 1 to 5% Zwittergent 3-12.
Purification of UspA1.
During the purification of UspA2, fractions enriched in UspA1 from the 0.5 mM phosphate buffer wash of the hydroxyapatite column were retained and stored at 4°C. The UspA1 enriched fractions saved from four separate UspA2 productions runs were pooled and concentrated approximately threefold by using the Amicon stirred cell with a YM-100 membrane under nitrogen pressure. The UspA1 concentrate was split into two 175-ml aliquots, and the buffer was exchanged by passage over a (∼550-ml) Sephadex G-25 column (50 by 280 mm) equilibrated with 10 mM PT. The buffer-exchanged material was loaded onto a 50 × 217 mm (∼425-ml) ceramic hydroxyapatite column (Bio-Rad) equilibrated with 10 mM PT. The column was washed with 450 ml of the equilibration buffer followed by 900 ml of 0.25 M NaPO4 (pH 7.0) containing 0.1% TX-100. The UspA1 was eluted with a linear NaPO4 gradient of 0.25 to 0.5 M NaPO4 (pH 7.0) containing 0.1% TX-100. The fractions containing UspA1 were identified by SDS-PAGE and pooled.
SDS-PAGE and Western blot analysis.
SDS-PAGE was carried out as described by Laemmli (15), using 4 to 20% (wt/vol) gradient acrylamide gels (Integrated Separation Systems [ISS], Natick, Mass.). Proteins were visualized by staining the gels with Coomassie brilliant blue R-250. Gels were scanned with a personal densitometer SI (Molecular Dynamics Inc., Sunnyvale, Calif.) and molecular weights were estimated by using the Fragment Analysis software (version 1.1; Molecular Dynamics) with prestained molecular weight markers (ISS) as standards. Transfer of proteins to polyvinylidene difluoride membranes was accomplished with a semidry electroblotter and electroblot buffers (ISS). The membranes were probed with protein-specific antisera or MAbs followed by goat anti-mouse alkaline phosphatase conjugate as the secondary antibody (BioSource International, Camarillo, Calif.). Western blots were developed with the 5-bromo-4-chloro-3-indolylphosphate–nitroblue tetrazolium (BCIP-NBT) phosphatase substrate system (Kirkegaard and Perry Laboratories, Gaithersburg, Md.).
Protein estimation.
Protein concentrations were estimated by the bicinchoninic assay (Pierce, Rockford, Ill.) with bovine serum albumin as the standard.
Enzymatic and chemical cleavages of UspA2 and UspA1. (i) CNBr cleavage.
Approximately 0.3 mg of the purified protein was precipitated with 90% (vol/vol) ethanol, and the pellet resuspended in 100 μl of 88% (vol/vol) formic acid containing 12 M urea. Following resuspension, 100 μl of 88% (vol/vol) formic acid containing 2 M CNBr (Sigma Chemical Co., St. Louis, Mo.) was added, and the mixture was incubated overnight at room temperature in the dark.
(ii) Trypsin and chymotrypsin cleavage.
Approximately 2 mg of the purified protein was precipitated with 90% (vol/vol) ethanol, and the pellet was resuspended in a total volume of 1 ml of phosphate-buffered saline (PBS) containing 0.1% TX-100. This preparation was added directly to a vial containing 25 μg of either trypsin or chymotrypsin (Boehringer Mannheim, Indianapolis, Ind.). The reaction mixture was incubated for 48 h at 37°C.
(iii) Endoproteinase Lys-C cleavage.
Approximately 2 mg of the purified protein was precipitated with 90% (vol/vol) ethanol, and the pellet was resuspended in a total volume of 1.0 ml of PBS containing 0.1% TX-100. This preparation was added directly to a vial containing 15 μg of endoproteinase Lys-C (Boehringer Mannheim). The reaction mixture was incubated for 48 h at 37°C.
(iv) Separation of peptides.
The above cleavage reaction mixtures were centrifuged in an Eppendorf centrifuge at 12,000 rpm for 5 min, and the supernatant was loaded directly onto a Vydac protein C4 high-performance liquid chromatography (HPLC) column (The Separations Group, Hesperia, Calif.). The solvents used were 0.1% (vol/vol) aqueous trifluoroacetic acid (TFA) (solvent A) and acetonitrile-H2O-TFA (80:20:0.1 [vol/vol/vol]) (solvent B) at a flow rate of 1.0 ml/min. Following the initial wash with solvent A, the peptides were eluted with a linear gradient of 0 to 100% solvent B and detected by determining the absorbance at 220 nm. Suitable fractions were collected, dried in a Speed-Vac concentrator (Jouan Inc., Winchester, Va.), and resuspended in distilled water. The fractions were analyzed by SDS-PAGE in 10 to 18% (wt/vol) acrylamide gradient gels (ISS) in a Tris-Tricine buffer system (22). The fractions containing a single peptide band were directly subjected to N-terminal sequence analysis. Fractions displaying multiple peptide bands by SDS-PAGE were electrophoretically transferred onto a polyvinylidene difluoride membrane as described above. The membrane was stained with Coomassie brilliant blue R-250, and the individual bands were excised for N-terminal sequence analysis (18).
Determination of subunit size by MALDI-TOF mass spectrometry.
Determination of molecular weight by matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometry (12) was done on a Lasermat model 2000 mass analyzer (Finnigan Mat, Hemel Hempstead, United Kingdom) with 3,5-dimethoxy-4-hydroxy-cinnamic acid as the matrix. Precipitation with 90% (vol/vol) ethanol was done with samples containing ≥0.1% (vol/vol) TX-100 to remove the detergent. The precipitated protein was resuspended in water.
Determination of aggregate sizes by gel filtration chromatography.
Approximately 1 mg of the purified protein was precipitated with 90% (vol/vol) ethanol, and the pellet was resuspended in a total volume of 1.0 ml of PBS containing 0.1% TX-100. Two hundred microliters of the preparation was applied to a Superose-6 HR 10/30 gel filtration column (10 by 30 mm; Pharmacia) equilibrated in PBS–0.1% TX-100 at a flow rate of 0.5 ml/min. The column was calibrated with the high-molecular-weight calibration kit (Pharmacia) that contains aldolase (molecular weight, 158,000), catalase (232,000), ferritin (440,000), thyroglobulin (669,000), and blue dextran (2,000,000).
Amino acid sequence analysis.
N-terminal sequence analysis was carried out using an model 477A protein/peptide sequencer equipped with an on-line phenylthiohydantoin (PTH) analyzer (model 120A; Applied Biosystems, Foster City, Calif.). The PTH derivatives were identified by reversed-phase HPLC using a Brownlee PTH C18 column (particle size, 5 μm; 2.1 mm [inside diameter] by 22 cm [length]; Applied Biosystems).
Immunizations.
Female BALB/c mice (Taconic Farms, Germantown, N.Y.) with ages of 6 to 8 weeks were immunized subcutaneously with two doses of UspA1 or UspA2 4 weeks apart. To prepare the vaccine, purified UspA1 or UspA2 was added to aluminum phosphate, and the mixture was rotated overnight at 4°C. 3-O-deacylated monophosphoryl lipid A (MPL; Ribi ImmunoChem Research, Hamilton, Mont.) was added just prior to administration. Each dose of vaccine contained 5 μg of purified protein, 100 μg of aluminum phosphate, and 50 μg of MPL resuspended in a 200-μl volume. Control mice were injected with 5 μg of the genetically detoxified diphtheria toxin (CRM197) with the same adjuvants. Serum samples were collected before the first vaccination and 2 weeks after the second immunization. Mice were housed in a specific-pathogen-free facility and provided water and food ad libitum.
MAbs.
MAbs 17C7, 11A6, and 17H4 were kindly provided by Eric Hansen. MAb 17C7 binds to both UspA1 and UspA2, while MAbs 11A6 and 17H4 are specific for UspA1 and UspA2, respectively (1). The lipooligosaccharide (LOS)-specific MAb Mcat73-11 was prepared as described previously (4).
ELISA procedures.
For the whole-cell enzyme-linked immunosorbent assay (ELISA), the bacteria were grown overnight on Mueller-Hinton agar and swabbed off the plate into PBS. The turbidity of the cells was adjusted to 0.10 at 600 nm, and 100 μl added to the wells of a 96-well F Immunoplate (Nunc Roskilde, Denmark). The cells were dried overnight at 37°C, sealed with a mylar plate sealer, and stored at 4°C until needed. On the day of the assay, the residual protein binding sites were blocked by the addition of 5% non-fat dry milk in PBS with 0.1% Tween 20 (BLOTTO) and incubation at 37°C for 1 h. The blocking solution was then removed, and 100 μl of serum was serially diluted in the wells with BLOTTO. The sera were allowed to incubate for 1 h at 37°C. The plate wells were soaked with 300 ml of PBS containing 0.1% Tween 20 for 30 s, washed three times for 5 s each with a SkanWasher 300 plate washer (Skatron Instruments AS, Lier, Norway), and then incubated for 1 h at 37°C with goat anti-mouse immunoglobulin G (IgG) conjugated to alkaline phosphatase (BioSource International) diluted 1:1,000 in BLOTTO. After being washed, the plates were developed at room temperature with 100 μl of 1-mg/ml p-nitrophenyl phosphate dissolved in diethanolamine buffer per well. Development was stopped by adding 50 μl of 3 N NaOH to each well. The absorbance of each well was read at 405 nm, and titers were calculated by linear regression. The titer is reported as the inverse of the dilution extrapolated to an absorption value of 0.10 U.
For the antigen-specific ELISAs, the proteins were diluted to a concentration of 5 μg/ml in a 50 mM sodium carbonate buffer (pH 9.8) containing 0.02% sodium azide (Sigma Chemical Co.). One hundred microliters was added to each well of a 96-well enzyme immunoassay/radioimmunoassay medium binding ELISA plate (Costar Corp., Cambridge, Mass.) and incubated for 16 h at 4°C. The plates were washed and subsequently treated as described for the whole-cell ELISA procedure.
Complement-dependent bactericidal assay.
This assay was performed similarly to that previously described (4). Relative killing was calculated as the percent reduction in the number of CFU in the sample relative to that in a sample in which heat-inactivated complement replaced active complement. The results are reported as the highest dilution of serum at which greater than 50% of the bacteria were killed.
Inhibition of bacterial adherence to HEp-2 cells.
A total of 5 × 104 HEp-2 cells in 300 μl of RPMI-10 was added to a sterile 8-well Lab-Tek chamber slide (Nunc) and incubated overnight in an incubator containing 5% CO2 to obtain a monolayer of cells on the slide. The slide was washed with PBS and incubated with 300 μl of bacterial suspension (A550 of 0.5) or with a bacterial suspension that had been incubated with antisera (1:100) at 37°C for 1 h. The slides were then washed with PBS and stained with quick stain (Difco) following the manufacture’s instructions. The slide was viewed and photographed by using a light microscope equipped with a camera (Nikon Microphot-SA, Nikon, Tokyo, Japan).
Protein binding to fibronectin and vitronectin.
The binding of purified UspA1 and UspA2 to fibronectin was examined by dot blot. Human plasma fibronectin (Sigma Chemical Co.) was spotted onto a nitrocellulose membrane and allowed to absorb, followed by incubation of the membrane in BLOTTO for 1 h at room temperature. The blot was then washed with PBS and incubated with purified UspA1 or UspA2 (2 μg/ml in BLOTTO) overnight at 4°C. After three washes with PBS, the membrane was incubated with MAb 17C7 diluted in BLOTTO for 2 h at room temperature, washed, and then incubated with secondary antibody (goat anti-mouse immunoglobulin conjugated to alkaline phosphatase [Bio-Rad]) diluted 1:2,000 in BLOTTO for 2 h at room temperature. The membrane was developed with BCIP-NBT as described above.
Binding of UspA1 and UspA2 to vitronectin was examined by spotting purified UspA1 and UspA2 onto a nitrocellulose membrane. After the proteins had been absorbed into the membrane, it was blocked with BLOTTO for 1 h and incubated with human plasma vitronectin (1 μg/ml in BLOTTO; GIBCO BRL, Grand Island, N.Y.) for 4 h at room temperature. After the membrane was washed, bound human vitronectin was detected by incubation first with rabbit anti-human vitronectin serum (GIBCO BRL) and then with goat anti-rabbit IgG-alkaline phosphatase conjugate overnight at 4°C. The bound conjugate antibody was detected with BCIP-NBT as described above.
Interaction of purified proteins with HEp-2 cells.
Each well of a 96-well cell culture plate (Costar) was seeded with 5 × 104 HEp-2 cells in 0.2 ml of RPMI containing 10% fetal calf serum, and the plate was incubated overnight in a 37°C incubator containing 5% CO2. Purified UspA1 or UspA2 (1 to 1,000 ng/well in BLOTTO) was added, and the wells were incubated at 37°C for 2 h. The plate was washed with PBS, incubated with a 1:1 mix of mouse antiserum to UspA1 and UspA2 diluted 1:1,000 in PBS containing 5% dry milk. The plate was washed and then incubated with rabbit anti-mouse IgG conjugated to horseradish peroxidase (1:5,000 in PBS containing 5% dry milk) at room temperature for 1 h. Finally, the plate was washed and developed with a substrate solution containing 2,2′-azino-bis-(3-ethyl-benzthiazoline-6-sulfonic acid) at 0.3 mg/ml in citrate buffer (pH 4.0) containing 0.03% hydrogen peroxide (Kirkegaard and Perry Laboratories). Whole bacteria of strain O35E were included as a positive control. The highest concentration of the bacteria had an optical density at 550 nm of 1.0 and was diluted in threefold dilutions, the same as for the proteins.
Murine model of M. catarrhalis pulmonary clearance.
Groups of 10 BALB/c mice were challenged by instilling the bacteria into their lungs. The degree of pulmonary clearance was assessed 6 h after challenge as described previously (4). The reported clearance values and statistical comparison were relative to a set of 10 mice contemporaneously immunized with CRM197. The data were analyzed by the nonparametric Wilcoxon rank sum test. A probability (P) equal to or less than 0.05 was considered significant.
RESULTS
Purification of UspA1 and UspA2.
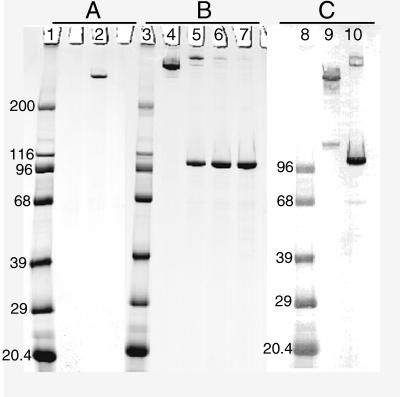
We developed a large-scale, high yield process for extracting and purifying UspA2 from M. catarrhalis cells. The method consisted of three critical steps. First, the UspA2 protein was extracted from the bacteria with 0.03 M THT (pH 8.0). Second, the cell extract was applied to a TMAE column, and the UspA2 protein was eluted with NaCl. Finally, the enriched fractions from the TMAE chromatography were applied to a ceramic hydroxyapatite column, and the UspA2 was eluted with a linear 0.1 to 0.2 mM sodium phosphate gradient. A yield of 250 mg of purified UspA2 was typically obtained from ∼400 g (wet weight) of M. catarrhalis O35E cells. A single band was seen for the UspA2 in SDS-PAGE gels by Coomassie blue staining. It corresponded to a molecular weight of ∼240,000 and contained greater than 95% of the protein based on scanning densitometry (Fig. 1A). A second band reacting with the 17C7 MAb at approximately 125,000, possibly a dimer, could be detected in the UspA2 preparation by Western blotting but not as clearly by Coomassie blue staining (Fig. 1C).
FIG. 1.
SDS-PAGE and Western blots of purified proteins. (A) Coomassie blue-stained gel of purified UspA2 heated for 10 min at 100°C (lane 2). (B) Coomassie blue-stained gel of purified UspA1 prepared either without heating of sample (lane 4) or with heating for 3 min at 100°C (lane 5), for 5 min at 100°C (lane 6), or heated for 10 min at 100°C (lane 7). (C) Western blot of purified UspA2 (lane 9) and purified UspA1 (lane 10) probed with the 17C7 MAb. Both proteins were heated for 10 min during sample preparation. Lanes: 1, 3, and 8, molecular size markers (in kilodaltons).
A method for the purification of the UspA1 protein was also developed. This protein copurified with UspA2 through the initial extraction and TMAE chromatography steps. Following hydroxyapatite chromatography, however, UspA1 remained bound to the column and had to be eluted at the higher salt concentration of 500 mM NaPO4. The partially purified UspA1 preparation obtained in this step was reapplied and eluted from the hydroxyapatite column with a linear sodium phosphate gradient. A total of 80 mg of purified UspA1 was isolated from ∼1.6 kg (wet weight) of M. catarrhalis O35E cells. UspA1 purified by this method migrated at three different apparent sizes by SDS-PAGE depending on the method of sample preparation. Unheated samples exhibited a single band at ∼280,000, whereas samples heated at 100°C for 3 min resulted in an apparent molecular weight shift to ∼350,000. Prolonged heating at 100°C resulted in a shift of the 350,000-molecular-weight band to one at 100,000 (Fig. 1B). Following heating of the sample for 7 min at 100°C, the band at 100,000 contained greater than 95% of the protein based on scanning densitometry of the Coomassie-stained gel. In contrast, UspA2 migrated at 240,000 by SDS-PAGE even when the preparation was heated at 100°C for 10 min. The Western blotting results (Fig. 1C) indicated that the protein preparations were substantially free of each other.
The preparations were examined for the presence of other contaminants, particularly LOS, by Western blotting and other assays. LOS, a common contaminant of proteins prepared from gram-negative bacteria, has been shown to elicit bactericidal activity toward M. catarrhalis (9). The preparations were found to be free of LOS by several methods. Western blotting of bacterial lysates probed with the sera elicited by the UspA1 or UspA2 preparations in mice did not detect material in the low-molecular-size range where M. catarrhalis LOS normally migrates. It was possible to estimate the amount of LOS in the preparations by probing Western blots of the purified proteins with the LOS-specific MAb Mcat73-11. For this, the SDS-PAGE gels were loaded with concentrates of the proteins along with different concentrations of purified LOS as standards. The amount of LOS detected by this method was less than 10 ng per μg of protein in the preparations (data not shown). Finally, the UspA2 preparation passed the rabbit pyrogenicity test required by the American Food and Drug Administration at a dose of 6 μg of protein per kg. Thus, the preparations were substantially free of LOS and other contaminants.
Molecular weight determinations.
MALDI-TOF mass spectrometric analysis for determination of the molecular weight of UspA2 with 3,5-dimethoxy-4-hydroxy-cinnamic acid matrix in the presence of 70% (vol/vol) aqueous acetonitrile and 0.1% TFA resulted in the identification of a predominant species with an average molecular mass of 59,518 Da. In addition to the expected [M+H]+ and [M+2H]2+ molecular ions, the [2M+H]+ and [3M+H]+ ions were also observed. The two latter ions were consistent with the dimer and the trimer species. Using similar conditions, we were unable to determine the mass of UspA1.
To determine the molecular sizes of the purified proteins in solution, UspA1 and UspA2 were independently run on a Superose-6 HR 10/30 gel filtration column (optimal separation range, 5,000 to 5,000,000) calibrated with molecular weight standards. Purified UspA1 exhibited a native molecular weight of 1,150,000, and UspA2 exhibited a molecular weight of 830,000. These weights, however, may be affected by the presence of TX-100.
N-terminal sequence analysis of internal UspA1 and UspA2 peptides.
All attempts to determine the N-terminal sequences of both UspA1 and UspA2 proved unsuccessful. This result suggested that the N-termini of both proteins were blocked. Thus, to confirm that the primary sequences of purified UspA1 and purified UspA2 corresponded to that deduced from their respective gene sequences, internal peptide fragments were generated and subjected to N-terminal sequence analysis. The N-terminal sequences obtained for the fragments generated from the digestion of the UspA2 and UspA1 proteins matched sequences within the primary amino acid sequence deduced from the respective gene sequences (Tables 1 and 2). The UspA2 fragments indicated by an asterisk in Table 1 exhibited sequence similarity with residues 505 to 515 and 605 to 614, respectively, of the sequence deduced from the UspA1 gene. The UspA1 fragment indicated by the asterisk in Table 2 exhibited sequence similarity with residues 278 to 294 of the UspA2 primary sequence. These common sequences corresponded with the domains within UspA1 and UspA2 that share 93% sequence identity (2). The remainder of the sequences were unique to the respective proteins.
TABLE 1.
N-terminal sequences of peptides derived from cleavage of UspA2
| UspA2 peptide sequencea | Matchb (Residue no.) | Cleavage agent |
|---|---|---|
| LLAEQQLNG | 92–100 | Trypsin |
| ALESNVEEGL | 216–225 | Lys-C |
| 245–254 | ||
| 274–283 | ||
| ALESNVEEGLLDLS | 274–288 | Trypsin |
| 505–515* | ||
| AKASAANTDR | 378–387 | Chymotrypsin |
| 605–614* | ||
| AATAADAITKNGN | 439–450 | Chymotrypsin |
| SITDLGTKVDGFDGR | 458–472 | Lys-C |
| VDALXTKVNALDXKVN | 473–488 | Trypsin |
| AAQAALSGLFVPYSVGKFNATAALGGYGSK | 506–535 | CNBr |
Underlined residues denote mismatch with the nucleotide-derived amino acid sequence (2).
An asterisk indicates that the sequence also matches that sequence of UspA1. No asterisk indicates a match only with the nucleotide-derived amino acid sequence of UspA2.
TABLE 2.
N-terminal sequences of peptides derived from cleavage of UspA1
| UspA1 peptide sequencea | Matchb (residue no.) | Cleavage agent |
|---|---|---|
| LENNVEEPXLNLS | 456–468 | Lys-C |
| DQKADI | 473–478 | Trypsin |
| NNVEEGLLDLSGRLIDQK | 504–521 | Lys-C |
| 278–294* | ||
| VAEGFEIF | 690–697 | Trypsin |
| AGIATNKQELILQNDRLNRI | 701–720 | Lys-C |
X denotes an unidentified amino acid residue; underlining denotes a mismatch with the nucleotide-derived amino acid sequence (2).
An asterisk indicates that the sequence also matches that sequence of UspA2; no asterisk indicates a match only with the nucleotide-derived amino acid sequence of UspA1.
Reactivity of Mabs with UspA1 and UspA2.
The Western blot analysis of purified UspA1 and UspA2 revealed that both proteins reacted strongly with the MAb 17C7 described by Helminen et al. (11) (Fig. 1). MAb 11A6 reacted with UspA1 but not UspA2 and MAb 17H4 reacted with UspA2 but not UspA1 as previously demonstrated by Aebi et al. (1) using mutants that did not express the respective proteins (data not shown). All three MAbs bound to whole bacterial cells of strain O35E by whole cell ELISA.
Immunogenicity and antibody cross-reactivity.
Antisera to the purified UspA1 and UspA2 proteins were generated in mice. The titers of antigen-specific antibodies (IgG and IgM) as well as the cross-reactive antibodies in these sera were determined by ELISA with each of the purified proteins (Table 3). Both proteins elicited antibody titers that were significantly greater against themselves than against the heterologous protein. Thus, the reactivities of the polyclonal antibodies as well as the MAbs indicate that the proteins possessed both shared and unique B-cell epitopes.
TABLE 3.
Cross-reactivity of antibodies to UspA1 and UspA2 proteins
| Antiserum targeta | Geometric mean ELISA titerb to:
|
|
|---|---|---|
| UspA1 | UspA2 | |
| UspA1 | 740,642c | 10,748c |
| UspA2 | 19,120d | 37,615d |
The preparation of the sera is described in the text.
ELISA geometric mean titers are for total IgG and IgM antibodies in sera from 10 mice.
The difference in titers of the anti-UspA1 with the two purified proteins was statistically different by the Wilcoxon signed rank test (P = 0.0002).
The difference in titers of the anti-UspA2 with the two purified proteins was statistically different by the Wilcoxon signed rank test (P = 0.01).
Antibody reactivity to whole bacterial cells and bactericidal activity.
Antisera to UspA1 and UspA2 were assayed by whole cell ELISA against the homologous O35E strain and several heterologous isolates (Table 4). The reactivities of the antibodies to UspA1 and UspA2 were strongest with the O35E strain. The reactivities of the sera toward the heterologous isolates indicated that they bound antibodies elicited by both UspA1 and UspA2.
TABLE 4.
ELISA and complement-mediated bactericidal titers to whole bacterial cells of multiple isolates of M. catarrhalis elicited by purified UspA1 and purified UspA2
| Isolate | Whole cell ELISAa
|
Bactericidal titerb
|
||
|---|---|---|---|---|
| Anti-UspA1 | Anti-UspA2 | Anti-UspA1 | Anti-UspA2 | |
| O35E | 195,261 | 133,492 | 400 | 800 |
| 430-345 | 12,693 | 18,217 | 400 | 400 |
| 1230-359 | 7,873 | 13,772 | 400 | 400 |
| TTA24 | 14,341 | 7,770 | 800 | 800 |
The titer was determined for a pool of sera from 10 mice. The titer of the serum drawn before the first immunization was <50 for all isolates.
Bactericidal titers were determined as the inverse of the highest serum dilution killing >50% of the bacteria. The titers for the sera from mice immunized contemporaneously with CRM197 were <100.
The bactericidal activities of the antisera to UspA1 and UspA2 were determined against O35E and other isolates (Table 4). Sera to both proteins had bactericidal titers ranging from 400 to 800 against O35E and the other tested disease isolates. Anti-CRM197 serum, the negative control, and sera drawn before immunization had titers of <100 against all strains.
Inhibition of bacterial adherence to epithelial cells.
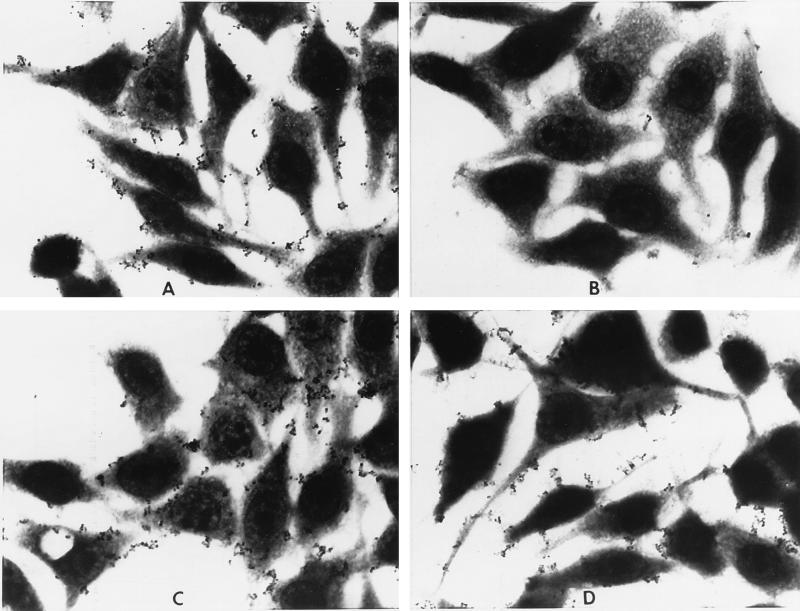
Adherence of the O35E strain to HEp-2 cell monolayers cultured in Lab-Tek chamber slides was observed by light microscopy. Preincubation of the bacteria with saline (Fig. 2A), a normal mouse serum (Fig. 2C), or an antiserum to the UspA2 (Fig. 2D) did not interfere with the attachment of the O35E bacterium. Each of the sera was tested at a 1:100 dilution. The bacteria were observed attached to the surface of the HEp-2 cells, as well as in the interstitial spaces between the cells. Few bacteria appeared to be unassociated with the HEp-2 cells. In contrast, preincubation with an antiserum to the UspA1 resulted in a nearly complete reduction of bacterial attachment to the HEp-2 cells (Fig. 2B).
FIG. 2.
Capacity of antisera to UspA1 to block bacterial attachment to HEp-2 cells. O35E bacteria were preincubated with saline (A), normal mouse serum (C), anti-UspA1 serum (B), or anti-UspA2 serum (D). All sera were tested at a 1:100 dilution. Magnification, ×400.
Interaction of purified proteins with HEp-2 cells.
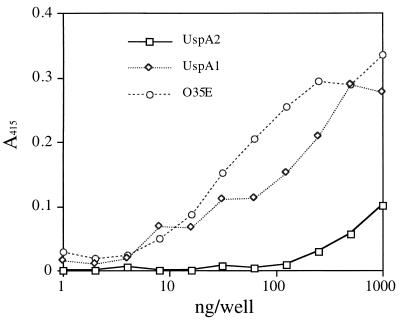
Purified UspA1 and UspA2 were tested for their ability to interact with HEp-2 cell monolayer in a 96-well plate by ELISA. Binding of the proteins to HEp-2 cells was detected with a 1:1 mix of the mouse antisera to UspA1 and UspA2. Purified UspA1 bound to HEp-2 cells at concentrations above 10 ng/well. A weak binding of UspA2 to HEp-2 cells was detected at concentrations above 100 ng/well (Fig. 3). The O35E bacterial isolate also bound to the HEp-2 cells, and this isolate was used as a positive control.
FIG. 3.
Interaction of purified UspA1 and UspA2 with HEp-2 cells determined by ELISA. HEp-2 cell monolayers cultured in a 96-well plate were incubated with serially diluted UspA1 or UspA2. Bacterial strain O35E was used as the positive control. The dilution of the bacteria was analogous to that of the proteins, beginning with a suspension with an A550 of 1.0. The bound proteins or attached bacteria were detected with a 1:1 pool of antisera to UspA1 and UspA2 as described in Materials and Methods.
Interaction of UspA1 and UspA2 with fibronectin and vitronectin.
The purified proteins were assayed for their abilities to interact with fibronectin and vitronectin by dot blot assays. Human plasma fibronectin immobilized on a nitrocellulose membrane bound purified UspA1 but not UspA2 (Fig. 4B and C), while UspA2 immobilized on the nitrocellulose membrane was capable of binding vitronectin (Fig. 4A). Vitronectin binding by UspA1 was also detected, but the reactivity was 1,000-fold weaker than that seen for UspA2. Collagen (type IV), porcine mucin (type III), fetuin, and heparin were also tested for interaction with purified UspA1 and purified UspA2, but they did not exhibit detectable binding (results not shown).
FIG. 4.
Interaction of proteins with fibronectin and vitronectin determined by dot blot. (A) Binding of vitronectin to UspA1 and UspA2. For this blot, the purified proteins were spotted onto the membrane, blocked, and incubated with human plasma vitronectin. The bound vitronectin was detected with a rabbit antivitronectin conjugate. (B) Binding of fibronectin to UspA1. Purified fibronectin was spotted onto the membrane, blocked, and incubated with purified UspA1. The bound UspA1 was then detected with MAb 17C7. (C) Binding of fibronectin to UspA2. This procedure was done as described for panel B except with UspA2. The amount of protein applied at each spot is indicated on the left.
Pulmonary challenge.
Immunized mice were given a pulmonary challenge with the homologous O35E strain or the heterologous TTA24 strain. Relative to that in the control mice immunized with CRM197, enhanced clearance of both strains was observed regardless of whether the mice were immunized with UspA1 or UspA2 (Table 5). Although the heterologous TTA24 strain appeared to be cleared more effectively, no statistical difference (P > 0.05) was seen between the mice immunized with UspA1 and with UspA2.
TABLE 5.
Pulmonary clearance of M. catarrhalis by mice immunized with purified UspA1 and UspA2
| Study (challenge strain) | Immunogen | Mean ± SEMa (CFU [105]/lung) | % Clearance | P |
|---|---|---|---|---|
| 1 (O35E) | UspA1 | 3.2 ± 1.8 | 49.0 | 0.013 |
| UspA2 | 4.3 ± 1.5 | 31.8 | 0.05 | |
| CRM197 | 6.3 ± 1.3 | 0 | ||
| 2 (TTA24) | UspA1 | 1.5 ± 0.9 | 54.6 | 0.02 |
| UspA2 | 1.1 ± 0.5 | 66.6 | 0.0003 | |
| CRM197 | 3.3 ± 0.7 | 0 |
SEM, standard error of the mean.
The challenge method is described in the text. There were 10 mice per group. The numbers are the percentage of bacteria cleared from the immunized mice compared to the percentage in control mice, which were immunized with CRM197.
DISCUSSION
Previous UspA purification attempts yielded preparations containing multiple high-molecular-weight protein bands by SDS-PAGE and Western blotting. Because each of the bands reacted with the UspA-specific MAb 17C7, it was thought they represented multiple forms of a single UspA protein (4). However, recent reports have demonstrated that there are two distinct genes encoding two different proteins that share an epitope recognized by the 17C7 MAb (1, 2). The present study shows that the proteins UspA1 and UspA2 can be separated from one another. Once separated, the isolated proteins were shown to have different SDS-PAGE mobility characteristics, different reactivities with a set of monoclonal antibodies, and many different internal peptide sequences. Yet, the results from the peptide analyses were also consistent with the proteins sharing a portion of their peptide sequences, including the MAb 17C7 epitope (2).
The separation of the proteins from one another allowed us to further demonstrate that these proteins possess different biochemical, functional, and immunological characteristics. The gel filtration studies indicate that in solution, the purified proteins are homopolymers of their respective subunits held together by strong noncovalent forces. The noncovalent nature of the interaction is indicated by the facts that UspA2 lacks any cysteines and treatment of both proteins with reducing agents did not alter their mobilities by SDS-PAGE. Both gene sequences encode leucine zipper motifs that might mediate intermolecular interactions (20) between monomers. Even so, it was surprising that the noncovalent bonds of both proteins were strong enough to resist not only dissociation by the conditions normally used to prepare samples for SDS-PAGE but also high concentrations of chaotropic agents such as urea (14). By SDS-PAGE, the dominant UspA2 oligomeric form migrated with an apparent molecular weight of 240,000 while a far smaller portion migrated at about 125,000, which was best detected by Western blot analysis. The mobility of UspA1, however, varied depending on how long the sample was heated. The smallest form was about 100,000, the size of the gene product missing from the uspA1 deletion mutant reported by Aebi et al. (2), but is different from the size of 88,000 Da predicted from the gene sequence. In solution, both proteins formed larger aggregates than those seen by SDS-PAGE. Their molecular weights, as measured by gel filtration chromatography, were 1,150,000 and 830,000 for UspA1 and UspA2, respectively. If the proteins behave this way in vivo, UspA1 and UspA2 likely occur as large molecular complexes on the bacterial surface.
The results of the N-terminal amino acid sequence analyses of the UspA2- and UspA1-derived peptides (Tables 1 and 2) are in agreement with the protein sequences derived from the respective gene sequences. This finding confirmed that the purified UspA1 and UspA2 proteins were the products of the respective uspA1 and uspA2 genes (2). Further, the experimental and theoretical amino acid compositions of UspA1 and UspA2 (2) were generally consistent (data not shown). There is, however, a discrepancy between the molecular weight of 59,518 determined by mass spectrometry and the molecular weight of 62,483 indicated from the gene sequence for UspA2 (2). This discrepancy suggests that this protein either undergoes posttranslational processing or proteolytic degradation. Further work is needed to elucidate the structures of both proteins.
The data are consistent with both proteins being exposed on the bacterial surface. That they are both exposed was shown by Aebi et al. (1) using MAbs 11A6 and 17H4 that were originally obtained by screening against the purified proteins. Surface exposure of the proteins is probably important for the two proteins’ functions. One function for UspA1 appears to be mediation of adherence to host tissues (1). The relevance of binding to HEp-2 cells is that these epithelial cells are derived from the larynx, a common site of M. catarrhalis colonization (23). Our preliminary data suggest that the target for UspA1 binding is fibronectin. Fibronectin has been reported to be a host receptor for other pathogens (17, 28). Examination of the UspA1 gene sequence, however, failed to reveal any obvious similarity to the known fibronectin binding motifs reported for gram-positive organisms (28). Thus, UspA1 probably plays a role in host adherence, possibly via cell-associated fibronectin.
The function of UspA2 has been reported to be associated with sensitivity to normal human serum (1). That it is not an adhesin is indicated by the facts that antibodies to it do not block adherence to the HEp-2 or Chang cell lines and that the purified protein did not bind to the cells. Our results, however, suggest that UspA2 binds vitronectin. While binding of pathogens to vitronectin has been linked to cell adherence and invasion (6, 7, 16), van Dijk and his coworkers reported that vitronectin binding by M. catarrhalis may subvert host defenses (27). The soluble form of vitronectin is also known as S-protein in the complement system, and it regulates the formation of the membrane attack complex (25). van Dijk et al. suggest that the binding of vitronectin to the M. catarrhalis surface inhibits the formation of the membrane attack complex, rendering the bacteria resistant to the complement-dependent killing activity of the sera. They have also described two types of human isolates. One that binds vitronectin and is resistant to the lytic activity of the serum, and the other that does not bind vitronectin and is serum sensitive (13). It must be noted, however, that vitronectin, like all the extracellular matrix proteins, has many forms and serves multiple functions in the host (21, 24). Thus, the interaction of both UspA1 and UspA2 with the extracellular matrix proteins fibronectin and vitronectin may serve the bacterium in ways other than subverting host defenses or as receptors for bacterial adhesion and invasion.
Our primary interest in these proteins is their potential as vaccine antigens. Even though the two proteins share epitopes and sequences, they have different biochemical characteristics and likely serve different biological functions. The results of the immunological studies of mice indicate that both proteins are good vaccine candidates. Mice immunized with either UspA1 or UspA2 developed high antibody titers toward the homologous and heterologous bacterial isolates. The sera from these mice had complement-dependent bactericidal activity toward heterologous isolates. In addition, immunized mice exhibited enhanced pulmonary clearance of both the homologous isolate and a heterologous isolate regardless of which protein was used for the immunization. We also examined the immunogenicity of a mixture of the two proteins, but this combination did not significantly improve the immune response compared with those seen for the proteins alone (results not shown). It is important that antibodies elicited by the proteins were cross-reactive. This finding was anticipated since both react with the 17C7 MAb and share an amino acid sequence (2). Based on these results, both proteins appear to be good vaccine candidates.
ACKNOWLEDGMENTS
We thank Terri Mininni for preparing the Mcat73-11 MAb, Ben Metcalf for his examination of the MAbs by Western blotting, and Denise Hahn for technical assistance in developing the adhesion assay. We thank Bruce Green and James Cowell for valuable discussions on the biochemical and immunological analysis of the proteins and the preparation of the manuscript. We are grateful to Eric Hansen for providing bacterial isolates and the other monoclonal antibodies.
REFERENCES
- 1.Aebi C, Lafontaine E R, Cope L D, Latimer J L, Lumbley S L, McCracken G H, Hansen E J. Phenotypic effect of isogenic uspA1 and uspA2 mutations on Moraxella catarrhalis O35E. Infect Immun. 1998;66:3113–3119. doi: 10.1128/iai.66.7.3113-3119.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aebi C, Maciver I, Latimer J L, Cope L D, Stevens M K, Thomas S E, McCracken G H, Hansen E J. A protective epitope of Moraxella catarrhalis is encoded by two different genes. Infect Immun. 1997;65:4367–4377. doi: 10.1128/iai.65.11.4367-4377.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyle F M, Georghiou P R, Tilse M H, McCormack J G. Branhamella (Moraxella) catarrhalis: pathogenic significance in respiratory infections. Med J Aust. 1991;154:592–596. doi: 10.5694/j.1326-5377.1991.tb121219.x. [DOI] [PubMed] [Google Scholar]
- 4.Chen D, McMichael J C, VanDerMeid K R, Hahn D, Mininni T, Cowell J, Eldridge J. Evaluation of purified UspA from Moraxella catarrhalis as a vaccine in a murine model after active immunization. Infect Immun. 1996;64:1900–1905. doi: 10.1128/iai.64.6.1900-1905.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davies B I, Maesen F P V. The epidemiology of respiratory tract pathogens in Southern Netherlands. Eur Respir J. 1988;1:415–420. [PubMed] [Google Scholar]
- 6.Dehio M, Gómez-Duarte O G, Dehio C, Meyer T F. Vitronectin-dependent invation of epithelial cells by Neisseria gonorrhoeae involves αv intrgrin receptors. FEBS Lett. 1998;424:84–88. doi: 10.1016/s0014-5793(98)00144-6. [DOI] [PubMed] [Google Scholar]
- 7.Gómez-Duarte O G, Dehio M, Guzman C A, Chhatwal G S, Dehio C, Meyer T F. Binding of vitronectin to Opa-expressing Neisseria gonorrhoeae mediates invasion of HeLa cells. Infect Immun. 1997;65:3857–3866. doi: 10.1128/iai.65.9.3857-3866.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gottfarb P, Brauner A. Children with persistent cough—outcome with treatment and role of Moraxella catarrhalis. Scand J Infect Dis. 1994;26:545–551. doi: 10.3109/00365549409011812. [DOI] [PubMed] [Google Scholar]
- 9.Gu X-X, Chen J, Barenkamp S J, Robbins J B, Tsai C-M, Lim D J, Battey J. Synthesis and characterization of lipooligosaccharide-based conjugates as vaccine candidates for Moraxella (Branhamella) catarralis. Infect Immun. 1998;66:1891–1897. doi: 10.1128/iai.66.5.1891-1897.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hager H, Verghese A, Alverez S, Berk S L. Branhamella catarrhalis respiratory infections. Rev Infect Dis. 1987;9:1140–1149. doi: 10.1093/clinids/9.6.1140. [DOI] [PubMed] [Google Scholar]
- 11.Helminen M E, Maciver I, Latimer J L, Klesney-Tait J, Cope L D, Paris M, McCracken G H, Hansen E J. A large, antigenically conserved protein on the surface of Moraxella catarrhalis is a target for protective antibodies. J Infect Dis. 1994;170:867–872. doi: 10.1093/infdis/170.4.867. [DOI] [PubMed] [Google Scholar]
- 12.Hillenkamp F, Karas M. Mass spectrometry of peptides and proteins by matrix-assisted ultraviolet laser desorption/ionization. Methods Enzymol. 1990;193:280–295. doi: 10.1016/0076-6879(90)93420-p. [DOI] [PubMed] [Google Scholar]
- 13.Hol C, Verduin C M, van Dijke E, Verhoef J, van Dijk H. Complement resistance in Branhamella (Moraxella) catarrhalis. Lancet. 1993;341:1281. doi: 10.1016/0140-6736(93)91185-o. [DOI] [PubMed] [Google Scholar]
- 14.Klingman K L, Murphy T F. Purification and characterization of a high-molecular-weight outer membrane protein of Moraxella (Branhamella) catarrhalis. Infect Immun. 1994;62:1150–1155. doi: 10.1128/iai.62.4.1150-1155.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 16.Limper A H, Standing J E, Hoffman O A, Castro M, Neese L W. Vitronectin binds to Pneumocystis carnii and mediates organism attachment to cultured lung epithelial cells. Infect Immun. 1993;61:4302–4309. doi: 10.1128/iai.61.10.4302-4309.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ljungh Ä, Wadström T. Binding of extracellular matrix proteins by microbes. Methods Enzymol. 1995;253:501–514. doi: 10.1016/s0076-6879(95)53041-x. [DOI] [PubMed] [Google Scholar]
- 18.Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987;262:10035–10038. [PubMed] [Google Scholar]
- 19.Murphy T F. Branhamella catarrhalis: epidemiology, surface antigenic structure, and immune response. Microbiol Rev. 1996;60:267–279. doi: 10.1128/mr.60.2.267-279.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Shea E K, Klemm J D, Kim P S, Alber T. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science. 1991;254:539–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- 21.Preissner K T. Structure and biological role of vitronectin. Annu Rev Cell Biol. 1991;7:275–310. doi: 10.1146/annurev.cb.07.110191.001423. [DOI] [PubMed] [Google Scholar]
- 22.Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 23.Schalen L, Eliasson I, Fex S, Kamme C, Schalen C. Acute laryngitis in adults: results of erythromycin treatment. Acta Otolaryngol Suppl (Stockholm) 1992;492:55–57. doi: 10.3109/00016489209136810. [DOI] [PubMed] [Google Scholar]
- 24.Seiffert D. Constitutive and regulated expression of vitronectin. Histol Histopathol. 1997;12:787–797. [PubMed] [Google Scholar]
- 25.Su H R. S-protein/vitronectin interaction with the C5b and the C8 of the complement membrane attack complex. Int Arch Allergy Immunol. 1996;110:314–317. doi: 10.1159/000237322. [DOI] [PubMed] [Google Scholar]
- 26.van Cauwenberge, P. B., M. A. M. Vander, and K. J. Ingels. 1993. The microbiology of acute and chronic sinusitis and otitis media: a review. Eur Arch Otorhinolaryngol 250(Suppl. 1):S3–S6. [DOI] [PubMed]
- 27.Verduin C M, Jansze M, Hol C, Mollines T E, Verhoef J, van Dijk H. Differences in complement activation between complement-resistant and complement-sensitive Moraxella (Branhamella) catarrhalis strains occur at the level of membrane attack complex formation. Infect Immun. 1994;62:589–595. doi: 10.1128/iai.62.2.589-595.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Westerlund B, Korhonen I K. Bacterial protein binding to mammalian extracellular matrix. Mol Microbiol. 1993;9:687–694. doi: 10.1111/j.1365-2958.1993.tb01729.x. [DOI] [PubMed] [Google Scholar]