Beaumale et al report that C. elegans Katanin binding to microtubules is dictated by its p80-like (MEI-2) subunit via two microtubule-binding domains (MTBDs) and that MEI-2-mediated MT binding is essential for female meiotic spindle assembly.
Abstract
Microtubule-severing enzymes (MSEs), such as Katanin, Spastin, and Fidgetin play essential roles in cell division and neurogenesis. They damage the microtubule (MT) lattice, which can either destroy or amplify the MT cytoskeleton, depending on the cellular context. However, little is known about how they interact with their substrates. We have identified the microtubule-binding domains (MTBD) required for Katanin function in C. elegans. Katanin is a heterohexamer of dimers containing a catalytic subunit p60 and a regulatory subunit p80, both of which are essential for female meiotic spindle assembly. Here, we report that p80-like(MEI-2) dictates Katanin binding to MTs via two MTBDs composed of basic patches. Substituting these patches reduces Katanin binding to MTs, compromising its function in female meiotic-spindle assembly. Structural alignments of p80-like(MEI-2) with p80s from different species revealed that the MTBDs are evolutionarily conserved, even if the specific amino acids involved vary. Our findings highlight the critical importance of the regulatory subunit (p80) in providing MT binding to the Katanin complex.
Introduction
Microtubules (MTs) are dynamic cellular structures with critical roles in numerous cellular processes, including cell motility, intracellular transport, and cell division (Nogales, 2000). These functions depend on their dynamical behavior, which is controlled by microtubule-associated proteins (MAPs) (Bodakuntla et al., 2019). Among the MAPs, the microtubule-severing enzyme (MSE) emerges as a specialized subclass of enzymes, which directly target the lattice of microtubules to sever them (McNally and Roll-Mecak, 2018; Roll-Mecak and McNally, 2010; Sarbanes et al., 2022; Sharp and Ross, 2012).
Belonging to the AAA+ ATPase (ATPase associated with diverse cellular activities) family, MSEs encompass three conserved enzymes: Fidgetin, Spastin, and Katanin. These MSEs are characterized by the presence of an AAA+ domain, enabling protein oligomerization and ATP hydrolysis, which defines the functional properties of the family (Kuo and Howard, 2021; Lynn et al., 2021; McNally and Roll-Mecak, 2018; Roll-Mecak and McNally, 2010; Sarbanes et al., 2022; Sharp and Ross, 2012).
Recently, the role of MSEs, previously reported as microtubule-severing enzymes, has been re-evaluated (Vemu et al., 2018). Vemu et al. (2018) showed that MSEs use ATP hydrolysis to damage the tubulin lattice, creating a hole at the surface of the microtubules. Then, depending on the cellular context, MTs can either be repaired by the incorporation of GTP-tubulin, leading to the formation of stable microtubules, or further weakened, leading to MT severing and the formation of two new microtubule ends. These new ends in turn might depolymerize to dismantle the microtubule or be used as seeds to polymerize new microtubules depending on the GTP–tubulin availability (Kuo and Howard, 2021; Kuo et al., 2019; Vemu et al., 2018).
Despite this breakthrough, how MSEs primarily interact with microtubules is still poorly understood. The most documented example is for the human Spastin, for which a domain of about a hundred amino acids (called the microtubule-binding domain, MTBD) has been identified (White et al., 2007). Unfortunately, Spastin MTBD motif (Homo sapiens residue 270–328) is not resolved in the available Spastin structures (e.g., PDB 6PEK, starting at residue 323) (Han et al., 2020; Roll-Mecak and Vale, 2008), and sequence analysis does not allow the identification of a conserved MTBD motif using interspecies comparison. Likewise, an MTBD cannot be identified on Katanin or Fidgetin by sequence analysis. Finally, sequence analysis using known MT-binding motifs present in other MAPs, for example, Kinesins, Dyneins, or TOG domain proteins, did not provide clues about putative MTBD sequences. Thus, the exact molecular determinants allowing MSE•MT interaction need to be firmly established and might vary between the three MSEs.
In contrast to Spastin and Fidgetin, which are organized as homohexamers, Katanin is composed of two subunits: a p60 catalytic subunit containing the AAA+ domain (Walker A and B motif allowing the enzyme to bind and to hydrolyze ATP, respectively) and a p80 regulatory subunit previously shown to dictate Katanin cellular localization (Akhmanova and Steinmetz, 2019; Hartman et al., 1998; Hartman and Vale, 1999; McNally and Vale, 1993; McNally et al., 2000; Mishra-Gorur et al., 2014). The hetero-oligomeric organization of Katanin adds complexity to the identification of a MTBD, which might be present in either p60, p80, or formed by the interaction between the two subunits (Faltova et al., 2019; Jiang et al., 2017; Rezabkova et al., 2017).
Using structural approaches, Zehr et al. (2019) recently showed that the central pore formed by p60 AAA+ domain hexamerization engages the acidic C-terminal tail (CTT) of tubulin. This interaction is sensitive to posttranslational modifications and is required for microtubule severing, but whether it constitutes the primary binding site is not known (Bailey et al., 2015; Gadadhar et al., 2021; Garnham and Roll-Mecak, 2012; Genova et al., 2023; Lindsay et al., 2023). If this interaction constitutes the sole Katanin–microtubule interaction, it would imply that the Katanin hexamer would land on the microtubules directly engaging the CTT. Alternatively, other MT-binding domains might first dock the catalytic subunit on MT and position the central pore of the enzyme on the CTT exposed at the MT surface. Several observations support this latter hypothesis. First, Hartman and Vale (Hartman and Vale, 1999) observed that the N-terminal non-AAA region of p60 binds microtubules with an affinity intermediate between full-length p60 in ATPγS or ADP, consistent with the observations (i) that sea urchin p60 can sever microtubules without p80 and (ii) that the presence of the p80 subunit stimulates its MT-severing activity (Hartman et al., 1998). Second, the presence of the con80 domain or KATNBL1, which binds MT with a low affinity, drastically enhanced MT binding activity to the N-terminal domain of human p60 (McNally et al., 2000; McNally and McNally, 2011). In contrast to previous observations, we reported for Caenorhabditis elegans Katanin (MEI-1•MEI-2) that MEI-2 alone, but not MEI-1, binds microtubules, providing direct evidence that MEI-2 contains a bona fide MTBD (Joly et al., 2016). However, sequence analysis did not reveal any obvious MTBD motif. More recently, Zehr et al. (2019) identified a positively charged region in MEI-1 between the MIT and AAA domain that contributes to microtubule binding and severing activity in the presence of MEI-2. In addition, Szczesna et al. (2022) reported that the N terminal region of MEI-1 (non-AAA), in complex with MEI-2 C-terminal domain is sensitive to the structure of the C-terminal tails of tubulin (CTT) and to its level of posttranslational modification. All together, these observations suggest a complex mode of interaction between Katanins and the microtubules, and that this interaction might slightly differ between species.
Here, we dissected the precise contribution of the MEI-2 subunit to Katanin activity in C. elegans. In this system, the inactivation of each Katanin subunit (MEI-1 or MEI-2) results in a similar failure in meiotic spindle assembly causing embryonic lethality (Mains et al., 1990; Srayko et al., 2000). However, partial reduction of Katanin function causes the assembly of a longer meiotic spindle, giving rise to the extrusion of abnormally large polar bodies after cytokinesis (Gomes et al., 2013; Han et al., 2009; Joly et al., 2016; Mains et al., 1990; McNally et al., 2006; Srayko et al., 2000, 2006). As Katanin activity is essential for the assembly of the meiotic spindle, monitoring the formation and the size of the polar body provides a quantifiable readout to monitor Katanin activity in vivo.
Here, by combining in vitro approaches with structure–function analysis in vivo, we revealed that MEI-2 primarily binds MT through two microtubule-binding domains (MTBDs) composed of basic patches and thereby dictates MEI-1 binding to microtubules. Importantly, we show that reducing MEI-2-binding to microtubules by only a factor of two causes major defects in Katanin function in vivo, indicating that optimal MEI-2•MT interaction is critical for Katanin activity in vivo.
Taken together, our findings demonstrate that the p80 regulatory subunit MEI-2 provides MT binding to the Katanin complex, thereby promoting MT severing and Katanin function in vivo.
Results
p80-like(MEI-2) subunit dictates Katanin binding to microtubules
To establish which subunit promotes initial Katanin binding to microtubules, we tested whether MEI-1 or MEI-2 is responsible for this interaction.
To do so, we set up an in vitro interaction assay (Fig. 1 A) in which we mixed purified Katanin and microtubules in the presence of the protein crosslinker EDC (1-ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride), allowing us to resolve covalent complexes between Katanin and MTs by SDS-PAGE if a Katanin•MT interaction occurs. To identify the protein present in each complex and to reveal the identity of the crosslinked subunits to microtubules, we performed the crosslinking experiment using MEI-1 alone or by changing the size of MEI-1 or MEI-2 using different tags or truncation variants (Fig. 1, B and C).
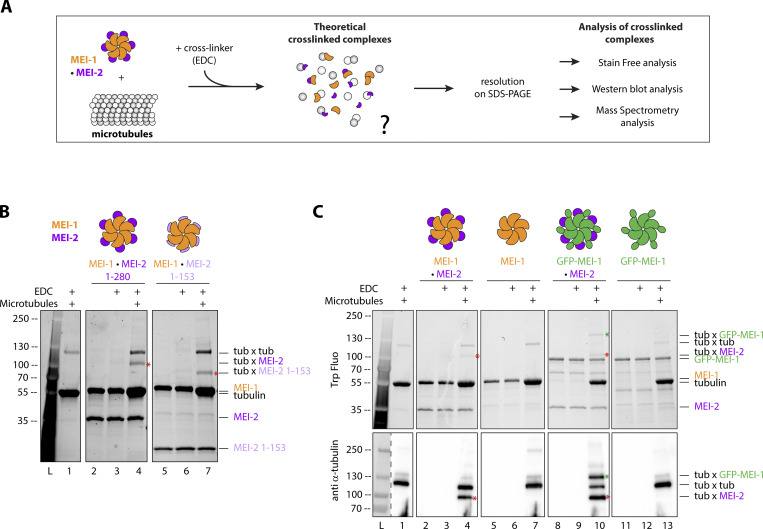
Figure 1.
Both Katanin subunits interact with microtubules. (A) Schematic of the in vitro crosslink assay. Briefly, purified proteins (KataninMEI-1-MEI-2 and microtubules) were mixed and incubated with EDC chemical crosslinker for 15 min. Samples were resolved on SDS PAGE and analyzed using stain-free (BioRad) based on tryptophan fluorescence labeling, Western blot, or mass spectrometry. (B) Crosslink assay between Katanin, composed of MEI-1 (orange) and MEI-2 full length (1–280) (purple) or N-terminal fragment (1–153) (light purple) and microtubules. Red stars indicate crosslinked complexes MEI-2*tubulin. SDS PAGE (BioRad) was analyzed by stain-free technique based on tryptophan fluorescence (Stain Free; Bio-Rad). (C) Crosslink assay between KataninMEI-1-MEI-2 or MEI-1 alone (orange) with or without GFP tag (green) and microtubules. Red stars indicate crosslinked complexes MEI-2*tubulin. Green star indicates the crosslinked complex GFP-MEI-1*tubulin. SDS PAGE was analyzed using tryptophan fluorescence (Stain Free; Bio-Rad) (upper panel) and by Western blot using anti-tubulin antibody (lower panel). Experiments were performed at least in triplicate and the maximal variation observed was <10%. Source data are available for this figure: SourceData F1.
First, we exposed Katanin to EDC and did not observe differences compared with the control condition in the absence of EDC (Fig. 1 B, compare lanes 2 and 3). From these results, we concluded that no crosslinked species were formed between MEI-1*MEI-1, MEI-1*MEI-2, or MEI-2*MEI-2, most likely due to the absence of compatible residues (K, E, D) at the level of these subunit interfaces. We then mixed Katanin with MTs in the presence of EDC and observed a specific band migrating around 100 kDa (red asterisk—Fig. 1 B compare lanes 4 to lanes 1, 2, and 3). This band was only detectable when Katanin was incubated with MTs. Based on its size (around 100 kDa), we suspected that this band is composed of MEI-2 (33 kDa) and tubulin (around 60 kDa). To test this hypothesis, we repeated the experiment with a shorter version of MEI-2, MEI-21–153 (25 versus 33 kDa for MEI-21–280), and observed that the crosslinked band was downshifted accordingly (75 kDa), demonstrating the presence of MEI-21–153 (red asterisks; Fig. 1 B compare lane 7 to 4). From these observations, we conclude that MEI-2 directly interacts with MTs.
In addition, we observed a band migrating around 120 kDa, which is similar in size to a band produced by the crosslink of tubulin moieties in the control experiment (tub × tub; Fig. 1 B, lane 1). However, the intensity of this band specifically increased in the presence of Katanin (Fig. 1 B, lane 4 and 7), suggesting some changes in the crosslinked species. Based on its size, we hypothesized that this band could correspond to different crosslinked complexes: either tubulin*tubulin, MEI-1*MEI-1, or MEI-1*tubulin. To reveal its composition, we modified the molecular weight of MEI-1 using a GFP tag (GFP-MEI-1: 80 versus 55 kDa for MEI-1). In this context, the crosslinked band was upshifted (around 140 kDa) when MTs were mixed with GFP-MEI-1•MEI-2 but not with GFP-MEI-1 alone (green asterisk; Fig. 1 C, compare lane 10 to 4–7 and 13). We confirmed the presence of tubulin in this upshifted band by Western blot (Fig. 1 C, bottom panel). From this observation, we conclude that MEI-1 interacts with microtubules, but only when MEI-2 is present.
Taken together, these results establish that MEI-2 interacts with MTs, allowing MEI–1•MT interaction to occur. These observations also suggest that MEI-2 brings MEI-1 in proximity to MTs to promote the interaction between the central pore of MEI-1 and the tubulin–CTT required for substrate remodeling (Zehr et al., 2019).
KataninMEI-1-MEI-2 binds to microtubules via two distinct MT-binding domains in MEI-2
We next aimed to identify the MT-interacting domain (MTBD) of MEI-2, promoting Katanin•MTs interaction.
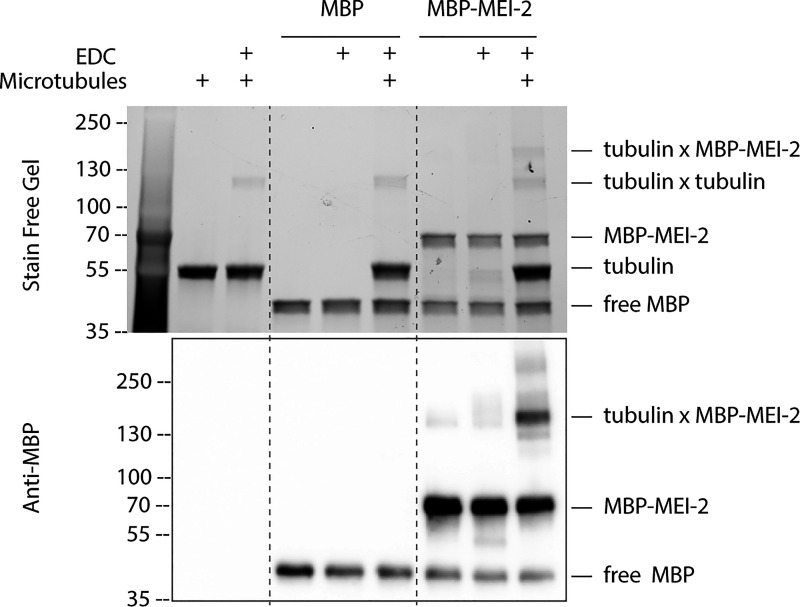
To this end, we produced and purified 20 different MEI-2 fragments spanning the N-terminal part, the central region, and the C-terminal part and tested their ability to interact with MT using EDC crosslink experiments (Fig. 2 A). We observed that the N-terminal (MEI-21–70) and the C-terminal part of MEI-2 (MEI-2241–280) crosslinked with microtubules while no crosslink could be detected using the central region of MEI-2 (MEI-270–240; Fig. 2 B compare lanes 5, 8, 11, and 14 and Fig. S2).
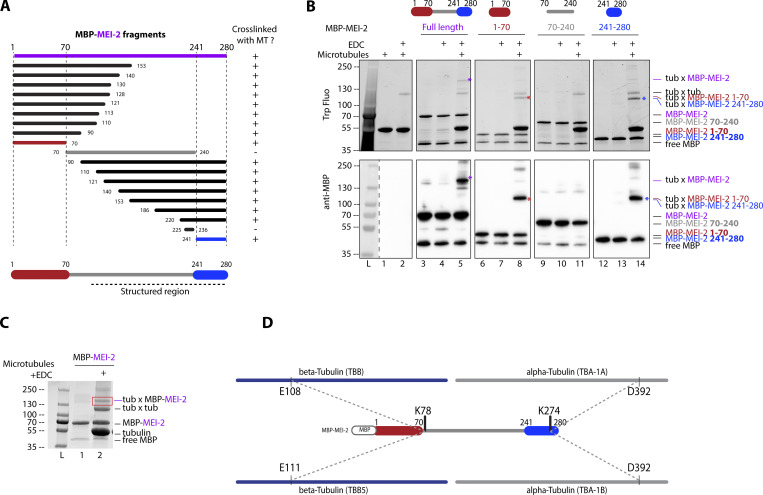
Figure 2.
Katanin binds to microtubules via two domains on MEI-2 subunit. (A) Schematic representation of all MEI-2 fragments tagged with MBP tested in crosslink with microtubules in vitro. Black fragments still crosslink with microtubules, whereas the gray fragment does not. MEI-2 1–70 (red) and MEI-2 241–280 (blue) were identified as two minimal MEI-2 fragments able to crosslink with microtubules. The dashed line corresponds to the putative structured region of MEI-2 based on the Mm Katnb1 fragment structure (Jiang et al., 2018). (B) Crosslink assay between MBP–MEI-2 full-length (purple) or fragments and microtubules. Colorful stars indicate crosslinked complexes MBP–MEI-2*tubulin or MBP-MEI-2 fragment*tubulin. SDS PAGE was analyzed using tryptophan fluorescence (Stain Free; Bio-Rad; upper panel) and by Western blot using anti-MBP antibody (lower panel). The absence of crosslink between MBP tag alone and microtubules is controlled in Fig. S2. Experiments were performed at least in triplicate and the maximal variation observed was <10%. (C) Red square corresponds to crosslink complex MBP–MEI-2*tubulin extracted from SDS PAGE to be analyzed by mass spectrometry. (D) Schematic representation of MEI-2, porcine alpha- and beta-tubulins crosslinked residues identified by mass-spectrometry (see Table S1). Two MEI-2 residues were identified as specifically crosslinked with alpha- or beta-tubulin. Source data are available for this figure: SourceData F2.
Figure S2.
MEI-2 specifically crosslinks with microtubules in vitro. Crosslink assay between MBP alone or MBP-MEI-2 and microtubules. SDS PAGE (BioRad) was analyzed by free staining based on tryptophan fluorescence (upper panel) and by Western blot using anti-MBP antibody (lower panel). Source data are available for this figure: SourceData FS2.
We thus conclude that MEI-2 binds to microtubules via two MTBDs located in MEI-21–70 (MTBD1) and MEI-2241–280 (MTBD2).
Next, we analyzed the MEI-2*tubulin crosslinked band (Fig. 2 C, red box) with mass spectrometry to map interacting sites on MEI-2 and MTs and to determine whether MEI-2 targets a specific site at the surface of MTs. We identified two crosslinked sites on MEI-2 (K78 and K274) targeting two distinct sites on tubulins (beta tubulin TBB E108 or TBB-5 E111, and alpha tubulin TBA-1 and TBA-2 D392, respectively; Fig. 2 D). Interestingly, the apparent molecular weight of the band extracted from SDS-PAGE corresponds to one MEI-2 moiety and one crosslinked tubulin (MEI-2*tubulin about 130 kDa—MBP-MEI-2 70 kDa + tubulin 60 kDa). We conclude that MEI-2 interacts with the globular domain of a single tubulin subunit at a time, and that each MEI-2 lysine binds specifically to either alpha or beta tubulin.
Taken together, these results indicate that MEI-2 directly interacts with microtubules via at least two distinct MTBDs, which target specific alpha or beta tubulin on their globular domain.
Basic residues of MEI-2 are involved in direct KataninMEI-1•MEI-2–microtubule interaction
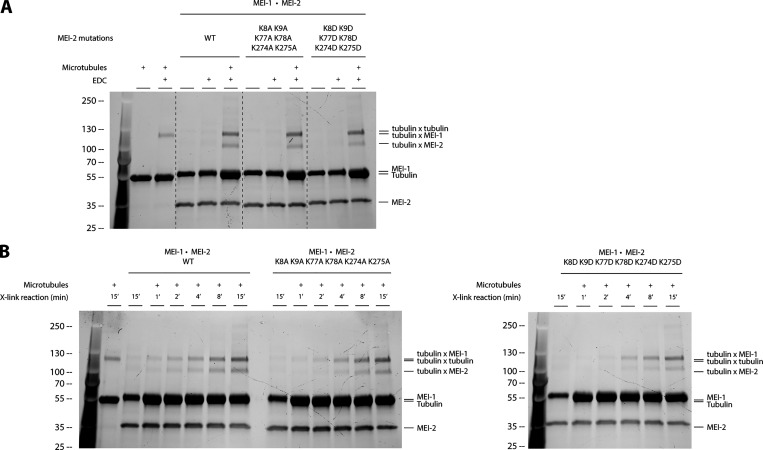
To further characterize Katanin•MT interaction, we aimed to identify key MEI-2 residues allowing this interaction. Guided by the crosslinking and mass spectrometry experiments (Fig. 2), we observed that the crosslinked lysines (K) identified in MEI-2 were organized in pairs (K77-K78, K274-K275) and that a KK pair was also present in MTBD1 (MEI-21–70; K8 K9). To test their importance, we replaced the positively charged KK pairs of both minimal MEI-2-interacting fragments identified (MEI-21–70 and MEI-2240–280) with either apolar (Alanine-A) or negatively charged (Aspartate-D) residues and tested their interaction with MTs using crosslink experiments (Fig. 3, A and B).
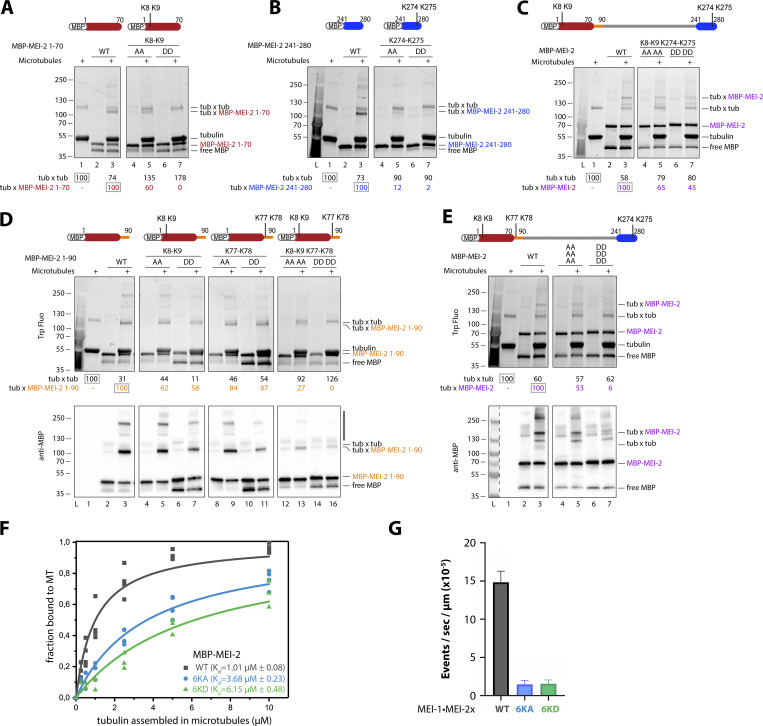
Figure 3.
Microtubule binding of MEI-2 depends on lysine patches. Crosslink assay between MBP-MEI-2 fragments and microtubules. (A–E) The lysines (KK) of interest were mutated in alanines (AA) or aspartates (DD) in minimal MBP-MEI-2 fragments (A) 1–70 (red) and (B) 241–280 (blue), in MBP-MEI-2 full length (C and E) or in MBP-MEI-2 fragment (D) 1–90 (orange). SDS PAGE (BioRad) was analyzed using tryptophan fluorescence (Stain Free; Bio-Rad; upper panel) and by Western blot using anti-MBP antibody (lower panel) for D and E. Quantification of Tubulin*tubulin and Tubulin*MBP-MEI-2 full length or fragment complexes were performed using ImageJ, and values are expressed as percentage of, respectively, tubulin alone and tubulin in the presence of MBP–MEI-2 full length or fragments WT (gray boxes). (F) Microtubule binding constant curves of MEI-2 using the copelleting approach. Different microtubule concentrations (as indicated) were incubated with purified MBP-MEI-2 WT or variant (6KA or 6KD, as indicated) for 20 min and spun down. Proteins present in the supernatant and the pellet fraction were analyzed using SDS-PAGE and quantified using ImageJ. This graph shows the amount of MBP-MEI-2 detected depending on the concentration of microtubule, and Kd was estimated using Origin software. At least three independent experiments were performed. (G) Microtubule severing activities of MEI-1 WT•MEI-2 WT or variant (6KA or 6KD, as indicated). Severing rates were calculated from three independent experiments with the following total number of microtubules: 206 for WT, 114 for 6KA, and 183 for 6KD. Error bars correspond to SEM. Source data are available for this figure: SourceData F3.
While crosslinked bands were observed in the context of WT fragments, substituting K8-K9 or K274-K275 reduced the amount of MEI-2*tubulin crosslink formed, with a more pronounced effect observed with aspartate substitutions compared with alanine (Fig. 3 A, compare lane 7 to 5 and 3 and Fig. 3 B, compare lane 7 to 5 and 3).
We then introduced these substitutions into full-length MEI-2 (MEI-2FL K8A-K9A K274A-K275A and K8D-K9D K274D-K275D). We observed that even with aspartate substitutions at all four positions, which almost abolished the interaction in the context of the fragments, the characteristic MEI-2*tubulin band was still formed using MEI-2FL, though in reduced amounts (Fig. 3 C, lane 7), suggesting the presence of other microtubule-interacting motif(s).
Based on the mass spectrometry results revealing a crosslink of the K77–K78 pair, which is only present in MEI-2FL but not in MTBD1 and MTBD2, we hypothesized that these residues played a role in the interaction with MTs. The presence of the K77–K78 pair in the MEI-270–240 fragment was not sufficient to observe stable MEI-2•MT interaction (Fig. 2 B, lane 11) while its presence in the MEI-2FL K8D-K9D K274D-K275D (Fig. 3 C, lane 7) strongly suggested that K77–K78 depends on its environment to interact with microtubules. We tested this hypothesis by extending the minimal N-terminal fragment MEI-21–70 to MEI-21–90 to include the K77–K78 pair identified by mass spectrometry. While the substitution of this motif alone was not sufficient to completely abolish the interaction with MT (Fig. 3 D, lanes 9–11, respectively 84% and 87%), in combination with K8–K9, the MEI-21–90*tubulin band was not detectable (Fig. 3 D, lane 16 -0%).
We conclude that the K77–K78 pair is playing an essential role in the MEI-2•MT interaction but that its function is sensitive to the flanking sequence environment.
We then substituted the three lysine pairs in full-length MEI-2. As expected, the simultaneous substitution of the six lysines by alanine (K8A-K9A K77A-K78A K274A-K275A; Fig. 3 E, compare lane 5 [53%] to 3 [100%]) or aspartate (K8D-K9D K77D-K78D K274D-K275D; Fig. 3 E compare lane 7 [6%] to 3 [100%]) strongly reduced the MEI-2*tubulin crosslinked band, with a more pronounced effect observed with aspartate substitutions. We observed similar results when MEI-2 was in complex with MEI-1 (Katanin—Fig. S1).
Figure S1.
The KataninMEI-1●MEI-2 affinity for microtubules is affected by the charge of residues. (A) Crosslink assay between Katanin composed of MEI-1 and MEI-2 WT, 6KA or 6KD, and microtubules in vitro. SDS PAGE (BioRad) was analyzed by free staining based on tryptophan fluorescence. (B) Time course of crosslink reaction between KataninMEI-1●MEI-2 WT, 6KA, or 6KD and microtubules showing the accumulation of crosslinked complex MEI-2●tubuline over time. SDS PAGE (BioRad) was analyzed by free staining based on tryptophan fluorescence. Source data are available for this figure: SourceData FS1.
From these results, we conclude that the three positively charged KK motifs independently contribute to MEI-2•microtubule interaction as substituting only one patch is not sufficient to drastically reduce MT binding in vitro.
To determine whether the substitution of the six lysines identified reduces the affinity of MEI-2 for the microtubules, we performed MT copelleting assays in the presence of MEI-2 WT or variants (6KA or 6KD). As expected, we observed the highest affinity for MT for MEI-2 WT with a Kd of about 1 µM (Fig. 3 F), which is in a similar range to the previously reported Kd of human p80 (c15orf29) for MTs of about 2 µM (McNally and McNally, 2011), strongly suggesting common properties of the p80s from different species. The substitution of the six lysines identified reduced the affinity of MEI-2 for microtubules with a Kd of 3.68 µM observed for 6KA and a Kd of 6.15 µM for 6KD (Fig. 3 F), leading to a drastic reduction of microtubule severing activity for both of these variants compared with WT (about 10-fold reduction, Fig. 3 G).
Thus, we conclude that the six lysines identified play a direct role in the interaction with microtubules and that their substitution is affecting the affinity of MEI-2 for the microtubules, resulting in the decrease of microtubule severing activity.
MEI-2 basic patches are critical for KataninMEI-1•MEI-2 function in vivo
Having identified key residues in MEI-2 necessary for the Katanin•MT interaction in vitro, we next asked whether these residues contribute to Katanin function in vivo. As mentioned earlier, loss of Katanin function leads to severe defects in meiotic spindle assembly resulting in embryonic lethality (Clark-Maguire and Mains, 1994a, 1994b; Mains et al., 1990). However, partial loss of Katanin activity is not lethal but often causes the assembly of a longer meiotic spindle, which is accompanied by the extrusion of a larger polar body (Clandinin and Mains, 1993; Gomes et al., 2013; Han et al., 2009; Mains et al., 1990; McNally et al., 2014). The system is, therefore, very sensitive and allows monitoring of even minor defects in Katanin activity.
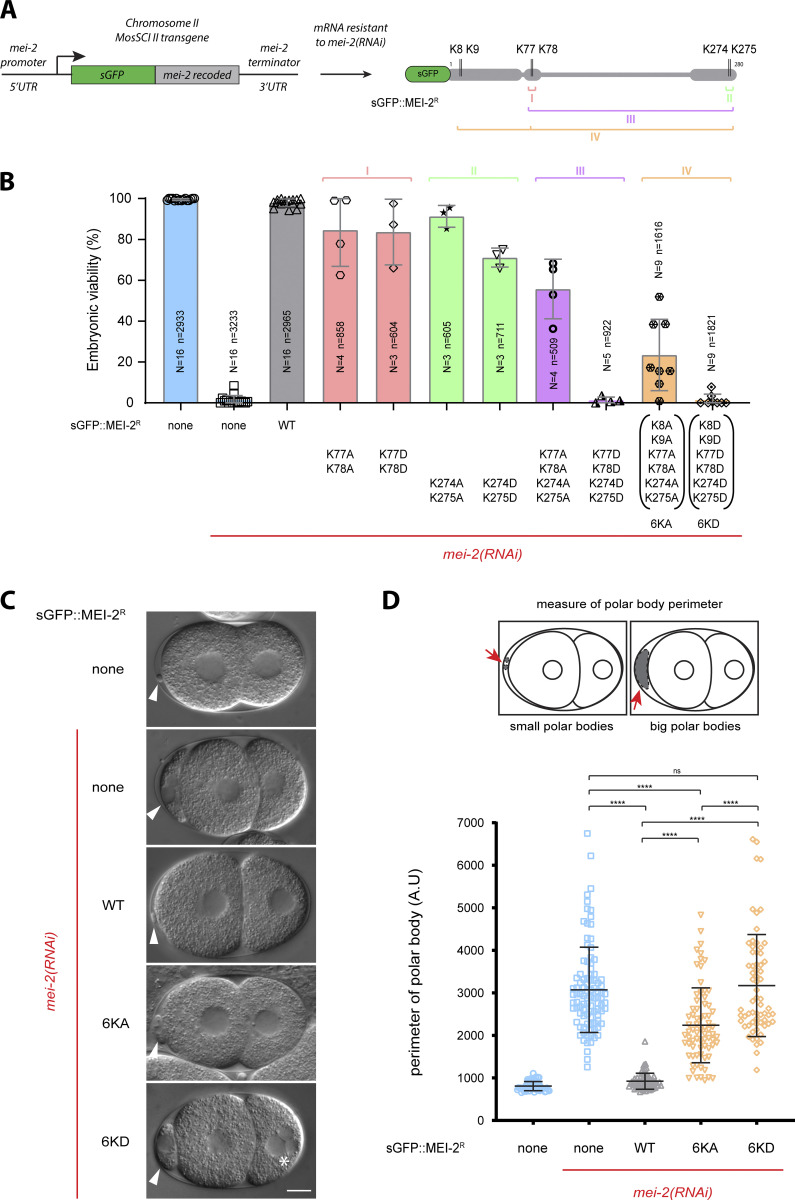
To analyze the role of MEI-2 MTBDs in vivo, we generated worm lines expressing sGFP::mei-2 Recoded (sGFP::mei-2R—RNAi-resistant transgene) wild-type or with different combinations of MTBD lysines substituted by alanine or aspartate (K77A-K78A), (K77D-K78D), (K274A-K275A), (K274D-K275D), or in combination (K77A-K78A K274A-K275A), (K77D-K78D K274D-K275D), 6KA (K8A-K9A K77A-K78A K274A-K275A), 6KD (K8D-K9D K77D-K78D K274D-K275D) (Fig. 4 A).
Figure 4.
Charge-dependent interaction MEI-2●microtubules is required for KataninMEI-1●MEI-2 activity during female meiosis. (A) Schematic representation of MosSCI transgene expressing sGFP::mei-2R (recoded) under the control of mei-2 promoter and terminator. Recoding of mei-2 allows mei-2 mRNA resistance to mei-2(RNAi). The schematics also represent sGFP::MEI-2R recombinant protein expressed in worms and mutated lysines. (B) Viability of embryos expressing sGFP::MEI-2R WT and mutants exposed to mei-2(RNAi). Error bars indicate SEM. N is the number of independent experiments, and n is the number of embryos. (C) DIC images of two-cell stage embryos expressing sGFP::MEI-2R WT, 6KA, and 6KD mutants exposed to mei-2(RNAi). White arrows indicate polar bodies and the white asterisk indicates multinuclei (under mei-2 RNAi, multinuclei observed for N2 n = 27/104, sGFP::MEI-2R WT n = 0/80, sGFP::MEI-2R 6KA n = 8/61 and sGFP::MEI-2R 6KD n = 23/62). Scale bars: 10 μm. (D) Measure of polar bodies perimeter of early embryos expressing sGFP::MEI-2R WT, 6KA, and 6KD mutants exposed to mei-2(RNAi). Error bars indicate SEM.
We then tested the ability of these transgenes to rescue the lethality and large polar body phenotype typically observed upon endogenous mei-2 depletion by RNAi (Fig. 4, B and C). sGFP::mei-2R WT transgene was able to fully rescue the lethality and the large polar body phenotypes induced by RNAi depletion of the endogenous mei-2 (Fig. 4, B and C). Likewise, sGFP::mei-2R with single pair substitutions (K77A-K78A), (K77D-K78D), (K274A-K275A), and (K274D-K275D) had only a moderate effect on the embryo viability (90–70% viability), and most of these transgenes rescued the lethality associated with endogenous mei-2 inactivation (Fig. 4 B). A more pronounced lethality phenotype was, however, observed for embryos expressing MEI-2 containing aspartate substitutions (Fig. 4 B).
Interestingly, the combination of two KK pair substitutions by alanine (K77A-K78A K274A-K275A) drastically reduces the viability of the embryo to 50% while the corresponding aspartate substitutions (K77D-K78D K274D-K275D) did not support embryo viability (0% viability) (Fig. 4 B). This result strongly suggests that the basic patches act synergistically to promote Katanin binding to microtubules in vivo.
As our in vitro experiments indicated that combining substitution of the basic patches is necessary to profoundly affect MEI-2 binding to microtubules, we tested sGFP::mei-2R transgenes with all three KK pairs substituted. Both GFP::MEI-2R 6KA and 6KD were expressed at normal levels in the germline, with an expression pattern indistinguishable from the wild-type (Fig. S3). However, embryos expressing these transgenes as the sole MEI-2 source were mainly unviable (Fig. 4 B). Notably, we observed a more drastic reduction of the embryo viability for embryos expressing GFP::MEI-2R 6KA (15% viability) than for GFP::MEI-2R 4KA (K77A-K78A K274A-K275A) (50% viability). Interestingly, and in agreement with in vitro observations showing less microtubule binding for MEI-2 6KD compared with 6KA (Fig. 3 E), the phenotype observed for the 6KD (0% viability) is more pronounced than for 6KA (15% viability; Fig. 4 B).
Figure S3.
Control of sGFP::MEI-2R expression in worms. (A) Representative spinning disk images of adult worms control (N2) or expressing sGFP::MEI-2R WT, 6KA, and 6KD exposed to mei-2(RNAi). sGFP fluorescence (gray) shows that the sGFP::MEI-2R WT, 6KA, and 6KD are expressed at the similar level in the oocytes cytoplasm in gonads. (B) Quantification of the GFP fluorescence using ImageJ software Graph representing the intensity of fluorescence by unit of surface (A.U.).
These results suggest that the observed embryonic lethality is due to a defect in Katanin’s activity and not merely due to the absence of Katanin. In addition, the fact that substitutions of lysine patches with acidic residues cause more severe phenotypes (lethality, Fig. 4 B) than substitutions with apolar residues suggests that acid residues cause a repulsion effect, more severely compromising Katanin binding to microtubules (as observed in vitro in Fig. 3 F).
As Katanin is essential for female meiosis, we suspected that the embryo lethality was due to a defect in meiotic spindle assembly. To test this hypothesis, we monitored the size of the polar bodies (Fig. 4, C and D) in WT, 6KA, and 6KD lines upon endogenous mei-2 depletion by RNAi. For both strains (6KA and 6KD), we observed 100% of large polar bodies (Fig. 4 C). Strikingly, even though both strains exhibit large polar bodies, embryonic viability differs (15% versus 0%, respectively, for sGFP::MEI-2R 6KA and sGFP::MEI-2R 6KD), suggesting that for the strain containing sGFP::MEI-2R 6KA, Katanin activity is severely reduced but not abolished. This reduced activity most likely results in the formation of altered meiotic spindles that are sufficient to achieve correct chromosome segregation. By measuring the perimeter of polar bodies of each embryo (Fig. 4 D), we noticed that polar bodies in sGFP::MEI-2R 6KD embryos are significantly bigger than those in sGFP::MEI-2R 6KA embryos. Moreover, we observed the presence of multiple nuclei with a much higher frequency in sGFP::MEI-2R 6KD mitotic embryos (Fig. 4 C, 6KD, white asterisk n = 23/62) than in sGFP::MEI-2R 6KA (n = 8/61), most likely as a consequence of a more severe chromosome segregation defects during meiosis.
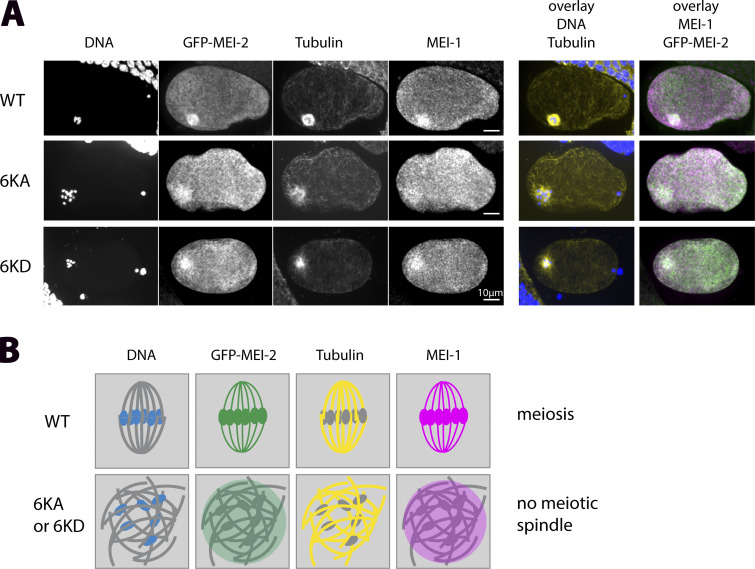
To directly visualize the meiotic spindles and colocalize MEI-1, MEI-2, chromosomes and MTs in GFP-MEI-2R WT, GFP-MEI-2R 6KA, and GFP-MEI-2R 6KD, we used indirect immunofluorescence (Fig. 5). In contrast to the WT situation, where two meiotic spindle poles were observed, decorated by MEI-1 and MEI-2, a mass of chromosomes surrounded by MTs is formed in both GFP-MEI-2R 6KA and GFP-MEI-2R 6KD mutant embryos (Fig. 5). Notably, MEI-1 and MEI-2 localized in this area, suggesting that the defects in meiotic spindle formation are not merely due to an absence of Katanin, but rather, a defect in Katanin activity due to reduced MT binding ability.
Figure 5.
Lack of microtubule-binding does not affect KataninMEI-1●MEI-2 localization during female meiosis. (A) Representative spinning disk images of fixed meiotic embryos expressing sGFP::MEI-2R WT, 6KA, and 6KD mutants exposed to mei-2(RNAi). Embryos (n = 5/5) were immunostained with anti-GFP (green), anti-tubulin (yellow), anti-MEI-1 (magenta) antibodies and colored with DAPI (blue). (B) Schematic representation of localization of the different colors at the meiotic spindle region for WT and variants.
From these observations, we conclude that MEI-2 6KA and 6KD substitutions do not affect the assembly or cellular localization of Katanin but only their activity by preventing Katanin optimal binding to microtubules.
Discussion
In this study, we demonstrate that MEI-2 promotes Katanin binding to microtubules. Using protein fragmentation, site-directed mutagenesis, and EDC crosslinking experiments, we revealed the existence of multiple microtubule-binding sites on MEI-2. By coupling the EDC crosslinker with mass spectrometry, we identified precisely targeted sites at the surface of the microtubule in the globular domain of alpha- and beta-tubulins. By introducing Katanin variants into C. elegans and monitoring polar body formation as a readout, we confirmed the importance of the identified MTBDs for Katanin activity in vitro and in vivo. Interestingly, based on our biochemical characterization of Katanin•MTs interaction, we were able to modulate Katanin activity in vivo by using Katanin variants harboring different affinities for microtubules.
Our study also revealed that after the initial binding of Katanin on MTs via MEI-2, MEI-1 subunit is brought in proximity to the microtubules and directly contacts them. We speculate that this microtubule proximity allows MEI-1 to engage in a productive interaction with the microtubule to damage it. From the structure of MEI-1 AAA+ domain associated with the C-terminal acidic tubulin tail (Zehr et al., 2019), we propose that this interaction might occur between the p60 hexamer central pore residues and the C-terminal exposed tail of tubulin to allow tubulin remodeling in an ATP-hydrolysis dependent manner, leading to microtubule damage.
Conservation of the fold
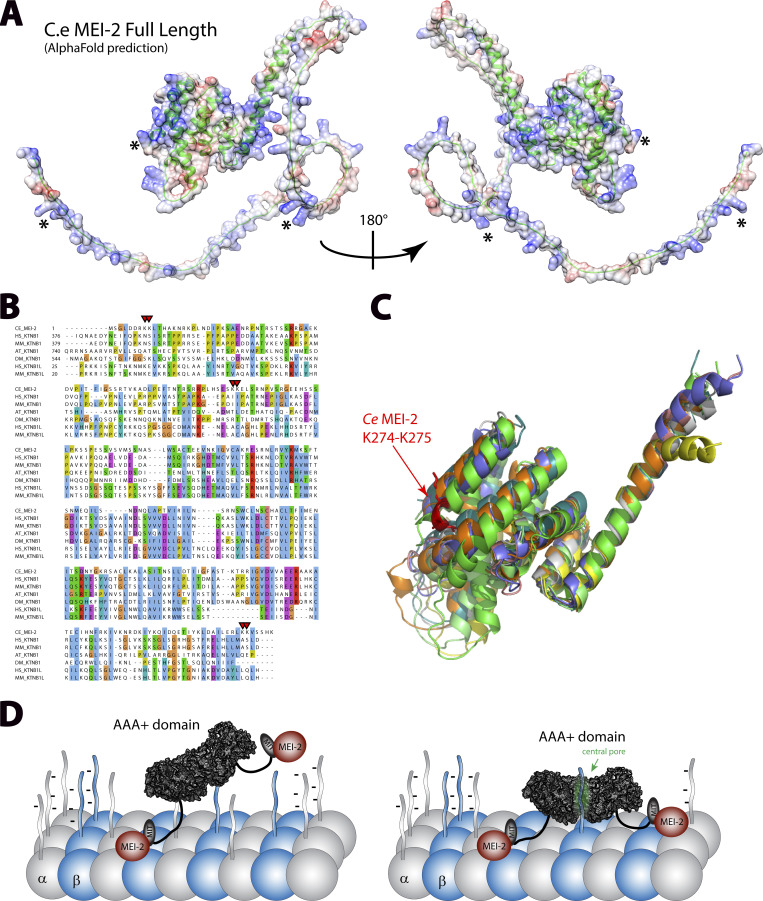
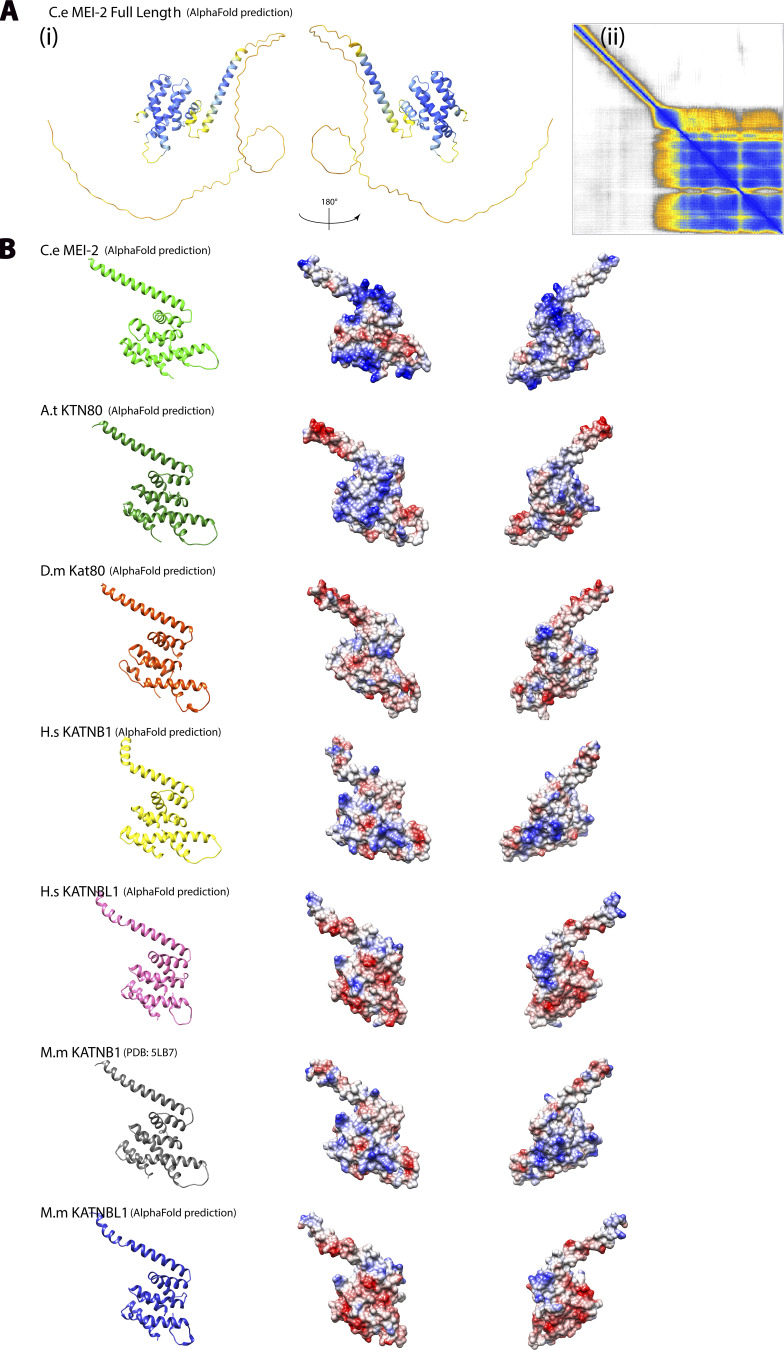
Having precisely identified MTBDs that are required for MEI-2 (Fig. 6 A) to interact with the microtubules, we asked whether these motifs were conserved within the p80s. Although protein sequence alignment did not reveal an obvious conserved motif (Fig. 6 B) nor the conservation of Lysine pairs identified in our study, structural modeling of p80 and p80-like from C. elegans, H. sapiens, Mus musculus, Drosophila melanogaster, and Arabidopsis thaliana using alphaFold2 (Fig. 6, A and C; and Fig. S4) revealed that the C. elegans MEI-2 had two distinct domains: a highly unstructured N-terminal domain (containing two of the lysine pairs) and a folded C-terminal domain (containing one of the lysine pair—Fig. 6 A). Structural alignment of the C-terminal domain from several p80s shows the clear conservation of the organization of this region corresponding to MEI-2 MTBD2 (Fig. 6 C). From this observation, we propose that the motif identified in MEI-2 might also be present in p80 subunits from other species implying that the role of p80 to initiate Katanin•MT interactions could be conserved. Consistent with this hypothesis, previous work on Katanin from sea urchins, mice, and humans have reported that the presence of p80 drastically increases Katanin affinity for microtubule as well as microtubule severing activity (Hartman et al., 1998; McNally et al., 2000; McNally and McNally, 2011).
Figure 6.
p80 structural alignment and Katanin•MT interacting model. (A) Structural model of MEI-2 obtained using AlphaFold2 prediction website. The carbon backbone is represented using cartoon mode (in green), and the surface color shows the electrostatic potential calculated using chimera electrostatic potential Coulombic function. * corresponds to the locations of KK pair residues identified in this study. (B) Protein sequences alignment of p80 and p80-like Katanin subunits from different species. CE: C. elegans; HS: H. sapiens; MM: M. musculus; AT: A. thaliana; DM: D. melanogaster. (C) Structural alignment of the C-terminal part of p80 and p80-like Katanin subunits from different species. Except for MM KATNB1, which is the X-ray structure obtained in complex with p60 fragment (PDB: 5LB7) (Jiang et al., 2017), structures shown are alpha-fold prediction aligned on MEI-2 structural prediction. The region shown is limited to the “structured region” of MEI-2 (aa 105–280). Single-protein structures are presented in Fig. S4. (D) Working model of microtubule binding of Katanin essential for microtubule severing activity.
Figure S4.
Conservation of structural organization of Katanin p80 subunits. (A) (i) Alpha Fold model of MEI-2 full-length corresponding to Fig. 6 A but colored using AlphaFold confidence color code. (ii) Error map corresponding to the structure prediction (i). (B) Except for MM KATNB1, which is the x-ray structure obtained in complex with p60 fragment (PDB: 5LB7) (Jiang et al., 2017), AlphaFold structural models of C-terminal part of p80 and p80-like subunits in different species based on protein sequences presented in Fig. 6. C. elegans C.e MEI-2; Homo sapiens H.s KATNB1; Homo sapiens H.s KATNBL1; Mus musculus M.m KATNB1; Mus musculus M.m KATNBL1; Drosophila melanogaster D.m Kat80; Arabidopsis thaliana A.t KTN80.
Adaptation of Katanins to different targets
One explanation for the conservation of the global organization but not of the protein sequence could be the differences between tubulin sequences and isoforms from the different species. We speculate that the sequences corresponding to the MTBD present in the different p80s could be used to recognize specific tubulin isoforms in a specific cellular context, leading to some Katanin•MT interaction specificity.
Interestingly, even after identifying precise MTBD into MEI-2, we could identify conserved motifs within the other p80s. Nevertheless, alpha-fold modeling strongly suggests that the global structure of the domain is conserved and that the p80s contain positively charged subdomains, which might be directly involved in the interaction with the negatively charged microtubule surface (Fig. 6 A and Fig. S4). This lack of sequence conservation could result from the natural variation of tubulin isoforms between different species. To adapt to these variations, p80 and microtubule surface sequences coevolved, ensuring optimal activity of the enzyme in a given organism.
Interestingly, most in vitro experiments were performed with microtubules assembled from pig or cow tubulin and MAPs from other species. We can anticipate that even if we observe some specific activities, the interaction between MAPs and the microtubule might be suboptimal. Notably, the average number of protofilaments constituting the microtubule is reduced in C. elegans (about 11–15 protofilaments) relative to vertebrate microtubules (13–15 protofilaments). In the case of 13 protofilaments, tubulin protofilaments align parallel to the microtubule-long axes. In contrast, non-13 protofilament microtubules exhibit a helical twist, more or less pronounced depending on the number of protofilaments constituting the microtubules, most likely impacting MAPs’ binding and activities (Amos and Schlieper, 2005; Chaaban and Brouhard, 2017; Chrétien and Wade, 1991; Ti et al., 2018; Tilney et al., 1973).
Multiple roles of MEI-2•tubulin interaction
We observed that MEI-2 directly interacts with the globular region of the tubulins. This encounter then promotes the interaction between MEI-1 and the tubulin CTT. If our results demonstrate that MEI-2 promotes Katanin•MT interaction, it may also stimulate the catalytic activity of the Katanin enzyme.
Indeed, while single MEI-2 basic patch substitutions did not lead to a significant difference in the binding of MEI-2 to the microtubules in vitro, they had a clear impact on the enzymatic activity in vivo, as revealed by the appearance of large polar bodies. In addition, Lu et al. (2004) have previously reported that a single mutation in the globular region of alpha-tubulin (TBA-2, sb27-E194K) drastically reduced Katanin’s ability to sever the MTs, suggesting that this region of tubulin is involved in the Katanin•MT contacts and impacts Katanin’s activity. From these observations, we propose that the MEI-2•microtubule interaction could have two roles: (i) to bring the catalytic MEI-1 subunit close to the tubulin CTT and (ii) to regulate the enzymatic activity of Katanin. We speculate that the level of Katanin activity is finely tuned by the nature of the amino acids of the globular domain of tubulin engaged in the interaction with MEI-2. Thus, even if changing the amino acids critical for this interaction will not lead to the disassociation of the Katanin from the microtubule, it might lead to changes in microtubule severing activity. Notably, the nature of the tubulin sequence changes depending on the tubulin isoform and tissue localization (Nishida et al., 2021). Katanin is required for many cellular processes in different cell types (McNally and Roll-Mecak, 2018; Sarbanes et al., 2022). Thus, modulating the Katanin’s microtubule severing activity via this interaction might allow different levels of Katanin activity depending on the cell type (e.g., cilia versus neurons), allowing the adaptation of the same enzyme to different cellular contexts. MEI-2•microtubule interaction might not only bring Katanin to the microtubule lattice, but the nature of the interaction between tubulin and Katanin might modulate the activity of the enzyme.
A model for Katanin•microtubule interaction leading to MT severing
From our results, we propose the following Katanin•MT interaction model (Fig. 6 D). We and others have previously shown that Katanin (MEI-1•MEI-2) is assembled as a hexamer of dimers (Joly et al., 2016; Sarbanes et al., 2022; Sharp and Ross, 2012; Zehr et al., 2017). We propose that one MEI-2 makes initial contact with the globular region of the tubulin (from crosslink and mass spectrometry results). Then, a second interaction between a second MEI-2 present in the Katanin oligomer promotes the stabilization and the proper positioning of the MEI-1 catalytic ring at the surface of the microtubule (based on the observation of crosslinks between MEI-1 and tubulin in the presence of MEI-2). Consequently, the MEI-1 hexamer’s central pore is positioned in close proximity to the charged CTT of tubulin exposed at the surface of the microtubule, allowing its engagement in the central pore. This model is entirely consistent with previous results showing that (i) the presence of MEI-2 is required for Katanin activity, (ii) the charged CTT stimulates the ATPase activity of the catalytic ring, and (iii) that the presence of p80 enhances the microtubule-severing activity in vitro (Hartman et al., 1998; Hartman and Vale, 1999; Jiang et al., 2017; Joly et al., 2016, 2020; McNally et al., 2000; McNally and McNally, 2011; Rezabkova et al., 2017; Vemu et al., 2018; Zehr et al., 2019). From this respect, we propose the following speculative model concerning microtubule-severing mechanism: (i) binding of MT by MEI-2 bringing MEI-1 in proximity to the microtubule lattice, (ii) tubulin CTT engagement into the central pore formed by MEI-1 hexamer leading to ATP hydrolysis, (iii) triggering conformational changes relayed by MEI-2 to the microtubule, and (iv) extraction of tubulin dimers damaging the microtubule lattice.
A deep understanding of Katanin function and regulation will help the design and engineering of tunable Katanin in space and time. In that respect, Opto-Katanin, an artificial enzyme tunable by light was recently developed (Meiring et al., 2022). Notably, the authors had to fuse the p60 subunit to the MT-binding domain of EB3 (Meiring et al., 2022). They concluded that p60 needs a good anchor, binding individual microtubules, and that a direct interaction of p60 with the CTT may be disadvantageous in their system. This observation is consistent with the major role played by p80, which anchors Katanin to the microtubule.
P80 mutations and disease
Indirect observations support the idea of a significant role of p80 in Katanin function. Indeed, there is an increase in observation of p80 mutation leading to major cellular defects linked to human diseases such as congenital cortical malformations, sterility, and cancer (Hu et al., 2014; Mishra-Gorur et al., 2014). Nevertheless, how these mutations affect the enzyme’s activity is not understood. As p80 has been reported to interact with different partners directing Katanin localization in vivo (Akhmanova and Hoogenraad, 2015; Atherton et al., 2019; Hartman et al., 1998; Hartman and Vale, 1999; Jiang et al., 2014, 2017, 2018; McNally and Roll-Mecak, 2018; Roll-Mecak and McNally, 2010; Sarbanes et al., 2022), one hypothesis could be that the mutations affect the direct interaction with partners and Katanin localization. A second hypothesis emanating from our study is that the mutations might affect the interaction between the Katanin and the microtubules, thus affecting directly the activity of the enzyme. To better understand how these mutations affect the enzyme’s activity, it is thus of prime importance to directly test whether they are affecting the affinity of the enzyme for the microtubules.
Materials and methods
Plasmids
The plasmids used in this study are described in Table S2.
Protein purification
Katanin (6xHis-MEI-1/Strep-MEI-2 or 6xHis-GFP-MEI-1/Strep-MEI-2) was produced in BL21 bacteria transformed with plasmids available in Table S2. Katanins were purified as described in Joly et al. (2016) and (2020). Briefly, Katanin production was induced with 0.5 mM of isopropyl thio-β-D-galactoside (IPTG) for 4–5 h at 24°C. After centrifugation, bacteria were resuspended in 50 mM Tris-HCl pH 8.0, 500 mM NaCl, and 5% glycerol supplemented with protease inhibitor cocktail (Roche) and broken by sonication. The soluble fraction was loaded onto 2 × 1 ml StrepTrap HP (GE-Healthcare), and after washing, proteins were eluted using 5 mM desthiobiotin in the purification buffer. The eluate was then loaded onto a 2 × 1 ml HiTrap Chelating HP column (GE-Healthcare) precharged with Nickel. After washing, proteins were eluted using imidazole in a purification buffer. Before storage, the buffer was exchanged to 50 mM Tris-HCl pH 8.0, 500 mM NaCl, and 5% glycerol using a G25 column, and the proteins were snap-frozen in liquid nitrogen and stored at −80°C.
MBP-MEI-2 variants or fragments were produced in BL21 bacteria transformed with plasmids available in Table S2. Briefly, protein production was induced with 0.5 mM final concentration of isopropyl thio-β-D-galactoside (IPTG) for 5 h at 24°C. After centrifugation, cells were resuspended in lysis buffer containing (50 mM Tris-HCl, pH 8.0, 500 mM NaCl, and 5% glycerol) and broken by sonication. The supernatant was loaded onto MBP-Trap columns (Cytiva), and after washing, the protein was eluted with 15 mM maltose. The buffer was exchanged using a G25 column for a storage buffer containing 50 mM Tris-HCl pH 8.0, 200 mM NaCl, and 5% glycerol. Then, proteins were frozen in liquid nitrogen and stored at −80°C before using for experiments.
Nematode strains and culture conditions
C. elegans strains were cultured and maintained using standard procedures (Brenner, 1974). Transgenic worms expressing sGFP::MEI-2R transgenes under the control of mei-2 promoter and terminator were generated by MosSCI (Mos1- mediated single-copy gene insertion) using the strain EG6699 (ttTi5605 II; unc-119 [ed3] III; ox|Ex1578) (Frøkjaer-Jensen et al., 2008). Correct insertion of the transgene into chromosome II was verified by PCR. C. elegans strains and plasmids used in this study are listed and described in Table 1 and Table S2, respectively.
Table 1.
C. elegans strains and plasmids used in this study
| Transgenic strains | Genotypes | Proteins expressed | References |
|---|---|---|---|
| wLP 903 | leaSi41[Pmei-2::sgfp::mei-2 WT recoded + unc-119(+)]II; unc-113(ed3) III | sGFP::MEI-2R WT | This study |
| wLP 980 | leaSi42[Pmei-2::sgfp::mei-2 K77A K78A recoded + unc-119(+)]II; unc-113(ed3) III | sGFP::MEI-2R K77A K78A | This study |
| wLP 982 | leaSi43[Pmei-2::sgfp::mei-2 K77D K78D K274D K275D recoded + unc-119(+)]II; unc-113(ed3) III | sGFP::MEI-2R K77D K78D K274D K275D | This study |
| wLP 1045 | leaSi44[Pmei-2::sgfp::mei-2 K77D K78D recoded + unc-119(+)]II; unc-113(ed3) III | sGFP::MEI-2R K77D K78D | This study |
| wLP 1047 | leaSi45[Pmei-2::sgfp::mei-2 K274A K275A recoded + unc-119(+)]II; unc-113(ed3) III | sGFP::MEI-2R K274A K275A | This study |
| wLP 1050 | leaSi46[Pmei-2::sgfp::mei-2 K274D K275D recoded + unc-119(+)]II; unc-113(ed3) III | sGFP::MEI-2R K274D K275D | This study |
| wLP 1067 | leaSi47[Pmei-2::sgfp::mei-2 K77A K78A K274A K275A recoded + unc-119(+)]II; unc-113(ed3) III | sGFP::MEI-2R K77A K78A K274A K275A | This study |
| wLP 1087 | leaSi48[Pmei-2::sgfp::mei-2 K246A K249A recoded + unc-119(+)]II; unc-113(ed3) III | sGFP::MEI-2R K246A K249A | This study |
| wLP 1092 | leaSi49[Pmei-2::sgfp::mei-2 K246D K249D recoded + unc-119(+)]II; unc-113(ed3) III | sGFP::MEI-2R K246D K249D | This study |
| wLP 1127 | leaSi50[Pmei-2::sgfp::mei-2 K8A K9A K77A K78A K274A K275A recoded + unc-119(+)]II; unc-113(ed3) III | sGFP::MEI-2R K8A K9A K77A K78A K274A K275A | This study |
| wLP 1130 | leaSi51[Pmei-2::sgfp::mei-2 K8D K9D K77D K78D K274D K275D recoded + unc-119(+)]II; unc-113(ed3) III | sGFP::MEI-2R K8D K9D K77D K78D K274D K275D | This study |
RNAi experiments
RNAi induced by feeding worm bacteria was used to deplete mei-2 mRNA in C. elegans strains expressing sGFP::MEI-2recoded, resistant to mei-2(RNAi). mei-2 RNAi targets the last exon of mei-2 (5′-AATATCACGAGCGACAATTATGGGAAGCGATCGGCTTGCTTGAAGGCTCTTGCATCAATTACCAACAGTTTACTGGACACAATCATCGGATTTGCATCCACCAAAACTCGGCGAATTGGAGTCGATGTGGTAGCTGAAGAGCGAGCTGCAAAAGCAACAGAGTGCATTCACAATTTTCGCAAAATAGTTAAAAATCGGGACAAAATCTACAAACAGATTGACCAGGAGACAATTTACAAACTCGATGCCATTCTCGAGCGATTGAAAAAAGTTTCCAGCCATAAGTAA-3′). 2 mM IPTG was added to the saturating culture of HT115 bacteria transformed with L4440 plasmid expressing control (empty vector) or mei-2 dsRNA. Bacteria were seeded on NGM plates complemented with 2 mM IPTG. L3 larvae were exposed to bacteria expressing the dsRNA and maintained at 23°C for 24 h. After 24 h of RNAi exposition, phenotypes of embryos contained in adult worms were analyzed by microscopy (DIC or fluorescence), and embryo development was evaluated by viability assay.
Embryonic viability assay/progeny test
Embryonic viability assay was achieved with adult worms exposed to mei-2(RNAi) for 24 h at 23°C. Five adults were transferred on a new plate for 5 h at 23°C. Worms were removed from the plates, the laid eggs were counted, and plates were maintained at 23°C for 16 h. 16 h later, the number of hatched and dead embryos was counted to evaluate the embryonic viability score as a percentage of the initial embryo count. The graphs were generated by using GraphPad Prism 8.0.2.
Immunofluorescence and imaging
Immunofluorescence analysis was performed on adult worms exposed to mei-2(RNAi) for 24 h at 23°C. 20 worms were dissected in 5 µl of Shelton buffer on slides previously coated with subbing-solution (0.4% gelatin, 0.04% Chromium [III] potassium sulfate dodecahydrate, 0.1% poly-L-lysine). A 24 × 32 mm coverslip was placed onto the drop and the slide was frozen on a block precooled on dry ice. After 20 min, the coverslip was flipped and the slide was incubated in −20°C methanol for 20 min. Slides were rehydrated in PBS twice for 5 min. Samples were blocked for 1 h at room temperature with 100 µl of 2% BSA in PBS. Then, slides were incubated overnight at 4°C with 100 μl primary antibodies in 2% BSA in PBS in a wet chamber. Goat anti-GFP antibody (polyclonal, 039600-101-215; Rockland) was used at a dilution of 1:300. Mouse anti-tubulin antibody (monoclonal, DM1A; Sigma-Aldrich) was used at a dilution of 1:400. Affinity-purified rabbit anti-MEI-1 antibody (Pintard et al., 2003) was used at a dilution of 1:300. Slides were washed with PBS 0.05% Tween-20 and incubated with 100 µl of secondary antibodies in 2% BSA in PBS for 45 min at room temperature. Donkey anti-goat, Alexa Fluor 488 (polyclonal, A-11055; Invitrogen); donkey anti-mouse, Alexa Fluor 568 (polyclonal, A-10037; Invitrogen); and donkey anti-rabbit, Alexa Fluor 667 (polyclonal, A-31573; Invitrogen) secondary antibodies were used at a dilution of 1:1,000. After washing with PBS, samples were mounted using a Vectashield mounting medium containing DAPI between the coverslip and slide. Coverslips were sealed with nail coat and slides were stored at 4°C. Fixed embryos were imaged using a spinning disk confocal X1 microscope with a 63× objective. Captured images were processed using ImageJ and Photoshop.
Imaging of dissected embryos by DIC
Imaging of polar bodies phenotype of early embryos was performed using a DIC microscope. Embryos were obtained by dissecting adult worms with a needle in 5 µl of M9 in a coverslip. Then, the coverslip was placed on a 3% agarose pad pre-prepared on a slide. DIC images were acquired by an Axiocam Hamamatsu ICcI camera (Hamamatsu Photonics) mounted on a Zeiss AxioImager A1 microscope equipped with a Plan Neofluar 100×/1.3-NA objective (Zeiss), and the acquisition system was controlled by Axiovision software (Zeiss). Captured images were processed using ImageJ and Photoshop.
Measurement of polar body perimeter
The polar bodies were measured using ImageJ on DIC images of early embryos after mei-2(RNAi) exposition. The graphs were generated by using GraphPad Prism 8.0.2.
Imaging of whole worms
Imaging of full worms was performed using a Spinning Disk W1 microscope. Adult worms were anesthetized in 5 µl of levamisole on a coverslip placed on a pre-prepared 3% agarose pad on a slide. Live imaging was performed at 23°C using a spinning disc confocal head (CSU-W1; Yokogawa Corporation of America) mounted on a DMI8 inverted microscope (Leica) equipped with 491- and 561-nm lasers (491-nm 150 mW; DPSS Laser 561 nm 150 mW) and a complementary metal-oxide semiconductor (Orca-Flash 4 V2+; Hamamatsu). Acquisition parameters were controlled by MetaMorph software (Molecular Devices). In all cases, a 40×, HC PL APO 40×/1.30 Oil CS2 (506358; Leica) lens was used, and ∼10 z-sections were collected at 0.5-µm intervals. Captured images were processed using ImageJ and Photoshop.
Taxol-stabilized microtubules
Microtubules were polymerized in vitro from purified porcine tubulin as previously described (Joly et al., 2016). After centrifugation at 90,000 g for 10 min, microtubules were resuspended in 80 mM PIPES-KOH, pH 6.8, 1 mM MgCl2, 1 mM EGTA, and 10 µM Taxol and kept at room temperature before use.
Microtubule crosslinking assay
In vitro crosslinking assay between stabilized microtubules and Katanin (6xHis-MEI-1•Strep-MEI-2) or MEI-2 (MBP-MEI-2) was performed at room temperature in 46.4 mM PIPES-KOH, pH = 6.8, 0.580 mM MgCl2, 0.580 mM EGTA, 16 mM Tris-HCl, pH 8.0, 160 mM NaCl, 1.6% glycerol, 7.8 µM taxol-stabilized microtubules, and 3.52 µM Katanin. The crosslinking reaction was initiated by adding 5 mM EDC (1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride). EDC specifically allows the crosslink between lysine (K) and aspartate (D) or glutamate (E). After 15 min, the reaction was stopped by adding Laemmli and was boiled at 95°C. Crosslinked complexes between bound proteins were identified and analyzed by running samples on SDS-PAGE gel (stained free-BioRad gel 4–20% acrylamide) for stain-free, Coomassie blue staining, or Western blot analysis.
Microtubule binding assay (copelleting assay)
Microtubule-binding reactions were carried out in binding buffer 80 mM PIPES-KOH, pH 6.8, 1 mM MgCl2, 1 mM EGTA, 10 mM Tris-HCl pH 8.0, 50 mM NaCl, and 1% glycerol. Purified MBP-MEI-2 WT or variants (1 µM) were incubated in the presence of indicated concentration of taxol-stabilized microtubules at 25°C for 15 min. The binding reactions were spun down at 100,000 g for 10 min. Supernatants and resuspended pellets were analyzed using gradient SDS-PAGE 4–20% and revealed using Stained-free (Biorad) technology. Quantification was performed using ImageJ and analysis using Origin software.
Microtubule severing assay
Microtubule severing activities of MEI-1 WT•MEI-2 WT or variant (6KA or 6KD, as indicated) were performed using TIRF-based microscopy at 25°C. Briefly, docetaxol-stabilized rhodamin-labeled microtubules (assembled from porcine tubulin) were adsorbed on a glass coverslip into BRB80 buffer complemented with 10 µM docetaxol, 5 mM DTT, 10 mM MgCl2, and 2 mM ATP. After 5 min incubation, the microtubules were exposed to a Katanin mix containing BRB80 with 10 µM docetaxol, 5 mM DTT, 10 mM MgCl2, and 200 nM of the different Katanin (as indicated). Images were immediately recorded using a TIRF home-built microscope at a frequency of one image every 2 s for a total of 10 min with an exposure time of 100 ms by frame.
Each movie was analyzed using ImageJ software. Briefly, each individual microtubule was hand-picked, and kymographs were generated using Multi-Kymograph ImageJ Pluggin. We defined the severing event as the number of severing observed by second and by micrometer of microtubule. Three independent experiments were performed for each Katanin (WT and variants). The graph was generated using Prism. Error bars represent the SEM.
Western blotting and analysis
Western blot analysis was performed using a nitrocellulose blotting membrane (0.45 µm NC, 10600002; Amersham Protrana) according to standard procedures (Sambrook et al., 1989). Mouse anti-MBP antibody (monoclonal, E8032S; NEB) was used at the dilution of 1:5,000, and rabbit anti-alpha tubulin (polyclonal, Ab18251; Abcam) was used at the dilution of 1:1,000. HRP-conjugated anti-mouse (A9917; Sigma-Aldrich) and anti-rabbit antibodies (A0545; Sigma-Aldrich) were used at 1:3,000, and the signal was detected with chemiluminescence (ECL-Millipore).
Mass spectrometry analysis
Briefly, mass spectrometry analysis was performed on complexes separated by SDS-PAGE, gel extracted, and digested using multienzymatic digestion with trypsin and AIP. Thus, the samples were analyzed using LC-MS/MS on the Qex2 instrument. Identification of the protein and sequence coverage were performed using Peaks Studio and detection of crosslinked peptides with Mass Spec Studio.
Online supplemental material
Fig. S1 shows that the KataninMEI-1●MEI-2 affinity for microtubules is affected by the charge of residues. Fig. S2 shows MEI-2 crosslinks with microtubules in vitro. Fig. S3 shows sGFP::MEI-2R expression in worms. Fig. S4 shows the conservation of Katanin p80 subunits’ structural organization. Table S1 shows peptides identified using mass spectrometry. Table S2 lists plasmids used in this study.
Supplementary Material
shows peptides identified using mass spectrometry.
lists plasmids used in this study.
is the source file for Fig. 1.
is the source file for Fig. 2.
is the source file for Fig. 3.
is the source file for Fig. S1.
is the source file for Fig. S2.
Acknowledgments
We thank H. Wioland, C. Leduc, A. Jegou, and G. Romet-Lemonne for their help with TIRF microscopy, suggestions, and discussions. We thank P. Moussounda for her help with media preparation. We thank C. Gaucher, N. Baouz, J. Lopez, and L. Gaichet for administrative and technical support. We acknowledge the ImagoSeine core facility of the Institut Jacques Monod, member of IBiSA and the France-BioImaging (ANR-10-INBS-04) infrastructure and the Institut Jacques Monod “Structural and Functional proteomic platform.” We thank R. Karess, J. Morvan, B. Ossareh-Nazari, and G. Velez Aguilera for their critical reading of the manuscript and valuable comments.
Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). We also thank WormBase. E. Beaumale was supported by a PhD fellowship from the Ministry of Research and by the Fondation ARC pour la recherche sur le cancer. N. Joly is supported by an ARC project Fellowship. This work has received two supports under the program “Investissement d’Avenir” launched by the French Government and implemented by Agence Nationale de la Recherche (ANR), with the reference «ANR-18-IdEx-0001» as part of its program «Emergence»-KataRep and «Dynamic». Work in the laboratory of LP is supported by the French National Research Agency under grant no. ANR-17-CE13-0011-01/01, by la Ligue Nationale Contre le Cancer (Programme équipe labélisée). L. Pintard team is also supported by the Labex “Who am I?” Laboratory of Excellence No. ANR-11-LABX-0071, the French Government through its Investments for the Future program operated by the French National Research Agency (ANR) under Grant no. ANR-11-IDEX-0005-01.
Author contributions: E. Beaumale and N. Joly designed the study. E. Beaumale and N. Joly generated DNA constructs, protein purifications, and crosslinking assays. E. Beaumale, L. Van Hove, and L. Pintard generated worm strains and performed immunostaining localization. E. Beaumale and N. Joly performed spinning disk experiments. E. Beaumale performed all remaining experiments and data analysis with N. Joly. E. Beaumale, L. Pintard, and N. Joly assembling the figures. E. Beaumale, L. Pintard, and N. Joly wrote the manuscript with input from all coauthors.
References
- Akhmanova, A., and Hoogenraad C.C.. 2015. Microtubule minus-end-targeting proteins. Curr. Biol. 25:R162–R171. 10.1016/j.cub.2014.12.027 [DOI] [PubMed] [Google Scholar]
- Akhmanova, A., and Steinmetz M.O.. 2019. Microtubule minus-end regulation at a glance. J. Cell Sci. 132:jcs227850. 10.1242/jcs.227850 [DOI] [PubMed] [Google Scholar]
- Amos, L.A., and Schlieper D.. 2005. Microtubules and maps. Adv. Protein Chem. 71:257–298. 10.1016/S0065-3233(04)71007-4 [DOI] [PubMed] [Google Scholar]
- Atherton, J., Luo Y., Xiang S., Yang C., Rai A., Jiang K., Stangier M., Vemu A., Cook A.D., Wang S., et al. 2019. Structural determinants of microtubule minus end preference in CAMSAP CKK domains. Nat. Commun. 10:5236. 10.1038/s41467-019-13247-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, M.E., Sackett D.L., and Ross J.L.. 2015. Katanin severing and binding microtubules are inhibited by tubulin carboxy tails. Biophys. J. 109:2546–2561. 10.1016/j.bpj.2015.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodakuntla, S., Jijumon A.S., Villablanca C., Gonzalez-Billault C., and Janke C.. 2019. Microtubule-associated proteins: Structuring the cytoskeleton. Trends Cell Biol. 29:804–819. 10.1016/j.tcb.2019.07.004 [DOI] [PubMed] [Google Scholar]
- Brenner, S. 1974. The genetics of Caenorhabditis elegans. Genetics. 77:71–94. 10.1093/genetics/77.1.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaaban, S., and Brouhard G.J.. 2017. A microtubule bestiary: Structural diversity in tubulin polymers. Mol. Biol. Cell. 28:2924–2931. 10.1091/mbc.e16-05-0271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrétien, D., and Wade R.H.. 1991. New data on the microtubule surface lattice. Biol. Cell. 71:161–174. 10.1016/0248-4900(91)90062-R [DOI] [PubMed] [Google Scholar]
- Clandinin, T.R., and Mains P.E.. 1993. Genetic studies of mei-1 gene activity during the transition from meiosis to mitosis in Caenorhabditis elegans. Genetics. 134:199–210. 10.1093/genetics/134.1.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Maguire, S., and Mains P.E.. 1994a. Localization of the mei-1 gene product of Caenorhaditis elegans, a meiotic-specific spindle component. J. Cell Biol. 126:199–209. 10.1083/jcb.126.1.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Maguire, S., and Mains P.E.. 1994b. mei-1, a gene required for meiotic spindle formation in Caenorhabditis elegans, is a member of a family of ATPases. Genetics. 136:533–546. 10.1093/genetics/136.2.533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faltova, L., Jiang K., Frey D., Wu Y., Capitani G., Prota A.E., Akhmanova A., Steinmetz M.O., and Kammerer R.A.. 2019. Crystal structure of a heterotetrameric katanin p60:p80 complex. Structure. 27:1375–1383.e3. 10.1016/j.str.2019.07.002 [DOI] [PubMed] [Google Scholar]
- Frøkjaer-Jensen, C., Davis M.W., Hopkins C.E., Newman B.J., Thummel J.M., Olesen S.P., Grunnet M., and Jorgensen E.M.. 2008. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 40:1375–1383. 10.1038/ng.248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadadhar, S., Alvarez Viar G., Hansen J.N., Gong A., Kostarev A., Ialy-Radio C., Leboucher S., Whitfield M., Ziyyat A., Touré A., et al. 2021. Tubulin glycylation controls axonemal dynein activity, flagellar beat, and male fertility. Science. 371:eabd4914. 10.1126/science.abd4914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnham, C.P., and Roll-Mecak A.. 2012. The chemical complexity of cellular microtubules: Tubulin post-translational modification enzymes and their roles in tuning microtubule functions. Cytoskeleton. 69:442–463. 10.1002/cm.21027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genova, M., Grycova L., Puttrich V., Magiera M.M., Lansky Z., Janke C., and Braun M.. 2023. Tubulin polyglutamylation differentially regulates microtubule-interacting proteins. EMBO J. 42:e112101. 10.15252/embj.2022112101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes, J.E., Tavernier N., Richaudeau B., Formstecher E., Boulin T., Mains P.E., Dumont J., and Pintard L.. 2013. Microtubule severing by the katanin complex is activated by PPFR-1-dependent MEI-1 dephosphorylation. J. Cell Biol. 202:431–439. 10.1083/jcb.201304174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, H., Schubert H.L., McCullough J., Monroe N., Purdy M.D., Yeager M., Sundquist W.I., and Hill C.P.. 2020. Structure of spastin bound to a glutamate-rich peptide implies a hand-over-hand mechanism of substrate translocation. J. Biol. Chem. 295:435–443. 10.1074/jbc.AC119.009890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, X., Gomes J.E., Birmingham C.L., Pintard L., Sugimoto A., and Mains P.E.. 2009. The role of protein phosphatase 4 in regulating microtubule severing in the Caenorhabditis elegans embryo. Genetics. 181:933–943. 10.1534/genetics.108.096016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman, J.J., Mahr J., McNally K., Okawa K., Iwamatsu A., Thomas S., Cheesman S., Heuser J., Vale R.D., and McNally F.J.. 1998. Katanin, a microtubule-severing protein, is a novel AAA ATPase that targets to the centrosome using a WD40-containing subunit. Cell. 93:277–287. 10.1016/S0092-8674(00)81578-0 [DOI] [PubMed] [Google Scholar]
- Hartman, J.J., and Vale R.D.. 1999. Microtubule disassembly by ATP-dependent oligomerization of the AAA enzyme katanin. Science. 286:782–785. 10.1126/science.286.5440.782 [DOI] [PubMed] [Google Scholar]
- Hu, W.F., Pomp O., Ben-Omran T., Kodani A., Henke K., Mochida G.H., Yu T.W., Woodworth M.B., Bonnard C., Raj G.S., et al. 2014. Katanin p80 regulates human cortical development by limiting centriole and cilia number. Neuron. 84:1240–1257. 10.1016/j.neuron.2014.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, K., Faltova L., Hua S., Capitani G., Prota A.E., Landgraf C., Volkmer R., Kammerer R.A., Steinmetz M.O., and Akhmanova A.. 2018. Structural basis of formation of the microtubule minus-end-regulating CAMSAP-katanin complex. Structure. 26:375–382.e4. 10.1016/j.str.2017.12.017 [DOI] [PubMed] [Google Scholar]
- Jiang, K., Hua S., Mohan R., Grigoriev I., Yau K.W., Liu Q., Katrukha E.A., Altelaar A.F.M., Heck A.J.R., Hoogenraad C.C., and Akhmanova A.. 2014. Microtubule minus-end stabilization by polymerization-driven CAMSAP deposition. Dev. Cell. 28:295–309. 10.1016/j.devcel.2014.01.001 [DOI] [PubMed] [Google Scholar]
- Jiang, K., Rezabkova L., Hua S., Liu Q., Capitani G., Altelaar A.F.M., Heck A.J.R., Kammerer R.A., Steinmetz M.O., and Akhmanova A.. 2017. Microtubule minus-end regulation at spindle poles by an ASPM-katanin complex. Nat. Cell Biol. 19:480–492. 10.1038/ncb3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joly, N., Beaumale E., Van Hove L., Martino L., and Pintard L.. 2020. Phosphorylation of the microtubule-severing AAA+ enzyme Katanin regulates C. elegans embryo development. J. Cell Biol. 219:e201912037. 10.1083/jcb.201912037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joly, N., Martino L., Gigant E., Dumont J., and Pintard L.. 2016. Microtubule-severing activity of the AAA+ ATPase Katanin is essential for female meiotic spindle assembly. Development. 143:3604–3614. [DOI] [PubMed] [Google Scholar]
- Kuo, Y.W., and Howard J.. 2021. Cutting, amplifying, and aligning microtubules with severing enzymes. Trends Cell Biol. 31:50–61. 10.1016/j.tcb.2020.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo, Y.W., Trottier O., Mahamdeh M., and Howard J.. 2019. Spastin is a dual-function enzyme that severs microtubules and promotes their regrowth to increase the number and mass of microtubules. Proc. Natl. Acad. Sci. USA. 116:5533–5541. 10.1073/pnas.1818824116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay, K.A., Abdelhamid N., Kahawatte S., Dima R.I., Sackett D.L., Finegan T.M., and Ross J.L.. 2023. A tale of 12 tails: Katanin severing activity affected by carboxy-terminal tail sequences. Biomolecules. 13:620. 10.3390/biom13040620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, C., Srayko M., and Mains P.E.. 2004. The Caenorhabditis elegans microtubule-severing complex MEI-1/MEI-2 katanin interacts differently with two superficially redundant beta-tubulin isotypes. Mol. Biol. Cell. 15:142–150. 10.1091/mbc.e03-06-0418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn, N.A., Martinez E., Nguyen H., and Torres J.Z.. 2021. The mammalian family of katanin microtubule-severing enzymes. Front. Cell Dev. Biol. 9:692040. 10.3389/fcell.2021.692040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mains, P.E., Kemphues K.J., Sprunger S.A., Sulston I.A., and Wood W.B.. 1990. Mutations affecting the meiotic and mitotic divisions of the early Caenorhabditis elegans embryo. Genetics. 126:593–605. 10.1093/genetics/126.3.593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally, F.J., and Roll-Mecak A.. 2018. Microtubule-severing enzymes: From cellular functions to molecular mechanism. J. Cell Biol. 217:4057–4069. 10.1083/jcb.201612104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally, F.J., and Vale R.D.. 1993. Identification of katanin, an ATPase that severs and disassembles stable microtubules. Cell. 75:419–429. 10.1016/0092-8674(93)90377-3 [DOI] [PubMed] [Google Scholar]
- McNally, K., Audhya A., Oegema K., and McNally F.J.. 2006. Katanin controls mitotic and meiotic spindle length. J. Cell Biol. 175:881–891. 10.1083/jcb.200608117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally, K., Berg E., Cortes D.B., Hernandez V., Mains P.E., and McNally F.J.. 2014. Katanin maintains meiotic metaphase chromosome alignment and spindle structure in vivo and has multiple effects on microtubules in vitro. Mol. Biol. Cell. 25:1037–1049. 10.1091/mbc.e13-12-0764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally, K.P., Bazirgan O.A., and McNally F.J.. 2000. Two domains of p80 katanin regulate microtubule severing and spindle pole targeting by p60 katanin. J. Cell Sci. 113:1623–1633. 10.1242/jcs.113.9.1623 [DOI] [PubMed] [Google Scholar]
- McNally, K.P., and McNally F.J.. 2011. The spindle assembly function of Caenorhabditis elegans katanin does not require microtubule-severing activity. Mol. Biol. Cell. 22:1550–1560. 10.1091/mbc.e10-12-0951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiring, J.C.M., Grigoriev I., Nijenhuis W., Kapitein L.C., and Akhmanova A.. 2022. Opto-katanin, an optogenetic tool for localized, microtubule disassembly. Curr. Biol. 32:4660–4674.e6. 10.1016/j.cub.2022.09.010 [DOI] [PubMed] [Google Scholar]
- Mishra-Gorur, K., Çağlayan A.O., Schaffer A.E., Chabu C., Henegariu O., Vonhoff F., Akgümüş G.T., Nishimura S., Han W., Tu S., et al. 2014. Mutations in KATNB1 cause complex cerebral malformations by disrupting asymmetrically dividing neural progenitors. Neuron. 84:1226–1239. 10.1016/j.neuron.2014.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida, K., Tsuchiya K., Obinata H., Onodera S., Honda Y., Lai Y.C., Haruta N., and Sugimoto A.. 2021. Expression patterns and levels of all tubulin isotypes analyzed in GFP knock-in C. elegans strains. Cell Struct. Funct. 46:51–64. 10.1247/csf.21022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales, E. 2000. Structural insights into microtubule function. Annu. Rev. Biochem. 69:277–302. 10.1146/annurev.biochem.69.1.277 [DOI] [PubMed] [Google Scholar]
- Pintard, L., Willis J.H., Willems A., Johnson J.L., Srayko M., Kurz T., Glaser S., Mains P.E., Tyers M., Bowerman B., and Peter M.. 2003. The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature. 425:311–316. 10.1038/nature01959 [DOI] [PubMed] [Google Scholar]
- Rezabkova, L., Jiang K., Capitani G., Prota A.E., Akhmanova A., Steinmetz M.O., and Kammerer R.A.. 2017. Structural basis of katanin p60:p80 complex formation. Sci. Rep. 7:14893. 10.1038/s41598-017-14194-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roll-Mecak, A., and McNally F.J.. 2010. Microtubule-severing enzymes. Curr. Opin. Cell Biol. 22:96–103. 10.1016/j.ceb.2009.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roll-Mecak, A., and Vale R.D.. 2008. Structural basis of microtubule severing by the hereditary spastic paraplegia protein spastin. Nature. 451:363–367. 10.1038/nature06482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch E.F., and Maniatis T.. 1989. Molecular Cloning. A Laboratory Manual. Second edition. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Sarbanes, S.L., Zehr E.A., and Roll-Mecak A.. 2022. Microtubule-severing enzymes. Curr. Biol. 32:R992–R997. 10.1016/j.cub.2022.08.046 [DOI] [PubMed] [Google Scholar]
- Sharp, D.J., and Ross J.L.. 2012. Microtubule-severing enzymes at the cutting edge. J. Cell Sci. 125:2561–2569. 10.1242/jcs.101139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srayko, M., Buster D.W., Bazirgan O.A., McNally F.J., and Mains P.E.. 2000. MEI-1/MEI-2 katanin-like microtubule severing activity is required for Caenorhabditis elegans meiosis. Genes Dev. 14:1072–1084. 10.1101/gad.14.9.1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srayko, M., O’toole E.T., Hyman A.A., and Müller-Reichert T.. 2006. Katanin disrupts the microtubule lattice and increases polymer number in C. elegans meiosis. Curr. Biol. 16:1944–1949. 10.1016/j.cub.2006.08.029 [DOI] [PubMed] [Google Scholar]
- Szczesna, E., Zehr E.A., Cummings S.W., Szyk A., Mahalingan K.K., Li Y., and Roll-Mecak A.. 2022. Combinatorial and antagonistic effects of tubulin glutamylation and glycylation on katanin microtubule severing. Dev. Cell. 57:2497–2513.e6. 10.1016/j.devcel.2022.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ti, S.-C., Alushin G.M., and Kapoor T.M.. 2018. Human β-tubulin isotypes can regulate microtubule protofilament number and stability. Dev. Cell. 47:175–190.e5. 10.1016/j.devcel.2018.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney, L.G., Bryan J., Bush D.J., Fujiwara K., Mooseker M.S., Murphy D.B., and Snyder D.H.. 1973. Microtubules: Evidence for 13 protofilaments. J. Cell Biol. 59:267–275. 10.1083/jcb.59.2.267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemu, A., Szczesna E., Zehr E.A., Spector J.O., Grigorieff N., Deaconescu A.M., and Roll-Mecak A.. 2018. Severing enzymes amplify microtubule arrays through lattice GTP-tubulin incorporation. Science. 361:eaau1504. 10.1126/science.aau1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, S.R., Evans K.J., Lary J., Cole J.L., and Lauring B.. 2007. Recognition of C-terminal amino acids in tubulin by pore loops in Spastin is important for microtubule severing. J. Cell Biol. 176:995–1005. 10.1083/jcb.200610072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehr, E., Szyk A., Piszczek G., Szczesna E., Zuo X., and Roll-Mecak A.. 2017. Katanin spiral and ring structures shed light on power stroke for microtubule severing. Nat. Struct. Mol. Biol. 24:717–725. 10.1038/nsmb.3448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehr, E.A., Szyk A., Szczesna E., and Roll-Mecak A.. 2019. Katanin grips the β-tubulin tail through an electropositive double spiral to sever microtubules. Dev. Cell. 52:118-131.e6. 10.1016/j.devcel.2019.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows peptides identified using mass spectrometry.
lists plasmids used in this study.
is the source file for Fig. 1.
is the source file for Fig. 2.
is the source file for Fig. 3.
is the source file for Fig. S1.
is the source file for Fig. S2.