Abstract
Actinomyces naeslundii genospecies 1 and 2 bind to acidic proline-rich proteins (APRPs) and statherin via type 1 fimbriae and to β-linked galactosamine (GalNAcβ) structures via type 2 fimbriae. In addition, A. naeslundii displays two types of binding specificity for both APRPs-statherin and GalNAcβ, while Actinomyces odontolyticus binds to unknown structures. To study the molecular basis for these binding specificities, DNA fragments spanning the entire or central portions of fimP (type 1) and fimA (type 2) fimbrial subunit genes were amplified by PCR from strains of genospecies 1 and 2 and hybridized with DNA from two independent collections of oral Actinomyces isolates. Isolates of genospecies 1 and 2 and A. odontolyticus, but no other Actinomyces species, were positive for hybridization with fimP and fimA full-length probes irrespective of binding to APRPs and statherin, GalNAcβ, or unknown structures. Isolates of genospecies 1 and 2, with deviating patterns of GalNAcβ1-3Galα-O-ethyl-inhibitable coaggregation with Streptococcus oralis Ss34 and MPB1, were distinguished by a fimA central probe from genospecies 1 and 2, respectively. Furthermore, isolates of genospecies 1 and 2 displaying preferential binding to APRPs over statherin were positive with a fimP central probe, while a genospecies 2 strain with the opposite binding preference was not. The sequences of fimP and fimA central gene segments were highly conserved among isolates with the same, but diversified between those with a variant, binding specificity. In conclusion, A. naeslundii exhibits variant fimP and fimA genes corresponding to diverse APRP and GalNAcβ specificities, respectively, while A. odontolyticus has a genetically related but distinct adhesin binding specificity.
A key primary event in bacterial colonization of the host is adherence mediated by adhesins which recognize either carbohydrate or peptide structures (15, 21, 22, 26). Adhesins also participate in adherence-associated events, such as bacterial uptake into cells and host cell signaling (10, 23). Moreover, multiple adhesin families and heterologous specificities among each adhesin family lead to specific animal and tissue tropisms (39, 42, 44, 45).
Actinomyces naeslundii genospecies 1 and 2 and Actinomyces odontolyticus are common oral species displaying specific tissue and animal host tropisms (12–14, 25, 32, 34, 38). The intraoral tropism of A. naeslundii genospecies 1 and 2 involves two antigenically and functionally distinct fimbriae, type 1 and type 2 (8, 9). The amino acid sequences of the type 1 and type 2 major fimbrial subunits as deduced from the corresponding fimP and fimA genes of strains T14V (genospecies 2) (48) and ATCC 12104T (genospecies 1) (11, 49), respectively, have 34% sequence identity (50). Recently, the biogenesis and function of type 1 fimbriae were found to involve six genes (open reading frames 1 through 6), in addition to the fimP gene (51). Moreover, the adhesive capacity of type 2 fimbriae has been associated with a 95-kDa protein or protein complex, although the genetic basis for this is unclear (27). The corresponding genes for adhesion of A. odontolyticus to oral surfaces remain unknown (18).
Type 1 fimbriae, present mainly on genospecies 2 (8, 39), mediate attachment to salivary acidic proline-rich proteins (APRPs) and statherin adsorbed onto hydroxyapatite surfaces (15, 16). Allelic (e.g., PRP-1, PRP-2, Db, Pa, and PIF) and posttranslational (e.g., PRP-3 and PRP-4) variants of APRPs and four statherin variants (3, 20) and variant binding patterns of Actinomyces to APRPs and statherin (18, 41) have been demonstrated. The genospecies 2 strains LY7 and ATCC 19246, both expressing type 1 fimbriae (16, 47) and isolated from human sites, display preferential binding to APRPs and statherin, respectively (40, 41). Type 2 fimbriae, expressed by both genospecies (8, 39), mediate binding to β1-3-linked galactose or galactosamine structures (referred to as GalNAcβ specificity) (33, 43) in cell surface glycolipids and glycoproteins (5, 40), salivary glycoproteins (41), and streptococcal capsular polysaccharides (1). However, the genospecies 1 strain ATCC 12104T and the genospecies 2 strain LY7 display different binding patterns to a panel of saccharides containing β-linked galactose or galactosamine structures (39, 40). Consequently, these two strains display different lactose- and GalNAcβ1-3Galα-O-ethyl-inhibitable adherence patterns to epithelial cells (5, 39–41), polymorphonuclear leukocytes (36, 39), and streptococci (7, 28, 29, 39, 40). The adherence (or coaggregation) of Actinomyces-Streptococcus involves heterogeneous interaction modes (coaggregation groups A through F) which are either inhibitable or unaffected in the presence of lactose (28, 29). However, the genetic basis for the different types of APRP and GalNAcβ specificity is unknown.
Recently, we surveyed the expression of APRP and GalNAcβ binding specificities among a collection of Actinomyces isolates from defined individual and tissue sites of the human oral cavity (18, 19, 39). The survey revealed (i) different types of GalNAcβ specificity, as measured by coaggregation with streptococci, in genospecies 1 (type 2:1 specificity) and genospecies 2 (type 2:2 specificity); (ii) different prevalence rates of APRP binding, but preferential binding to APRPs over statherin, for isolates of genospecies 1 and 2; (iii) tongue isolates of A. odontolyticus expressing fimbriae and coaggregating with streptococci in a fashion that was not inhibited in the presence of GalNAcβ1-3Galα-O-ethyl.
The aim of this study was to generate full-length and central DNA fragments specific for the fimP (type 1) and fimA (type 2) fimbrial subunit genes and to use these fragments as probes in DNA-DNA hybridization assays with our collection of Actinomyces isolates and representative strains of the Actinomyces coaggregation groups A through F. Using this approach, we identified genetic variation among the fimP and fimA fimbrial subunit genes that correlated with the different types of APRP and GalNAcβ specificity displayed by Actinomyces species. Furthermore, we provide evidence that A. odontolyticus may exhibit a genetically related but functionally distinct adhesin binding specificity.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and characterization.
Strains of Actinomyces were isolated from different intraoral sites and subjects and characterized as previously described (18, 19). Representative reference strains of coaggregation group A (A. naeslundii T14V), group B (A. naeslundii PK19), group C (A. naeslundii PK947), group D (A. naeslundii PK606), group E (A. odontolyticus PK984), and group F (A. naeslundii PK1259) were obtained from P. Kolenbrander, National Institutes of Health, Bethesda, Md. A. naeslundii LY7 was obtained from R. J. Gibbons, Forsyth Dental Center, Boston, Mass. A. naeslundii ATCC 19246 and ATCC 12104T were obtained from the National Bacteriology Laboratory, Stockholm, Sweden. Strains LY7 and ATCC 19246 and reference strains for coaggregation groups A through F were characterized by multivariate statistical analysis of phenotypic characteristics, serotyping, and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as previously described (18). Streptococcus oralis MPB1 was obtained from the Department of Cariology, Göteborg University, Göteborg, Sweden, and S. oralis Ss34 was obtained from P. Kolenbrander. Escherichia coli HB101 and K-12 and Actinomyces ATCC 10048T, ATCC 12102T, ATCC 35568T, ATCC 23860T, and ATCC 49285T were all obtained from the Culture Collection, Göteborg University (CCUG). All Actinomyces and streptococcal strains were cultured at 37°C for 24 h in 5% CO2–10% H2 in nitrogen on Columbia II agar base plates (Becton Dickinson and Co., Cockeysville, Md.) supplemented with 30 ml of a human erythrocyte suspension per liter. E. coli strains were cultured on Luria-Bertani (LB) agar plates at 37°C for 24 h. Bacteria were metabolically labeled by mixing 10 μl of [35S]methionine (10 mCi/ml; Amersham, Little Chalfont, United Kingdom) with bacteria suspended in 100 μl of 10 mM phosphate-buffered saline (pH 7.2) prior to growth for 24 h.
Chromosomal DNA isolation.
Chromosomal DNA was isolated essentially as described previously (6), except for the following minor modifications: (i) the concentration of lysozyme was increased to 20 mg/ml; (ii) bacterial lysis was induced by the addition of 60 μl of 10% SDS and 150 μl of pronase (25 mg/ml; Sigma Chemical Co., St. Louis, Mo.) prior to overnight incubation at 37°C; and (iii) after lysis, DNA was extracted with an equal volume of phenol, followed by chloroform-isoamyl alcohol (24:1, vol/vol), and precipitated with 2 volumes of ethanol (95%)–1 M NaCl at −20°C. Following treatment with 0.5 mg of RNase (Boehringer GmbH, Mannheim, Germany) at 37°C for 45 min, DNA was finally recovered with 375 μl of cold ethanol (95%) in the presence of 50 μl of 7.5 M ammonium acetate for 30 min at −20°C.
Oligonucleotide primer pairs.
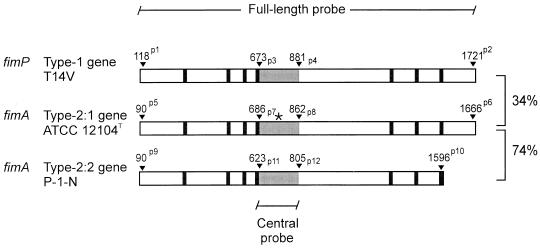
The PCR primer sequences used to generate DNA probes specific for fimP (type 1), fimA (type 2:1), and fimA (type 2:2) fimbrial subunit genes were as follows: P1, 5′-ACA GCA ATG CAC TCC CTC AA-3′, and P2, 5′-TG CTT GGC AAC GTG ACG GC-3′ (used to generate a 1,603-bp type 1 full-length probe); P3, 5′-ACC CTC TCC GGT GTG GAC AA-3′, and P4, 5′-ACC TCG TTC TGA CCG ACG AT-3′ (a 208-bp type 1 central probe); P5, 5′-G AAG TAC AAC ACC AGC ACG C-3′, and P6, 5′-GAG GTC CCG GTT CCG CTT-3′ (a 1,576-bp type 2:1 full-length probe); P7, 5′-AGA AGA TCG AAG TCG CCA AGA-3′, and P8, 5′-CTG TAG CCG TCA CCT GCT TCA-3′ (a 176-bp type 2:1 central probe); P9, 5′-G AAG TAC AAC ACC AGC ACG C-3′, and P10, 5′-CTT GGC ACC AGT GAG GGG-3′ (a 1,506-bp portion of the type 2:2 fimbrial subunit gene); and P11, 5′-AGG CCA TCA GCG TTG AGA AGA-3′, and P12, 5′-AGA CCT CAG TGG CGG TCA-3′ (a 182-bp type 2:2 central probe). The locations of these primer sequences with respect to the type 1 and type 2 fimbrial subunit genes are indicated in Fig. 1.
FIG. 1.
Schematic illustration of the fimP and fimA fimbrial subunit genes from A. naeslundii. DNA probes representing full-length and central (gray area) gene segments specific for different adhesin specificities were PCR amplified by using synthetic oligonucleotide primers specific for fimP from A. naeslundii T14V (genospecies 2), fimA from ATCC 12104T (genospecies 1), and fimA from P-1-N (genospecies 2, strain CCUG 33910). The nucleotide positions of all primers are indicated by arrowheads and labeled p1 through p12 (see Materials and Methods). Primers set as superscripts to position numbers were used to amplify full-length gene segments; primers set as subscripts to position numbers were used to amplify central gene segments. The central DNA probes were generated from a region of the gene sequences having comparably low homology and being devoid of highly conserved proline-containing segments. The overall nucleotide sequence identities between the three fimbrial subunit genes are indicated on the right. An asterisk marks the position of the BamHI cleavage site in the type 2:1 fimbrial subunit gene.
PCR.
The PCRs were performed in 50-μl reaction mixtures containing 100 ng of DNA; 50 ng of each oligonucleotide primer; 2 U of Taq polymerase (Boehringer); 100 μM each dATP, dGTP, dCTP, and dTTP (deoxynucleoside triphosphate [dNTP] mixture; Boehringer); and 1× PCR buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 1.5 mM MgCl2). DNA fragments were labeled with digoxigenin by replacing the dNTP mixture with the digoxigenin dNTP labeling mixture (Boehringer) in the second PCR amplification. Chromosomal DNA isolated from A. naeslundii T14V (type 1), ATCC 12104T (type 2:1), and P-1-N (type 2:2) was used as the template for probe construction. The full-length gene fragments were amplified by using an initial cycle with denaturation at 94°C for 5 min, annealing at 59°C for 150 s, and extension at 72°C for 150 s, followed by a repeat of 33 cycles with denaturation at 94°C for 75 s, annealing at 59°C for 150 s, extension at 72°C for 150 s, and a final hold at 72°C for 7 min. The type 1 and type 2:1 central gene segments were amplified by using an initial cycle with denaturation at 94°C for 4 min, annealing at 63°C for 2 min, and extension at 72°C for 2 min, followed by a repeat of 29 cycles with denaturation at 94°C for 75 s, primer annealing at 63°C for 75 s, primer extension at 72°C for 75 s, and a final hold at 72°C for 7 min. The type 2:2 central gene segments were amplified in the same way except that the annealing temperature was decreased to 50°C.
Slot blot hybridization assay.
Chromosomal DNA (3 μg) was transferred to nylon membranes (Hybond N+; Amersham) with a slot blot manifold filter system (Milliblot-S; Millipore, Västra Frölunda, Sweden) as described previously (2). Membranes were prehybridized in 5× SSC (0.75 M sodium chloride, 0.075 M sodium citrate)–0.02% N-lauroylsarcosine–0.1% SDS–1% blocking reagent (Boehringer) at 80°C for 5 h (high-stringency conditions). Following overnight hybridization in prehybridization solution containing probe DNA, the membranes were washed twice in 2× SSC–0.1% SDS for 5 min at room temperature and twice in 0.5× SSC–0.1% SDS (for type 1 and type 2:1 full-length and central probes) or 0.3× SSC–0.1% SDS (for type 2:2 central probe) for 15 min at 80°C. Additional rounds of hybridization and stringency washes were also performed under the same conditions except that the temperature was lowered from 80 to 50°C (low-stringency conditions). Hybridization was detected by using a digoxigenin DNA detection kit, as recommended by the manufacturer (Boehringer), scanned in a densitometer (GS-700 imaging densitometer; Bio-Rad, Hercules, Calif.), and analyzed with Molecular Analyst software (Bio-Rad). The degree of hybridization was scored from 0 to 6 according to the following densitometric values: score of 0 = a densitometric value of <0.01, 1 = 0.01 to <0.04, 2 = 0.04 to <0.10, 3 = 0.10 to <0.16, 4 = 0.16 to <0.22, 5 = 0.22 to <0.27, and 6 = ≥0.27.
Southern blot analysis.
Chromosomal DNA (8 μg) was digested with BamHI or PstI, and DNA fragments were separated on 0.7% agarose gels. DNA was transferred to nylon membranes (Hybond N+; Amersham) as described previously (35). Membranes were prehybridized in 5× SSC–0.02% N-lauroylsarcosine–0.1% SDS–1% blocking reagent (Boehringer) at 80°C for 5 h (high-stringency conditions). Following overnight hybridization in prehybridization solution containing probe DNA, the membranes were washed twice in 2× SSC–0.1% SDS for 5 min at room temperature and twice in 0.5× SSC–0.1% SDS (for type 1 and type 2:1 full-length and central probes) or 0.3× SSC–0.1% SDS (for type 2:2 central probe) for 15 min at 80°C. Hybridization was detected by using a digoxigenin DNA detection kit, as recommended by the manufacturer (Boehringer).
Subcloning and nucleotide sequencing.
PCR-amplified DNA fragments were purified with the Jetpure PCR purification kit (Saveen, Malmö, Sweden). Type 1 and type 2 full-length and central DNA fragments were cloned directly into the pGEM-T vector system (Promega, Madison, Wis.) in accordance with the manufacturer’s instructions. The ligation mixture was transformed into E. coli JM109 competent cells, and appropriate transformants were selected on LB agar plates containing ampicillin (50 μg/ml), 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (40 μg/mL; X-Gal), and isopropyl-β-d-thiogalactopyranoside (0.1 mM; IPTG). Plasmid DNA was isolated by miniprep DNA purification systems (Wizard; Promega) and sequenced by using the Pharmacia (Uppsala, Sweden) T7 sequencing system (37) and analyzed in 8 M urea–6% polyacrylamide gels.
Computer analysis.
DNA and deduced amino acid sequence comparisons were performed by using the Sequence Analysis software package, version 8.1, from the Genetics Computer Group, University of Wisconsin, Madison.
Coaggregation.
The coaggregation of streptococci (3 × 109 cells/ml) and Actinomyces cells (3 × 109 cells/ml), both suspended in coaggregation buffer (1.0 mM Tris-HCl, 0.1 mM Ca2+, 0.1 mM Mg2+, 0.02% NaN3), was determined by visual inspection after mixing the cells as described previously (18).
Isolation of APRPs and statherin.
Freshly collected parotid saliva from one individual homozygous for allelic APRP variants (PRP-1 and PIF-s) was separated on a DEAE-Sephacel column (15 by 1.6 cm; Pharmacia) by using a linear gradient of 25 mM to 1.0 M NaCl in 50 mM Tris-HCl (pH 8.0). The peak containing APRPs and statherin was concentrated by ultrafiltration on a Centriprep 10 concentrator (Amicon Inc., Beverly, Mass.) and separated by gel filtration (HiLoad 26/60 Superdex S-200 Prepgrade; Pharmacia) in 20 mM Tris-HCl–0.5 M NaCl (pH 8.0). Each of the resolved PRP-1–PIF-s, PRP-3–PIF-f, and statherin proteins were then finally purified on a Macroprep high Q column (15 by 1.6 cm; Bio-Rad) by using a linear gradient of 25 mM to 1.0 M NaCl in 50 mM Tris-HCl (pH 8.0). The purity and identity of APRPs and statherin were confirmed by SDS-PAGE, native alkaline electrophoresis, NH2-terminal amino acid sequencing, and bacterial binding properties.
Hydroxyapatite assay.
The adherence of [35S]methionine-labeled bacteria (6 × 104 cpm/ml; 5 × 108 cells/ml) to purified proteins (5.0 μg/ml) adsorbed onto hydroxyapatite beads (BDH Chemicals Ltd., Poole, United Kingdom) was measured as described previously (16, 41).
RESULTS
Genetically related Actinomyces adhesin specificities.
To determine the genetic relatedness of different binding specificities in A. naeslundii and A. odontolyticus, DNA fragments which span the known fimP and fimA fimbrial subunit genes of strains T14V (genospecies 2) and ATCC 12104T (genospecies 1), respectively, were amplified by PCR (Fig. 1) and hybridized with DNA from Actinomyces isolates with defined binding specificities (Table 1). Isolates of A. naeslundii were positive for hybridization with the fimP and fimA full-length probes, irrespective of genospecies 1 or 2, type of APRP and GalNAcβ specificity, or tissue origin (teeth or buccal mucosa) (Table 1 and see Table 3). Isolates of genospecies 1 displayed stronger hybridization signals with the fimA probe than genospecies 2 did, while the opposite was true for the fimP probe. Isolates of A. odontolyticus, which binds to unknown structures, were positive with both probes, whereas Actinomyces israelii, Actinomyces meyeri, Actinomyces gerencseriae, Actinomyces georgiea, and E. coli were negative. Thus, A. naeslundii and A. odontolyticus may express a family of genetically related but distinct adhesin specificities.
TABLE 1.
Hybridization of full-length and central fimP (type 1) and fimA (type 2) gene probes with chromosomal DNA from Actinomyces strains of defined binding specificities
| Speciesa and strain | Originb | Degree of hybridizationc
|
Binding specificityd
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
fimP fimbrial gene
|
fimA fimbrial gene
|
|||||||||
| Full-length probe | Central probe | Full-length probe | Central probes
|
APRPs | GalNAcβ
|
Unknown structure | ||||
| 2:1 | 2:2 | 2:1 | 2:2 | |||||||
| A. naeslundii genospecies 1 | ||||||||||
| B-2-G | Buccae | 2 | 0 | 6 | 4 | 0 | − | + | − | |
| B-1-L | Buccae | 3 | 0 | 6 | 3 | 0 | − | + | − | |
| P-4-N | Plaque | 2 | 0 | 5 | 4 | 0 | − | + | − | |
| Pn-1-GA | Plaque | 2 | 0 | 6 | 4 | 0 | − | + | − | |
| Pn-22-E | Plaque | 3 | 0 | 6 | 4 | 0 | − | + | − | |
| P-5-L | Plaque | 2 | 0 | 6 | 4 | 0 | − | + | − | |
| P-10-A | Plaque | 2 | 0 | 6 | 4 | 0 | − | + | − | |
| A. naeslundii genospecies 2 | ||||||||||
| B-3-N | Buccae | 6 | 6 | 5 | 0 | 3 | + | − | + | |
| B-10-N | Buccae | 6 | 4 | 5 | 0 | 3 | + | − | + | |
| B-3-K | Buccae | 6 | 4 | 6 | 0 | 3 | + | − | + | |
| B-10-K | Buccae | 6 | 5 | 5 | 0 | 2 | + | − | + | |
| B-1-G | Buccae | 6 | 2 | 4 | 0 | 2 | + | − | + | |
| B-3-L | Buccae | 5 | 4 | 5 | 0 | 2 | + | − | + | |
| B-10-L | Buccae | 6 | 5 | 5 | 0 | 2 | + | − | + | |
| B-1-A | Buccae | 6 | 4 | 5 | 0 | 4 | + | − | + | |
| P-1-N | Plaque | 6 | 5 | 4 | 0 | 3 | + | − | + | |
| P-6-K | Plaque | 5 | 5 | 4 | 0 | 3 | + | − | + | |
| P-1-G | Plaque | 6 | 3 | 6 | 0 | 4 | + | − | + | |
| P-1-L | Plaque | 4 | 4 | 5 | 0 | 2 | + | − | + | |
| P-1-A | Plaque | 6 | 3 | 5 | 0 | 4 | + | − | + | |
| A. odontolyticus | ||||||||||
| B-4-L | Buccae | 4 | 4 | 5 | 0 | 2 | + | − | + | |
| Pn-13-N | Plaque | 3 | 0 | 3 | 0 | 0 | + | − | + | |
| T-5-G | Tongue | 3 | 0 | 2 | 0 | 0 | + | − | − | + |
| T-1-G | Tongue | 3 | 0 | 2 | 0 | 0 | − | − | − | + |
| T-21-N | Tongue | 3 | 0 | 2 | 0 | 0 | − | − | − | + |
| T-25-N | Tongue | 3 | 0 | 2 | 0 | 0 | − | − | − | + |
| T-26-N | Tongue | 3 | 0 | 2 | 0 | 0 | − | − | − | + |
| T-27-N | Tongue | 3 | 0 | 2 | 0 | 0 | − | − | − | + |
| T-1-K | Tongue | 3 | 0 | 2 | 0 | 0 | − | − | − | + |
| T-3-K | Tongue | 3 | 0 | 2 | 0 | 0 | − | − | − | + |
| T-10-K | Tongue | 2 | 0 | 1 | 0 | 0 | − | − | − | + |
| A. israelii | ||||||||||
| ATCC 10048T | 0 | 0 | 0 | 0 | 0 | |||||
| ATCC 12102T | 0 | 0 | 0 | 0 | 0 | |||||
| A. meyeri ATCC 35568T | 0 | 0 | 0 | 0 | 0 | |||||
| A. gerencseriae ATCC 23860T | 0 | 0 | 0 | 0 | 0 | |||||
| A. georgiae ATCC 49285T | 0 | 0 | 0 | 0 | 0 | |||||
| Escherichia coli | ||||||||||
| K12 | 0 | 0 | 0 | 0 | 0 | |||||
| HB101 | 0 | 0 | 0 | 0 | 0 | |||||
Strains were isolated from several individuals (last suffix in strain designations for A. naeslundii genospecies 1 and 2 and A. odontolyticus). The isolates were identified by multivariate statistical analysis of phenotypic characteristics, serological reactions, and protein banding patterns of cell extracts analyzed by SDS-PAGE (18).
Strains were isolated from the buccal mucosa (buccae), teeth (plaque), and tongue of several individuals.
The type 1 DNA probes were generated from the fimP fimbrial subunit gene of A. naeslundii T14V, and the type 2:1 and type 2:2 DNA probes were generated from the variant fimA fimbrial subunit genes of A. naeslundii ATCC 12104T (genospecies 1) and P-1-N (genospecies 2), respectively. The probes were labeled with digoxigenin and used in slot blot hybridization with chromosomal DNA from Actinomyces species under high-stringency conditions. The degree of hybridization was scored from 0 to 6 according to the following densitometric values: 0 = <0.01, 1 = 0.01 to <0.04, 2 = 0.04 to <0.10, 3 = 0.10 to <0.16, 4 = 0.16 to <0.22, 5 = 0.22 to <0.27, and 6 = ≥0.27. Essentially identical results, though analyzed for only half of the strains, were obtained under low-stringency conditions.
APRP specificity denotes binding of Actinomyces strains to APRP-1 and APRP-3 and statherin adsorbed onto hydroxyapatite beads (18, 19). Preferential binding of all strains was in the order of APRP-1 > APRP-3 > statherin. GalNAcβ specificity denotes hemagglutination and aggregation of Actinomyces strains with an established collection of streptococcal strains in a GalNAcβ1-3Galα-O-ethyl-inhibitable fashion (18, 19). The GalNAcβ specificities, types 2:1 and 2:2, denote different aggregation patterns with S. oralis Ss34 and MPB1: type 2:1 coaggregates with both strains, whereas type 2:2 coaggregates only with Ss34 (18, 19). Binding specificity to unknown structures denotes aggregation of Actinomyces with streptococci that was not inhibitable by the presence of GalNAcβ1-3Galα-O-ethyl (18, 19). +, positive result; −, negative result.
TABLE 3.
Hybridization of full-length and central fimP (type 1) and fimA (type 2) gene probes with chromosomal DNA from an independent reference collection of Actinomyces strains and with DNA from a single strain characterized by a statherin binding pattern
| Strain or coaggregation group (strain) | Speciesa | Degree of hybridization tob:
|
Binding specificityc
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
fimP fimbrial gene
|
fimA fimbrial gene
|
||||||||||
| Full-length probe | Central probe | Full-length probe | Central probes
|
APRPs
|
GalNAcβ
|
Unknown structure | |||||
| 2:1 | 2:2 | APRP-1 | Stath. | 2:1 | 2:2 | ||||||
| Reference strainsb | |||||||||||
| LY7 | A. naeslundii genospecies 2 | 6 | 6 | 4 | 0 | 3 | + | (+) | − | + | − |
| ATCC 12104T | A. naeslundii genospecies 1 | 3 | 0 | 6 | 6 | 0 | − | − | + | − | − |
| Coaggregation groupsd | |||||||||||
| A (T14V) | A. naeslundii genospecies 2 | 6 | 6 | 4 | 0 | 3 | + | (+) | − | + | − |
| B (PK19) | A. naeslundii genospecies 1 | 3 | 0 | 6 | 5 | 0 | − | − | + | − | − |
| C (PK947) | A. naeslundii genospecies 1 | 6 | 5 | 6 | 4 | 0 | − | − | + | − | − |
| D (PK606) | A. naeslundii genospecies 1 | 5 | 5 | 6 | 4 | 0 | − | − | + | − | − |
| E (PK984) | A. odontolyticus | 2 | 0 | 5 | 0 | 0 | − | − | − | − | + |
| F (PK1259) | A. naeslundii genospecies 2 | 4 | 3 | 4 | 0 | 1 | + | (+) | − | + | − |
| Statherin binding straine ATCC 19246 | A. naeslundii genospecies 2 | 6 | 0 | 5 | 0 | 0 | (+) | + | − | − | − |
The isolates were identified by multivariate statistical analyses of phenotypic characteristics, serological reactions, and proteins banding patterns of cell extracts analyzed by SDS-PAGE (18).
The type 1 DNA probes were generated from the fimP fimbrial subunit gene of A. naeslundii T14V, and the type 2:1 and type 2:2 DNA probes were generated from the fimA type 2 fimbrial genes of A. naeslundii ATCC 12104T and A. naeslundii P-1-N (CCUG 33910), respectively. The probes were labeled with digoxigenin and used in slot blot hybridization with chromosomal DNA from isolates of Actinomyces spp. under high-stringency conditions. The degree of hybridization was scored from 0 to 6 according to the following densitometric values: 0 = <0.01, 1 = 0.01 to <0.04, 2 × 0.04 to <0.10, 3 = 0.10 to <0.16, 4 = 0.16 to <0.22, 5 = 0.22 to <0.27, and 6 = ≥0.27. Essentially identical results were obtained under low-stringency conditions.
APRP or statherin (Stath.) specificity denotes binding to acidic proline-rich proteins (APRPs) or statherin adsorbed onto hydroxyapatite beads, respectively (18, 19). A negative binding result (−) means complete absence of visible binding, and a positive binding result [+ and (+)] indicates that more than 15% of bacteria adhered to the protein-treated beads. A strong positive binding result, +, indicates markedly more binding than a low positive result, (+). GalNAcβ specificity denotes hemagglutination and aggregation of Actinomyces strains with an established collection of streptococcal strains in a GalNAcβ1-3Galα-O-ethyl-inhibitable fashion (18, 19). The different GalNAcβ specificities, types 2:1 and 2:2, denote different aggregation patterns with S. oralis Ss34 and MPB1: type 2:1 coaggregates with both strains, while type 2:2 coaggregates only with Ss34 (18, 19). Binding specificity to unknown structures denotes aggregation with streptococci that was not inhibitable by GalNAcβ1-3Galα-O-ethyl (18, 19).
The coaggregation groups A to F include strains showing adherence properties typically found in an independent reference collection of Actinomyces strains with a mainly subgingival plaque origin (28).
Statherin binding denotes a preferential binding to statherin over APRPs adsorbed onto hydroxyapatite beads (41).
A. naeslundii genospecies 1 exhibits a variant fimA gene associated with type 2:1 GalNAcβ specificity.
To investigate the genetic basis for different types of GalNAcβ specificity, a DNA probe which spans the central segment of the fimA gene of strain ATCC 12104T (genospecies 1) was amplified by PCR (Fig. 1) and hybridized with DNA from isolates of A. naeslundii with different types of GalNAcβ specificity (Tables 1 and 2). Isolates of A. naeslundii genospecies 1, which display GalNAcβ1-3Galα-O-ethyl-inhibitable coaggregation with S. oralis Ss34 and MPB1 (type 2:1 specificity), were positive with the probe irrespective of tooth (18 isolates) or buccal mucosa (2 isolates) origin. In contrast, isolates of A. naeslundii genospecies 2, which harbor APRP and another GalNAcβ specificity, were negative with the probe (63 of 63 isolates). Thus, A. naeslundii genospecies 1 appears to encode a variant fimA gene that confers type 2:1 GalNAcβ specificity.
TABLE 2.
Hybridization of central fimP (type 1) and fimA (type 2) gene probes with chromosomal DNA from strains of A. naeslundii and A. odontolyticus with defined binding specificities and tissue origin
| Species and site of isolationa | No. of isolates hybridized withb:
|
No. of isolates with:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
fimP central DNA probe
|
fimA central DNA probes
|
APRP specificityc
|
GalNAcβ specificityd
|
||||||||
| Pos | Neg | Type 2:1
|
Type 2:2
|
Pos | Neg | Type 2:1 | Type 2:2 | Neg | |||
| Pos | Neg | Pos | Neg | ||||||||
| A. naeslundii genospecies 1 (n = 20) | |||||||||||
| Tooth (n = 18) | 6 | 12 | 18 | 18 | 5 | 13 | 18 | ||||
| Buccal mucosa (n = 2) | 2 | 2 | 2 | 2 | 2 | ||||||
| Tongue (n = 0) | |||||||||||
| A. naeslundii genospecies 2 (n = 63) | |||||||||||
| Tooth (n = 41) | 40 | 1 | 41 | 41 | 41 | 41 | |||||
| Buccal mucosa (n = 1) | 21 | 21 | 21 | 21 | 21 | ||||||
| Tongue (n = 1) | 1 | 1 | 1 | 1 | 1 | ||||||
| A. odontolyticus (n = 19) | |||||||||||
| Tooth (n = 1) | 1 | 1 | 1 | 1 | 1 | ||||||
| Buccal mucosa (n = 1) | 1 | 1 | 1 | 1 | 1 | ||||||
| Tongue (n = 17) | 17 | 17 | 17 | 1 | 16 | 17 | |||||
The isolates were identified by multivariate statistical analysis of phenotypic characteristics, serology, and protein banding patterns of cell extracts analyzed with SDS-PAGE (18). Strains isolated from eight individuals were grouped based on tissue origin (tooth, buccal mucosa, or tongue) or binding specificity.
The central type 1 DNA probe was generated from the fimP fimbrial subunit gene of A. naeslundii T14V, and the central type 2:1 and type 2:2 DNA probes were generated from the variant fimA fimbrial subunit genes of A. naeslundii ATCC 12104T and P-1-N (CCUG 33910), respectively. The probes were labeled with digoxigenin and used in slot blot hybridization with chromosomal DNA from Actinomyces species under high-stringency conditions. The degree of hybridization was scored from 0 to 6 according to the following densitometric values; 0 = <0.01, 1 = 0.01 to <0.04, 2 = 0.04 to <0.10, 3 = 0.10 to <0.16, 4 = 0.16 to <0.22, 5 = 0.22 to <0.27, and 6 = ≥0.27. Positive signals (Pos) denote hybridization reactions with scores of 2 to 6 and negative signals (Neg) denote hybridization reactions with scores of 0 to 1. The majority of strains showed hybridization reactions with scores of 3 to 4. Essentially identical results, though analyzed for only half of the strains, were obtained under low-stringency conditions.
APRP specificity denotes binding to APRPs adsorbed onto hydroxyapatite beads (18, 19). All APRP-binding isolates showed preferential binding in the order of APRP-1 > APRP-3 > statherin (18, 19).
GalNAcβ specificity denotes hemagglutination and aggregation of Actinomyces strains with a collection of streptococcal strains in a GalNAcβ1-3Galα-O-ethyl-inhibitable fashion (18, 19). The different GalNAcβ specificities, types 2:1 and 2:2, denote different aggregation patterns with S. oralis Ss34 and MPB1: type 2:1 coaggregates with both strains, while type 2:2 coaggregates only with Ss34 (18, 19). Negative GalNAcβ binding is characterized as aggregation of Actinomyces with streptococci that was not inhibited in the presence of GalNAcβ1-3Galα-O-ethyl (18, 19).
A. naeslundii genospecies 2 exhibits a variant fimA gene associated with type 2:2 GalNAcβ specificity.
To investigate the molecular basis for the variant GalNAcβ specificity of genospecies 2, essentially the entire fimA gene from the genospecies 2 strain P-1-N (CCUG 33910) was amplified by PCR, cloned, and sequenced (Fig. 1). This gene segment showed 74% nucleotide sequence identity to the known fimA gene of the genospecies 1 strain ATCC 12104T (data not shown).
A DNA probe was generated by PCR from the central region of the novel fimA gene and hybridized with DNA from the Actinomyces isolates (Tables 1 and 2). Isolates of genospecies 2, which display GalNAcβ1-3Galα-O-ethyl-inhibitable coaggregation with S. oralis Ss34 but not with strain MPB1 (type 2:2 specificity), were positive with this probe irrespective of tooth (41 isolates), buccal mucosa (21 isolates), or tongue (1 isolate) origin (63 of 63 isolates). In contrast, isolates of genospecies 1, which display type 2:1 GalNAcβ specificity, were negative with the probe (20 of 20 isolates). Thus, A. naeslundii genospecies 2 exhibits a variant fimA gene that is responsible for a type 2:2 GalNAcβ specificity.
Variant fimP genes correspond to different types of type 1 APRP specificity.
To characterize the molecular basis for different types of APRP specificity, a DNA probe which spans the central segment of the fimP gene was generated by PCR from the genospecies 2 strain T14V (Fig. 1). When used in hybridization assays, this central DNA probe hybridized with DNA from virtually all A. naeslundii isolates characterized by a preferential binding to APRPs over statherin (67 of 68 isolates), irrespective of genospecies (62 genospecies 2 isolates and 5 genospecies 1 isolates) or tissue origin (45 tooth, 21 buccal mucosa, and 1 tongue isolate). In contrast, the genospecies 2 strain ATCC 19246, displaying preferential binding to statherin over APRPs, was positive with the fimP full-length, but not with the central, probe (Table 3). Thus, A. naeslundii genospecies 2 may contain variant fimP genes that correspond to different types of APRP specificity.
APRP and GalNAcβ specificity in A. odontolyticus.
Since we have demonstrated that genetic diversity within the fimP and fimA fimbrial structural genes correlates with different types of binding specificity, we aimed to use hybridization analysis with specific probes to characterize APRP and GalNAcβ specificity in A. odontolyticus (Tables 1 and 2). Isolates of A. odontolyticus were positive with both fimP and fimA full-length probes (19 of 19 isolates), irrespective of binding to unknown structures (17 tongue isolates), APRP and GalNAcβ (1 buccal mucosa and 1 tooth isolate), or to APRP alone (1 tongue isolate). However, only the isolate from buccal mucosa with APRP and type 2:2 GalNAcβ specificity was positive with the fimP and fimA central probes. These data suggest a further genetic heterogeneity in fimbrial subunit genes in A. odontolyticus.
Adhesin specificities among an independent reference collection of Actinomyces strains.
We next characterized the binding properties present in an independent reference collection of Actinomyces strains, i.e., Actinomyces coaggregation groups A through F, which represent the different adherence properties of isolates of largely subgingival origin (28). Representative strains of groups A through F bound to APRPs, GalNAcβ, and unknown structures, and DNA from these isolates hybridized to the fimP and fimA full-length probes (Table 3).
The group B, C, and D strains, belonging to A. naeslundii genospecies 1, displayed a type 2:1 GalNAcβ specificity and were positive with the fimA type 2:1 central probe (Table 3). Although binding to APRPs was not distinct, the group C and D strains, but not the group B strain, were positive with the fimP central probe. The group A and F strains, belonging to A. naeslundii genospecies 2, displayed a type 2:2 GalNAcβ specificity and were positive with the fimA type 2:2 central probe. These strains also bound more efficiently to APRP-1 than to statherin and were positive with the fimP central probe. The group E strain, belonging to A. odontolyticus, displayed binding to unknown structures and was positive only with the fimP and fimA full-length probes. Thus, the same family of genetically related but distinct adhesin specificities are present in an independent reference collection of Actinomyces strains.
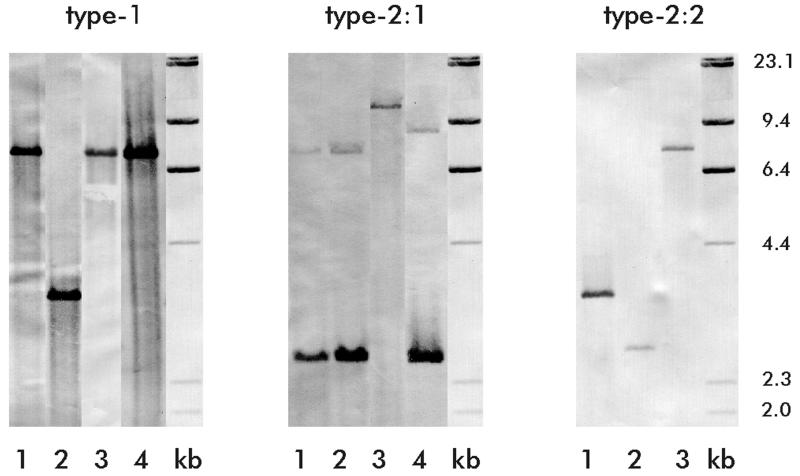
Single type 1 and type 2 fimbrial gene copies.
To exclude the possibility of multiple fimP and fimA genes contributing to genetic diversity, Southern blot hybridizations were used to demonstrate single fimP and fimA gene copies among randomly selected isolates (Fig. 2). The type 1 and type 2:2 central probes hybridized to single BamHI or PstI DNA fragments of chromosomal DNA from several isolates of A. naeslundii genospecies 2. The type 2:1 central probe hybridized to two BamHI DNA fragments of chromosomal DNA from four isolates of A. naeslundii genospecies 1. This is expected since a BamHI restriction site is located within the central region of the fimA gene of the genospecies 1 strain ATCC 12104T (Fig. 1).
FIG. 2.
The type 1 and type 2 fimbrial subunits are encoded by single gene copies in Actinomyces isolates. Southern blot hybridization of chromosomal DNA from A. naeslundii isolates with the fimP type 1 central DNA probe (left panel: lane 1, strain B-1-K; lane 2, P-1-K; lane 3, T14V; and lane 4, LY7), the fimA type 2:1 central DNA probe (middle panel: lane 1, strain Pn-22-E; lane 2, Pn-6-GA; lane 3, P-5-N; and lane 4, ATCC 12104T), and the fimA type 2:2 central DNA probe (right panel: lane 1, strain P-1-K; lane 2, P-1-N; and lane 3, LY7). Chromosomal DNA was digested with BamHI prior to hybridization with either type 1 or type 2:1 DNA probes and with PstI prior to hybridization with the type 2:2 DNA probe. The sizes (in kilobases) of HindIII-digested bacteriophage lambda DNA standards (lanes kb) are indicated on the right.
The variant fimA and fimP genes conferring a given binding specificity are highly conserved within each Actinomyces genospecies.
To gain further insights into the genetic diversity of fimA and fimP genes, central fimA and fimP gene fragments from isolates of A. naeslundii were amplified by PCR, cloned, and sequenced (Fig. 3). The fimP genes from five isolates of A. naeslundii genospecies 2, which bind preferentially to APRPs over statherin, displayed central gene segments with 98 to 100% nucleotide and deduced amino acid sequence identity to the known fimP gene. Similarly, the fimA genes from six isolates of A. naeslundii genospecies 1, which display type 2:1 GalNAcβ specificity, displayed central gene segments with 90 to 100% nucleotide and deduced amino acid sequence identity to the known fimA gene. The fimA genes of three isolates of genospecies 2, which display type 2:2 GalNAcβ specificity, had central gene segments with 92 to 100% nucleotide and deduced amino acid identity to each other. The fimP and fimA type 2:1 central gene segments had 42 to 51% nucleotide sequence identity and 28 to 43% deduced amino acid sequence identity; the fimP and fimA type 2:2 central gene segments had 44 to 50% nucleotide sequence identity and 26 to 38% deduced amino acid sequence identity. In contrast, the nucleotide and deduced amino acid sequence identities between the fimA type 2:1 and type 2:2 central segments were 70 to 75% and 71 to 81%, respectively. These data reveal a direct correlation between genetic diversity among fimP and fimA genes and the binding specificity mediated by the protein product encoded by these genes.
FIG. 3.
Alignment of the deduced amino acid sequences (in single-letter code) of central segments of the fimP and fimA type 2:1 and type 2:2 fimbrial subunit genes (see Fig. 1). Identical amino acids are indicated by gray shading, and those conserved in structure are indicated by a colon (:). Amino acids in boldface type are variable in fimbrial subunit sequences corresponding to the same binding specificity. The type 1 sequence data were collected from five isolates of genospecies 2 (strains B-10-K, B-1-K, P-1-K, P-3-K, and LY7), the type 2:1 sequence data are from six strains of genospecies 1 (strains P-3-N, P-5-N, Pn-1-GA, Pn-6-N, Pn-20-E, and Pn-22-E), and the type 2:2 sequence data are from a comparison of three strains of genospecies 2 (strains P-1-N, P-1-K, and LY7).
DISCUSSION
In the present study, we have used two independent Actinomyces reference collections of supra- and subgingival origin to extend our previous demonstration of two types of both APRP and GalNAcβ specificity in A. naeslundii and a yet-uncharacterized binding specificity in A. odontolyticus. We show that variant fimP and fimA fimbrial subunit genes confer the two types of APRP and GalNAcβ specificity and that a genetically related fimbrial adhesin may account for the binding of A. odontolyticus to unknown structures. Thus, structural variation in fimbria-associated proteins conferring adhesin specificity may account for the many tissues and animal hosts being colonized by Actinomyces species.
Our findings provide the following evidence that genetic variation in the fimP and fimA genes of A. naeslundii corresponds to different types of both APRP and GalNAcβ specificity. First, isolates with different types of APRP and GalNAcβ specificity were distinguished by DNA probes spanning the central portions of the fimA and fimP genes, while full-length probes were positive for hybridization with the isolates irrespective of type of binding specificity. Second, inverse hybridization signals with fimA and fimP full-length probes occurred between isolates with differing binding specificities (i.e., genospecies 1 versus 2). Third, the sequences of the fimA and fimP central gene segments were highly conserved among isolates with the same, but diversified between those with a variant, binding specificity. Furthermore, the different binding specificities observed were not due to a contribution of multiple genes, since randomly selected isolates contained only single fimA and fimP gene copies.
The present finding of variant fimA and fimP genes corresponding to different types of both APRP and GalNAcβ specificity may suggest the presence of a binding domain in the fimbrial subunit. This is reminiscent of the E. coli K88 and K99 major fimbrial subunits, both of which contain a domain for binding to host carbohydrate or protein structures (4, 24). Many bacterial adhesins constitute minor proteins distinct from the major structural subunit (21, 22), and similar to the situation for the minor Sfa adhesin of S fimbriae (17), structural variations in the major subunit could modulate the specificity of a separate, as-yet-uncharacterized adhesin in Actinomyces. Both extensive structural differences, as observed for PapG adhesins recognizing different glycolipid conformations (21, 22, 31, 44, 45), and single amino acid substitutions, similar to those described for different influenza virus binding specificities (46), could confer different binding specificities to Actinomyces species.
The binding of tongue isolates of A. odontolyticus to unknown structures in coaggregation with streptococci (18) may reflect a genetically related but functionally distinct fimbrial adhesin. Evidence for this is that all tongue isolates of A. odontolyticus, which produce fimbriae (18, 47), were positive with the full-length but not the central fimP and fimA probes. The fact that three A. odontolyticus isolates expressing APRP and GalNAcβ specificity were positive with the full-length probes while only one of those was positive with the central probes provides further arguments for a genetic variability among fimbrial subunit genes in A. odontolyticus.
The present association of variant fimbrial subunit genes with different types of APRP and GalNAcβ specificity may explain the intraoral tissue and animal tropisms of Actinomyces species (40, 41). For example, the different GalNAcβ1-3Galα-O-ethyl-inhibitable adherence patterns of genospecies 1 (ATCC 12104T) and genospecies 2 (LY7) to oral epithelial cells (40) may suggest variation in GalNAcβ specificity as a contributing factor to the relative dominance of genospecies 2 on the buccal mucosa and that of genospecies 1 on the tongue (12, 32). Furthermore, the differing appearances of genospecies 1 and 2 in plaque formation and their distinct patterns of coaggregation with streptococci irrespective of tissue origin (teeth or buccal mucosa) may suggest variation in GalNAcβ binding specificity as a tool to establish specific ecological niches (28). Although the role of variation in APRP specificity is less well understood, APRPs and statherin are polymorphic proteins displaying allelic and posttranslational variants. Therefore, the weak binding of some isolates (group C and D strains and one tooth isolate) to the allelic APRP-1 variant but positive fimP hybridization signals may suggest either binding to other APRP variants or a repressed fimbrial gene expression. Nonetheless, in line with the absence of preferential binding to statherin of strain 19246, which originated from a human case of actinomycosis, in both Actinomyces collections, statherin binding seems to be a characteristic of Actinomyces isolates from the oral cavity of rats and hamsters (30).
ACKNOWLEDGMENTS
This work was supported by grants from the Swedish Medical Research Council (10906), the Swedish Board for Technological Development (513), the Swedish Medical Society (748), and the Swedish Dental Society.
We thank B. Carlsson and J. Carlsson for kind assistance.
REFERENCES
- 1.Abeygunawardana C, Bush C A, Cisar J O. Complete structure of the cell surface polysaccharide of Streptococcus oralis ATCC 10557: a receptor for lectin-mediated interbacterial adherence. Biochemistry. 1991;30:6528–6540. doi: 10.1021/bi00240a025. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. Vol. 1. New York, N.Y: Greene Publishing Associates; 1995. [Google Scholar]
- 3.Azen E A, Maeda N. Molecular genetics of human salivary proteins and their polymorphism. Adv Hum Genet. 1988;17:141–199. doi: 10.1007/978-1-4613-0987-1_5. [DOI] [PubMed] [Google Scholar]
- 4.Bakker D, Willemsen P T J, Simons L H, van Zijderveld F G, de Graaf F K. Characterization of the antigenic and adhesive properties of FaeG, the major subunit of K88 fimbriae. Mol Microbiol. 1992;6:247–255. doi: 10.1111/j.1365-2958.1992.tb02006.x. [DOI] [PubMed] [Google Scholar]
- 5.Brennan M J, Cisar J O, Vatter A E, Sandberg A L. Lectin-dependent attachment of Actinomyces naeslundii to receptors on epithelial cells. Infect Immun. 1984;46:459–464. doi: 10.1128/iai.46.2.459-464.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chassy B M, Giuffrida A. Method for the lysis of gram-positive, asporogenous bacteria with lysozyme. Appl Environ Microbiol. 1980;39:153–158. doi: 10.1128/aem.39.1.153-158.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cisar J O, Kolenbrander P E, McIntire F C. Specificity of coaggregation reactions between human oral streptococci and strains of Actinomyces viscosus and Actinomyces naeslundii. Infect Immun. 1979;24:742–752. doi: 10.1128/iai.24.3.742-752.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cisar J O, Sandberg A L, Mergenhagen S E. The function and distribution of different fimbriae on strains of Actinomyces viscosus and Actinomyces naeslundii. J Dent Res. 1984;63:393–396. doi: 10.1177/00220345840630030701. [DOI] [PubMed] [Google Scholar]
- 9.Cisar J O, Vatter A E, Clark W B, Curl S H, Hurst-Calderone S, Sandberg A L. Mutants of Actinomyces viscosus T14V lacking type 1, type 2, or both types of fimbriae. Infect Immun. 1988;56:2984–2989. doi: 10.1128/iai.56.11.2984-2989.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cossart P, Boquet P, Normark S, Rappuoli R. Cellular microbiology emerging. Science. 1996;271:315–316. doi: 10.1126/science.271.5247.315. [DOI] [PubMed] [Google Scholar]
- 11.Donkersloot J A, Cisar J O, Wax M E, Harr R J, Chassy B M. Expression of Actinomyces viscosus antigens in Escherichia coli: cloning of a structural gene (fimA) for type 2 fimbriae. J Bacteriol. 1985;162:1075–1078. doi: 10.1128/jb.162.3.1075-1078.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellen R P. Establishment and distribution of Actinomyces viscosus and Actinomyces naeslundii in the human oral cavity. Infect Immun. 1976;14:1119–1124. doi: 10.1128/iai.14.5.1119-1124.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellen R P. Oral colonization by gram-positive bacteria significant to periodontal disease. In: Genco R J, Mergenhagen S E, editors. Host-parasite interactions in peridontal diseases. Washington, D.C: American Society for Microbiology; 1982. pp. 98–111. [Google Scholar]
- 14.Ellen R P, Segal D N, Grove D A. Relative proportions of Actinomyces viscosus and Actinomyces naeslundii in dental plaques collected from single sites. J Dent Res. 1978;57:550. doi: 10.1177/00220345780570040201. [DOI] [PubMed] [Google Scholar]
- 15.Gibbons R J. Bacterial adhesion to oral tissues: a model for infectious diseases. J Dent Res. 1989;68:750–760. doi: 10.1177/00220345890680050101. [DOI] [PubMed] [Google Scholar]
- 16.Gibbons R J, Hay D I. Human salivary acidic proline-rich proteins and statherin promote the attachment of Actinomyces viscosus LY7 to apatitic surfaces. Infect Immun. 1988;56:439–445. doi: 10.1128/iai.56.2.439-445.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hacker J, Kestler H, Hoschützky H, Jann K, Lottspeich F, Korhonen T K. Cloning and characterization of the S fimbrial adhesion II complex of an Escherichia coli O18:K1 meningitis isolate. Infect Immun. 1993;61:544–550. doi: 10.1128/iai.61.2.544-550.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hallberg, K., K.-J. Hammarström, E. Falsen, G. Dahlén, R. J. Gibbons, D. I. Hay, and N. Strömberg. Actinomyces naeslundii genospecies 1 and 2 express different binding specificities to N-acetyl-β-d-galactosamine, whereas Actinomyces odontolyticus expresses a different binding specificity in colonizing the human mouth. Oral Microbiol. Immunol., in press. [DOI] [PubMed]
- 19.Hallberg K, Holm C, Hammarström K-J, Kalfas S, Strömberg N. Ribotype diversity of Actinomyces with similar intraoral tropism but different types of N-acetyl-β-d-galactosamine binding specificity. Oral Microbiol Immunol. 1998;13:188–192. doi: 10.1111/j.1399-302x.1998.tb00731.x. [DOI] [PubMed] [Google Scholar]
- 20.Hay D I, Ahern J M, Schluckebier S K, Schlesinger D H. Human salivary acidic proline-rich polymorphism and biosynthesis studied by high-performance liquid chromatography. J Dent Res. 1994;73:1717–1726. doi: 10.1177/00220345940730110701. [DOI] [PubMed] [Google Scholar]
- 21.Hultgren S J, Abraham S, Caparon M, Falk P, St. Geme III J W, Normark S. Pilus and nonpilus bacterial adhesins: assembly and function in cell recognition. Cell. 1993;73:887–901. doi: 10.1016/0092-8674(93)90269-v. [DOI] [PubMed] [Google Scholar]
- 22.Hultgren S J, Normark S, Abraham S N. Chaperone-assisted assembly and molecular architecture of adhesive pili. Annu Rev Microbiol. 1991;45:383–415. doi: 10.1146/annurev.mi.45.100191.002123. [DOI] [PubMed] [Google Scholar]
- 23.Isberg R R. Discrimination between intracellular uptake and surface adhesion of bacterial pathogens. Science. 1991;252:934–938. doi: 10.1126/science.1674624. [DOI] [PubMed] [Google Scholar]
- 24.Jacobs A A C, Simons B H, de Graaf F K. The role of lysine-132 and arginine-136 in the receptor-binding domain of the K99 fibrillar subunit. EMBO J. 1987;6:1805–1808. doi: 10.1002/j.1460-2075.1987.tb02434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson J L, Moore L V H, Kaneko B, Moore W E C. Actinomyces georgiae sp. nov., Actinomyces gerencseriae sp. nov., designation of two genospecies of Actinomyces naeslundii, and inclusion of A. naeslundii serotypes II and III and Actinomyces viscosus serotypes II in A. naeslundii genospecies 2. Int J Syst Bacteriol. 1990;40:273–286. doi: 10.1099/00207713-40-3-273. [DOI] [PubMed] [Google Scholar]
- 26.Karlsson K A. Animal glycosphingolipids as membrane attachment sites for bacteria. Annu Rev Biochem. 1989;58:309–350. doi: 10.1146/annurev.bi.58.070189.001521. [DOI] [PubMed] [Google Scholar]
- 27.Klier C M, Kolenbrander P E, Roble A G, Marco M L, Cross S, Handley P S. Identification of a 95 kDa putative adhesin from Actinomyces serovar WVA963 strain PK1259 that is distinct from type 2 fimbrial subunits. Microbiology. 1997;143:835–846. doi: 10.1099/00221287-143-3-835. [DOI] [PubMed] [Google Scholar]
- 28.Kolenbrander P E. Intergeneric coaggregation among human oral bacteria and ecology of dental plaque. Annu Rev Microbiol. 1988;42:627–656. doi: 10.1146/annurev.mi.42.100188.003211. [DOI] [PubMed] [Google Scholar]
- 29.Kolenbrander P E, Williams B L. Lactose-reversible coaggregation between oral actinomycetes and Streptococcus sanguis. Infect Immun. 1981;33:95–102. doi: 10.1128/iai.33.1.95-102.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li T, Johansson I, Hay D, Strömberg N. Actinomyces naeslundii display structurally variant type-1 fimbrial subunit genes with ThrPhe and ProGln recognition motifs in tooth-adsorbed salivary proteins. Caries Res. 1998;32:299. . (Abstract.) [Google Scholar]
- 31.Marklund B-I, Tennent J M, Garcia E, Hamers A, Båga M, Lindberg F, Gaastra W, Normark S. Horizontal gene transfer of the Escherichia coli pap and prs pili operons as a mechanism for the development of tissue-specific adhesive properties. Mol Microbiol. 1992;6:2225–2242. doi: 10.1111/j.1365-2958.1992.tb01399.x. [DOI] [PubMed] [Google Scholar]
- 32.Marsh P, Martin M. Acquisition, adherence, distribution and functions of the oral flora. Dental plaque. In: Marsh P, Martin M, editors. Oral microbiology. 3rd ed. London, United Kingdom: Chapman & Hall, Ltd.; 1992. pp. 56–132. [Google Scholar]
- 33.McIntire F C, Crosby L K, Barlow J J, Matta K L. Structural preferences of β-galactoside-reactive lectins on Actinomyces viscosus T14V and Actinomyces naeslundii WVU45. Infect Immun. 1983;41:848–850. doi: 10.1128/iai.41.2.848-850.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ritz H L. Microbial population shifts in developing human dental plaque. Arch Oral Biol. 1967;12:1561–1568. doi: 10.1016/0003-9969(67)90190-2. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 36.Sandberg A L, Ruhl S, Joralmon R A, Brennan M J, Sutphin M J, Cisar J O. Putative glycoprotein and glycolipid polymorphonuclear leucocyte receptors for the Actinomyces naeslundii WVU45 fimbrial lectin. Infect Immun. 1995;63:2625–2631. doi: 10.1128/iai.63.7.2625-2631.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Socransky S S, Manganiello A D, Propas D, Oram V, van Houte J. Bacteriological studies of developing supragingival dental plaque. J Periodontal Res. 1977;12:90–106. doi: 10.1111/j.1600-0765.1977.tb00112.x. [DOI] [PubMed] [Google Scholar]
- 39.Strömberg N, Ahlfors S, Borén T, Bratt P, Hallberg K, Hammarström K-J, Holm C, Johansson I, Järvholm M, Kihlberg J, Li T, Ryberg M, Zand G. Anti-adhesion and diagnostic strategies for oro-intestinal bacterial pathogens. Adv Exp Med Biol. 1996;408:9–24. doi: 10.1007/978-1-4613-0415-9_2. [DOI] [PubMed] [Google Scholar]
- 40.Strömberg N, Borén T. Actinomyces tissue specificity may depend on differences in receptor specificity for GalNAcβ-containing glycoconjugates. Infect Immun. 1992;60:3268–3277. doi: 10.1128/iai.60.8.3268-3277.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strömberg N, Borén T, Carlén A, Olsson J. Salivary receptors for GalNAcβ-sensitive adherence of Actinomyces spp.: evidence for heterogeneous GalNAcβ and proline-rich protein receptor properties. Infect Immun. 1992;60:3278–3286. doi: 10.1128/iai.60.8.3278-3286.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strömberg N, Hultgren S, Russel D G, Normark S. Microbial attachment, molecular mechanisms. Encycl Microbiol. 1992;3:143–158. [Google Scholar]
- 43.Strömberg N, Karlsson K-A. Characterization of the binding of Actinomyces naeslundii (ATCC 12104) and Actinomyces viscosus (ATCC 19246) to glycosphingolipids, using a solid-phase overlay approach. J Biol Chem. 1990;265:11251–11258. [PubMed] [Google Scholar]
- 44.Strömberg N, Marklund B-I, Lund B, Ilver D, Hamers A, Gastra W, Karlsson K-A, Normark S. Host-specificity of uropathogenic Escherichia coli depends on differences in binding specificity to Galα1-4Gal-containing isoreceptors. EMBO J. 1990;9:2001–2010. doi: 10.1002/j.1460-2075.1990.tb08328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strömberg N, Nyholm P-G, Pascher I, Normark S. Saccharide orientation at the cell surface affects glycolipid receptor function. Proc Natl Acad Sci USA. 1991;88:9340–9344. doi: 10.1073/pnas.88.20.9340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weis W, Brown J H, Cusack S, Paulson J C, Skehel J J, Wiley D C. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature. 1988;333:426–431. doi: 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- 47.Yeung M K. Conservation of an Actinomyces viscosus T14V type 1 fimbrial subunit homolog among divergent groups of Actinomyces spp. Infect Immun. 1992;60:1047–1054. doi: 10.1128/iai.60.3.1047-1054.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yeung M K, Chassy B M, Cisar J O. Cloning and expression of a type 1 fimbrial subunit of Actinomyces viscosus T14V. J Bacteriol. 1987;169:1678–1683. doi: 10.1128/jb.169.4.1678-1683.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yeung M K, Cisar J O. Cloning and nucleotide sequence of a gene for Actinomyces naeslundii WVU45 type 2 fimbriae. J Bacteriol. 1988;170:3803–3809. doi: 10.1128/jb.170.9.3803-3809.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yeung M K, Cisar J O. Sequence homology between the subunits of two immunologically and functionally distinct types of fimbriae of Actinomyces spp. J Bacteriol. 1990;172:2462–2468. doi: 10.1128/jb.172.5.2462-2468.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yeung M K, Ragsdale P A. Synthesis and function of Actinomyces naeslundii T14V type 1 fimbriae require the expression of additional fimbria-associated genes. Infect Immun. 1997;65:2629–2639. doi: 10.1128/iai.65.7.2629-2639.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]