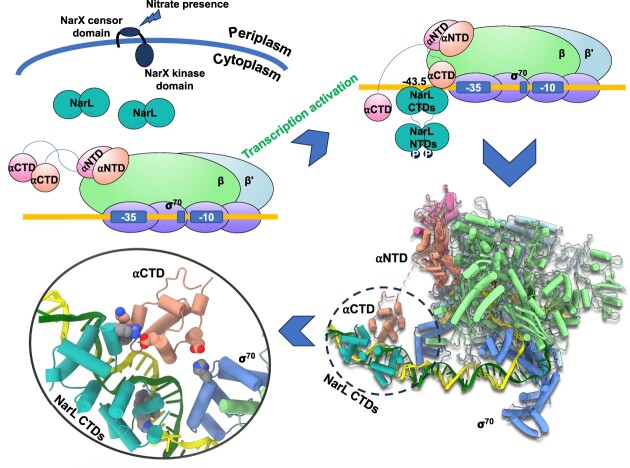
Abstract
Transcription activation is a crucial step of regulation during transcription initiation and a classic check point in response to different stimuli and stress factors. The Escherichia coli NarL is a nitrate-responsive global transcription factor that controls the expression of nearly 100 genes. However, the molecular mechanism of NarL-mediated transcription activation is not well defined. Here we present a cryo-EM structure of NarL-dependent transcription activation complex (TAC) assembled on the yeaR promoter at 3.2 Å resolution. Our structure shows that the NarL dimer binds at the −43.5 site of the promoter DNA with its C-terminal domain (CTD) not only binding to the DNA but also making interactions with RNA polymerase subunit alpha CTD (αCTD). The key role of these NarL-mediated interactions in transcription activation was further confirmed by in vivo and in vitro transcription assays. Additionally, the NarL dimer binds DNA in a different plane from that observed in the structure of class II TACs. Unlike the canonical class II activation mechanism, NarL does not interact with σ4, while RNAP αCTD is bound to DNA on the opposite side of NarL. Our findings provide a structural basis for detailed mechanistic understanding of NarL-dependent transcription activation on yeaR promoter and reveal a potentially novel mechanism of transcription activation.
Graphical Abstract
Graphical Abstract.
Introduction
Transcription is the first and essential step of gene expression and regulation within cells. It is initiated in response to different stimuli and stresses (1,2). In bacteria, transcription is controlled by a diverse network of transcription factors (TFs) and being precisely activated or repressed in response to different external signals (1,3,4). During simple transcription initiation, RNA polymerase (RNAP) holoenzyme (α2ββ'ωσ) binds to the conservative promoter elements (–35 element and –10 element) to form a closed promoter complex (RPc) and then isomerize to an open promoter complex (RPo) for starting RNA synthesis (5–9). Transcription from promoters controlled under specific conditions or where DNA lacks optimal recognition elements or could not accommodate optimal RNAP recognition by itself, is initiated with the help of specific TFs named activators and the entire process is called transcription activation (2,10–13). It has been well known that canonical transcription activation could be completed mainly in two ways: (i) Recruitment mechanism, in which the activator guides RNAP holoenzyme to the promoter region and/or assists the RPc or RPo formation (10–12,14,15); (ii) Promoter twisting mechanism, in which the activator bends and twists the DNA to facilitate correct recognition by RNAP holoenzyme (16–18). Canonical recruitment mechanisms during bacterial transcription activation have been reported on the classic example, catabolite activator protein (CAP), and involve different sets of protein/protein and protein/DNA interactions depending on the specific context of the promoter (10–12,14,15,19,20). In the canonical class I activation mode, CAP dimer binds DNA at the −61.5 site of the lac promoter and interacts with the C-terminal domain of alpha subunit (αCTD) to recruit RNAP holoenzyme (11,14,19–24), while in the canonical class II activation mode, CAP dimer binds at the −41.5 site of the gal promoter DNA, which overlaps the −35 element (12,15,19,20,23,25). In the latter activation mode, CAP simultaneously interacts with DNA, αNTD, αCTD, the β flap, and domain 4 of σ70 subunit (σ4) to create a diverse interaction network, allowing the recruitment of the RNAP and remodeling and stabilization of RPc (10,12,15,19,20,23,26). The transcription activation mechanisms for the promoters where TF binding site is located in a non-canonical position that differs from the canonical class I (−61.5) and class II (−41.5) begin to be revealed by recent cryo-EM studies (21,22,26,27), however, transcription activation by many TFs binding at such positions, including NarL, are still lacking structural understanding.
The Escherichia coli global transcription regulator NarL controls transcription of a large set of genes involved in nitrate respiration during anaerobiosis and regulates gene expression in response to nitrate and nitrite ions (28). While transcription of the majority of NarL-activated genes also requires the oxygen-responsive transcription activator Fnr for full activation (29), previous studies identified two promoters, yeaR and ogt, that can be fully activated solely by NarL (30–32). Ogt promoter controls the expression of an O6-alkylguanine-DNA-alkyltransferase, an important DNA repair enzyme, and yeaR controls the expression of proteins of unknown function or possibly involved in tellurite resistance (30,33,34). NarL is a typical response-regulator of the two-component signal transduction system (33–35). Two inner membrane-bound sensor kinases, NarX and NarQ, are activated by nitrate or nitrite ions (36–38) and phosphorylate NarL at the residue D59 (34). This phosphorylation leads to a structural rearrangement and dimerization of NarL with subsequent DNA binding and promoter activation. NarL belongs to the FixJ/NarL family, in which members are defined by a similar fold of N-terminal signal receiver domain (39–42), dimerization pattern, and a LuxR-like DNA-binding domain (43–45). NarL consists of two domains, N-terminal receiver domain (RD) and C-terminal effector DNA binding domain (DBD), which are joined by a linker (34,46,47). DNA binding is regulated by the phosphorylation state of the RD. In the nonphosphorylated state, RD blocks the access of DNA to DBD (48), while phosphorylation disrupts the RD-DBD interdomain interface, allowing the binding of DNA to DBD (46).
NarL binding sites are organized as a palindromic repeats, where two 7-base pair (bp) elements are separated by 2 bp, known as the ‘7-2-7’ sequence (34). The yeaR promoter contains one ‘7-2-7’ sequence located just upstream of the promoter −35 element (centered at -43.5 site) and its full activation requires the binding of only one NarL dimer (30). By contrast, ogt promoter contains two ‘7-2-7’ sequences (positioned at −44.5 and −77.5 sites, respectively) and requires tandem binding of NarL to both sites for full activation (31). Although the genetics of the NarX/NarL system has been extensively characterized (28,30–32,36,43,49) and the crystal structures of unphosphorylated NarL (34) or NarL DBD in complex with DNA (46–48) have been reported, the molecular mechanism of NarL-dependent transcription activation remains unclear and necessities a high-resolution structure of the intact NarL-dependent transcription activation complex (TAC).
To this end, we assembled NarL-TAC on a yeaR promoter and determined its structure using cryo-EM at 3.2 Å resolution. In this structure, we observed extensive interactions between NarL DBD/promoter DNA, and αCTD/promoter DNA, less extensive interactions between NarL/αCTD, but no TF or αCTD/σ4 interactions, which are different from those shown in the canonical class II activation mechanism. In addition, mutagenesis analysis combined with in vivo and in vitro DNA binding and transcription assays revealed the role of two NarL residues, Lys174 and Arg178, at the NarL/αCTD interface in transcription activation. In summary, our study provides the structural basis to understand the detailed molecule mechanism of NarL-dependent transcription activation and suggests a mode of noncanonical class-II transcription activation.
Materials and methods
Preparation of protein samples
E. coli RNAP and σ70 were prepared as previously (11,50–52). The αCTD deleted RNAP was expressed using recombinant pVS10 constructed by deleting the region coding for amino acids 248–330 of the α subunit. The DNA fragment encoding NarL was synthesized using the standard colony PCR approach, using the K12 E. coli strain as a source of template DNA. Specifically designed DNA primers, flanked with the NdeI and XhoI restriction sites, were ordered from Integrated DNA technologies (Supplementary Table S1). The PCR product was gel purified, digested with NdeI and XhoI restriction enzymes and cloned into pET21a vector. Mutations in the NarL coding region were introduced by Quickchange site-directed mutagenesis kit (Stratagene). His6-tagged NarL protein was overexpressed in E. coli BL21 (DE3) (ThermoFisher Scientific) at 37°C for 6 h. Induction was performed at OD600 in a range between 0.6 and 0.8 using 1 mM final concentration of Isopropyl β-d-1-thiogalactopyranoside (IPTG). Cells were collected, flash frozen, and stored at −80°C until needed. For His-tag affinity purification cell pellet was resuspended in lysis buffer (20 mM Tris HCl pH 8.0, 150 mM NaCl, 5 mM Imidazole with Protease Inhibitor Cocktail (cOmplete™)) sonicated for 40 cycles: 30 second pulse, 1 min rest on ice at 60% power output. Lysate was centrifuged twice at 14K RPM for 20 and 15 min respectively and supernatant was further filtered through 0.22 μM filter (VWR International). Protein-containing solution was applied onto the 5ml HP HisTrap column (Cytiva) preequilibrated with Buffer A (20mM Tris–HCl pH 8.0, 150 mM NaCl, 5 mM Imidazole). Protein was eluted over the 100 ml Imidazole gradient (5–500 mM). Peak fractions were analyzed with SDS-PAGE. Target fractions were pulled together and diluted with Buffer B (20 mM Tris–HCl pH 8.0) up to 150 ml to reduce Imidazole and NaCl concentration to the minimum due to the next purification step. Diluted fractions were applied to 5 ml Q HP column (Cytiva) preequilibrated in buffer B, to further purify NarL. Protein was eluted over the 100 ml NaCl gradient (0–1 M). Protein-containing fractions were pulled together and concentrated using centrifugal filter unit (MilliporeSigma™ Amicon™ Ultra-15 MWCO 10 kDa) to 12 mg/ml and 260/280 ratio 0.67. Concentrated protein was aliquoted, flash frozen, and stored at −80°C.
NarL phosphorylation
To achieve the active form, NarL protein was phosphorylated using acetyl phosphate (Lithium potassium acetyl phosphate, Sigma Aldrich) by following the procedure described earlier (38). In brief, reaction was performed in 30 μl volume. Acetyl phosphate and NarL were mixed in the reaction buffer (5 mM Tris–HCl pH 7.5, and 10 mM MgCl2) to the final concentration of 10 mM and 80 μM, respectively. Reaction was incubated at 37°C for 45 min.
Preparation of yeaR promoter DNA scaffold
The design of the template DNA (tDNA) and nontemplate DNA (ntDNA) strands was modified based on the sequence published earlier (30) to accommodate discriminator region within the transcription bubble to help to form the stable open promoter complex (Figure 1A, Supplementary Table S1). DNA fragments were ordered from Integrated DNA technology and resuspended to the final concentration of 1mM. Stock solutions of both ntDNA and tDNA were mixed at 1:1 molar ratio, heated to 100°C and annealed together through the temperature gradient (100–12°C) using a thermocycler.
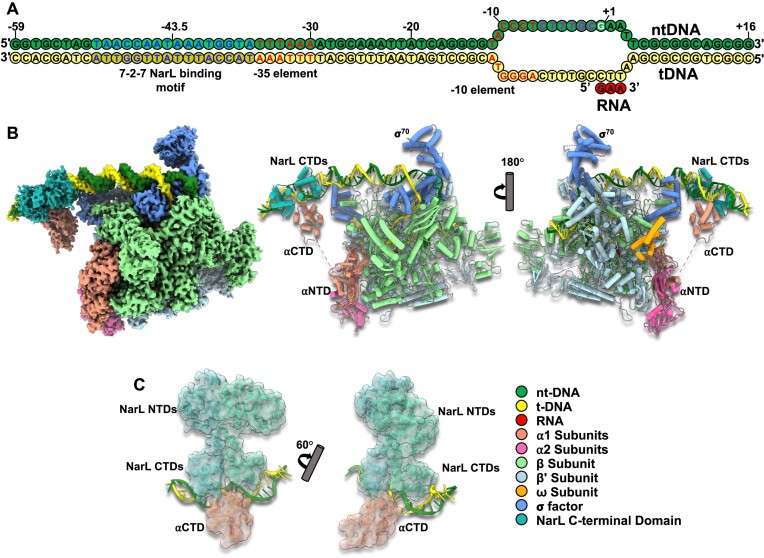
Figure 1.
The cryo-EM structures of E. coli NarL-TAC. (A) Schematic representation of yeaR promoter scaffold: forest green, non-template DNA (ntDNA); yellow, template DNA (tDNA); red, de novo RNA transcript. Transcription start site (TSS), discriminator region, −35 and −10 promoter elements, and NarL binding site are highlighted in white, magenta, red and blue, respectively. (B) Overview of cryo-EM reconstruction map and model of the E. coli NarL-TAC at overall resolution 3.2 Å. Individual subunits are labeled with different colors. (C) The docked NarL/DNA binding site with the locally refined map at resolution 4.4 Å. NarL is colored in light sea green. NarL-TAC, NarL-dependent transcription activation complex; αNTD, amino-terminal domain of the alpha subunit; αCTD, carboxyl-terminal domain of the alpha subunit.
NarL-TAC assembly and purification
NarL-TAC was assembled as follows: Core RNAP (α2ββ'ω) was mixed with the annealed yeaR promoted DNA scaffold and free NTPs (GTP and ATP), then incubated at 37°C for 1 min. Obtained complex was mixed with σ70 and further incubated at 37°C for 1 min. Lastly, phosphorylated NarL was added to the mixture and incubated at 37°C for 8 minutes. Complex components were mixed at the final 1:2:2.4 molar ratio of RNAP holoenzyme/promoter DNA/Phosphorylated NarL respectively. Free NTPs for the de novo RNA synthesis were added to the final concentration of 0.2 mM. Assembly reaction was performed in the 500 μl volume, loaded onto Superose® 6 Increase 10/300 GL size exclusion column (Cytiva). Elution fractions were analyzed using SDS-PAGE (Supplementary Figure S1A). Fractions containing all complex components and having a 260/280 ratio of 1.4 and higher are pulled together and concentrated to ∼30 μM and used for the cryo-EM grids preparation.
NarL-TAC negative staining
To confirm sample quality and absence of aggregation, negative staining EM was performed to examine the sample (Supplementary Figure S1B). 3 μl of nonconcentrated fraction were applied onto freshly glow-discharged 200-mesh carbon-coated copper grids (EM Science), incubated for 1 min with following blotting of liquid excess. Grids were washed with water 3 times and then stained with 0.75% uranyl formate solution for 15 s. Excess liquid is blotted, and grids are air dried. Imaging is performed using Biotwin Tecnai Spirit 120 kV LaB6 electron microscope (ThermoFisher Scientific) with 4K Gatan CCD at 68K magnification.
Cryo-EM grid preparation and data acquisition
The purified NarL-TAC complex (4 μl at ∼30 μM), supplemented with 8 mM CHAPSO immediately before grid preparation, was applied to freshly glow-discharged Quantifoil R1.2/1.3 300-mesh copper grids (EM Sciences) and then blotted for 4 s at 22°C under 100% chamber humidity and plunge-frozen in liquid ethane using a Vitrobot Mark IV (FEI). Data were collected at the Hormel Institute, University of Minnesota using Latitude-S (Gatan) on a Titan Krios electron microscope (ThermoFisher Scientific) equipped with a K3 direct electron detector with a Biocontinuum energy filter (Gatan) in CDS mode. The movies were collected at a nominal magnification of 130 000× (corresponding to 0.664 Å per pixel), slit width of 20 eV, a dose rate of 21 e–/Å2/s, and a total dose of 42 e−/Å2 for K3 detector. The statistics of cryo-EM data collection are summarized in Supplementary Table S2.
Image processing
Cryo-EM data were processed using cryoSPARC v4.0.3 (53), and the procedure is outlined in (Supplementary Figure S2). In brief, dose-fractionated movies were subjected to Patch motion correction with MotionCor2 (54) and Patch CTF estimation with CTFFIND-4.1.13 (55). Particles were picked using both Blob picker and Template picker accompanied by removing duplicate particles. Multiple rounds of 2D classifications were applied to remove junk particles. Particles extracted from the good 2D classes were used for Ab-initio reconstruction of four maps and then for the heterogeneous refinements, followed by further homogeneous, non-uniform, and CTF refinements to generate the 3.16 Å map. Particles in the good 3D class (247,809) were then imported into RELION-4.0 (56) using the csparc2star.py module (57) and subjected to signal subtraction to keep only the NarL dimer binding region in RELION-4.0, followed by masked 3D classification. Particles in the good classes (104,500) were selected to perform further local refinements with signal subtraction in cryoSPARC v4.0.3 to generate a 4.37 Å map for improving the densities of the NarL N-terminal domain (NTD). In the meantime, these particles were also reverted to original particles and subjected to non-uniform refinement that resulted in a 3.23 Å map of the whole NarL-TAC complex. The star2bild.py module (57) was performed to generate an angular distribution diagram, suggesting no preferred orientation issue. Map resolutions were determined by gold-standard Fourier shell correlation (FSC) at 0.143 between the two half-maps. Local resolution variation was estimated from the two half-maps in cryoSPARC v4.0.3.
Cryo-EM model building and refinement
Initial model building of the NarL-TAC complex was performed in Coot-0.8.9 (58) using PDB IDs: 6B6H, 3K4G, 1ZG5 or 1A04 as starting models. The E. coli holoenzyme and the corresponding bound DNA template were docked using the cryo-EM structure of E. coli class I TAC (PDB ID:6B6H) in Chimera v1.15 (59) and rebuilt in Coot-0.8.9 (58) using the 3.2 Å whole NarL-TAC map. Then several rounds of refinement in Phenix-1.16 (60) and manual building in Coot-0.8.9 were performed until the final reliable models were obtained. The de novo RNA transcript (GAA) is clearly shown on the map and unambiguously built. The density of partial DNA bubble region is poor due to DNA scrunch but remained for an intact model representation. NarL DBD dimer and its bound DNA were modeled using the structure of NarL-DNA complex (PDB ID:1ZG5). The α-CTD was modeled using the structure of E. coli α-CTD (PDB ID:3K4G). The NarL dimer region was also docked using the 4.4 Å local refinement map with the model of the apo NarL structure (PDB ID:1A04) for the better density of NarL NTD. The final models have good stereochemistry by evaluation in MolProbity (61). The statistics of cryo-EM data collection, 3D reconstruction, and model refinement were shown in Supplementary Table S2. Figures were generated using UCSF Chimera X v0.93 (62).
Contact area analysis
The contact area between σ4 domain (residues 535–612) and –35 promoter region was analyzed using the command: ‘measure buriedarea obj1 with obj2’ in UCSF ChimeraX v0.93 (62).
Sequence alignment analysis
Sequence alignments were performed using online tools, Clustal Omega, https://www.ebi.ac.uk/Tools/msa/clustalo/ (63) and ESPript 3.0 https://espript.ibcp.fr/ESPript/ESPript/index.php (64).
In vitro transcription assay
In vitro transcription assays were performed as previously described (65). Briefly, different concentration of phosphorylated NarL protein was incubated with assembled RNAP holoenzyme (100 nM RNAP core with 300 nM σ70) in 5 μl TB buffer (20 mM Tris–HCl, pH 7.9, 50 mM NaCl, 10 mM KCl, 5 mM MgSO4, 1 mM DTT, 0.1 mM EDTA, 5% glycerol) for 10 min at 37°C. A 123-bp fluorescein labeled yeaR promoter fragment was amplified from E. coli genomic DNA and added into the RNAP mixture at 15 nM for another 10 min incubation. Transcription was initiated by the addition of 250 μM CTP, GTP and ATP, 15 μM UTP and 1 μCi of [α-32P]UTP. The reactions were carried out at 37 °C for 10 min and stopped by 1 volume of 95% formamide solution. RNA products were incubated at 70 °C for 5 min and analyzed on denaturing 16% polyacrylamide gels. Gels were scanned by Amersham Typhoon scanner (GE Healthcare).
DNA-binding analysis
The electrophoretic mobility shift assays (EMSA) using phosphorylated NarL were performed as previously described (66) with slight modification. Briefly, different amounts of NarL protein were incubated with fluorescein labeled yeaR promoter fragment in TB buffer. After incubation at 37°C for 30 min, samples were loaded on 6% native 0.5× TBE-PAGE. Gels were scanned by Amersham Typhoon scanner (GE Healthcare).
In vivo test of NarL-yeaR regulation
To confirm the regulation of NarL to yeaR transcription, we constructed strains expressing K174A or R178A mutated NarL based on a K12 MG1655 strain with deletion of lacZ gene (67) using a CRISPR-Cas9 system (68). After that, we constructed a lacZ reporter plasmid in fusion with yeaR promoter using the ClonExpress II One Step Cloning Kit (Vazyme) as described previously (66). For determining β-galactosidase the yeaRp::lacZ plasmid was transformed into relevant strains, which were then grown overnight aerobically at 37 °C with and without 1% KNO3 in M9 minimal medium supplemented with 0.5% glucose, 0.5% casamino acids, 0.1 mM Na2MoO4, 0.1 mM NasSeO3, and 50 μg/ml kanamycin (69), β-galactosidase activities were measured to indicate the activities of yeaR promoter in different strains (66). RNA of strains under the same conditions was extracted using TRIzol reagent as described in the manufacturer's protocol (Invitrogen, USA). Reverse transcription-quantitative PCR (RT-qPCR) (65) was performed to quantify the mRNA levels of yeaR gene and 16S rRNA in these strains (primers were listed in Supplementary Table S1). The copy number of 16S rRNA in each strain was used for normalizing the relative mRNA level of yeaR gene, and the relative mRNA level of yeaR gene in E. coli wild-type strain was then normalized to 1. Three biological repetitions were performed, and each repetition contains two technical replicates. The raw data for RT-qPCR assay were summarized in Supplementary Table S3.
Quantification and statistical analysis
RNAs from in vitro transcription assays were quantified by ImageQuant-TL software. RNA bands shown in each figure quantified together. Data are shown as mean ± SD from three experiments. The β-galactosidase activity data were obtained from three colonies performed in duplicates for each strain and data are shown as mean ± SD. Statistical analyses were performed using the unpaired Student's t-test (two-tailed) between each of two groups.
Results
Overall structure of NarL-TAC
The cryo-EM structure of the intact E. coli NarL-TAC consisting of a NarL dimer, σ70-RNAP holoenzyme, and a complete NarL-specific promoter yeaR with a de novo synthesized RNA transcript (GAA) was determined at an overall resolution of 3.2 Å. The complex was reconstituted on a synthetic DNA scaffold that contains a region (−59 to + 1) corresponding to the original yeaR promoter (30), a pre-opened discriminator region (−6 to −1) to facilitate formation of stable open complex, and a designed downstream region (+2 to +16) to further stabilize the TAC (Supplementary Table S1) (11,16,65,66,70). The yeaR promoter regulation region contains the −35 element, −10 element, and the NarL-binding site corresponding to the ‘7-2-7 site’ (TAACCAATAAATGGTA) centered at position −43.5 (Figure 1A). The 3.2 Å cryo-EM map shows well-defined density for all major components of the NarL-TAC excluding NarL NTD and supports reliable model building (Figure 1B and Supplementary Figure S3). The density for the NarL NTD was further improved by local refinement of the NarL binding region and was sufficient for the docking of the NarL dimer (Figure 1C). Apparently, the NarL NTD is far from RNAP β and σ subunits (22 and 40 Å distance, respectively) (Supplementary Figure S4). The following structural analyses will focus on the structure built on the NarL-TAC map excluding NarL NTD.
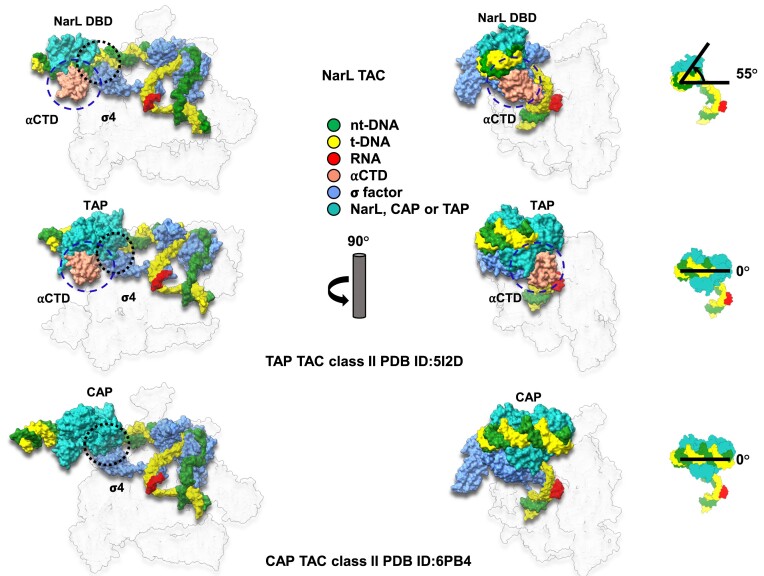
A comparison of our NarL-TAC structure with class II CAP TAC (PDB ID: 6PB4) (12) and class II TAP TAC (PDB ID: 5I2D) (10) demonstrated transcription activation on yeaR promoter by NarL belongs to class II activation mechanism. However, the position of the NarL DBD is rotated by ∼55° in relation to the position of canonical class II CAP or TAP DBDs (Figure 2) that places it in a different plane in relation to RNAP holoenzyme, allowing the formation of αCTD/DNA interactions similar to those present in class I TAC (11) (see below), but not observed in class II TAC. Moreover, while the activator-σ4 interaction is important for class II activation, NarL does not interact with the σ4 domain although they are spatially close to each other. All these structural observations suggest that NarL-dependent transcription activation on yeaR promoter adopts a noncanonical class II activation mode.
Figure 2.
Comparison of E. coli NarL-TAC and E. coli class II CAP-TAC. Two complexes (surface representations) are compared in the relationship of transcription factor binding position and interaction with σ70 domain 4 (σ4). Interaction regions are encircled with black dash. When setting CAP binding position as a starting point, NarL binds DNA within a different plane being rotated ∼55° anticlockwise. CAP interacts with σ4 while NarL does not, although it is spatially close to σ4. The subunit color codes are the same as shown in Figure 1B except for CAP dimer in light sea green and others are colored in gray and transparent.
NarL/DNA interactions
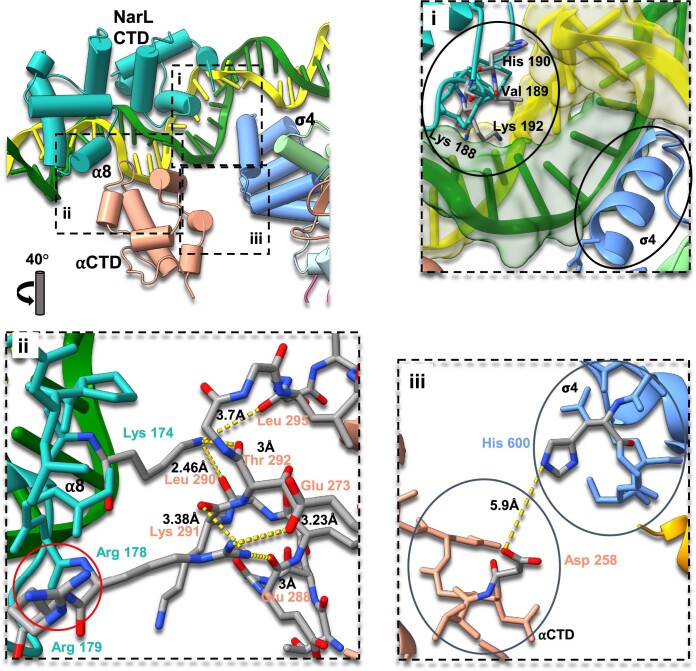
Our observations are in good agreement with the previous crystal structure of NarL-DBD/DNA complex where NarL primarily interacts with DNA major grooves via a specific ‘7-2-7’ sequence. Using the crystal structure of NarL DBD/DNA complex (PDB ID: 1ZG5) (46) as a reference, we modeled this region to resolve NarL DBD/DNA interaction around −43.5 site and confirmed that NarL utilizes the same conserved DNA-binding mechanism shared by other members of the NarL/FixJ family (39,43). Helices 7, 8, 9 and 10 (α7–α10) form a luxR-type DNA-binding HTH domain with α9 inserting into the major groove of DNA and forming extensive base-specific interactions with the DNA through Lys188, Val189, and Lys192 (Figure 3i). Sequence alignment with two other members of NarL family, TraR and RcsB, reveals that the DBD is generally conserved but has a significant sequence variation within α9 that is involved in sequence-specific DNA recognition (Supplementary Figure S5A and B). Structural alignment of NarL-TAC, NarL DBD/DNA (PDB ID: 1ZG5) (46), TraR/DNA (PDB ID: 1H0M) (44), and RcsB/DNA (PDB ID: 5W43) (45) suggests the high structural similarity of the DBD domains and conservation of the DNA binding mechanism within the family (Supplementary Figure S5C and D).
Figure 3.
NarL CTD–αCTD–DNA interactions within the E. coli NarL-TAC. Cartoon view of the NarL CTD binding region with specific residues depicted as sticks. The color scheme is the same as Figure 1. (i) Zoom view of NarL/DNA interface. NarL/DNA and σ4/DNA regions are encircled. A typical HTH domain of NarL where Lys188, Val189, and Lys192 of α9 contact the base of the major groove and His190 interacts with a DNA backbone. NarL does not interact with σ4 and affects σ4/DNA interactions. (ii) Zoom view of NarL/αCTD interface. NarL Lys174 forms hydrogen bonds with αCTD Leu290, Tre292 and Leu295, stabilizing the loop involving αCTD/DNA interactions. NarL Arg178 forms hydrogen bonds with αCTD Glu273, Lys291 and Glu288. NarL Arg179 is highlighted with a red circle. (iii) Zoom view of αCTD-σ4 interface. The closest side chain distance is around 5.9 Å, suggesting no direct interaction between them.
NarL/αCTD interactions
NarL directly interacts with αCTD through the NarL α8 and the 287 determinant of αCTD (E. coli residues 285-289, 315, 317, 318) (19,20,71,72). This kind of TF-αCTD interaction is normally observed from canonical class I TACs (11,21,22), nevertheless TFs in class I TACs could potentially interact with the second αCTD through its 265 determinant (21). TFs in class II TACs could interact with either 265 determinant (E. coli residues 265, 268, 294, 296, 298, 299, 302) or 287 determinant of single αCTD copy (10,12,21,22,26). NarL Arg178 side chain forms three hydrogen bonds with the side chain of Glu273 and the main chains of Lys291 and Glu288 of αCTD. In addition, positively charged side chain of NarL Lys174 is hydrogen bonded with the main chain oxygens of αCTD Leu290, Thr292 and Leu295, likely contributing to stabilizing the loop involving αCTD/DNA interactions (Figure 3). While Arg179 in the NarL α8 was previously suggested to be involved in the NarL–αCTD interaction on the ogt promoter (32,43), such interaction is not observed in our structure. Previous studies showed that mutation of either Arg178 or Arg179 compromises the ability of NarL to activate modified promoter ogt1052. Moreover, it was assumed that Arg178 directly interacts with the backbone of Glu273 but not with the side chain (32). In our study with the yeaR promoter, while Arg178 forms contacts with three αCTD residues, partially confirming and extending the results presented earlier (32), Arg179 side chain is facing the opposite direction and is too far to interact with αCTD (Figure 3ii). This difference in Arg179 orientations could be resulted from different promoters used in the assemblies. Also, it is worth noting that other residues such as Pro172 and Met175 that are located on the NarL/αCTD interface could potentially contribute hydrophobic interactions. The distance between two prolines: NarL Pro172 and αCTD Pro293 (∼ 4.1 Å) could be considered close for interactions, but spatial orientation of the side chains does not resemble common hydrophobic interactions as π-stacking or T-shaped π-stacking (73). The distance between NarL Met175 side chain and αCTD Pro293 side chain is around 4.5 Å suggesting possible interactions, but an earlier study showed that mutation of this residue doesn’t significantly affect transcription activation (43).
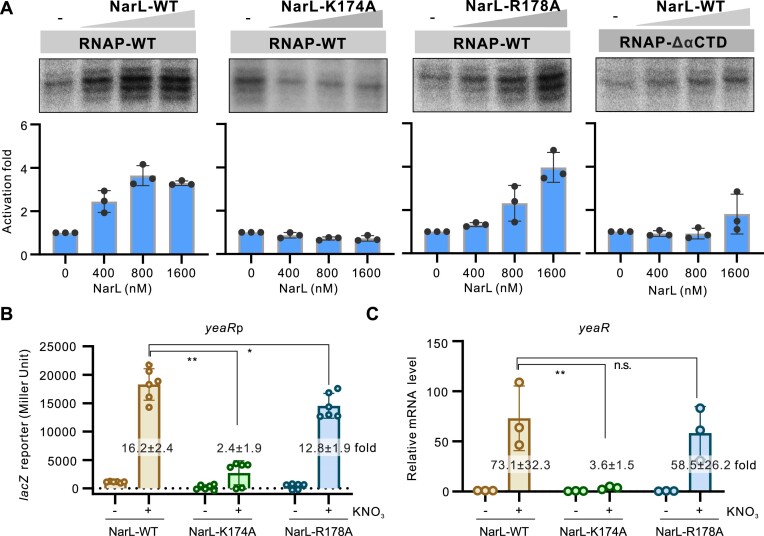
To confirm the roles of NarL–RNAP interaction in NarL-dependent transcription, we purified Lys174 or Arg178 mutated NarL protein and performed in vitro transcription assay on yeaR promoter. As expected, comparing with wild type NarL protein, mutation of either Arg178 or Lys174 obviously decreased the activation rates of NarL protein (Figure 4). Furthermore, we also constructed E. coli strain with a point mutation of Arg178 or Lys174 in narL coding region, and the yeaR promoter-lacZ reporter plasmid to test the in vivo influences of these two residues. Both the lacZ reporter and the yeaR mRNA quantification assays showed that mutation of Lys174 obviously decreased the activation of NarL to yeaR transcription in the presence of the signal factor KNO3, and mutation of Arg178 showed relatively less influence that could be neglected (Figure 4). The observed modest role of Arg178 in NarL-dependent transcription activation may explain the variance of Arg178-associated interactions. Both these two residues are not close to the DNA binding region (Figure 3ii). Consistently, either of these mutations influenced the DNA binding of NarL protein in EMSA assay (Supplementary Figure S6). These data suggest that the interaction between NarL and RNAP is important for NarL-activated transcription. Consistently, deletion of the αCTD in RNAP obviously reduced the NarL-activated transcription on yeaR promoter (Figure 4), highlighting its importance for the activation.
Figure 4.
Roles of key residues of NarL involved in interacting with RNAP in transcription activation. (A) Roles of NarL–RNAP interaction in NarL-activated transcription. In vitro transcription using purified E. coli RNAP with wild-type (WT) or mutated NarL protein on yeaR promoter was applied. The Lys174 residue was mutated to alanine and named as K174A. The Arg178 residue was mutated to alanine and named as R178A. The αCTD deleted RNAP (RNAP–ΔαCTD) was also purified and applied in this assay. RNA products were quantified from three experiments and are shown as mean ± SD in the bottom panel. (B) Activities of yeaRp in E. coli strain expressing WT or mutated NarL protein. The yeaRp was fused with lacZ reporter gene and the expression of lacZ was measured by β-galactosidase test. Bacteria were grown to late logarithmic growth phase at 37°C with or without 1% KNO3. Individual values of biological replicates (n = 6) are shown as dots, and the mean ± SD values are displayed as error bars. ** P < 0.01.* P < 0.05. (C) Transcriptional level of yeaR gene in E. coli strain expressing wild type or mutated NarL protein under the same conditions as in panel B. The transcriptional levels were analyzed by RT-qPCR assay. The mRNA levels of 16S rRNA in each strain was used for normalization, and the relative level of yeaR mRNA in E. coli WT strain without KNO3 induction was normalized to 1. In panels B and C, the activation fold by WT or mutated NarL to yeaRp activity in the presence of 1% KNO3 compared with WT strain without KNO3 was indicated in each column.
αCTD/DNA interactions
The αCTD binds to the minor groove around -43.5 site between the two NarL monomers. However, in the canonical class II activation models (10,12,26), TFs mainly bind at the major groove at –41.5 site, and also occupy the nearby minor groove for the potential αCTD binding as observed in our NarL-TAC structure. Structural comparison of αCTD-associated complexes suggests that αCTD/DNA interaction is highly conserved and is mediated through its 265 determinant (Supplementary Figure S7A) (19,20,23,74). As a result, αCTD binds to DNA through interactions between the positively charged elements of Arg265, Asn294, Gly296 and Lys298 and the negatively charged backbones of tDNA bases A –44, T –43 and T –42 (Supplementary Figure S7B).
The relative orientation among NarL, σ4, and αCTD
In the canonical class II activation mode, CAP interacts with the 596 determinant of σ4 (E. coli residues 593–603), but αCTD does not contact σ4 (12,15,19–22). In this structure, due to the ∼55° rotation of DNA binding position, NarL does not interact with σ4 (Figures 2 and 3). Interestingly, in the canonical class I activation mode, αCTD makes contact with the 596 determinant of σ4 through its 261 determinant (E. coli residues 257–259, 261) (11,19,20,24,72). In the present structure, although the position of αCTD 261 determinant is in the correct place and orientation facing to the σ4, the distances between His600 of σ4 and Asp258 of αCTD are longer than 5 Å and should not be considered direct interaction (Figure 3iii).
NarL activation by phosphorylation
Since NarL is a representative of two component signal transduction system it requires phosphorylation to drive transcription activation (34,37,38,46). Superimposition of the phosphorylated NarL in this structure with the apo unphosphorylated NarL (PDB ID: 1A04) based on their CTDs demonstrates that NarL NTD makes a significant turn of around 180° after phosphorylation to expose the dimerization helix α10 and main DNA binding helix α9 (Supplementary Figure S8), confirming the previous NMR and EPR Spectroscopy studies (75,76). In the apo NarL structure, α9 and α10 are blocked by its NTD. Similarly, helix α8, which is buried in the interdomain interface within unphosphorylated NarL (75), is involved in the NarL/αCTD interaction in this structure (Supplementary Figure S8A and B). Based on our observation and the previous studies (75,76), it is likely that phosphorylation of the conserved Asp59 would affect the structural elements around it (loops 2), then triggering further changes within the loops 1 on the NarL interdomain surface, and finally leading to a rotation of the NTD that disrupts interdomain interactions and exposes important interaction determinants (Supplementary Figure S8C).
Discussion
NarL α8 and its role in the NarL/αCTD interaction
The NarL/αCTD interface shown in this NarL–TAC on the yeaR promoter displays that α8 in one of the NarL subunits is the main region that interacts with RNAP (Figure 3). Previous study on the ogt promoter, in which NarL binds at the −44.5 site, revealed that mutations of residues Arg178 and Arg179 on α8 slightly impair transcription, suggesting their modest roles in transcription activation on the ogt promoter (32). By contrast, our study shows that while Arg178, which interacts with Glu273 side chain and the main chains of Lys291 and Glu288, also participates in the NarL/αCTD interaction, Arg179 is facing a different direction and is not involved in the NarL/αCTD interaction. Instead, another residue in NarL α8, Lys174, significantly contributes to the NarL/αCTD interface by interacting with the main chains of Leu290, Thr292 and Leu295. Further mutational study demonstrated that Arg178 has only modest effect on the transcription activation similar to the previously reported (32), but mutation of Lys174 almost abolishes activation. Thus, we speculate that residues in α8 could potentially form multiple patterns of interactions with αCTD, with the specificity of these interactions being defined by the binding position on the promoter. It is also plausible that the potential rotation of the binding position around the DNA axis and the presence of α8 as a main interaction partner for αCTD make NarL a very versatile TF that can utilize different activation mechanisms. This kind of a small TF/αCTD interface where limited residues are involved in RNAP recruitment and transcription activation is different from the extensive contact in the canonical class II CAP-TAC (12), Interestingly, it is similar to the one observed from the previous RamA TAC structure (PDB: 7BEG), in which the small TF/αCTD interface emphasizing RamA residue H31 is critical for recruiting RNAP and activating transcription (22).
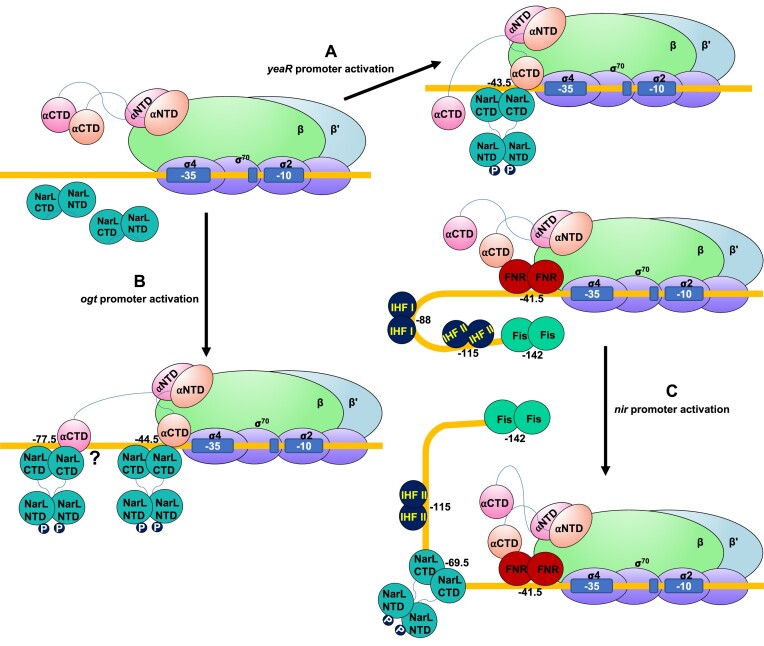
Proposed models for transcription activation involving NarL
As a global transcription regulator, NarL is involved in the activation of a large set of genes. Due to the different positions of NarL binding on the promoters and cooperative activation modes on certain promoters, NarL could potentially utilize at least three distinct mechanisms of transcription activation (Figure 5). During transcription activation on the yeaR promoter (Figure 5A), NarX senses nitrate ions and phosphorylates NarL. Upon phosphorylation, NarL undergoes significant conformational changes, with its NTD repositioned to the opposite side of its CTD, to expose its dimerization interface and DNA binding sites for promoter binding. NarL dimer binds at −43.5 site (30,31) and recruits RNAP through the interactions with αCTD. The NarL/αCTD interaction likely stabilizes αCTD/DNA binding. The unique feature of NarL-mediated transcription activation mechanism, namely the absence of NarL–σ4 interaction, distinguishes it from the canonical class II activation mode, which is characterized by TF–σ4 interaction.
Figure 5.
Proposed models of NarL-dependent transcription activation. (A) Activation mode on yeaR promoter. yeaR promoter is a NarL-dependent promoter, on which transcription is activated by an ‘unusual’ class II mechanism. NarL is phosphorylated by sensor kinase NarX with major repositioning of its NTD, binds DNA with help in recruiting RNAP, and thereby facilitates transcription initiation. (B) Activation mode on ogt promoter. NarL adopts a cooperative mode. NarL is phosphorylated by sensor kinase NarX and binds at positions −44.5 and −77.5 to activate transcription on the ogt promoter. (C) Activation mode on nir promoter. NarL utilizes a cooperative anti-repression mechanism. NarL is phosphorylated by sensor kinase NarX to bind promoter DNA at position −69.5, interferes with the binding site of IHF I, and removes Fis repression, allowing FNR to activate transcription.
In addition to recruiting RNAP, NarL may also apply other mechanisms for activating transcription. In the absence of NarL/αCTD interaction, we also observed a certain degree of transcription activation by high concentration of NarL (Figure 4), suggesting the presence of another element within the transcription activation. The comparison of σ4/DNA contact area of NarL-TAC and two canonical CAP-TACs shows that the buried surface area between σ4 (residues 535–612) and –35 element of this NarL–TAC (636 Å2) is significantly larger than that in class II CAP–TAC (PDB: 6PB4, 297 Å2), but similar to the one of class I CAP–TAC (PDB: 6b6h, 661 Å2). Therefore, the enhancement of recognition of the intrinsically weak –35 element of the yeaR promoter facilitated by changes within DNA topology upon NarL binding may play an auxiliary role in this regulatory process.
Previous DNase I footprinting and mutational analysis suggested that phosphorylated NarL dimers bind to the tandem repeat sites at −77.5 and −44.5 positions and presumably utilize both class I and class II modes to cooperatively activate transcription on the ogt promoter (32) (Figure 5B), although further structural evidences are needed to confirm this model. In addition, it was also shown that modification of the ogt promoter region to possess a closer to consensus NarL binding site sequence or reposition it to a different place allowed full promoter activation by only a single NarL dimer (32,77). This finding suggests a potentially universal and novel activation mechanism by NarL at different promoters and binding sites and has important implications for synthetic biology applications.
Similarly, those DNase I footprinting and mutational analysis also indicated that NarL potentially utilizes a completely different mechanism when activating transcription on the nir promoter (78). NarL has its binding site at position −69.5, which clashes with the IHF (integration host factor) binding site at−88 position. Phosphorylated NarL dimer displaces IHF and potentially affects DNA conformation, leading to the removal of Fis (factor for inversion stimulation) inhibition and transcription activation by FNR on this promoter (Figure 5C). This regulation mechanism involves four different transcription factors and could be classified as cooperative activation by anti-repression. On yeaR promoter, the Lys174 mutated NarL showed no activation in vitro but still slightly activated transcription in vivo (Figure 4), suggesting some other factors in E. coli may also be involved in the NarL-yeaR regulatory process. The structural bases of this highly cooperative activation mode, which may involve not only anti-repression role of NarL, could significantly improve the mechanistic understanding of the entire transcription regulation network.
In summary, we determined a 3.2 Å resolution of NarL-dependent TAC on a yeaR promoter and proposed the molecular mechanism of transcription activation. The NarL dimer binds to−43.5 site, which is also recognized by αCTD, and recruits RNAP via interaction with αCTD. Such activation mode is distinct from the canonical class II activation mechanism. NarL-dependent transcription activation is also an important target for further structural studies due to the involvement of different modes on the gene expression activation from different promoters and high biotechnological significance due to its regulation by inorganic nitrate ions (77).
Supplementary Material
Acknowledgements
We thank the staff at the cryo-EM facility and instrument core facility in the Hormel Institute, University of Minnesota, which is funded by the Hormel Foundation, for providing help. We also thank the Core Facility and Technical Support of Wuhan Institute of Virology for help in radioactive tests.
Author contributions: B.L. and Y.H. initiated, designed and supervised the experiments. D.K. performed protein sample preparations and assembly of the complexes used in the structure determination. D.K. and B.L. performed cryo-EM grid preparation, screening, optimization, image processing, map reconstruction, and model building and refinement. B.L. conducted high throughput data collection on Titan Krios. L.H. and W.Z. constructed mutations and purified proteins. L.H. performed in vitro and in vivo biochemical assays. All authors contributed to the analysis of the data and the interpretation of the results. D.K., Y.Y., Y.H. and B.L. wrote the manuscript with contributions from the other authors.
Contributor Information
Dmytro Kompaniiets, Section of Transcription & Gene Regulation, The Hormel Institute, University of Minnesota, Austin, MN 55912, USA.
Lina He, State Key Laboratory of Virology, Wuhan Institute of Virology, Center for Biosafety Mega-Science, Chinese Academy of Sciences, Wuhan 430071, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Dong Wang, Section of Transcription & Gene Regulation, The Hormel Institute, University of Minnesota, Austin, MN 55912, USA.
Wei Zhou, State Key Laboratory of Virology, Wuhan Institute of Virology, Center for Biosafety Mega-Science, Chinese Academy of Sciences, Wuhan 430071, China.
Yang Yang, Roy J. Carver Department of Biochemistry, Biophysics and Molecular Biology, Iowa State University, Ames, IA 50011, USA.
Yangbo Hu, State Key Laboratory of Virology, Wuhan Institute of Virology, Center for Biosafety Mega-Science, Chinese Academy of Sciences, Wuhan 430071, China; Hubei JiangXia Laboratory, Wuhan 430071, China.
Bin Liu, Section of Transcription & Gene Regulation, The Hormel Institute, University of Minnesota, Austin, MN 55912, USA.
Data availability
The cryo-EM density maps for NarL–RNAP TAC complex reported in this paper have been deposited in Electron Microscopy Data Bank under accession number EMD-41856. The atomic coordinates for the atomic model are deposited in Protein Data Bank under the accession number 8U3B.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
The Hormel Institute, University of Minnesota (to B.L.); Young Top-notch Talent Cultivation Program of Hubei Province (to Y.H.). Funding for open access charge: Hormel Institute, University of Minnesota.
Conflict of interest statement. None declared.
References
- 1. Browning D.F., Butala M., Busby S.J.W.. Bacterial transcription factors: regulation by pick “N” mix. J. Mol. Biol. 2019; 431:4067–4077. [DOI] [PubMed] [Google Scholar]
- 2. Browning D.F., Busby S.J.W.. Local and global regulation of transcription initiation in bacteria. Nat. Rev. Microbiol. 2016; 14:638–650. [DOI] [PubMed] [Google Scholar]
- 3. Libis V., Delépine B., Faulon J.L.. Sensing new chemicals with bacterial transcription factors. Curr. Opin. Microbiol. 2016; 33:105–112. [DOI] [PubMed] [Google Scholar]
- 4. Philips S.J., Canalizo-Hernandez M., Yildirim I., Schatz G.C., Mondragón A., O’Halloran T.V.. TRANSCRIPTION. Allosteric transcriptional regulation via changes in the overall topology of the core promoter. Science. 2015; 349:877–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boyaci H., Chen J., Jansen R., Darst S.A., Campbell E.A.. Structures of an RNA polymerase promoter melting intermediate elucidate DNA unwinding. Nature. 2019; 565:382–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Feklistov A., Darst S.A.. Structural basis for promoter-10 element recognition by the bacterial RNA polymerase σ subunit. Cell. 2011; 147:1257–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bae B., Feklistov A., Lass-Napiorkowska A., Landick R., Darst S.A.. Structure of a bacterial RNA polymerase holoenzyme open promoter complex. eLife. 2015; 4:e08504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen J., Boyaci H., Campbell E.A.. Diverse and unified mechanisms of transcription initiation in bacteria. Nat. Rev. Microbiol. 2021; 19:95–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hu Y., Liu B.. Roles of zinc-binding domain of bacterial RNA polymerase in transcription. Trends Biochem. Sci. 2022; 47:710–724. [DOI] [PubMed] [Google Scholar]
- 10. Feng Y., Zhang Y., Ebright R.H.. Structural basis of transcription activation. Science. 2016; 352:1330–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu B., Hong C., Huang R.K., Yu Z., Steitz T.A.. Structural basis of bacterial transcription activation. Science. 2017; 358:947–951. [DOI] [PubMed] [Google Scholar]
- 12. Shi W., Jiang Y., Deng Y., Dong Z., Liu B.. Visualization of two architectures in class-II CAP-dependent transcription activation. PLoS Biol. 2020; 18:e3000706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ye F., Gao F., Liu X., Buck M., Zhang X.. Mechanisms of DNA opening revealed in AAA+ transcription complex structures. Sci. Adv. 2022; 8:eadd3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ebright R.H. Transcription activation at Class I CAP-dependent promoters. Mol. Microbiol. 1993; 8:797–802. [DOI] [PubMed] [Google Scholar]
- 15. Busby S., Ebright R.H.. Transcription activation at Class II CAP-dependent promoters. Mol. Microbiol. 1997; 23:853–859. [DOI] [PubMed] [Google Scholar]
- 16. Yang Y., Liu C., Zhou W., Shi W., Chen M., Zhang B., Schatz D.G., Hu Y., Liu B.. Structural visualization of transcription activated by a multidrug-sensing MerR family regulator. Nat. Commun. 2021; 12:2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brown N.L., Stoyanov J.V., Kidd S.P., Hobman J.L.. The MerR family of transcriptional regulators. FEMS Microbiol. Rev. 2003; 27:145–163. [DOI] [PubMed] [Google Scholar]
- 18. Ansari A.Z., Bradner J.E., O’Halloran T.V.. DNA-bend modulation in a repressor-to-activator switching mechanism. Nature. 1995; 374:371–375. [DOI] [PubMed] [Google Scholar]
- 19. Lawson C.L., Swigon D., Murakami K.S., Darst S.A., Berman H.M., Ebright R.H.. Catabolite activator protein: DNA binding and transcription activation. Curr. Opin. Struct. Biol. 2004; 14:10–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Busby S., Ebright R.H.. Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 1999; 293:199–213. [DOI] [PubMed] [Google Scholar]
- 21. Shi J., Wang L., Wen A., Wang F., Zhang Y., Yu L., Li F., Jin Y., Feng Z., Li J.et al.. Structural basis of three different transcription activation strategies adopted by a single regulator SoxS. Nucleic Acids Res. 2022; 50:11359–11373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hao M., Ye F., Jovanovic M., Kotta-Loizou I., Xu Q., Qin X., Buck M., Zhang X., Wang M.. Structures of Class I and Class II Transcription Complexes Reveal the Molecular Basis of RamA-Dependent Transcription Activation. Adv Sci (Weinh). 2022; 9:e2103669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Benoff B., Yang H., Lawson C.L., Parkinson G., Liu J., Blatter E., Ebright Y.W., Berman H.M., Ebright R.H.. Structural basis of transcription activation: the CAP-alpha CTD-DNA complex. Science. 2002; 297:1562–1566. [DOI] [PubMed] [Google Scholar]
- 24. Hudson B.P., Quispe J., Lara-Gonzalez S., Kim Y., Berman H.M., Arnold E., Ebright R.H., Lawson C.L.. Three-dimensional EM structure of an intact activator-dependent transcription initiation complex. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:19830–19835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhou Y., Merkel T.J., Ebright R.H.. Characterization of the activating region of Escherichia coli catabolite gene activator protein (CAP) II. Role at class I and class II CAP-dependent promoters. J. Mol. Biol. 1994; 243:603–610. [DOI] [PubMed] [Google Scholar]
- 26. Shi J., Wang F., Li F., Wang L., Xiong Y., Wen A., Jin Y., Jin S., Gao F., Feng Z.et al.. Structural basis of transcription activation by Rob, a pleiotropic AraC/XylS family regulator. Nucleic Acids Res. 2022; 50:5974–5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yang X., Wang Y., Liu G., Deng Z., Lin S., Zheng J.. Structural basis of Streptomyces transcription activation by zinc uptake regulator. Nucleic Acids Res. 2022; 50:8363–8376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Constantinidou C., Hobman J.L., Griffiths L., Patel M.D., Penn C.W., Cole J.A., Overton T.W.. A reassessment of the FNR regulon and transcriptomic analysis of the effects of nitrate, nitrite, NarXL, and NarQP as Escherichia coli K12 adapts from aerobic to anaerobic growth. J. Biol. Chem. 2006; 281:4802–4815. [DOI] [PubMed] [Google Scholar]
- 29. Ramos Cruz, Boursier H., Moszer L., Kunst I., Danchin F., Glaser P. Anaerobic transcription activation in Bacillus subtilis: identification of distinct FNR-dependent and -independent regulatory mechanisms. EMBO J. 1995; 14:5984–5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lin H.Y., Bledsoe P.J., Stewart V.. Activation of yeaR-yoaG operon transcription by the nitrate-responsive regulator NarL is independent of oxygen- responsive regulator Fnr in Escherichia coli K-12. J. Bacteriol. 2007; 189:7539–7548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Squire D.J., Xu M., Cole J.A., Busby S.J., Browning D.F.. Competition between NarL-dependent activation and Fis-dependent repression controls expression from the Escherichia coli yeaR and ogt promoters. Biochem. J. 2009; 420:249–257. [DOI] [PubMed] [Google Scholar]
- 32. Ruanto P., Chismon D.L., Hothersall J., Godfrey R.E., Lee D.J., Busby S.J.W., Browning D.F.. Activation by NarL at the Escherichia coli ogt promoter. Biochem. J. 2020; 477:2807–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gao R., Bouillet S., Stock A.M.. Structural basis of response regulator function. Annu. Rev. Microbiol. 2019; 73:175–197. [DOI] [PubMed] [Google Scholar]
- 34. Baikalov I., Schröder I., Kaczor-Grzeskowiak M., Grzeskowiak K., Gunsalus R.P., Dickerson R.E.. Structure of the Escherichia coli response regulator NarL. Biochemistry. 1996; 35:11053–11061. [DOI] [PubMed] [Google Scholar]
- 35. Darwin A.J., Stewart V.. Lin E.C.C., Lynch A.S.. Regulation of Gene Expression in Escherichia coli. 1996; Boston, MA: Springer US; 343–359. [Google Scholar]
- 36. Huynh T.N., Chen L.L., Stewart V.. Sensor-response regulator interactions in a cross-regulated signal transduction network. Microbiology (Reading). 2015; 161:1504–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stewart V. Nitrate- and nitrite-responsive sensors NarX and NarQ of proteobacteria. Biochem. Soc. Trans. 2003; 31:1–10. [DOI] [PubMed] [Google Scholar]
- 38. Schröder I., Wolin C.D., Cavicchioli R., Gunsalus R.P.. Phosphorylation and dephosphorylation of the NarQ, NarX, and NarL proteins of the nitrate-dependent two-component regulatory system of Escherichia coli. J. Bacteriol. 1994; 176:4985–4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Galperin Michael Y. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J. Bacteriol. 2006; 188:4169–4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schnell R., Agren D., Schneider G.. 1.9 A structure of the signal receiver domain of the putative response regulator NarL from Mycobacterium tuberculosis. Acta Crystallogr. Sect. F. 2008; 64:1096–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Park A.K., Moon J.H., Oh J.S., Lee K.S., Chi Y.M.. Crystal structure of the response regulator spr1814 from Streptococcus pneumoniae reveals unique interdomain contacts among NarL family proteins. Biochem. Biophys. Res. Commun. 2013; 434:65–69. [DOI] [PubMed] [Google Scholar]
- 42. Katsir G., Jarvis M., Phillips M., Ma Z., Gunsalus R.P.. The Escherichia coli NarL receiver domain regulates transcription through promoter specific functions. BMC Microbiol. 2015; 15:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lin A.V., Stewart V.. Functional roles for the GerE-family carboxyl-terminal domains of nitrate response regulators NarL and NarP of Escherichia coli K-12. Microbiology (Reading). 2010; 156:2933–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vannini A., Volpari C., Gargioli C., Muraglia E., Cortese R., De Francesco R., Neddermann P., Di Marco S.. The crystal structure of the quorum sensing protein TraR bound to its autoinducer and target DNA. EMBO J. 2002; 21:4393–4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Filippova Ekaterina V., Zemaitaitis B., Aung T., Wolfe Alan J., Anderson Wayne F.. Structural basis for DNA recognition by the two-component response regulator RcsB. mBio. 2018; 9:e01993-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Maris A.E., Kaczor-Grzeskowiak M., Ma Z., Kopka M.L., Gunsalus R.P., Dickerson R.E.. Primary and secondary modes of DNA recognition by the NarL two-component response regulator. Biochemistry. 2005; 44:14538–14552. [DOI] [PubMed] [Google Scholar]
- 47. Baikalov I., Schröder I., Kaczor-Grzeskowiak M., Cascio D., Gunsalus R.P., Dickerson R.E.. NarL dimerization? Suggestive evidence from a new crystal form. Biochemistry. 1998; 37:3665–3676. [DOI] [PubMed] [Google Scholar]
- 48. Maris A.E., Sawaya M.R., Kaczor-Grzeskowiak M., Jarvis M.R., Bearson S.M., Kopka M.L., Schröder I., Gunsalus R.P., Dickerson R.E.. Dimerization allows DNA target site recognition by the NarL response regulator. Nat. Struct. Biol. 2002; 9:771–778. [DOI] [PubMed] [Google Scholar]
- 49. Browning D.F., Cole J.A., Busby S.J.. Suppression of FNR-dependent transcription activation at the Escherichia coli nir promoter by Fis, IHF and H-NS: modulation of transcription initiation by a complex nucleo-protein assembly. Mol. Microbiol. 2000; 37:1258–1269. [DOI] [PubMed] [Google Scholar]
- 50. Liu B., Zuo Y., Steitz T.A.. Structural basis for transcription reactivation by RapA. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:2006–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shi W., Zhou W., Chen M., Yang Y., Hu Y., Liu B.. Structural basis for activation of Swi2/Snf2 ATPase RapA by RNA polymerase. Nucleic Acids Res. 2021; 49:10707–10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liu B., Steitz T.A.. Structural insights into NusG regulating transcription elongation. Nucleic Acids Res. 2017; 45:968–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A.. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017; 14:290–296. [DOI] [PubMed] [Google Scholar]
- 54. Rubinstein J.L., Brubaker M.A.. Alignment of cryo-EM movies of individual particles by optimization of image translations. J. Struct. Biol. 2015; 192:188–195. [DOI] [PubMed] [Google Scholar]
- 55. Rohou A., Grigorieff N.. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015; 192:216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zivanov J., Nakane T., Scheres S.H.W.. Estimation of high-order aberrations and anisotropic magnification from cryo-EM data sets in RELION-3.1. IUCrJ. 2020; 7:253–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Asarnow D., Palovcak E., Cheng Y.. 2019; asarnow/pyem: UCSF pyem v0.5.
- 58. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta. Crystallogr. D Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 59. Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E.. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004; 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 60. Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W.et al.. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta. Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chen V.B., Arendall W.B., Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C.. MolProbity: all-atom structure validation for macromolecular crystallography. Acta. Crystallogr. D Biol. Crystallogr. 2010; 66:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Goddard T.D., Huang C.C., Meng E.C., Pettersen E.F., Couch G.S., Morris J.H., Ferrin T.E.. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 2018; 27:14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sievers F., Higgins D.G.. Clustal omega. Curr. Protoc. Bioinformatics. 2014; 48:3.13.1–3.13.16. [DOI] [PubMed] [Google Scholar]
- 64. Robert X., Gouet P.. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014; 42:W320–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Shi W., Zhou W., Zhang B., Huang S., Jiang Y., Schammel A., Hu Y., Liu B.. Structural basis of bacterial sigma(28) -mediated transcription reveals roles of the RNA polymerase zinc-binding domain. EMBO J. 2020; 39:e104389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Shi W., Zhang B., Jiang Y., Liu C., Zhou W., Chen M., Yang Y., Hu Y., Liu B.. Structural basis of copper-efflux-regulator-dependent transcription activation. iScience. 2021; 24:102449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hu Y., Wang Z., Feng L., Chen Z., Mao C., Zhu Y., Chen S.. sigma(E) -dependent activation of RbpA controls transcription of the furA-katG operon in response to oxidative stress in mycobacteria. Mol. Microbiol. 2016; 102:107–120. [DOI] [PubMed] [Google Scholar]
- 68. Jiang Y., Chen B., Duan C., Sun B., Yang J., Yang S.. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl. Environ. Microb. 2015; 81:2506–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Dong X.R., Li S.F., DeMoss J.A.. Upstream sequence elements required for NarL-mediated activation of transcription from the narGHJI promoter of Escherichia coli. J. Biol. Chem. 1992; 267:14122–14128. [PubMed] [Google Scholar]
- 70. Liu B., Zuo Y., Steitz T.A.. Structures of E. coli sigmaS-transcription initiation complexes provide new insights into polymerase mechanism. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:4051–4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Savery N.J., Lloyd G.S., Kainz M., Gaal T., Ross W., Ebright R.H., Gourse R.L., Busby S.J.. Transcription activation at Class II CRP-dependent promoters: identification of determinants in the C-terminal domain of the RNA polymerase alpha subunit. EMBO J. 1998; 17:3439–3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Savery N.J., Lloyd G.S., Busby S.J., Thomas M.S., Ebright R.H., Gourse R.L.. Determinants of the C-terminal domain of the Escherichia coli RNA polymerase alpha subunit important for transcription at class I cyclic AMP receptor protein-dependent promoters. J. Bacteriol. 2002; 184:2273–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zondlo N.J. Aromatic-proline interactions: electronically tunable CH/pi interactions. Acc. Chem. Res. 2013; 46:1039–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Gaal T., Ross W., Blatter E.E., Tang H., Jia X., Krishnan V.V., Assa-Munt N., Ebright R.H., Gourse R.L.. DNA-binding determinants of the alpha subunit of RNA polymerase: novel DNA-binding domain architecture. Genes Dev. 1996; 10:16–26. [DOI] [PubMed] [Google Scholar]
- 75. Eldridge A.M., Kang H.S., Johnson E., Gunsalus R., Dahlquist F.W.. Effect of phosphorylation on the interdomain interaction of the response regulator, NarL. Biochemistry. 2002; 41:15173–15180. [DOI] [PubMed] [Google Scholar]
- 76. Zhang J.H., Xiao G., Gunsalus R.P., Hubbell W.L.. Phosphorylation triggers domain separation in the DNA binding response regulator NarL. Biochemistry. 2003; 42:2552–2559. [DOI] [PubMed] [Google Scholar]
- 77. Hothersall J., Lai S., Zhang N., Godfrey R.E., Ruanto P., Bischoff S., Robinson C., Overton T.W., Busby S.J.W., Browning D.F.. Inexpensive protein overexpression driven by the NarL transcription activator protein. Biotechnol. Bioeng. 2022; 119:1614–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Browning D.F., Cole J.A., Busby S.J.W.. Transcription activation by remodelling of a nucleoprotein assembly: the role of NarL at the FNR-dependent Escherichia coli nir promoter. Mol. Microbiol. 2004; 53:203–215. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM density maps for NarL–RNAP TAC complex reported in this paper have been deposited in Electron Microscopy Data Bank under accession number EMD-41856. The atomic coordinates for the atomic model are deposited in Protein Data Bank under the accession number 8U3B.