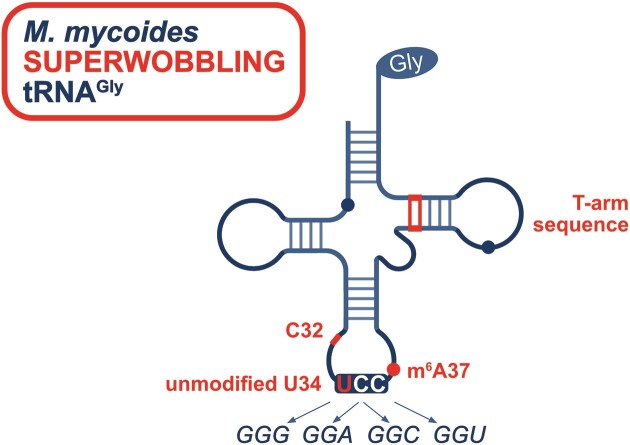
Abstract
tRNA superwobbling, used by certain bacteria and organelles, is an intriguing decoding concept in which a single tRNA isoacceptor is used to decode all synonymous codons of a four-fold degenerate codon box. While Escherichia coli relies on three tRNAGly isoacceptors to decode the four glycine codons (GGN), Mycoplasma mycoides requires only a single tRNAGly. Both organisms express tRNAGly with the anticodon UCC, which are remarkably similar in sequence but different in their decoding ability. By systematically introducing mutations and altering the number and type of tRNA modifications using chemically synthesized tRNAs, we elucidated the contribution of individual nucleotides and chemical groups to decoding by the E. coli and M. mycoides tRNAGly. The tRNA sequence was identified as the key factor for superwobbling, revealing the T-arm sequence as a novel pivotal element. In addition, the presence of tRNA modifications, although not essential for providing superwobbling, was shown to delicately fine-tune and balance the decoding of synonymous codons. This emphasizes that the tRNA sequence and its modifications together form an intricate system of high complexity that is indispensable for accurate and efficient decoding.
Graphical Abstract
Graphical Abstract.
Introduction
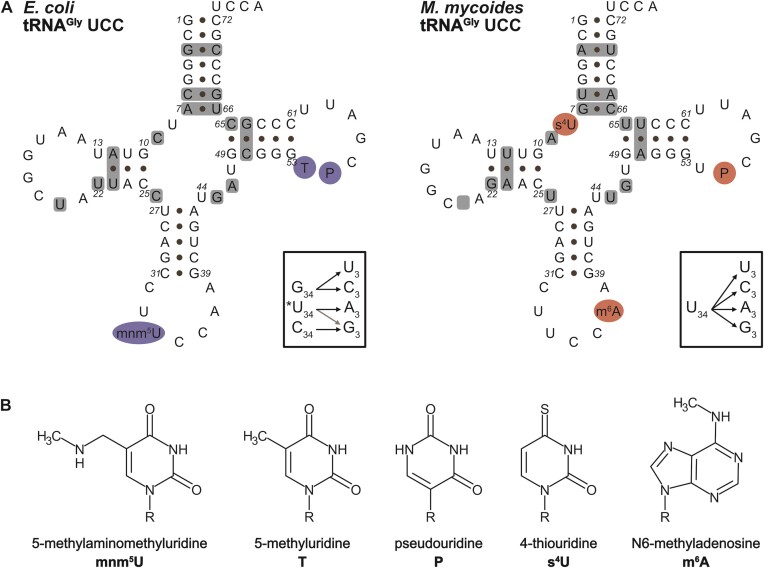
Translation is a central process common to all living organisms. Although the basic concept and mechanism is shared by all known translation systems, numerous differences and variations have evolved (1,2). One particularly intriguing aspect that varies significantly between different organisms is the decoding strategies used to translate the genetic information into protein. While the genetic code is highly conserved across species, the number of tRNA types required to decode all sense codons varies widely. For example, eukaryotes require between 41 and 55 tRNA types (3), while bacteria use 28–46 (4). Mammalian mitochondria have further reduced the number of tRNA types required for translation to a total of 22 (5,6). The reason that different numbers of tRNA types can decode all sense codons lies in the nature of the codon/anticodon interaction. During the decoding process, only the first and second nucleotide of the codon are strictly required to form canonical Watson–Crick (W/C) base pairs with the nucleotides at position 36 and 35 of the tRNA, respectively. This is not the case for the interaction between the third nucleotide of the codon and the tRNA nucleotide at position 34, the first anticodon position. This so-called wobble position (7) tolerates different base pairing geometries, such as G–U wobble base pairs and other non-W/C base pairs (8–10). These interactions are often facilitated or stabilized by the presence of tRNA modifications, making them critical elements for precise and efficient translation. More than 100 different RNA nucleoside derivatives have been identified within tRNAs throughout all kingdoms of life (11). Such modifications can be broadly categorized into body and anticodon loop modifications. Body modifications are primarily thought to contribute to the tRNA structure, folding and stability. Anticodon loop modifications are mainly considered to modulate decoding during translation either directly (via modification at nucleotide 34) or indirectly (via modifications at nucleotides 32, 37, 38) by promoting interactions that enable or stabilize non-W/C base pairs, thereby increasing the selectivity and accuracy of translation (8,10,12–14). This functional variability is reflected in their chemical composition, ranging from simple 2’O-methylations, to complex modifications like wybutosine (W) present at nucleotide 37 or 5-carboxymethoxyuridine (cmo5U) and derivatives present at nucleotide 34 both requiring multiple steps and modifying enzymes for their biosynthesis (11,15,16). Escherichia coli and Mycoplasma mycoides are representatives of bacteria that use quite different decoding strategies. The well-studied model organism E. coli expresses 41 tRNA types encoded by 87 tRNA genes (17). On the contrary, M. mycoides, belonging to the class of Mollicutes, which are the smallest and simplest self-replicating bacteria depends on the expression of only 28 different tRNA types encoded by 30 tRNA genes (17–19). In order to compensate for the small number of tRNA types, Mycoplasma species as well as mitochondria and chloroplasts employ a remarkable decoding concept (20–22). For example, whereas E. coli decodes the four-fold degenerate codon boxes for glycine (GGN), alanine (GCN), valine (GUN), proline (CCN), serine (UCN) and leucine (CUN) by two to three tRNA isoacceptors, M. mycoides employs only a single tRNA (17). It has been postulated that for this to occur, these M. mycoides tRNAs must either exhibit four-way wobble pairing capability (20) or lack engagement with the third codon position, as suggested by the ‘two-out-of-three’ hypothesis (23). All these tRNAs overcoming the standard rules of decoding (7) by extended wobbling, so-called superwobbling tRNAs, share a common feature, which is an unmodified uridine at position 34 (5,24). No additional common identity element has yet been identified. The four-fold degenerate codon box glycine is one example for the different decoding concepts employed by E. coli and M. mycoides. In E. coli, the four synonymous glycine codons GGA, GGC, GGG and GGU are decoded by three tRNA isoacceptors, namely tRNAGly CCC, GCC and UCC. In M. mycoides, the tRNAGly UCC is the only tRNA responsible for decoding of all the four glycine codons. The comparison of the tRNAGly UCC of both bacteria revealed that the sequences are largely shared between the two tRNAs (Figure 1A). In particular, the complete nucleotide sequence within the anticodon stem–loop is identical. However, the modifications in the body and in the anticodon loop are different. The E. coli tRNAGly UCC harbors 5-methylaminomethyluridine (mnm5U) at position 34, as well as 5-methyluridine (T) and pseudouridine (P) within the T-loop at positions 54 and 55, respectively (Figure 1). In contrast, the tRNAGly UCC of M. mycoides carries 4-thiouridine (s4U) at position 8, N6-methyladenosine (m6A) at position 37 and P at position 55 but not the usual T at position 54 (Figure 1). The body modifications s4U8, T54 and P55 are primarily considered to stabilize the L-shaped tRNA structure (25,26). The s4U8 modification is a commonly present modification in tRNAs, and, interestingly, it is also present in the E. coli tRNAGly CCC and GCC isoacceptors in combination with dihydrouridine (D) within the D-arm (27) (Supplementary Figure S1). Structure analysis of a functional tRNA showed stacking of the 4-thio group on the residue at position 13 and the formation of a W/C or Hoogsteen base pair of s4U8 with the nucleotide at position 14 (Supplementary Figure S2) (28). T54 was identified to form an intraloop reverse Hoogsteen base pair with A58 (29) and to stabilize the tRNA structure by increasing the melting temperature of the tRNA (31). P55 is involved in tRNA fold formation (25) due to efficient base stacking ability and base pairing with the G18 within the D-loop (29). mnm5U34 and m6A37 within the anticodon loop are thought to modulate the decoding capability of the tRNA (32–34). mnm5U34 was proposed to stabilize decoding of purine-ending codons, which is especially important for split codon box tRNAs (8,35). m6A37, as well as other modified As at position 37, have been proposed to stabilize the codon/anticodon interaction, which is particularly critical in the case of a weak A–U or U–A base pair between nucleotides anticodon first position and codon third position (13), and to contribute to the preorganization of the anticodon loop (34).
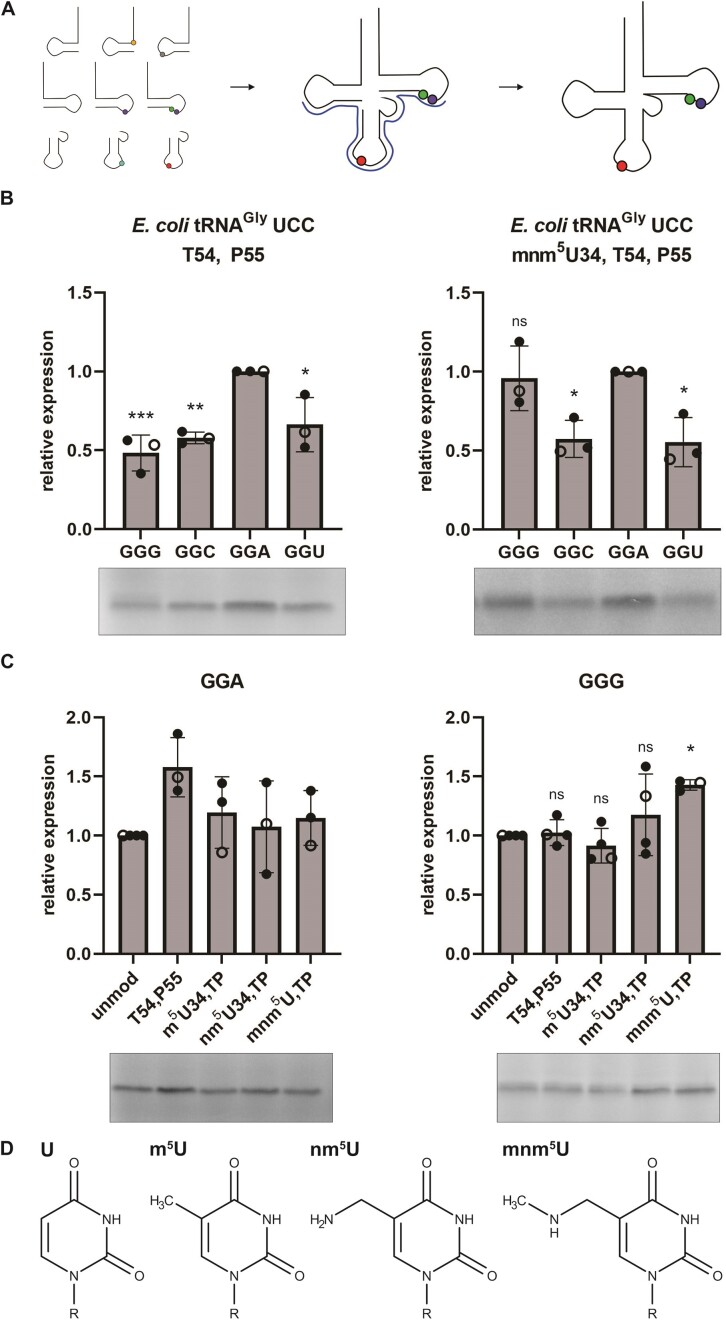
Figure 1.
tRNA sequences and modifications. (A) Secondary structure of tRNAGly UCC from E. coli and M. mycoides. Decoding of the four synonymous glycine codons (GGN) in E. coli requires three tRNAGly isoacceptors with the anticodons CCC, GCC, and *UCC (left box; ‘*’ modified U). In contrast, the M. mycoides tRNAGly with the anticodon UCC is capable of decoding the four GGN codon (right box). Grey shaded boxes in the tRNA structures indicate the sequence differences between the E. coli and M. mycoides tRNAGly UCC. The D-arm of the M. mycoides tRNA is one nucleotide smaller compared to E. coli (indicated by an empty grey box). The modified positions are highlighted in blue (E. coli) or red (M. mycoides). (B) The chemical structures of the indicated tRNA modifications decorating the tRNAs are depicted.
Since M. mycoides tRNAGly UCC, in contrast to E. coli tRNAGlyUCC, can decode all four synonymous codons, we wondered to what extent specific sequence elements or tRNA modifications contribute to the difference in their decoding capacity. Therefore, we established an in vitro translation approach based on splinted ligation (Supplementary Figure S3) (36–38). This allowed us to generate differentially modified tRNAs from a set of synthetic RNA oligonucleotides carrying different modifications at different positions. Thus, the contribution of a single modification, but also of a combined set of modifications, on translation can be assessed. We generated tRNAGly UCC from E. coli and M. mycoides varying in their sequence and modifications and determined to what extent their decoding capacity was affected. We thoroughly characterized these tRNAs and identified the contribution of different sequence elements and modifications to tRNA superwobbling. This not only broadens our understanding of how modifications and sequence affect the decoding, but also reveals the basis for alternative decoding strategies employed by organisms with reduced genome size.
Materials and methods
Plasmids
To generate minimal mRNAs encoding the peptide MRFGRFGEGRFGRF* (1719.98 Da), a short coding sequence (5′ – ATG AGA TTY GGN AGA TTY GGN GAA GGN AGA TTY GGN AGA TTY TAA – 3′; N indicates any of the four canonical bases, and Y indicates pyrimidines), was cloned downstream of the T7 promotor replacing the folA gene of the DHFR control plasmid (NEB). Cloning was performed using the Q5® Site-Directed Mutagenesis Kit (NEB, E0554S) according to manufacturer's protocol. Primers were designed using the NEBaseChanger™ tool and purchased from Integrated DNA Technologies (IDT). Plasmids were harvested and purified using the Monarch® Plasmid Miniprep Kit (NEB, T1010). The sequence of each construct was verified by Sanger Sequencing (Eurofins Genomics, Mix2Seq Kit NightXpress).
The firefly luciferase (fLuc) gene (Supplementary Figure S4) was introduced into the multiple cloning site of a pUC19 plasmid according to (39).
tRNA transcription
tRNA sequences were obtained from the tRNA Leipzig Database (http://trnadb.bioinf.uni-leipzig.de/) (40). DNA templates for transcription were generated by Phusion Green High-Fidelity DNA Polymerase (F534L, Thermo Scientific™) based on overlapping primers (41) purchased from IDT (Supplementary Table S1). The forward primer harbored a BamHI sequence, T7 promotor, as well as the 5′ part of the respective tRNA sequence. The reverse primer encoded the 3′ part of the tRNA sequence. In order to ensure a defined 3′ end of the DNA template, the two first 5′ nucleotides of the reverse primer were 2'O-methylated (42). Transcription by the HiScribe™ T7 High Yield RNA Synthesis Kit (E2040S, NEB) was performed according to manufacturer's protocol for small RNAs. The template was removed by a DNase treatment using 1 U of DNase 1 (Thermo Scientific™, EN0521) for 30 min at 37°C. The RNA was purified using the Monarch® RNA Cleanup Kit (50 μg) (NEB, T2040). The concentration was determined using a photo spectrometer (NanoDrop® ND-1000).
RNase H mediated selective tRNA depletion of E. coli tRNA bulk
Native E. coli tRNA bulk included in the PURExpress® Δ(aa, tRNA) Kit (NEB, E6840S) was depleted for up to three tRNAGly isoacceptors by RNase H. The protocol for the selective depletion of tRNAGly CCC, GCC and UCC by RNase H digest in presence of DNA antisense oligonucleotides was adapted from (43). For the depletion, 100 μg tRNA bulk were mixed with 400 pmol of antisense DNA (Supplementary Table S2) in 200 mM NaCl and 100 mM Tris–HCl (pH 8.0) in a volume of 60 μl. The solution was denatured for 2 min at 80°C, cooled down to 22°C (0.1°C/s) and incubated for 5 min at 22°C. Subsequently, 10x RNase H buffer (f.c. 1×), DTT (f.c. 10 μM) and 20 U RNase H (NEB, M0297S) were added to a final volume of 80 μl and incubated for 1 h at 37°C in a thermo cycler. Afterwards, the antisense DNA was removed by the addition of RQ1 DNase (Promega, M6101) and incubation for 45 min at 37°C. The amount of DNase was dependent on the number of tRNAs to be depleted (10 U, 15 U or 20 U for one, two or three antisense DNA oligonucleotides, respectively). The RNA was purified using the Monarch® RNA Cleanup Kit (500 μg) (NEB, T2050S). The concentration was determined using photo spectrometry (NanoDrop® ND-1000) and the concentration was adjusted to 10 μg/μl.
Chemical synthesis of RNA
RNA solid-phase synthesis
RNA synthesis was performed on an H6 GeneWorld DNA/RNA automated synthesizer (K&A, Laborgeraete GbR, Germany) on a 1.0 μmol scale or on an ABI 391 PCR-MATE (Applied Biosystems) on a 1.4 μmol scale using standard phosphoramidite chemistry and optimized in-house written synthetic cycles. Standard 2′-O-TOM or 2′-O-TBDMS and acetyl protected nucleoside phosphoramidite building blocks (ChemGenes), 2′-O-TBS 1000 Å CPG solid support (ChemGenes) and in-house synthesized S4-cyanoethyl-protected 4-thiouridine as well as N-trifluoroacetyl (TFA) protected 5-aminomethyluridine (nm5U) and 5-methylaminomethyluridine (mnm5U) phosphoramidites were used. On the K&A synthesizer, detritylation, coupling, capping and oxidation reagents were dichloroacetic acid/1,2-dichloroethane (4/96), phosphoramidite/acetonitrile (100 mM) and benzylthiotetrazole/acetonitrile (300 mM), Cap A/Cap B (1/1) (Cap A: 4-(dimethylamino)pyridine/acetonitrile (500 mM), Cap B: acetic anhydride/sym-collidine/acetonitrile (2/3/5)) and iodine (20 mM) in tetrahydrofuran/pyridine/H2O (35/10/5). On the ABI synthesizer, detritylation and capping solutions were exchanged to 5%-dichloroacetic acid in anhydrous toluene, Cap A: acetic anhydride/lutidine/tetrahydrofuran 1/1/8 (v/v/v) and Cap B: tetrahydrofuran/N-methylimidazole 86/14 (v/v), respectively. Solutions of phosphoramidites and tetrazole were dried over activated molecular sieves (3 Å) overnight.
Deprotection and purification of RNA
For deprotection of unmodified RNA, the solid support was mixed with aqueous methylamine (40%, 0.50 ml) and aqueous ammonia (28%, 0.50 ml) for 20 min at 65°C. The supernatant was removed and the solid support was washed three times with H2O/THF (1.0 ml; 1/1). Combined supernatant and washings were evaporated to dryness and the residue was dissolved either in a solution of tetrabutylammonium fluoride in tetrahydrofuran (1.0 M, 1.5 ml) in the case of 2′-O-TOM or in a mixture of 300 μl anhydrous dimethylsulfoxide and 375 μl triethylamine trihydrofluoride for 2′-O-TBDMS protecting groups. The solution was subsequently incubated for 16 h at 37°C for the complete removal of the 2′-O-silyl protecting groups. Then, the reaction was quenched by addition of triethylammonium acetate/H2O (1.0 M, 1.5 ml, pH 7.4). Eventually present tetrahydrofuran was removed under reduced pressure and the sample was desalted by size-exclusion column chromatography (GE Healthcare, HiPrep™ 26/10 Desalting; Sephadex G25) eluting with H2O; collected fractions were evaporated and the RNA dissolved in H2O (1 ml).
For deprotection of amino modified oligonucleotides, the solid support in the synthesis cartridge was pretreated with 40 ml of 10% diethylamine in anhydrous acetonitrile at a flow rate of 2 ml min−1 using two syringes. After washing with 40 ml acetonitrile the solid support was dried in vacuo and the deprotection was continued according to the standard procedure described above.
Oligonucleotides modified with 4-thiouridine were treated with 1,8-diazabicyclo[5.4.0]undec-7-en (DBU) in acetonitrile (1.0 M, 2 ml) for 3 h at room temperature. The resin was washed three times with acetonitrile (0.5 ml), dried in vacuo and incubated with a mixture of aqueous methylamine (40%, 0.50 ml), aqueous ammonia (28%, 0.50 ml) and dithiothreitol (50 mM) for 20 min at 65°C. The supernatant was removed and the solid support was washed three times with H2O/ethanol (1.0 ml; 1/1). Combined supernatant and washings were evaporated to dryness and the residue was dissolved in a mixture of dry DMSO (200 μl) and triethylamine trihydrofluoride (275 μl) and heated for 2.5 h at 65°C. After cooling down to room temperature, sodium acetate (3.0 M, 25 μl) and ethanol (1.0 ml) were added and the RNA was stored at 20°C overnight. After centrifugation (13 000 rpm, 20 min, 4°C), the supernatant was discarded and the pellet was washed twice with ethanol (0.5 ml). The pellet was then dried in vacuo and dissolved in H2O (1 ml).
The crude RNA was purified by anion exchange chromatography (Thermo Scientific Ultimate 3000 HPLC System) on a semipreparative Dionex DNAPac® PA-100 column (9 mm × 250 mm) at 80°C with a flow rate of 2 ml/min (eluent A: 20 mM NaClO4, 25 mM Tris·HCl, pH 8.0, 20 v/v % acetonitrile; eluent B: 600 mM NaClO4, 25 mM Tris·HCl, pH 8.0, 20% v/v acetonitrile). Fractions containing RNA were evaporated and the residue redissolved in 0.1 M triethylammonium bicarbonate solution (10–20 ml), loaded on a C18 SepPak Plus® cartridge (Waters/Millipore), washed with H2O, and then eluted with acetonitrile/H2O (1/1). Crude and purified RNA were analyzed by anion exchange chromatography (Thermo Scientific Ultimate 3000 HPLC System) on a Dionex DNAPac® PA-100 column (4 mm x 250 mm) at 80°C with a flow rate of 1 ml/min. For RNA up to 15 nucleotides in length, a gradient of 0–40% B in 30 min was applied; for longer RNA a gradient of 0–60% B was applied; eluent A: 20 mM NaClO4, 25 mM Tris·HCl, pH 8.0, 20% v/v acetonitrile; eluent B: 600 mM NaClO4, 25 mM Tris·HCl, pH 8.0, 20% v/v acetonitrile. HPLC traces were recorded at UV absorption by 260 nm.
The synthesis yield was determined via UV-photometrical analysis on an Implen NanoPhotometer 7122 V2.0.0 and the mass was verified using ion pair reversed phase LC-ESI-MS analysis on a Finnigan LCQ Advantage Max mass spectrometer equipped with an electrospray ionization (ESI) interface coupled to a Thermo Scientific Dionex Ultimate 3000 HPLC system. The sample was applied to a Thermo Scientific DNAPacTM RP column (2.1 × 50 mm) and eluted using an increasing gradient from Buffer A: 8.6 mM triethylamine 100 mM hexafluoriisopropanol in ddH2O (miliQ) to Buffer B: MeOH.
Splinted RNA ligation
Specifically modified tRNAs were generated via splinted ligation of up to three synthetic RNA oligonucleotides (Supplementary Table S3) using reverse complement DNA splinter (Supplementary Table S2). The enzymatic ligation protocol was adapted from (37,44). For one ligation usually 1 nmol of each RNA fragment and the respective DNA splint were combined in a final volume of 70 μl and denatured at 90°C for 2 min. Subsequently, the solution was passively cooled down to room temperature over 15 min. For the ligation reaction 10 μl 10× T4 DNA ligase buffer, 10 μl 50% PEG4000, and 50 U T4 DNA ligase (Thermo Scientific) were added and incubated for 3 h at 37°C. RQ1 DNase (Promega, M6101) (5 U/reaction) was added and incubated 30 min at 37°C. The ligation was denatured for 2 min at 95°C containing 0.5× volume formamide before loading on a denaturing 10% polyacrylamide gel (29:1; 7 M urea). The ligated product was detected by UV-shadowing. The respective band was cut out and fragmented into small pieces and subsequently collected in a fresh Eppendorf tube. The sample was overlaid with RNA gel elution buffer containing 300 mM NaCl, 0.2% SDS, 60 mM NaOAc (pH 5.2) according to (45). The sample was shaken overnight at 4°C following an incubation at 60°C for 1 h. The samples were cooled on ice before purification using the Monarch® RNA Cleanup Kit (10 μg) (NEB, T2030L). The concentration was determined using photo spectrometry (NanoDrop® ND-1000).
As an alternative to polyacrylamide gel electrophoresis, analysis and purification of the ligation products were performed by anion-exchange HPLC (for conditions see section ‘Deprotection and purification of RNA’). After incubation at 37°C for 3 h, the ligation reaction was extracted twice with phenol/chloroform/isoamyl alcohol solution (25/24/1 v/v/v) and then twice with chloroform to remove any remaining phenol from the mixture. The solution was concentrated in vacuo and purified by AE-HPLC on an analytical column. The ligated products were obtained with high purity, that was confirmed by AE-HPLC analysis.
In vitro translation
All in vitro translations were performed using the PURExpress® Δ(aa, tRNA) Kit (NEB, E6840S). For in vitro translation 10 pmol tRNAfMet CAU, 30 pmol tRNAPhe GAA, 30 pmol tRNAArg UCU transcripts and 15 pmol of the transcribed or modified tRNAGly were prepared in a PCR tube and lyophilized in a Concentrator Plus (Eppendorf). Due to a specific contamination of the PURExpress® Δ(aa, tRNA) Kit with native tRNAGlu, the addition of an in vitro transcribed tRNAGlu was dispensable. Lyophilized tRNAs were dissolved in the reaction mixture containing 50 ng of plasmid template, 1.25 μl Solution A, 1.875 μl Solution B, 0.63 μl amino acid mix, 0.5 μl [35S]-Met/Cys (Hartmann Analytic; 10 mCi/ml) in a final volume of 6.25 μl. The reactions were incubated for 2 h at 37°C. The translation was stopped on ice by adding 2 μl of 4× Laemmli buffer (8% SDS, 40% glycerol, 350 mM Tris pH 8.8, 400 mM β-mercaptoethanol, 0.1% bromophenol blue). The peptide yields were determined by a Tricine-SDS-PAGE with a 16% separating (6 M urea), a 10% spacing and 4% stacking gel (46). Afterwards, the gel was shaken for approximately 5 min each in staining (50% ethanol, 5% acetic acid) and destaining (20% ethanol, 7.5% acetic acid) solution. The gel was dried on a Whatman paper for 1 h at 70°C in a slab gel dryer (Cleaver Scientific), exposed to a phosphor screen overnight and scanned by Typhoon™ FLA 9500 (GE Healthcare). The bands were quantified using the ImageQuantTL software.
For the luciferase assays, 15 pmol tRNA transcript or synthetic tRNAGly isoacceptors were lyophilized in PCR tubes in a Concentrator Plus (Eppendorf). When all E. coli tRNAGly isoacceptors were employed simulaneously, 7.5 pmol, 3.25 pmol and 3.25 pmol tRNAGly GCC, CCC and UCC transcripts were used, respectively. 10.7 μg tRNA bulk or depleted tRNA bulk were added together with 36 ng DNA template, 0.9 μl Solution A, 1.36 μl Solution B, and 0.45 μl amino acid mix. The reactions containing a total volume of 4.5 μl were incubated for 2 h at 37°C. To determine the luciferase activity 25 μl of the luciferase assay reagent (Promega, E1483) was prepared in a clear bottom, black 96 well plate (Greiner Bio-one). 2 μl of the translation reaction was pipetted to the wall of the well and all reactions were started simultaneously by short centrifugation at 1000×g. The luminescence of two technical replicates per sample was measured in a BMG Labtech FLUOstar Omega plate reader with a detection time of 9.5 s and a gain of 3000.
Statistical analysis
The quantitative data from the in vitro translation assays were subjected to a two-step statistical analysis using R. First, an ANOVA comparison was performed. In case the ANOVA model was significant (P-value < 0.05), the statistical analysis was continued with Dunnett's post-hoc test. Dunnett's test was used to determine which normalized sample means were significantly different from the reference sample mean. For fLuc assays, the ‘3× depletion’ shown in the graphs served as a quality control sample and was therefore not included in the statistical analysis.
Results
tRNAGly UCC from M. mycoides and E. coli (Figure 1) are very similar in their sequence differing only in some nucleotides within the tRNA body. The sequence alignments (Supplementary Figure S1) revealed an identical anticodon stem-loop (nucleotide 27 to 43), while the acceptor stem is GC-rich in E. coli compared to M. mycoides, in which two of the first six G–C pairs from E. coli are changed into A-U pairs in M. mycoides. However, there is one A–U pair in E. coli which is replaced by a G–C pair in M. mycoides located at the end of the acceptor stem (position 7–66). The sequences of the T-loops are identical, but the T-stem contains only G–C in E. coli while it contains a G-U and A-U pair at the beginning of the T-stem in M. mycoides. More variations are found in the D-arm. The D-loop is one nucleotide smaller in M. mycoides but otherwise the sequences are identical. Further, there is a rare C9 in E. coli while M. mycoides harbors the more common A9. In the common tertiary structure of tRNAs, the base pair 12-23 makes a triple interaction with residue 9 (26,47,48) leading probably to A9/A23 non-W/C base pair in M. mycoides containing the base triplet A9, U12–A23. In bacterial sequences, all tRNAGly with C9 have an A12–U23 pair (17) of which the potential contact between C9 and U23 is not known and needs to be verified. An additional difference in the D-stem is the U13/U22 in E. coli replaced by a U13–G22 in M. mycoides. In the variable loop, E. coli harbors a GA while M. mycoides a UG sequence. To identify the sequence elements and the modifications responsible for this difference in the decoding capability of these tRNAs, we generated tRNAGly variants that differ in their sequence and modifications.
Decoding of synonymous glycine codons by E. coli and M. mycoides tRNAGly isoacceptors
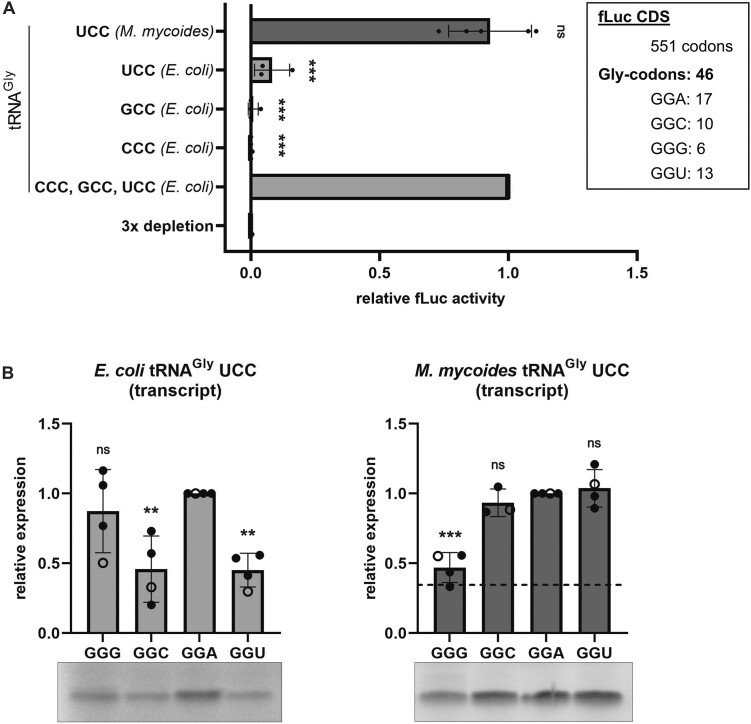
Previous studies have shown that the tRNAGly UCC of M. mycoides is compatible with the E. coli translation machinery (49,50). This allows to test the decoding strategy of superwobbling tRNAs of Mycoplasma in well-established E. coli translation systems. In order to avoid the tRNAs to be modified by E. coli enyzmes (51,52), we employed a defined recombinant in vitro translation system that lacks all modifying enzymes (53). The first question addressed was whether an in vitro transcribed and thus unmodified M. mycoides tRNAGly UCC is still able to decode all four synonymous glycine codons or whether the modified nucleotides are a prerequisite for superwobbling. In addition, the decoding capacity of the unmodified E. coli tRNA was determined, since it contains the same anticodon stem-loop sequence as the M. mycoides tRNA and an unmodified U34, which is a characteristic feature for extended wobbling.
To determine the decoding ability of these transcribed tRNAs, the native E. coli tRNAGly isoacceptors within tRNA bulk from E. coli were substituted with the tRNAs of interest and tested in in vitro translation of the fLuc reporter mRNA containing all four synonymous glycine codons in multiple copies (Figure 2A, Supplementary Figure S4). Therefore, the native E. coli tRNA pool was depleted of the three tRNAGly isoacceptors using RNase H (Supplementary Figure S5A). Long antisense DNA oligonucleotides (Supplementary Table S2) were hybridized to the E. coli tRNAGly isoacceptors and subsequently digested with RNase H, which is specific for RNA-DNA duplexes. Due to the high abundance of tRNAGly GCC compared to the other two isoacceptors (four gene copies instead of one) (54,55), a single depletion step targeting all three isoacceptors simultaneously was not sufficient. Therefore, a second depletion targeting tRNAGly GCC was required to further deplete residual amounts of native tRNA (Supplementary Figure S5B). This pool of tRNAs lacking all three tRNAGly isoacceptors, was subsequently used in the PURExpress Δ(aa, tRNA) translation kit in combination with a fLuc assay. This recombinant PURExpress system was designed to allow the separate addition of tRNAs and amino acids and is therefore an ideal system for testing tRNA pools of different compositions (53,56). The RNase H treated tRNA pool did not provide any luciferase activity, indicating that the tRNAGly isoacceptors were successfully eliminated. However, translation could be efficiently rescued by the simultaneous addition of the three in vitro transcribed E. coli tRNAGly isoacceptors with the anticodons GCC, CCC and UCC. Interestingly, the addition of E. coli tRNAGly UCC alone provided only small amounts of luciferase signal. In contrast, the M. mycoides tRNAGly UCC resulted in comparable luciferase activities as the three E. coli tRNAGly isoacceptors combined, revealing its superwobbling capability even in the absence of all its tRNA modifications (Figure 2A).
Figure 2.
Decoding of synonymous glycine codons by unmodified tRNAGly isoacceptors. (A) In vitro translation of the firefly luciferase (fLuc) gene with E. coli tRNA bulk depleted in the tRNAGly isoacceptors was rescued with tRNAGly isoacceptor transcripts from E. coli (light grey) and M. mycoides (dark grey). The glycine codon composition of the fLuc coding sequence (CDS) is shown in the upper right (box). Datapoints represent technical duplicates of individual experiments and fLuc activity was normalized to the sample using all three E. coli tRNAGly isoacceptor transcripts. (B) Decoding of specific glycine codons by the E. coli and M. mycoides tRNA transcripts within a short mRNA. Each datapoint represents the quantification of [35S]-Met labelled peptides on autoradiographs. Open circles indicate quantifications from the representative gels. Peptide yields resulting from the sequence containing GGA codons were used as reference. The dashed line indicates the peptide yield resulting from translation of the GGA harbouring mRNA by E. coli tRNAGlyUCC transcript. (A, B) Mean of individual datapoints with standard deviation is shown. Significance was tested by ANOVA ((A) < 2e-16; (B) 0.00337 (left); 1.85e-05 (right)) followed by Dunnett's test (0.001 ‘***’, 0.01 ‘**’, 0.05 ‘*’, >0.05 ‘ns’).
To understand how the tRNAGly UCC from E. coli and M. mycoides differ in their ability to decode the synonymous glycine codons, we tested each glycine codon individually. Therefore, a defined translation assay employing a minimal mRNA encoding for a 14 amino acid long peptide was established. To determine the decoding capability of the tRNAs for individual synonymous glycine codons, four variations of the coding sequences were generated, differing only in the third nucleotide of the glycine codons (Supplementary Figure S6). Thereby, we were able to observe differences in the efficiency of each tRNA to decode the individual synonymous codons (Figure 2B), which cannot be determined by using the fLuc assay as the coding sequence contains all four codons. Whereas, the unmodified E. coli tRNAGly UCC was most efficient in decoding GGA and GGG codons, the M. mycoides tRNA showed lowest decoding efficiency for GGG codons. However, the M. mycoides tRNA generally provided higher protein yields than the E. coli tRNAGly UCC, even when decoding the mRNA harboring the least favored GGG codons.
Contribution of tRNA sequence to superwobbling
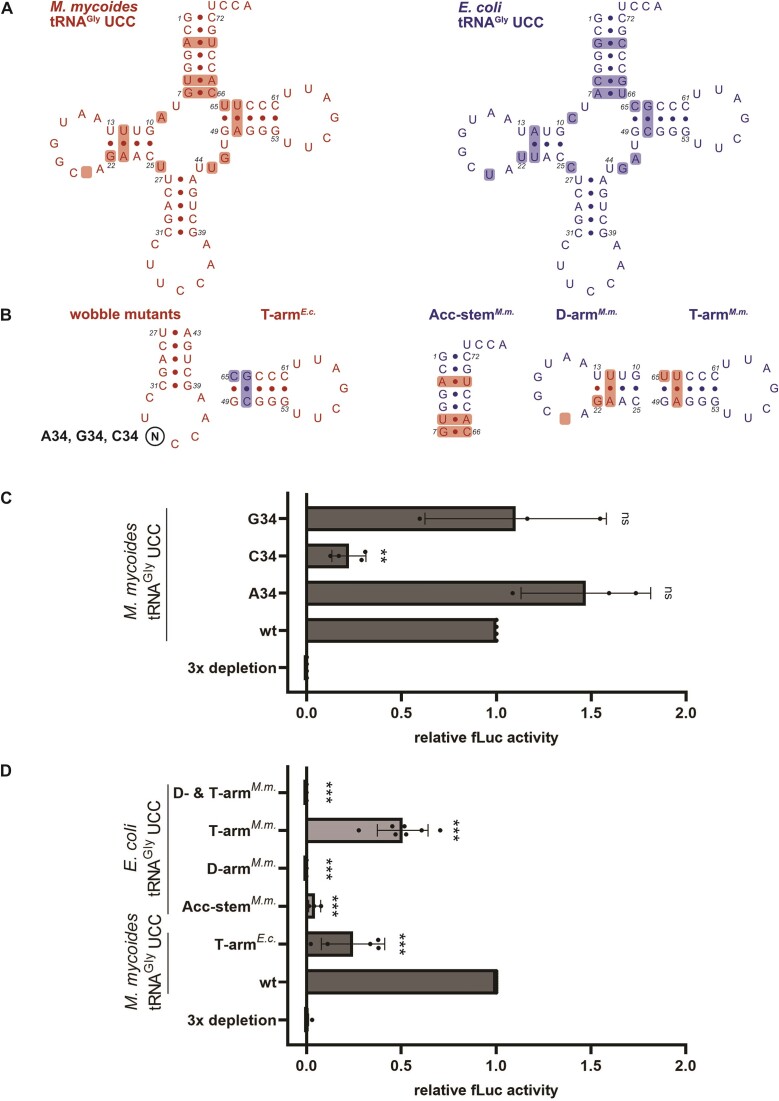
The observation that the in vitro transcribed M. mycoides tRNAGly UCC lacking all tRNA modifications, was able to compensate for the loss of all three E. coli tRNAGly isoacceptors suggests that its superwobbling ability is not dependent on the modifications and might be mainly based on the tRNA sequence (Figure 1). To address which nucleotides or sequence elements enable superwobbling, several tRNA mutants of E. coli and M. mycoides were created (Figure 3A, B).
Figure 3.
Impact of tRNA sequence on decoding of synonymous glycine codons. (A) The secondary structure of the unmodified wild-type (wt) M. mycoides (M.m.; red) and E. coli (E.c.; blue) tRNAGly UCC sequences are shown, with sequence differences highlighted. (B) Single nucleotides (wobble mutants) or sequence elements (acceptor (Acc)-stem, D-arm, T-arm) were exchanged between the M. mycoides and the E. coli tRNAGly. (C) M. mycoides tRNA mutated at position 34 and (D) other E. coli and M. mycoides tRNA mutants were tested for their ability to sustain fLuc translation in absence of the native E. coli tRNA isoacceptors. Datapoints represent technical duplicates of individual experiments. The fLuc activity was normalized to the wt M. mycoides tRNA sample. (C, D) Mean of individual datapoints with standard deviation is shown. Significance was tested by ANOVA ((C) 0.000675; (D) <2e-16) followed by Dunnett's test (0.001 ‘***’, 0.01 ‘**’, 0.05 ‘*’, >0.05 ‘ns’).
As the unmodified U34 is the only common feature between all superwobbling tRNAs of mitochondria, chloroplasts and Mycoplasma species, we determined the importance of this U34 on decoding of the synonymous glycine codons. We mutated the M. mycoides tRNAGly UCC at the wobble position resulting in tRNAs carrying an ACC, CCC or GCC anticodon and tested for their aminoacylation (Supplementary Figure S7A). Remarkably, the ACC and GCC anticodon carrying tRNAs were able to efficiently compensate for the depletion of the three tRNAGly isoacceptors, whereas the C34 mutant could only partially sustain translation (Figure 3C). This suggests that U34 is not strictly required for superwobbling in the context of the M. mycoides tRNAGly UCC sequence, nor does it seem to be the sole determinant, as the unmodified U34 within the E. coli sequence did not increase the decoding capacity.
Since the anticodon stem-loop sequence of E. coli and M. mycoides tRNAGly UCC are identical, the contribution of the T-arm, D-arm and the acceptor stem were determined. Therefore, the respective sequence elements from the M. mycoides tRNA were introduced into the E. coli tRNAGly UCC (Figure 3A, B). After efficient aminoacylation of the mutants by the E. coli glycyl-tRNA synthetase (GlyRS) has been confirmed (Supplementary Figure S7B), they were tested for their potential to compensate for the depletion of the three E. coli tRNAGly isoacceptors during translation. Indeed, the introduction of the M. mycoides T-arm, consisting of the mutation C65U and the substitution of the base pair C50-G64 for A50-U64, resulted in a partial rescue of translation by the mutant E. coli tRNA. The exchange of the acceptor stem or the D-arm did not equip the E. coli tRNA with the ability to compensate for the loss of the tRNAGly isoacceptors. Also, the combined introduction of the T- and D-arm did not enable translation of the luciferase mRNA (Figure 3D). Interestingly, even the capability to decode the cognate GGA codon was reduced by the D-arm muntant and completely abolished by the mutant with combined D- and T-arm (Supplementary Figure S7C). Since the T-arm sequence of M. mycoides was a key element to improve the superwobbling ability of the E. coli tRNA, we wondered how the T-arm sequence of E. coli would affect the M. mycoides tRNA. Indeed, a decreased luciferase activity was detected (Figure 3D), underlining the importance of the T-arm for the extended decoding capability of the M. mycoides tRNA.
Contribution of tRNA modifications in E. coli tRNAGly UCC to decoding
tRNAs are the most abundantly modified RNA molecules and the importance of these modifications is undisputable. Although, tRNAGly isoacceptors are in general only sparsely modified in bacteria (27,40), their modifications are still considered to have indispensable functions for tRNA structure and efficient decoding. Especially, modifications at the wobble position are postulated to modulate decoding efficiency and accuracy of the respective tRNAs. The E. coli tRNAGly UCC harbors three modifications (Figure 1): T54 and P55 within the T-arm and the wobble modification mnm5U34. This wobble modification was postulated to improve base pairing to G at the third codon position (57,58).
To systematically determine the impact of the modifications on decoding, we generated site-specifically modified tRNAs independent of any modifying enzyme. Thereby, chemically synthesized RNA oligonucleotides were ligated via splinted ligation to result in full-length modified tRNAs (Figure 4A). This approach enabled the site-specific and quantitative introduction of specific modifications or combinations thereof into tRNAs. As first step, only the conserved T-arm modifications T54 and P55 were inserted but U34 was left unmodified. Already the presence of the T-arm modifications significantly modulated decoding of glycine codons (Figure 4B). Whereas the unmodified tRNAs decoded the mRNAs harboring GGA and GGG codons similarly efficient (Figure 2B), the T-arm modifications seemed to be disadvantageous for GGG decoding (Figure 4B). By adding mnm5U at position 34, resulting in a fully-modified tRNAGly, the decoding of GGG was significantly improved again, consistent with its proposed role for enhancing the decoding of G-ending codons (9,58) (Figure 4B). These results indicate that the full set of modifications is needed to provide balanced decoding of GGA and GGG, revealing a significant crosstalk between the body and anticodon modifications.
Figure 4.
The effect of tRNA modifications on decoding by E. coli tRNAGly UCC. (A) Schematic depiction of splinted ligation of differently modified chemically synthesized RNA oligonucleotides to generate full-length site-specifically modified tRNAs. (B) Decoding efficiency of the four synonymous glycine codons within a small mRNA by partly or fully modified E. coli tRNAGly UCC. [35S]-Met labelled peptides were quantified and normalized to the peptide yields resulting from the GGA containing mRNA. (C) Translation efficiency of tRNAs harboring different modifications depicted in (D) at position 34. Small mRNAs either containing GGA or GGG codons were in vitro translated in presence of [35S]-Met and the respective modified E. coli tRNAGly UCC. Peptide yields were normalized to the sample of unmodified tRNAGly UCC. Open circles indicate datapoints shown in the representative gels. (B, C) Mean of individual datapoints with standard deviation is shown. Significance was tested by ANOVA ((B) 0.00158 (left); 0.00677 (right); (C) not significant (left); 0.0204 (right) followed by Dunnett's test (0.001 ‘***’ 0.01 ‘**’ 0.05 ‘*’ >0.05 ‘ns’).
The chemical assembly used here to introduce and to test modifications not only allows the introduction of natural modifications but also other non-standard RNA derivative. Next, we tried to reveal whether the complete mnm5U modification is needed for modulating decoding. Therefore, the metabolic precursor nm5U and also m5U (T) (Figure 4D) were inserted at the wobble position of the E. coli tRNAGly UCC harboring the T-arm modifications. Remarkably, all tested modifications reduced the efficiency of decoding mRNAs harboring GGA glycine codons. Already the presence of a methylation slightly reduced peptide yields. In respect to the GGG codon, only mnm5U was able to improve its decoding, whereas the precursor nm5U or m5U did not change the translation activity (Figure 4C).
Contribution of tRNA modifications in M. mycoides tRNAGly UCC to decoding
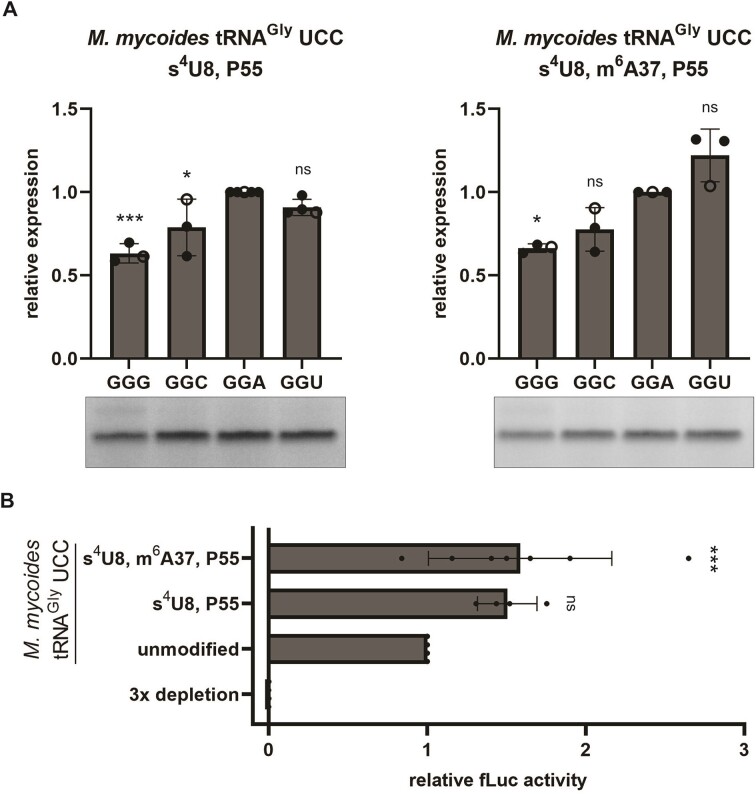
Like the E. coli tRNA also the M. mycoides tRNAGly UCC carries only a small number of tRNA modifications (Figure 1). To assess the impact of the tRNA body modifications, s4U at position 8 and P at position 55 were introduced into the M. mycoides tRNA. This partially modified tRNA showed comparable decoding as the unmodified tRNA transcript (Figures 2B and 5A). The additional introduction of the m6A modification at position 37 resulted in an enhanced translation of the GGU containing mRNA (Figure 5A).
Figure 5.
The effect of tRNA modifications on decoding by M. mycoides tRNAGly UCC. (A) The M. mycoides tRNAGly UCC carrying different sets of modifications were tested for their capability to decode the synonymous glycine codons. [35S]-Met labelled peptides were quantified and normalized to the peptide yields resulting from the GGA containing mRNA. Open circles indicate datapoints shown in the representative gels. (B) Depletion of the three E. coli tRNAGly isoacceptors within E. coli tRNA bulk was rescued with the differently modified M. myocides tRNAGly. The fLuc activities were normalized to the unmodified tRNA sample. (A, B) Mean of individual datapoints with standard deviation is shown. Significance was tested by ANOVA ((A) 0.000424 (left); 0.000782 (right); (B) 0.000722) followed by Dunnett's test (0.001 ‘***’, 0.01 ‘**’, 0.05 ‘*’, >0.05 ‘ns’).
In addition to translation of the short mRNAs, we employed the luciferase reporter gene assay to determine the ability of the differently modified tRNAs to decode all glycine codons within the luciferase reporter mRNA. Therefore, tRNAs without modifications, tRNAs harboring only the body modifications s4U8 and P55, and tRNAs with the complete set of modifications were compared in their potential to provide translation in absence of the native E. coli tRNAGly isoacceptors. Thereby, we observed that the body modifications lead to increased luciferase activities, which was not further improved by m6A at position 37 (Figure 5B). This finding indicates that the modifications are not essential to provide the ability to decode all four synonymous glycine codons, but they modulate the efficiency by potentially stabilizing and prestructuring the tRNA in a native fold for an overall better translation performance.
m6A at position 37 enables superwobbling of E. coli tRNAGly UCC
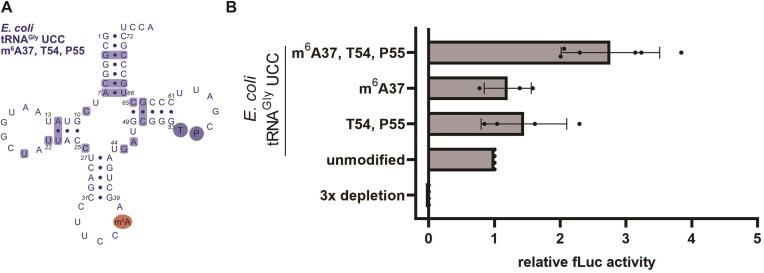
Although M. mycoides tRNA was highly efficient in decoding all four glycine codons even in absence of its modifications, the presence of them was still beneficial. All superwobbling tRNAs from M. capricolum, a Mollicutes closely related to M. mycoides, of which the sequences and modifications of the complete set of tRNAs are known, harbor either m6A or m1G at position 37 (18,24). As m6A at position 37 was postulated to significantly impact decoding through a structural preorganization of the anticodon loop (34), this modification might contribute to the superwobbling capability of a tRNA. Therefore, m6A was inserted in E. coli tRNAGly UCC in combination with the conserved E. coli T-arm modifications (Figure 6A) and tested for its ability to compensate for the loss of the E. coli tRNAGly isoacceptors. As controls also a tRNA carrying only the T-arm modifications or only the m6A modification were tested alongside in the luciferase reporter assay. Remarkably, m6A did provide the E. coli tRNA with improved ability to decode all four glycine codons, but interestingly only in combination with the T-arm modifications, T54 and P55 (Figure 6B). This observation suggests the importance of tRNA modifications for structural integrity and preorganization of the anticodon loop to enable efficient protein synthesis.
Figure 6.
The effect of m6A37 on decoding. (A) Secondary structure of the hybrid modified E. coli tRNAGly UCC carrying the native T-arm modifications T54 and P55 (shaded in blue) and the M. mycoides tRNA modification m6A at position 37 (shaded in red). (B) Depletion of the three E. coli tRNAGly isoacceptors within E. coli tRNA bulk was rescued with the differently modified E. coli tRNAGly UCC. Datapoints represent measurements of technical duplicates of individual experiments. The fLuc activity was normalized to the unmodified tRNA sample. Mean of individual datapoints with standard deviation is shown. Significance was tested by ANOVA (not significant).
Discussion
tRNAs play a crucial role during protein synthesis by enabling decoding of the codons within mRNAs and the integration of the corresponding amino acid into the nascent peptide chains. Different sequence elements and modification patterns have evolved to allow the multiple steps that tRNAs go through in order to provide the ribosome with an adequate supply of amino acids (59,60). To be delivered to the A-site, the tRNA needs to undergo aminoacylation and subsequently form a ternary complex with the GTPase EF-Tu and GTP. At the ribosomal A-site, the anticodon of the tRNA forms an interaction with the mRNA codon. Following decoding, the aminoacyl-tRNA fully accommodates into the A-site, enabling peptide bond formation and translocation to the P-site (61,62). This path of a tRNA during translation, is conserved in all organisms. However, tRNAs vary in their sequence, modifications, and decoding concepts. Remarkable decoding strategies based on superwobbling tRNAs decoding four-fold degenerate codon boxes, have been evolved by certain bacteria and organelles. This resulted in a significantly reduced number of tRNAs required to decode all sense codons (18). To better understand the requirements for extended wobbling, we compared the superwobbling tRNAGly UCC from M. mycoides with the tRNAGly UCC from E. coli using a novel approach based on the ligation of chemically synthesized RNA oligonucleotides.
We generated and tested unmodified and variations of modified M. mycoides and E. coli tRNAGlyUCC for their ability to decode all four synonymous glycine codons. The results show that the modifications of the M. mycoides tRNAGly are not essential to provide superwobbling, which led to the conclusion that its tRNA sequence is the primary prerequisite for extended wobbling (Figure 2). The unmodified U34 and C32, within the M. mycoides tRNAGly, were previously identified as crucial elements for its superwobbling ability (49,51,63). However, U34 was not essential for superwobbling by M. mycoides tRNAGly. Mutants with A34 or G34 were both highly efficient in decoding all four synonymous glycine codons (Figure 3C). G34 is commonly used in bacterial tRNAs and a loss of decoding specificity has not been described yet (13,40,64). In contrast, A34 is not found in any of the tRNAs except for tRNAArg, where A34 undergoes A-to-I editing (64,65) and A34 has already been shown to base pair with all four nucleotides (52). However, at position 32 the mutation of C to U was detrimental to superwobbling (Supplementary Figure S8) (49,51). The presence of U32 allows the formation of a base pair with A38, thereby affecting the preorganization of the tRNA anticodon loop (49,51,66). The interaction between the nucleotides at position 32 and 38 has been identified to influence the binding affinity to the ribosomal A-site and to balance the different strengths of codon/anticodon interactions to achieve uniform binding (67–71). Since the sequence of the anticodon stem-loop of M. mycoides and E. coli is identical, it was unexpected that the unmodified E. coli tRNAGly UCC was not able to efficiently decode the four synonymous glycine codons (Figure 2). Only the additional insertion of the M. mycoides T-arm into the E. coli tRNA extended the decoding capability and thereby revealed an additional sequence element important for superwobbling (Figure 3D). The T-stem sequence has been shown to determine the binding affinity to EF-Tu in accordance with the thermodynamic contributions of the tRNA body and its esterified amino acid (72,73), potentially supporting extended decoding. However, tRNAs from different bacteria within an anticodon class have similar predicted mean ΔG° values for EF-Tu binding despite sequence variations in their T-stems (74), arguing against a major impact of EF-Tu.
An alternative explanation could be that the T-arm sequence modulates the structure or folding of the M. mycoides and E. coli tRNA. The tRNA sequence is primarily responsible for folding into a native and functional tRNA. However, based on the sequence, the partition function will exhibit varied populations, with some sequences adopting numerous folding states while others having a limited number. Among those states, the proportion of native folds is critical for function. Different folding preferences between the tRNAs were calculated from secondary structure predictions, resulting in the correct secondary cloverleaf structure for M. mycoides tRNA, but not for E. coli (Supplementary Figure S9) (75). UV melting analyses had found application for tRNAs already before (76–79) and can provide an insight into tRNA folding. Indeed, these experiments show differences between the two tRNAs in absence and in presence of Mg2+ ions implicating different folding (Supplementary Figure S10). As an alternative approach, NMR melting curves were performed which, in agreement with UV melting, show a higher stability (but not necessarily of the native folding state) of the E. coli tRNA compared to the M. mycoides tRNA (Supplementary Figure S11). However, due to the complexity of the tRNA structure, the melting curves obtained did not allow conclusions to be drawn about specific structural differences.
Folding of tRNAs is assisted by the presence of modifications. By introducing modifications separately into the tRNAs, the individual contribution of the body and anticodon loop modifications to decoding could be determined. Thereby, we observed a crosstalk between the modifications as well as a modulatory effect on the decoding efficiency of the tRNAs for specific codons. Decoding of GGA, GGC and GGU by the E. coli tRNAGly UCC (Figure 4) was enhanced by the introduction of the T-arm modifications T54 and P55. Modifications in the T-loop support the folding of the T-arm into its native state with the long-range tertiary contacts with the D-loop (25,29–31). The additional introduction of mnm5U balanced decoding of GGG and GGA codons by slightly impairing GGA but enhancing GGG. Uhlenbeck and colleagues have shown that the binding affinity of different tRNAs to their cognate codons in the ribosome is nearly uniform through a delicate interplay of amino acid identity, tRNA sequence and its modifications (80,81). Our results also indicate that the E. coli tRNAGly UCC provides the balanced and fine-tuned decoding of GGA and GGG codons only in presence of the full set of native modifications. The indispensability of T-arm modifications in conjunction with mature mnm5U, rather than precursor nm5U or m5U, highlights the sensitivity of decoding to even subtle chemical variations within modified nucleotides. This underscores the intricate and delicately balanced network of interactions governing this process. Also, in the superwobbling M. mycoides tRNAGly modifications contributed to the overall translation efficiency (Figure 5). While the s4U8 and P55 body modifications alone already increased the overall translation efficiency, they did not modulate the preference for specific codons. Only the full set including m6A37 slightly improved the decoding of GGU codons, resulting in a preference for A- and U-ending codons, which reflects the codon usage in M. mycoides (17).
Although our study on M. mycoides tRNAGly revealed the sequence as key prerequisite for efficient recognition of all four synonymous codons, it did not exclude a contribution of the modifications to superwobbling. Therefore, we aimed to confer the E. coli tRNAGly with superwobbling ability by the introduction of the M. mycoides tRNA anticodon loop modification. m6A at position 37 has previously been described as important for the preorganization (82), for limiting the flexibility (34) and for the prevention of a collapsed anticodon loop (29,83,84). Consequently, we hypothesized an increased binding affinity of the tRNA to the ribosomal A-site in presence of m6A37 to enable extended wobbling. Remarkably, m6A in the E. coli tRNA improved the its ability to decode all four synonymous codons (Figure 6). However, this beneficial effect of the m6A modification on four-way wobbling became apparent only when combined with the T-arm modifications, which might additionally support correct folding of the tRNA. The requirement for m6A37, T54 and P55 to arm the E. coli tRNA for efficient superwobbling again implies the importance of modification in stabilizing and prestructuring the entire tRNA molecule to achieve an optimized conformation for efficient participation in translation.
By employing chemically synthesized tRNAs a complete characterization of tRNA sequence and modifications on decoding can be performed. In the present study, the modifications of the M. mycoides tRNAGly were shown not to be essential for superwobbling. Instead, the tRNA sequence itself was identified as the primary prerequisite revealing the T-arm sequence as novel important element contributing to superwobbling. However, modifications modulate and improve decoding by being part of a delicate set of interactions required for efficient and accurate translation, possibly by facilitating the folding and prestructuring of the tRNA molecule. Our results highlight that the tRNA sequence and its modifications form a fully integrated system extremely complex to disentangle experimentally, requiring the need for novel approaches to study their impact on translation systematically.
Supplementary Material
Acknowledgements
We thank Nina Clementi and Maximilian Kohl for helpful discussions and Claudia Lamina for the assistance with the statistical analysis. Alexander Hüttenhofer and all lab members of the Institute of Genomics and RNomics for valuable input and discussions.
Author contributions: M.K., K.B., K.E., R.P. and F.S.J. performed the experiments, M.K. and M.E. designed the experiments, M.K., M.E., K.B., C.K., R.M. and E.W. analyzed the data. M.K., M.E. and E.W. wrote the manuscript. M.E. provided the concept and funding of the study. All authors contributed to the final version of the manuscript.
Contributor Information
Maria Kompatscher, Institute of Genomics and RNomics, Biocenter, Medical University of Innsbruck, Innrain 80-82, 6020 Innsbruck, Austria.
Karolina Bartosik, Institute of Organic Chemistry, Center for Molecular Biosciences Innsbruck, University of Innsbruck, Innrain 80-82, 6020 Innsbruck, Austria.
Kevin Erharter, Institute of Organic Chemistry, Center for Molecular Biosciences Innsbruck, University of Innsbruck, Innrain 80-82, 6020 Innsbruck, Austria.
Raphael Plangger, Institute of Organic Chemistry, Center for Molecular Biosciences Innsbruck, University of Innsbruck, Innrain 80-82, 6020 Innsbruck, Austria.
Fabian Sebastian Juen, Institute of Organic Chemistry, Center for Molecular Biosciences Innsbruck, University of Innsbruck, Innrain 80-82, 6020 Innsbruck, Austria.
Christoph Kreutz, Institute of Organic Chemistry, Center for Molecular Biosciences Innsbruck, University of Innsbruck, Innrain 80-82, 6020 Innsbruck, Austria.
Ronald Micura, Institute of Organic Chemistry, Center for Molecular Biosciences Innsbruck, University of Innsbruck, Innrain 80-82, 6020 Innsbruck, Austria.
Eric Westhof, Université de Strasbourg, Institut de Biologie Moléculaire et Cellulaire, Architecture et Réactivité de l’ARN, CNRS UPR 9002, 2, allée Konrad Roentgen, F-67084 Strasbourg, France.
Matthias D Erlacher, Institute of Genomics and RNomics, Biocenter, Medical University of Innsbruck, Innrain 80-82, 6020 Innsbruck, Austria.
Data availability
The data that support the findings of this study are available from the corresponding author upon request.
Supplementary data
Supplementary Data are available at NAR online.
Funding
Austrian Science Fund FWF [P34132-B, F8004 to M.E.; P31961 and F8011 to R.M.; P30370, P32773 and I52870 to C.K]; Austrian Research Promotion Agency FFG [858017, West-Austrian BioNMR to C.K and R.M]. Funding for open access charge: Austrian Science Fund FWF.
Conflict of interest statement. None declared.
References
- 1. Kyrpides N.C., Woese C.R.. Universally conserved translation initiation factors. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ganoza M.C., Kiel M.C., Aoki H.. Evolutionary Conservation of Reactions in Translation. Microbiol. Mol. Biol. Rev. 2002; 66:460–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Goodenbour J.M., Pan T.. Diversity of tRNA genes in eukaryotes. Nucleic Acids Res. 2006; 34:6137–6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ayan G.B., Park H.J., Gallie J.. The birth of a bacterial trna gene by large-scale, tandem duplication events. eLife. 2020; 9:e57947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Suzuki T., Yashiro Y., Kikuchi I., Ishigami Y., Saito H., Matsuzawa I., Okada S., Mito M., Iwasaki S., Ma D.et al.. Complete chemical structures of human mitochondrial tRNAs. Nat. Commun. 2020; 11:4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Suzuki T., Nagao A., Suzuki T.. Human mitochondrial trnas: biogenesis, function, structural aspects, and diseases. Annu. Rev. Genet. 2011; 45:299–329. [DOI] [PubMed] [Google Scholar]
- 7. Crick F.H.C. Codon-anticodon pairing: the wobble hypothesis. J. Mol. Biol. 1966; 19:548–555. [DOI] [PubMed] [Google Scholar]
- 8. Rozov A., Demeshkina N., Khusainov I., Westhof E., Yusupov M., Yusupova G.. Novel base-pairing interactions at the tRNA wobble position crucial for accurate reading of the genetic code. Nat. Commun. 2016; 7:10457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Westhof E., Yusupov M., Yusupova G.. The multiple flavors of GoU pairs in RNA. J. Mol. Recognit. 2019; 32:e2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Agris P.F., Eruysal E.R., Narendran A., Väre V.Y.P., Vangaveti S., Ranganathan S.V. Celebrating wobble decoding: half a century and still much is new. RNA Biol. 2018; 15:537–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McCown P.J., Ruszkowska A., Kunkler C.N., Breger K., Hulewicz J.P., Wang M.C., Springer N.A., Brown J.A.. Naturally occurring modified ribonucleosides. Wiley Interdiscip. Rev. RNA. 2020; 11:e1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Suzuki T. The expanding world of tRNA modifications and their disease relevance. Nat. Rev. Mol. Cell Biol. 2021; 22:375–392. [DOI] [PubMed] [Google Scholar]
- 13. Grosjean H., Westhof E.. An integrated, structure- and energy-based view of the genetic code. Nucleic Acids Res. 2016; 44:8020–8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Prabhakar P.S., Takyi N.A., Wetmore S.D.. Posttranscriptional modifications at the 37th position in the anticodon stem-loop of tRNA: structural insights from MD simulations. RNA. 2021; 27:202–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sakai Y., Kimura S., Suzuki T.. Dual pathways of tRNA hydroxylation ensure efficient translation by expanding decoding capability. Nat. Commun. 2019; 10:2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang W., Foo M., Eren A.M., Pan T.. tRNA modification dynamics from individual organisms to metaepitranscriptomics of microbiomes. Mol. Cell. 2022; 82:891–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chan P.P., Lin B.Y., Mak A.J., Lowe T.M.. TRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 2021; 49:9077–9096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. De Crécy-Lagard V., Marck C., Brochier-Armanet C., Grosjean H.. Comparative RNomics and Modomics in mollicutes: prediction of gene function and evolutionary implications. IUBMB Life. 2007; 59:634–658. [DOI] [PubMed] [Google Scholar]
- 19. Grosjean H., Breton M., Sirand-Pugnet P., Tardy F., Thiaucourt F., Citti C., Barré A., Yoshizawa S., Fourmy D., de Crécy-Lagard V.et al.. Predicting the Minimal Translation Apparatus: lessons from the Reductive Evolution of Mollicutes. PLoS Genet. 2014; 10:e1004363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Osawa S., Jukes T.H., Watanabe K., Muto A.. Recent evidence for evolution of the genetic code. Microbiol. Rev. 1992; 56:229–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Alkatib S., Scharff L.B., Rogalski M., Fleischmann T.T., Matthes A., Seeger S., Schöttler M.A., Ruf S., Bock R.. The Contributions of Wobbling and Superwobbling to the Reading of the Genetic Code. PLoS Genet. 2012; 8:e1003076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rogalski M., Karcher D., Bock R.. Superwobbling facilitates translation with reduced tRNA sets. Nat. Struct. Mol. Biol. 2008; 15:192–198. [DOI] [PubMed] [Google Scholar]
- 23. Lagerkvist U. ‘Two out of three’: an alternative method for codon reading. Proc. Natl. Acad. Sci. U.S.A. 1978; 75:1759–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Andachi Y., Yamao F., Muto A., Osawa S.. Codon recognition patterns as deduced from sequences of the complete set of transfer RNA species in Mycoplasma capricolum. Resemblance to mitochondria. J. Mol. Biol. 1989; 209:37–54. [DOI] [PubMed] [Google Scholar]
- 25. Biedenbänder T., De Jesus V., Schmidt-Dengler M., Helm M., Corzilius B., Fürtig B.. RNA modifications stabilize the tertiary structure of tRNAfMet by locally increasing conformational dynamics. Nucleic Acids Res. 2022; 50:2334–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Quigley G.J., Wang A.H.J., Seeman N.C., Suddath F.L., Rich A., Sussman J.L., Kim S.H.. Hydrogen bonding in yeast phenylalanine transfer RNA. Proc. Natl. Acad. Sci. U.S.A. 1975; 72:4866–4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Boccaletto P., Stefaniak F., Ray A., Cappannini A., Mukherjee S., Purta E., Kurkowska M., Shirvanizadeh N., Destefanis E., Groza P.et al.. MODOMICS: a database of RNA modification pathways. 2021 update. Nucleic Acids Res. 2022; 50:D231–D235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rozov A., Demeshkina N., Westhof E., Yusupov M., Yusupova G.. Structural insights into the translational infidelity mechanism. Nat. Commun. 2015; 6:7251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lorenz C., Lünse C.E., Mörl M.. tRNA Modifications: impact on Structure and Thermal Adaptation. Biomolecules. 2017; 7:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nobles K.N., Yarian C.S., Liu G., Guenther R.H., Agris P.F.. Highly conserved modified nucleosides influence Mg2+-dependent tRNA folding. Nucleic Acids Res. 2002; 30:4751–4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sengupta R., Vainauskas S., Yarian C., Sochacka E., Malkiewicz A., Guenther R.H., Koshlap K.M., Agris P.F.. Modified constructs of the tRNA TΨC domain to probe substrate conformational requirements of m1A58 and m5U54 tRNA methyltransferases. Nucleic Acids Res. 2000; 28:1374–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Duechler M., Leszczyńska G., Sochacka E., Nawrot B.. Nucleoside modifications in the regulation of gene expression: focus on tRNA. Cell. Mol. Life Sci. 2016; 73:3075–3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Agris P.F., Narendran A., Sarachan K., Väre V.Y.P., Eruysal E.. The importance of being modified: the role of RNA modifications in translational fidelity. Enzymes. 2017; 41:1–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Agris P.F. Bringing order to translation: the contributions of transfer RNA anticodon-domain modifications. EMBO Rep. 2008; 9:629–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Takai K., Yokoyama S.. Roles of 5-substituents of tRNA wobble uridines in the recognition of purine-ending codons. Nucleic Acids Res. 2003; 31:6383–6391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kurschat W.C., Müller J., Wombacher R., Helm M.. Optimizing splinted ligation of highly structured small RNAs. RNA. 2005; 11:1909–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lang K., Micura R.. The preparation of site-specifically modified riboswitch domains as an example for enzymatic ligation of chemically synthesized RNA fragments. Nat. Protoc. 2008; 3:1457–1466. [DOI] [PubMed] [Google Scholar]
- 38. Graber D., Moroder H., Steger J., Trappl K., Polacek N., Micura R.. Reliable semi-synthesis of hydrolysis-resistant 3′-peptidyl-tRNA conjugates containing genuine tRNA modifications. Nucleic Acids Res. 2010; 38:6796–6802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kohl M.P., Kompatscher M., Clementi N., Holl L., Erlacher M.D.. Initiation at AUGUG and GUGUG sequences can lead to translation of overlapping reading frames in E. coli. Nucleic Acids Res. 2023; 51:271–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jühling F., Mörl M., Hartmann R.K., Sprinzl M., Stadler P.F., Pütz J.. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009; 37:D159–D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Horton R.M., Cai Z., Ho S.N., Pease L.R.. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. BioTechniques. 1990; 8:528–535. [PubMed] [Google Scholar]
- 42. Kao C., Zheng M., Rüdisser S.. A simple and efficient method to reduce nontemplated nucleotide addition at the 3′ terminus of RNAs transcribed by T7 RNA polymerase. RNA. 1999; 5:1268–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Van Goethem A., Yigit N., Everaert C., Moreno-Smith M., Mus L.M., Barbieri E., Speleman F., Mestdagh P., Shohet J., Van Maerken T.et al.. Depletion of tRNA-halves enables effective small RNA sequencing of low-input murine serum samples. Sci. Rep. 2016; 6:37876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vušurović N., Altman R.B., Terry D.S., Micura R., Blanchard S.C.. Pseudoknot formation seeds the twister ribozyme cleavage reaction coordinate. J. Am. Chem. Soc. 2017; 139:8186–8193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hoernes T.P., Hüttenhofer A., Erlacher M.D.. mRNA modifications: dynamic regulators of gene expression?. RNA Biol. 2016; 13:760–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schägger H. Tricine–SDS-PAGE. Nat. Protoc. 2006; 1:16–22. [DOI] [PubMed] [Google Scholar]
- 47. Quigley G.J., Rich A.. Structural domains of transfer RNA molecules. Science (80-.). 1976; 194:796–806. [DOI] [PubMed] [Google Scholar]
- 48. Kim S.H., Sussman J.L., Suddath F.L., Quigley G.J., McPherson A., Wang A.H., Seeman N.C., Rich A.. The general structure of transfer RNA molecules. Proc. Natl. Acad. Sci. U.S.A. 1974; 71:4970–4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Claesson C., Lustig F., Borén T., Simonsson C., Barciszewska M., Lagerkvist U.. Glycine codon discrimination and the nucleotide in position 32 of the anticodon loop. J. Mol. Biol. 1995; 247:191–196. [DOI] [PubMed] [Google Scholar]
- 50. Samuelsson T., Guindy Y.S., Lustig F., Borén T., Lagerkvist U.. Apparent lack of discrimination in the reading of certain codons in Mycoplasma mycoides. Proc. Natl. Acad. Sci. U.S.A. 1987; 84:3166–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lustig F., Borén T.E., Claesson C., Simonsson C., Barciszewska M.Z., Lagerkvist U.. The nucleotide in position 32 of the tRNA anticodon loop determines ability of anticodon UCC to discriminate among glycine codons. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:3343–3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Borén T., Elias P., Samuelsson T., Claesson C., Barciszewska M., Gehrke C.W., Kuo K.C., Lustig F.. Undiscriminating codon reading with adenosine in the wobble position. J. Mol. Biol. 1993; 230:739–749. [DOI] [PubMed] [Google Scholar]
- 53. Shimizu Y., Inoue A., Tomari Y., Suzuki T., Yokogawa T., Nishikawa K., Ueda T.. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 2001; 19:751–755. [DOI] [PubMed] [Google Scholar]
- 54. Dong H., Nilsson L., Kurland C.G.. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J. Mol. Biol. 1996; 260:649–663. [DOI] [PubMed] [Google Scholar]
- 55. Komine Y., Adachi T., Inokuchi H., Ozeki H.. Genomic organization and physical mapping of the transfer RNA genes in Escherichia coli K12. J. Mol. Biol. 1990; 212:579–598. [DOI] [PubMed] [Google Scholar]
- 56. Li J., Tang M., Qi H.. Codon-reduced protein synthesis with manipulating tRNA components in cell-free system. Front. Bioeng. Biotechnol. 2022; 10:891808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Westhof E., Yusupov M., Yusupova G.. Recognition of Watson-Crick base pairs: constraints and limits due to geometric selection and tautomerism. F1000Prime Rep. 2014; 6:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kurata S., Weixlbaumer A., Ohtsuki T., Shimazaki T., Wada T., Kirino Y., Takai K., Watanabe K., Ramakrishnan V., Suzuki T.. Modified uridines with C5-methylene substituents at the first position of the tRNA anticodon stabilize U.G wobble pairing during decoding. J. Biol. Chem. 2008; 283:18801–18811. [DOI] [PubMed] [Google Scholar]
- 59. Giegé R., Eriani G.. The tRNA identity landscape for aminoacylation and beyond. Nucleic Acids Res. 2023; 51:1528–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Beuning P.J., Musier-Forsyth K.. Transfer RNA recognition by aminoacyl-tRNA synthetases. Biopolymers. 1999; 52:1–28. [DOI] [PubMed] [Google Scholar]
- 61. Rodnina M.V. Translation in prokaryotes. Cold Spring Harb. Perspect. Biol. 2018; 10:a032664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rodnina M.V., Fischer N., Maracci C., Stark H.. Ribosome dynamics during decoding. Philos. Trans. R. Soc. B. Biol. Sci. 2017; 372:20160182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lustig F., Borén T., Guindy Y.S., Elias P., Samuelsson T., Gehrke C.W., Kuo K.C., Lagerkvist U.. Codon discrimination and anticodon structural context. Proc. Natl. Acad. Sci. U.S.A. 1989; 86:6873–6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Grosjean H., de Crécy-Lagard V., Marck C.. Deciphering synonymous codons in the three domains of life: co-evolution with specific tRNA modification enzymes. FEBS Lett. 2010; 584:252–264. [DOI] [PubMed] [Google Scholar]
- 65. Ehrlich R., Davyt M., López I., Chalar C., Marín M.. On the track of the missing tRNA genes: a source of non-canonical functions?. Front. Mol. Biosci. 2021; 8:643701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Pernod K., Schaeffer L., Chicher J., Hok E., Rick C., Geslain R., Eriani G., Westhof E., Ryckelynck M., Martin F.. The nature of the purine at position 34 in tRNAs of 4-codon boxes is correlated with nucleotides at positions 32 and 38 to maintain decoding fidelity. Nucleic Acids Res. 2020; 48:6170–6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Olejniczak M., Uhlenbeck O.C.. tRNA residues that have coevolved with their anticodon to ensure uniform and accurate codon recognition. Biochimie. 2006; 88:943–950. [DOI] [PubMed] [Google Scholar]
- 68. Olejniczak M., Dale T., Fahlman R.P., Uhlenbeck O.C.. Idiosyncratic tuning of tRNAs to achieve uniform ribosome binding. Nat. Struct. Mol. Biol. 2005; 12:788–793. [DOI] [PubMed] [Google Scholar]
- 69. Murakami H., Ohta A., Suga H.. Bases in the anticodon loop of tRNAAlaGGC prevent misreading. Nat. Struct. Mol. Biol. 2009; 16:353–358. [DOI] [PubMed] [Google Scholar]
- 70. Ledoux S., Olejniczak M., Uhlenbeck O.C.. A sequence element that tunes Escherichia coli tRNA(Ala)(GGC) to ensure accurate decoding. Nat. Struct. Mol. Biol. 2009; 16:359–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Auffinger P., Westhof E.. Singly and bifurcated hydrogen-bonded base-pairs in tRNA anticodon hairpins and ribozymes. J. Mol. Biol. 1999; 292:467–483. [DOI] [PubMed] [Google Scholar]
- 72. Iwane Y., Kimura H., Katoh T., Suga H.. Uniform affinity-tuning of N-methyl-aminoacyl-tRNAs to EF-Tu enhances their multiple incorporation. Nucleic Acids Res. 2021; 49:10807–10817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Schrader J.M., Chapman S.J., Uhlenbeck O.C.. Understanding the sequence specificity of tRNA binding to elongation factor Tu using tRNA mutagenesis. J. Mol. Biol. 2009; 386:1255–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Schrader J.M., Uhlenbeck O.C.. Is the sequence-specific binding of aminoacyl-tRNAs by EF-Tu universal among bacteria?. Nucleic Acids Res. 2011; 39:9746–9758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003; 31:3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Serebrov V., Vassilenko K., Kholod N., Gross H.J., Kisselev L.. Mg2+ binding and structural stability of mature and in vitro synthesized unmodified Eschierichia coli tRNAPhe. Nucleic Acids Res. 1998; 26:2723–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Stein A., Crothers D.M.. Conformational changes of transfer RNA. The role of magnesium(II). Biochemistry. 1976; 15:160–168. [DOI] [PubMed] [Google Scholar]
- 78. Schroeder S.J., Turner D.H.. Optical Melting Measurements of Nucleic Acid Thermodynamics. 2009; 1st edn.Elsevier Inc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Mustoe A.M., Liu X., Lin P.J., Al-Hashimi H.M., Fierke C.A., Brooks C.L.. Noncanonical secondary structure stabilizes mitochondrial tRNASer(UCN) by reducing the entropic cost of tertiary folding. J. Am. Chem. Soc. 2015; 137:3592–3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Fahlman R.P., Dale T., Uhlenbeck O.C.. Uniform binding of aminoacylated transfer RNAs to the ribosomal A and P sites. Mol. Cell. 2004; 16:799–805. [DOI] [PubMed] [Google Scholar]
- 81. Ledoux S., Uhlenbeck O.C.. Different aa-tRNAs are selected uniformly on the ribosome. Mol. Cell. 2008; 31:114–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kilpatrick M., Walker R.. The nucleotide sequence of glycine tRNA from Mycoplasma mycoides sp. capri. Neuroscience. 1980; 8:2783–2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Denmon A.P., Wang J., Nikonowicz E.P.. Conformation effects of base modification on the anticodon stem-loop of Bacillus subtilis tRNATyr. J. Mol. Biol. 2011; 412:285–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Chang A.T., Nikonowicz E.P.. Solution nuclear magnetic resonance analyses of the anticodon arms of proteinogenic and nonproteinogenic tRNAGly. Biochemistry. 2012; 51:3662–3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request.