Abstract
During infection, the Legionnaires’ disease bacterium, Legionella pneumophila, survives and multiplies within a specialized phagosome that is near neutral pH and does not fuse with host lysosomes. In order to understand the molecular basis of this organism’s ability to control its intracellular fate, we have isolated and characterized a group of transposon-generated mutants which were unable to kill macrophages and were subsequently found to be defective in intracellular multiplication. These mutations define a set of 20 genes (19 icm [for intracellular multiplication] genes and dotA [for defect in organelle trafficking]). In this report, we describe a quantitative assay for phagosome-lysosome fusion (PLF) and its use to measure the levels of PLF in cells that have been infected with either wild-type L. pneumophila or one of several mutants defective in different icm genes or dotA. By using quantitative confocal fluorescence microscopy, PLF could be scored on a per-bacterium basis by determining the extent to which fluorescein-labeled L. pneumophila colocalized with host lysosomes prelabeled with rhodamine-dextran. Remarkably, mutations in the six genes that were studied resulted in maximal levels of PLF as quickly as 30 min following infection. These results indicate that several, and possibly all, of the icm and dotA gene products act at an early step during phagosome establishment to determine whether L. pneumophila-containing phagosomes will fuse with lysosomes. Although not ruled out, subsequent activity of these gene products may not be necessary for successful intracellular replication.
Legionella pneumophila (the Legionnaires’ disease bacterium) is a gram-negative bacterium that grows within a wide range of phagocytic host cells, ranging from freshwater amoebae to mammalian macrophages (Mφs). Although the cytological events that occur during infection of human Mφs have been described in great detail, there is little information about the molecular basis for the ability of this organism to survive and replicate within host cells. Infection of human Mφs by L. pneumophila appears to occur by a sequential multistep process (for reviews, see references 25, 31, and 39). Following attachment to complement receptors, the bacteria are engulfed either by a single pseudopod that coils around the bacterium (20) or by conventional phagocytosis (35). In the former case, the multilayered membrane of the coiled pseudopod resolves to form a phagosome. During this phagosome formation, a sorting of plasma membrane proteins occurs so that some markers are excluded from the phagosomal membrane while others are included (7, 8). The phagosome containing live wild-type L. pneumophila does not acidify below pH 6, nor does it fuse with host lysosomes (19, 21). Instead, this Legionella-specific phagosome (LSP) goes through a series of intracellular trafficking events, associating sequentially with smooth vesicles, mitochondria, and ribosomes (18). After 4 to 6 h postinfection, the LSP becomes ribosome studded and seems to be surrounded by rough endoplasmic reticulum, as indicated by the presence of the endoplasmic reticulum luminal marker BiP (18, 42). At approximately 10 h postinfection, L. pneumophila begins to multiply inside the LSP. Eventually, the host cell is lysed, releasing L. pneumophila which can then initiate new rounds of infection.
Genetic analysis has provided some information about the bacterial genes that are required for the ability of L. pneumophila to specifically replicate within human Mφs. Two regions within the L. pneumophila genome have been identified which encode a total of 20 genes (19 icm genes and dotA) that are dispensable for growth on bacteriologic media but are required for intracellular replication and host cell killing (2, 4, 29, 34, 37, 38). These genes are likely to encode functions that are important for the intracellular events described above.
Two models can be considered for how the icm and dotA genes may act during these intracellular events. In a sequential model, subsets of the genes would be expressed temporally so that L. pneumophila may direct its phagosome to maintain nearly neutral pH; prevent phagosome-lysosome fusion (PLF); associate sequentially with smooth vesicles, mitochondria, and ribosomes; and promote bacterial replication. Alternatively, in a concerted model, all the genetic information in the bacteria is expressed prior to infection so that the incoming L. pneumophila can form a specialized phagosome that possesses all the properties needed for intracellular life. Subsequent events would then be dictated by the features of the LSP that are established during its formation or shortly thereafter. These models should be distinguishable by analysis of the icm and dotA mutants that fail to replicate within Mφs, since each mutant should be defective in some step in infection.
In this paper we describe measurements of PLF for cells infected with either wild-type L. pneumophila or strains with mutations in five different icm genes and the dotA gene. We find that in contrast to the cells infected with wild-type L. pneumophila, cells infected with the mutants exhibited maximal levels of PLF as rapidly as 30 min following infection. Indeed, the levels of PLF of cells infected with the mutants were indistinguishable from the levels of PLF observed in cells infected with paraformaldehyde (Para)-killed bacteria, indicating that the mutants had no measurable ability to prevent PLF. These results are consistent with a concerted model of gene expression in which many or all of the icm and dotA gene products act together either during phagosome formation or shortly thereafter to produce an LSP compartment that does not fuse with lysosomes. For L. pneumophila, it appears that early events in Mφ infection, phagosome formation, and/or establishment determine its intracellular fate.
MATERIALS AND METHODS
Reagents.
5,6-Carboxyfluorescein succinimidyl ester (FSE) (catalog no. C-1311) and tetramethylrhodamine dextran (Rh-dextran) (70,000 molecular weight [MW], lysine fixable) (catalog no. D-1818) were purchased from Molecular Probes (Seattle, Wash.). Also purchased were RPMI 1640 medium lacking l-glutamine (RPMI) (JRH Biosciences, Lenexa, Kans.), l-glutamine (Gln) (Mediatech, Washington, D.C.), and agarose (IBI, New Haven, Conn.). Normal human serum (NHS) was obtained from healthy male volunteers and stored in 5-ml aliquots at −80°C. Fetal calf serum (FCS) (catalog no. F-2442), poly-d-lysine hydrobromide (200,000 MW) (catalog no. P-1149), and all other reagents and chemicals and were purchased from Sigma (St. Louis, Mo.). FCS was incubated at 56°C for 30 min to inactivate the complement. Phosphate-buffered saline (PBS) was prepared as 137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, and 1.4 mM KH2PO4 (pH 7.2). M63 salts contain 22.0 mM KH2PO4, 40.2 mM K2HPO4, 14.6 mM (NH4)2SO4, and 500 nM FeSO4 (pH 6.5).
Bacterial strains.
All bacteria were derived from L. pneumophila Philadelphia-1 and are described in Table 1.
TABLE 1.
Strains of L. pneumophilaa
| Strainb | Relative LD50 | Phenotype and genotype | Refer- ence(s) |
|---|---|---|---|
| JR32 | 1 | Mak+ | 37 |
| 25D | >500,000 | Mak− | 17 |
| LELA1802 | >500,000 | Mak−icmX::Tn903dIIlacZ | 34, 37 |
| LELA1984 | 13,000 | Mak−icmE::Tn903dIIlacZ | 34, 37 |
| LELA2474 | 10,000 | Mak−icmU::Tn903dIIlacZ | 34, 37 |
| LELA2955 | >500,000 | Mak−icmX::Tn903dIIlacZ | 34, 37 |
| LELA3118 | >500,000 | Mak−dotA::Tn903dIIlacZ | 34, 37 |
| LELA3150 | 32,000 | Mak−icmB::Tn903dIIlacZ | 34, 37 |
| LELA3278 | 14,000 | Mak−icmR::Tn903dIIlacZ | 37, 38 |
| LELA4004 | >500,000 | Mak−icmX::Tn903dIIlacZ | 34, 37 |
Relative LD50s, phenotypes, genotypes, and strain designations are taken from reference 37. Relative LD50s relate to the number of L. pneumophila organisms that are needed to kill one-half of a monolayer of Mφ-like HL-60 cells. Defects in macrophage killing are designated Mak−.
All LELA strains are derived from JR32 and contain one Tn903dIIlacZ insertion in the indicated gene.
Bacterial viability assay.
Propidium iodide (PI) is known to intercalate with DNA (23) in the nuclei of dead cells (28). Its exclusion by L. pneumophila was used as a rapid measurement of bacterial viability. A 40-μl suspension of L. pneumophila at approximately 109 bacteria per ml in M63 salts was added to 40 μl of a 100-μg/ml solution of PI in water. After a 15-min incubation at room temperature, 1.0 ml of M63 salts was added. The solution was centrifuged, the supernatant was removed, and the bacteria were resuspended in 1.0 ml of M63 salts. Bacteria were then visualized by epifluorescence and phase-contrast microscopy to determine the number of red fluorescent bacteria and the total number of bacteria, respectively (Fig. 1H, I, and J). The number of red fluorescent bacteria divided by the total number of bacteria was taken as a measure of cell death.
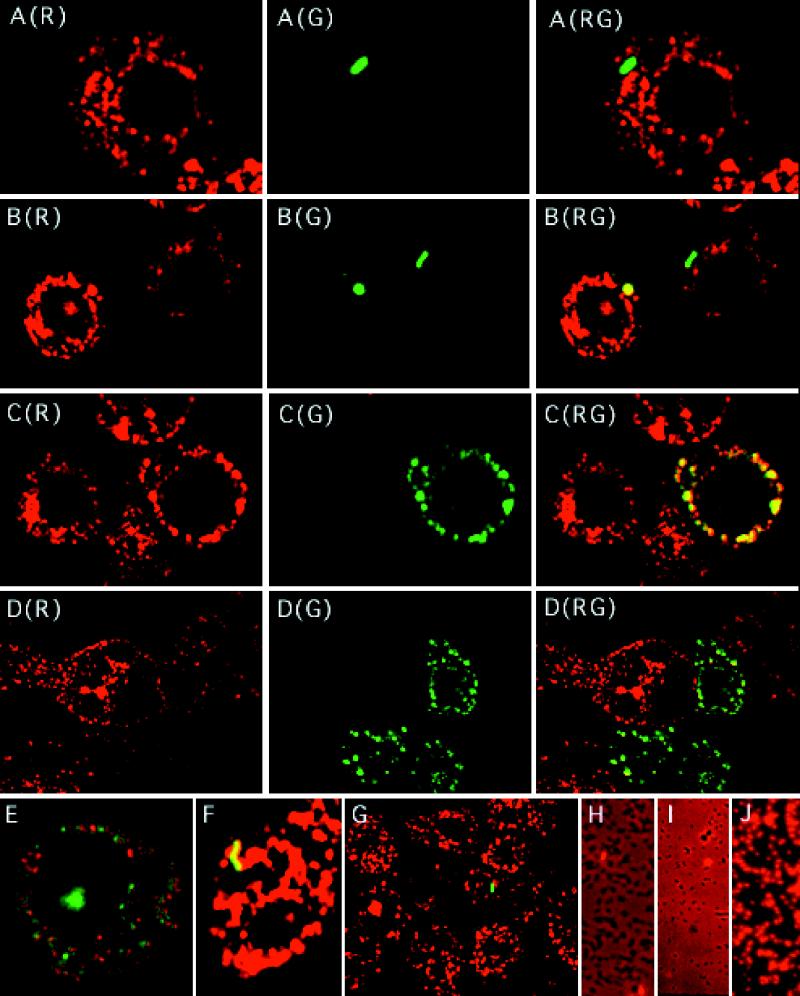
FIG. 1.
Fluorescence images of PLF in macrophage-like U937 cells (A to G) and L. pneumophila stained with PI (H to J). (A to G) U937 cells were incubated with Rh-dextran (70,000 MW, lysine fixable) for 40 h, washed, and then incubated for 1.0 h in RPMI medium to chase the Rh-dextran into lysosomes. U937 cells were then infected with various strains of FL-labeled L. pneumophila at a final concentration of 5 × 107 per ml for 0.5 h at 37°C. The differentiated U937 cells were washed again, covered with agarose, and then fixed with 4% Para in PBS immediately or after an additional 5.5 h of incubation at 37°C. Shown are confocal microscopy images of infected U937 cells. (A) Wild-type strain JR32 after 6 h in a phagosome which has not fused with host lysosomes (R575/530 = 0.178, compared to FL-labeled L. pneumophila RFL = 0.319 ± 0.058); (B) LELA2955 (icmX) after 0.5 h in one phagosome which has fused (R575/530 = 1.18 at the edge of one Mφ-like cell) while another phagosome has not fused (R575/530 = 0.243) (compare both values to RFL = 0.299 ± 0.045); (C) LELA1984 (icmE) after 6 h in a phagosome that fused with lysosomes and whose contents have been degraded and dispersed to form Mv-Mφ in which the FL is colocalized with Rh (of 22 defined spots, all had R575/530 values of >0.88, compared to FL-labeled L. pneumophila RFL = 0.264 ± 0.049); (D) LELA3118 (dotA) after 6 h in phagosomes that have fused with lysosomes and whose contents have been degraded and dispersed to form Mv Mφs whose FL was occasionally colocalized with Rh (greater than one-half of the punctate green vesicles in each cell have associated R575/530 values of less than the τ value [RFL + four SDs]); (E) LELA3118 (dotA) after 6 h in a phagosome which has fused and whose contents have been degraded to form an Mv Mφ containing discrete green punctate vesicles (R575/530 values were less than the τ value) among Rh-dextran-containing lysosomes; (F) LELA1984 (icmE) after 6 h in a phagosome which has fused with a lysosome and is completely filled with Rh-dextran (R575/530 = 1.80, compared to RFL = 0.264 ± 0.049); and (G) wild-type JR32 after 6 h in a phagosome which has not fused (R575/530 = 0.317, compared to RFL = 0.235 ± 0.053) and in a typical field of sparsely infected macrophages. Red indicates lysosomes containing Rh-dextran, green indicates FI-labeled whole bacteria or their remnants, and yellow indicates colocalization of the two markers. Panels A through D display single images of cross-talk-corrected I575-B (R) and I530-B (G) as well as their composite images (RG). Composite images of cross-talk-corrected I575-B and I530-B are shown in panels E through G. (H to J) L. pneumophila JR32 was grown for 2 days on ABCYE medium, suspended in phosphate buffer, and either not labeled (H), FL labeled and incubated overnight on ice (I), or FL labeled, incubated overnight on ice, and then incubated for 5 min in phosphate buffer containing 25% ethanol and 25% acetone (J). Bacteria were then incubated with PI, washed, and photographed under epifluorescence and phase-contrast microscopy with a Nikon Labophot UFX camera system. Shown are double exposures of L. pneumophila, which appears as dark rods. Red indicates nonviable L. pneumophila that stains positively with PI and represents 2, 2, and 100% of the bacteria in panels H, I, and J, respectively.
Preparation of FL-labeled L. pneumophila.
Two- and 3-day-old cultures of L. pneumophila strains were scraped from ACES [N-(2-acetamido)-2-aminoethanesulfonic acid)-buffered charcoal-yeast extract (ABCYE) agar plates (11) and then suspended and washed three times in 100 mM potassium phosphate (pH 8.0). Right before use, solutions containing 50, 16.7, 5.6, and 1.9 mg of FSE per ml were made in dimethyl sulfoxide. The FSE solution (10 μl for each concentration) was added to approximately 1010 bacteria in 1.0 ml of 100 mM potassium phosphate (pH 8.0). The bacterium-FSE mixtures, in 1.6-ml plastic centrifuge tubes, were periodically inverted throughout a 20-min incubation at room temperature. After being labeled, the bacteria were washed five times in M63 salts and stored in the same solution overnight on ice.
Epifluorescence was used to determine which bacteria were suitably labeled. The fluorescein (FL) fluorescence of L. pneumophila from each of the labeling reactions was visually compared to that of other FL-labeled L. pneumophila that had previously gave an adequate intensity of FL fluorescence in the PLF assay described below.
On the following day, the viability of the selected FL-labeled bacteria was determined on the basis of PI exclusion. Only those bacteria that were greater than 97% viable were suspended, at 108 per ml, in RPMI with 2 mM Gln and 10% NHS and used to infect monolayers of differentiated U937 cells.
Properties of FL-labeled L. pneumophila.
The derivatization of L. pneumophila with FI did not reduce its viability or cytopathogenicity. All of the FL-derivatized rod-shaped bacteria observed by phase-contrast microscopy emitted green fluorescence (data not shown) and formed colonies on ABCYE medium (1.08 ± 0.17 CFU per particle) to the same extent as non-FL-labeled L. pneumophila (0.94 ± 0.24 CFU per particle). The percentage of L. pneumophila that failed to exclude the dye PI after FL labeling (1.9% ± 1.3%) was not appreciably different than that before labeling (2.3% ± 1.4%), indicating that the labeling procedure affected neither the integrity of the bacterial membrane nor viability (Fig. 1H, I, and J). The FL labeling also did not alter the viability of any of the mutant L. pneumophila strains as judged by PI exclusion.
Labeling of L. pneumophila with FL did not alter its ability to carry out productive infections in Mφ-like cells. Wild-type FL-labeled L. pneumophila JR32 killed monolayers of differentiated HL-60 and U937 cells (data not shown) like its non-FL-labeled counterpart as measured in a cytotoxicity assay (30). Moreover, wild-type FL-labeled L. pneumophila JR32 multiplied in monolayers of differentiated U937 and HL-60 cells with the same growth kinetics and to the same extent as nonlabeled bacteria (data not shown) as measured in a growth assay (45). Finally, both FL-labeled and nonlabeled wild-type L. pneumophila JR32 began to multiply at 10 h postinfection (4 h after the last PLF time point was taken) as determined by a synchronous infection growth assay (45). Thus, FL-labeled L. pneumophila is apparently able to enter, survive in, and multiply in differentiated U937 cells like the nonlabeled bacteria.
Preparation of FL-labeled Para-killed L. pneumophila.
FL-labeled L. pneumophila JR32 was washed three times and suspended in 0.5 ml of 100 mM potassium phosphate (pH 8.0). A 0.5-ml solution of 4% Para was mixed with the bacteria, and the solution was incubated for 30 min at room temperature. The reaction was quenched by adding 50 μl of 1 M NH4Cl. The bacteria were then resuspended in PBS containing 50 mM NH4Cl and incubated at room temperature for an additional 5 min. Finally, the bacteria were washed three times in M63 salts before being suspended at 108 per ml in RPMI with 2 mM Gln and 10% NHS. Para-treated JR32 (JR32-Para) was nonviable (less than 10−9 CFU per bacterial particle) and permeable to PI (data not shown).
Cell culture.
The human leukemia cell lines HL-60 (9) and U937 (41) were maintained in RPMI supplemented with 2 mM Gln and 10% heat-inactivated FCS at 37°C under 5% CO2–95% air. For L. pneumophila cytotoxicity, growth, and synchronous growth assays, HL-60 and U937 cells were differentiated into Mφ-like cells by incubating them for 2 days with 10 ng of phorbol 12-myristate 13-acetate per ml in RPMI with 2 mM Gln and 10% NHS (15). Adherent cells were washed three times with RPMI containing 2 mM Gln and then incubated in RPMI with 2 mM Gln and 10% NHS prior to infection.
Preparation, infection, and fixation of U937 cells for confocal microscopy.
For the PLF assay, monolayers of differentiated U937 cells were made by harvesting the cells at 1 × 106 per ml and resuspending the cells at 7.5 × 105 per ml in RPMI with 2 mM Gln, 10% NHS, 10 ng of phorbol 12-myristate 13-acetate per ml, and 1 mg of Rh-dextran per ml. The cell suspension (0.2 ml) was placed on a poly-d-lysine-coated glass coverslip (22 by 22 mm; no. 1, Gold Seal; Becton-Dickinson Labware) that had been first mounted on the bottom of a 35- by 10-mm culture dish (Corning 25000) with a 12-mm-diameter hole punched through the bottom. The coverslips were affixed to the plastic plates by using a mixture of paraffin and petroleum jelly at a 3-to-1 ratio. After a 40-h incubation under 5% CO2–95% air at 37°C, adherent cells were washed three times with RPMI containing 2 mM Gln at 37°C and then incubated in 100 μl of RPMI with 2 mM Gln and 10% NHS for 1 h under 5% CO2–95% air at 37°C.
The kinetics of PLF were measured after infecting U937 cells for 30 min. The differentiated U937 cells were infected by the addition of 100 μl of FL-labeled bacteria at 1.0 × 108 per ml in RPMI with 2 mM Gln and 10% NHS to produce a final concentration of 5 × 107 bacteria per ml. After a 0.5-h incubation under 5% CO2–95% air at 37°C, nonadherent bacteria were removed by washing the U937 monolayers three times with RPMI containing 2 mM Gln at 37°C. The infected U937 cells were then immobilized by adding 200 μl of RPMI with 2 mM Gln, 10% NHS, and 0.8% agarose at approximately 40°C over the monolayer. The overlay prevented the loss of infected U937 cells that tended to detach from glass coverslips after prolonged incubations and also limited the free diffusion of any remaining extracellular L. pneumophila as demonstrated in plaque assays (12, 30).
For those infected U937 cells that were to have an additional incubation at 37°C under 5% CO2–95% air, approximately 0.5 ml of RPMI with 2 mM Gln and 10% NHS was placed on top of the agarose plug to prevent its desiccation. To fix the cells for microscopy, any medium in the culture dish was first aspirated. Approximately 3 ml of an ice-cold solution of PBS containing 4% Para was then added to the culture dish, and the cells were incubated for at least 12 h at 4°C. Finally, the Para-PBS solution was aspirated and replaced by 3 ml of PBS.
Preparation of FL-labeled L. pneumophila for confocal microscopy.
A 20-μl solution of FL-labeled L. pneumophila at 1010 per ml in RPMI with 2 mM Gln and 10% NHS was mixed with 200 μl of 2.0% agarose at approximately 40°C. The bacterial suspension was placed on coverslips previously mounted on the bottom of a culture dish with a hole punched out. Bacteria in the solidified agar were fixed and handled like the infected U937 cells described above.
Image acquisition.
Fixed, infected U937 cells were examined by using a laser scanning confocal microscope (MRC 600; Bio-Rad Microscience, Cambridge, Mass.) on an inverted microscope (Axiovert; Zeiss, Oberkochen, Germany) with a 63× (numerical aperture, 1.4) Zeiss Plan-Apo infinity-corrected objective. Light from a 25-mW argon laser was passed through a discriminating filter (488 DF 10) to illuminate infected U937 cells. Emitted fluorescence was then passed through a dichroic reflector (DR 510 LP) and split into two beams by another dichroic reflector (DR 560 LP). One beam was passed through a discriminating filter (530 DF 30) and the other was passed through a long-pass filter (EF 575 LP) to simultaneously generate dual fluorescence images, I530 and I575, respectively. Images were recorded in fields where the FL fluorescence emanated from within U937 cells. This was verified by focusing up and down during image collection to ensure that the FL fluorescence was surrounded by punctate Rh fluorescence produced by the Rh-dextran that labeled the lysosomes. The average from eight scans was used to produce the final digitized images.
Determination of the distribution of L. pneumophila in infected U937 cells.
Monolayers of differentiated U937 cells whose lysosomes were prelabeled with Rh-dextran were infected with twofold increasing concentrations (ranging from 4 × 106 to 1 × 109 bacteria per ml [final concentration]) of FL-labeled wild-type L. pneumophila JR32. After a 0.5-h incubation, monolayers were washed three times with RPMI containing 2 mM Gln to remove nonadherent bacteria, fixed with 4% Para in PBS, and then examined by confocal microscopy. The observed number of internalized L. pneumophila organisms per Mφ was determined by adding the number of fluorescent FL-labeled L. pneumophila organisms among the punctate Rh fluorescence that delineated the extent of the Mφ’s cytoplasm. The expected number of internalized bacteria per Mφ was calculated by multiplying the total number of Mφs examined by the Poisson probability distribution function P(n) = (mn/n!)(e−m), where n equals the number of L. pneumophila organisms internalized per Mφ and m equals the total number of observed intracellular L. pneumophila organisms divided by the total number of Mφs examined. The distribution of L. pneumophila within the Mφs matched a Poisson distribution as determined by chi-square analysis to measure fit (P > 0.35 in one infection and P > 0.99 in the other eight infections). At the concentration of FL-labeled L. pneumophila used in our PLF assay, approximately 7% of the Mφs are calculated to contain two or more bacteria. Thus, in any given experiment, 10% or fewer of the Mφs with internalized FL-labeled L. pneumophila will be erroneously scored as fused when multiply infected Mφs (7%) and degraded dead bacteria (maximally 3% as determined by PI exclusion) are taken into account.
Image analysis.
Image processing was done with an image processor (Gould-Vicom IP8000; VICOM Visual Computing, Freemont, Calif.) run on a microVAX minicomputer (Digital Equipment Corporation, Maynard, Mass.). Digitized I530 and I575 had the background (B) subtracted to obtain background-corrected images, I530-B and I575-B, respectively, as described previously (32, 33). To resolve FI-labeled bacteria or any intracellular compartments containing FI, an automated procedure was used to remove dim pixels in the I530-B whose brightness was less than 40% of the brightest pixels found in the same spot (32). Spots defined in the I530-B were used to create a mask of the same areas in the I575-B. Pixels within these areas that have nonzero intensity values comprised the final spots used for analysis. The intensity values of the pixels within each trimmed spot of the I575-B were summed and divided by the sum of the corresponding pixel intensity values in the I530-B to obtain a ratiometric (R575/530) value for each spot. Spots were excluded from analysis if they were composed of fewer than 20 pixels, since the reliability of R575/530 values decreased rapidly as the size of the spot diminished below this number of pixels (data not shown). Detector saturation was avoided by excluding from analysis spots in either the I530 or the I575 that contained a pixel whose value was greater than 240 (of a maximum of 255).
Scoring of PLF.
PLF was scored for both intact and degraded bacteria. For intact bacteria within Mφs, the R575/530 values of spots corresponding to FL-labeled L. pneumophila were used to determine whether PLF had occurred. Filter sets were chosen so that FI fluorescence would register in both the I530 and the I575. Thus, a mean ratiometric value of FL fluorescence (RFI) and a standard deviation (SD) could be calculated simply by obtaining R575/530 values of hundreds of FL-labeled L. pneumophila. The RFL value depended on the acquisition settings and was measured in each experiment. Since cellular autofluorescence was negligible, an increase in the R575/530 value of a spot that defined a bacterium was due to the presence of coincident lysosomal Rh-dextran. Because the R575/530 values assigned to FL-labeled L. pneumophila fit a normal distribution (see Fig. 2A), a confidence value could be calculated based on the SD. Fusion was scored when the R575/530 value exceeded the high stringency value of four SDs above the RFL value (τ), thus ensuring that a negligible number (fewer than 0.01%) of spots that corresponded to nonfused FL-labeled L. pneumophila will have, by statistical chance, an R575/530 value that is higher. Studies with unlabeled cells demonstrated that the increase in R575/530 values associated with FL-labeled L. pneumophila was due to its colocalization with Rh-dextran, whose fluorescence is registered almost exclusively in the pixels comprising the I575.
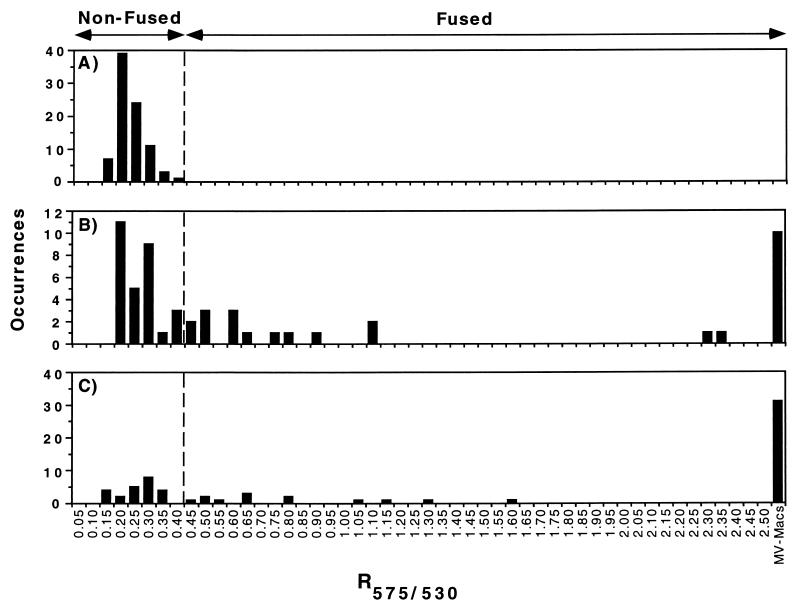
FIG. 2.
Histograms of the R575/530 values obtained from one PLF experiment with FL-labeled L. pneumophila 25D (avirulent) alone (A) or within Rh-dextran-labeled U937 cells after an incubation of 0.5 h, where 38% had fused to lysosomes (B), and 6 h, where 64% had fused (C). The calculated τ value (RFL + four SDs) is indicated by the dotted vertical lines. FL-labeled L. pneumophila organisms with associated R575/530 values greater than the τ value were counted as fused, while those with values equal to or less than τ were counted as nonfused. Mv Mφs (MV-Macs) displaying many FL-containing compartments, presumably arising from the degradation of an FL-labeled bacterium, could be counted as a fusion event, since the number of Mφs that were multiply infected was low (calculated to be 7%; see Materials and Methods).
FL-labeled L. pneumophila bacteria that were degraded after their phagosome fused with lysosomes could also be accurately counted based on statistical inference for the bacterial population infecting U937 cells and the retention of FL fluorescence within these Mφs. First, the number of infecting L. pneumophila organisms within Mφs was distributed in a Poisson fashion. The small fraction of Mφs infected (typically 10%) in a given PLF experiment relates to a low probability (7%) of an infected Mφ containing two or more L. pneumophila organisms. Thus, any Mφs containing more than one punctate vesicle of FL have a high (93%) probability of arising from a single bacterium which had been degraded. Second, the FL fluorescence from degraded bacteria was retained within the Mφ and did not spread to surrounding cells (Fig. 1C and D) (see below). Typically, many punctate vesicles were formed after FL-labeled L. pneumophila was degraded (Fig. 1C, D, and E) (see below). Mφs containing these FL-containing compartments were called multivesiculated (Mv) Mφs and appeared yellow [Fig. 1C(RG)] or green [Fig. 1D(RG)], depending on how much Rh-dextran colocalized with these compartments.
By using the method described above, PLF was scored for individual bacteria that gained entry into U937 Mφs. The fraction of L. pneumophila that had fused with lysosomes at any given time after infection was calculated as (fused L. pneumophila + Mv Mφs)/(fused L. pneumophila + Mv Mφs + nonfused L. pneumophila).
Statistical analysis.
The probabilities and statistical significance for chi-square analysis, paired two-tailed Student’s t test, SD, normal distribution, and Poisson distribution were calculated as described by Remington and Schork (36). Probability (P) values of less than 0.05 or 0.001 were judged significant or highly significant, respectively.
RESULTS
Parameters affecting colocalization of phagosomal and lysosomal markers as an indicator of PLF.
To compare the abilities of wild-type and mutant L. pneumophila strains to inhibit PLF, a quantitative colocalization assay was developed that measures the presence of a lysosomal marker within phagosomes containing individual bacteria. For two reasons, confocal fluorescence microscopy was used to determine colocalization. First, phagosomes and lysosomes containing different fluorophores can be viewed within a thin focal plane. Second, digitized images can be acquired and used for quantification. To mark phagosomes, L. pneumophila was covalently labeled with the fluorescent dye carboxy-FL. The FL-labeled L. pneumophila bacteria were then internalized by U937-derived Mφs. The lysosomes of these Mφs were preloaded with Rh-dextran for 40 h prior to infection. The Rh-dextran is taken up by fluid phase endocytosis and delivered to lysosomes (14). PLF could then be scored by determining the fraction of phagosomes containing FL-labeled L. pneumophila that had colocalized with lysosomes bearing Rh-dextran. To make the scoring of FL and Rh colocalization objective and to make its detection more sensitive, fluorescence images were digitized and processed to quantify each fluorophore. Thus, small amounts of Rh-dextran added incrementally to phagosomes containing FL-labeled L. pneumophila upon transient fusions with lysosomes could be measured (10).
Before PLF could be measured on a per-bacterium basis, several parameters needed to be established: (i) all of the L. pneumophila organisms were labeled with FL, (ii) the labeling did not kill L. pneumophila or interfere with the ability of the wild type to multiply within and kill Mφs, (iii) a low multiplicity of infection was used to minimize multiple infections so that Mφs containing many discrete FL-containing compartments could be scored as a single PLF event (such Mφs arise when an FL-labeled bacterium within a phagolysosome is degraded and fragments are distributed throughout the cell [see below]), (iv) the infection was performed for a discrete period of time (30 min) so that kinetics of PLF could be monitored, and (v) measurements were made before the internalized bacteria multiplied, which would have interfered with counting the initial number of bacteria that had been taken up by the Mφ (see Materials and Methods for a detailed description of each parameter).
Colocalization and dispersal of FL-labeled bacterial fragments as a measure of PLF for intact and digested FL-labeled L. pneumophila.
To characterize the behavior of the FL-labeled L. pneumophila and the Rh-labeled dextran within Mφs, we took advantage of the fluorescence emission of FL, which is greater at 530 nm than at 575 nm. Images of fixed U937 cells, in which only a few cells (approximately 10%) were infected (Fig. 1G shows for a representative field), were acquired both at 530 nm (I530), which is near the FL emission maximum, and at 575 nm (I575), which is near the Rh emission maximum but which also contains some FL fluorescence. Thus, FL-labeled L. pneumophila organisms that are observed will occupy areas at identical pixel locations in each image. Consequently, a ratiometric (R575/530) value can be assigned to these areas or spots by simply dividing the total fluorescence power of one spot identified in the processed I575 by the total fluorescence power of the corresponding spot found in the processed I530 (see “Image analysis” in Materials and Methods). In each experiment, a standard mean ratiometric value for fluorescein (RFL) was determined by averaging the R575/530 values of many FL-labeled L. pneumophila bacteria. A threshold value, τ, which equals the RFL value of FL plus four SDs, was then calculated (Fig. 2A). The τ value established the criterion used to score PLF. When there was no colocalization (i.e., R575/530 ≤ τ), the phagosomes containing L. pneumophila were scored as not fused [Fig. 1A(RG) and G]. In contrast, when the two fluorescent markers colocalized (i.e., R575/530 > τ), phagosomes containing FL-labeled L. pneumophila were scored as fused [Fig. 1B(RG) and F]. The stringent τ value also guaranteed that fewer than 0.01% of the spots will have, by statistical chance, an R575/530 value that is greater than τ and be scored as a fusion event.
Although colocalization of FL-labeled L. pneumophila and Rh-dextran might be due either to the bacteria binding residual Rh-dextran that may have been present during its internalization or to the uptake of Mφ plasma membrane components with trace amounts of Rh-dextran, these possibilities seem unlikely. FL-labeled L. pneumophila that internalized in the presence of 1 mg of extracellular Rh-dextran per ml formed phagosomes with associated R575/530 values (0.538 ± 0.069) that were almost identical to the RFL values (0.529 ± 0.064) corresponding to isolated FL-labeled L. pneumophila. These results indicate that very little extracellular or plasma membrane-bound Rh-dextran is taken into the phagosome, and this therefore cannot account for the increase in R575/530 values seen in our colocalization experiments. We conclude that colocalization (i.e., R575/530 > τ) is indicative of PLF (Fig. 2).
In the process of collecting images, we observed many instances of Mv Mφs that contained a large number of FL-containing compartments. We infer that these represent the products of bacterial digestion following PLF [Fig. 1C(G), D(G), and E]. The Mv Mφ appearance ranged from yellow to green depending on the amount of Rh-dextran that colocalized with these FL-containing compartments [Fig. 1C(RG) and D(RG), respectively]. Infrequently, an Mv Mφ containing both green and yellow compartments was seen 6 h after infection (Fig. 1E). The Mv Mφs probably represent the products of a single PLF event and not multiple infection of an Mφ, for two reasons. First, these Mφs occur at frequencies much greater than the fraction of Mφs expected to be multiply infected (see Materials and Methods). Second, the number of FL-containing compartments found within these Mφs greatly exceeded the number of FL-labeled L. pneumophila organisms found in any Mφ that had been infected with even a 20-fold-greater concentration of bacteria (data not shown).
Mv Mφs did not arise from the uptake of extracellular FL that may have been released into the medium during the initial incubation with FL-labeled L. pneumophila. No low basal level of FL fluorescence was ever detected throughout the Mφ monolayers [Fig. 1A(G), B(G), C(G), and D(G)]. FL fluorescence also remained contained within Mφs that had degraded the FL-labeled L. pneumophila, since the FL fluorescence was never seen to spread into adjacent cells (Fig. 1C and D).
The Mv Mφs observed at 6 h postinfection also were not formed by the multiplication of FL-labeled L. pneumophila within the host cells. Replication of FL-labeled wild-type L. pneumophila JR32 in synchronously infected Mφs was not detectable until 10 h postinfection, a full two L. pneumophila intracellular generation times (2 h per generation) beyond the 6-h time point at which PLF was measured (data not shown). Thus, by objectively quantifying fusion in Mφs infected at a low multiplicity of bacteria, PLF could be measured on a per-bacterium basis. The total Mv Mφs plus intact internalized bacteria that had associated R575/530 values greater than the τ value represented the number of bacteria whose phagosomes fused with lysosomes. Histograms of PLF were then generated and used to calculate the fraction of individual FL-labeled L. pneumophila organisms that had either fused to a lysosome or fused and been degraded (Fig. 2B and C) (see Materials and Methods).
Kinetics of PLF for wild-type, mutant, and killed L. pneumophila.
In order to find out if measuring PLF by these criteria yielded results similar to those obtained by others with electron microscopy, we compared levels of PLF for three types of L. pneumophila as a function of time after infection: live wild-type strain JR32; live avirulent mutant 25D, which is known to be defective in preventing PLF; and Para-killed JR32 (17). Initial time course experiments indicated that measurements made 0.5 and 6 h after infection would suffice to differentiate the three samples (data not shown). The fractions of bacteria that fused at these times were calculated from histograms (Fig. 2), and the PLF assay was then repeated so that meaningful statistical comparisons could be made.
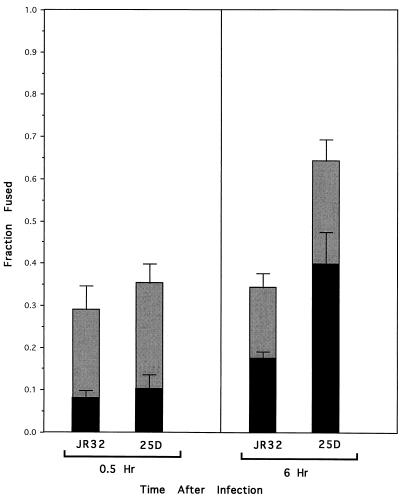
The fraction of phagosomes containing FL-labeled JR32 that fused with lysosomes 0.5 h after infection was 29%, significantly above the 10% background expected in the assay due to dead bacteria and multiple infections (Fig. 3) (see Materials and Methods). Approximately the same low level of fusion was also found when PLF was measured 5.5 h later (P = 0.28 by Students’ t test). Thus, 29% of phagosomes containing live wild-type L. pneumophila fuse with lysosomes early after infection. However, once the phagosome containing JR32 is established, no additional fusion seems to occur (Fig. 3).
FIG. 3.
Comparison of abilities of wild-type L. pneumophila JR32 and the avirulent mutant 25D to prevent PLF (gray bars) and form Mv Mφs (black bars) during an infection of U937 cells. Values represent the means from five experiments in which at least 50 Mφs containing intracellular FL fluorescence were scored per experiment at each of the indicated times after infection. Error bars indicate the standard errors of the means.
Interestingly, the fraction of phagosomes containing 25D that fused with lysosomes 0.5 h after infection was only slightly higher than that found for JR32 (Fig. 3), and the difference was not statistically significant (P = 0.090). Thus, at early times after infection, the majority of JR32 and 25D seem to form phagosomes that do not appreciably fuse with lysosomes.
However, at 6 h after infection, the fraction of phagosomes containing 25D that fused was significantly greater than the fraction of fused phagosomes containing either 25D at 0.5 h or JR32 at 6 h postinfection (P < 0.001 in both cases). The fraction of phagosomes containing 25D that fused did not appreciably increase with longer incubation times, indicating that maximal fusion was reached approximately 6 h after infection (data not shown). Bacteria in these fused phagosomes seem to be progressively degraded (Fig. 3). At 16 h postinfection, the fraction of Mv Mφs represented nearly all of the PLF events scored for 25D at 6 h postinfection (data not shown). This not only supports the idea that the maximal number of fused 25D phagosomes was reached at 6 h after infection but also supports the idea that PLF precedes Mv Mφs formation. Thus, in contrast to phagosomes containing JR32, phagosomes bearing 25D initially resist PLF but then lose this ability with time.
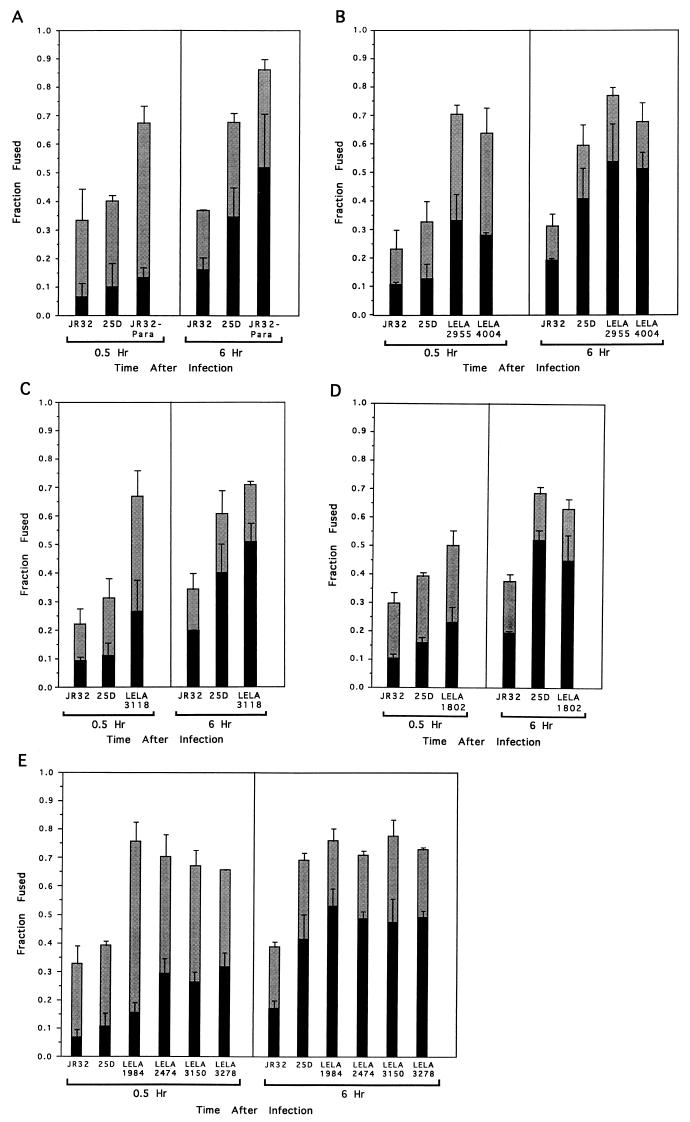
The levels of PLF were also measured for cells infected with FL-labeled L. pneumophila killed with Para (JR32-Para). Unlike phagosomes containing living wild-type JR32 and mutant 25D, phagosomes containing JR32-Para rapidly fused with lysosomes during the first 0.5 h of incubation (Fig. 4A). The high fraction (0.86 ± 0.04) of phagosomes containing JR32-Para that fused with lysosomes 6 h after infection was not statistically different (P = 0.31) from the fraction fused 0.5 h after infection (0.67 ± 0.06), confirming the complete inability of nonviable L. pneumophila to prevent PLF immediately after infection (19).
FIG. 4.
Comparison of JR32-Para (A) and the transposon-generated L. pneumophila mutants LELA2955 (icmX) and LELA4004 (icmX) (B), LELA3118 (dotA) (C), LELA1802 (icmX) (D), and LELA1984 (icmE), LELA2474 (icmU), LELA3150 (icmB), and LELA3278 (icmR) (E) with wild-type JR32 and the avirulent mutant 25D for the ability to prevent PLF (gray bars) and form Mv Mφs (black bars) in U937 cells. Values in each panel represent the means from three experiments in which at least 50 Mφs containing intracellular FL fluorescence were scored per experiment at each of the indicated times after infection. Error bars indicate the standard errors of the means.
Among these measurements, the largest fraction of phagosomes (0.86) that fused with lysosomes was that found for JR32-Para at 6 h after infection. Assuming that all phagosomes containing dead bacteria fuse with lysosomes, the remaining fraction of phagosomes containing apparently intact bacteria (0.14) may have fused with lysosomes containing little or no Rh-dextran. This is consistent with the fraction (approximately 0.10) of Mv-Mφs containing predominately green vesicles (Fig. 1D) that comprised the total fusion events of all strains of L. pneumophila (except JR32) at 6 h postinfection (data not shown). The high τ value used as a stringent criterion to measure fusion demands that a significant amount of Rh-dextran be colocalized with the FL-labeled L. pneumophila. With 0.86 as an upper limit for the fraction fused, essentially all of the phagosomes containing 25D seemed to have been fused with lysosomes 6 h after infection, since the fraction fused for 25D was not statistically different than that for JR32-Para (P = 0.23). Thus, the results obtained with this assay are in basic agreement with those obtained by using a completely different experimental system (17). In addition, we have obtained new information about the time dependence of fusion events that occur between phagosomes containing either wild-type L. pneumophila or the mutant 25D in Mφ-like cells that are mainly singly infected.
Phagosomes containing icm and dot mutants rapidly fuse with lysosomes.
As described in the introduction, we have characterized a total of 20 genes (icm and dotA) whose products are required for both intracellular multiplication and host cell killing. In order to find out whether these genes play a role in determining the ability of L. pneumophila to prevent PLF, we measured the levels of PLF for eight different mutants carrying insertion mutations in six of these genes. Instead of the low levels of PLF displayed by JR32 and 25D early after infection, seven of the eight mutants analyzed were found in phagosomes that rapidly fused with host lysosomes. Indeed, for L. pneumophila LELA2955 (icmX), LELA4004 (icmX), LELA3118 (dotA), LELA1984 (icmE), LELA2474 (icmU), LELA3150 (icmB), and LELA3278 (icmR), the fraction of PLF after 0.5 h of infection was as high as that for JR32-Para, ranging from 0.64 to 0.76 (Fig. 4B, C, and E). The fraction of PLF for these mutants at 0.5 h postinfection was significantly higher than the PLF observed for either JR32 or 25D (P < 0.05).
Like for the nonviable JR32-Para, the fraction of fused phagosomes did not increase appreciably with an additional 5.5 h of incubation (P values ranged between 0.09 and 0.97), indicating that all of the phagosomes containing these mutants fused within 0.5 h after being internalized. Moreover, the average fractions of fused phagosomes for these mutants after infection (0.68 ± 0.04 at 0.5 h and 0.73 ± 0.04 at 6 h) are similar to those for JR32-Para (0.67 ± 0.06 at 0.5 h and 0.86 ± 0.04 at 6 h), indicating a complete lack of ability to prevent fusion with lysosomes.
Early fusion with degradative lysosomal compartments is also suggested by the formation of Mv Mφs (Fig. 4). More Mv Mφs were found 0.5 h after infection with FL-labeled L. pneumophila LELA2955 (icmX), LELA4004 (icmX), LELA3118 (dotA), LELA1984 (icmE), LELA2474 (icmU), LELA3150 (icmB), and LELA3278 (icmR) than were found 0.5 h after infection with FL-labeled L. pneumophila 25D or JR32. With a 5.5-h longer incubation, the Mv Mφs comprised greater than 50% of the total fraction fused, a value similar to that found for JR32-Para.
Although seven of the eight mutants tested have no ability to prevent PLF within 0.5 h of infection, mutant LELA1802 (icmX) seems to maintain a marginal ability to prevent PLF (Fig. 4D). At 0.5 h after infection, Phagosomes containing FL-labeled LELA1802 (icmX) colocalized with lysosomes to a significantly higher degree (P = 0.0011) than phagosomes containing wild-type JR32, indicating that LELA1802 is defective in preventing PLF. However, although phagosomes containing LELA1802 (icmX) also colocalize with lysosomes more than phagosomes containing 25D after 0.5 h of infection, the difference was not statistically significant (P = 0.11), indicating that they may have a partial ability to prevent PLF, like 25D. At 6 h after infection, the fractions of phagosomes containing LELA1802 (icmX) and 25D that fused with lysosomes, 0.63 ± 0.04 and 0.68 ± 0.02, respectively, were statistically indistinguishable (P = 0.42) and similar in magnitude to that for JR32-Para (0.64 ± 0.04). Thus, LELA1802 (icmX) is also unable to inhibit PLF within 6 h but may possess a slight ability to prevent PLF early in infection.
These results indicate that strains with null mutations in six of the genes all exhibit the same phenotype, a total lack of ability to inhibit PLF. The fact that phagosomes containing these mutants are all fused with lysosomes as rapidly as 30 min following infection indicates that the decision about whether each phagosome will or will not fuse with lysosomes is determined during the initial encounter between the bacteria and the host cell. In addition, at least the six gene products tested here (those of icmX, dotA, icmB, icmR, icmU, and icmE) are essential for determining this key aspect of the intracellular fate of L. pneumophila.
DISCUSSION
Infection of host cells by L. pneumophila results in the formation of a specialized compartment (LSP) that is essential for intracellular bacterial survival and growth. Lack of fusion between the LSP and lysosomes is of paramount importance for L. pneumophila, since mutants that are unable to prevent PLF are unable to multiply intracellularly, do not kill host cells, and do not cause disease in animals (18, 19, 29). The experiments reported in this paper distinguish three types of L. pneumophila-containing phagosomes based on their ability to fuse with lysosomes. The first type consists of those that do not measurably fuse with lysosomes. These are exemplified by the 67% of the phagosomes containing wild-type L. pneumophila JR32 that never colocalized with the Rh-dextran marking the lysosomal compartments, even 6 h after infection. The second type, which initially resist fusion but eventually do fuse with lysosomes between 0.5 and 6 h postinfection consists of those exemplified by mutants 25D and LELA1802 (icmX). The third type contains strains that have no capacity to prevent PLF; these phagosomes have completely fused to lysosomes at some point prior to 0.5 h postinfection. These are exemplified by the phagosomes containing either Para-killed wild-type JR32 or the remaining transposon-generated mutant L. pneumophila strains.
Our results demonstrate that the ability to prevent PLF during or right after phagocytosis seems to require at least the products of the icmX, icmE, icmR, icmB, icmU, and dotA genes of L. pneumophila (Table 1). Mutants containing transposon insertions in these genes are as defective as non-viable JR32-Para in preventing PLF within 30 min of bacterial uptake. Since de novo protein synthesis is not required to prevent PLF, expression of the dotA and icm genes must occur before the phagocytosis of L. pneumophila (22). In this regard, the 29% of viable FL-labeled wild-type L. pneumophila JR32 organisms whose phagosomes fail to resist PLF may not sufficiently express the putative dotA and icm gene products. Such population heterogeneity may explain why L. pneumophila grown in Acanthamoeba castellani is more invasive for epithelial cells (6), and why coinoculation of L. pneumophila and Hartmannella vermiformis increases growth of L. pneumophila within the lungs of mice (5). Both types of cocultivations may enrich for L. pneumophila populations that sufficiently express the information encoded by the dotA and icm loci. Consequently, all the L. pneumophila would be able to prevent PLF and thus display increased invasiveness and bacterial multiplication, both of which are linked to increased virulence.
Within 30 min of infection, all dotA and icm L. pneumophila mutants tested here were found mainly in phagolysosomes. PLF seems to be permanently inhibited only by wild-type L. pneumophila that makes and maintains a proper LSP. Once formed, the LSP may then have all the determinants necessary to specify its subsequent intracellular trafficking. Thus, our results are most consistent with a concerted model of gene expression and do not support a model in which different icm gene products act at different stages of the infection process. If this had been the case, some of the mutants might have retained the ability to prevent PLF and been defective at a later step. While we cannot rigorously exclude the possibility that we inadvertently chose only those mutants that were defective in preventing PLF and that other mutants would retain this ability, we feel that this is unlikely because the mutations represent different regions within the two different loci containing the icm and dotA genes. Our model predicts that null mutations in all of the icm genes should cause the same rapid, high-level PLF phenotype.
Evidence for sequential gene activation, however, has recently been obtained (43). Based on finding distinct intracellular fates of bacterial mutants defective for intracellular growth, Swanson and Isberg concluded that “corresponding gene products are likely to act in different bacterial pathways” (43). These differing conclusions may be due to a variety of experimental differences, including differences in the viability of the mutants, differences due to the microscopy techniques used, and, finally, differences in the nature of the mutants analyzed. The mutants studied by Swanson and Isberg are likely the result of missense mutations generated by ethyl methanesulfonate mutagenesis and may display complex phenotypes as a result of partial activities of the mutated gene products (43). Indeed, some of the mutants they studied are reminiscent of 25D and LELA1802, which exhibit a partial ability to prevent PLF. Since approximately 6 h is needed for lysosomes to completely fuse with phagosomes containing 25D, it may have time to display special intracellular trafficking properties like the mutants studied by Swanson and Isberg. In contrast, the mutants analyzed in our study all contained a single, well-defined transposon insertion. The transposon insertion is the only mutation in the mutants, since moving the mutation to a fresh genetic background produces the same phenotypes (37).
Only one of the mutants that contained a well-defined mutation (transposon insertion) and was defective in killing Mφs, LELA1802 (icmX), seems to temporarily resist PLF at 0.5 h postinfection. Oddly, the transposon inserted in the icmX gene of LELA1802 is located between the other two transposon insertions found in LELA4004 and LELA2955. It seems that the truncated icmX gene product made only in LELA1802 confers enough activity to allow its phagosome to resist PLF. Nevertheless, this activity cannot be very strong, since within 6 h after infection, phagosomes containing LELA1802 fuse with host lysosomes. The phenotype exhibited by LELA1802 suggests that the activity of one icm gene (icmX) may be needed after phagosome formation. Moreover, the fact that the mutants containing icm and dotA disruptions are placed in phagosomes that readily fuse with lysosomes does not exclude the possibility that these genes function at a later time in the infection. Obviously, further experimentation on the activity of the dotA and icm genes during infection is needed.
Occasionally, Mv Mφs containing both green and yellow compartments were seen 6 h after infection (Fig. 1E). These Mv Mφs may have arisen after FL-labeled L. pneumophila fused with lysosomal compartments lacking Rh-dextran. The degraded bacterial contents may then have been distributed through a lysosomal network devoid of Rh-dextran. Alternatively, lower-molecular-weight molecules of FL (and its derivatives), thought to be liberated by the digestion of FL-labeled L. pneumophila in a phagolysosome, may partition from high-molecular-weight molecules like Rh-dextran (3, 44). The partitioning would form compartments containing only FL fluorescence despite the presence of lysosomes containing Rh-dextran.
Phagosomes bearing mutants LELA1984 (icmE), LELA3278 (icmR), LELA3150 (icmB), and LELA2474 (icmU), which displayed some ability to kill Mφs (50% lethal doses [LD50s] of less than 500,000 relative to that for wild-type strain JR32), seem to fuse as readily to lysosomes as phagosomes containing LELA1802 (icmX), LELA2955 (icmX), LELA3118 (dotA), and LELA4004 (icmX), whose Mφ killing ability is below the level of detection (relative LD50s of greater than 500,000) (Table 1). This indicates that L. pneumophila may have a killing mechanism which is independent of its ability to survive and multiply within Mφs. This killing ability, which seems to be present only when large numbers of L. pneumophila organisms are incubated with Mφ-like cells, may be due to a reported cytotoxin of L. pneumophila (13, 24).
Previously, investigators used electron microscopy to measure PLF by scoring the frequency at which recognizable bacteria colocalized with the lysosomal marker acid phosphatase or electron-dense particles that were allowed to accumulate in lysosomes before infection (19, 40). There are limitations in both of these methods. Since the bacteria are subjectively identified, underestimates of PLF will occur when lysosomal degradation obscures their morphological features. Indeed, complete degradation would eliminate that population of bacteria whose phagosomes fused with lysosomes. Additionally, PLF studies usually employed host cells that were highly infected with bacteria, which may make counting the degraded bacteria difficult.
The pathogen L. pneumophila subverts many host defense mechanisms in order to gain access to an environment conducive for its survival and growth. The fact that this environment is a phagosome within human macrophages presents the additional challenge of avoiding fusion to lysosomes, a trait shared by other intracellular pathogens (1, 16, 27). Although the molecular basis for PLF inhibition and regulation is unknown, bacterial entry seems to play an important role in determining the fate of intracellular pathogens. For example, Toxoplasma gondii and L. pneumophila, when coated with antibodies and allowed to infect cells expressing the Mφ-lymphocyte Fc receptor, will form phagosomes that fuse to lysosomes (19, 26). The icm and dotA genes of L. pneumophila therefore appear to control the properties displayed by the LSP and act early during its formation and/or its establishment.
ACKNOWLEDGMENTS
We are indebted to Mike Hillmeyer for assistance in computer programming and maintenance. For helpful discussion, we thank John Presley, Moira Lawson, Nadia Khelef, Cynthia Panagiotidis, and David Figurski. For unpublished sequence information, we thank Mary Purcell and Laura Hales. We also thank Carmen Rodriguez for technical assistance.
This work was supported by National Research Service Award AI-08299 (to L.A.W.) and National Institutes of Health grants AI-2354 (to H.A.S.) and DK27083 (to F.R.M.).
REFERENCES
- 1.Armstrong J A, Hart P D. Phagosome-lysosome interactions in cultured macrophages infected with virulent tubercle bacilli. J Exp Med. 1975;142:1–16. doi: 10.1084/jem.142.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger K H, Isberg R R. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol Microbiol. 1993;7:7–19. doi: 10.1111/j.1365-2958.1993.tb01092.x. [DOI] [PubMed] [Google Scholar]
- 3.Berthiaume E P, Medina C, Swanson J A. Molecular size-fractionation during endocytosis in macrophages. J Cell Biol. 1995;129:989–998. doi: 10.1083/jcb.129.4.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brand B C, Sadosky A B, Shuman H A. The Legionella pneumophila icm locus: a set of genes required for intracellular multiplication in human macrophages. Mol Microbiol. 1994;14:797–808. doi: 10.1111/j.1365-2958.1994.tb01316.x. [DOI] [PubMed] [Google Scholar]
- 5.Brieland J, McClain M, Heath L, Chrisp C, Huffnagle G, LeGendre M, Hurley M, Fantone J, Engleberg C. Coinoculation with Hartmannella vermiformis enhances replicative Legionella pneumophila lung infection in a murine model of Legionnaires’ disease. Infect Immun. 1996;64:2449–2456. doi: 10.1128/iai.64.7.2449-2456.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cirillo J D, Falkow S, Tompkins L S. Growth of Legionella pneumophila in Acanthamoeba castellanii enhances invasion. Infect Immun. 1994;62:3254–3261. doi: 10.1128/iai.62.8.3254-3261.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clemens D L, Horwitz M A. Hypoexpression of major histocompatibility complex molecules on Legionella pneumophila phagosomes and phagolysosomes. Infect Immun. 1993;61:2803–2812. doi: 10.1128/iai.61.7.2803-2812.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clemens D L, Horwitz M A. Membrane sorting during phagocytosis: selective exclusion of major histocompatibility complex molecules but not complement receptor CR3 during conventional and coiling phagocytosis. J Exp Med. 1992;175:1317–1326. doi: 10.1084/jem.175.5.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins S J, Ruscetti F W, Gallagher R E, Gallo R C. Terminal differentiation of human promyelocytic leukemia cells induced by dimethyl sulfoxide and other polar compounds. Proc Natl Acad Sci USA. 1978;75:2458–2462. doi: 10.1073/pnas.75.5.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desjardins M, Huber L A, Parton R G, Griffiths G. Biogenesis of phagolysosomes proceeds through a sequential series of interactions with the endocytic apparatus. J Cell Biol. 1994;124:677–688. doi: 10.1083/jcb.124.5.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feeley J C, Gorman G W, Weaver R E, Mackel D C, Smith H W. Primary isolation media for Legionnaires’ disease bacterium. J Clin Microbiol. 1978;8:320–325. doi: 10.1128/jcm.8.3.320-325.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernandez R C, Lee S H, Haldane D, Sumarah R, Rozee K R. Plaque assay for virulent Legionella pneumophila. J Clin Microbiol. 1989;27:1961–1964. doi: 10.1128/jcm.27.9.1961-1964.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedman R L, Iglewski B H, Miller R D. Identification of a cytotoxin produced by Legionella pneumophila. Infect Immun. 1980;29:271–274. doi: 10.1128/iai.29.1.271-274.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geisow M J. Fluorescein conjugates as indicators of subcellular pH. Exp Cell Res. 1984;150:29–35. doi: 10.1016/0014-4827(84)90698-0. [DOI] [PubMed] [Google Scholar]
- 15.Hass R, Bartels H, Topley N, Hadman M, Kohler L, Goppelt-Strube M, Resch K. TPA-induced differentiation and adhesion of U937 cells: changes in ultrastructure, cytoskeletal organization and expression of cell surface antigens. Eur J Cell Biol. 1989;48:282–293. [PubMed] [Google Scholar]
- 16.Heinzen R A, Scidmore M A, Rockey D D, Hackstadt T. Differential interactions with endocytic and exocytic pathways distinguish parasitophorous vacuoles of Coxiella burnetii and Chlamydia trachomatis. Infect Immun. 1996;64:796–809. doi: 10.1128/iai.64.3.796-809.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horwitz M A. Characterization of avirulent mutant Legionella pneumophila that survive but do not multiply within human monocytes. J Exp Med. 1987;166:1310–1328. doi: 10.1084/jem.166.5.1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horwitz M A. Formation of a novel phagosome by the Legionnaires’ disease bacterium (Legionella pneumophila) in human monocytes. J Exp Med. 1983;158:1319–1331. doi: 10.1084/jem.158.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horwitz M A. The Legionnaires’ disease bacterium (Legionella pneumophila) inhibits phagosome-lysosome fusion in human monocytes. J Exp Med. 1983;158:2108–2126. doi: 10.1084/jem.158.6.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horwitz M A. Phagocytosis of the Legionnaires’ disease bacterium (Legionella pneumophila) occurs by a novel mechanism: engulfment within a pseudopod coil. Cell. 1984;36:27–33. doi: 10.1016/0092-8674(84)90070-9. [DOI] [PubMed] [Google Scholar]
- 21.Horwitz M A, Maxfield F R. Legionella pneumophila inhibits acidification of its phagosome in human monocytes. J Cell Biol. 1984;99:1936–1943. doi: 10.1083/jcb.99.6.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horwitz M A, Silverstein S C. Intracellular multiplication of Legionnaires’ disease bacteria (Legionella pneumophila) in human monocytes is reversibly inhibited by erythromycin and rifampin. J Clin Invest. 1983;71:15–26. doi: 10.1172/JCI110744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hudson B, Upholt W B, Devinny J, Vinograd J. The use of an ethidium analogue in the dye-buoyant density procedure for the isolation of closed circular DNA: the variation of the superhelix density of mitochondrial DNA. Proc Natl Acad Sci USA. 1969;62:813–820. doi: 10.1073/pnas.62.3.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Husmann L K, Johnson W. Cytotoxicity of extracellular Legionella pneumophila. Infect Immun. 1994;62:2111–2114. doi: 10.1128/iai.62.5.2111-2114.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Isberg R R. Intracellular trafficking of Legionella pneumophila within phagocytic cells. In: Miller V L, Kaper J B, Portnoy D A, Isberg R R, editors. Molecular genetics of bacterial pathogenesis. Washington, D.C: American Society for Microbiology; 1994. pp. 263–278. [Google Scholar]
- 26.Joiner K A, Fuhrman S A, Miettinen H M, Kasper L H, Mellman I. Toxoplasma gondii: fusion competence of parasitophorous vacuoles in Fc receptor-transfected fibroblasts. Science. 1990;249:641–646. doi: 10.1126/science.2200126. [DOI] [PubMed] [Google Scholar]
- 27.Jones T C, Hirsch J G. The interaction between Toxoplasm gondii and mammalian cells. II. The absence of lysosomal fusion with phagocytic vacuoles containing living parasites. J Exp Med. 1972;136:1173–1194. doi: 10.1084/jem.136.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krishan A. Rapid flow cytofluorometric analysis of mammalian cell cycle by propidium iodide staining. J Cell Biol. 1975;66:188–193. doi: 10.1083/jcb.66.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marra A, Blander S J, Horwitz M A, Shuman H A. Identification of a Legionella pneumophila locus required for intracellular multiplication in human macrophages. Proc Natl Acad Sci USA. 1992;89:9607–9611. doi: 10.1073/pnas.89.20.9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marra A, Horwitz M A, Shuman H A. The HL-60 model for the interaction of human macrophages with the Legionnaires’ disease bacterium. J Immunol. 1990;144:2738–2744. [PubMed] [Google Scholar]
- 31.Marra A, Shuman H A. Genetics of Legionella pneumophila virulence. Annu Rev Genet. 1992;26:51–69. doi: 10.1146/annurev.ge.26.120192.000411. [DOI] [PubMed] [Google Scholar]
- 32.Maxfield F R. Measurement of vacuolar pH and cytoplasmic calcium in living cells using fluorescence microscopy. Methods Enzymol. 1989;173:745–771. doi: 10.1016/s0076-6879(89)73048-2. [DOI] [PubMed] [Google Scholar]
- 33.Maxfield F R, Dunn K W. Studies of endocytosis using image intensification fluorescence microscopy and digital image analysis. In: Herman B, Jacobson K, editors. Digitized video microscopy. New York, N.Y: Alan R. Liss, Inc.; 1989. pp. 387–417. [Google Scholar]
- 34.Purcell M, Shuman H. The Legionella pneumophila icmGCDJBF genes are required for killing of human macrophages. Infect Immun. 1998;66:2245–2255. doi: 10.1128/iai.66.5.2245-2255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rechnitzer C, Blom J. Engulfment of the Philadelphia strain of Legionella pneumophila within pseudopod coils in human phagocytes. Comparison with other Legionella strains and species. APMIS. 1989;97:105–114. [PubMed] [Google Scholar]
- 36.Remington R D, Schork A. Statistics with applications to the biological and health sciences. Englewood Cliffs, N.J: Prentice-Hall, Inc.; 1985. [Google Scholar]
- 37.Sadosky A B, Wiater L A, Shuman H A. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect Immun. 1993;61:5361–5373. doi: 10.1128/iai.61.12.5361-5373.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Segal G, Shuman H A. Characterization of a new region required for macrophage killing by Legionella pneumophila. Infect Immun. 1997;65:5057–5066. doi: 10.1128/iai.65.12.5057-5066.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shuman H A, Horwitz M A. Legionella pneumophila invasion of mononuclear phagocytes. Curr Top Microbiol Immunol. 1996;209:99–112. doi: 10.1007/978-3-642-85216-9_6. [DOI] [PubMed] [Google Scholar]
- 40.Steinberg T H, Swanson J A. Measurement of phagosome-lysosome fusion and phagosomal pH. Methods Enzymol. 1994;236:147–160. doi: 10.1016/0076-6879(94)36014-6. [DOI] [PubMed] [Google Scholar]
- 41.Sundstrom C, Nilsson K. Establishment and characterization of a human histiocytic lymphoma cell line (U-937) Int J Cancer. 1976;17:565–577. doi: 10.1002/ijc.2910170504. [DOI] [PubMed] [Google Scholar]
- 42.Swanson M S, Isberg R R. Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect Immun. 1995;63:3609–3620. doi: 10.1128/iai.63.9.3609-3620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swanson M S, Isberg R R. Identification of Legionella pneumophila mutants that have aberrant intracellular fates. Infect Immun. 1996;64:2585–2594. doi: 10.1128/iai.64.7.2585-2594.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y L, Goren M B. Differential and sequential delivery of fluorescent lysosomal probes into phagosomes in mouse peritoneal macrophages. J Cell Biol. 1987;104:1749–1754. doi: 10.1083/jcb.104.6.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wiater L A, Sadosky A B, Shuman H A. Mutagenesis of Legionella pneumophila using Tn903dII lacZ: identification of a growth-phase-regulated pigmentation gene. Mol Microbiol. 1994;11:641–653. doi: 10.1111/j.1365-2958.1994.tb00343.x. [DOI] [PubMed] [Google Scholar]