Abstract

Highly reflective assemblies of purine, pteridine, and flavin crystals are used in the coloration and visual systems of many different animals. However, structure determination of biogenic crystals by single-crystal XRD is challenging due to the submicrometer size and beam sensitivity of the crystals, and powder XRD is inhibited due to the small volumes of powders, crystalline impurity phases, and significant preferred orientation. Consequently, the crystal structures of many biogenic materials remain unknown. Herein, we demonstrate that the 3D electron diffraction (3D ED) technique provides a powerful alternative approach, reporting the successful structure determination of biogenic guanine crystals (from spider integument, fish scales, and scallop eyes) from 3D ED data confirmed by analysis of powder XRD data. The results show that all biogenic guanine crystals studied are the previously known β-polymorph. This study highlights the considerable potential of 3D ED for elucidating the structures of biogenic molecular crystals in the nanometer-to-micrometer size range. This opens up an important opportunity in the development of organic biomineralization, for which structural knowledge is critical for understanding the optical functions of biogenic materials and their possible applications as sustainable, biocompatible optical materials.
Short abstract
This study demonstrates the successful structure determination of biogenic guanine crystals from three biological origins directly from 3D ED. This method overcomes problems arising from crystal beam sensitivity, multiple crystal domains, sample contamination, small sample volumes, and polymorphic transformations encountered with single-crystal and powder XRD.
Assemblies of high refractive index molecular crystals give rise to many different optical phenomena in animals.1−3 Guanine is the most widespread biogenic molecular crystal, responsible for the silvery reflectance of fish scales,4−6 the brilliant iridescence of copepods,7 the mirrored eyes of scallops,8,9 and many more.10−13 Initially, one crystal structure of anhydrous guanine—the α-polymorph—was recognized,14 and it was originally thought that the biogenic crystals adopt this structure.5 However, Hirsch et al. showed that biogenic guanine crystals adopt a different structure—the β-polymorph.15
The nanometer-to-micrometer size of biogenic molecular crystals prevents structure determination from single-crystal X-ray diffraction (SC-XRD). Furthermore, powder XRD is challenging due to the small quantities of biogenic crystals, the presence of crystalline impurity phases, significant line broadening due to effects of small particle size and/or strain, and the likelihood of a significant degree of preferred orientation in the powder sample due to the anisotropic crystal morphologies that are typical for biogenic materials.
To circumvent these challenges in the structure determination of biogenic guanine, a multitechnique approach was employed15 in which DFT-calculated structures were compared with powder XRD data for the biogenic materials, leading to the structure determination of the β-polymorph from powder XRD data. The crystal structures of both the α-polymorph (P21/c) and the β-polymorph (which we describe here in space group P21/c) comprise H-bonded sheets (bc-plane), which are π-stacked along the a-axis. The α and β polymorphs have the same H-bonding connectivity within the sheets but differ primarily in the relative offset of adjacent sheets parallel to the bc-plane. Similar multitechnique approaches have been used to solve the crystal structures of biogenic isoxanthopterin19,20 and 7,8-dihydroxanthopterin.21
Since structure determination of biogenic crystals from SC-XRD or powder XRD data is challenging, we were motivated to apply three-dimensional electron diffraction (3D ED; also called MicroED) for direct structure determination of biogenic crystals. There is currently growing interest in the use of 3D ED data to determine the structures of molecular crystals;22,23 however, to the best of our knowledge, this method has not yet been applied to biominerals. 3D ED is conceptually analogous to SC-XRD, involving sampling of all reflections in the tilt range of the goniometer.24 However, 3D ED offers a distinct advantage since the strong interaction with electrons allows data collection from submicrometer-sized crystals. The beam sensitivity of biogenic molecular crystals and their susceptibility to undergo damage upon extraction from the organism present major challenges to structure determination.2,16−18 Compared to conventional zone-axis ED, continuous rotation during data collection minimizes the effects of beam damage and reduces dynamical effects.25 3D ED data collection times are further reduced by using radiation hard hybrid detectors that are very sensitive and have negligible readout times. Compared to powder XRD, only minute quantities of crystals are required for 3D ED, meaning that gentle extraction and cleaning methods can be applied to the biogenic crystals. Furthermore, since individual crystals can be selected and measured using a small beam, contaminants can also be spatially avoided. Together, these advantages make continuous rotation 3D ED ideal for radiation-sensitive crystals of small molecules,22,26−29 proteins30−32 and now also biogenic materials.
Herein, we demonstrate the successful structure determination of biogenic guanine crystals directly from 3D ED data, focusing on samples from three biological origins: spider integument, scallop eyes, and fish scales. White widow spiders use 300–500 nm thick prismatic guanine crystals (usually found as doublets) in specialized skin cells to scatter light diffusely and produce their characteristic white coloration (Figure 1a).33,34 Scallops have dozens to hundreds of eyes lining the mantle tissue, which use concave mirrors, constructed from guanine crystals, to generate images;9 each 70 nm thick square crystal is made up of three twin domains (Figure 1b).35 In fish scales, disordered stacks of 25 nm thick elongated hexagonal crystals produce silvery reflectance (Figure 1c).5
Figure 1.
Morphologies of biogenic guanine crystals imaged by TEM. (a) Prismatic guanine crystal doublets extracted from iridophore cells in the integument of the white widow spider (Latrodectus pallidus). Top left: L. pallidus. Top right: higher magnification optical micrograph of the white iridophore cells. (b) Square-plate guanine crystals extracted from the image-forming mirror of scallop eyes. Top left: adult scallop, Pecten maximus. Top right: optical micrograph of part of the mantle tissue with three visible eyes. (c) Elongated hexagonal plate crystals extracted from salmon (Salmo salar) scales. Top left: salmon skin. Top right: optical micrograph of one scale at higher magnification showing the iridescent iridophore cells. In the TEM images in parts a–c, the (100) crystal face was assigned by zone-axis ED carried out during imaging.
Results and Discussion
For the 3D ED measurements, guanine crystals were extracted from the biological tissue (Section S1; Supporting Information) and deposited on a TEM grid. Continuous rotation 3D ED data were collected on an ELDICO ED-1 electron diffractometer at room temperature. For each measurement, the data collection time was 120 s, and the electron dose was 0.01 electrons Å–2 s–1 (see Section S2). Since no significant beam damage was detected during the measurement, data could be collected over a large tilt range (−60° to +60°).25 3D ED data for guanine crystals from spider and fish samples were recorded with a diffraction resolution of ca. 0.67 Å (in each case, three data sets from different crystals were recorded). The degree of crystallinity of the scallop samples was lower, and the highest diffraction resolution of the 3D ED data was ca. 0.80 Å. The 3D ED data were indexed, integrated, and merged using the APEX4 software package (Bruker AXS Inc. 2022). The 3D ED data gave a unit cell and space group consistent with the reported structure15 of the β-polymorph of guanine: P21/b, a = 3.59 Å, b = 9.72 Å, c = 18.34 Å, and γ = 119.5°. While previous work15 on the β-polymorph used the nonstandard P21/b space group setting (with unique c-axis), we utilize the standard P21/c setting (with unique b-axis) in the present work. In both space groups, the H-bonded layers lie parallel to the bc-plane. The (0kl), (h0l) and (hk0) reciprocal lattice planes in the 3D ED data for the spider sample are shown in Figure 2. Systematic absences in the (0kl) plane [(0k0), k = 2n; (00l), l = 2n], the (hk0) plane [(0k0), k = 2n] and the (h0l) plane [l = 2n] are consistent with space group P21/c.
Figure 2.
Reconstructed precession images from the 3D ED data recorded for a guanine crystal from a spider showing the reciprocal lattice planes reconstructed from the experimental data. In addition, the gray panels correspond to simulated planes (see Section S2) for the crystal structure determined for the spider sample from the 3D ED data in P21/c: (a) (0kl), (b) (h0l), and (c) (hk0). The inset in part a shows the crystal from which the data set was collected (scale bar 1 μm).
From the 3D ED data recorded in the present work, there was no evidence for the presence of the α-polymorph of guanine in any of the crystals studied. However, for each 3D ED data set, we identified between two and five domains that are orientationally distinct but have equivalent unit cells. These multiple domains could be manually selected in the reconstructed 3D reciprocal lattice and separated easily using the APEX4 software. Consequently, for each data set, structure solution was carried out considering only the major domain. The ability to differentiate between individual crystal domains is extremely useful for biogenic crystals, which often exist as mesocrystals13,34,36−38 or twinned crystals.35
Given the plate-like morphology of the biogenic crystals (Figure 1b,c), for which the large face is assigned as (100),15 the crystals on the grid typically exhibit preferred orientation (lying parallel to their (100) face), which prevents the collection of some diffraction information around the a*-axis. However, the completeness (>80%) of the 3D ED data was sufficient to allow successful structure determination. Structure solution from the 3D ED data was carried out in parallel using both the dual-space algorithm in SHELXT39,40 and the direct-space genetic algorithm (GA) in the program EAGER, which was originally developed for structure solution from powder XRD data41−46 and has recently been adapted for 3D ED data.47,48 Structure refinement from the 3D ED data was carried out using the full-matrix least-squares technique in SHELXL.39,40
The structure of the β-polymorph of guanine determined from 3D ED data is essentially the same as that reported by Hirsch et al.,15 but with higher structural quality, evidenced inter alia by the successful location of H atoms in the structure refinement. The crystal structure comprises π-stacking of H-bonded layers (distance between aromatic centroids ca. 3.4 Å; perpendicular distance between adjacent layers ca. 3.2 Å). Within the H-bonded layer, each molecule interacts with four neighboring molecules by a total of eight H-bonds (Figure 3a–c). The H-bonded layers in the β-polymorph are not strictly planar; although the molecular planes are essentially parallel to the mean plane of the H-bonded layer, alternate molecules are displaced slightly above and below this mean plane on moving along the b-axis. In contrast, for the α-polymorph of guanine, the position of each molecule relative to the mean plane of the H-bonded layer is the same and the plane of each molecule is tilted slightly from the mean plane of the layer.
Figure 3.
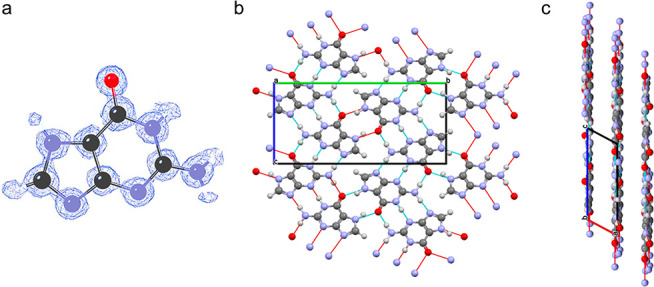
(a) Electron density map following structure refinement from 3D ED data of guanine crystals from the spider sample showing the successful identification of the hydrogen atom positions, (b) the H-bonded layer parallel to bc-plane of the crystal structure of the β-polymorph, and (c) view along the b-axis showing the H-bonded layers.
In the structure solution calculations from individual 3D ED data sets, both using the direct-space GA technique and traditional methods, all non-H atoms were found directly in the structure solution. In addition, the structure solutions obtained from direct-space GA calculations contain the positions of H atoms (Figure S3). For the structure solutions obtained by traditional methods, the H atoms were identified subsequently in electron density maps after structure refinement (Figure 3a). For the fish and spider samples, the structure was refined from the 3D ED data with anisotropic atomic displacement parameters (ADPs) following merging of data sets from three individual crystals. The final R-factor was R1 = 0.177 (for 1188 reflections) for the fish sample and R1 = 0.241 (for 1085 reflections) for the spider sample. For the scallop sample, 3D ED data sets from individual crystals were also merged to increase the completeness (to ∼80%), but the poor resolution did not allow a sensible refinement of anisotropic ADPs. The structure was instead refined with isotropic ADPs for non-H atoms with a riding model for H atoms (R1 = 0.374 for 558 reflections). The lower resolution and intensities of diffraction from the scallop crystals may result from the presence of multiple crystalline domains, which reduce the coherence length of the crystallites, resulting in a higher R1 value; nevertheless, the crystal structure was successfully solved from the 3D ED data.
As the accuracy of unit cell parameters determined from 3D ED data is not as high as that from powder XRD data, the unit cell parameters were determined independently from powder XRD data by profile fitting using the Pawley method, specifically for the fish and spider samples and for a synthetic sample of the β-polymorph of guanine. Powder XRD data were not recorded for the scallop sample as only a very small amount of polycrystalline sample can be obtained from this organism, which further illustrates the utility of 3D ED for many biological systems. The unit cell parameters determined from the 3D ED data and powder XRD are shown in Table 1. In addition, a full structure refinement was carried out from the powder XRD data for the synthetic sample by using the Rietveld refinement technique (Section S6.3). The initial structural model was taken from the structure solution obtained from the EAGER calculations on the 3D ED data for the spider sample but using the unit cell from Pawley fitting of the powder XRD data. The final Rietveld refinement included preferred orientation correction using the March–Dollase method,49,50 which established that the preferred orientation direction corresponds to the (100) plane in the P21/c space group setting.51 Significantly, this plane corresponds to the large face of the plate-like crystal morphology of the β polymorph, as shown in Figure 1.
Table 1. Unit Cell Parameters Determined from 3D ED Data and Powder XRD Data for samples of β guanine from Hirsch et al.15 and this studya.
| Space
group P21/c |
Space
group P21/b |
|||||||
|---|---|---|---|---|---|---|---|---|
| 3D ED |
Powder
XRD |
Hirsch
et al.15 |
||||||
| Sample | Spider | Fish | Scallop | Spider | Fish | Synthetic | Synthetic Powder XRD | Calculated |
| a/Å | 3.615(22) | 3.630(8) | 3.616(12) | 3.6310(1) | 3.6381(1) | 3.6317(1) | 3.629 | 3.5938 |
| b/Å | 18.39(11) | 18.34(4) | 18.43(5) | 18.4396(2) | 18.4227(2) | 18.4214(11) | 9.813 | 9.7195 |
| c/Å | 9.791(50) | 9.803(19) | 9.790(28) | 9.8145(4) | 9.8172(4) | 9.8138(10) | 18.427 | 18.3401 |
| β/° | 118.02(16) | 117.94(6) | 117.82(9) | 118.048(1) | 117.930(1) | 117.945(4) | 118.04 (γ) | 119.493 (γ) |
| V/Å3 | 574.6(60) | 576.6(23) | 576.9(33) | 579.95(1) | 581.33(1) | 580.00(3) | 557.6 | 557.6 |
Unit cells in the columns headed “3D ED” were obtained by indexing the 3D ED data. Unit cells in the columns headed “powder XRD” were determined by Pawley fitting (spider and fish samples) and Rietveld refinement (synthetic sample) of the powder XRD data. The final structure refinement calculations for the fish, spider, and scallop samples used the unit cells determined from powder XRD data together with the diffraction intensities from 3D ED data, as discussed in the text.
Comparison of the crystal structures of the three biogenic materials refined from 3D ED data and the synthetic material refined from powder XRD data reveals that these structures are essentially identical (Figure 4). One measure of structural similarity is to determine the average position of each atom in the asymmetric unit in the four structures and then to compare (by means of a calculated root-mean-squared deviation (RMSD)) the atomic positions in each structure relative to the average atomic positions. The values of RMSD calculated by this method are 0.063 Å (fish), 0.077 Å (spider), 0.106 Å (scallop), and 0.071 Å (synthetic). Clearly, the small values of RMSD indicate that there are no significant differences between these structures. This conclusion is corroborated by results from DFT-D geometry optimization calculations (Section S7), for which the calculated energies of the four structures are within 0.75 kJ mol–1 of each other following geometry optimization with fixed (experimental) unit cell, and within 0.05 kJ mol–1 of each other following geometry optimization with relaxation of the unit cell. For the four structures resulting from DFT-D geometry optimization with unit cell relaxation, the values of RMSD relative to the average atomic positions (as described above) are very small: 0.022 Å (fish), 0.018 Å (spider), 0.028 Å (scallop), and 0.017 Å (synthetic). Thus, the four structures determined from the experimental 3D ED data and powder XRD data represent essentially identical structures corresponding to the same local minimum on the energy hypersurface.
Figure 4.

Overlay of the relative position of the guanine molecule in the asymmetric unit of the crystal structures determined from 3D ED data for biogenic fish (cyan), spider (yellow), and scallop (magenta) samples and from powder XRD data for the synthetic sample of the β polymorph (green).
Concluding Remarks
Our studies represent the first reported crystal structure determination of biogenic molecular crystals directly from 3D ED data, specifically the crystal structures of biogenic guanine crystals from spider integument, scallop eyes, and fish scales. In each case, the structure closely matches the published structure of the β-polymorph of guanine, reported previously15 from DFT calculations and Rietveld refinement from powder XRD data for both synthetic and spider samples. Our work demonstrates the utility of 3D ED for structure determination of submicrometer biogenic crystals, which circumvents the inherent challenges associated with SC-XRD or powder XRD of biogenic crystals including the small crystal size, beam sensitivity, small sample amounts, presence of crystalline impurity phases, line broadening due to small particle size and/or strain, and significant preferred orientation due to anisotropic crystal morphologies. Moreover, we note that the positions of H atoms were established in our structure determinations (both from 3D ED data and powder XRD data), representing an improvement in the quality of the β-guanine crystal structure compared to the structure published previously.15 Our results confirm that biogenic guanine crystals from fish, spiders, and scallops correspond to the β-polymorph,15 rather than the α-polymorph or the theoretically predicted orthorhombic γ-polymorph (which was postulated in Hirsch et al.15). The observation of small quantities of the α-polymorph in powder XRD data recorded for the spider sample (Figure S5) likely derive from partial transformation of the β-polymorph to the thermodynamically stable α-polymorph5,10,15 during the harsher extraction methods used in preparing samples for powder XRD. Interestingly, while the biogenic guanine crystals obtained from spider, scallop, and fish samples exhibit differences in crystal morphology (Figure 1), we have shown here that their crystal structures are essentially identical (Table 1; CCDC deposition number 2304787, and see Supporting Information). Even in the potentially challenging case of biogenic crystals that comprise multiple domains (as observed for spider and scallop samples), successful structure determination from 3D ED data was achieved by appropriate analysis of crystal twinning. Moreover, while biogenic guanine crystals from spider, fish and scallop samples are known to contain some amounts of other purine dopants such as hypoxanthine,10 the crystal structures remain remarkably unaffected. This is in full agreement with the results of Pinsk et al., which showed that hypoxanthine is included into the guanine host structure as a homogeneous solid solution with the crystal structure of the β-polymorph of guanine.10 Until now, the inability to directly determine the structures of biogenic crystals has limited the rate of progress in organic biomineralization. We anticipate that 3D ED will play a major role in the future exploration of recently discovered uncharacterized molecular biocrystals.16,52,53
Acknowledgments
We thank Dr. Alexander Upcher and Dr. Vladimir Ezersky from the Ilse Katz Institute for Nanoscale Science & Technology at Ben-Gurion University of the Negev for many fruitful discussions. We acknowledge Dr. Lee Shelly for photographing the scallop, and Dr. Barak Ratzker for preparing illustrations used in the Table of Contents graphic. Funding was provided by an ERC Starting Grant (Grant No. 852948, “CRYSTALEYES”), a HFSP grant (Grant No. RGP0037/2022) and an ISF grant (Grant No.1565/22) awarded to B.A.P. In addition, B.A.P. is the Nahum Guzik Presidential Recruit and the recipient of the 2019 Azrieli Faculty Fellowship. A.W. is a recipient of an Azrieli Graduate Student Fellowship 2022/23. K.D.M.H., B.M.K., and C.E.H. are grateful to Cardiff University for support.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.cgd.3c01290.
Detailed materials and methods sections for extraction of crystals (S1); 3D ED measurements and data processing (S2); periodic DFT-D geometry optimization calculations (S3); space group transformation from P21/n to P21/c (S4); structure determination from 3D ED (S5), including traditional structure solution and refinement (S5.1) and direct-space structure solution using the genetic algorithm technique (S5.2); powder XRD measurements (S6.1); profile fitting and unit cell determination from powder XRD data (S6.2); structure refinement of β-polymorph from powder XRD by Rietveld refinement (S6.3); DFT-D geometry optimization of crystal structures from 3D ED and powder XRD data (S7) (PDF)
Crystal structures of guanine from the fish (CIF), scallop (CIF), and synthetic samples (CIF)
The authors declare no competing financial interest.
Supplementary Material
References
- Gur D.; Palmer B. A.; Weiner S.; Addadi L. Light Manipulation by Guanine Crystals in Organisms: Biogenic Scatterers, Mirrors, Multilayer Reflectors and Photonic Crystals. Adv. Funct. Mater. 2017, 27 (6), 1603514. 10.1002/adfm.201603514. [DOI] [Google Scholar]
- Palmer B. A.; Gur D.; Weiner S.; Addadi L.; Oron D. The Organic Crystalline Materials of Vision: Structure–Function Considerations from the Nanometer to the Millimeter Scale. Adv. Mater. 2018, 30 (41), 1800006. 10.1002/adma.201800006. [DOI] [PubMed] [Google Scholar]
- Wagner A.; Wen Q.; Pinsk N.; Palmer B. A. Functional Molecular Crystals in Biology. Isr. J. Chem. 2021, 61 (9), 668–678. 10.1002/ijch.202100069. [DOI] [Google Scholar]
- Jordan T. M.; Partridge J. C.; Roberts N. W. Non-Polarizing Broadband Multilayer Reflectors in Fish. Nat. Photonics 2012, 6 (11), 759–763. 10.1038/nphoton.2012.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy-Lior A.; Pokroy B.; Levavi-Sivan B.; Leiserowitz L.; Weiner S.; Addadi L. Biogenic Guanine Crystals from the Skin of Fish May Be Designed to Enhance Light Reflectance. Cryst. Growth Des. 2008, 8 (2), 507–511. 10.1021/cg0704753. [DOI] [Google Scholar]
- Gur D.; Leshem B.; Oron D.; Weiner S.; Addadi L. The Structural Basis for Enhanced Silver Reflectance in Koi Fish Scale and Skin. J. Am. Chem. Soc. 2014, 136 (49), 17236–17242. 10.1021/ja509340c. [DOI] [PubMed] [Google Scholar]
- Gur D.; Leshem B.; Farstey V.; Oron D.; Addadi L.; Weiner S. Light-Induced Color Change in the Sapphirinid Copepods: Tunable Photonic Crystals. Adv. Funct. Mater. 2016, 26 (9), 1393–1399. 10.1002/adfm.201504339. [DOI] [Google Scholar]
- Land M. F. A Multilayer Interference Reflector in the Eye of the Scallop, Pecten Maximus. J. Exp. Biol. 1966, 45 (3), 433–447. 10.1242/jeb.45.3.433. [DOI] [Google Scholar]
- Palmer B. A.; Taylor G. J.; Brumfeld V.; Gur D.; Shemesh M.; Elad N.; Osherov A.; Oron D.; Weiner S.; Addadi L. The Image-Forming Mirror in the Eye of the Scallop. Science 2017, 358 (6367), 1172–1175. 10.1126/science.aam9506. [DOI] [PubMed] [Google Scholar]
- Pinsk N.; Wagner A.; Cohen L.; Smalley C. J. H.; Hughes C. E.; Zhang G.; Pavan M. J.; Casati N.; Jantschke A.; Goobes G.; Harris K. D. M.; Palmer B. A. Biogenic Guanine Crystals Are Solid Solutions of Guanine and Other Purine Metabolites. J. Am. Chem. Soc. 2022, 144 (11), 5180–5189. 10.1021/jacs.2c00724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur D.; Palmer B. A.; Leshem B.; Oron D.; Fratzl P.; Weiner S.; Addadi L. The Mechanism of Color Change in the Neon Tetra Fish: A Light-Induced Tunable Photonic Crystal Array. Angew. Chemie - Int. Ed. 2015, 54 (42), 12426–12430. 10.1002/anie.201502268. [DOI] [PubMed] [Google Scholar]
- Gur D.; Nicolas J. D.; Brumfeld V.; Bar-Elli O.; Oron D.; Levkowitz G. The Dual Functional Reflecting Iris of the Zebrafish. Adv. Sci. 2018, 5 (8), 1800338. 10.1002/advs.201800338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G.; Yallapragada V. J.; Halperin T.; Wagner A.; Shemesh M.; Upcher A.; Pinkas I.; McClelland H. L. O.; Hawlena D.; Palmer B. A. Lizards Exploit the Changing Optics of Developing Chromatophore Cells to Switch Defensive Colors during Ontogeny. Proc. Natl. Acad. Sci. U. S. A. 2023, 120, e2215193120. 10.1073/pnas.2215193120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guille K.; Clegg W. Anhydrous Guanine: A Synchrotron Study. Acta Crystallogr. Sect. C Cryst. Struct. Commun. 2006, 62 (8), o515–o517. 10.1107/S0108270106026011. [DOI] [PubMed] [Google Scholar]
- Hirsch A.; Gur D.; Polishchuk I.; Levy D.; Pokroy B.; Cruz-Cabeza A. J.; Addadi L.; Kronik L.; Leiserowitz L. “Guanigma”: The Revised Structure of Biogenic Anhydrous Guanine. Chem. Mater. 2015, 27 (24), 8289–8297. 10.1021/acs.chemmater.5b03549. [DOI] [Google Scholar]
- Friedman O.; Böhm A.; Rechav K.; Pinkas I.; Brumfeld V.; Pass G.; Weiner S.; Addadi L. Structural Organization of Xanthine Crystals in the Median Ocellus of a Member of the Ancestral Insect Group Archaeognatha. J. Struct. Biol. 2022, 214 (1), 107834. 10.1016/j.jsb.2022.107834. [DOI] [PubMed] [Google Scholar]
- Rez P.; Aoki T.; March K.; Gur D.; Krivanek O. L.; Dellby N.; Lovejoy T. C.; Wolf S. G.; Cohen H. Damage-Free Vibrational Spectroscopy of Biological Materials in the Electron Microscope. Nat. Commun. 2016, 7, 10945. 10.1038/ncomms10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biran I.; Houben L.; Weissman H.; Hildebrand M.; Kronik L.; Rybtchinski B. Real-Space Crystal Structure Analysis by Low-Dose Focal-Series TEM Imaging of Organic Materials with Near-Atomic Resolution. Adv. Mater. 2022, 34, 2202088. 10.1002/adma.202202088. [DOI] [PubMed] [Google Scholar]
- Palmer B. A.; Hirsch A.; Brumfeld V.; Aflalo E. D.; Pinkas I.; Sagi A.; Rosenne S.; Oron D.; Leiserowitz L.; Kronik L.; Weiner S.; Addadi L. Optically Functional Isoxanthopterin Crystals in the Mirrored Eyes of Decapod Crustaceans. Proc. Natl. Acad. Sci. U. S. A. 2018, 115 (10), 2299–2304. 10.1073/pnas.1722531115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch A.; Palmer B. A.; Ramasubramaniam A.; Williams P. A.; Harris K. D. M.; Pokroy B.; Weiner S.; Addadi L.; Leiserowitz L.; Kronik L. Structure and Morphology of Light-Reflecting Synthetic and Biogenic Polymorphs of Isoxanthopterin: A Comparison. Chem. Mater. 2019, 31 (12), 4479–4489. 10.1021/acs.chemmater.9b01039. [DOI] [Google Scholar]
- Zhang G.; Hirsch A.; Shmul G.; Avram L.; Elad N.; Brumfeld V.; Pinkas I.; Feldman Y.; Ben Asher R.; Palmer B. A.; Kronik L.; Leiserowitz L.; Weiner S.; Addadi L. Guanine and 7,8-Dihydroxanthopterin Reflecting Crystals in the Zander Fish Eye: Crystal Locations, Compositions, and Structures. J. Am. Chem. Soc. 2019, 141 (50), 19736–19745. 10.1021/jacs.9b08849. [DOI] [PubMed] [Google Scholar]
- Saha A.; Nia S. S.; Rodríguez J. A. Electron Diffraction of 3D Molecular Crystals. Chem. Rev. 2022, 122 (17), 13883–13914. 10.1021/acs.chemrev.1c00879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karothu D. P.; Alhaddad Z.; Göb C. R.; Schürmann C. J.; Bücker R.; Naumov P. The Elusive Structure of Levocetirizine Dihydrochloride Determined by Electron Diffraction. Angew. Chemie - Int. Ed. 2023, 62, e202303761. 10.1002/anie.202303761. [DOI] [PubMed] [Google Scholar]
- Huang Z.; Willhammar T.; Zou X. Three-Dimensional Electron Diffraction for Porous Crystalline Materials: Structural Determination and Beyond. Chem. Sci. 2021, 12 (4), 1206–1219. 10.1039/D0SC05731B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemmi M.; Mugnaioli E.; Gorelik T. E.; Kolb U.; Palatinus L.; Boullay P.; Hovmöller S.; Abrahams J. P. 3D Electron Diffraction: The Nanocrystallography Revolution. ACS Cent. Sci. 2019, 5 (8), 1315–1329. 10.1021/acscentsci.9b00394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Genderen E.; Clabbers M. T. B.; Das P. P.; Stewart A.; Nederlof I.; Barentsen K. C.; Portillo Q.; Pannu N. S.; Nicolopoulos S.; Gruene T.; Abrahams J. P. Ab Initio Structure Determination of Nanocrystals of Organic Pharmaceutical Compounds by Electron Diffraction at Room Temperature Using a Timepix Quantum Area Direct Electron Detector. Acta Crystallogr. Sect. A Found. Adv. 2016, 72, 236–242. 10.1107/S2053273315022500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruene T.; Wennmacher J. T. C.; Zaubitzer C.; Holstein J. J.; Heidler J.; Fecteau-Lefebvre A.; De Carlo S.; Müller E.; Goldie K. N.; Regeni I.; Li T.; Santiso-Quinones G.; Steinfeld G.; Handschin S.; van Genderen E.; van Bokhoven J. A.; Clever G. H.; Pantelic R. Rapid Structure Determination of Microcrystalline Molecular Compounds Using Electron Diffraction. Angew. Chemie - Int. Ed. 2018, 57 (50), 16313–16317. 10.1002/anie.201811318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. G.; Martynowycz M. W.; Hattne J.; Fulton T. J.; Stoltz B. M.; Rodriguez J. A.; Nelson H. M.; Gonen T. The CryoEM Method MicroED as a Powerful Tool for Small Molecule Structure Determination. ACS Cent. Sci. 2018, 4 (11), 1587–1592. 10.1021/acscentsci.8b00760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson Grape E.; Rooth V.; Nero M.; Willhammar T.; Inge A. K. Structure of the Active Pharmaceutical Ingredient Bismuth Subsalicylate. Nat. Commun. 2022, 13, 1984. 10.1038/s41467-022-29566-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nannenga B. L.; Shi D.; Leslie A. G. W.; Gonen T. High-Resolution Structure Determination by Continuous-Rotation Data Collection in MicroED. Nat. Methods 2014, 11 (9), 927–930. 10.1038/nmeth.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi D.; Nannenga B. L.; De La Cruz M. J.; Liu J.; Sawtelle S.; Calero G.; Reyes F. E.; Hattne J.; Gonen T. The Collection of MicroED Data for Macromolecular Crystallography. Nat. Protoc. 2016, 11 (5), 895–904. 10.1038/nprot.2016.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clabbers M. T. B.; Van Genderen E.; Wan W.; Wiegers E. L.; Gruene T.; Abrahams J. P. Protein Structure Determination by Electron Diffraction Using a Single Three-Dimensional Nanocrystal. Acta Crystallogr. Sect. D Struct. Biol. 2017, 73 (9), 738–748. 10.1107/S2059798317010348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy-Lior A.; Shimoni E.; Schwartz O.; Gavish-Regev E.; Oron D.; Oxford G.; Weiner S.; Addadi L. Guanine-Based Biogenic Photonic-Crystal Arrays in Fish and Spiders. Adv. Funct. Mater. 2010, 20 (2), 320–329. 10.1002/adfm.200901437. [DOI] [Google Scholar]
- Wagner A.; Ezersky V.; Maria R.; Upcher A.; Lemcoff T.; Aflalo E. D.; Lubin Y.; Palmer B. A. The Non-Classical Crystallization Mechanism of a Composite Biogenic Guanine Crystals. Adv. Mater. 2022, 34, 2202242. 10.1002/adma.202202242. [DOI] [PubMed] [Google Scholar]
- Hirsch A.; Palmer B. A.; Elad N.; Gur D.; Weiner S.; Addadi L.; Kronik L.; Leiserowitz L. Biologically Controlled Morphology and Twinning in Guanine Crystals. Angew. Chemie - Int. Ed. 2017, 56 (32), 9420–9424. 10.1002/anie.201704801. [DOI] [PubMed] [Google Scholar]
- Levy-Lior A.; Shimoni E.; Schwartz O.; Gavish-Regev E.; Oron D.; Oxford G.; Weiner S.; Addadi L. Guanine-Based Biogenic Photonic-Crystal Arrays in Fish and Spiders. Adv. Funct. Mater. 2010, 20 (2), 320–329. 10.1002/adfm.200901437. [DOI] [Google Scholar]
- Eyal Z.; Deis R.; Varsano N.; Dezorella N.; Rechav K.; Houben L.; Gur D. Plate-like Guanine Biocrystals Form via Templated Nucleation of Crystal Leaflets on Preassembled Scaffolds. J. Am. Chem. Soc. 2022, 144 (49), 22440–22445. 10.1021/jacs.2c11136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A.; Upcher A.; Maria R.; Magnesen T.; Zelinger E.; Raposo G.; Palmer B. A. Macromolecular Sheets Direct the Morphology and Orientation of Plate-like Biogenic Guanine Crystals. Nat. Commun. 2023, 14, 589. 10.1038/s41467-023-35894-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick G. M. SHELXT - Integrated Space-Group and Crystal-Structure Determination. Acta Crystallogr. Sect. A Found. Crystallogr. 2015, 71 (1), 3–8. 10.1107/S2053273314026370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick G. M. A Short History of SHELX. Acta Crystallogr. Sect. A Found. Crystallogr. 2008, 64 (1), 112–122. 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- Kariuki B. M.; Serrano-González H.; Johnston R. L.; Harris K. D. M. The Application of a Genetic Algorithm for Solving Crystal Structures from Powder Diffraction Data. Chem. Phys. Lett. 1997, 280 (3–4), 189–195. 10.1016/S0009-2614(97)01156-1. [DOI] [Google Scholar]
- Cheung E. Y.; McCabe E. E.; Harris K. D. M.; Johnston R. L.; Tedesco E.; Raja K. M. P.; Balaram P. C–H···O Hydrogen Bond Mediated Chain Reversal in a Peptide Containing a γ-Amino Acid Residue, Determined Directly from Powder X-ray Diffraction Data. Angew. Chemie - Int. Ed. 2002, 41 (3), 494–496. . [DOI] [PubMed] [Google Scholar]
- Harris K. D. M.; Habershon S.; Cheung E. Y.; Johnston R. L. Developments in Direct-Space Techniques for Structure Solution from Powder Diffraction Data. Z. Kristallogr. 2004, 219, 838–846. 10.1524/zkri.219.12.838.55856. [DOI] [Google Scholar]
- Martí-Rujas J.; Meazza L.; Lim G. K.; Terraneo G.; Pilati T.; Harris K. D. M.; Metrangolo P.; Resnati G. An Adaptable and Dynamically Porous Organic Salt Traps Unique Tetrahalide Dianions. Angew. Chemie - Int. Ed. 2013, 52 (50), 13444–13448. 10.1002/anie.201307552. [DOI] [PubMed] [Google Scholar]
- Williams P. A.; Hughes C. E.; Harris K. D. M. L-lysine: Exploiting Powder X-ray Diffraction to Complete the Set of Crystal Structures of the 20 Directly Encoded Proteinogenic Amino Acids. Angew. Chemie - Int. Ed. 2015, 54 (13), 3973–3977. 10.1002/anie.201411520. [DOI] [PubMed] [Google Scholar]
- Watts A. E.; Maruyoshi K.; Hughes C. E.; Brown S. P.; Harris K. D. M. Combining the Advantages of Powder X-ray Diffraction and NMR Crystallography in Structure Determination of the Pharmaceutical Material Cimetidine Hydrochloride. Cryst. Growth Des. 2016, 16 (4), 1798–1804. 10.1021/acs.cgd.6b00016. [DOI] [Google Scholar]
- Sun T.; Hughes C. E.; Guo L.; Wei L.; Harris K. D. M.; Zhang Y. B.; Ma Y. Direct-Space Structure Determination of Covalent Organic Frameworks from 3D Electron Diffraction Data. Angew. Chemie - Int. Ed. 2020, 59 (50), 22638–22644. 10.1002/anie.202009922. [DOI] [PubMed] [Google Scholar]
- Smalley C. J. H.; Hoskyns H. E.; Hughes C. E.; Johnstone D. N.; Willhammar T.; Young M. T.; Pickard C. J.; Logsdail A. J.; Midgley P. A.; Harris K. D. M. A Structure Determination Protocol Based on Combined Analysis of 3D-ED Data, Powder XRD Data, Solid-State NMR Data and DFT-D Calculations Reveals the Structure of a New Polymorph of L-tyrosine. Chem. Sci. 2022, 13, 5277–5288. 10.1039/D1SC06467C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- March A. Mathematical Theory on Regulation According to the Particle Shape in Affine Deformation. Z. Kristallogr. 1932, 81, 285–297. 10.1524/zkri.1932.81.1.285. [DOI] [Google Scholar]
- Dollase W. A. Correction of Intensities of Preferred Orientation in Powder Diffractometry: Application of the March Model. J. Appl. Crystallogr. 1986, 19 (4), 267–272. 10.1107/S0021889886089458. [DOI] [Google Scholar]
- Wittig N. K.; Christensen T. E. K.; Grünewald T. A.; Birkedal H. Vase-like β-Polymorph Guanine Crystal Aggregates Formed at the Air-Water Interface. ACS Mater. Lett. 2020, 2 (5), 446–452. 10.1021/acsmaterialslett.9b00447. [DOI] [Google Scholar]
- Pilátová J.; Pánek T.; Oborník M.; Čepička I.; Mojzeš P. Revisiting Biocrystallization: Purine Crystalline Inclusions are Widespread in Eukaryotes. ISME J. 2022, 16 (9), 2290–2294. 10.1038/s41396-022-01264-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavan M. E.; Movilla F.; Pavan E. E.; Di Salvo F.; López N. I.; Pettinari M. J. Guanine Crystal Formation by Bacteria. BMC Biol. 2023, 21, 66. 10.1186/s12915-023-01572-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.




