Abstract
Purpose
This study investigated a three-dimensional indicator in spectral-domain optical coherence tomography (SD-OCT) and established phenotype-genotype correlation in X-linked retinoschisis (XLRS).
Methods
Thirty-seven patients with XLRS underwent comprehensive ophthalmic examinations, including visual acuity (VA), fundus examination, electroretinogram (ERG), and SD-OCT. SD-OCT parameters of central foveal thickness (CFT), cyst cavity volume (CCV), and photoreceptor outer segment length were assessed. CCV was defined as the sum of the areas of cyst cavities in uential B-scans, measured automatically by self-developed software (OCT–CCSEG). Structural changes of the protein associated with missense variants were quantified by molecular dynamics (MD). The correlation between genotype and phenotype was analyzed.
Results
Twenty-seven different RS1 variants were identified, including a novel variant c.336_337insT(p.L113Sfs*8). The average age of onset was 14.76 ± 15.75 years, and the mean VA was 0.84 ± 0.43 logMAR. The mean CCV was 1.69 ± 1.87 mm3, correlating significantly with CFT (R = 0.66; P < 0.01). In the genotype–phenotype analysis of missense variants, CCV significantly correlated with the structural effect on the protein of mutational changes referred to as wild type, including root-mean-square deviation (R = 0.34; P = 0.04), solvent accessible surface area (R = 0.38; P = 0.02), and surface hydrophobic area (R = 0.37; P = 0.03). The amplitude of scotopic 3.0 ERG a-waves and b-waves significantly correlated with the percentage change of the β-strand in the secondary structure (a-wave: R = −0.58, P < 0.01; b-wave: R = −0.53, P < 0.01).
Conclusions
CCV is a promising indicator to quantify the structural disorganization of XLRS retina. The OCT–CCSEG software calculated CCV automatically, potentially facilitating prognosis assessment and development of personalized treatment. Moreover, MD-involved genotype–phenotype analysis suggests an association between protein structural alterations and XLRS severity measured by CCV and ERG.
Keywords: X-linked retinoschisis, RS1, OCT, molecular dynamics
X-linked retinoschisis (XLRS; OMIM 312700) is an X-linked recessive inherited retinal disorder with an estimated prevalence ranging from 1 in 5000 to 1 in 25,000.1–4 Patients with XLRS typically present with visual acuity loss, characterized by bilateral foveal schisis arising from the splitting of the inner retinal layers and a negative electroretinogram (ERG) waveform with significant attenuation of the b-wave amplitude.2,5–8 XLRS is caused by variants in the RS1 gene, which encodes an adhesive protein, retinoschisin (RS1), which is essential for the structural and functional integrity of the retina.
The phenotype of XLRS exhibits considerable heterogeneity, presenting practical challenges in disease management. Prior studies have found that cystoid changes in retinoschisis can involve various retinal layers, extending from the retinal nerve fiber layer to the nuclear layer, with the extent of the schisis showing significant variability.9–11 This highlights the necessity to identify appropriate clinical parameters that accurately reflect the effect of the disease on retinal structural changes. For the assessment of structural retina changes and prognosis of XLRS, spectral-domain optical coherence tomography (SD-OCT) is an effective approach to examine the multiple properties of retinal schisis and provide quantifiable indicators of structural changes. However, existing studies have mainly focused on one- or two-dimensional SD-OCT indicators, such as total retinal thickness, central foveal thickness (CFT), and area of the macular schisis cavity (AMS).12,13 These indicators have limitations with regard to reflecting the actual structural disorganization. Thus, we were motivated to consider three-dimensional (3D) volumetric parameters in SD-OCT, which potentially offer a more precise evaluation of retinal structural characteristics in patients with XLRS.
In addition to the diverse degree of structural lesions in the XLRS retina, considerable variability is observed in retinal function among XLRS patients, as manifested primarily in the ERG a-wave, b-wave, and the b-wave/a-wave ratio. To explain the broad clinical heterogeneity of XLRS, several studies have sought to establish the genotype–phenotype correlation of the disease. Pioneer studies mainly aimed to correlate clinical features, such as visual acuity, foveal pathology, complications, and ERG b-wave amplitude, with variant types but failed to establish a clear correlation.8,14–17 Recent studies have provided insight into the molecular mechanisms involved. Research conducted by Sergeev et al.18,19 correlated molecular models of RS1 obtained from molecular dynamics (MD) simulations with retinal function and suggested an association between the structural alteration of protein and the severity of XLRS as determined by ERG b-wave/a-wave ratios. In addition, Sudha's research investigated the association between the molecular characterization of the equilibrated structure of the mutant RS1 protein and its secretion profile.20 These studies evaluated the impact of a variant at the atomic level, shedding new light on the genotype–phenotype analysis of XLRS.
In this study, we aimed to analyze the clinical and genetic characteristics of 37 Chinese XLRS patients. Our findings revealed a promising method for obtaining 3D volumetric parameters using SD-OCT that can be applied along with other indicators to further investigate the correlation between phenotype and genotype in XLRS. Moreover, we gained a novel perspective on the genotype–phenotype correlation analysis of XLRS from molecular dynamic insights.
Methods
Recruitment of Subjects
Thirty-seven patients with molecularly confirmed XLRS were recruited from the Department of Ophthalmology, Peking Union Medical College Hospital (PUMCH), Beijing, China, and Beijing Mei'ermu Hospital, Beijing, China. Informed consent was collected from them or their legal guardians before enrollment. The PUMCH review board approved this study (no. JS-2059), which was conducted in accordance with the Guidance on Sample Collection of Human Genetic Diseases by the Ministry of Public Health of China, as well as the tenets of the Declaration of Helsinki.
Clinical Evaluations
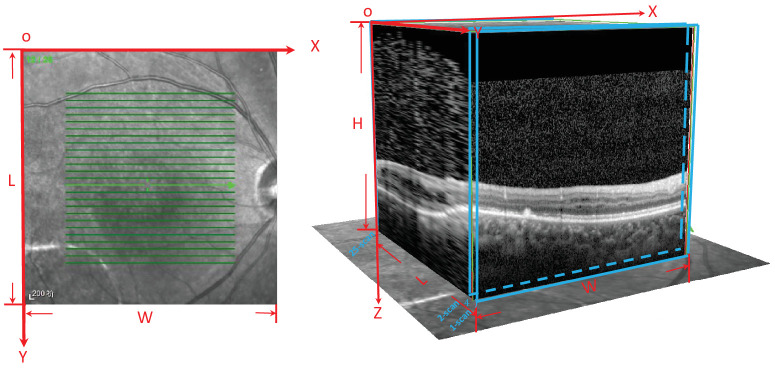
Detailed medical and family histories were recorded for each patient. Patients underwent comprehensive ophthalmological examinations, including visual acuity, slit-lamp biomicroscopy, dilated indirect ophthalmoscopy, fundus photography, fundus autofluorescence photography (SPECTRALIS; Heidelberg Engineering, Heidelberg, Germany), SD-OCT (SPECTRALIS; Heidelberg Engineering), and full-field standard ERG (RETIport; an-vision, Salt Lake City, UT, USA). VA was assessed using the Snellen chart and converted to logMAR units. For OCT image acquisition, a skilled technician performed an SD-OCT examination on all of the patients with XLRS. The scanning area was 5.6 × 5.6 mm in in the XY plane, centered at the fovea, generating a set of 25 horizontal cross-sectional B-scan images. Each OCT scan was 5.6 × 1.9 mm in the XZ plane (Fig. 1). ERG was performed with corneal ERG-Jet contact lens electrodes (Fabrinal, Neuchatel, Switzerland) or DTL electrodes (Roland Consult Stasche & Finger GmbH, Brandenburg ad Havel, Germany). The test protocol followed the standards of the International Society for Clinical Electrophysiology of Vision.
Figure 1.
Illustration of the SD-OCT scanning pattern. The scanning pattern area was 5.6 × 5.6 mm in in the XY plane, centered at the fovea, which generated a set of 25 horizontal cross-sectional B-scan images. Each OCT scan was 5.6 × 1.9 mm in the XZ plane.
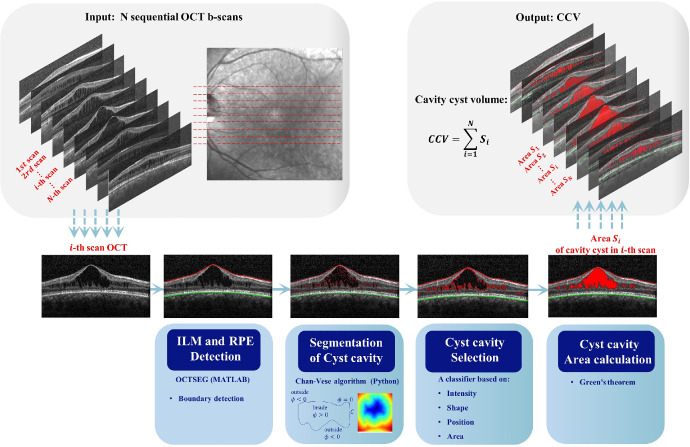
To further assess SD-OCT imaging biomarkers, the images were processed through ImageJ (National Institutes of Health, Bethesda, MD, USA) to calculate the CFT and photoreceptor outer segment (PROS) length. The PROS length was determined as the perpendicular distance between the posterior surface of the inner segment (IS)/outer segment (OS) junction and the anterior surface of the retinal pigment epithelium (RPE), taken at the foveal center on a horizontal central SD-OCT image (Supplementary Fig. S1). The retinal layers with foveoschisis were documented. To accurately quantify the macular cyst cavities in SD-OCT images, we defined and calculated cyst cavity volume (CCV) as the sum of the areas of cyst cavities across 25 sequential horizontal cross-sectional B-scans, multiplied by the distance between these scans. Mathematically, the CCV can be approximated by:
where Δl represents the interscan distance. It was measured automatically by a self-developed software system, OCT cyst cavity segmentation (OCT–CCSEG), using Python. Specifically, the system consists of five steps: (1) detecting the retinal layers, inner limiting membrane (ILM), and RPE by OCT segmentation with MATLAB software OCTSEG (MathWorks, Natick, MA, USA)21; (2) initially segmenting candidate cyst cavities by identification of the contours of all dark regions between the ILM and RPE using the Chan–Vese algorithm; (3) selecting cyst cavities from the candidates based on their intensity, shape, and position; (4) calculating the cyst cavity area in each scan; and (5) computing CCV from 25 SD-OCT scans. The entire framework is depicted in Figure 2.
Figure 2.
The CCV automated calculation system OCT–CCSEG. The input is a group of OCT scans, and the output is CCV. The system includes five modules: image boundaries detection module, image segmentation module, filter module, cavity area calculation module, and cavity volume calculation module.
Genetic Analysis of RS1 Variants
The procedure and variant filtering were conducted as previously described.22 Targeted panel sequencing of 256 known retinal disease genes and whole exome sequencing were used to identify pathogenic variants. Briefly, the genomic DNA of all probands and their available relatives was isolated from peripheral leukocytes and captured via the SeqCap EZ Choice XL Library (Roche NimbleGen, Pleasanton, CA, USA), then sequenced on the Illumina HiSeq platform (Illumina, San Diego, CA, USA). The raw data were analyzed using NextGENe 2.3.4 software, and the reads were compared to the reference sequence of hg19 from the UCSC Genome Browser (University of California Santa Cruz Genomics Institute, Santa Cruz, CA, USA). A comparison was conducted using the 1000 Genomes Project database, dbSNP database, and Exome Aggregation Consortium database to exclude nonpathogenic polymorphisms.
Pathogenicity of the missense variants and nucleotide conservation were accessed through the VarCards database.23 In silico algorithms, including SIFT,24 PolyPhen2,25 MutationTaster,26 MutationAssessor,27 FATHMM,28 PROVEAN,29 VEST3,30 MetaSVM,31 MetaLR,31 M-CAP,32 CADD,33 DANN,34 fathmm-MKL,35 GenoCanyon,36 and REVEL37 were applied to predict the pathogenicity of the missense variants. Nucleotide conservation was analyzed using GERP++,38 phastCons,39 phyloP,40 and SiPhy.41 The pathogenicity and conservation of small deletions, insertions, and nonsense variants were assessed by Phylop, PhastCons, and MutationTaster. The potential effects of splicing variants were evaluated by the Human Splicing Finder42 and SpliceAI To identify regions in the RS1 protein that are sensitive to mutations, in silico saturation mutagenesis analysis was performed on the wild-type RS1 protein using Rhapsody.43 Putative pathogenic RS1 variants (NM_000330.3) were verified by Sanger sequencing and segregation analysis when family members were available. Variants were classified according to the standards and guidelines of the American College of Medical Genetics and Genomics (ACMG), and the nomenclature of all variants was adjusted to the Human Genome Variation Society guidelines.44,45 The protein model of human RS1 (PDB-3JD6) was applied to visualize the distribution of the variants within RS1 on the protein structure. The rendering of the protein structure and annotation of interesting regions were achieved by the PyMOL Molecular Graphics System 2.0 (Schrödinger, LLC, New York, NY, USA).
Molecular Dynamics Simulation and Trajectory Analysis
The structural stability and properties of the wild-type and RS1 mutant proteins were analyzed through MD simulations. We applied the monomeric form of PDB-3JD646 as the initial model for wild-type RS1. The structures of the mutant RS1 were generated by Discovery Studio software (Accelrys, San Diego, CA, USA) with the RS1 wild-type structure as a template. The predicted structures were confirmed through a Ramachandran plot that showed no residues in the disallowed region (Supplementary Fig. S2, Supplementary Table S1). Prior to the MD simulation, we performed an energy minimization for the structures to remove any steric conflicts using the Smart Minimizer tool in Discovery Studio. The MD simulations were executed in four steps, including simulation setup, energy minimization, equilibration, and production simulation, using CHARMM force field and Gromacs 2022.1 software.47 The parameters used in the MD simulations are detailed in Supplementary File S1.
The trajectory files of the MD simulations were processed by GROMACS utilities, including gmx rmsd, gmx sasa, and gmx hbond, to analyze root-mean-square deviation (RMSD), solvent-accessible surface area (SASA), and hydrogen bond. The secondary structure was determined through DSSP48 and virtualized in UCSF Chimera.49 The hydrophilic and hydrophobic areas were estimated by the software BioLuminate (Schrödinger), and the free energy changes in the protein due to missense variants were evaluated through the FoldX force field.50 Alteration in the structural property of a mutant protein in comparison to the wild type was computed and is represented by Δ (delta). For example, the ΔRMSD of a given variant signifies the deviation in RMSD between the mutant protein and the wild type. The figures were generated using the Python Matplotlib 3.7.0 package.
Statistical Analysis
The statistical analysis in the study was performed by Python. The normality of the data was assessed by the D'Agostino or Pearson algorithm. The t-test was performed to determine the difference in phenotype features between patients and normal controls. Results with P < 0.05 were considered statistically significant. The missing observations were ignored, and analysis was done on the variables present. The time-to-event analysis was performed to analyze the impact of age on VA. The non-numerical VA was transformed into logMAR values, such as hand movements to 2.7 logMAR. Based on the visual impairment criteria of the World Health Organization, we divided VA into four levels: blindness (level 1, VA < 1.30 logMAR), severe visual impairment (level 2, 0.30 logMAR ≤ VA< 1.00 logMAR), moderate visual impairment (level 3, 1.00 logMAR ≤ VA < 0.48 logMAR), and mild visual impairment (level 4, 0.48 logMAR ≤ VA < 0.30 logMAR). The Kaplan–Meier survival analysis was applied to estimate the age at these visual impairment levels, and a spline regression model was used to account for censored data. The ordinal logistic regression was used to explore the annual decline/growth ratio for VA, a-wave amplitude, b-wave amplitude, and the b-wave/a-wave ratio. In the genotype–phenotype analysis, patients were categorized into two groups based on the type of variants they harbored18: missense variants and null variants (nonsense, insertion, small deletion, splicing, and gross deletions).
Results
Clinical Characteristics
The clinical characteristics of the 37 Chinese patients are summarized in Table 1, including age of onset, VA, SD-OCT indicators (CFT, CCV, and PROS), and ERG parameters. Detailed clinical information for each enrolled patient is presented in Supplementary Table S2. Representative fundus and SD-OCT images and ERG recordings of patients with XLRS are shown in Figure 3. The mean ± SD age of onset of the 37 patients in our cohort was 14.76 ± 15.75 years (interquartile range [IQR], 5.0–16.0; range, 1.0–58.0), and the mean VA was 0.84 ± 0.43 logMAR (IQR, 0.52–1.3; range, 0.10–2.70). To quantify the structural disorganization of the XLRS retina, SD-OCT was conducted for 34 patients (64 eyes). The mean CFT, PROS, and CCV were calculated to be 0.456 ± 0.235 mm (IQR, 0.317–0.551; range, 0.1–1.321), 21.62 ± 7.13 µm (IQR, 17.69–25.32; range, 0.33–42.58), and 1.69 ± 1.87 mm3 (IQR, 3.36–7.99; range, 0–52.18), respectively.
Table 1.
Clinical Observations of Patients (N = 37)
| Counts (Patients, Eyes) | Mean | SD | Minimum | 25th Percentiles | 50th Percentiles | 75th Percentiles | Maximum | |
|---|---|---|---|---|---|---|---|---|
| Onset age (y) | 37/37 | 14.76 | 15.64 | 1.00 | 5.00 | 7.00 | 16.00 | 58.00 |
| VA (logMAR) | 74/74 | 0.84 | 0.43 | 0.10 | 0.52 | 0.82 | 1.03 | 2.70 |
| Scotopic 3.0 ERG | ||||||||
| a-wave | 73/74 | 190.90 | 75.30 | 13.40 | 157.00 | 200.00 | 247.80 | 354.50 |
| b-wave | 73/74 | 191.16 | 86.99 | 13.70 | 127.00 | 184.90 | 246.60 | 382.80 |
| b-wave/a-wave ratio | 73/74 | 1.02 | 0.25 | 0.38 | 0.85 | 1.01 | 1.19 | 1.56 |
| Photopic 3.0 ERG | ||||||||
| a-wave | 70/74 | 33.60 | 12.09 | 2.90 | 25.83 | 35.35 | 42.80 | 56.60 |
| b-wave | 70/74 | 61.50 | 26.61 | 8.72 | 41.08 | 60.60 | 81.90 | 120.10 |
| b-wave/a-wave ratio | 70/74 | 2.08 | 1.34 | 0.23 | 1.44 | 1.83 | 2.17 | 8.01 |
| CFT | 64/74 | 0.46 | 0.23 | 0.10 | 0.32 | 0.43 | 0.55 | 1.32 |
| PROS | 64/74 | 21.62 | 7.13 | 0.32 | 17.69 | 22.94 | 25.32 | 42.58 |
| CCV | 64/74 | 1.69 | 1.87 | 0.01 | 0.78 | 1.25 | 1.86 | 12.16 |
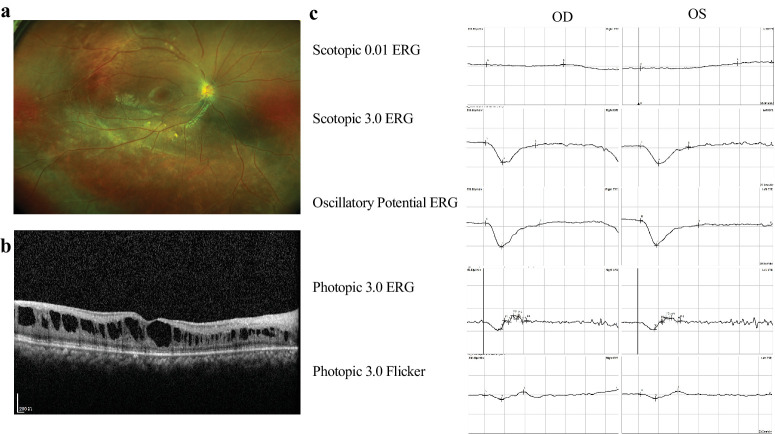
Figure 3.
Representative clinical images of XLRS patients. (a) Fundus displaying a spoke-wheel–like schisis at the macula. (b) OCT showing splitting of the inner retinal layers. (c) ERG exhibiting reduced waveforms of rod and cone responses and a negative ERG waveform on scotopic 3.0 ERG.
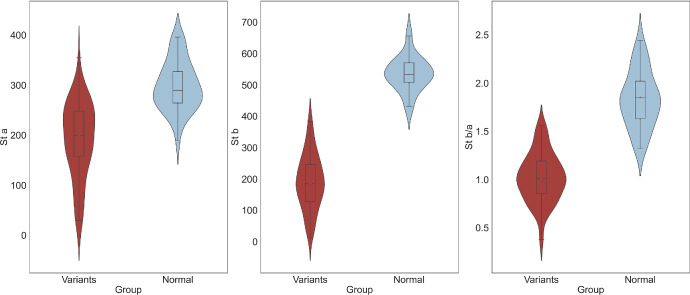
ERGs were obtained for all patients in this cohort, and the parameters for the scotopic and photopic 3.0 stimuli are shown in Table 1. The scotopic ERG a-wave amplitude, b-wave amplitude, and b-wave/a-wave ratio were significantly lower in our XLRS cohort compared to a control group of 40 normal individuals (P < 0.001) (Fig. 4). It was worth noting that significant correlations were identified between the VA and the a-wave and b-wave amplitudes, as well as the b-wave/a-wave ratio for scotopic ERGs (a-wave: R = −0.30, P = 0.01; b-wave: R = −0.43, P < 0.01; b-wave/a-wave ratio: R = −0.36, P < 0.01). To delve deeper into these associations, we defined the better eye as the eye with better VA. In the results from the better eye, we discovered further significant correlations. Specifically, the better eye VA was significantly correlated with the amplitude of the b-wave (R = −0.39; P = 0.03) and the b-wave/a-wave ratio (R = −0.40; P = 0.03) of the better eye. When gene variants were categorized into two classes (null variants and missense variants), no significant differences were observed between these two groups in terms of ERG parameters and VA.
Figure 4.
Comparison of ERG in patients and the control group. The violin and box plots illustrate scotopic 3.0 a-wave amplitude, scotopic 3.0 b-wave amplitude, and 3.0 scotopic b-wave/a-wave ratio in different groups.
Age Impact on VA
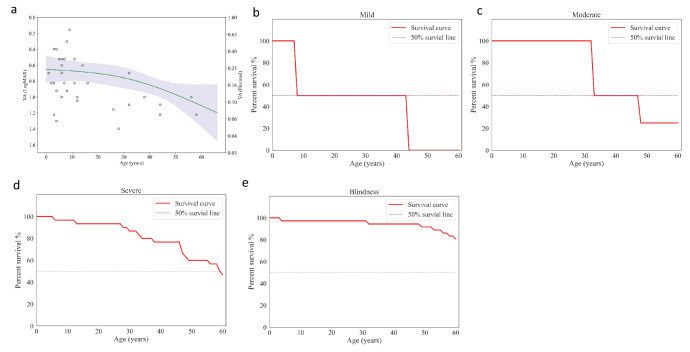
The VA of the patients in the study demonstrated a decreasing trend with increasing age, with an acceleration of this trend observed at older ages (Fig. 5a). Specifically, the trend was characterized by an average annual rate of 0.002 logMAR/y under the age of 10, 0.003 logMAR/y between 10 and 20 years of age, and 0.010 logMAR/y above 20 years old. In the survival analysis, the median ages to reach mild, moderate, and severe impairments were 8 years, 33 years, and 59 years, respectively (Figs. 5b–5d). In the analysis of blindness level, we found that 19.9% of patients were predicted to reach blindness at the age of 60 years (Fig. 5e).
Figure 5.
Age impact on VA. (a) The spline regression model of VA and age. (b–e) Kaplan–Meier survival analysis of different visual function indicators for age: survival curves for best-corrected visual acuity (BCVA) of 0.30 logMAR (b); survival curves for BCVA of 0.48 logMAR (c); survival curves for BCVA of 1.00 logMAR (d); and survival curves for BCVA of 1.30 logMAR (e).
Properties and Qualification of Cysts
The retinal layers with foveoschisis were summarized from the SD-OCT images of 64 eyes. Cysts were observed in the ganglion cell layer (GCL), inner plexiform layer (IPL), inner nuclear layer (INL), outer plexiform layer (OPL), and outer nuclear layer (ONL) with prevalences of 10.9%, 1.6%, 100%, 21.9%, and 31.2%, respectively. Pearson's correlation analysis was conducted to investigate the association among the OCT indicators CFT, CCV, and PROS. In the entire cohort, we found that CCV significantly correlated with CFT (R = 0.307; P = 0.014) and PROS (R = −0.25; P = 0.045). Our data indicated that CCV was not correlated with the b-wave/a-wave ratio in scotopic ERGs (R = −0.016; P = 0.900). Additionally, no statistical difference was found in CCV, CFT, and PROS between the patients with missense variants and those with null variants. The distribution of CCV versus age is shown in Supplementary Figure S3.
Genetic Characteristics
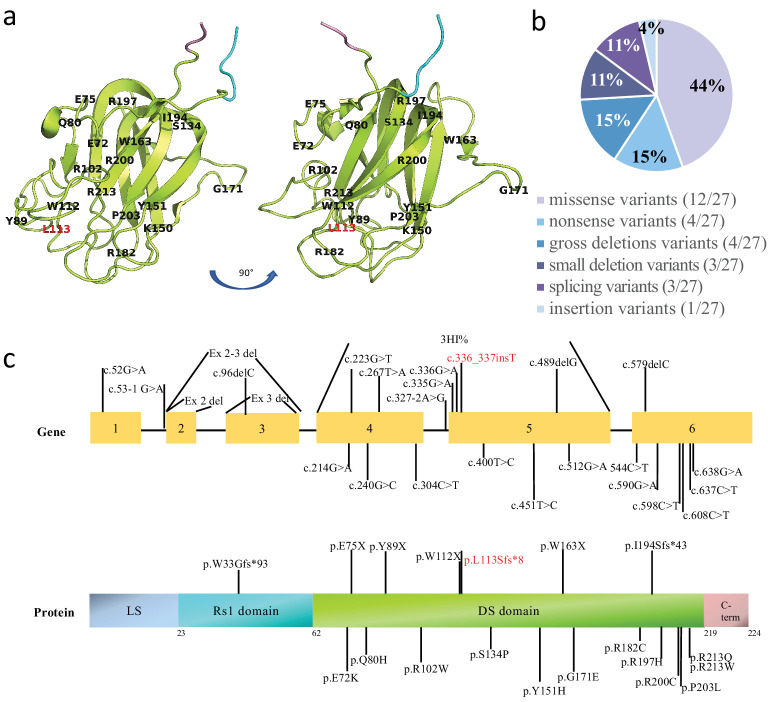
In this study, 37 patients with genetically confirmed XLRS were enrolled. A total of 27 different pathogenic variants were identified, including one novel variant, c.336_337insT(p.L113Sfs*8). The variants were categorized as 12 missense, four nonsense, four gross deletions, three small deletions, three splicings, and one insertion variants. Most of the variants were located in the discoidin domain (21/27), followed by the RS1 domain (4/27) and the N-terminal leader sequence (2/27). Table 2 presents a comprehensive list of the variants, their associated protein changes, variant types, and protein domains. The genomic location of the variants in the protein structure model of RS1 (PDB-3JD6) and the distribution of the variants among protein domain structure are illustrated in Figure 6. The most common variants were c.637C>T (four patients, 10.8%) and c.214G>A (four patients, 10.8%), followed by c.336_337insT (two patients, 5.2%), c.451T>C (two patients, 5.2%), c.590G>A (two patients, 5.2%), and c.608G>A (two patients, 5.2%). The remaining 21 variants were identified in one patient each. Most variants were located in exons 4, 5, and 6.
Table 2.
Genotype Summary of Patients
| Variant cDNA Change | Variant Protein Change | Variant Type | Domain | Cases, n |
|---|---|---|---|---|
| c.52G>A | — | Splicing | LS | 1 |
| c.53-1G>A | — | Splicing | LS | 1 |
| c.96delC | p.W33Gfs*93 | Small deletion | RS1 domain | 1 |
| c.214G>A | p.E72K | Missense | Discoidin domain | 4 |
| c.223G>T | p.E75X | Nonsense | Discoidin domain | 1 |
| c.240G>C | p.Q80H | Missense | Discoidin domain | 1 |
| c.267T>A | p.Y89X | Nonsense | Discoidin domain | 1 |
| c.304C>T | p.R102W | Missense | Discoidin domain | 1 |
| c.327-2A>G | — | Splicing | Discoidin domain | 1 |
| c.335G>A | p.W112X | Nonsense | Discoidin domain | 1 |
| c.336G>A | p.W112X | Nonsense | Discoidin domain | 1 |
| c.336_337insT | p.L113Sfs*8 | Insertions | Discoidin domain | 2 |
| c.400T>C | p.S134P | Missense | Discoidin domain | 1 |
| c.451T>C | p.Y151H | Missense | Discoidin domain | 2 |
| c.489delG | p.W163X | Small deletion | Discoidin domain | 1 |
| c.512G>A | p.G171E | Missense | Discoidin domain | 1 |
| c.544C>T | p.R182C | Missense | Discoidin domain | 1 |
| c.579delC | p.I194Sfs*43 | Small deletion | Discoidin domain | 1 |
| c.590G>A | p.R197H | Missense | Discoidin domain | 2 |
| c.598C>T | p.R200C | Missense | Discoidin domain | 1 |
| c.608C>T | p.P203L | Missense | Discoidin domain | 2 |
| c.637C>T | p.R213W | Missense | Discoidin domain | 4 |
| c.638G>A | p.R213Q | Missense | Discoidin domain | 1 |
| Exon 2 del | — | Deletion | RS1 domain | 1 |
| Exon 2-3 del | — | Deletion | RS1 domain | 1 |
| Exon 3 del | — | Deletion | RS1 domain | 1 |
| Exon 4-5 del | — | Deletion | Discoidin domain | 1 |
LS, leader sequence.
Figure 6.
Distribution of reported variants within RS1. (a) The genomic location of the variants in the homology model of RS1 (PDB-3JD6). The RS1 domain is shown in cyan, the DS domain in lemon, and the C-terminal in pink. (b) The spectrum of the 27 RS1 variants. (c) The distribution of the variants; the frequency of variants in each exon is indicated in the bar plot. The locations of variants in the gene and the protein are marked by lines.
In Silico Analysis
In silico analysis was performed on missense variants, small deletion variants, insertion variants, nonsense variants, and splicing variants in our cohort using various algorithms. The majority of the in silico algorithms, such as PolyPhen2, FATHMM, PROVEAN, VEST3, MetaSVM, MetaLR, M-CAP, CADD, DANN, fathmm-MKL, GenoCanyon, and REVEL, indicated that the 12 missense variants in our cohort were damaging or probably damaging to protein structure and could cause disease. The evaluation of the conservation of these missense variants was performed using GERP, phyloP, PhastCons, and SiPhy, which confirmed their conservation. Evaluation of small deletions, insertions (including the novel variant c.336_337insT), and nonsense variants was performed using phyloP, phastCons, and MutationTaster, revealing that these variants were conserved by phyloP and phastCons and predicted to be disease causing by MutationTaster. The HSF Pro platform (HSF and MaxEnt) and SpliceAI were employed to evaluate the potential effects of the splicing variants. MaxEnt and HSF analysis indicated that variant c.53-1G>A may cause a broken wild-type acceptor site. SpliceAI predicted that this variant could cause acceptor loss within the pre-mRNA position of −1 bp, with a probability of 0.69. Similarly, variant c.327-2A>G was predicted to cause a broken wild-type acceptor site by MaxEnt and HSF. SpliceAI analysis suggested that this variant may cause acceptor loss within the pre-mRNA position of −2 bp, with a high probability of 0.99. For additional information on the results of the in silico analysis, refer to Supplementary Tables S3 to S5. Remarkably, Vijayasarathy et al.51 previously discussed the variant c.52G>A, which involved the terminal nucleotide of exon 1 and was within the 5′ donor splice site of intron 1. The c.52G>A variant disrupted base pairing between the splice site and small nuclear RNA, thereby reducing normal splicing efficiency.
To investigate the hot region of variants in RS1, we applied the in silico algorithm Rhapsody, which has the benefit of predicting the effect of substituting any amino acid residue in a protein with any of the other 19 amino acids, enabling in silico site-directed mutagenesis analysis. Consequently, we identified residue changes that may be poorly tolerated, which can provide valuable insights for future research. Through protein-wide, site-directed mutagenesis, we identified the top-scoring variants in RS1 with a Rhapsody score of at least 0.86. Our analysis revealed that the highest scoring variants were located in eight locations in the protein, indicating that these crucial residues have the potential to cause disease when mutated (Supplementary Fig. S4).
Molecular Dynamics Analysis
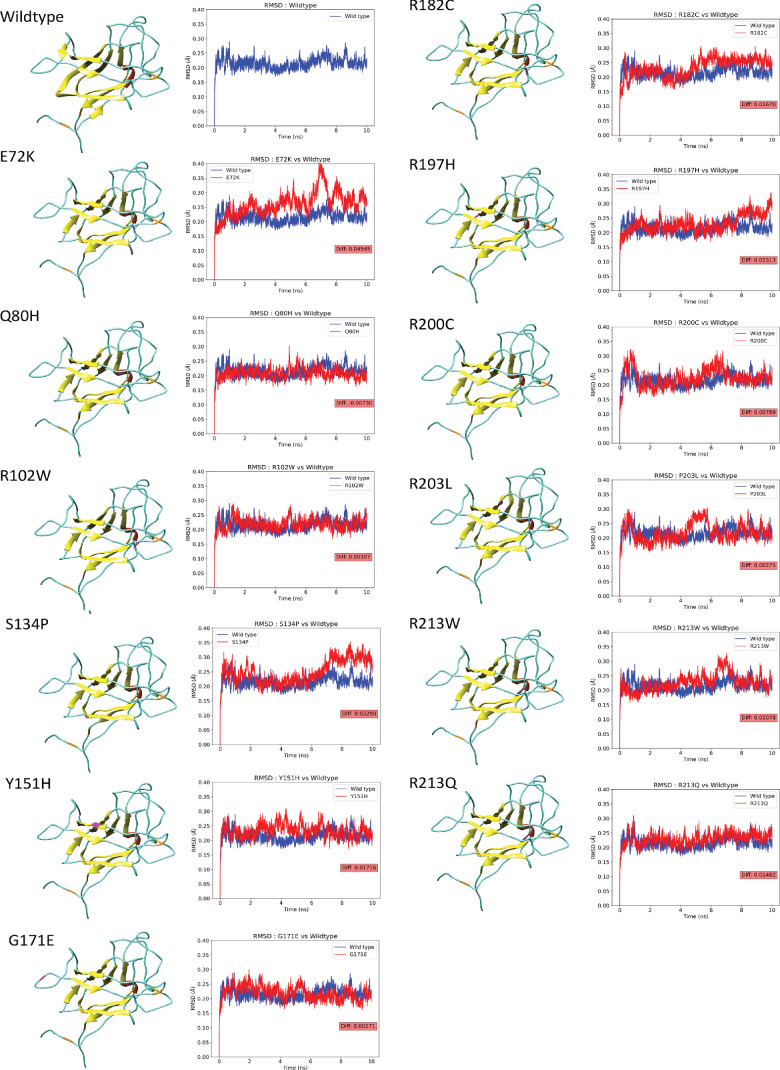
Trajectory data obtained from MD simulations were utilized to analyze the impact of missense variants on the RS1 protein structure. The stability of the protein was evaluated by the RMSDs of the backbone atoms, which determined the extent of their alteration or displacement during the simulation. We computed RMSDs for both the wild-type and mutant RS1. The RMSD of the wild-type protein was 0.215 Å. The variants E72K (0.260 Å), R102W (0.218 Å), S134P (0.248 Å), Y151H (0.233 Å), G171E (0.218 Å), R182C (0.232 Å), R197H (0.231 Å), R200C (0.223 Å), P203L (0.218 Å), R213W (0.226 Å), and R213Q (0.230 Å) showed higher deviations compared to the wild type, suggesting a decrease in stability due to increased backbone flexibility. The variant Q80H (0.208 Å) represented the lower deviations, indicating increased rigidity in the backbone. These RMSDs of mutants measure the effect of variants on the structure of the protein. The 3D structures of the wild-type and mutant proteins of RS1 in the MD simulation are displayed in Figure 7, and the RMSD and ΔRMSD values for both wild type and variants are provided in Table 2.
Figure 7.
Structure and RMSD graph of wild-type and variant forms of RS1 in MD simulation. In the 3D structure, helices are shown in red, strand in yellow, coli in cyan, disulfide bonds in orange, and variant spots in magenta. The RMSD graph shows the deviation of the backbone in the wild-type (blue) and variant (red) forms of RS1 during the MD simulation. Their difference is represented in the red box.
An analysis was performed to assess the impact of the variants on the secondary structure of the protein during the MD simulation. An increase in the proportion of α-helical structures was found in all missense variants compared to the wild type. Except for R213Q, the proportion of β-strands was decreased in all missense variants. The percentages of α-helical strands and β-strands in the secondary structure, as well as their changes compared with the wild type, are listed in Table 3. Notably, all missense variants maintained two intact disulfide bonds, which are crucial for protein folding and stability (Table 3). These changes in secondary structure impact the overall stability and function of the protein.
Table 3.
Structure Properties of Variant Proteins
| Variants | Wild-Type | E72K | Q80H | R102W | S134P | Y151H | G171E | R182C | R197H | R200C | P203L | R213W | R213Q |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| hbnumsp | 427 | 401 | 396 | 395 | 394 | 391 | 416 | 397 | 399 | 391 | 391 | 379 | 388 |
| hbnumsp diff | 0.00% | −6.09% | −7.26% | −7.49% | −7.73% | −8.43% | −2.58% | −7.03% | −6.56% | −8.43% | −8.43% | −11.24% | −9.13% |
| hbnumpp | 100 | 105 | 104 | 116 | 106 | 99 | 100 | 103 | 108 | 101 | 101 | 100 | 102 |
| hbnumpp diff | 0.00% | 5.00% | 4.00% | 16.00% | 6.00% | −1.00% | 0.00% | 3.00% | 8.00% | 1.00% | 1.00% | 0.00% | 2.00% |
| SASA | 99.26 | 104.36 | 97.3 | 98.98 | 104.68 | 97.98 | 100.46 | 94.23 | 100.09 | 97.21 | 98.42 | 97.75 | 100.7 |
| SASA diff | 0.00% | 5.13% | −1.98% | −0.28% | 5.46% | −1.29% | 1.21% | −5.08% | 0.84% | −2.07% | −0.85% | −1.52% | 1.45% |
| Hydrophilic area | 9215.928 | 9141.156 | 8827.822 | 9097.777 | 9138.324 | 9009.037 | 9411.617 | 8974.289 | 8930.672 | 8719.049 | 9201.594 | 8859.148 | 9176.891 |
| Hydrophilic area diff | 0.00% | −0.81% | −4.21% | −1.28% | −0.84% | −2.24% | 2.12% | −2.62% | −3.10% | −5.39% | −0.16% | −3.87% | −0.42% |
| Hydrophobic area | 759.629 | 924.825 | 810.795 | 808.761 | 953.833 | 740.211 | 812.356 | 642.217 | 794.462 | 694.482 | 669.083 | 709.807 | 801.754 |
| Hydrophobic area diff | 0.00% | 21.75% | 6.74% | 6.47% | 25.57% | −2.56% | 6.94% | −15.46% | 4.59% | −8.58% | −11.92% | −6.56% | 5.55% |
| RMSD | 0.22 | 0.26 | 0.21 | 0.22 | 0.25 | 0.23 | 0.22 | 0.23 | 0.23 | 0.22 | 0.22 | 0.23 | 0.23 |
| RMSD diff | 0.00 | 0.05 | −0.01 | 0.00 | 0.03 | 0.02 | 0.00 | 0.02 | 0.02 | 0.01 | 0.00 | 0.01 | 0.01 |
| Delta_delta_G | −0.27 | 1.16 | 3.08 | 0.49 | 2.79 | 3.32 | 0.06 | 3.44 | 1.36 | 1.32 | 25.41 | 3.76 | −2.21 |
| Grantham distance | 0.00 | 56.00 | 24.00 | 101.00 | 74.00 | 83.00 | 98.00 | 180.00 | 29.00 | 180.00 | 98.00 | 101.00 | 43.00 |
| Residue accessibility | 0.00 | 104.50 | 27.12 | 41.61 | 8.82 | 1.00 | 58.43 | 88.16 | 61.88 | 39.72 | 0.12 | 0.00 | 0.00 |
| Alpha | 0.00 | 5.38 | 5.98 | 3.59 | 5.98 | 5.38 | 3.59 | 2.39 | 4.19 | 4.19 | 1.79 | 0.00 | 4.19 |
| Beta | 38.92 | 33.53 | 37.12 | 35.32 | 35.32 | 36.52 | 35.92 | 32.33 | 31.13 | 35.92 | 38.32 | 33.53 | 40.71 |
| Other | 61.07 | 61.07 | 56.88 | 61.07 | 58.68 | 58.08 | 60.47 | 65.26 | 64.67 | 59.88 | 59.88 | 66.46 | 55.08 |
| Beta diff | 0.00 | −0.14 | −0.05 | −0.09 | −0.09 | −0.06 | −0.08 | −0.17 | −0.20 | −0.08 | −0.02 | −0.14 | 0.05 |
| Disulfide bond | 2 (Cys63-Cys219, Cys 110-Cys142) | ||||||||||||
diff, difference.
Additionally, the chemical properties of the mutant protein were assessed to gain further insight into their impact. The parameters that were calculated included the number of hydrogen bonds within the protein and between the protein and solvent, SASA, and hydrophilic and hydrophobic areas of the protein and their differences compared to the wild type (Table 3). We found that the variants had a greater impact on the hydrophobic area than the hydrophilic area. Changes in the hydrophobic and hydrophilic areas of the protein surface can affect its structure and function, including cell adhesion, secretion, and other intermolecular interactions. This could potentially result in more severe clinical symptoms for patients with vision problems.
Genotype and Phenotype Correlation for Missense Variants
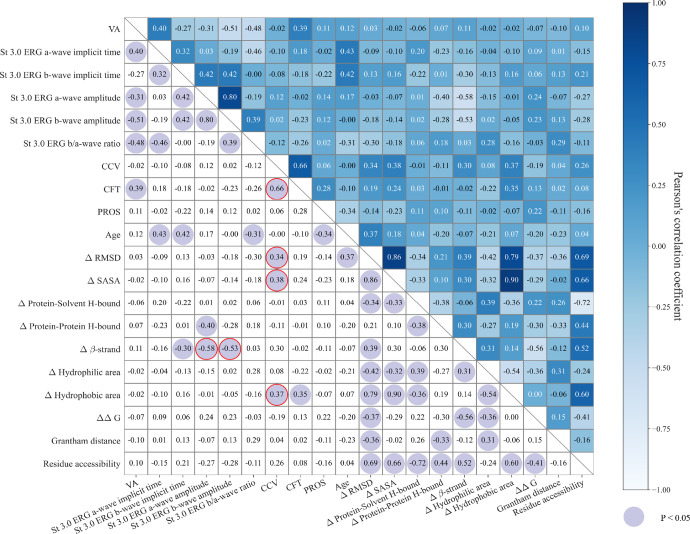
The correlation matrices and P values for the phenotype and genotype for patients with missense variants are shown in Figure 8. The analysis revealed that the CCV exhibited significant correlations with the structural effect on the protein of mutational changes referred to as wild type, including ΔRMSD (R = 0.34; P = 0.04), ΔSASA (R = 0.38; P = 0.02), and Δ surface hydrophobic area (R = 0.37; P = 0.03). Conversely, CFT showed weaker correlations or non-significant relationships with these three genotype features with correlation coefficients of 0.340 (P = 0.046), 0.383 (P = 0.023), and 0.371 (P = 0.028), respectively. Also, the amplitudes of scotopic 3.0 ERG a-waves and b-waves were significantly correlated with the percentage change of the β strand in the secondary structure (a-wave Δβ-strand: R = −0.58, P < 0.01; b-wave Δβ-strand: R = −0.53, P < 0.01). However, the scotopic 3.0 ERG a-wave/b-wave ratio was not correlated with the Δβ-strand (R = 0.03; P = 0.85).
Figure 8.
Correlation matrix of genotype and phenotype. The value in each cell is the Pearson's correlation coefficient of two variables in the corresponding row and column. The cells with purple circles are significant results (P < 0.05).
Discussion
This study identified the clinical and genetic features of an XLRS patient cohort and established a link between the phenotype and genotype in XLRS from the molecular dynamic insight. The phenotype of XLRS patients is known for its heterogeneity, which was also significant in our cohort. In terms of the structural disorganization observed from SD-OCT images, cysts were found in the GCL, IPL, INL, OPL, and ONL with prevalences of 10.9%, 1.6%, 100%, 21.9%, and 31.2%, respectively. Moreover, the degree of schisis differed extensively among different patients. This heterogeneity posed a challenge for disease management; thus, searching for the appropriate clinical parameter to reflect the severity of effect of the disease on structural changes of the retina is worth investigating. In this study, we explored a potential OCT indicator to quantify the structural disorganization of XLRS and further applied it to compare the retinal structural changes in patients with XLRS with different genotypes.
Previous OCT studies of XLRS focused on CFT, but this indicator is limited to the depiction of a single OCT scan at a one-dimensional level.12,13 Lin et al.52 proposed utilizing the AMS as a parameter in OCT and explored its correlation with VA in patients with XLRS. The measurement of the AMS was performed through manual annotation and selection, a process that is time consuming and labor intensive. Also, evaluating the AMS remained at the two-dimensional level when describing the cyst cavity and was limited as far as reproducing the real cyst cavity. To quantify the 3D structure of the cyst cavity directly, we defined the total CCV as the sum of the total area of the 25 sequential B-scans of the OCT. However, a challenge in its calculation is the heavy workload of manually annotating images. To the best of our knowledge, there is still no commercial and open-source software packages that support the CCV automated computing. Given the current software vacancies, we self-developed an automated CCV computing platform OCT–CCSEG.
The OCT–CCSEG platform involves three major steps. Initially, we used the open-source program OCTSEG21 to detect the boundaries of ILM and RPE in OCT. The feasibility and open-source nature of this program made it a suitable choice. Next, we applied the Chan–Vese algorithm, which is commonly used in medical imaging to segment the brain, heart, and trachea,53–55 to identify the dark region between the two boundaries. The robustness of this algorithm against noise was particularly beneficial for detecting cysts. Last, we employed a filter based on regional features such as intensity, shape, and position to eliminate non-cyst regions, which further improves the accuracy of our system. We have validated the accuracy, flexibility, and stability of our system in OCT images for human patients and mouse models (data not shown). The results of our experiments reinforced the robustness of our approach and instilled confidence in its reliable application. Our system also automates all processes, reducing manual labor and increasing efficiency in data analysis. Moreover, the results obtained from the 3D measurements are more accurate and provide a more precise depiction of changes in the cyst cavity compared to other existing OCT biomarkers.
Furthermore, the OCT–CCSEG system demonstrates versatility in its potential functions. Current natural history and gene therapy studies of XLRS lack clinical indicators to understand and quantify the disease progression and prognosis. Previous OCT studies of XLRS focused on total retinal thickness. However, retinal thickness can be influenced by two factors: thinning caused by the outer retinal atrophy or thickening caused by the presence of cystic cavities. When the total retinal thickness is reduced, it is difficult to determine whether this is due to a reduction in cyst cavity volume or photoreceptor loss. To monitor the disease progression in structural changes, CCV could serve as a more precise indicator. Moreover, the automation and rapid calculation capabilities of our platform facilitate frequent and efficient assessments of changes in CCV throughout disease progression. Considering the significant association between CCV and clinical features of genotype in our patients, our platform may assist in selecting therapeutic windows and endpoints in XLRS intervention trials.
In addition to its application in XLRS, OCT–CCSEG has potential applications in other retinal diseases due to the high demand for retinal volume indicators. This platform can measure retinal volume in OCT. Specifically, the initial step of the platform entails detecting the ILM and RPE layers in OCT. By calculating the region between these two boundaries, the total volume can be determined, and the difference between this total volume and the cyst volume can accurately measure the volume of the retina. The precise measurements of retinal volume render it an invaluable diagnostic tool for diseases that affect retinal volume. Furthermore, this platform holds potential for application in diseases that impact the microstructure of the retina, such as exudates, cysts, and fluid. These diseases include age-related macular degeneration, retinal vein occlusion, diabetic macular edema, ocular inflammation, and diabetic retinopathy.
In addition to clinical heterogeneity, gene heterogeneity is highly evident in our cohort. A total of 27 different pathogenic variants were confirmed, including one novel variant. These variants were classified into 12 missense, four nonsense, four gross deletions, three small deletions, three splicings, and one insertion variants. Existing literature mostly agrees on the absence of a clear genotype–phenotype correlation in XLRS. Therefore, it is impossible to draw prognostic conclusions in newly diagnosed patients based solely on the molecular result. Previous studies compared missense variants and null variants with respect to various indicators, including ERG indicators (a-wave amplitude, b-wave amplitude, and b-wave/a-wave ratio), CFT and PROS in OCT, and VA.16,56,57 However, our study did not find any significant differences between missense variants and null variants in terms of the ERG indicators CFT and PROS, CCV in OCT, or VA. This suggests that simply categorizing the variants as missense and null may not be sufficient to fully describe their impact on the phenotype. Inspired by studies that approached the problem from the perspective of protein structure change,18–20 we conducted MD simulations for all missense variants to gain a wealth of structural information about the mutant protein, such as RMSD, SASA, surface hydrophobic area, and percentage of β-strand in the secondary structure. These MD metrics measure the impact of variants on protein structure from different perspectives, providing a comprehensive representation of the genotype. For the phenotype, we also included PROS length in our analysis, which has been found to have a strong correlation with VA in patients with XLRS.56,57
Our genotype–phenotype analysis of missense variants revealed that CCV was significantly correlated with mutation-induced structural alterations, such as ΔRMSD, ΔSASA, and Δ hydrophobic area. In contrast, the widely used metric of CFT showed a weaker correlation or failed to show significance with these structural alterations. This finding suggests that CCV could potentially serve as an alternative indicator for OCT in genotype–phenotype correlation analyses. Although PROS length was found to have a strong correlation with VA in earlier studies,56,57 our analysis found that the correlations of PROS length with most genotype and phenotype features were not significant. Furthermore, the correlation between the amplitude of the scotopic a-wave and b-wave with the percentage change of the β-strand in the secondary structure suggested that a change in the secondary structure in the protein might result in a reduction of the amplitude of a-waves and the b-waves in scotopic ERGs.
Conclusions
We analyzed the clinical and genetic features of 37 Chinese patients with XLRS. In this process, we proposed that CCV could be a promising indicator to quantify the structural disorganization of the XLRS retina in three dimensions. To automate the calculation of CCV, we designed a self-developed software system, OCT–CCSEG. This system was applied to compare the retinal structural changes of patients with different genotypes in our cohort. The results from the MD-involved genotype–phenotype analysis of missense variants implied an association between the predicted protein structural alteration and the severity of XLRS as measured by the CCV and ERG. Moreover, CCV has the potential to serve as a valuable indicator of disease progression and may provide a foundation for determining appropriate therapeutic windows and endpoints in XLRS intervention trials.
Supplementary Material
Acknowledgments
The authors thank all of the patients and their family members for participating in the study.
Supported by grants from Beijing National Natural Science Foundation (L222083), CAMS Innovation Fund for Medical Sciences (CIFMS 2022-I2M-C&T-B-024), Fundamental Research Funds for the Central Universities (3332022007) and National Natural Science Foundation of China (82201192).
Disclosure: X. Wei, None; H. Li, None; T. Zhu, None; Z. Sun, None; R. Sui, None
References
- 1. Sieving PA, MacDonald IM, Hoang S, et al.. X-linked congenital retinoschisis. In: GeneReviews. Seattle, WA: University of Washington, Seattle; 1993. [PubMed] [Google Scholar]
- 2. Tantri A, Vrabec TR, Cu-Unjieng A, Frost A, Annesley WH Jr, Donoso LA. X-linked retinoschisis: a clinical and molecular genetic review. Surv Ophthalmol. 2004; 49: 214–230. [DOI] [PubMed] [Google Scholar]
- 3. George N, Yates J, Moore A. X linked retinoschisis. Br J Ophthalmol. 1995; 79: 697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sieving PA, Traboulsi E.. Juvenile Retinoschisis. Genetic Diseases of the Eye. New York: Oxford University Press; 1998: 347–355. [Google Scholar]
- 5. Apushkin M, Fishman G.. Fundus findings and longitudinal study of visual acuity loss in patients with X-linked retinoschisis. Invest Ophthalmol Vis Sci. 2004; 45: 4026–4026. [DOI] [PubMed] [Google Scholar]
- 6. Forsius H. Visual acuity in 183 cases of X-chromosomal retinoschisis. Can J Ophthalmol. 1973; 8: 385–393. [PubMed] [Google Scholar]
- 7. George N, Yates J, Moore A.. Clinical features in affected males with X-linked retinoschisis. Arch Ophthalmol. 1996; 114: 274–280. [DOI] [PubMed] [Google Scholar]
- 8. Pimenides D, George N, Yates J, et al.. X-linked retinoschisis: clinical phenotype and RS1 genotype in 86 UK patients. J Med Genet. 2005; 42: e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gerth C, Zawadzki RJ, Werner JS, Héon E.. Retinal morphological changes of patients with X-linked retinoschisis evaluated by Fourier-domain optical coherence tomography. Arch Ophthalmol. 2008; 126: 807–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gregori NZ, Berrocal AM, Gregori G, et al.. Macular spectral-domain optical coherence tomography in patients with X linked retinoschisis. Br J Ophthalmol. 2009; 93: 373–378. [DOI] [PubMed] [Google Scholar]
- 11. Yu J, Ni Y, Keane PA, Jiang C, Wang W, Xu G.. Foveomacular schisis in juvenile X-linked retinoschisis: an optical coherence tomography study. Am J Ophthalmol. 2010; 149: 973–978.e2. [DOI] [PubMed] [Google Scholar]
- 12. Apushkin MA, Fishman GA, Janowicz MJ.. Correlation of optical coherence tomography findings with visual acuity and macular lesions in patients with X-linked retinoschisis. Ophthalmology. 2005; 112: 495–501. [DOI] [PubMed] [Google Scholar]
- 13. Jeffrey BG, Cukras CA, Vitale S, Turriff A, Bowles K, Sieving PA.. Test–retest intervisit variability of functional and structural parameters in X-linked retinoschisis. Transl Vis Sci Technol. 2014; 3: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Riveiro-Alvarez R, Trujillo-Tiebas M-J, Gimenez-Pardo A, et al.. Correlation of genetic and clinical findings in Spanish patients with X-linked juvenile retinoschisis. Invest Ophthalmol Vis Sci. 2009; 50: 4342–4350. [DOI] [PubMed] [Google Scholar]
- 15. Hewitt AW, FitzGerald LM, Scotter LW, Mulhall LE, McKay JD, Mackey DA.. Genotypic and phenotypic spectrum of X-linked retinoschisis in Australia. Clin Exp Ophthalmol. 2005; 33: 233–239. [DOI] [PubMed] [Google Scholar]
- 16. Vincent A, Robson AG, Neveu MM, et al.. A phenotype–genotype correlation study of X-linked retinoschisis. Ophthalmology. 2013; 120: 1454–1464. [DOI] [PubMed] [Google Scholar]
- 17. Fahim AT, Ali N, Blachley T, Michaelides M.. Peripheral fundus findings in X-linked retinoschisis. Br J Ophthalmol. 2017; 101: 1555–1559. [DOI] [PubMed] [Google Scholar]
- 18. Sergeev Y, Caruso R, Meltzer M, Smaoui N, MacDonald I, Sieving P.. Molecular modeling of retinoschisin with functional analysis of pathogenic mutations from human X-linked retinoschisis. Hum Mol Genet. 2010; 19: 1302–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sergeev YV, Vitale S, Sieving PA, et al.. Molecular modeling indicates distinct classes of missense variants with mild and severe XLRS phenotypes. Hum Mol Genet. 2013; 22: 4756–4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sudha D, Neriyanuri S, Sachidanandam R, et al.. Understanding variable disease severity in X-linked retinoschisis: does RS1 secretory mechanism determine disease severity? PLoS One. 2018; 13(5): e0198086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mayer M. OCTSEG (Optical Coherence Tomography Segmentation and Evaluation GUI). Natick, MA: MathWorks; 2023. [Google Scholar]
- 22. Wei X, Li H, Wu S, Zhu T, Sui R.. Genetic analysis and clinical features of three Chinese patients with Oguchi disease. Doc Ophthalmol. 2023; 146: 17–32. [DOI] [PubMed] [Google Scholar]
- 23. Li J, Shi L, Zhang K, et al.. VarCards: an integrated genetic and clinical database for coding variants in the human genome. Nucleic Acids Res. 2018; 46: D1039–D1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sim NL, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC.. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012; 40: W452–W457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Adzhubei IA, Schmidt S, Peshkin L, et al.. A method and server for predicting damaging missense mutations. Nat Methods. 2010; 7: 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schwarz JM, Cooper DN, Schuelke M, Seelow D.. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014; 11: 361–362. [DOI] [PubMed] [Google Scholar]
- 27. Reva B, Antipin Y, Sander C.. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011; 39: e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shihab HA, Gough J, Cooper DN, et al.. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum Mutat. 2013; 34: 57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Choi Y, Sims GE, Murphy S, Miller JR, Chan AP.. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012; 7: e46688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Carter H, Douville C, Stenson PD, Cooper DN, Karchin R.. Identifying Mendelian disease genes with the variant effect scoring tool. BMC Genomics. 2013; 14(suppl 3): S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dong C, Wei P, Jian X, et al.. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum Mol Genet. 2015; 24: 2125–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jagadeesh KA, Wenger AM, Berger MJ, et al.. M-CAP eliminates a majority of variants of uncertain significance in clinical exomes at high sensitivity. Nat Genet. 2016; 48: 1581–1586. [DOI] [PubMed] [Google Scholar]
- 33. Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J.. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014; 46: 310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Quang D, Chen Y, Xie X.. DANN: a deep learning approach for annotating the pathogenicity of genetic variants. Bioinformatics. 2015; 31: 761–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shihab HA, Rogers MF, Gough J, et al.. An integrative approach to predicting the functional effects of non-coding and coding sequence variation. Bioinformatics. 2015; 31: 1536–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lu Q, Hu Y, Sun J, Cheng Y, Cheung KH, Zhao H.. A statistical framework to predict functional non-coding regions in the human genome through integrated analysis of annotation data. Sci Rep. 2015; 5: 10576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ioannidis NM, Rothstein JH, Pejaver V, et al.. REVEL: an ensemble method for predicting the pathogenicity of rare missense variants. Am J Hum Genet. 2016; 99: 877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S.. Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput Biol. 2010; 6: e1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Siepel A, Bejerano G, Pedersen JS, et al.. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005; 15: 1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Siepel A, Pollard KS, Haussler D.. New Methods for Detecting Lineage-Specific Selection. Berlin: Springer; 2006: 190–205. [Google Scholar]
- 41. Garber M, Guttman M, Clamp M, Zody MC, Friedman N, Xie X.. Identifying novel constrained elements by exploiting biased substitution patterns. Bioinformatics. 2009; 25: i54–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Desmet FO, Hamroun D, Lalande M, Collod-Béroud G, Claustres M, Béroud C.. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009; 37: e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ponzoni L, Peñaherrera DA, Oltvai ZN, Bahar I.. Rhapsody: predicting the pathogenicity of human missense variants. Bioinformatics. 2020; 36: 3084–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Richards S, Aziz N, Bale S, et al.. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015; 17: 405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. den Dunnen JT, Dalgleish R, Maglott DR, et al.. HGVS recommendations for the description of sequence variants: 2016 update. Human Mutat. 2016; 37: 564–569. [DOI] [PubMed] [Google Scholar]
- 46. Tolun G, Vijayasarathy C, Huang R, et al.. Paired octamer rings of retinoschisin suggest a junctional model for cell–cell adhesion in the retina. Proc Natl Acad Sci USA. 2016; 113: 5287–5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ.. GROMACS: fast, flexible, and free. J Comput Chem. 2005; 26: 1701–1718. [DOI] [PubMed] [Google Scholar]
- 48. Kabsch W, Sander C.. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983; 22: 2577–2637. [DOI] [PubMed] [Google Scholar]
- 49. Pettersen EF, Goddard TD, Huang CC, et al.. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004; 25: 1605–1612. [DOI] [PubMed] [Google Scholar]
- 50. Schymkowitz J, Borg J, Stricher F, Nys R, Rousseau F, Serrano L.. The FoldX web server: an online force field. Nucleic Acids Res. 2005; 33: W382–W388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vijayasarathy C, Sui R, Zeng Y, et al.. Molecular mechanisms leading to null-protein product from retinoschisin (RS1) signal-sequence mutants in X-linked retinoschisis (XLRS) disease. Hum Mutat. 2010; 31: 1251–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lin Z, Zang S, Maman Lawali DJA, et al.. Investigation of correlations between optical coherence tomography biomarkers and visual acuity in X-linked retinoschisis. Front Med (Lausanne). 2021; 8: 734888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rousseau O, Bourgault Y.. Heart segmentation with an iterative Chan-Vese algorithm. Available at: https://hal.science/hal-00403627v2/file/HeartSegmentation.pdf. Accessed January 30, 2024.
- 54. Huang Q, Zhou Y, Tao L, et al.. A Chan-Vese model based on the Markov chain for unsupervised medical image segmentation. Tsinghua Sci Technol. 2021; 26: 833–844. [Google Scholar]
- 55. Zhang N, Zhang J, Shi R.. An Improved Chan-Vese model for medical image segmentation. In: Proceedings of the 2008 International Conference on Computer Science and Software Engineering. Piscataway, NJ: Institute of Electrical and Electronics Engineers; 2008: 864–867. [Google Scholar]
- 56. Kwon HJ, Kim YN, Min CH, et al.. Macular microvasculature in X-linked retinoschisis: optical coherence tomography and optical coherence tomography angiography study. Retina. 2022; 42: 1939–1949. [DOI] [PubMed] [Google Scholar]
- 57. Yang HS, Lee JB, Yoon YH, Lee JY.. Correlation between spectral-domain OCT findings and visual acuity in X-linked retinoschisis. Invest Ophthalmol Vis Sci. 2014; 55: 3029–3036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.