Abstract
The concept of control is central to understanding and applications of biological network models. Some of their key structural features relate to control functions, through gene regulation, signaling, or metabolic mechanisms, and computational models need to encode these. Applications of models often focus on model-based control, such as in biomedicine or metabolic engineering. This paper presents an approach to model-based control that exploits two common features of biological networks, namely their modular structure and canalizing features of their regulatory mechanisms. The paper focuses on intracellular regulatory networks, represented by Boolean network models. A main result of this paper is that control strategies can be identified by focusing on one module at a time. This paper also presents a criterion based on canalizing features of the regulatory rules to identify modules that do not contribute to network control and can be excluded. For even moderately sized networks, finding global control inputs is computationally very challenging. The modular approach presented here leads to a highly efficient approach to solving this problem. This approach is applied to a published Boolean network model of blood cancer large granular lymphocyte (T-LGL) leukemia to identify a minimal control set that achieves a desired control objective.
Keywords: Boolean networks, modularity, control, canalization, gene regulatory networks
1. Introduction
With the availability of more experimental data and information about the structure of biological networks, computational modeling can capture increasingly complex features of biological networks [1, 2]. However, the increased size and complexity of dynamic network models also poses challenges in understanding and applying their structure as a tool for model-based control, important for a range of applications [3, 4]. This is our focus here. To narrow the scope of the problems we address we limit ourselves to intracellular networks represented by Boolean network (BN) models. BNs are widely used in molecular systems biology to capture the coarse-grained dynamics of a variety of regulatory networks [5]. They have been shown to provide a good approximation of the dynamics of continuous processes [6].
For the commonly-used modeling framework of ordinary differential equations, there is a well-developed theory of optimal control, which is largely absent for other modeling frameworks, such as Boolean networks or agent-based models, both frequently used in systems biology and biomedicine. Furthermore, control inputs, in many cases, are of a binary nature, such as gene knockouts or the blocking of mechanisms. For BNs, there is no readily available mathematical theory that could be used for control, leaving sampling and simulation. As networks get larger, with hundreds [7] or even thousands of nodes [8], this leaves few computational tools to identify control inputs for achieving preselected objectives, such as moving a network from one phenotype (e.g., cancer) to another (e.g., normal). One approach is to reduce the system in a way that the reduced system maintains relevant dynamical properties such as its attractors [9, 10]. This allows the control methods to be applied to the reduced system, and the same controls can then be used for the original system.
An approach that has not been used so far, to our knowledge, is to exploit the modular structure of many biological systems to identify control strategies by focusing on one module at a time. Modularity refers to the division of the system into separate units, or modules, that each have a specific function [11, 12]. Modularity is a fundamental property of biological systems that is essential for the evolution of new functions and the development of robustness [13, 14]. In [15], we developed a mathematical theory of modularity for Boolean network models and showed that one can identify network-level control inputs at the modular level. That is, we obtain global control inputs by identifying them at the local, modular level and assembling them to global control. This enables network control for much larger networks than would otherwise be computationally feasible. In this paper, we develop this approach into a mathematical theory of biological network control.
We further propose to use another property of biological networks, represented through Boolean network models. Almost all Boolean rules that describe the dynamics of over 120 published, expert-curated biological Boolean network models have the property that they exhibit some degree of canalization [16]. A Boolean function is canalizing if it has one or more variables that, when they take on a particular value, they determine the value of the function, irrespective of the values of all the other variables. As an example, any variable in a conjunctive rule (e.g., ) determines the value of the entire rule, when it takes on the value 0. We derive a criterion for Boolean network models whose Boolean functions are all canalizing, that can be used to exclude certain modules from needing to be considered for the identification of controls.
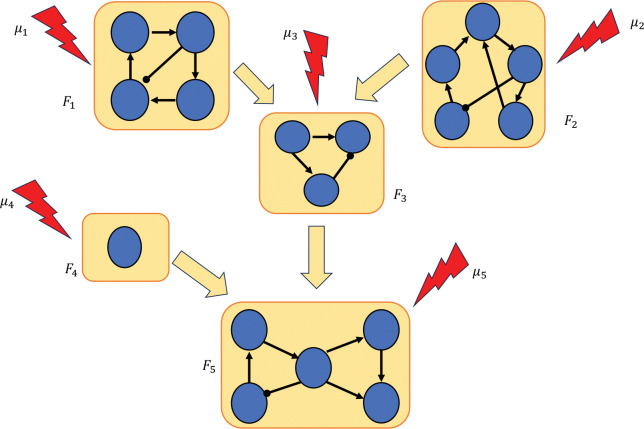
Our approach to control via modularity is summarized in Figure 1. We decompose the network into its constituent modules, then apply control methods to each module to identify a control target for the entire network. We show that by combining the controls of the modules, we can control the entire network. In the last part of the paper, we present theoretical results that exploit the canalizing properties of the regulatory functions to exclude certain modules from the control search. Finally, we demonstrate our approach by applying it to a published model of the blood cancer large granular lymphocyte (T-LGL) leukemia [17].
Figure 1:
Control via modularity. First, the network is decomposed into its constituent modules: . Then, controls are identified for each module. Combining the controls of the modules yields a set of controls for the whole network.
2. Background
We first describe Boolean networks and how to decompose a network into modules. In a BN, each gene is represented by a node that can be in one of two states: ON or OFF. Time is discretized as well, and the state of a gene at the next time step is determined by a Boolean function that takes as input the current states of a subset of the nodes in the BN. The dependence of a gene on the state of another gene can be graphically represented by a directed edge, and the wiring diagram contains all such dependencies.
2.1. Boolean Networks
Boolean networks can be seen as discrete dynamical systems. Specifically, consider variables each of which can take values in , where is the field with two elements, 0 and 1, where arithmetic is performed modulo 2. Then, a synchronously updated Boolean network is a function , where each coordinate function describes how the future value of variable depends on the present values of all variables. All variables are updated at the same time (synchronously).
The wiring diagram of a Boolean network is the directed graph with vertices and an edge from to if depends on . That is, if there exists such that , where is the th unit vector.
Example 2.1.
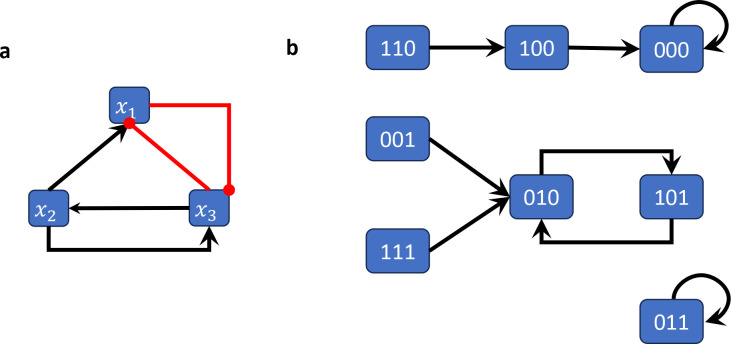
Figure 2a shows the wiring diagram of the Boolean network given by
Figure 2:
Wiring diagram and state space of the Boolean network in Example 2.1–2.8. (a) The wiring diagram encodes the dependency between variables. (b) The state space is a directed graph with edges between all states and their images. This graph therefore encodes all possible trajectories.
2.2. Dynamics of Boolean networks
Another directed graph associated with a BN is the state space. It describes all possible transition of the BN from one time step to another. The attractors of a BN are sets of states from which there is no escape as the system evolves. An attractor with a single state is also called a steady state (or fixed point). In mathematical models of intracellular regulatory networks, the attractors of the model are often associated with the possible phenotypes of the cell. This idea can be traced back to Waddington 18 and Kauffman 19. For example, in a model of cancer cells, the steady states of the model correspond to proliferative, apoptotic, or growth-arrest phenotypes 20. Mathematically, a phenotype is associated with a group of attractors where a subset of the system’s variables have the same states. These shared states are then used as biomarkers that indicate diverse hallmarks of the system.
There are two ways to describe the dynamics of a Boolean network , (i) as trajectories for all possible initial conditions, or (ii) as a directed graph with nodes in . Although the first description is less compact, it will allow us to formalize the dynamics of coupled networks.
Definition 2.2.
A trajectory of a Boolean network is a sequence of elements of such that for all .
Example 2.3.
For the network in the example above, , there are possible initial states giving rise to the following trajectories (commas and parenthesis for states are omitted for brevity).
We can see that and are periodic trajectories with period 2. Similarly, and are periodic with period 1. All other trajectories eventually reach one of these 4 states.
When seen as trajectories, and are different, but they can both be encoded by the fact that and . Similarly, and can be encoded by the equalities and . This alternative, more compact way of encoding the dynamics of a Boolean network is the standard approach, which we formalize next.
Definition 2.4.
The state space of a (synchronously updated) Boolean network is a directed graph with vertices in and an edge from to if .
Example 2.5.
Figure 2b shows the state space of the (synchronously updated) Boolean network from Example 2.1.
From the state space, one can easily obtain all periodic points, which form the attractors of the network.
Definition 2.6.
The space of attractors for a Boolean network is the collection of all minimal subsets satisfying .
The subset of sets of exact size 1 consists of all steady states (also known as fixed points) of .
-
The subset of sets of exact size consists of all cycles of length of .
Equivalently, an attractor of length is an ordered set with elements, , such that .
Remark 2.7.
In the case of steady states, the attractor may be denoted simply by .
Example 2.8.
The Boolean network from Example 2.1 has 2 steady states (i.e., attractors of length 1) and one cycle of length 2, which can be easily derived from its state space representation (Figure 2b).
2.3. Modules
Before defining modules we will describe the restriction of a Boolean network to a subset of its variables using the following example. For a formal definition of restrictions, please see [15].
Example 2.9.
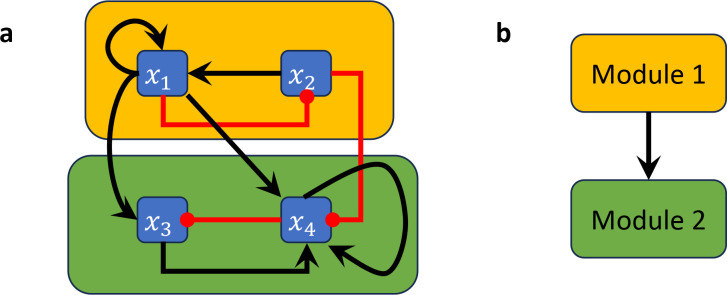
Consider the Boolean network
with wiring diagram in Figure 3a. The restriction of this network to is the 2-variable network , which forms the first module (indicated by the amber box in Figure 3a), while the restriction of to is the 2-variable network , which forms the second module (indicated by the green module in Figure 3a). Note that the wiring diagram of is always a subgraph of the wiring diagram of , irrespective of the choice of .
Figure 3:
Boolean network decomposition into modules. (a) Wiring diagram of a non-strongly connected Boolean network where the non-trivial modules are highlighted by amber and green boxes. (b) Directed acyclic graph describing the corresponding connections between the nontrivial modules.
Definition 2.10.
The wiring diagram of a Boolean network is strongly connected if every pair of nodes is connected by a directed path. That is, for each pair of nodes , in the wiring diagram with there exists a directed path from to (and vice versa). In particular, a one-node wiring diagram is strongly connected by definition.
Remark 2.11.
The wiring diagram of any Boolean network is either strongly connected or it is composed of a collection of strongly connected components where connections between different component move in only one direction.
Let be a Boolean network and let be the strongly connected components of its wiring diagram, with denoting the set of variables in strongly connected component . Then, the modules of are defined by .
Definition 2.12.
Let be the strongly connected components of the wiring diagram of a Boolean network . By setting if there exists at least one edge from a vertex in to a vertex in , we obtain a (directed) acyclic graph
which describes the connections between the strongly connected components of .
Example 2.13.
For the Boolean network from Example 2.9, the wiring diagram has two strongly connected components and with variables and (Figure 3a), connected according to the directed acyclic graph . The two modules of are given by the restriction of to and , that is, and (Figure 3b). Note that the module , i.e., the restriction of to , is simply the projection of onto the variables because does not receive feedback from the other component (i.e., because ).
3. Control via Modularity
In this section, we apply the modular decomposition theory described in the previous section and in [15] to make the control problem of Boolean networks more tractable. We show how the decomposition into modules can be used to obtain controls for each module, which can then be combined to obtain a control for the entire network. In this context, two types of control actions are generally considered: edge controls and node controls. For each type of control, one can consider deletions or constant expressions as defined below. The motivation for considering these control actions is that they represent the common interventions that can be implemented in practice. For instance, edge deletions can be achieved by the use of therapeutic drugs that target specific gene interactions, whereas node deletions represent the blocking of effects of products of genes associated to these nodes; see [21, 22].
Once the modules have been identified, different methods for phenotype control (that is, control of the attractor space) can be used to identify controls in these networks. Some of these methods employ stable motifs 23, feedback vertex sets 24, as well as algebraic approaches 25, 26, 27. For our examples below, we will use the methods defined in 23, 25, 24 to find controls for the simple networks.
A Boolean network with control is a Boolean network , where is a set that denotes all possible controls, defined below. The case of no control coincides with the original Boolean network, that is, . Given a control , the dynamics are given by . See [25] for additional details and examples of how to encode control edges and nodes in a Boolean network.
Definition 3.1 (Edge Control).
Consider the edge in the wiring diagram . The function
| (1) |
where is a constant in , encodes the control of the edge , since for each possible value of we have the following control settings:
If , . That is, the control is not active.
If , . In this case, the control is active, and the action represents the removal of the edge when , and the constant expression of the edge if . We use to denote that the control is active.
This definition can be easily extended for the control of many edges, so that we obtain , where is the number of edges in the wiring diagram. Each coordinate, , of in encodes the control of an edge .
Definition 3.2 (Node Control).
Consider the node in the wiring diagram . The function
| (2) |
encodes the control (knock-out or constant expression) of the node , since for each possible value of we have the following control settings:
For , , . That is, the control is not active.
For , , . This action represents the knock-out of the node .
For , , . This action represents the constant expression of the node .
For , , . This action changes the Boolean function to its negative value.
Definition 3.3.
A control stabilizes a network at an attractor when the resulting network after applying to (denoted here as ) has as its only attractor.
For a Boolean network , we let denote the set of its attractors. Whenever the Boolean network is decomposable into multiple constituent modules , we write where the semi-product operation indicates the coupling of the subnetworks, as described in 15. Furthermore, from the decomposition theory described in [15, the attractors of are of the form where is an attractor of the subnetwork, for . The following theorem takes advantage of the modular structure of the network to find controls one module at a time.
Theorem 3.4.
Given a decomposable network , if is a control that stabilizes in (whether is an existing attractor or a new one) and is a control that stabilizes in (whether is an existing attractor or a new one), then is a control that stabilizes in provided that either or is a steady state.
Proof.
Let be the resulting network after applying the control . Thus, the dynamics of is , that is . Similarly, the dynamics of is . That is, . Then,
Thus,
For the last equality we used the fact that the product of a steady state and a cycle (or vice versa) will result in only one attractor for the combined network. The former is not always true in general because multiplying two attractors (of length greater than 1) might result in several attractors for the composed network due to the attractors starting at different states.
It follows that there is only one attractor of and that attractor is . Thus, is stabilized by and we have .▫
Theorem 3.4 shows how the modular structure can be used to identify controls that stabilize the network in any desired state. In particular, we can use the modular structure of a network to find controls that stabilize a network at an existing attractor, which is often the case in biological control applications. We state this fact in the following corollary.
Corollary 3.5.
Given a decomposable network , let be an attractor of , where and and at least or is a steady state. If is a control that stabilizes in and is a control that stabilizes in , then is a control that stabilizes in .
Remark 3.6.
In Theorem 3.4 we required one of the stabilized attractors to be a steady state in order to be able to combine the controls from the modules. We can remove this requirement from Theorem 3.4 by using the following definition of stabilization for non-autonomous networks, which will guarantee that and can be combined in a unique way, resulting in a unique attractor of the whole network.
Definition 3.7.
A non-autonomous Boolean network is defined by
where and is a sequence with elements in . We call this type of network non-autonomous because its dynamics will depend on . We use to denote this non-autonomous network.
A state is a steady state of if for all . Similarly, an ordered set with elements, , is an attractor of length of if , , , Note that in general is not necessarily of period and may even not be periodic.
If for some network (that is, it does not depend on ) for all , then , and this definition of attractors coincides with the classical definition of attractors for (autonomous) Boolean networks (Definition 2.6).
Definition 3.8.
Consider a controlled non-autonomous network given by , where is a trajectory representation of an attractor of an upstream network. We say that a control stabilizes this network, (defined as in Definition 3.7), at an attractor when the resulting network after applying (denoted here as ) has as its unique attractor. For non-autonomous networks the definition of unique attractor requires that has a unique periodic trajectory up to shifting of (which is automatically satisfied if or is a steady state).
Example 3.9.
Consider, again, the network , which can be decomposed into , with and . Suppose we want to stabilize in 0110 (which is not an attractor of ). Note that the non-autonomous network and .
Consider the control . That is, the control is the combined action of setting the input from to to 1 and the input from to to 0. The control stabilizes at 01, which is not an original attractor of . Let . Note that the space of attractors for is .
Now consider the control . That is, the control is the combined action of setting the input from to to 1 and the input from to to 0. This control stabilizes at , which is not an original attractor of .
Finally, the control stabilizes at . Note that is a new attractor for .
Theorem 3.4 uses the modular structure of a Boolean network to identify controls that stabilize the network in any desired attractor. In biological applications, the attractors typically correspond to distinct biological phenotypes (defined more rigorously in the next section) and a common question is how to force a network to always transition to only one of these phenotypes. For example, cancer biologists may use an appropriate Boolean network model with the two phenotypes proliferation and apoptosis to identify drug targets (i.e., edge or node controls), which force the system to always undergo apoptosis. The following example illustrates this specific control aspect, described in Corollary 3.5
Example 3.10.
Consider again the network from Example 3.9 with and . Suppose we want to stabilize in 1111, which is an attractor of (but not the only one). Note that the non-autonomous network and . Let .
The edge control (that is, the control that constantly expresses the edge from to ) stabilizes at . The space of attractors for is then . Note that would be an alternative control.
The edge control (that is, the control that constantly expresses the edge from to ) stabilizes at . Again, note that would be an alternative control.
Now, the control stabilizes at .
4. Control via Modularity and Canalization
In addition to using the modular structure of the network, we can take advantage of the canalizing structure of the regulatory functions to identify contol targets.
We first review some concepts and definitions, and introduce the concept of canalization.
Definition 4.1.
A Boolean function is essential in the variable if there exists an such that
where is the ith unit vector. In that case, we also say depends on .
Definition 4.2.
A Boolean function is canalizing if there exists a variable , a Boolean function and such that
In that case, we say that canalizes (to ) and call the canalizing input (of ) and the canalized output.
Definition 4.3.
A Boolean function is nested canalizing with respect to the permutation , inputs and outputs , if
The last line ensures that actually depends on all variables.
We restate the following stratification theorem for reference.
Theorem 4.4([28]).
Every Boolean function can be uniquely written as
| (3) |
where each is a nonconstant extended monomial, is the core polynomial of , and is the canalizing depth. Each appears in exactly one of , and the only restrictions are the following “exceptional cases”:
1. If and , then ;
2. If and and , then .
When is not canalizing (i.e., when ), we simply have .
From Equation 3, we can directly derive an important summary statistic of canalizing functions.
Definition 4.5.
Given a Boolean function represented as in Equation 3, we call the extended monomials the layers of and define, as in [29], the layer structure as the vector , which describes the number of variables in each layer. Note that is nested canalizing if and only if .
Example 4.6.
The Boolean functions and are nested canalizing. consists of three layers with layer structure (1, 1, 2), while possesses only two layers and layer structure (1, 3).
While finding the layer structure of a Boolean function is an NP-hard problem, there exist several algorithmic implementations [30].
A phenotype is associated with a group of attractors where a subset of the system’s variables have a shared state. The states of the shared attractors will be called markers of the phenotype.
Suppose is a decomposable network, and that there is a phenotype that depends on variables in only (that is, all markers of the phenotype are part of ), and that we wish to control the phenotype through . The most straightforward approach is to set the variables that the phenotype depends on to the appropriate values that result in the desired phenotype. However, such intervention may not be experimentally possible. Instead, we can exploit the canalizing properties of the functions corresponding to the nodes connecting the modules and to identify control targets.
Lemma 4.7.
Suppose is a decomposable network. Suppose further that only one node with update function is regulated by nodes in . If is canalizing with layers, let be the lowest (i.e., most important) layer of , which contains nodes from . If all regulators of from appear in the core polynomial, we set . Then, setting to its canalizing value decouples the systems and , as long as appears in a layer .
Proof.
The lemma is a direct consequence of Theorem 4.4. If receives its canalizing input and is in a more important layer of than all variables in , then none of these variables can affect anymore. Thus, controlling to receive its canalizing input eliminates the link between and . □
Theorem 4.8.
Suppose is a decomposable network. If for some ,
only one node with update function is regulated by nodes in , and
is a canalizing function, which possesses none of the variables from in its most important layer, and
the phenotype of interest depends only on variables in and modules that are not “downstream” of in the directed acyclic graph of (see Defintion 2.12), then the module can be excluded from the control search by setting any node to its canalizing input, as long as this node appears in a more dominant layer of than all variables of .
Proof.
By Lemma 4.7, setting to its canalizing value results in decoupling and . will no longer have any effect on , and thus, due to condition (iii), on the phenotype of interest. can therefore be removed from the control search.▫
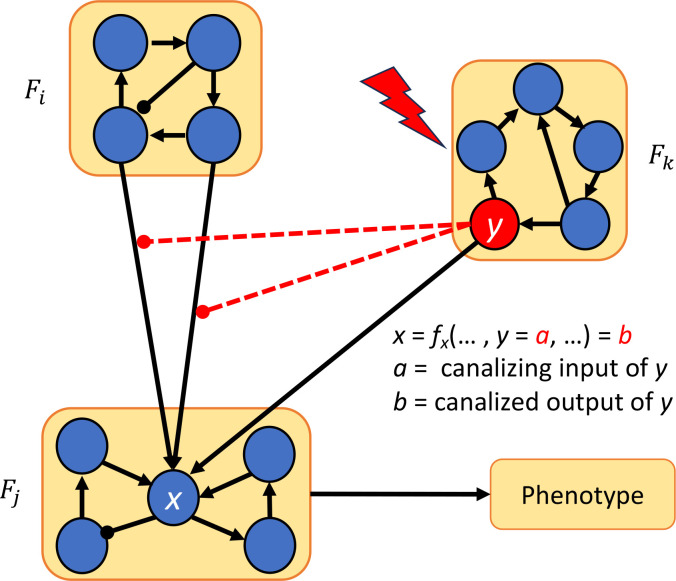
Theorem 4.8 is illustrated in Fig. 4 Note that node can be in or some other module as in the figure.
Figure 4:
Control via modularity and canalization. Once the network is decomposed into modules , we could override the effect of module by the using another module ( in this case) whose variables are inputs of that are located in a higher canalizing hierarchy than the layers containing the variables of .
Remark 4.9.
The method in Theorem 4.8 can be extended to the case when and are connected via multiple nodes. In that case decoupling is achieved through the same procedure presented above, applied to each node in that is regulated by nodes in .
In Theorem 4.8, we assumed that none of the variables of are in the most dominant layer in the update rules of variables in . If some of the variables of are in the most dominant layer, we can still remove module from the control search using an edge control, as shown in the following theorem.
Theorem 4.10.
Suppose is a decomposable network. If for some ,
only one node with update function is regulated by nodes in , and
is a canalizing function with some variables from in its most important layer, and
the phenotype of interest depends only on variables in and modules that are not “downstream” of in the directed acyclic graph of (see Defintion 2.12), then the module can be excluded from the control search by applying an edge control to any input in the most dominant layer of .
Proof.
Let such that , and that is located in the most dominant layer . Then, setting to its canalizing value results in decoupling the subnetworks and . Thus, will no longer have any effect on and thus it can be removed from the control search. □
Remark 4.11.
The method can be extended to the case when and are connected via multiple nodes. In that case decoupling is achieved through the same procedure presented above applied to each node in with regulators from .
To showcase these methods, we will now decompose a published Boolean network model into its modules, and then identify the minimal set of controls for the entire network by exploiting the canalizing structure of the regulatory functions within the modules. The identified set of controls will force the entire system into a desired attractor.
Example 4.12.
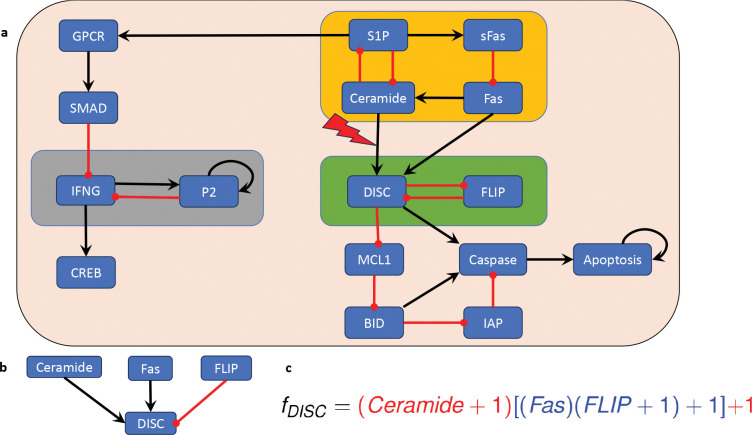
We consider a Boolean network model for the blood cancer large granular lymphocyte (T-LGL) leukemia, which was published in [17]. T-LGL leukemia is a clonal hematological disorder characterized by persistent increases of large granular lymphocytes in the absence of reactive cause [31]. The wiring diagram of this model is depicted in Figure 5a. This network has 16 nodes and three nontrivial modules (highlighted by the amber, green, and gray boxes in Figure 5a). The control objective here is to identify control targets that lead the system to programmed cell death. In other words, we aim to direct the system into an attractor that has the marker apoptosis ON.
Figure 5:
(a) Wiring diagram of the T-LGL model, published in [17], which describes the mechanisms that regulate apoptosis. The non-trivial modules are highlighted by amber, green, and gray boxes. (b) The regulatory inputs of the node DISC. (c) Writing the regulatory function corresponding to node DISC in its standard monomial form (Theorem 4.4) reveals its canalizing structure.
Since the model has three nontrivial modules, the approach described in Section 3 would require us to identify control targets for three modules. However, an exploitation of the canalizing structure and common sense reveals that we do not need to control every module to ensure apoptosis, the desired control objective. First, irrespective of canalization, the module highlighted in gray in Figure 5a does not affect the phenotype apoptosis. Therefore, we can focus on the modules “upstream” of apoptosis (i.e., the amber and green modules in Figure 5a).
In this case, we will apply Theorem 4.10 to identify control targets for this model. We note that the edges from the upstream module (amber box in Figure 5a) to the downstream module (green box in Figure 5a) all end in the node DISC. Therefore, we will investigate the canalizing properties of the regulatory function of DISC (see Figure 5b),
Using the approach described in [30], we find that has two canalizing layers, and and its layering structure is given by (see Figure 5c)
We note that the only variable in the most important canalizing layer, Ceramide, is in the upstream module. Thus, we can decouple the modules via an edge control on the connection between the upstream and downstream modules. That is, the constant expression of the edge from Ceramide to DISC will decouple the two modules and will lead to constant expression of DISC. We can check that this control is effective at stabilizing the system in the desired attractor and the control set obviously has minimal size.
In summary, in this example we used an edge control to decouple the upstream and downstream modules and then identified a control target in the downstream module which contains the markers of the phenotype of interest.
5. Conclusion
Model-based control is a mainstay of industrial engineering, and there is a well-developed mathematical theory of optimal control that can be applied to models consisting of systems of ordinary differential equations. While this model type is also commonly used in biology, for instance in biochemical network modeling or epidemiology and ecology, there are many biological systems that are more suitably modeled in other ways. Boolean network models provide a way to encode regulatory rules in networks that can be used to capture qualitative properties of biological networks, when it is unfeasible or unnecessary to determine kinetic information. While they are intuitive to build, they have the drawback that there is very little mathematical theory available that can be used for model analysis, beyond simulation approaches. And for large networks, simulation quickly becomes ineffective.
The results in this paper, building on those in [15], can be considered as a contribution to a mathematical control theory for Boolean networks, incorporating key features of biological networks. There are many open problems that remain, and we hope that this work will inspire additional developments.
Our concrete contributions here are as follows. The modularization method makes the control search far more efficient and allows us to combine controls at the module level obtained with different control methods. For example, methods based on computational algebra [25, 27] can identify controllers that can create new (desired) steady states, which other methods cannot. Feedback vertex set [32, 24] is a structure-based method that identifies a subset of nodes whose removal makes the graph acyclic. Stable motifs [23] are based on identifying strongly connected subgraphs in the extended graph representation of the Boolean network. Other control methods include [33, 34, 35]. We can use any combination of these methods to identify the controls in each module.
6. Acknowledgments
Author Matthew Wheeler was supported by The American Association of Immunologists through an Intersect Fellowship for Computational Scientists and Immunologists. This work was further supported by the Simons foundation [grant numbers 712537 (to C.K.), 850896 (to D.M.), 516088 (to A.V.)]; the National Institute of Health [grant number 1 R01 HL169974-01 (to R.L.)]; and the Defense Advanced Research Projects Agency [grant number HR00112220038 (to R.L.)]. The authors also thank the Banff International Research Station for support through its Focused Research Group program during the week of May 29, 2022 (22frg001), which was of great help in framing initial ideas of this paper.
References
- [1].Aguilar Boris, Gibbs David L, Reiss David J, McConnell Mark, Danziger Samuel A, Dervan Andrew, Trotter Matthew, Bassett Douglas, Hershberg Robert, Ratushny Alexander V, et al. A generalizable data-driven multicellular model of pancreatic ductal adenocarcinoma. GigaScience, 9(7):giaa075, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Plaugher Daniel and Murrugarra David. Modeling the pancreatic cancer microenvironment in search of control targets. Bulletin of Mathematical Biology, 83(11):1–26, 2021. [DOI] [PubMed] [Google Scholar]
- [3].Rozum Jordan and Albert Réka. Leveraging network structure in nonlinear control. NPJ systems biology and applications, 8(1):36, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Plaugher Daniel and Murrugarra David. Phenotype control techniques for boolean gene regulatory networks. Bulletin of Mathematical Biology, 85(10):1–36, 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Schwab Julian D, Kühlwein Silke D, Ikonomi Nensi, Kühl Michael, and Kestler Hans A. Concepts in boolean network modeling: What do they all mean? Computational and structural biotechnology journal, 18:571–582, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Veliz-Cuba Alan, Arthur Joseph, Hochstetler Laura, Klomps Victoria, and Korpi Erikka. On the relationship of steady states of continuous and discrete models arising from biology. Bulletin of mathematical biology, 74:2779–2792, 2012. [DOI] [PubMed] [Google Scholar]
- [7].Singh Vidisha, Naldi Aurelien, Soliman Sylvain, and Niarakis Anna. A large-scale boolean model of the rheumatoid arthritis fibroblast-like synoviocytes predicts drug synergies in the arthritic joint. NPJ systems biology and applications, 9, 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Aghamiri Sara Sadat, Singh Vidisha, Naldi Aurélien, Helikar Tomáš, Soliman Sylvain, and Niarakis Anna. Automated inference of boolean models from molecular interaction maps using casq. Bioinformatics, 36(16):4473–4482, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Veliz-Cuba A.. Reduction of Boolean network models. Journal of Theoretical Biology, 289:167–172, 2011. [DOI] [PubMed] [Google Scholar]
- [10].Saadatpour Assieh, Albert Réka, and Reluga Timothy. A reduction method for boolean network models proven to conserve attractors. SIAM Journal on Applied Dynamical Systems, 12:1997–2011, 01 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kashtan Nadav and Alon Uri. Spontaneous evolution of modularity and network motifs. Proceedings of the National Academy of Sciences, 102(39):13773–13778, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hartwell Leland H, Hopfield John J, Leibler Stanislas, and Murray Andrew W. From molecular to modular cell biology. Nature, 402(Suppl 6761):C47–C52, 1999. [DOI] [PubMed] [Google Scholar]
- [13].Kitano Hiroaki. Biological robustness. Nature Reviews Genetics, 5(11):826–837, 2004. [DOI] [PubMed] [Google Scholar]
- [14].Lorenz Dirk M, Jeng Alice, and Deem Michael W. The emergence of modularity in biological systems. Physics of life reviews, 8(2):129–160, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kadelka Claus, Wheeler Matthew, Alan Veliz-Cuba David Murrugarra, and Laubenbacher Reinhard. Modularity of biological systems: a link between structure and function. Journal of the Royal Society Interface, 20(207):20230505, 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kadelka Claus, Butrie Taras-Michael, Hilton Evan, Kinseth Jack, Schmidt Addison, and Serdarevic Haris. A meta-analysis of boolean network models reveals design principles of gene regulatory networks. Science Advances, 10(2):eadj0822, 2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Saadatpour Assieh, Wang Rui-Sheng, Liao Aijun, Liu Xin, Loughran Thomas P, Albert István, and Albert Réka. Dynamical and structural analysis of a t cell survival network identifies novel candidate therapeutic targets for large granular lymphocyte leukemia. PLoS Comput Biol, 7(11):e1002267, Nov 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Conrad Hal Waddington. The strategy of the genes. Routledge, 2014. [Google Scholar]
- [19].Kauffman S A. Metabolic stability and epigenesis in randomly constructed genetic nets. J Theor Biol, 22(3):437–67, Mar 1969. [DOI] [PubMed] [Google Scholar]
- [20].Plaugher Daniel, Aguilar Boris, and Murrugarra David. Uncovering potential interventions for pancreatic cancer patients via mathematical modeling. Journal of theoretical biology, 548:111197, 2022. [DOI] [PubMed] [Google Scholar]
- [21].Choi Minsoo, Shi Jue, Jung Sung Hoon, Chen Xi, and Cho Kwang-Hyun. Attractor landscape analysis reveals feedback loops in the p53 network that control the cellular response to dna damage. Science signaling, 5(251):ra83–ra83, 2012. [DOI] [PubMed] [Google Scholar]
- [22].Wooten David J, Zañudo Jorge Gómez Tejeda, Murrugarra David, Perry Austin M, Dongari-Bagtzoglou Anna, Laubenbacher Reinhard, Nobile Clarissa J, and Albert Réka. Mathematical modeling of the candida albicans yeast to hyphal transition reveals novel control strategies. PLoS computational biology, 17(3):e1008690, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zanudo Jorge GT and Albert Réka. Cell fate reprogramming by control of intracellular network dynamics. PLoS computational biology, 11(4):e1004193, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zañudo Jorge Gomez Tejeda, Yang Gang, and Albert Réka. Structure-based control of complex networks with nonlinear dynamics. Proceedings of the National Academy of Sciences, 114(28):7234–7239, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Murrugarra David, Veliz-Cuba Alan, Aguilar Boris, and Laubenbacher Reinhard. Identification of control targets in boolean molecular network models via computational algebra. BMC systems biology, 10:1–11, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Murrugarra D and Dimitrova ES. Molecular network control through boolean canalization. eurasip j. Bioinformatics Syst. Biol, 9, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Vieira Luis Sordo, Laubenbacher Reinhard C, and Murrugarra David. Control of intracellular molecular networks using algebraic methods. Bulletin of mathematical biology, 82(1):2, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].He Qijun and Macauley Matthew. Stratification and enumeration of boolean functions by canalizing depth. Physica D: Nonlinear Phenomena, 314:1–8, 2016. [Google Scholar]
- [29].Kadelka Claus, Kuipers Jack, and Laubenbacher Reinhard. The influence of canalization on the robustness of boolean networks. Physica D: Nonlinear Phenomena, 353:39–47, 2017. [Google Scholar]
- [30].Dimitrova Elena, Stigler Brandilyn, Kadelka Claus, and Murrugarra David. Revealing the canalizing structure of boolean functions: Algorithms and applications. Automatica, 146:110630, 2022. [Google Scholar]
- [31].Rashid Anila, Khurshid Mohammad, and Ahmed Arsalan. T-cell large granular lymphocytic leukemia: 4 cases. Blood research, 49(3):203–205, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Mochizuki Atsushi, Fiedler Bernold, Kurosawa Gen, and Saito Daisuke. Dynamics and control at feedback vertex sets. ii: A faithful monitor to determine the diversity of molecular activities in regulatory networks. Journal of theoretical biology, 335:130–146, 2013. [DOI] [PubMed] [Google Scholar]
- [33].Choo Sang-Mok, Ban Byunghyun, Joo Jae Il, and Cho Kwang-Hyun. The phenotype control kernel of a biomolecular regulatory network. BMC systems biology, 12(1):1–15, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Cifuentes-Fontanals Laura, Tonello Elisa, and Siebert Heike. Control in boolean networks with model checking. Frontiers in Applied Mathematics and Statistics, 8, 2022. [Google Scholar]
- [35].Borriello Enrico and Daniels Bryan C. The basis of easy controllability in boolean networks. Nature communications, 12(1):1–15, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]