Abstract
Biological systems are inherently noisy; however, they produce highly stereotyped tissue morphology. Drosophila pupal wings show a highly stereotypic folding through uniform expansion and subsequent buckling of wing epithelium within a surrounding cuticle sac. The folding pattern produced by buckling is generally stochastic; it is thus unclear how buckling leads to stereotypic tissue folding of the wings. We found that the extracellular matrix (ECM) protein, Dumpy, guides the position and direction of buckling-induced folds. Dumpy anchors the wing epithelium to the overlying cuticle at specific tissue positions. Tissue-wide alterations of Dumpy deposition and degradation yielded different buckling patterns. In summary, we propose that spatiotemporal ECM remodeling shapes stereotyped tissue folding through dynamic interactions between the epithelium and its external structures.
Remodeling of extracellular matrix protein, Dumpy, guides epithelial tissue folding during Drosophila wing development.
INTRODUCTION
In origami, a single sheet of paper is folded in a sequence to create a stereotypic shape. Similar folded structures are found everywhere in life, including leaves in buds, insect wings in pupae, and internal organs such as the brain and gastrointestinal tract. Intriguingly, many of the folded structures are highly stereotyped among individuals. These biological folded structures are often formed during development through a physical process called buckling, which generates folds of elastic sheets in response to compressive forces (1, 2). Because most of the internal organs develop not in free space but under the physical constraints of the surrounding structures [i.e., neighboring tissues and extracellular matrices (ECMs)], tissues need to buckle to relax the compressive stresses imposed by their surroundings. However, unlike origami, folds generated by buckling are generally difficult to predict (2) because they are influenced by various physical interactions with their surroundings. Therefore, how tissues achieve stereotypic morphology counteracting the stochasticity of buckling remains an important question.
Here, we demonstrate that the core factor in regulating the stereotypic folding is an apical ECM protein Dumpy (Dpy). We found that the spatiotemporal regulation of Dpy “deposition” and “destruction” ensures the stereotypic three-dimensional (3D) tissue shapes of Drosophila wing. While it is widely acknowledged that patterned cell behaviors, such as cell shape changes and cell rearrangement, can profoundly affect tissue morphology, our findings highlight the role of external cues provided by ECM remodeling in shaping 3D tissue structures.
RESULTS
Stereotypic folding of Drosophila pupal wing epithelia emerges within a cuticle sac
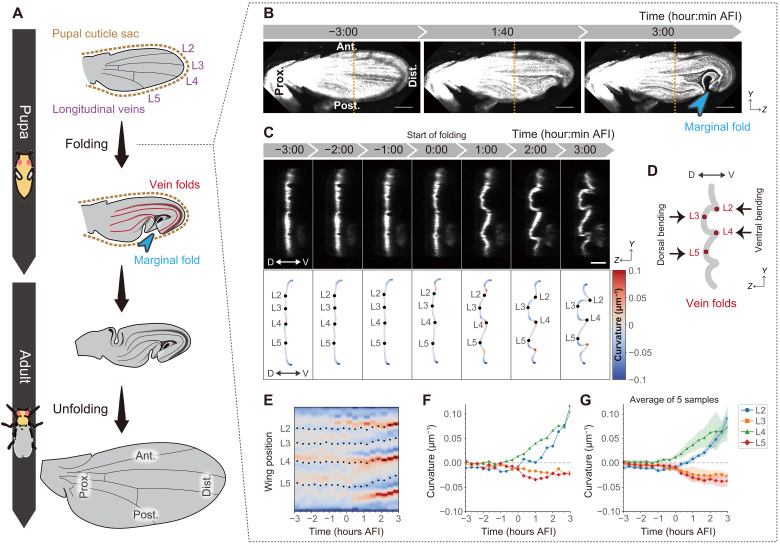
To investigate the mechanisms underlying the production of highly stereotyped tissue morphology, we studied the wing folding of Drosophila pupae. The wing folding process is considered to be caused by buckling with an almost uniform expansion of wing epithelia (fig. S1) in the confined space of the pupal cuticle (brown dotted lines in Fig. 1A) (3–6). The folding pattern is highly reproducible (fig. S2). We thus focused on the two types of major stereotypic folds, one along four longitudinal veins (L2 to L5) (“vein folds,” red lines in Fig. 1A) and the other perpendicular to the margin of the wing blade formed by bending the entire wing in the posterior direction (“marginal fold,” blue arrowhead in Fig. 1A). To observe the sequential dynamics of the folding, we performed time-lapse imaging of the pupa (Fig. 1, B and C, and movie S1). In movie S1, wing expansion and the formation of the vein folds initiated from about 39:40 hours after puparium formation (APF), which we define as the timing of folding initiation. The wing expansion was not due to cell division but due to cell surface expansion through cell flattening as reported previously (fig. S3 and movie S2) (4–6). Meanwhile, from about 1.7 hours after folding initiation (AFI), the marginal fold starts forming at the distal-posterior region (blue arrowhead in Fig. 1B). In addition, laser perturbation experiments support that the wings are under tensile conditions before folding, but after initiating the wing expansion, they become less tensile and rather compressive (fig. S4).
Fig. 1. Emergence of stereotypic folding within a cuticle sac.
(A) Schematics of wing morphogenesis. (B) Maximum projections of confocal time-lapse images in a control wing labeling dorsal cells. (C) Top: Anterior-posterior cross sections along the orange dotted lines in (B). Bottom: Color-coded tissue curvature with longitudinal veins (black dots). (D) Schematics of folded pupal wing representing vein folds on anterior-posterior cross-sectional view. (E to G) (E and F) Tissue curvature measurement from a control wing in (C). (E) Spatiotemporal color map of curvature with longitudinal veins (black dots). The color range is identical to the color bar shown in (C). (F and G) Time evolution of the average local curvature around each longitudinal vein for a pupa (F) and the mean with SD for the five different pupae shown in fig. S5B (G). Scale bars, 100 μm (B) and 50 μm (C). AFI, after folding initiation.
We first focused on the formation of the vein folds that occurs initially. Among the four vein folds, the L2 and L4 vein folds bent ventrally (“ventral bending”), and the L3 and L5 vein folds bent dorsally (“dorsal bending”) (Fig. 1, C, top, and D). To map the folding dynamics, we calculated the wing surface curvature along the anterior-posterior axis (Fig. 1C, bottom). The profiles of the local curvature of five different pupae reproducibly show alternating positive and negative curvatures (Fig. 1, C to G, and fig. S5, A and B). These observations indicate that wings are stereotypically folded along veins.
Dpy along veins regulates the stereotypic buckling direction by connecting the wing epithelium and the pupal cuticle along veins
Because the position and direction of folds produced by buckling are generally stochastic (2), there must be unknown mechanisms simultaneously acting to fix the positions and directions of folds in wings. In several developmental processes, such as ventral furrow formation of Drosophila gastrula and neural tube formation of vertebrates, epithelial tissues locally generate intrinsic forces using actomyosin for stereotypic folding (7, 8). To test whether the actomyosin force is responsible for the stereotypic folding of wings, we knocked down the myosin II regulatory light chain encoded by spaghetti squash (sqh) (9); however, the wing folding pattern was not affected (fig. S6 and movies S3 and S4), suggesting that the local differences in cell behaviors driven by actomyosin hardly contribute to determining the folding positions in the pupal wing.
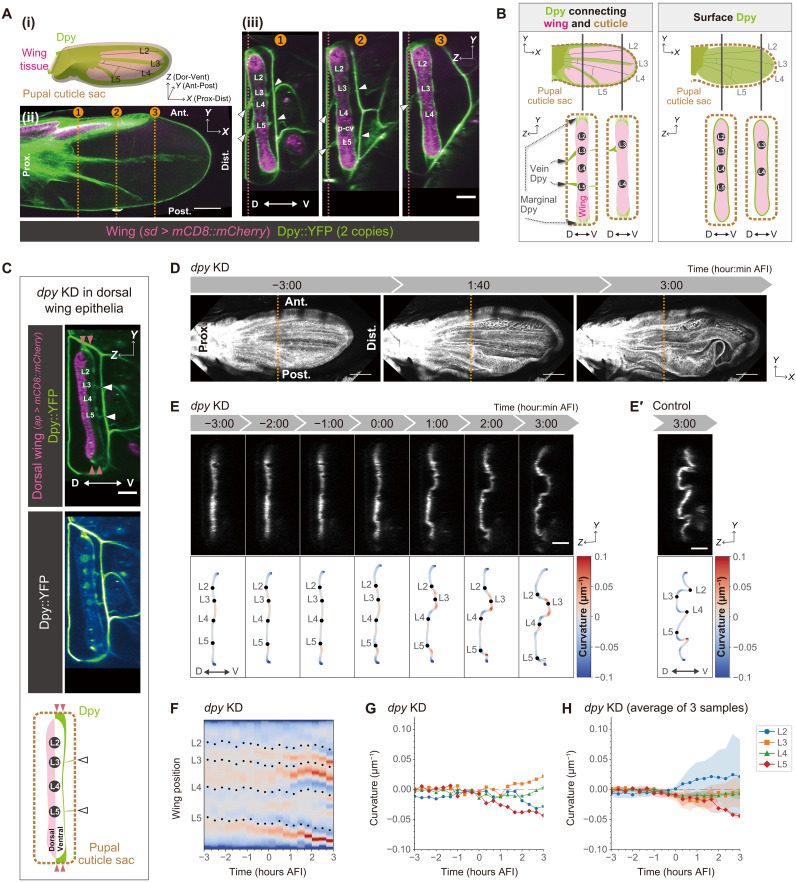
We then investigated whether the interactions with the extrinsic environment might control the folding position. We focused on Dpy, a fibrous apical ECM protein that mediates the attachment of the epithelium to the surrounding cuticle in pupa (10–13). We imaged prefolded wings of flies harboring the protein-trapped dpy allele (Dpy::YFP) (14, 15) using a multiphoton laser scanning microscope (MP microscope) (Fig. 2A, fig. S7, and movie S5): We found that Dpy covers the apical cell surface of the wing epithelium (Fig. 2B, right, “Surface Dpy”) and that Dpy is also present on the pupal cuticle. In addition, as reported previously (10, 11), a dense accumulation of fibrous Dpy connecting the wing epithelium and the pupal cuticle was found at the wing margin (“Marginal Dpy”), hinge region, and along the vicinity of L3 and L5 veins (“Vein Dpy”) (Fig. 2B, left, “Dpy connecting wing and cuticle”). The specific accumulation pattern of the fibrous Dpy is not solely determined by the difference in the expression level (fig. S8, A and B) but by the genetic and molecular mechanisms underlying vein pattern formation (fig. S9, A to D).
Fig. 2. Dumpy (Dpy)regulates the stereotypic buckling direction by connecting the wing epithelium and the pupal cuticle along veins.
(A) (i) Schematics of a prefolded wing. (ii and iii) Multiphoton laser scanning (MP) microscope snapshot images of a prefolded control wing, which expresses endogenous Dpy::YFP and mCD8::mCherry induced by sd-Gal4 [32.5 hours after puparium formation (APF)]. (ii) XY view of a slice at the red dotted lines in (iii). (iii) Anterior-posterior cross sections along the orange dotted lines in (ii). White arrowheads indicate the position of the vein Dpy. (B) Schematics showing the two different types of Dpy localization before folding. (C) MP microscope snapshot images (top and middle) and schematics (bottom) of a wing with depleted dorsal Dpy. Marginal Dpy (red arrowheads), vein Dpy (white arrowheads), and surface Dpy remains on the ventral side. (D) Maximum projections of confocal time-lapse images of a wing with depleted dorsal Dpy. (E) Top: Anterior-posterior cross sections along the orange dotted lines in (D). Bottom: Color-coded tissue curvature with longitudinal veins (black dots). (E′) For comparison with the dpy knockdown results (E), control results from Fig. 1C are presented. (F to H) (F and G) Tissue curvature measurement from the wing in (E). (F) Spatiotemporal color map of curvature with longitudinal veins (black dots). The color range is identical to the color bar shown in (C). (G and H) Time evolution of the average local curvature around each longitudinal vein for a pupa (G) and the mean with SD for the three different pupae (samples #1, #3, and #5 shown in fig. S5D) that allowed curvature measurement over the entire −3 to 3 hours after folding initiation (AFI) (H). Scale bars, 100 μm [A (ii) and D], 50 μm [A (iii), C, E, and E′)].
We then noticed that the vein Dpy corresponded to the positions of dorsal bending folds (Fig. 1, C to G; fig. S10; and movie S6). The vein Dpy was present both on the dorsal and ventral sides; however, its localization pattern was different. On the dorsal side, the vein Dpy were continuously localized proximally-distally, whereas on the ventral side, it was hardly detectable at the distal half of the wing (Fig. 2A (iii), cross section 3). On the basis of these observations, we hypothesized that at the positions along L3 and L5 veins, the wing is more strongly anchored to the cuticle dorsally than ventrally, resulting in the dorsally biased stereotypic bending along L3 and L5 veins during buckling. To test this hypothesis, we depleted Dpy only from the dorsal wing by RNAi knockdown (KD) (Fig. 2, C to H; fig. S11; and movie S7). In the example shown in Fig. 2E, L2 and L4 veins are bent to the dorsal side and L3 vein to the ventral side, in the opposite directions to the control shown in Fig. 2E′. Moreover, the folding position and direction varied among individual wings (Fig. 2F and fig. S5C), and the curvature of most of the veins was close to zero (Fig. 2, G and H, and fig. S5D). These results indicate that Dpy depletion from the dorsal wing epithelium randomized the pattern of vein folds. In contrast, Dpy depletion from the ventral wing epithelium exhibited a lesser impact on the folding pattern (fig. S12). Together, we conclude that dorsal Dpy, but not ventral Dpy, plays a pivotal role in determining the position and direction of buckling by anchoring the wing epithelium to the cuticle at specific positions in the tissue. The importance of Dpy anchorage is further supported by the observation that Dpy overexpression (OE), which does not result in considerable changes in the Dpy anchorage pattern (fig. S8, A and B), leads to the stereotypic folding pattern (fig. S8, C and D, and movies S8 and S9). In addition, although the mutant wing lacking veins lost most of the vein Dpy, residual Dpy-anchoring regions still form dorsal folds (fig. S9, E and F, and movie S10). These results suggest that the Dpy anchorage, rather than differential expression of Dpy or mechanical/biochemical properties of the vein cells, instructs the pattern of buckling.
Dpy degradation is indispensable for the marginal fold formation
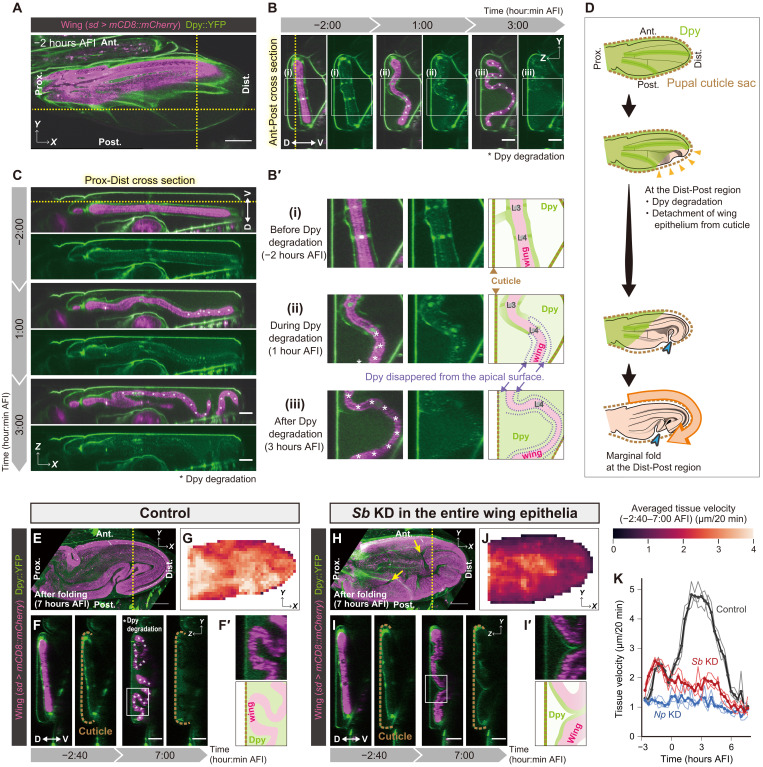
We next investigated how the stereotypic marginal fold at the distal-posterior region emerges. Then, we closely observed the spatiotemporal dynamics of Dpy distribution in a control wing (Fig. 3, A to D, and movie S11) and noticed that Dpy::YFP fluorescent signals on the wing tissue gradually disappeared during folding [see Dpy::YFP signal nearby asterisks in Fig. 3 (B, B′, and C) and see also purple dotted lines in Fig. 3B′], regardless of its localization type (vein/marginal Dpy or surface Dpy) (fig. S13, see Fig. 2B for the type of Dpy). Accordingly, the Dpy::YFP fluorescent signal increased at the extracellular space between the cuticle sac and wing tissue (fig. S14), suggesting that Dpy was disassembled and released into the extracellular space during folding. The Dpy disappearance starts at the distal-posterior region [see the position of Dpy degradation shown by asterisks in Fig. 3 (B and C)] before forming the marginal fold at the same distal-posterior region (fig. S13D). This implies that the distal-posterior edge of the wing margin started to get detached from the pupal cuticle first (yellow arrowheads in Fig. 3D) and consequently formed the marginal fold (blue arrowhead in Fig. 3D).
Fig. 3. Dumpy (Dpy) degradation is indispensable for the marginal fold formation.
(A to C) Multiphoton laser scanning (MP) microscope time-lapse images of a control wing expressing mCD8::mCherry induced by sd-Gal4 and endogenous Dpy::YFP. (A) XY view of a slice at the yellow dotted lines in (B) and (C). (B and C) Cross sections along the lines in (A). (B′) Magnified views of rectangles in (B) and their schematics. (D) Schematics of Dpy degradation and the position of the marginal fold. (E to I) Confocal time-lapse imaging of Dpy::YFP and Particle image velocimetry (PIV) analysis in control (E to G) and Stubble (Sb) knockdown (H to J). (E and H) Maximum projections of postfolded wings [7 hours after folding initiation (AFI)]. Yellow arrows in (H) indicate ectopic folds. (F and I) Anterior-posterior cross sections along the yellow dotted lines in (E) and (H). (F′ and I′) Magnified views of rectangles in (F) and (I) and their schematics. (G and J) Tissue velocity averaged in each grid throughout the folding process (−2:40 to 7:00 AFI). (K) Temporal dynamics of tissue velocity. N = 3 wings for each genotype (thin lines) and the average (thick lines). Asterisks in (B), (B′), (C), and (F) indicate the wing position where Dpy::YFP signal disappeared from the nearby apical surface. Scale bars, 100 μm (A, E, and H) and 50 μm (B, C, F, and I).
To examine the impact of Dpy degradation on the marginal fold formation, we focused on Stubble (Sb) or Notopleural (Np), both of which encode transmembrane proteases that are responsible for degrading Dpy (16–19). When we depleted Sb or Np throughout the wing, the Dpy localization pattern before folding was similar to that in controls (fig. S15); however, we observed abnormalities after folding. The surface and vein/marginal Dpy proteins remained in their original positions even after the folding was initiated (Fig. 3, E to I; figs. S16 and S17; and movies S12 and S13), and the Dpy::YFP fluorescent signal at the extracellular space did not increase (fig. S14), indicating that Sb or Np depletion prevents Dpy disassembly. In terms of the folding process, the Sb- or Np-depleted wings initiated expansion and the first formation of the vein folds was in a similar fashion to controls (movie S12), suggesting that blocking Dpy degradation does not affect the initiation of buckling along veins. However, they failed to eventually form any marginal fold (Fig. 3H and fig. S16A), suggesting that a persistent Dpy anchorage prevents cells from sliding along the wing margin. Further reflecting these abnormalities, their tissue velocities were lower during folding (Fig. 3K, fig. S18A, and movie S14), especially at the wing margin associated with persistent Dpy anchorage (Fig. 3, G and J, and figs. S16B and S18, B and C). Moreover, the Sb- or Np-depleted wings formed ectopic folds in the intervein region, some of which crossed over the veins (yellow arrows in Fig. 3H and fig. S16A). In addition, in the Sb- or Np-depleted wings, cell area reached after folding was smaller than that in control wings (fig. S19), suggesting that the persistent Dpy structure also interferes with cell flattening. Together, Dpy degradation is indispensable to proceed toward the complete formation of the marginal fold as well as full flattening.
Patterned Dpy degradation specifies the marginal fold position
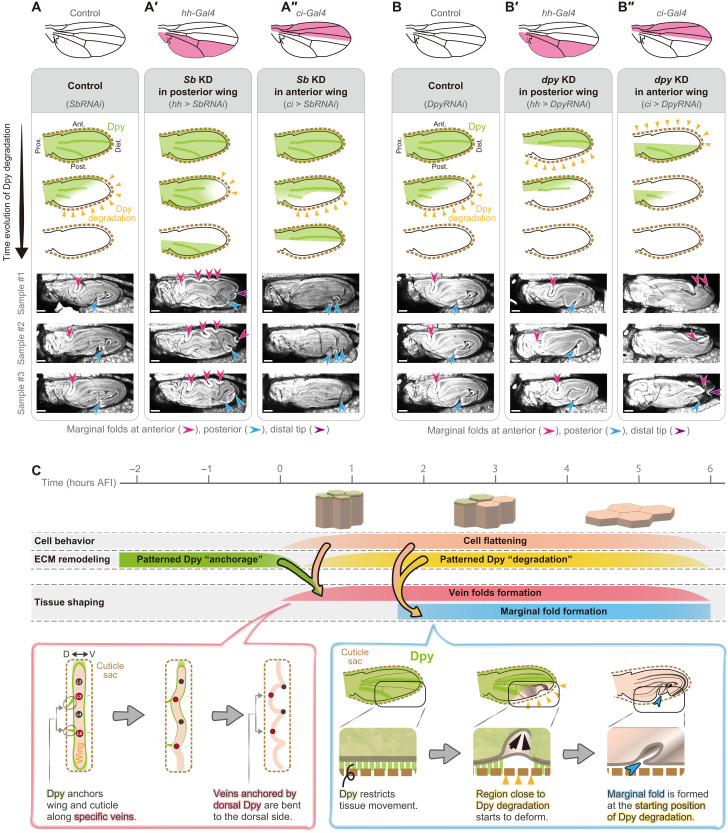
To test whether the starting position of the Dpy degradation determines the position of the marginal fold, we sought to alter the Dpy degradation pattern by knocking down Sb in a region-specific manner and examined whether ectopic folds are formed (Fig. 4, A to A″, and fig. S20). Sb depletion in the posterior compartment allows anterior compartment-specific Dpy degradation, which can be regarded as Dpy degradation from the anterior compartment (Fig. 4A′). In this situation, multiple ectopic folds were formed along the anterior margin (Fig. 4A′ and fig. S20, E to H). In contrast, Sb depletion in the anterior compartment led to distal-posterior marginal folds similar to those seen in controls (Fig. 4A″ and fig. S20, I and J). Silencing dpy expression in such a region-specific manner further supports our hypothesis (Fig. 4, B to B″, and fig. S21): Dpy depletion in the anterior compartment, which could create a situation of premature anterior Dpy degradation before posterior Dpy degradation, resulted in ectopic marginal folds at the distal-anterior position (Fig. 4B″ and fig. S21, G and H). On the other hand, Dpy depletion in the posterior compartment resulted in a distal-posterior marginal fold, located at almost the same position as that of the control (Fig. 4B′ and fig. S21, C to F). Together, we conclude that spatial coordination of the Dpy degradation specifies the marginal fold position and eventually shapes the higher-order 3D folding.
Fig. 4. Patterned Dumpy (Dpy) degradation specifies the marginal fold position.
(A to A″ and B to B″) Top: Schematics showing the region expressing Gal4 (magenta). Middle: Expected dynamics of Dpy degradation. Yellow arrowheads indicate the starting position of Dpy degradation. Bottom: Maximum projections of confocal snapshot images of GAP43::GFP-expressing postfolded wings in control (A and B), localized silencing of Stubble (Sb) at posterior or anterior region (A′ and A″), localized silencing of dpy at posterior or anterior region (B′ and B″). Arrowheads represent folds along the wing margin. Scale bars, 100 μm. (C) A model for shaping stereotypic tissue folding.
DISCUSSION
We have revealed that the spatiotemporally coordinated deposition and destruction of the ECM protein (Dpy) guide the position and direction of buckling (Fig. 4C). Our results demonstrate that although cell populations show spatially homogeneous cellular behaviors (i.e., cell flattening), they can yield stereotypic tissue buckling morphology through the positional information encoded by ECM remodeling.
We observed that Dpy, which anchors the wing tissue and cuticle, is constructed at specific positions (Fig. 2, A and B, left). The position of Dpy anchorage is likely controlled posttranslationally, because Dpy overexpression did not result in the ectopic formation of Dpy anchorage (fig. S8). The construction of the Dpy matrix involves multiple processes, including synthesis, secretion (12, 20), assembly into polymers (12), and association of the cuticle with the apical cell surface (21). Although the signaling pathway regulating Dpy anchoring remains unknown, the loss of vein Dpy in the mutant wing lacking veins (fig. S9) suggests that positional information and signaling pathways related to wing vein patterning may contribute to the spatial differences in Dpy structure formation.
We found that Dpy degradation initiates in the distal-posterior region and that the spatial pattern of degradation controls the position of the marginal fold (Figs. 3 and 4). The degradation of Dpy is regulated by the action of Sb and Np apical transmembrane proteases, suggesting that the spatial pattern of Np/Sb expression or localization to the apical surface may control the degradation pattern of Dpy. Alternatively, another possible mechanism for the propagation pattern of Dpy degradation could be attributed to the spatial pattern of proteolytic activation of the proteases, because Np and Sb are expressed as catalytically inactive zymogens and require proteolytic cleavage to become active (16, 18). In addition, regional differences in the physical properties of Dpy and its sensitivity to the proteases could contribute to the propagation pattern of Dpy degradation. It has been reported that Dpy undergoes filamentous conversion in response to increasing tension during indirect flight muscle development (12). Considering that Dpy filaments in wing tissue are formed under anisotropic proximal-distal–oriented tension caused by hinge contraction (10, 11, 22, 23), the extent of Dpy filamentous conversion may vary regionally within the tissue depending on the applied stress. It would be interesting to investigate the regional difference in the physical properties of Dpy filaments within a tissue and their relevance to the propagation pattern of Dpy degradation.
Our findings are unique as they reveal the potential of external cues in generating stereotypic 3D tissue shapes within a spatially confined environment. This ECM-based mechanism should confer a potential means to generate diverse and controllable 3D tissue shaping in parallel with cell-intrinsic genetic programming. Future application of this ECM modification to tissue engineering would pave the way for manufacturing precisely folded tissues in any desired manner.
MATERIALS AND METHODS
Table 1. Fly strains used in this paper.
A list of the fly strains used in this paper, including their respective sources or references, identifiers, and additional information.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Drosophila melanogaster | w1118 | Bloomington Drosophila Stock Center | BDSC: 6326 FlyBase: FBal0018186 | |
| D. melanogaster | ap-GAL4 | Kyoto Stock Center | DGRC: 106798 FlyBase: FBti0002785 | FlyBase symbol: P{GawB}apmd544 |
| D. melanogaster | UAS-Dcr2 | Bloomington Drosophila Stock Center | BDSC: 24651 FlyBase: FBti0100276 | FlyBase symbol: P{UAS-Dcr-2.D}10 |
| D. melanogaster | UAS-DsRed | Bloomington Drosophila Stock Center | BDSC: 6282 FlyBase: FBti0018001 | FlyBase symbol: P{UAS-AUG-DsRed}A |
| D. melanogaster | sd-Gal4 | Bloomington Drosophila Stock Center | BDSC: 8609 FlyBase: FBti0004225 | FlyBase symbol: P{GawB}sdSG29.1 |
| D. melanogaster | Dpy::YFP | Kyoto Stock Center | DGRC: 115238 FlyBase: FBti0143891 | FlyBase symbol: PBac{681.P.FSVS-1}dpyCPTI001769 |
| D. melanogaster | UAS-mCD8::mCherry | Bloomington Drosophila Stock Center | BDSC: 27391 FlyBase: FBti0115768 | FlyBase symbol: P{UAS-mCD8.ChRFP}2 |
| D. melanogaster | UAS-mCD8::mCherry | Bloomington Drosophila Stock Center | BDSC: 27392 FlyBase: FBti0115769 | FlyBase symbol: P{UAS-mCD8.ChRFP}3 |
| D. melanogaster | UAS-dpyRNAi | Vienna Drosophila Resource Center | VDRC: 44029 FlyBase: FBti0091762 | FlyBase symbol: P{GD4443}v44029 |
| D. melanogaster | UAS-SbRNAi | Vienna Drosophila Resource Center | VDRC: 1613 FlyBase: FBti0092159 | FlyBase symbol: P{GD473}v1613 |
| D. melanogaster | UAS-NpRNAi | Vienna Drosophila Resource Center | VDRC: 105297 FlyBase: FBti0121532 | FlyBase symbol: P{KK109020}VIE-260B |
| D. melanogaster | UAS-NpRNAi | Vienna Drosophila Resource Center | VDRC: 23381 FlyBase: FBti0083222 | FlyBase symbol: P{GD13443}v23381 |
| D. melanogaster | Ubi-GAP43::GFP | This study | ||
| D. melanogaster | Ubi-GAP43::tdTomato | This study | ||
| D. melanogaster | ci-Gal4 | (24) | FlyBase: FBal0193963 | FlyBase symbol: P{ci-GAL4.C} |
| D. melanogaster | hh-Gal4 | (25) | FlyBase: FBti0017278 | FlyBase symbol: P{GAL4}hhGal4 |
| D. melanogaster | E-cadherin::GFP | Bloomington Drosophila Stock Center | BDSC: 60584 FlyBase: FBti0168565 | FlyBase symbol: TI{TI}shg[GFP] |
| D. melanogaster | Act > CD2 > GAL4 | Bloomington Drosophila Stock Center | BDSC: 4780 FlyBase: FBti0012408 | FlyBase symbol: P{GAL4-Act5C(FRT.CD2).P}S |
| D. melanogaster | sqh::eGFP | (26) | FlyBase: FBti0206965 | FlyBase symbol: TI{TI}sqh[EGFP.29B] |
| D. melanogaster | Tub-Gal80ts | Bloomington Drosophila Stock Center | BDSC: 7019 FlyBase: FBti0027796 | FlyBase symbol: P{tubP-GAL80ts}20 |
| D. melanogaster | UAS-Nslmb-vhhGFP4 | Bloomington Drosophila Stock Center | BDSC: 38421 FlyBase: FBti0147362 | FlyBase symbol: P{UAS-Nslmb-vhhGFP4}3 |
| D. melanogaster | ap-lexA | Bloomington Drosophila Stock Center | BDSC: 54268 FlyBase: FBti0155833 | FlyBase symbol: P{GMR42A06-lexA}attP40 |
| D. melanogaster | LexAop-GAL80 | Bloomington Drosophila Stock Center | BDSC: 32215 FlyBase: FBti0131978 | FlyBase symbol: P{8XLexAop2-IVS-GAL80-WPRE}su(Hw)attP1 |
| D. melanogaster | en-Gal4 | Bloomington Drosophila Stock Center | BDSC: 30564 FlyBase: FBti0003572 | FlyBase symbol: P{en2.4-GAL4}e16E |
| D. melanogaster | dpy-gRNA | Bloomington Drosophila Stock Center | BDSC: 80265 FlyBase: FBti0201086 | FlyBase symbol: P{TOE.GS02831}attP40 |
| D. melanogaster | UAS-3xFLAG.dCas9.VPR | Bloomington Drosophila Stock Center | BDSC: 67049 FlyBase: FBti0184669 | FlyBase symbol: P{UAS-3xFLAG.dCas9.VPR}attP2 |
| D. melanogaster | rhove-1, vn1 | (27) | FlyBase: FBal0017855, FBal0018018 | FlyBase symbol: rhove-1, vn1 |
Experimental genotypes
Figure 1
(B, C, and E to G) ap-GAL4/+; UAS-Dcr2/UAS-DsRed
Figure 2
(A) sd-Gal4/+; Dpy::YFP, UAS-mCD8::mCherry/Dpy::YFP
(C) ap-Gal4, Dpy::YFP/UAS-dpyRNAi; UAS-mCD8::mCherry/UAS-Dcr2
(D, E, F, G, and H) ap-GAL4/UAS-dpyRNAi; UAS-Dcr2/UAS-DsRed
(E′) ap-GAL4/+; UAS-Dcr2/UAS-DsRed
Figure 3
(A, B, B′, C, E, F, F′, G, and K Control) sd-Gal4/+; Dpy::YFP, UAS-mCD8::mCherry/+
(H, I, I′, J, and K Sb KD) sd-Gal4/+; Dpy::YFP, UAS-mCD8::mCherry/+; UAS-SbRNAi/+
(K Np KD) sd-Gal4/+; Dpy::YFP, UAS-mCD8::mCherry/UAS-NpRNAi
Figure 4
(A) Ubi-GAP43::GFP/+; UAS-SbRNAi/+
(A′) Ubi-GAP43::GFP/+; UAS-SbRNAi/hh-Gal4
(A″) Ubi-GAP43::GFP/+; UAS-SbRNAi/ci-Gal4
(B) Ubi-GAP43::GFP, UAS-dpyRNAi/+
(B′) Ubi-GAP43::GFP, UAS-dpyRNAi/+; hh-Gal4/+
(B″) Ubi-GAP43::GFP, UAS-dpyRNAi/+; ci-Gal4/+
Figure S1
(A Control, B Control, and C Control) ap-GAL4/+; UAS-Dcr2/UAS-DsRed
(A dpy KD, B dpy KD, and C dpy KD) ap-GAL4/UAS-dpyRNAi; UAS-Dcr2/UAS-DsRed
Figure S2
w1118
Figure S3
(B to D) y, w, hs-flp; E-Cadherin::GFP; Act > CD2 > GAL4, UAS-mCD8::mCherry
Figure S4
(A and B) E-Cadherin::GFP
Figure S5
(A and B) ap-GAL4/+; UAS-Dcr2/UAS-DsRed
(C and D) ap-GAL4/UAS-dpyRNAi; UAS-Dcr2/UAS-DsRed
Figure S6
(A to D and I bottom) y, w, sqh::eGFP/Y; Tub-Gal80ts/+; UAS-Nslmb-vhhGFP4, UAS-mCD8::mCherry/ci-Gal4
(E to H and J bottom) y, w, sqh::eGFP/Y; Tub-Gal80ts/+; UAS-Nslmb-vhhGFP4, UAS-mCD8::mCherry/hh-Gal4
(I top) y, w, sqh::eGFP/+; Tub-Gal80ts/+; UAS-Nslmb-vhhGFP4, UAS-mCD8::mCherry/ci-Gal4
(I middle) y, w, sqh::eGFP/Y; Tub-Gal80ts/+; ci-Gal4/TM3, Sb ftz-lacZ
(J top) y, w, sqh::eGFP/+; Tub-Gal80ts/+; UAS-Nslmb-vhhGFP4, UAS-mCD8::mCherry/hh-Gal4
(J middle) y, w, sqh::eGFP/Y; Tub-Gal80ts/+; hh-Gal4/TM3, Sb ftz-lacZ
Figure S7
(A to C) sd-Gal4/+; Dpy::YFP, UAS-mCD8::mCherry/Dpy::YFP
Figure S8
(A Control and C Control) Dpy::YFP, UAS-mCD8::mCherry/dpy-gRNA; ci-Gal4/+
(A Dpy OE and C Dpy OE) Dpy::YFP, UAS-mCD8::mCherry/dpy-gRNA; ci-Gal4/UAS-3xFLAG.dCas9.VPR
(B Control and D Control) Dpy::YFP, UAS-mCD8::mCherry/dpy-gRNA; hh-Gal4/+
(B Dpy OE and D Dpy OE) Dpy::YFP, UAS-mCD8::mCherry/dpy-gRNA; hh-Gal4/UAS-3xFLAG.dCas9.VPR
Figure S9
(A) y, w, hs-flp; E-Cadherin::GFP; Act > CD2 > GAL4, UAS-mCD8::mCherry
(B Control) rhove-1, vn1/+
(B Mutant lacking veins) rhove-1, vn1/rhove-1, vn1
(C and E) Dpy::YFP, Ubi-GAP43::tdTomato/+; rhove-1, vn1/+
(D and F) Dpy::YFP, Ubi-GAP43:: tdTomato/+; rhove-1, vn1/rhove-1, vn1
Figure S10
sd-Gal4/+; Dpy::YFP, UAS-mCD8::mCherry/+
Figure S11
(A) ap-Gal4, Dpy::YFP/+; UAS-mCD8::mCherry/UAS-Dcr2
(B) ap-Gal4, Dpy::YFP/UAS-dpyRNAi; UAS-mCD8::mCherry/UAS-Dcr2
Figure S12
(A) sd-Gal4/+; ap-lexA, UAS-mCD8::mCherry/Dpy::YFP; UAS-Dcr2, LexAop-GAL80/+
(B to E) sd-Gal4/+; ap-lexA, UAS-mCD8::mCherry/+; UAS-Dcr2, LexAop-GAL80/+
(F) sd-Gal4/+; ap-lexA, UAS-mCD8::mCherry/Dpy::YFP, UAS-dpyRNAi; UAS-Dcr2, LexAop-GAL80/+
(G to J) sd-Gal4/+; ap-lexA, UAS-mCD8::mCherry/UAS-dpyRNAi; UAS-Dcr2, LexAop-GAL80/+
Figure S13
(A to C, C′, D, and D′) sd-Gal4/+; Dpy::YFP, UAS-mCD8::mCherry/+
Figure S14
(A Control and B Control) sd-Gal4/+; Dpy::YFP, UAS-mCD8::mCherry/+
(A Sb KD and B Sb KD ) sd-Gal4/+; Dpy::YFP, UAS-mCD8::mCherry/+; UAS-SbRNAi/+
(A Np KD and B Np KD ) sd-Gal4/+; Dpy::YFP, UAS-mCD8::mCherry/UAS-NpRNAi
Figure S15
(A) sd-Gal4/+; Dpy::YFP, UAS-mCD8::mCherry/+; UAS-SbRNAi/+
(B) sd-Gal4/+; Dpy::YFP, UAS-mCD8::mCherry/UAS-NpRNAi
Figure S16
(A to C and C′) sd-Gal4/+; Dpy::YFP, UAS-mCD8::mCherry/UAS-NpRNAi
Figure S17
(A) sd-Gal4/+; Dpy::YFP, UAS-mCD8::mCherry/+
(B) sd-Gal4/+; Dpy::YFP, UAS-mCD8::mCherry/+; UAS-SbRNAi/+
(C) sd-Gal4/+; Dpy::YFP, UAS-mCD8::mCherry/UAS-NpRNAi
Figure S18
(A Control, B Control, and C Control) sd-Gal4/+; Dpy::YFP, UAS-mCD8::mCherry/+
(A Sb KD, B Sb KD, and C Sb KD ) sd-Gal4/+; Dpy::YFP, UAS-mCD8::mCherry/+; UAS-SbRNAi/+
(A Np KD, B Np KD, and C Np KD ) sd-Gal4/+; Dpy::YFP, UAS-mCD8::mCherry/UAS-NpRNAi
Figure S19
(A Control and B Control) sd-Gal4/+; UAS-mCD8::mCherry/E-Cadherin::GFP
(A Sb KD and B Sb KD ) sd-Gal4/+; UAS-mCD8::mCherry/E-Cadherin::GFP; UAS-SbRNAi/+
(A Np KD and B Np KD ) sd-Gal4/+; UAS-mCD8::mCherry/E-Cadherin::GFP; UAS-NpRNAi/+
(A dpy KD and B dpy KD) sd-Gal4/+; UAS-dpyRNAi/E-Cadherin::GFP; UAS-mCD8::mCherry/UAS-Dcr2
Figure S20
(B) Ubi-GAP43::GFP/+; UAS-SbRNAi/+
(C) sd-Gal4/+; Ubi-GAP43::GFP/+; +/UAS-DsRed
(D) sd-Gal4/+; Ubi-GAP43::GFP/+; UAS-SbRNAi/+
(E) Ubi-GAP43::GFP/+; hh-Gal4/UAS-DsRed
(F) Ubi-GAP43::GFP/+; UAS-SbRNAi/hh-Gal4
(G) Ubi-GAP43::GFP/en-Gal4; +/UAS-DsRed
(H) Ubi-GAP43::GFP/en-Gal4; UAS-SbRNAi/+
(I) Ubi-GAP43::GFP/+; ci-Gal4/UAS-DsRed
(J) Ubi-GAP43::GFP/+; UAS-SbRNAi/ci-Gal4
Figure S21
(B) Ubi-GAP43::GFP, UAS-dpyRNAi/+
(C) Ubi-GAP43::GFP/+; hh-Gal4/UAS-DsRed
(D) Ubi-GAP43::GFP, UAS-dpyRNAi/+; hh-Gal4/+
(E) Ubi-GAP43::GFP/en-Gal4; +/UAS-DsRed
(F) Ubi-GAP43::GFP, UAS-dpyRNAi/en-Gal4
(G) Ubi-GAP43::GFP/+; ci-Gal4/UAS-DsRed
(H) Ubi-GAP43::GFP, UAS-dpyRNAi/+; ci-Gal4/+
Movie S1
ap-GAL4/+; UAS-Dcr2/UAS-DsRed
Movie S2
y, w, hs-flp; E-Cadherin::GFP; Act > CD2 > GAL4, UAS-mCD8::mCherry
Movie S3
y, w, sqh::eGFP/Y; Tub-Gal80ts/+; UAS-Nslmb-vhhGFP4, UAS-mCD8::mCherry/ci-Gal4
Movie S4
y, w, sqh::eGFP/Y; Tub-Gal80ts/+; UAS-Nslmb-vhhGFP4, UAS-mCD8::mCherry/hh-Gal4
Movie S5
sd-Gal4/+; Dpy::YFP, UAS-mCD8::mCherry/Dpy::YFP
Movie S6
sd-Gal4/+; Dpy::YFP, UAS-mCD8::mCherry/+
Movie S7
ap-GAL4/UAS-dpyRNAi; UAS-Dcr2/UAS-DsRed
Movie S8
(Control) Dpy::YFP, UAS-mCD8::mCherry/dpy-gRNA; ci-Gal4/+
(Dpy OE) Dpy::YFP, UAS-mCD8::mCherry/dpy-gRNA; ci-Gal4/UAS-3xFLAG.dCas9.VPR
Movie S9
(Control) Dpy::YFP, UAS-mCD8::mCherry/dpy-gRNA; hh-Gal4/+
(Dpy OE) Dpy::YFP, UAS-mCD8::mCherry/dpy-gRNA; hh-Gal4/UAS-3xFLAG.dCas9.VPR
Movie S10
(Control) Dpy::YFP, Ubi-GAP43::tdTomato/+; rhove-1, vn1/+
(Mutant lacking veins) Dpy::YFP, Ubi-GAP43:: tdTomato/+; rhove-1, vn1/rhove-1, vn1
Movie S11
sd-Gal4/+; Dpy::YFP, UAS-mCD8::mCherry/+
Movie S12
(Control) sd-Gal4/+; Dpy::YFP, UAS-mCD8::mCherry/+
(Sb KD) sd-Gal4/+; Dpy::YFP, UAS-mCD8::mCherry/+; UAS-SbRNAi/+
(Np KD) sd-Gal4/+; Dpy::YFP, UAS-mCD8::mCherry/UAS-NpRNAi
Movie S13
(Control) sd-Gal4/+; Dpy::YFP, UAS-mCD8::mCherry/+
(Sb KD) sd-Gal4/+; Dpy::YFP, UAS-mCD8::mCherry/+; UAS-SbRNAi/+
(Np KD) sd-Gal4/+; Dpy::YFP, UAS-mCD8::mCherry/UAS-NpRNAi
Movie S14
(Control) sd-Gal4/+; Dpy::YFP, UAS-mCD8::mCherry/+
(Sb KD) sd-Gal4/+; Dpy::YFP, UAS-mCD8::mCherry/+; UAS-SbRNAi/+
(Np KD) sd-Gal4/+; Dpy::YFP, UAS-mCD8::mCherry/UAS-NpRNAi
Husbandry conditions
Fly stocks were kept in vials at 18° or 25°C with a standard corn-sugar-yeast food and transferred every 4 or 3 weeks, respectively. For cross experiments, flies were flipped every 3 to 4 days, and vials containing offspring were raised at 18°, 25°, or 29°C until dissection or imaging.
Live imaging
Pupae for live imaging were prepared as previously described with some modifications (28, 29): White prepupae (WPP) were collected and kept at 25°C (unless otherwise noted) until they reached the appropriate stage. Staged pupae were washed with water, dried on KimWipe, and then mounted on a glass slide using double-sided tape with the right wing on top. The pupal case was carefully removed with forceps so as not to cause an influx of hemolymph into the wings. Filter paper (ADVANTEC 02103020) wetted with water was placed around the pupa to avoid desiccation. A bank of silicone oil compounds (Shin-Etsu Chemical Co. Ltd., HIVAC-G) was placed around the dissected pupa and covered with a coverslip. The coverslip was carefully placed parallel to the dorsal wing cuticle to avoid shear forces, which cause abnormal wing packing. We note that dissection before folding for time-lapse imaging reproducibly formed the stereotypic vein folds and marginal fold at the distal-posterior region, however, sometimes caused ectopic folds at the anterior wing margin. Therefore, we dissected wings after the folding was completed and confirmed that the positions of marginal fold in control wings are reproducible among individuals [fig. S20B (n = 10 of 10 samples) and fig. S21B (n = 10 of 10 samples)]. For Figs. 2C and 3 (E to K) and figs. S4, S6, S8, S9 (C to F), S11, S13, and S16 to S19, exposed wings were covered with a small drop of halocarbon oil (a 4:1 mixture of halocarbon oil 700 and 27, Sigma-Aldrich). Images were obtained at room temperature (at 22° to 25°C) using a Zeiss LSM 710 upright laser confocal microscope with a 20× objective (EC Plan-Neofluar 20×/0.50 M27), an LSM 800 inverted laser confocal microscope with a 10× objective (Plan-Apochromat 10×/0.45 M27), a 20× objective (Plan-Apochromat 20×/0.8 M27), and a silicon oil immersion objective (LD LCI Plan-Apochromat 40×/1.2 Imm Korr DIC M27) or an Olympus FV1000MPE-IX83 MP microscope (Spectra-Physics InSight DeepSee Laser set to 930-nm wavelength) with a silicon oil immersion objective (UPLSAPO30XS). For the time-lapse recording, each wing was imaged every 20 min except for fig. S19 (every 1 hour). For the protease knockdown experiment in the entire wings (Fig. 3, H to K, and figs. S14 to S17, B and C, and S18 to S20D), we excluded pupae whose prefolded wings are abnormal contour shape (small and round) from the live-imaging analysis because the abnormal shape makes it difficult to evaluate the impact of the proteases on wing folding.
Image processing
All images were processed using Fiji software (https://fiji.sc/). The range of intensity was adjusted with the “setMinAndMax(min, max)” function by setting the minimum and maximum displayed pixel values. For Figs. 1 (B, C, and E to G), 2 (A and C to H), and 3 (A to C and E to K) and figs. S1 (A to C, adult), S2, S5 to S9 (C to F), and S11 to S18, images were rotated once to bring the anterior side on top and the distal side to the right. In case the imaging was started before the wings were folded, the image was rotated so that the line connecting the intersection of L2 and L3 veins and the distal end point of L3 vein is horizontal to the xy axis of the image. Stack projection views were generated by applying maximum intensity z-projection. Cross sections of a stack were generated by using the “Image/Stacks/Reslice” command. For Figs. 2 (A and C) and 3 (A to C) and figs. S1 (A to C, adult), S7, S11, and S12 (A and F), tiles spanning a wing were stitched together using Pairwise Stitching or Grid/Collection Stitching Plugin. For images captured by an MP microscope (Olympus FV1000MPE-IX83), the stitched images were smoothed by using the “Process/Smooth” command. The positions of veins are inferred from the absence of a fluorescent signal within the tissue. For 3D view images rotating around the y axis in movie S13, Fiji 3D Viewer plugin was used.
Characterization and curvature measurement of folds along four longitudinal veins
Cross sections of a stack of time-lapse images were generated along the anterior-posterior axis that passes through the intersection of the L4 longitudinal vein and the posterior cross vein at 38 hours APF [for Figs. 1 (C and E to G) and 2 (E to H) and figs. S5 and S12 (C to E and H to J)] or at 7 hours after the temperature shift (for fig. S6, A to H). Wing surface outlines were extracted based on DsRed, mCD8::mCherry, or Sqh::eGFP signals using Fiji and analyzed using custom-written code in Python. In Fiji, the wing surface contour was manually traced from the cross-sectional images (YZT images) using the segmented line tool, interpolated using the “Edit/Selection/Interpolate” command, and acquired a series of wing surface contour points (x and y). The pixel width used for interpolation for each image was adjusted to ensure uniform μm width (0.31 μm) for all images, to compensate for differences in image resolution and to ensure that curvature could be quantified on the same scale for all images. The position of longitudinal veins (L2 to L5) was determined from the original stacked images (XYZT images) and projected to the corresponding position in the cross-sectional images because it was difficult to recognize the vein positions in cross-sectional images of wings labeling only dorsal or ventral wing epithelia. In Python, we fit a third-order polynomial to a sliding window along the wing surface contour points (x and y) and measured the first and second derivatives of position with respect to distance along the contour, from which we calculated local signed curvature k using the following formula
In our analysis, the curve bending to the ventral side is defined as positive. We evaluated the curvature at a series of 200 points equally spaced in distance along the contour. Of the 200 points that constitute the wing surface, the points closest to the vein coordinates were defined as the locations of the vein. The curvature values were color-coded, and the vein positions were represented by black dots on the corresponding coordinates in sequential time series images [Figs. 1C and 2 (E and E′) and figs. S5 (A and C), S6 (A, B, E, and F), and S12 (C, D, H, and I)] or on the spatiotemporal heatmaps [Figs. 1E and 2F and figs. S5 (A and C), S6 (B and F), and S12 (D and I)]. Curvature values of 11 points in the vicinity of the vein were averaged and plotted as a function of time [Figs. 1F and 2G and figs. S5 (B and D), S6 (C and G), and S12 (E and J)]. The time evolution of the average local curvature around each longitudinal vein was averaged across different pupae and presented as a plot with the corresponding SD [Figs. 1G and 2H and figs. S6 (D and H) and S12 (E and J)]. The Python code for calculating curvature was downloaded from the Harvard Dataverse [Draper, W. E. Analysis code. Harvard Dataverse. doi:10.7910/DVN/CHOTF0 (2016).49.], which was published by Draper and Liphardt (30).
Particle image velocimetry analysis
Time-lapse images were rotated, maximum intensity z-projections were applied (see the “Image processing” section for details), and background signals from outside wing tissue were removed from the images by erasing the area outside the regions of interest (ROIs) that were manually located along the wing margin (“Edit/Clear Outside” command in Fiji). Particle image velocimetry (PIV) analysis was performed on the red fluorescent protein channels (sd-Gal4 > UAS-mCD8::mCherry) of the processed time-lapse images using time series particle image velocimetry (TPIV) customized version of version 3.3 (https://signaling.riken.jp/tools/imagej-plugins/490/, a Fiji plugin developed by Housei Wada). Parameters for TPIV were set as follows: window width and height (pixel): 50; overlap (%): 50; extended value: 3; subpixel value: 10; color method: absolute; max length limit: 16; Check “OverWrite,” “WithOriginalImage,” and “CorrectByMedian.” Briefly, images were split into 50 × 50–pixel (31.2 × 31.2 μm) interrogation windows, and the interrogation window overlap was set to 50% to calculate tissue velocities (respective displacement vectors between subsequent frames). Each tissue velocity was color-coded according to its magnitude by colorizing each vector into eight colors using the “Absolute” mode with 16 pixels (9.98 μm) as the maximum value (fig. S18A). The tissue velocity output data from TPIV were further analyzed with a custom-written code in Python and plotted using Python or Excel. For the temporal dynamics of tissue velocity (Fig. 3K), the magnitude of all the tissue velocities within the wing margin was averaged for each time frame and plotted as a function of time. For the spatial color maps of tissue velocity (Fig. 3, G and J, and figs. S16B and S18B), the temporal average of all velocity vector magnitudes throughout the folding process (−2:40 to 7:00 AFI) was calculated within wing margin in each grid [25 × 25 pixels (15.6 × 15.6 μm)], which was compartmentalized according to the location of the initial point of each vector. For the spatiotemporal tissue velocity (fig. S18C), we first spatially categorized the tissue velocity vectors within the wing margin according to the normalized distance to wing margin = floor(Dv-m/G), with Dv-m being the minimum distance from the initial point of each vector to the wing margin and G being the constant value representing the distance between adjacent vectors [25 × 25 pixels (15.6 × 15.6 μm)]. The spatiotemporal tissue velocity was calculated by averaging the tissue velocities for each spatial and temporal category, and a heatmap was plotted as a function of time (hours AFI) (x axis) and normalized distance to wing margin (y axis). Only if the number of PIV vectors belonging to the category was 10 or more were they displayed in the heatmap.
Plasmid construction and transgenesis for Ubi-GAP43::GFP and Ubi-GAP43::tdTomato strain
For Ubi-GAP43::GFP strain, GFP fused with an N-terminal 20–amino acid peptide of plasma membrane localization tag from growth-associated protein 43 (GAP43) gene was amplified by polymerase chain reaction (PCR) with PrimeSTAR (Takara Bio) and primers (5′-AACAGATCTGCGGCCGCAACATGCTGTGCTGTATGCGAAGAACCAAAC AGGTTGAAAAAAATGATGAGGACCAAAAGATTATGAGTAAAGGAGAAGAACTTTTC-3′ (underline corresponds to plasma membrane localization tag) and 5′-GGATTCCTCCACGGGGTACCTTATTTGTATAGTTCATCCATGCCATGTG-3′). For Ubi-GAP43::tdTomato strain, tdTomato fused with same N-terminal plasma membrane localization tag was synthesized and amplified by PCR with primers (5′-AACAGATCTGCGGCCGCAACATGCTGTGCTGTATGCGGCGC-3′ and 5′-GGATTCCTCCACGGGTTACTTATAGAGTTCATCCATG-3′). These PCR fragments were subcloned into pUbi-attB (31) with the In-Fusion PCR cloning kit (Clontech).With these plasmids, transgenic strains were generated by the phiC31 integrase-mediated transgene integration into an attP target site of the ZH-22A (32) for Ubi-GAP43::GFP and VK02 for Ubi-GAP43::tdTomato (33).
Observation of adult wings using a stereo microscope
For fig. S2, adult flies right after eclosion were immediately transferred to the freezer until they stopped moving. Images were taken using an Olympus SZ61 stereo microscope equipped with a LUMIXDMC-G7H digital camera. For fig. S9B, adult flies 1 day after eclosion were washed in 70% ethanol (EtOH), and their wings were cut off. The wings were then mounted on a glass slide in 80% glycerol in 1× phosphate-buffered saline (PBS). Images were taken using a Leica S8APO stereomicroscope equipped with an Olympus AIR01 digital camera. Images were adjusted using Fiji.
Quantification of wing tissue area
To prevent overcrowding, 15 females and 3 males were crossed in a vial at 25°C and transferred to a new vial with fresh food every 3 to 4 days. For the preparation of pupal wings, pupae before wing folding (28 to 34 hours APF) were dissected as described in the “Live imaging” section above. For the preparation of adult wings, adult flies that had eclosed at least 24 hours ago, which is sufficient time for wing expansion in the wild type, were transferred into 1.5-ml tubes or dishes (IWAKI nontreated 35-mm culture dishes 1000-035, IWAKI, Shizuoka, Japan) and moved to the freezer. Adult wings were cut off, washed in 70% EtOH, and mounted on a glass slide in 80% glycerol in 1× PBS. Both pupal and adult wings were observed at room temperature (25°C) using an LSM 800 inverted laser confocal microscope with Plan-Apochromat 10×/0.45 M27. We applied maximum intensity Z-projection. In the case of adult wings, tiles of the maximum intensity Z-projections spanning a wing were stitched together using the Pairwise Stitching Plugin in Fiji. The wing area was measured by manually tracing the wing outline from subcostal break to alula notch.
Comparison of planer contour shape of adult and pupal wing
For comparison of the planer contour shape of pupal and adult wings in the right of fig. S1A, the pupal wing image was colored orange, and the adult wing image was colored blue. Then, the images were made translucent, sized so that the pupal and adult wings were approximately the same size, rotated, and overlaid in Adobe Illustrator.
Quantification of cell area
For fig. S3 (B to D), larvae were incubated in a water bath at 34°C for 30 min to induce clones of mCherry-marked cells. Then, 1, 25, or 26 hours after heat shock, WPP were collected and kept at 25°C until the appropriate stage. For fig. S19, control (sd-Gal4 > UAS-mCD8::mCherry), Sb knockdown (sd-Gal4 > UAS-mCD8::mCherry, UAS-SbRNAi), Np knockdown (sd-Gal4 > UAS-mCD8::mCherry, UAS-NpRNAi), and dpy knockdown (sd-Gal4 > UAS-mCD8::mCherry, UAS-dpyRNAi, UAS-Dcr2) wings expressing E-cadherin::GFP were raised at 25°C. Time-lapse imaging was performed using an LSM 800 (equipped with LD LCI Plan-Apochromat 40×/1.2 Imm Korr DIC M27) as described above. Because it is difficult to measure the cell area in the deep folded regions, images cropped from the region where the tissue is relatively flat were used for cell area quantification. On the basis of the adherens junction marker E-cadherin::GFP, we segmented images and tracked cells at the intervein region using Fiji plug-in Tissue Analyzer (22, 34). The brief segmentation procedure is as follows:
1) Apply maximum intensity Z-projection to seven slices (1 μm per slice) containing an adherens junction marker E-cadherin::GFP, apply median filter (1-pixel radius), convert to red, green, blue (RGB) color, and save as image sequence (TIF format) with Fiji. For some samples, prior to applying the maximum intensity Z-projection, the background was subtracted using the "Subtract Background" tool with a 10-pixel (3.1 μm) rolling ball radius.
2) Automated segmentation images were obtained using Tissue Analyzer or the Python package EPySeg (35). In the case of Tissue Analyzer, select the channel of E-cadherin::GFP and click the “Detect bonds V3 (save watershed)” in the “Segmentation” tab. In the case of EPySeg, select a pretrained model (2D epithelial segmentation) in the “Model” tab and set the “Channel of interest” to the channel of E-cadherin::GFP and output segmentation images in “Tissue Analyzer mode” in the “Predict” tab. (All of the following procedures were conducted using Tissue Analyzer.)
3) Manually correct the mistakes of the automated segmentation and remove cells in and around the vein region (“Correction” tab).
4) Click “Finish all” and “Check finish all” in the “PostProcess” tab. The parameter ("4-way vertex vs bond cut-off") was set to 2.
5) Click “Autocenter based on 2D correction” in the “Recenter” tab.
6) Click “Track cells (dynamics tissue)” in the “Tracking” tab. For fig. S3 (C and D), mCherry-marked cells were used as landmarks to check for tracking errors (e.g., cell swapping errors and cell pairing errors), and if any errors were found, they were manually fixed from the edit button “Correct/edit cell tracks” to track the same cells through time. Click “Check track” and “Update track mask” to finalize cell tracking.
7) Click “Generate/Update database” in the “SQLite DB” tab. The parameter ("4-way vertex vs bond cut-off") was set to 2.
8) Click “Export cell data” in the “Plots” tab to save the area of cells in .csv format.
For fig. S3 (C and D), the same cells were tracked over time using the location of the clonally mCherry-marked cells as landmarks. Subsequently, in fig. S3C, the cell area for each individual cell was plotted as a function of time. In fig. S3D, the average and SD of the cell area were calculated for each group of cells compartmentalized by wing veins. For fig. S19B, the area of cells at the same time (hours AFI) was averaged and plotted as a function of time.
NSlmb-vhhGFP4–mediated degradation of Sqh::eGFP
To deplete Sqh from the stage right before wing folding, deGradFP (9) and Gal80ts (36) systems were used in combination. In Sqh::eGFP knock-in flies harboring Tub-Gal80ts, NSlmb-vhhGFP4 and mCD8::mCherry were overexpressed by ci-Gal4/hh-Gal4 driver with temperature shift from 18° to 29°C at 48 hours APF. Here, 48 hours APF at 18°C approximately corresponds to about 21 to 22 hours APF at 25°C (37). For fig. S6 (A to H), temperature-shifted pupae were kept at 29°C for about 4 hours in the incubator to select pupae with the necessary markers for observation using a fluorescence stereomicroscope, and time-lapse imaging of the degradation process of Sqh::eGFP was performed with 20-min interval as described above, except that the temperature was changed using a thermal controller (TOKAI HIT, Thermo Plate, TPi-SQH26FT). Note that the thermal controller was set to 31°C to keep samples between 29° and 30°C at room temperature (25°C). For fig. S6 (I and J), temperature-shifted pupae were kept at 29°C for 1 day in an incubator and then dissected to take snapshot images of folded wings. Female pupae harboring both Sqh::GFP and Sqh (endogenous non–GFP-fused Sqh, which is not subject to degradation) and male pupae without driving NSlmb-vhhGFP4–mediated knockdown were used as control, and male pupae driving NSlmb-vhhGFP4–mediated knockdown were used as Sqh knockdown. Female and male pupae were distinguished at the WPP stage by identifying the enlarged male gonads. Observed pupae were raised while mounted on the cover glass, and their sexes were reconfirmed at the adult stage.
Quantification of intensity
For figs. S13 (C′ and D′) and S17, we obtained the images using the same microscope setup. Images for quantifying the signal intensities were maximum projections [fig. S13, C′ and D′, and S17, A (i), B (ii), and C (iii)] or cross sections of a stack [fig. S17, A (iv), B (v), and C (vi)] processed with Fiji as described in the “Image processing” section. Intensity profiles along the straight line (line width: 10 pixels) were obtained using the “Analyze/Plot Profile” command in Fiji. For fig. S13 (C′ and D′), the intensity profiles were normalized by the maximum intensity value among −1:40 to 1:40 AFI (40-min increments).
For fig. S14, we obtained the images using the same microscope setup and generated cross sections of a stack processed with Fiji as described in the “Image processing” section. In the cross-sectional image, we measured the average intensity of Dpy::YFP within ROIs [10 × 10 pixels (6.2 × 6.2 μm)], positioned in the extracellular space between the dorsal cuticle and dorsal wing surface, excluding regions of vein, marginal, or surface Dpy. mCD8::mCherry signal in wing tissue was used to make sure the position of the wing surface.
Laser perturbation
Laser perturbation experiments were performed on an Olympus FV1000MPE-IX83 MP microscope (Spectra-Physics InSight DeepSee Laser set to 930-nm wavelength) with a silicon oil immersion objective (UPLSAPO30XS). The region performing liner perturbation was between L3 and L4 veins and near the upper portion of the posterior cross vein. Wings are mounted with the anterior side on top and the distal side to the right and set line ROIs oriented in a vertical direction (almost along the anterior-posterior axis of the wing). Ablation was performed in a single XY plane, where the ROI could be captured in one focal plane. “Stimulus Setting” of OLYMPUS FLUOVIEW software was set as follows: UseScanner: Main; ROI: Line; Laser line: 930; power level: 100%; StimulateStartSetting: Activation in Series; PreActivation: 10 frames. For the perturbation with lower laser power (fig. S4A), pixel dwell time was set to 10.0 μs/pixel, and the number of scans (activation time) was set to 20 frames. For the perturbation with higher laser power (fig. S4B), pixel dwell time was set to 20.0 μs/pixel and the number of scans (activation time) was set to 350 frames. Images of pre- and postablation were acquired at the same single plane at a frame interval of 1.109 s.
Acknowledgments
We thank the Kyoto and Bloomington Drosophila Stock Centers, the Vienna Drosophila Resource Center, M. Suzanne, R. A. Holmgren, T. Tabata, S. Ohsawa, Y. Hattori, K. Sugimura, and D. Umetsu for fly stocks; M. Hirohata, K. Ikeguchi, M. Miki, and A. Nakata for assistance with data analysis and experiments; H. Wada for the TPIV; W. Draper for the code to calculate curvature; B. Aigouy for the Tissue Analyzer and EPySeg; KULIC, K. Takakura, and H. Yukinaga for imaging with a MP microscope; S. Hayashi, T. Uemura, T. Usui, T. Harumoto, M. S. Kitazawa, S. Tsugawa, and D. Umetsu for comments on the manuscript; and members of the Kondo, Uemura, and Fujimoto laboratories for discussions. A.T. was a JSPS Research Fellow.
Funding: This work was supported by JSPS KAKENHI 19J00764 (to A.T.), 21H05779 (to A.T.), 17H06386 (to K.F.), and 16H06280 “ABiS”; a grant from the NIPPON Genetics (to A.T.); The Keihanshin Consortium for Fostering the Next Generation of Global Leaders in Research (K-CONNEX) established by the program of Building of Consortia for the Development of Human Resources in Science and Technology, MEXT (to T.K.); and Japan Science and Technology Agency JPMJCR2121 (to K.F.).
Author contributions: Conceptualization: A.T., K.F., and T.K. Formal analysis: A.T. Investigation: A.T. Funding acquisition: A.T., K.F., and T.K. Project administration: A.T. Supervision: T.K. Writing—original draft: A.T. and T.K. Writing—review and editing: K.F.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S21
Legends for movies S1 to S14
References
Other Supplementary Material for this manuscript includes the following:
Movies S1 to S14
REFERENCES AND NOTES
- 1.C. M. Nelson, On buckling morphogenesis. J. Biomech. Eng. 138, 021005 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.C. Collinet, T. Lecuit, Programmed and self-organized flow of information during morphogenesis. Nat. Rev. Mol. Cell Biol. 22, 245–265 (2021). [DOI] [PubMed] [Google Scholar]
- 3.C. H. Waddington, The genetic control of wing development in Drosophila. J. Genet. 41, 75–113 (1940). [Google Scholar]
- 4.D. Fristrom, M. Wilcox, J. Fristrom, The distribution of PS integrins, laminin A and F-actin during key stages in Drosophila wing development. Development 117, 509–523 (1993). [DOI] [PubMed] [Google Scholar]
- 5.F. Roch, C. R. Alonso, M. Akam, Drosophila miniature and dusky encode ZP proteins required for cytoskeletal reorganisation during wing morphogenesis. J. Cell Sci. 116, 1199–1207 (2003). [DOI] [PubMed] [Google Scholar]
- 6.M. C. Diaz de la Loza, B. J. Thompson, Forces shaping the Drosophila wing. Mech. Dev. 144, 23–32 (2017). [DOI] [PubMed] [Google Scholar]
- 7.T. Lecuit, P.-F. Lenne, Cell surface mechanics and the control of cell shape, tissue patterns and morphogenesis. Nat. Rev. Mol. Cell Biol. 8, 633–644 (2007). [DOI] [PubMed] [Google Scholar]
- 8.C.-P. Heisenberg, Y. Bellaïche, Forces in tissue morphogenesis and patterning. Cell 153, 948–962 (2013). [DOI] [PubMed] [Google Scholar]
- 9.E. Caussinus, O. Kanca, M. Affolter, Fluorescent fusion protein knockout mediated by anti-GFP nanobody. Nat. Struct. Mol. Biol. 19, 117–121 (2012). [DOI] [PubMed] [Google Scholar]
- 10.R. P. Ray, A. Matamoro-Vidal, P. S. Ribeiro, N. Tapon, D. Houle, I. Salazar-Ciudad, B. J. Thompson, Patterned anchorage to the apical extracellular matrix defines tissue shape in the developing appendages of Drosophila. Dev. Cell 34, 310–322 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.R. Etournay, M. Popović, M. Merkel, A. Nandi, C. Blasse, B. Aigouy, H. Brandl, G. Myers, G. Salbreux, F. Jülicher, S. Eaton, Interplay of cell dynamics and epithelial tension during morphogenesis of the Drosophila pupal wing. eLife 4, e07090 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.W.-C. Chu, S. Hayashi, Mechano-chemical enforcement of tendon apical ECM into nano-filaments during Drosophila flight muscle development. Curr. Biol. 31, 1366–1378.e7 (2021). [DOI] [PubMed] [Google Scholar]
- 13.S. J. Smith, L. A. Davidson, M. Rebeiz, Evolutionary expansion of apical extracellular matrix is required for the elongation of cells in a novel structure. eLife 9, e55965 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.C. M. Lye, H. W. Naylor, B. Sanson, Subcellular localisations of the CPTI collection of YFP-tagged proteins in Drosophila embryos. Development 141, 4006–4017 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.N. Lowe, J. S. Rees, J. Roote, E. Ryder, I. M. Armean, G. Johnson, E. Drummond, H. Spriggs, J. Drummond, J. P. Magbanua, H. Naylor, B. Sanson, R. Bastock, S. Huelsmann, V. Trovisco, M. Landgraf, S. Knowles-Barley, J. D. Armstrong, H. White-Cooper, C. Hansen, R. G. Phillips, K. S. Lilley, S. Russell, D. St Johnston, Analysis of the expression patterns, subcellular localisations and interaction partners of Drosophila proteins using a pigP protein trap library. Development 141, 3994–4005 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.L. F. Appel, M. Prout, R. Abu-Shumays, A. Hammonds, J. C. Garbe, D. Fristrom, J. Fristrom, The Drosophila Stubble-stubbloid gene encodes an apparent transmembrane serine protease required for epithelial morphogenesis. Proc. Natl. Acad. Sci. U.S.A. 90, 4937–4941 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.M.-C. Diaz-de-la-Loza, R. P. Ray, P. S. Ganguly, S. Alt, J. R. Davis, A. Hoppe, N. Tapon, G. Salbreux, B. J. Thompson, Apical and basal matrix remodeling control epithelial morphogenesis. Dev. Cell 46, 23–39.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.L. Drees, T. Königsmann, M. H. J. Jaspers, R. Pflanz, D. Riedel, R. Schuh, Conserved function of the matriptase-prostasin proteolytic cascade during epithelial morphogenesis. PLOS Genet. 15, e1007882 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.M.-C. Diaz-de-la-Loza, R. Loker, R. S. Mann, B. J. Thompson, Control of tissue morphogenesis by the HOX gene Ultrabithorax. Development 147, dev184564 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.L. D. Ríos-Barrera, S. Sigurbjörnsdóttir, M. Baer, M. Leptin, Dual function for Tango1 in secretion of bulky cargo and in ER-Golgi morphology. Proc. Natl. Acad. Sci. U.S.A. 114, E10389–E10398 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Y. Sakaidani, T. Nomura, A. Matsuura, M. Ito, E. Suzuki, K. Murakami, D. Nadano, T. Matsuda, K. Furukawa, T. Okajima, O-Linked-N-acetylglucosamine on extracellular protein domains mediates epithelial cell–matrix interactions. Nat. Commun. 2, 583 (2011). [DOI] [PubMed] [Google Scholar]
- 22.B. Aigouy, R. Farhadifar, D. B. Staple, A. Sagner, J.-C. Röper, F. Jülicher, S. Eaton, Cell flow reorients the axis of planar polarity in the wing epithelium of Drosophila. Cell 142, 773–786 (2010). [DOI] [PubMed] [Google Scholar]
- 23.K. V. Iyer, R. Piscitello-Gómez, J. Paijmans, F. Jülicher, S. Eaton, Epithelial viscoelasticity is regulated by mechanosensitive E-cadherin turnover. Curr. Biol. 29, 578–591.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 24.J. A. Croker, S. L. Ziegenhorn, R. A. Holmgren, Regulation of the Drosophila transcription factor, Cubitus interruptus, by two conserved domains. Dev. Biol. 291, 368–381 (2006). [DOI] [PubMed] [Google Scholar]
- 25.H. Tanimoto, S. Itoh, P. ten Dijke, T. Tabata, Hedgehog creates a gradient of DPP activity in Drosophila wing imaginal discs. Mol. Cell 5, 59–71 (2000). [DOI] [PubMed] [Google Scholar]
- 26.A. Proag, B. Monier, M. Suzanne, Physical and functional cell-matrix uncoupling in a developing tissue under tension. Development 146, dev172577 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.M. A. Sturtevant, E. Bier, Analysis of the genetic hierarchy guiding wing vein development in Drosophila. Development 121, 785–801 (1995). [DOI] [PubMed] [Google Scholar]
- 28.A. Koto, E. Kuranaga, M. Miura, Temporal regulation of Drosophila IAP1 determines caspase functions in sensory organ development. J. Cell Biol. 187, 219–231 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.A. Tsuboi, S. Ohsawa, D. Umetsu, Y. Sando, E. Kuranaga, T. Igaki, K. Fujimoto, Competition for space is controlled by apoptosis-induced change of local epithelial topology. Curr. Biol. 28, 2115–2128.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 30.W. Draper, J. Liphardt, Origins of chemoreceptor curvature sorting in Escherichia coli. Nat. Commun. 8, 14838 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.T. Kondo, S. Hayashi, Mitotic cell rounding accelerates epithelial invagination. Nature 494, 125–129 (2013). [DOI] [PubMed] [Google Scholar]
- 32.J. Bischof, R. K. Maeda, M. Hediger, F. Karch, K. Basler, An optimized transgenesis system for Drosophila using germ-line-specific ϕC31 integrases. Proc. Natl. Acad. Sci. U.S.A. 104, 3312–3317 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.K. J. T. T. Venken, Y. He, R. A. Hoskins, H. J. Bellen, P[acman]: A BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314, 1747–1751 (2006). [DOI] [PubMed] [Google Scholar]
- 34.B. Aigouy, D. Umetsu, S. Eaton, Segmentation and Quantitative Analysis of Epithelial Tissues, in Methods in molecular biology (2016); 10.1007/978-1-4939-6371-3_13, pp. 227–239. [DOI] [PubMed]
- 35.B. Aigouy, C. Cortes, S. Liu, B. Prud’Homme, EPySeg: A coding-free solution for automated segmentation of epithelia using deep learning. Development 147, dev194589 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.S. E. McGuire, P. T. Le, A. J. Osborn, K. Matsumoto, R. L. Davis, Spatiotemporal rescue of memory dysfunction in Drosophila. Science 302, 1765–1768 (2003). [DOI] [PubMed] [Google Scholar]
- 37.A.-K. Classen, B. Aigouy, A. Giangrande, S. Eaton, Imaging Drosophila Pupal Wing Morphogenesis, in Methods in molecular biology (Clifton, N.J.) (2008); 10.1007/978-1-59745-583-1_16, vol. 420, pp. 265–275. [DOI] [PubMed]
- 38.B. Ewen-Campen, D. Yang-Zhou, V. R. Fernandes, D. P. González, L.-P. Liu, R. Tao, X. Ren, J. Sun, Y. Hu, J. Zirin, S. E. Mohr, J.-Q. Ni, N. Perrimon, Optimized strategy for in vivo Cas9-activation in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 114, 9409–9414 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S21
Legends for movies S1 to S14
References
Movies S1 to S14