Abstract
Pattern recognition receptor–mediated innate immunity is critical for host defense against viruses. A growing number of coding and noncoding genes are found to encode microproteins. However, the landscape and functions of microproteins in responsive to virus infection remain uncharacterized. Here, we systematically identified microproteins that are responsive to vesicular stomatitis virus infection. A conserved and endoplasmic reticulum–localized membrane microprotein, MAVI1 (microprotein in antiviral immunity 1), was found to interact with mitochondrion-localized MAVS protein and inhibit MAVS aggregation and type I interferon signaling activation. The importance of MAVI1 was highlighted that viral infection was attenuated and survival rate was increased in Mavi1-knockout mice. A peptide inhibitor targeting the interaction between MAVI1 and MAVS activated the type I interferon signaling to defend viral infection. Our findings uncovered that microproteins play critical roles in regulating antiviral innate immune responses, and targeting microproteins might represent a therapeutic avenue for treating viral infection.
The critical role of a microprotein in antiviral innate immune responses was uncovered.
INTRODUCTION
As the first line of defense against microbial pathogens, such as RNA and DNA viruses, innate immunity can elicit robust antiviral responses (1). Upon viral infection, viral-derived RNA/DNA is recognized by pattern recognition receptors, which include Toll-like receptors (TLRs), retinoic acid–inducible gene I (RIG-I)–like receptors (RLRs), NOD-like receptors, C-type lectin receptors, and several other nucleic acid sensors (2, 3). TLR family protein of RNA sensors includes TLR3, TLR7, and TLR8, which are localized in the endosome (4), and RLRs family protein of RNA sensors includes RIG-I, Melanoma differentiation-associated protein 5 (MDA5) , and Laboratory of genetics and physiology 2 (LGP2), which are mainly localized in the cytosol of cells (5). Among them, TLR3 recognizes double-stranded RNA (dsRNA) in a sequentially independent manner (6), while TLR7 and TLR8 detect single-stranded RNA (7, 8). As for the RLRs, all of them are characterized by a C-terminal dsRNA-binding domain and a central Aspartic acid-Glutamic acid-Alanine-Aspartic acid (DEAD) box helicase/adenosine triphosphate binding domain (9). RIG-I prefers to recognizing short dsRNA (<300 bp) with 5′ triphosphate moiety as well as short fragments of Polyinosinic-polycytidylic acid [poly (I:C)], 5′ diphosphate, or circular RNA (10, 11), while MAD5 mainly binds lengthy and irregular dsRNA (>300 bp) (12). Upon RNA viral infection, the conformation of both RNA sensors changes and forms a tandem CARD (caspase activation and recruitment domain) tetramer in synchrony with translocation to the mitochondria (13). Subsequently, the CARDs of RIG-I and MDA5 are equipped with the K63 multiubiquitin chain by E3 ubiquitin ligases (14), which is required for exposing the CARD domains to bind to the downstream key adaptor protein MAVS on the surface of mitochondria (also known as IPS1, VISA, or Cardif) (15–19). MAVS then forms a functional prion-like aggregates structure using the mitochondrial outer membrane as scaffold (20), which further activates cytosolic TANK-binding kinase 1 (TBK1) and Inhibitor of nuclear factor kappa-B kinase (IKK) complex. Phosphorylation of Interferon regulatory factor 3 (IRF3) and IRF7 by TBK1 and nuclear factor κB (NF-κB) by IKK promotes the translocation of IRF3, IRF7, and NF-κB into the nucleus, inducing type I interferons (IFNs) and proinflammatory cytokines (21–23). The secreted proinflammatory cytokines promote neutrophil recruitment and activate macrophages to clear pathogens, while secreted interferons activate the Janus kinase/signal transducers and activators of transcription signaling to induce the expression of hundreds of interferon-stimulated genes (ISGs) (24–26). The activation of RLRs signaling is critical to limit the spread of widespread spectrum viral infections (27, 28). RLR-MAVS–mediated type I IFN and ISGs induction are orchestrated by a variety of different mechanisms (29–31).
With the completion of the first draft of the human genome in 2001, the number of annotated human genes increased markedly over the next decade (32). It spurred an era of elucidating the function of the noncoding portion of the genome. The number of their search for protein-coding genes at ~20,000 in the mid-2000s is far underestimated (33, 34). With the development of ribosome profiling (Ribo-seq) and mass spectrometry (MS) techniques, various RNA transcripts including microRNA, circRNA, small nuclear RNA (snRNA), Piwi-interacting RNA, and long noncoding RNA (lncRNA), previously considered to be noncoding, have been shown to be capable of encoding short peptides or proteins (35, 36). Proteins encoded by lncRNA are involved in various biological processes such as development (37), metabolism (38), calcium homeostasis (39), RNA decapping (40), stress signaling (41), cell death (42), and tumorigenesis (43), among others. Whether microproteins, particularly for those are membrane localized, are involved in RLR-MAVS–mediated antiviral immune responses remain unexplored.
Here, we systematically identified microproteins that are responsive to vesicular stomatitis virus (VSV) infection from both coding and noncoding RNAs through transcriptomic profiling in combination with Ribo-seq. One of these microproteins, which we named as MAVI1, contains a signal peptide (SP) and a transmembrane (TM) domain and is localized on ER membrane. MAVI1 attenuates RLR-mediated type I IFN signaling and antiviral immune responses by directly targeting MAVS protein on mitochondrion. Concordantly, Mavi1 deficiency in mice notably enhances innate immune responses against RNA viruses and improves mouse survival. Peptide inhibitor targeting MAVI1 and MAVS interaction is potent in activating the type I IFN signaling and defending viral infection.
RESULTS
A microprotein MAVI1 is down-regulated in response to RNA virus infection and suppresses the expression of IFNB1
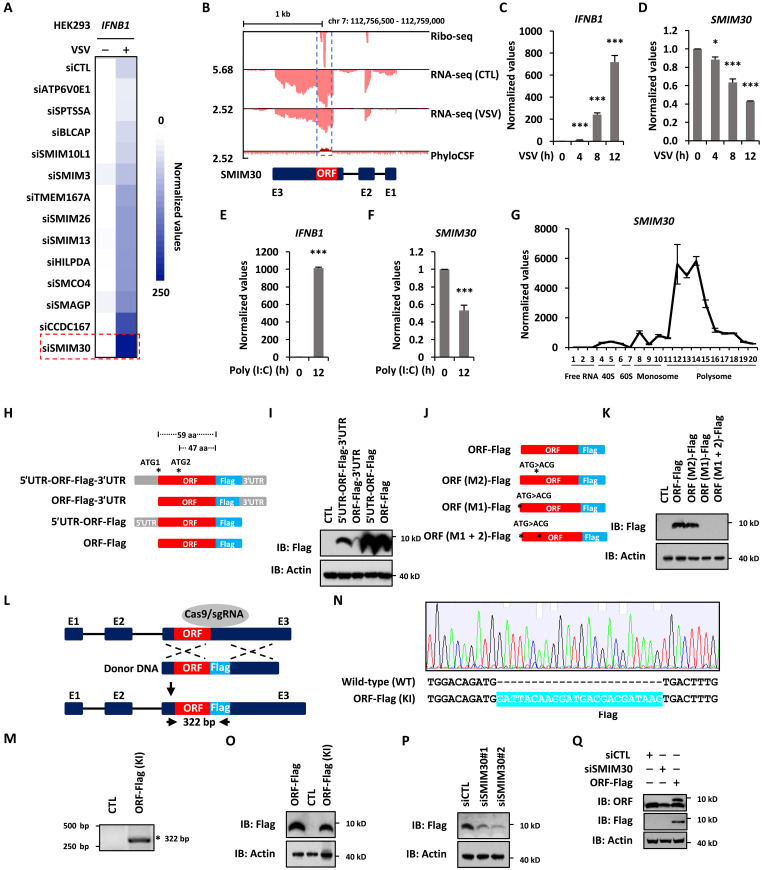
To explore the landscape of microproteins in response to RNA viral infection, we performed transcriptomic analysis in human embryonic kidney (HEK) 293 cells with or without VSV infection as well as analyzed Ribo-seq in HEK293 cells reported previously (44). Transcriptomic analysis results revealed that 1999 and 2318 genes were induced and repressed upon VSV infection, respectively [Fold change (FC) ≥ 1.5, q ≤ 0.05], and Ribo-seq analysis results revealed that there were 14,809 ORFs (open reading frames) predicted, including both known and de novo ORFs (fig. S1A and table S1). Among all the predicted ORFs, the transcripts of 881 and 1800 ORFs were induced and repressed by VSV, respectively (fig. S1A and table S1). In particular, 816 of these ORFs were predicted to have less than 100 amino acids, which were defined as short ORFs (sORFs) (fig. S1A and table S1). There were 77 sORFs from known coding genes, while the rest were de novo ones (fig. S1A and table S1). We noticed that a large number of protein products from these known sORFs, namely, microproteins, were predicted to contain TM domain (n = 13) (fig. S1A and table S1). We then performed a small-scale small interfering RNA (siRNA) screening against these 13 genes to examine their impact on VSV-induced IFNB1 expression (Fig. 1A). Knockdown of microprotein SMIM30 (45, 46), once considered as a noncoding RNA (LINC00998) (47, 48), exhibited the most marked effects on IFNB1 expression. We therefore focused on investigating whether and how SMIM30 regulates antiviral immune responses in the current study.
Fig. 1. A microprotein MAVI1 is down-regulated in response to RNA virus infection.
(A) HEK293 cells transfected with control siRNA (siCTL) or siRNA targeting genes as indicated were treated with or without VSV (2 × 106 p.f.u.) for 12 hours before examining IFNB1 expression. (B) Ribo-seq, RNA-seq, and PhyloCSF for SMIM30 are shown. (C and D) The expression of IFNB1 (C) and SMIM30 (D) were examined in HEK293 cells infected with VSV (mean ± SEM, * P < 0.05, ***P < 0.001). (E and F) The expression of IFNB1 (E) and SMIM30 (F) were examined in HEK293 cells transfected with Poly (I:C) (5 μg/ml) (mean ± SEM, ***P < 0.001). (G) HEK293 cells were subjected to polysome profiling, and the expression of SMIM30 was examined. (H) Schematic representation of Flag-tagged ORF with or without 5′ or 3′UTR. aa, amino acid. (I) HEK293 cells were transfected with constructs as shown in (H), followed by immunoblotting (IB) analysis. (J) Schematic representation of Flag-tagged ORF and its mutants with the first (M1), the second (M2), or both (M1 + M2) ATG mutated. (K) HEK293 cells were transfected with constructs as shown in (J), followed by IB analysis. (L) Schematic representation of Flag tag insertion at the end of ORF {ORF-Flag [knock-in (KI)]} in HEK293 cells. (M) Genomic DNA was extracted from ORF-Flag (–KI) cells, followed by PCR analysis. The expected size is 322 bp. (N) The PCR product was subjected to Sanger sequencing. The Flag tag inserted is highlighted in light blue. (O) HEK293 cells transfected with empty vector (CTL) or ORF-FLAG, or ORF-Flag (KI) HEK293 cells were subjected to IB analysis. (P) HEK293 cells transfected with siCTL, siSMIM30#1, and siSMIM30#2 were subjected to IB analysis. (Q) HEK293 cells transfected with siCTL, siSMIM30, or SMIM30-FLAG were subjected to IB analysis.
As described above, SMIM30 was found to be repressed by VSV as seen from RNA sequencing (RNA-seq) analysis (Fig. 1B). Repression of SMIM30 by VSV was confirmed in a time-dependent manner by reverse transcription quantitative polymerase chain reaction (RT-qPCR) analysis in both HEK293 (Fig. 1, C and D) and HeLa (fig. S1, B and C) cells. Similarly, poly (I:C), a synthetic analog of dsRNA used to mimic the replication intermediates presented in cells when infected with RNA viruses, repressed the expression of SMIM30 (Fig. 1, E and F). As SMIM30 was originally thought to be a lncRNA (LINC00998), we next examined whether SMIM30 is truly encoding a microprotein, and if so, what the nature of this microprotein. UCSC genome browser views from Ribo-seq analysis showed that SMIM30 has coding potential, which was supported by PhyloCSF prediction (Fig. 1B). The coding potential of SMIM30 was further demonstrated by polysome profiling assay in HEK293 cells, such that SMIM30 was largely associated with polysome fractions (Fig. 1G). As expected, YY1 and 7 SK snRNA were largely associated with polysome and ribosome-free fractions, respectively (fig. S1, D and E). To validate the coding capacity of the predicted ORF in SMIM30 and assess the impact of the corresponding 3′ and 5′ untranslated region (UTR) has on the translation efficiency of the predicted ORF, a series of constructs in which the predicted ORF was fused to a Flag-tag at its C-terminal end in the presence or absence of the 3′- or 5′UTR (Fig. 1H). These constructs expressed a microprotein at the predicted size (59 amino acids in length and less than 10 kDa in size) (Fig. 1I). Deletion of the 5′UTR markedly inhibited the translation efficiency, while deletion of the 3′UTR exhibited the opposite effects (Fig. 1I). The predicted ORF has an alternative translational start site 12 amino acids downstream of the first one (Fig. 1J). Mutation analysis of each translational start site (ATG to ACG) revealed that the first ATG is critical for translation (Fig. 1K). To further determine whether the predicted ORF is endogenously expressed in cells, we inserted a Flag tag right after the predicted ORF in the genome in HEK293 cells using CRISPR-Cas9–mediated homologous recombination (Fig. 1L). PCR-based genotyping (Fig. 1M) and Sanger sequencing (Fig. 1N) analyses verified the successful insertion of the Flag tag, which was further validated by immunoblotting analysis using anti-Flag antibody, yielding a microprotein of ~10 kDa (Fig. 1O). The expression of the endogenous Flag-tagged microprotein was attenuated when cells were transfected with siRNAs specifically targeting SMIM30 (Fig. 1P). To further validate the existence of the predicted microprotein, we generated a polyclonal antibody that specifically detects a microprotein, which was reduced when cells were transected with siRNA targeting SMIM30 (Fig. 1Q). Collectively, our data indicated that SMIM30, used to be considered as a lncRNA (LINC00998), encodes a microprotein, which is down-regulated in response to VSV infection. As we will demonstrate below that SIMIM30 plays a critical role in antiviral immunity, we renamed this microprotein as microprotein in antiviral immunity 1 (MAVI1).
MAVI1 negatively regulates antiviral immune responses (RNA virus) in vitro
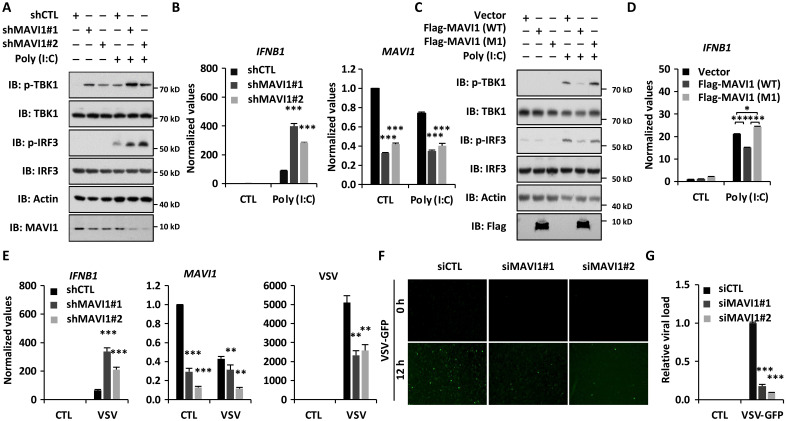
The repression of microprotein MAVI1 by RNA virus infection prompts us to examine the functional role of MAVI1 in RLR-mediated antiviral innate immune responses. HeLa cells were infected with lentivirus expressing control short hairpin RNA (shRNA) or two individual shRNAs targeting MAVI1 (shMAVI1 #1 and shMAVI1 #2) and treated with or without poly (I:C). Poly (I:C) treatment induced the phosphorylation of TBK1 and IRF3, which was further enhanced when knocking down of MAVI1 (Fig. 2A). Consequently, the expression of IFNB1 was markedly induced upon MAVI1 knockdown (Fig. 2B). Similarly, knockdown of MAVI1 by using siRNAs led to further induction of IFNB1 expression (fig. S2A). Overexpression of MAVI1, attenuated poly (I:C)–induced TBK1 and IRF3 phosphorylation (Fig. 2C) as well as IFNB1 expression (Fig. 2D), whereas MAVI1 (M1) with the start codon mutated failed to so, suggesting that the microprotein itself, but not the RNA transcript, played a repressive role in type I IFN signaling pathway. Furthermore, reintroduction of MAVI1 [wild type (WT)], but not MAVI1 (M1), abolished the effects of MAVI1 knockdown on type I IFN signaling (fig. S2B). In line with the above results, knockdown of MAVI1 further enhanced VSV-induced IFNB1 expression, which was concomitant with decreased viral replication (Fig. 2, E to G, and fig. S2C).
Fig. 2. MAVI1 negatively regulates RLR-mediated antiviral immune responses in vitro.
(A) Hela cells were infected with lentivirus expressing control shRNA (shCTL) or two individual shRNAs specifically targeting MVAI1 (shMVAI1#1 and shMVAI1#2) and treated with or without poly (I:C) (5 μg/ml, 6 hours), followed by immunoblotting (IB). (B) Hela cells were infected with lentivirus expressing shCTL, shMVAI1#1, or shMVAI1#2 and treated with or without poly (I:C) (5 μg/ml, 6 hours), followed by examination of IFNB1(left) and MAVI1 (right) expression (mean ± SEM, ***P < 0.001). (C) HeLa cells infected with lentivirus expressing empty vector, WT MAVI1 (Flag-MAVI1), or mutant MAVI1 with the first ATG mutated [Flag-MAVI1 (M1)] were subjected to IB analysis. (D) Cells as described in (C) were subjected to examination of IFNB1 expression (mean ± SEM, *P < 0.05, ***P < 0.001). (E) Hela cells were infected with lentivirus expressing shCTL, shMVAI1#1, shMVAI1#2 and treated with or without VSV (2 × 106 p.f.u., 12 hours), followed by examination of IFNB1, MAVI1, and VSV expression (mean ± SEM, **P < 0.01, ***P < 0.001). (F) Hela cells transfected with siCTL or siMAVI1 were treated with VSV-GFP for 12 hours, followed by microscopy imaging analysis. (G) The number of GFP-positive (GFP+) cells as shown in (F) were analyzed by ImageJ and normalized to siCTL-transfected cells.
MAVI1 negatively regulates antiviral immune responses (RNA virus) in vivo
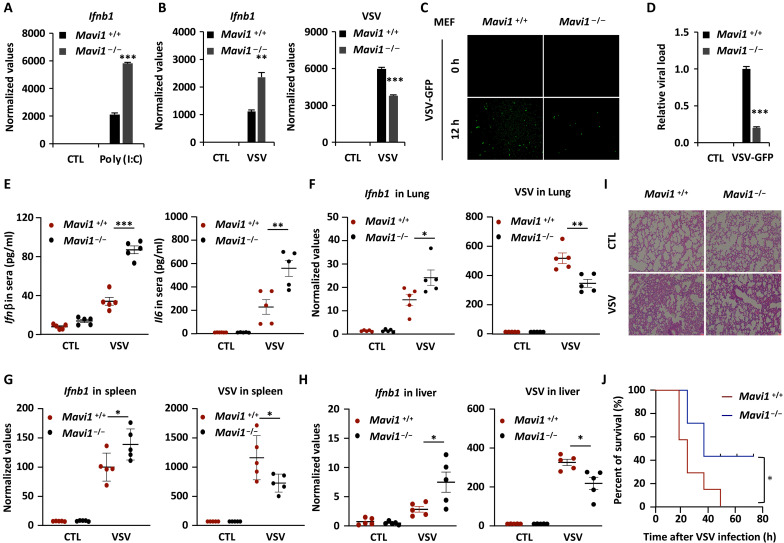
To further explore the role of MAVI1 in vivo, we generated Mavi1-deficient (Mavi1−/−) mice by CRISPR-Cas9 engineering (fig. S3, A and B). Mavi1−/− mice were born in accordance with Mendelian inheritance and gender ratios (fig. S3, C and D). There was no significant difference between WT (Mavi1+/+) and Mavi1−/− mice in terms of the weight of body and various organs (fig. S3, E and F). Mouse embryonic fibroblast (MEF) isolated from Mavi1−/− mice exhibited much stronger induction of Ifnb1 expression compared to those from Mavi1+/+ mice upon poly (I:C) transfection (Fig. 3A). Similarly, MEF from Mavi1−/− mice exhibited enhanced Ifnb1 expression when infected with VSV, which was concomitant with decreased viral replication (Fig. 3, B to D, and fig. S3G). Reintroduction of MAVI1 (WT), but not MAVI1 (M1), abolished the effects of Mavi1 knockout on VSV-induced Ifnb1 and VSV expression (fig. S3H). Furthermore, when mice were challenged with VSV, Mavi1−/− mice produced notably more cytokines including Infβ and Il6 in sera than those in Mavi1+/+ mice (Fig. 3E). The levels of Ifnb1 in spleen, lung, and liver tissues were also much higher in Mavi1−/− mice, which was concomitant with decreased viral burden (Fig. 3, F to H). Hematoxylin and eosin staining results showed that less infiltration of immune cells and less injury in the lung of Mavi1−/− mice upon VSV infection (Fig. 3I). Consequently, Mavi1−/− mice had higher survival rates compared to Mavi1+/+ mice when challenged with VSV infection VSV (Fig. 3J). Collectively, our data demonstrated that MAVI1 negatively regulates antiviral innate immune responses both in vitro and in vivo.
Fig. 3. MAVI1 negatively regulates RLR-mediated antiviral immune responses in vivo.
(A) WT (Mavi1+/+) and Mavi1 knockout (Mavi1−/−) MEFs treated with or without poly (I:C) (5 μg/ml, 6 hours) were subjected to examination of Ifnb1 expression (mean ± SEM, ***P < 0.001). (B) Mavi1+/+ and Mavi1−/− MEFs treated with or without VSV (2 × 106 p.f.u., 12 hours) were subjected to examination of the expression of genes as indicated (mean ± SEM, **P < 0.01, ***P < 0.001). (C) Mavi1+/+ and Mavi1−/− MEFs were treated with VSV-GFP for 12 hours, followed by microscopy imaging analysis. (D) The number of GFP+ cells as shown in (J) were analyzed by ImageJ and normalized to Mavi1+/+ MEFs without VSV-GFP treatment. (E) The levels of Infβ and Il6 in serum from Mavi1 +/+ or Mavi1−/− mice (n = 5) treated with or without VSV (5 × 108 p.f.u. per mouse) for 12 hours were examined by enzyme-linked immunosorbent assay (mean ± SEM, **P < 0.01, ***P < 0.001). (F to H) The levels of Infb and VSV from the lung (F), spleen (G), and liver (H) of mice as described in (L) were examined (mean ± SEM, *P < 0.05, **P < 0.01). (I) Lung tissue sections from mice as described in (M) were subjected to hematoxylin and eosin staining. (J) Survival curve of Mavi1 +/+ or Mavi1−/− mice (n = 7) treated with VSV (5 × 108 p.f.u. per mouse) is shown (mean ± SEM, *P < 0.05).
MAVI1 is an endoplasmic reticulum TM protein
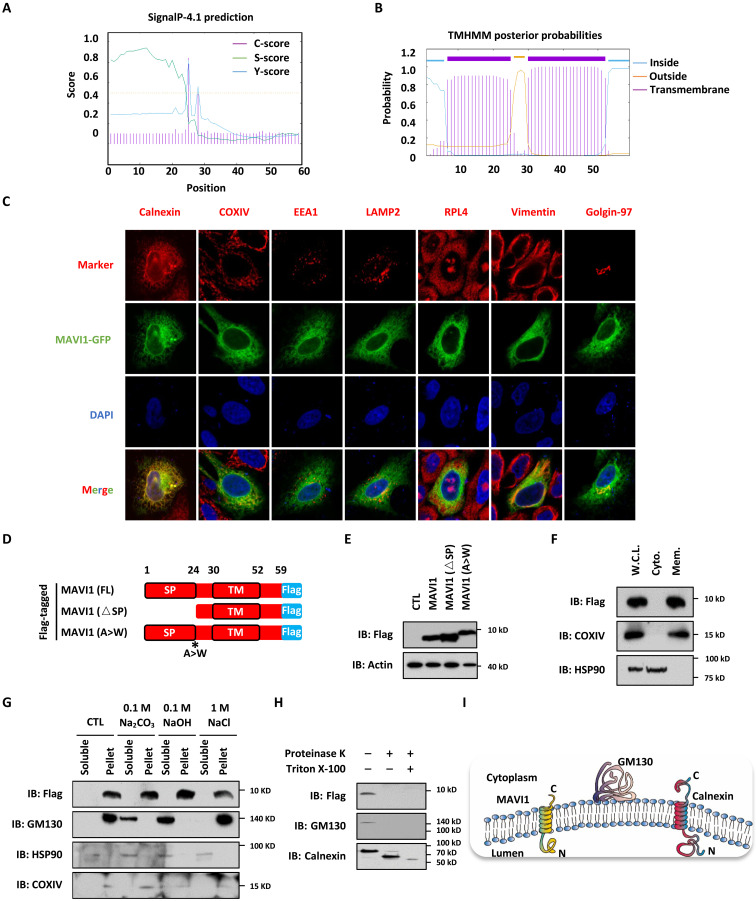
We next sought to investigate the molecular mechanisms underlying MAVI1 regulation of type I IFN signaling and antiviral immunity. SignaIP-4.1 and TMHMM predicted the presence of an SP (amino acids 1 to 24) and a TM domain (amino acids 30 to 52) in MAVI1 (Fig. 4, A and B). Immunofluorescence staining analysis results revealed that green fluorescent protein (GFP)–tagged MAVI1 largely colocalized with calnexin, a marker for endoplasmic reticulum (ER), and partly with COXIV and Golgin-97, which represent mitochondria and Golgi apparatus, respectively, in HEK293 cells (Fig. 4C). There was barely no colocalization with EEA1, LAMP2, RPL4, or Vimentin, which represents early endosomes, lysosomes, ribosome, and cytoskeleton, respectively (Fig. 4C). Cellular fractionation results also showed that MAVI1 was largely in the ER fraction, while a small portion was found in the mitochondria and Golgi apparatus fractions (fig. S4A). The colocalization of MAVI1 with ER was confirmed in several other cell lines, such as Hela, MCF7, Hep G2, A-498, and U87 cells (fig. S4B). To test whether the SP is required for MAVI1 targeting to ER, GFP-tagged MAVI1 with the SP-deleted (ΔSP) was constructed (fig. S4C). Immunofluorescence staining results showed that deletion of SP significantly impaired MAVI1 targeting to ER compared to WT MAVI1 (fig. S4D). Both WT and ΔSP expressed equally well (fig. S4E). The protein products from MAVI1 (WT) and MAVI1 (ΔSP) constructs were the same, indicating that the SP was removed after targeting (Fig. 4, D and E). To further support the removal of SP, mutation of Ala to Trp (A to W), a critical residue predicted to be essential for protease cleavage (49), caused a shift of the protein product (Fig. 4, D and E). The predicted TM domain in MAVI1 prompted us test whether MAVI1 is an ER membrane protein. Cellular fractionation analysis results revealed that MAVI1 was exclusively in the membrane fraction (Fig. 4F). Similar as the TM protein COXIV, MAVI1 remained in the pellet when the supernatant from homogenized cells were treated with Na2CO3, NaOH, or NaCl followed by centrifugation, while the outer membrane protein GM130 was released into the soluble fraction with Na2CO3 or NaOH treatment (Fig. 4G). Serving as a control, cytosolic protein HSP90 was detected in the soluble fraction both before and after treatment (Fig. 4G). To illustrate how MAVI1 is integrated with ER membrane, we transfected HEK293 cells with vector expressing MAVI1 with a Flag-tag at its C terminus. The membrane fraction was isolated and treated with or without proteinase K in the presence or absence of Triton X-100. MAVI1 was undetectable when we used anti-Flag antibody for immunoblotting after treated with proteinase K, indicating that the C terminus of MAVI 1 was exposed to the cytosol (Fig. 4H). As expected, the TM protein Calnexin was partially digested with proteinase K, while it nearly undetectable when Triton X-100 was added (Fig. 4H). The outer membrane protein GM130 was completely digested as long as proteinase K was added (Fig. 4H). Together, our results suggested that MAVI1 is an ER TM protein with its C terminus exposed to the cytosol of cells (Fig. 4I).
Fig. 4. MAVI1 is an ER membrane protein.
(A and B) The SP (1 to 24 amino acids) (A) and TM domain (30 to 52 aa) (B) in MAVI1 predicted by SignaIP-4.1and TMHMM website, respectively, are shown. (C) HEK293 cells transfected with GFP-tagged MAVI1 were stained with Calnexin (ER maker), COXIV (mitochondria maker), EEA1 (endosome maker), LAMP2 (lysosome maker), RPL4 (ribosome maker), Vimentin (cytoskeleton maker), or Golgin-97 (Golgi apparatus marker) (Red). Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) (blue). (D) Schematic representation of Flag-tagged, full length (FL) MAVI1, SP-deleted (△SP) MAVI1, and MAVI1 mutant with the SP cleavage site mutated (A > W) is shown. (E) HEK293 cells transfected with empty vector (CTL) or expression vectors as described in (D) were subjected to IB analysis. (F) HEK293 cells stably expressing Flag-tagged MAVI1 were subjected to cellular fraction, followed by IB analysis. W.C.L., whole cell lysate; Cyto., cytoplasmic fraction; Mem., membrane fraction. (G) HEK293 cells stably expressing Flag-tagged MAVI1 were subjected to membrane interaction assay, and the supernatant was treated with or without 0.1 M Na2CO3, 0.1 M NaOH, or 1 M NaCl. The resultant soluble solution and pellet were subjected to IB analysis. (H) HEK293 cells stably expressing Flag-tagged MAVI1 were subjected to cellular fractionation, and membrane fractions were treated with or without proteinase K in the presence or absence of Triton X-100, followed by IB analysis. (I) Membrane topology of calnexin, GM130, and MAVI1 proteins is shown.
MAVI1 interacts with MAVS
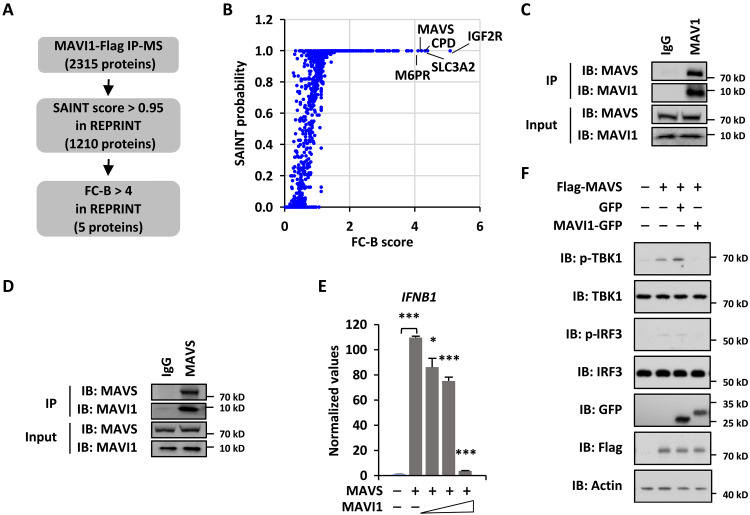
Next, to clarify the molecular targets of MAVI1 that are responsible for its role in antiviral immune responses, we analyzed MAVI1 interactome in HEK293 cells stably expressing Flag-tagged MAVI1 by immunoaffinity purification coupled with MS analysis. The false positives and common contaminates were removed by REPRINT (50), which led to the identification of five proteins, including IGF2R (insulin like growth factor 2 receptor), SLC3A2 (solute carrier family 3 member 2), CPD (carboxypeptidase D), MAVS (mitochondrial antiviral signaling protein, and M6PR (mannose-6-phosphate receptor), that potentially interact with MAVI1 (Fig. 5, A and B, and table S2). MAVS caught our attention due to the fact that it plays a vital role in host innate immune defense against RNA viruses by acting as the common adaptor of the RLR protein family of RNA sensors to activate downstream IRF3 and NF-κB and subsequently induce the expression of IFNs and ISGs (51). We therefore tested whether MAVI1 interacts with MAVS. Immunoprecipitation analysis using lysates from HEK293 cells stably expressing Flag-tagged MAVI1 confirmed that MAVI1 specifically interacted with MAVS (fig. S5A). Furthermore, MAVI1 interaction with MAVS was demonstrated by immunoprecipitation analysis in HEK293 cells overexpressing MAVI1 and MAVS proteins (fig. S5B). The interaction between MAVI1 and MAVS was also shown at endogenous level (Fig. 5C and 5D). We then isolated the ER, mitochondria, and mitochondria-associated ER membrane (MAM) fractions and found that MAVS and MAVI1 largely coexisted in the MAM fraction (fig. S5C). Furthermore, immunoprecipitation analysis using lysates from MAM fraction revealed that MAVI1 is associated with MAVS in MAM (fig. S5D). To support the interaction between MAVI1 and MAVS, overexpression MAVI1 suppressed MAVS-induced IFNB1 expression as well as TBK1 and IRF3 phosphorylation (Fig. 5, E and F). MAVI1 overexpression did not affect the protein level and subcellular localization of MAVS (fig. S5, E and F).
Fig. 5. MAVI1 interacts with MAVS.
(A) HEK293 cells stably expressing Flag-tagged MAVI1 were subjected to immunoprecipitation and mass spectrometry (IP-MS) analysis. (B) MAVI1-associated proteins identified were analyzed in REPRINT database to exclude common contaminants. Proteins with SAINT ≥ 0.95 and FC-B ≥ 4 were considered as specific binding. (C and D) HEK293 cells were subjected to IP analysis with anti-MAVI1 (C) or MAVS (D) antibody followed by IB analysis. (E) HEK293 cells transfected with or without MAVS in the presence or absence of MAVI1 were subjected to examine the expression of IFNB1 (mean ± SEM, *P < 0.05, ***P < 0.001). (F) HEK293 cells transfected with or without Flag-tagged MAVS in the presence of GFP or GFP-tagged MAVI1 were subjected to IB analysis.
MAVI1 interacts with MAVS through the TM domain
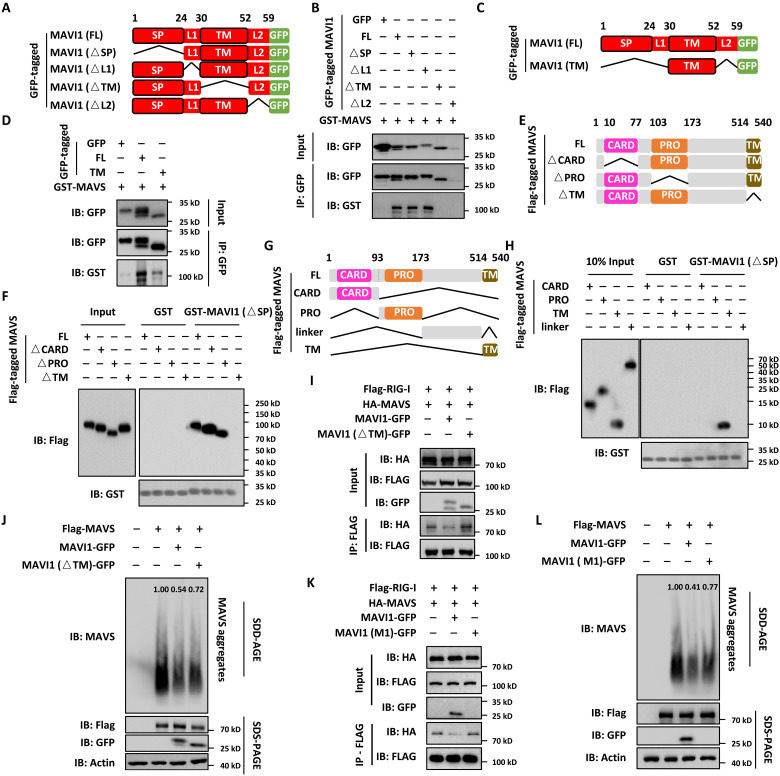
To further understand the interaction domain between MAVI1 and MAVS, a series of MAVI1 deletion constructs were transfected into HEK293 cells, purified, and coincubated with purified, bacterially expressed MAVS protein (Fig. 6A). Pull-down assay results revealed that TM deletion in MAVI1 (△TM) abolished its interaction with MAVS (Fig. 6B and fig. S6A). It should be noted that L2-deleted MAVI1 (△L2) was nearly undetectable (Fig. 6B). The TM domain of MAVI1 seemed to be able to be sufficient to interact with MAVS, although the affinity was much weaker than full-length MAVI1 (Fig. 6, C and D). Similarly, a series of MAVS deletion constructs were transfected into HEK293 cells, purified, and coincubated with purified, bacterially expressed MAVI1 (Fig. 6E). Pull-down assay results revealed that TM deletion in MAVS (△TM) abolished its interaction with MAVI1 (Fig. 6F and fig. S6B). To test whether TM domain in MAVS is sufficient for binding with MAVI1, different fragments containing the CARD domain, PRO domain, unknown region, and TM domain were cloned, transfected into HEK293 cells, purified, and coincubated with purified, bacterially expressed MAVI1. Pull-down assay results indicated that the TM domain, but not other fragments, interacted with MAVI1 (Fig. 6, G and H). The interaction with upstream sensor proteins such as RIG1 and subsequent aggregation of MAVS are critical for VSV-induced type I IFN signaling pathway. Immunoprecipitation analysis results indicated that the interaction between RIG-I and MAVS as well as was the aggregation of MAVS were largely interrupted in the presence of MAVI1, but not MAVI1 (△TM) or MAVI1 (M1) (Fig. 6, I to L). Together, our results indicated that MAVI1 directly interacts with MAVS to interrupt its interaction with RIG-I and its aggregation, suppressing antiviral innate immune responses.
Fig. 6. MAVI1 interacts with MAVS through the TM domain.
(A) Schematic representation of GFP-tagged, full length (FL), SP-deleted (△SP), linker region 1–deleted (△L1), TM domain–deleted (△L2), and linker region 2–deleted (△L2) MAVI1 is shown. (B) In vitro pull-down assay was performed by mixing GFP-tagged proteins purified from HEK293 cells as shown in (A) and purified, bacterially expressed GST-tagged MAVS. (C) Schematic representation of GFP-tagged MAVI1 (FL) and the TM domain only (TM) is shown. (D) In vitro pull-down assay was performed by mixing GFP-tagged proteins purified from HEK293 cells as shown in (C) and purified, bacterially expressed GST-tagged MAVS. (E) Schematic representation of Flag-tagged, MAVS (FL), caspase recruitment domain-deleted (△CARD), proline rich domain-deleted (△PRO), and TM domain–deleted (△TM) MAVS is shown. (F) In vitro pull-down assay was performed by mixing Flag-tagged proteins purified from HEK293 cells as shown in (E) and purified, bacterially expressed GST-tagged MAVS (△SP). (G) Schematic representation of Flag-tagged, MAVS (FL), CARD only (CARD), PRO only (PRO), unknown region only (unknown), and TM domain only (TM) is shown. (H) In vitro pull-down assay was performed by mixing Flag-tagged proteins purified from HEK293 cells as shown in (G) and purified, bacterially expressed GST-tagged MAVS (△SP). (I) HEK293 cells transfected with vectors as indicated were subjected to IP analysis. (J and L) Cell lysates from HEK293 cells transfected with vectors as indicated were resolved by SDD-AGE (top) and SDS-PAGE (bottom three) gel. The intensity of MAVS aggregates was quantified by ImageJ. (K) HEK293 cells transfected with vectors as indicated were subjected to IP analysis.
MAVI1 interaction with MAVS is critical for MAVI1 function in regulating antiviral innate immune responses
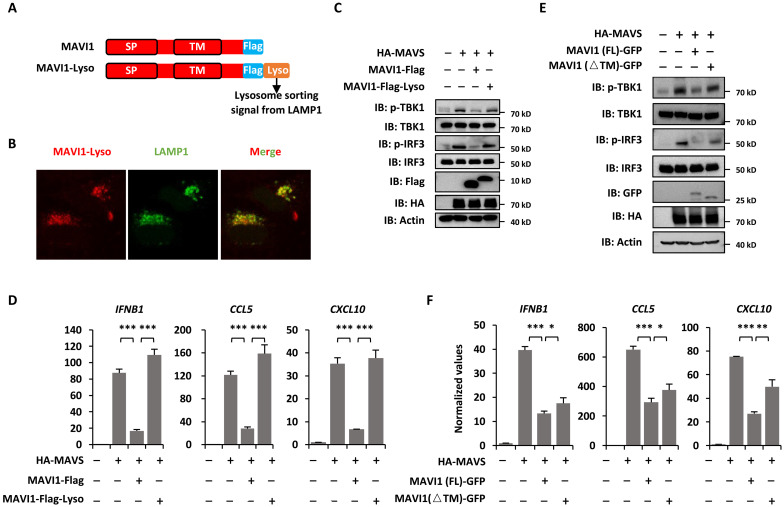
Contacts between the ERs and mitochondria are now recognized to be essential for diverse biological processes, such as calcium homeostasis (52, 53), cholesterol and ceramide biosynthesis (54), apoptosis (55), autophagy (56), and innate immune response (57). We next tested whether the interaction between ER-localized MAVI1 and mitochondrion-localized MAVS is critical for MAVI1 function in suppressing antiviral immune responses. First, MAVI1 fused to a lysosome sorting signal (MAVI1-Lyso) changed the cellular localization of MAVI1 from ER to lysosome as MAVI1-Lyso perfectly colocalized with LAMP1 as seen from immunofluorescence staining analysis (Fig. 7, A and B). Furthermore, MAVS-induced TBK1 and IRF3 phosphorylation as well as IFNB1, CCL5, and CXCL10 expression were attenuated in the presence of WT MAVI1, but not MAVI1-Lyso, indicating that ER localization of MAVI 1 is critical for its function in inhibiting type I IFN signaling (Fig. 7, C and D). As described above, TM domain of MAVI 1 is required for its interaction with MAVS. We then transfected HEK293 cells with MAVS in the presence or absence of MAVI 1 (FL) and MAVI 1 (△TM). MAVI 1 (△TM) was not as efficient as MAVI 1 (FL) in suppressing MAVS-induced TBK1 and IRF3 phosphorylation as well as IFNB1, CCL5, and CXCL10 expression (Fig. 7, E and F). In conclusion, our data demonstrated that the contact of ER-localized MAVI1 and mitochondrion-localized MAVS is critical for inhibiting type I IFN signaling pathway and antiviral immune responses.
Fig. 7. MAVI1 interaction with MAVS is critical for MAVI1 function in regulating antiviral innate immune responses.
(A) Schematic representation of Flag-tagged MAVI1 and Flag-tagged MAVI1 with the lysosome sorting signal from LAMP1 attached (MAVI1-Lyso) is shown. (B) HEK293 cells transfected with Flag-tagged MAVI1-lyso were subjected to immunofluorescence analysis using anti-Flag (red) and anti-LAMP1 antibody (green). (C) HEK293 cells transfected with vectors as indicated were subjected to IB analysis. (D) HEK293 cells as described in (C) were subjected to examination of IFNB1, CCL5, and CXCL10 expression (mean ± SEM, ***P < 0.001). (E) HEK293 cells transfected with vectors as indicated were subjected to IB analysis. (F) HEK293 cells as described in (E) were subjected to examination of IFNB1, CCL5, and CXCL10 expression (mean ± SEM, *P < 0.05, **P < 0.01, ***P < 0.001).
Peptide inhibitor targeting the interaction between MAVI 1 and MAVS (PiMAVI1) is potent in activating type I IFN signaling and antiviral immune responses
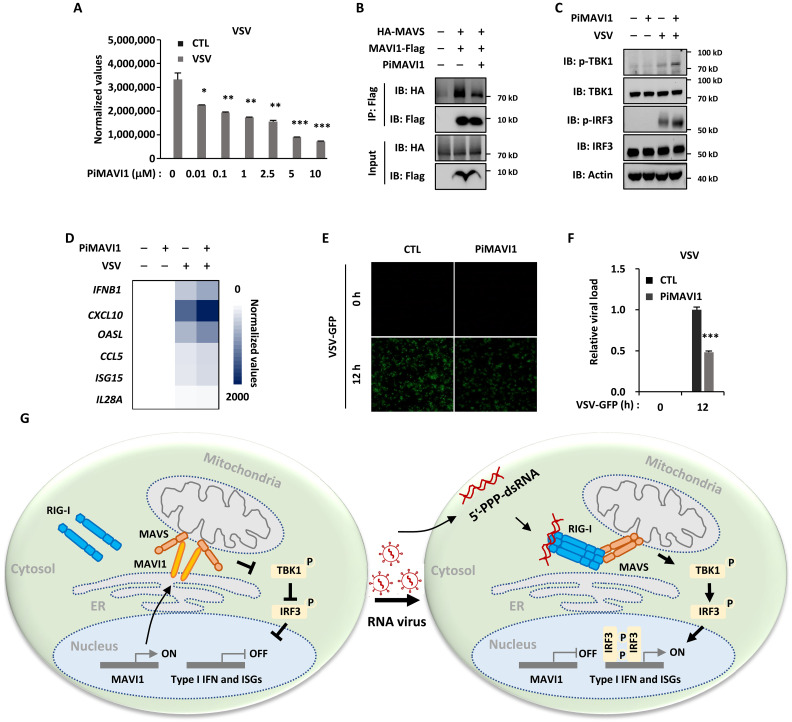
MAVI1 interacts with MAVS to suppress the antiviral immune responses prompted us to test whether interrupting such interaction will activate type I IFN signaling to defend against viral infection. As MAVI1 uses its TM domain to specifically interact MAVS, we designed and synthesized a short peptide with the membrane penetrating TAT sequence to mimic this region and tested its impact on MAVI1 and MAVS interaction, type I IFN signaling, and RNA viral replication. We first confirmed that such peptide conjugated with fluorescein isothiocyanate could enter into cells (fig. S7A). The cellular activity of this peptide in suppressing VSV viral replication as measured by VSV expression level was tested by treating HEK293 cells with increasing concentrations of this peptide (Fig. 8A). The best working concentration seemed to be around 10 μM without causing nonspecific cellular toxicity (Fig. 8A). We named this peptide as peptide inhibitor targeting MAVI1 and MAVS interaction (PiMAVI1). PiMAVI1 treatment suppressed the interaction between MAVI 1 and MAVS in HEK293 cells (Fig. 8B). Accordingly, PiMAVI1 treatment further enhanced VSV-induced phosphorylation of TBK1 and IRF3 as well as expression of IFNB1 and ISGs (Fig. 8, C and D). As a result, VSV replication was significantly impaired with PiMAVI1 treatment (Fig. 8, E and F, and fig. S7B). Together, our data demonstrated that targeting MAVI1 and MAVS interaction using PiMAVI1 is an effective way to activate type I IFN signaling pathway to defend against RNA viral infection.
Fig. 8. Peptide inhibitor targeting the interaction between MAVI1 and MAVS (PiMAVI1) is potent in activating type I IFN signaling and antiviral immune responses.
(A) HEK293 cells were pretreated with peptide inhibitor PiMAVI1 at concentrations as indicated for 8 hours and then infected with or without VSV treatment (2 × 106 p.f.u., 12 hours) were subjected to examination of VSV replication (mean ± SEM, *P < 0.05, **P < 0.01, ***P < 0.001). (B) HEK293 cells transfected with hemagglutinin (HA)–tagged MAVS and Flag-tagged MAVI1 and treated with or without PiMAVI1 (10 μM, 8 hours) were subjected to immunoprecipitation (IP) with anti-Flag antibody followed by immunoblotting (IB) analysis. (C and D) HEK293 cells pretreated with PiMAVI1 (10 μM, 8 hours) and then infected with VSV (12 hours) were subjected to IB (C) or RT-qPCR analysis (D). (E) HEK293 cells were pretreated with or without PiMAVI1 (10 μM, 8 hours) and then infected with or without VSV-GFP (2 × 106 p.f.u., 12 hours), followed by microscopy imaging analysis. (F) The number of GFP-positive (GFP+) cells as shown in (E) was analyzed by ImageJ and normalized to control cells. (G) The proposed model of MAVI1 function in antiviral innate immune responses. ER-localized TM protein MAVI1 contacts with MAVS protein on mitochondria to compete with RIG-I for binding to MAVS, resulting in the repression of type I IFN signaling and innate immune responses. Upon VSV infection, MAVI1 is down-regulated, and its inhibition on the interaction between RIG-I and MAVS is released, leading to the activation of type I IFN signaling and innate immune responses. Peptide inhibitor targeting the interaction between MAVI1 and MAVS is capable of disrupting MAVI1-MAVS interaction, activating type I IFN signaling, and enhancing host antiviral innate immunity.
DISCUSSION
Microproteins concealed within coding and noncoding genes remain highly underrepresented in genome annotations, which is due in large part to the difficulty in identifying functional sORFs in RNA transcripts. Although a number of genes that code for functional microproteins were identified and characterized with the rapid development of ribosome profiling, mass spectrum, and bioinformatics technology (58, 59), the physiological or pathological roles of the putatively large number of microproteins remain underexplored, in particular, those microproteins involved in antiviral innate immune response in host cells are poorly understood. Antiviral innate immune response is the first line of defense against viruses or microbial pathogens. Many diseases, such as severe acute respiratory syndrome (60), Middle East respiratory syndrome (61), severe fever with thrombocytopenia syndrome (62), and coronavirus disease 2019 (63), among others, are associated with infection with virus or microbial pathogens. Hence, it is meaningful to identify and explore the mechanisms of microproteins in the host defense against virus or microbial pathogens.
Transcriptomic combined with Ribo-seq analysis was performed in HEK293 cells, which revealed that more than 800 microproteins, both from coding and noncoding genes, are potentially responsive to VSV viral infection. Among these microproteins, 739 microproteins are de novo ones, which await future experimental validation. For those 77 known microproteins, membrane microproteins were highly enriched. It has been reported that about 30% of microproteins, significantly higher than proteins in the proteome, contain the alpha TM domain (64), suggesting that microproteins have a tendency of biofilm enrichment. Membrane microproteins have been reported to be involved in ER stress responses (41), muscle development (65), and antiviral immune responses (66). One of these microproteins, which we named as MAVI1, was found to be an ER TM protein. MAVI1, used to be recognized as a lncRNA, has been reported to function in its RNA form in cancer development (47, 48). However, its function in type I IFN signaling and antiviral immune responses reported here is largely dependent on its protein product. MAVI1 directly targets MAVS on mitochondria to interfere its interaction with the RNA sensor protein RIG-I. It has been reported that lactic acid inhibits the MAVS/RIG-1 interaction and MAVS aggregation by binding to the TM domain in MAVS (67). The mechanism of action is that lactic acid causes changes of the localization of MAVS in mitochondria. However, our results showed that MAVI1 has no obvious effects on the localization of MAVS. The underlying molecular mechanism through which MAVI1 inhibits the interaction between RIG-I and MAVS remains unknown, which will be an interesting question to pursue. The contact between different cellular organelles has been shown to be functionally important in a variety of cellular processes, such as the synthesis of the mitochondrial cytochrome c oxidase (68), the transmission of Ca2+ signals (69), formation of Golgi Ca2+ gradients (70), and the synthesis of phospho- and glycosphingolipids (71). Contact region between ER and mitochondria, known as MAM, was separated and identified by Vance (72) a few decades ago. Furthermore, MAM is involved in various biological functions such as phospholipid synthesis (73), cellular organelle-mediated inflammation (57), calcium homeostasis (53), autophagy (56), and mitochondrial fission (74). Except for MAVI1 in our screening, the role of remaining membrane microproteins in antiviral immunity is worthy of future investigation. For instance, CCDC167 was reported to be a regulator of cell cycle and served as a potential therapeutic target in breast cancer (75). Small Cell Adhesion Glycoprotein (SMAGP) was reported as a biomarker of cervical cancer development and progression (76).
As mentioned above, ER localization of MAVI1 and its interaction with MAVS are critical for inhibiting type I IFN signaling and antiviral innate immune responses. We designed a peptide inhibitor (Pi) that interferes with the interaction between MAVI1 and MAVS, which was able to activate type I IFN and ISG expression and defend viral infection. Further improvement of the specificity and efficacy of PiMAVI1 is needed in future study, such as replacing the hydrophobic noncritical amino acid Leu with Ala to improve it solubility and shortening from both ends to improve its stability.
In conclusion, an ER TM microprotein MAVI1 targets MAVS to suppress the type I IFN signaling and antiviral immune responses in host cells. Upon viral infection, MAVI 1 is down-regulated, leading to the activation of MAVS-mediated antiviral immune responses. Peptide inhibitor targeting MAVI1 and MAVS (PiMAVI1) interaction is potent in activating host antiviral immunity and defending viral infection. Our study uncovered that microproteins might represent a class of drug targets for antiviral innate immunity, and PiMAVI1 might represent a potential therapeutic avenue for treating diseases related to the type I IFN signaling in the clinic.
MATERIALS AND METHODS
Cell culture
HEK293T, HEK293, Hela, and BHK21 cells were purchased from the American Type Culture Collection. MEF cells were isolated from WT (Mavi1+/+) and Mavi1-deficient (Mavi1−/−) embryos (day 13.5). HEK293T, HEK293, Hela, BHK21, and MEF cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) Bioind with 10% (v/v) fetal bovine serum (FBS; BI) and 1% penicillin/streptomycin (Hyclone) at 37°C in 5% CO2.
Molecular cloning
Full-length human MAVI1 was amplified from cDNA of HEK293T cells and cloned into pBoBi vector with a Flag or GFP tag at its C terminus or pGEX6p1 vector with a GST tag at its N terminus. MAVI1 (M1), MAVI1 (M2), MAVI1 (M1 + 2), and MAVI1 (A > W) were generated by the overlap extension PCR method using PrimeSTAR HS DNA Polymerase from pBoBi-MAVI1 plasmid. MAVI1 truncations were cloned into pBoBi vector with a GFP tag at C terminus. Full-length human MAVS were amplified from cDNA of HEK293T cells and cloned into pBoBi vector with a Flag or hemagglutinin (HA) tag at its N terminus or pGEX6p1 vector with a GST tag at its N terminus. shRNAs targeting MAV1 were cloned into pLKO.1-TRC vector between Age I and Eco RI sites (targeting sequence: 5′-AAGAAGTAAGATGGCTGAAAT-3′ (shMAVI1#1); 5’-AATCTTAGGAAATCACCACTG-3′ (shMAVI1#2).
Peptide treatment, plasmid and siRNA transfection, and lenti virus packaging and infection
PiMAVI1 (IALLLGVVLSITGICACLGVYA-YGRKKRRQRRR) was synthesized by Genscript Inc. Plasmid and siRNA transfection were performed using polyethyleneimine (PEI; Polysciences) and Lipofectamine 2000 (Invitrogen), respectively, according to the manufacturer’s protocol. Targeting sequence were as follows: 5′-GGTTCCAAGAATCAGTAAA-3′ (siMAVI1#1); 5′-GGAAGTTCTTGCCCAGAAT-3′ (siMAVI1#2); 5′-TGCCTTGGTTCATCCCTAA-3′ (siATP6V0E1); 5′-CCATGCTGGTTTCCATTGT-3′ (siSPTSSA); 5′-CCATGTTCATGGGCTTCTA-3′ (siBLCAP); 5′-GGAAGACACTATATCCTAT-3′ (siSMIM10L1); 5′-CCAAGCACATCCTGGATAT-3′ (siSMIM3); 5′-TCAGCATCCTCTTCATACA-3′ (siTMEM167A); 5′-CCATGACTGGCTGCTGAAT-3′ (siSMIM26); 5′-TGCTGATGAAGGCCACAGA-3′ (siSMIM13); 5′-GTTGAACCTCTACCTGTTA-3′ (siHILPDA); 5′-AGACCTCCAAGGACAAGAA-3′ (siSMCO4); 5′-GCAGCGAGAAAGAGGAATA-3′ (siSMAGP); 5′-CTGTGGCCATCTTTATCCT-3′ (siCCDC167).
For lenti virus packaging, HEK293T cells were seeded in culture plates coated with poly-d-lysine [0.1% (w/v), Sigma-Aldrich, P7280] and transfected with lentiviral vectors together with packaging vectors, pMDL, VSVG, and REV, at a ratio of 10:5:3:2 using PEI for 48 hours according to the manufacturer’s protocol. Viruses were collected, filtered, and added in the presence of polybrene (10 μg/ml; Sigma-Aldrich, H9268), followed by centrifugation for 30 min at 1500g at 37°C. Medium was replaced 24 hours later.
RNA isolation and RT-qPCR
Total RNA was isolated using TRIzol (Invitrogen) following the manufacturer’s protocol. First-strand cDNA synthesis from total RNA was carried out using 5× HiScriptIIQ RT SuperMix (Vazyme), followed by qPCR using an AriaMx Real-Time PCR machine (Agilent Technologies). The sequence information for all primers used for qPCR analysis can be found in table S3.
RNA sequencing
Total RNA was isolated using the RNeasy Mini Kit (Qiagen) following the manufacturer’s protocol. Deoxyribonuclease I in column digestion was included to ensure the RNA quality. RNA library preparation was performed by using the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (E7420L, Illumina). Paired-end sequencing was performed at Amo-gene Biotech Co. Ltd. Sequencing reads were aligned to reference genome (hg38) by using STAR. Cuff-diff was used to quantify the expression of GENCODE V36 annotated genes with the option -M (reads aligned to repetitive regions were masked) and -u (multiple aligned reads were corrected using the “rescue method”). Coding genes with FPKM (fragments per kilobase per million mapped reads) larger than 0.5, either in control or VSV-treated sample, were included in the analysis.
Immunoblotting analysis
Cells were lysed in lysis buffer [50 mM tris-HCl (pH 7.9), 150 mM NaCl, 100 mM EDTA, 1% Triton X-100, and complete protease inhibitor cocktail (Roche)] on ice for 30 min. Sonication was performed 10 times in 30% power (Sonics) on ice before centrifugation at 12,000 rpm for 10 min at 4°C. The resultant supernatant was subjected to SDS–polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting analysis following standard protocols. Antibodies used were listed as follows: anti-IRF3 (4302 S, 1:1000), anti-p-IRF3 (4947 S, 1:1000), anti-TBK1(3504 S, 1:1000), and anti–p-TBK1 (5483 S, 1:1000) antibodies were purchased from Cell Singling Technologies; anti-COXIV (11242-1-AP; 1:1000), anti-Calnexin (10427-2-AP; 1:1000), anti-MAVS (14341-1-AP; 1:1000), anti-STING (19851-1-AP; 1:1000), anti-HA (51064-2-AP; 1:1000), anti-GFP (50430-2-AP; 1:1000), anti-GST (10000-O-AP; 1:1000), and anti-Actin (66009–1-IG, 1:1000) antibodies were purchased from Proteintech; anti-Flag (ab1804; 1:1000) antibody was purchased from Abcam; anti-HSP90 (D120009-0010; 1:1000) antibody was purchased from Sangon Biotech; anti-YY1 (SC-7341, 1:1000) antibody was purchased from Santa Cruz Biotechnology. To detect MAVI1, Tricine–SDS-PAGE was used, which is the preferred electrophoretic system for the resolution of proteins smaller than 30 kDa (77). Tricine–SDS-PAGE gel includes upper layer gel [(1 ml of AB-3 buffer: 48 g of acrylamide, 1.5 g of bisacrylamide in 100 ml of H2O; 3 ml of gel buffer: 3.0 M tris-HCL and 0.3% SDS (pH 8.45); 12 ml of H2O, 90 μl of 10% Ammonium persulphate (APS), and 9 μl Tetramethylethylenediamine (TEMED), intermediate layer gel (6 ml of AB-3 buffer, 10 ml of gel buffer, 3 g of glycerol, 30 ml of H2O, 150 μl of 10% APS, and 15 μl of TEMED), and under layer gel (10 ml of AB-3 buffer, 10 ml of gel buffer, 10.8 g of urea, 30 ml of H2O, 100 μl of 10% APS, and 10 μl of TEMED) was applied to cathode buffer [1 M tris-HCl (pH 8.9)] and anode buffer [1 M tris, 1 M tricine, and 1% SDS (pH 8.25)]. Electrophoresis was stopped when bromophenol blue had passed 2/3 of the gel, and transfer with transfer buffer (3.03 g of tris, 14.42 g of glycine, 1 liter of H2O, and 20% methanol) was carried out at 100 V for 70 min.
Immunofluorescence analysis
Cells were washed with phosphate-buffered saline (PBS) buffer for three times and then fixed with 4% paraformaldehyde in PBS buffer for 15 min and then permeabilized with 0.1% Triton X-100 in PBS buffer on ice for 10 min. After rinsing with PBS buffer for three times, blocking solution (5% bovine serum albumin in PBS buffer) was applied for 30 min and the corresponding primary antibody (anti-Calnexin (Proteintech, 10427-2-AP; 1:200 dilution), anti-COXIV (Proteintech, 11242-1-AP; 1:100), anti- EEA1 (home-made; 1:200 dilution), anti-LAMP2 (Santa Cruz Biotechnology, sc-18822; 1:200 dilution), anti-RPL4 (Santa Cruz Biotechnology, sc-100838; 1:200 dilution), anti-Vimentin (Santa Cruz Biotechnology, sc-373717; 1:200 dilution), or anti-Golgi 97 (Santa Cruz Biotechnology, sc-59820; 1:200 dilution) was added in blocking buffer at 4°C overnight. After washing with PBS buffer for five times, cells were incubated with secondary antibodies conjugated with Cy3 fluorescent dyes (Beyotime, a0521; 1:200) for 1 hour at room temperature and washed with PBS buffer for three times. 4′,6-Diamidino-2-phenylindole (1 mg/ml; Sigma-Aldrich) was then added for 5 min. Cells were then washed with PBS buffer for three times and mounted in Fluoromount-G (Southern Biotech). Images were recorded on a ZEISS Exciter 5 microscope (ZEISS).
Subcellular fractionation, membrane interaction assay, and proteinase K digestion
Subcellular fractionation, membrane interaction assay, and proteinase K digestion were performed as previously described (78). Briefly, cells were homogenized by passing through a 25G 5/8-gauge syringe (BD PrecisionGlide) in hypotonic buffer [20 mM Hepes (pH 7.9), 1.5 mM MgCl2, and 60 mM KCl] supplemented with protease inhibitor cocktail (Roche). Samples were then subjected to centrifugation at 1000g to remove nuclei and unbroken cells. The supernatant was centrifuged at 10,000g for 15 min at 4°C, and sediment fractions were collected as crude heavy membrane fraction. Cytosolic fractions were collected after centrifuged at 25,000g for 1 hour at 4°C. Alternatively, the supernatant was added with 0.1 M Na2CO3, 0.1 M NaOH, or 1 M NaCl. After incubation for 30 min on ice, the samples were centrifuged at 25,000g for 1 hour to separate the soluble and pellet fractions. Samples were analyzed by SDS-PAGE followed by immunoblotting.
Proteinase K (0.5 mg/ml; Thermo Fisher Scientific) was added to the freshly prepared membrane fraction in the absence or presence of 1% Triton X-100 in Tris sodium chloride EDTA solution (TNES) buffer [50 mM tris-Cl (pH 7.4), 150 mM NaCl, 5 mM EDTA, and 250 mM sucrose] without protease inhibitors. The mixtures were incubated on ice for 1 hour before termination by the addition of phenylmethylsulfonyl fluoride to a final concentration of 1 mM.
Immunoprecipitation analysis
Cells were lysed in lysis buffer [50 mM tris-HCl (pH 7.9), 150 mM NaCl, 100 mM EDTA, 1% Triton X-100, and complete protease inhibitor cocktail (Roche)] on ice for 30 min and then centrifuged at 12,000 rpm for 10 min at 4°C. The resultant supernatant was incubated with primary antibodies (anti-Flag M2 affinity gel, Sigma-Aldrich, A2220; MAVS antibody, Santa Cruz Biotechnology, sc-166583; MAVI1 antibody, homemade) in lysis buffer at 4°C overnight. After washing five times with lysis buffer, resins were boiled in SDS sample buffer [1% SDS, 5% glycerol, 50 mM dithiothreitol (DTT), 30 mM tris-HCl (pH 6.8), and 0.25% bromophenol blue] for 5 min, resolved by SDS-PAGE and analyzed by immunoblotting analysis.
Isolation of ER, mitochondria, golgi apparatus, and mitochondria-associated membrane fractionation
The ER, mitochondria (Mito.), and Golgi apparatus were isolated with an ER isolation kit (Solarbio, EX2690), mitochondria isolation kit (Solarbio, SM0020), and Golgi apparatus isolation kit (Solarbio, EX1360-100T) according to the manufacturer’s protocol. Mitochondria-associated membrane was isolated following the protocol as reported previously (79).
Enzyme-linked immunosorbent assay
IFN-β and interleukin-6 were measured with enzyme-linked immunosorbent assay kits (Dakewe) according to the manufacturer’s protocol.
Purification of MAVI1-associated proteins
To purify proteins associated with MAVI1, the cytosolic and nuclear fractions of HEK293 cells stably expressing pBobi-MAVI1-Flag were first separated, and nuclear extracts were then prepared in a buffer containing 50 mM tris-HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, and 1% Triton X-100, followed by affinity purification using anti-Flag M2 Affinity gel (Sigma-Aldrich, A2220). Immunoprecipitates were then washed with low-salt buffer containing 50 mM tris-HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, and 1% Triton X-100 for three times followed by high-salt buffer containing 50 mM tris-HCl (pH 7.4), 420 mM NaCl, 1 mM EDTA, and 1% Triton X-100 for two times before elution with 3× Flag peptides (Sigma-Aldrich, F4799). Eluates were then subjected to in-solution digestion following the protocol as described previously (80). MS experiments were performed on a nanoscale Ultra High Performance Liquid Chromatography (UHPLC) system (Thermo Fisher Scientific, EASY-nLC1200) connected to an Orbitrap Fusion Lumos equipped with a nanoelectrospray source (Thermo Fisher Scientific). The peptides were separated on a Reverse Phase High Performance Liquid Chromatography (RP-HPLC) analytical column (75 μm by 25 cm) packed with 2-μm C18 beads (Thermo Fisher Scientific, 164941) using a linear gradient ranging from 9 to 28% Acetonitrile (ACN) in 90 min and followed by a linear increase to 45% B in 20 min at a flow rate of 300 nl/min. The Orbitrap Fusion Lumos acquired data in a data-dependent manner alternating between full-scan MS and MS2 scans. The MS spectra [350 to 1500 mass/charge ratio (m/z)] were collected with 120,000 resolution, Automatic Gain Control (AGC) of 4 × 105, and 50-ms maximal injection time. Selected ions were sequentially fragmented in a 3-s cycle by High Energy Collision Dissociation (HCD) with 30% normalized collision energy, specified isolated windows 1.6 m/z, and 15,000 resolution. AGC of 5 × 104 and 40-ms maximal injection time were used. Dynamic exclusion was set to 30 s. Raw data were processed using Proteome Discoverer (version 2.2), and tandem mass spectrometry spectra were searched against the reviewed Swiss-Prot human proteome database.
Polysome profiling
Cells were incubated with cycloheximide (CHX) (Sigma-Aldrich) for 15 min and washed with cold PBS buffer with cycloheximide (PBS/CHX) once. Cells were then collected in cold PBS/CHX and spun down before adding lysis buffer [20 mM tris-HCl (pH 7.4), 150 mM NaCl, 5 mM MgCl2, 1% Triton X-100, 1% deoxycholate, 2.5 mM DTT, RNasin (200 U/ml), CHX (100 μg/ml), EDTA-free protease inhibitor (Roche), and α1-antitrypsin (EMD Biosciences)]. Cell lysates were spun down at 7500 rpm for 5 min at 4°C. The precipitates were added to 15 to 50% sucrose gradient medium [CHX (100 μg/ml), heparin (0.2 mg/ml), and 1 mM DTT], followed by centrifugation at 36,000 rpm for 3 hours at 4°C. The polyribosomal fraction profile was analyzed by fractionation with a fractionated collector. Absorbance was measured with an UA-6 absorbance detector (254 nm). RNA was isolated from fractions and subjected to RT-qPCR analysis as described above.
In vitro pull-down assay
HEK293T cells were transfected with Flag-tagged MAVS, GST-tagged MAVI1, and GFP-tagged MAVI1 for 48 hours, and proteins were purified by using anti–Flag-M2 affinity gel (Sigma-Aldrich, A2220), glutathione agarose beads (Thermo Fisher Scientific, 16102BID), and GFP magnetic beads (ABclonal, AE079), respectively, following the manufacturer’s protocols.
Purified, GST- or GFP-MAVI1 proteins immobilized on glutathione agarose beads or GFP magnetic beads were incubated with Flag- or GST-tagged MAVS proteins eluted with 3× Flag peptide (Sigma-Aldrich) or reduced glutathione at 4°C for overnight in reaction buffer [40 mM Hepes (pH 7.4), 2 mM EGTA, 2.5 mM MgCl2, and 0.3% CHAPS] and then washed with the same buffer for five times. Beads were boiled for 10 min in 1× SDS sample buffer and resolved by SDS-PAGE gel before immunoblotting analysis.
Plaque assay
BHK21 cells were plated in six-well plates (2 to 4 × 105 per well). The viruses were subjected to gradient dilution in DMEM culture medium with 2% FBS gradient on ice. Agra (1.5%) was mixed with DMEM with 2% FBS and kept at 42°C. Cells were washed with DMEM with 2% FBS once. Diluted viruses (500 μl) were then added and incubated at 37°C with 5% CO2 for 90 min. Viruses were then removed, and agarose/DMEM solution (3 ml) was added to each well and incubated at 37°C with 5% CO2 for 48 hours. Formaldehyde diluted with PBS (10%; 2 ml) was added into each well and incubated at room temperature for 30 min. Fixative was then removed, and the fixed cells were washed with PBS. Crystal violet solution (0.1%; 2 ml) was added to each well and incubated for 30 min. The staining solution was removed, and cells were thoroughly washed with running water, dry completely before counting the plaque, and determine the virus titer. Titer (plaque-forming units/milliliter) = number of plaques × dilution ratio.
MAVI1-Flag knock-in cells
sgRNA (targeting sequence: 5′-TGGACAGATGTGACTTTGAA-3′) targeting the MAVI1 genomic location was designed via http://crispr.genome-engineering.org/ and then cloned into the pSpCas9 (BB)-2A-puro-PX459 V2.0 vector. Donor DNA was amplified from genomic DNA using primer set as follows: forward primer: 5′-GCTCTAGAATGACCTCAGTTTCAACACAGTTG-3′; reverse primer: 5′-CGCGGATCCTTACTTATCGTCGTCATCCTTGTAATCCATCTGTCCATTTCTTTTTCGTGCA-3′. HEK293T cells were transfected with sgRNA expression vector together with the donor DNA for 48 hours before puromycin (1 μg/ml) was added. Single colonies were expanded for genomic DNA extraction and PCR amplification (forward primer: 5′-GGCTGGCCTGTTATTTGTCA-3′; reverse primer: 5′-CGCAAAGGTTTTCAGGGTGA-3′), followed by Sanger sequencing. Immunoblotting analysis was used to confirm the success knock-in of the Flag tag.
Mavi1-knockout (Mavi1−/−) mice
Mavi1-knockout mice (Mavi1−/−, C57BL/6) were generated by CRISPR-Cas9 genome editing technology using sgRNA (target sequence: 5′-GATCTTAGTCCTCGCTTCGCTGCTTTTGATCCT-3′) targeting mouse Mavi1. The deletion of Mavi1 was confirmed by PCR (forward primer: 5′-CTGGTCAGGGTGCAGTCTAA-3′; reverse primer: 5′-AAGTTCAGCCCTCCAGATCC-3′) and Sanger sequencing.
Virus infection in mice
C57BL/6 mice (8-week-old female), both WT and Mavi1−/−, were intraperitoneally injected with VSV [5 × 108 plaque forming unit (p.f.u.) per mouse] for 12 hours before serum collection. Spleen, liver, and lung tissues were collected for RT-qPCR analysis. Lung tissues were fixed in 10% phosphate-buffered formalin, embedded into paraffin, sectioned, stained with hematoxylin and eosin solution, and examined by light microscopy for histological changes. All animal experiments were conducted in accordance with a protocol approved by the Animal Care and Use Committee of Xiamen University. All animals were maintained in an animal room with 12-hour light/12-hour dark cycles and cared with free access to standard rodent chow and water in accordance with institutional guidelines.
Acknowledgments
We thank J.-h. Han for providing VSV and VSV-GFP. We thank Xiamen University Laboratory Animal Center for providing technical assistance for generating MAVI1-deficient mice.
Funding: This work was supported by National Natural Science Foundation of China (82125028, U22A20320, 31871319, 91953114, 81761128015, and 81861130370), National Key Research and Development Program of China (2020YFA0803600 and 2020YFA0112300), Natural Science Foundation of Fujian Province of China (2020J02004), and the Fundamental Research Funds for the Central University (20720190145 and 20720220003) to W.L. This work was also supported by National Natural Science Foundation of China (82203475) and China Postdoctoral Science Foundation (2022 M710705) to W.-j.L.
Author contributions: Conceptualization: W.L. Experimental design: W.-j.L., Y.Hu., and T.S. Performed experiments: Y.Hu., T.S., Y.L., W.-j.L., X.D., Y.S., and Q.Y. Methodology: Y.He, J.D., and T.R. Writing—original draft: W.L. and Y.Hu. Writing—review and editing: W.L. and Y.Hu.
Competing interests: The authors declare they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. RNA-seq data were deposited in the Gene Expression Omnibus database under accession GSE221218. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository with the dataset identifier PXD039375.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Legends for tables S1 to S3
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S3
REFERENCES AND NOTES
- 1.Liu X., Zhu C., Zha H., Tang J., Rong F., Chen X., Fan S., Xu C., du J., Zhu J., Wang J., Ouyang G., Yu G., Cai X., Chen Z., Xiao W., SIRT5 impairs aggregation and activation of the signaling adaptor MAVS through catalyzing lysine desuccinylation. EMBO J. 39, e103285 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cai C., Tang Y. D., Xu G., Zheng C., The crosstalk between viral RNA- and DNA-sensing mechanisms. Cell. Mol. Life Sci. 78, 7427–7434 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brubaker S. W., Bonham K. S., Zanoni I., Kagan J. C., Innate immune pattern recognition: A cell biological perspective. Annu. Rev. Immunol. 33, 257–290 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu G., Gack M. U., Distinct and orchestrated functions of RNA sensors in innate immunity. Immunity 53, 26–42 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tan X., Sun L., Chen J., Chen Z. J., Detection of microbial infections through innate immune sensing of nucleic acids. Annu. Rev. Microbiol. 72, 447–478 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Alexopoulou L., Holt A. C., Medzhitov R., Flavell R. A., Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 413, 732–738 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Heil F., Hemmi H., Hochrein H., Ampenberger F., Kirschning C., Akira S., Lipford G., Wagner H., Bauer S., Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science 303, 1526–1529 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Tanji H., Ohto U., Shibata T., Taoka M., Yamauchi Y., Isobe T., Miyake K., Shimizu T., Toll-like receptor 8 senses degradation products of single-stranded RNA. Nat. Struct. Mol. Biol. 22, 109–115 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Yoneyama M., Kikuchi M., Matsumoto K., Imaizumi T., Miyagishi M., Taira K., Foy E., Loo Y. M., Gale M. Jr., Akira S., Yonehara S., Kato A., Fujita T., Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 175, 2851–2858 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Goubau D., Schlee M., Deddouche S., Pruijssers A. J., Zillinger T., Goldeck M., Schuberth C., van der Veen A. G., Fujimura T., Rehwinkel J., Iskarpatyoti J. A., Barchet W., Ludwig J., Dermody T. S., Hartmann G., Reis e Sousa C., Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5′-diphosphates. Nature 514, 372–375 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y. G., Kim M. V., Chen X., Batista P. J., Aoyama S., Wilusz J. E., Iwasaki A., Chang H. Y., Sensing self and foreign circular RNAs by intron identity. Mol. Cell 67, 228–238.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Junior A. G. D., Sampaio N. G., Rehwinkel J., A balancing act: MDA5 in antiviral immunity and autoinflammation. Trends Microbiol. 27, 75–85 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rehwinkel J., Gack M. U., RIG-I-like receptors: Their regulation and roles in RNA sensing. Nat. Rev. Immunol. 20, 537–551 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peisley A., Wu B., Xu H., Chen Z. J., Hur S., Structural basis for ubiquitin-mediated antiviral signal activation by RIG-I. Nature 509, 110–114 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meylan E., Curran J., Hofmann K., Moradpour D., Binder M., Bartenschlager R., Tschopp J., Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437, 1167–1172 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Seth R. B., Sun L., Ea C. K., Chen Z. J., Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122, 669–682 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Xu L. G., Wang Y. Y., Han K. J., Li L. Y., Zhai Z., Shu H. B., VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell 19, 727–740 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Kawai T., Takahashi K., Sato S., Coban C., Kumar H., Kato H., Ishii K. J., Takeuchi O., Akira S., IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6, 981–988 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Chow K. T., Gale M. Jr., Loo Y. M., RIG-I and other RNA sensors in antiviral immunity. Annu. Rev. Immunol. 36, 667–694 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Hou F., Sun L., Zheng H., Skaug B., Jiang Q. X., Chen Z. J., MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell 146, 448–461 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Honda K., Takaoka A., Taniguchi T., Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity 25, 349–360 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Ivashkiv L. B., Donlin L. T., Regulation of type I interferon responses. Nat. Rev. Immunol. 14, 36–49 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamura T., Yanai H., Savitsky D., Taniguchi T., The IRF family transcription factors in immunity and oncogenesis. Annu. Rev. Immunol. 26, 535–584 (2008). [DOI] [PubMed] [Google Scholar]
- 24.MacMicking J. D., Interferon-inducible effector mechanisms in cell-autonomous immunity. Nat. Rev. Immunol. 12, 367–382 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schoggins J. W., Wilson S. J., Panis M., Murphy M. Y., Jones C. T., Bieniasz P., Rice C. M., Erratum: Corrigendum: A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 525, 144 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Stark G. R., Darnell J. E. Jr., The JAK-STAT pathway at twenty. Immunity 36, 503–514 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang P., Yang L., Cheng G., Yang G., Xu Z., You F., Sun Q., Lin R., Fikrig E., Sutton R. E., UBXN1 interferes with Rig-I-like receptor-mediated antiviral immune response by targeting MAVS. Cell Rep. 3, 1057–1070 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loo Y. M., Gale M. Jr., Immune signaling by RIG-I-like receptors. Immunity 34, 680–692 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen W., Han C., Xie B., Hu X., Yu Q., Shi L., Wang Q., Li D., Wang J., Zheng P., Liu Y., Cao X., Induction of Siglec-G by RNA viruses inhibits the innate immune response by promoting RIG-I degradation. Cell 152, 467–478 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Feng H., Lenarcic E. M., Yamane D., Wauthier E., Mo J., Guo H., McGivern D. R., González-López O., Misumi I., Reid L. M., Whitmire J. K., Ting J. P. Y., Duncan J. A., Moorman N. J., Lemon S. M., NLRX1 promotes immediate IRF1-directed antiviral responses by limiting dsRNA-activated translational inhibition mediated by PKR. Nat. Immunol. 18, 1299–1309 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin S., Tian S., Luo M., Xie W., Liu T., Duan T., Wu Y., Cui J., Tetherin suppresses type I interferon signaling by targeting MAVS for NDP52-mediated selective autophagic degradation in human cells. Mol. Cell 68, 308–322.e4 (2017). [DOI] [PubMed] [Google Scholar]
- 32.International Human Genome Sequencing Consortium , Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Leong A. Z.-X., Lee P. Y., Mohtar M. A., Syafruddin S. E., Pung Y.-F., Low T. Y., Short open reading frames (sORFs) and microproteins: An update on their identification and validation measures. J. Biomed. Sci. 29, 19 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gates A. J., Gysi D. M., Kellis M., Barabasi A. L., A wealth of discovery built on the Human Genome Project - by the numbers. Nature 590, 212–215 (2021). [DOI] [PubMed] [Google Scholar]
- 35.Ingolia N. T., Brar G. A., Stern-Ginossar N., Harris M. S., Talhouarne G. J. S., Jackson S. E., Wills M. R., Weissman J. S., Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep. 8, 1365–1379 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu S., Zhang J., Lian X., Sun L., Meng K., Chen Y., Sun Z., Yin X., Li Y., Zhao J., Wang T., Zhang G., He Q. Y., A hidden human proteome encoded by 'non-coding' genes. Nucleic Acids Res. 47, 8111–8125 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kondo T., Plaza S., Zanet J., Benrabah E., Valenti P., Hashimoto Y., Kobayashi S., Payre F., Kageyama Y., Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis. Science 329, 336–339 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Chugunova A., Loseva E., Mazin P., Mitina A., Navalayeu T., Bilan D., Vishnyakova P., Marey M., Golovina A., Serebryakova M., Pletnev P., Rubtsova M., Mair W., Vanyushkina A., Khaitovich P., Belousov V., Vysokikh M., Sergiev P., Dontsova O., LINC00116 codes for a mitochondrial peptide linking respiration and lipid metabolism. Proc. Natl. Acad. Sci. U.S.A. 116, 4940–4945 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nelson B. R., Makarewich C. A., Anderson D. M., Winders B. R., Troupes C. D., Wu F., Reese A. L., McAnally J. R., Chen X., Kavalali E. T., Cannon S. C., Houser S. R., Bassel-Duby R., Olson E. N., A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science 351, 271–275 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.D'Lima N. G., Ma J., Winkler L., Chu Q., Loh K. H., Corpuz E. O., Budnik B. A., Lykke-Andersen J., Saghatelian A., Slavoff S. A., A human microprotein that interacts with the mRNA decapping complex. Nat. Chem. Biol. 13, 174–180 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chu Q., Martinez T. F., Novak S. W., Donaldson C. J., Tan D., Vaughan J. M., Chang T., Diedrich J. K., Andrade L., Kim A., Zhang T., Manor U., Saghatelian A., Regulation of the ER stress response by a mitochondrial microprotein. Nat. Commun. 10, 4883 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo B., Zhai D., Cabezas E., Welsh K., Nouraini S., Satterthwait A. C., Reed J. C., Humanin peptide suppresses apoptosis by interfering with Bax activation. Nature 423, 456–461 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Meng N., Chen M., Chen D., Chen X. H., Wang J. Z., Zhu S., He Y. T., Zhang X. L., Lu R. X., Yan G. R., Small protein hidden in lncRNA LOC90024 promotes "Cancerous" RNA splicing and tumorigenesis. Adv. Sci. 7, 1903233 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang P., He D., Xu Y., Hou J., Pan B. F., Wang Y., Liu T., Davis C. M., Ehli E. A., Tan L., Zhou F., Hu J., Yu Y., Chen X., Nguyen T. M., Rosen J. M., Hawke D. H., Ji Z., Chen Y., Genome-wide identification and differential analysis of translational initiation. Nat. Commun. 8, 1749 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pang Y., Liu Z., Han H., Wang B., Li W., Mao C., Liu S., Peptide SMIM30 promotes HCC development by inducing SRC/YES1 membrane anchoring and MAPK pathway activation. J. Hepatol. 73, 1155–1169 (2020). [DOI] [PubMed] [Google Scholar]
- 46.Yang J.-E., Zhong W.-J., Li J.-F., Lin Y.-Y., Liu F.-T., Tian H., Chen Y.-J., Luo X.-Y., Zhuang S.-M., LINC00998-encoded micropeptide SMIM30 promotes the G1/S transition of cell cycle by regulating cytosolic calcium level. Mol. Oncol. 17, 901–916 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cai H., Yu Y., Ni X., Li C., Hu Y., Wang J., Chen F., Xi S., Chen Z., LncRNA LINC00998 inhibits the malignant glioma phenotype via the CBX3-mediated c-Met/Akt/mTOR axis. Cell Death Dis. 11, 1032 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fang X., Pan X., Mai H., Yuan X., Liu S., Wen F., LINC00998 functions as a novel tumor suppressor in acute myeloid leukemia via regulating the ZFP36 ring finger protein/mammalian target of rapamycin complex 2 axis. Bioengineered 12, 10363–10372 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blobel G., Dobberstein B., Transfer of proteins across membranes. II. Reconstitution of functional rough microsomes from heterologous components. J. Cell Biol. 67, 852–862 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mellacheruvu D., Wright Z., Couzens A. L., Lambert J. P., St-Denis N. A., Li T., Miteva Y. V., Hauri S., Sardiu M. E., Low T. Y., Halim V. A., Bagshaw R. D., Hubner N. C., al-Hakim A., Bouchard A., Faubert D., Fermin D., Dunham W. H., Goudreault M., Lin Z. Y., Badillo B. G., Pawson T., Durocher D., Coulombe B., Aebersold R., Superti-Furga G., Colinge J., Heck A. J. R., Choi H., Gstaiger M., Mohammed S., Cristea I. M., Bennett K. L., Washburn M. P., Raught B., Ewing R. M., Gingras A. C., Nesvizhskii A. I., The CRAPome: A contaminant repository for affinity purification-mass spectrometry data. Nat. Methods 10, 730–736 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun Q., Sun L., Liu H. H., Chen X., Seth R. B., Forman J., Chen Z. J., The specific and essential role of MAVS in antiviral innate immune responses. Immunity 24, 633–642 (2006). [DOI] [PubMed] [Google Scholar]
- 52.Hayashi T., Rizzuto R., Hajnoczky G., Su T. P., MAM: More than just a housekeeper. Trends Cell Biol. 19, 81–88 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Szabadkai G., Bianchi K., Várnai P., De Stefani D., Wieckowski M. R., Cavagna D., Nagy A. I., Balla T., Rizzuto R., Chaperone-mediated coupling of endoplasmic reticulum and mitochondrial Ca2+ channels. J. Cell Biol. 175, 901–911 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bionda C., Portoukalian J., Schmitt D., Rodriguez-Lafrasse C., Ardail D., Subcellular compartmentalization of ceramide metabolism: MAM (mitochondria-associated membrane) and/or mitochondria? Biochem. J. 382, 527–533 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Giorgi C., Ito K., Lin H. K., Santangelo C., Wieckowski M. R., Lebiedzinska M., Bononi A., Bonora M., Duszynski J., Bernardi R., Rizzuto R., Tacchetti C., Pinton P., Pandolfi P. P., PML regulates apoptosis at endoplasmic reticulum by modulating calcium release. Science 330, 1247–1251 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hamasaki M., Furuta N., Matsuda A., Nezu A., Yamamoto A., Fujita N., Oomori H., Noda T., Haraguchi T., Hiraoka Y., Amano A., Yoshimori T., Autophagosomes form at ER-mitochondria contact sites. Nature 495, 389–393 (2013). [DOI] [PubMed] [Google Scholar]
- 57.Thoudam T., Jeon J. H., Ha C. M., Lee I. K., Role of mitochondria-associated endoplasmic reticulum membrane in inflammation-mediated metabolic diseases. Mediators Inflamm. 2016, 1851420 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Crappe J., Ndah E., Koch A., Steyaert S., Gawron D., De Keulenaer S., De Meester E., De Meyer T., Van Criekinge W., Van Damme P., Menschaert G., PROTEOFORMER: Deep proteome coverage through ribosome profiling and MS integration. Nucleic Acids Res. 43, e29 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Olexiouk V., Crappé J., Verbruggen S., Verhegen K., Martens L., Menschaert G., sORFs.org: A repository of small ORFs identified by ribosome profiling. Nucleic Acids Res. 44, D324–D329 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rodriguez-Morales A. J., Bonilla-Aldana D. K., Balbin-Ramon G. J., Rabaan A. A., Sah R., Paniz-Mondolfi A., Pagliano P., Esposito S., History is repeating itself: Probable zoonotic spillover as the cause of the 2019 novel Coronavirus Epidemic. Infez. Med. 28, 3–5 (2020). [PubMed] [Google Scholar]
- 61.Baharoon S., Memish Z. A., MERS-CoV as an emerging respiratory illness: A review of prevention methods. Travel Med. Infect. Dis. 32, 101520, 101520 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rodriguez-Morales A. J., Ramírez-Jaramillo V., Patiño-Barbosa A. M., Bedoya-Arias H. A., Henao-San Martin V., Murillo-García D. R., Cardona-Ospina J. A., Lagos-Grisales G. J., Severe fever with thrombocytopenia syndrome - A bibliometric analysis of an emerging priority disease. Travel Med. Infect. Dis. 23, 97–98 (2018). [DOI] [PubMed] [Google Scholar]
- 63.Ahmad T., Khan M., Haroon, Musa T. H., Nasir S., Hui J., Bonilla-Aldana D. K., Rodriguez-Morales A. J., COVID-19: Zoonotic aspects. Travel Med. Infect. Dis. 36, 101607 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Aspden J. L., Eyre-Walker Y. C., Phillips R. J., Amin U., Mumtaz M. A. S., Brocard M., Couso J. P., Extensive translation of small Open Reading Frames revealed by Poly-Ribo-Seq. eLife 3, e03528 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Anderson D. M., Anderson K. M., Chang C. L., Makarewich C. A., Nelson B. R., McAnally J. R., Kasaragod P., Shelton J. M., Liou J., Bassel-Duby R., Olson E. N., A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 160, 595–606 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moore C. B., Bergstralh D. T., Duncan J. A., Lei Y., Morrison T. E., Zimmermann A. G., Accavitti-Loper M. A., Madden V. J., Sun L., Ye Z., Lich J. D., Heise M. T., Chen Z., Ting J. P. Y., NLRX1 is a regulator of mitochondrial antiviral immunity. Nature 451, 573–577 (2008). [DOI] [PubMed] [Google Scholar]
- 67.Zhang W., Wang G., Xu Z. G., Tu H., Hu F., Dai J., Chang Y., Chen Y., Lu Y., Zeng H., Cai Z., Han F., Xu C., Jin G., Sun L., Pan B. S., Lai S. W., Hsu C. C., Xu J., Chen Z. Z., Li H. Y., Seth P., Hu J., Zhang X., Li H., Lin H. K., Lactate is a natural suppressor of RLR signaling by targeting MAVS. Cell 178, 176–189.e15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Parimoo S., Rao N., Padmanaban G., Cytochrome c oxidase is preferentially synthesized in the rough endoplasmic reticulum-mitochondrion complex in rat liver. Biochem. J. 208, 505–507 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Csordás G., Renken C., Várnai P., Walter L., Weaver D., Buttle K. F., Balla T., Mannella C. A., Hajnóczky G., Structural and functional features and significance of the physical linkage between ER and mitochondria. J. Cell Biol. 174, 915–921 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dolman N. J., Gerasimenko J. V., Gerasimenko O. V., Voronina S. G., Petersen O. H., Tepikin A. V., Stable Golgi-mitochondria complexes and formation of Golgi Ca(2+) gradients in pancreatic acinar cells. J. Biol. Chem. 280, 15794–15799 (2005). [DOI] [PubMed] [Google Scholar]
- 71.Voelker D. R., Bridging gaps in phospholipid transport. Trends Biochem. Sci. 30, 396–404 (2005). [DOI] [PubMed] [Google Scholar]
- 72.Vance J. E., Phospholipid synthesis in a membrane fraction associated with mitochondria. J. Biol. Chem. 265, 7248–7256 (1990). [PubMed] [Google Scholar]
- 73.Li Z., Agellon L. B., Allen T. M., Umeda M., Jewell L., Mason A., Vance D. E., The ratio of phosphatidylcholine to phosphatidylethanolamine influences membrane integrity and steatohepatitis. Cell Metab. 3, 321–331 (2006). [DOI] [PubMed] [Google Scholar]
- 74.Lewis S. C., Uchiyama L. F., Nunnari J., ER-mitochondria contacts couple mtDNA synthesis with mitochondrial division in human cells. Science 353, aaf5549 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen P. S., Hsu H. P., Phan N. N., Yen M. C., Chen F. W., Liu Y. W., Lin F. P., Feng S. Y., Cheng T. L., Yeh P. H., Omar H. A., Sun Z., Jiang J. Z., Chan Y. S., Lai M. D., Wang C. Y., Hung J. H., CCDC167 as a potential therapeutic target and regulator of cell cycle-related networks in breast cancer. Aging 13, 4157–4181 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jia Y., Li H., Liu G., Song F., SMAGP a novel biomarker of cervical cancer development and progression. Onco. Targets. Ther. 11, 6925–6935 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schagger H., Tricine-SDS-PAGE. Nat. Protoc. 1, 16–22 (2006). [DOI] [PubMed] [Google Scholar]
- 78.Matsumoto A., Pasut A., Matsumoto M., Yamashita R., Fung J., Monteleone E., Saghatelian A., Nakayama K. I., Clohessy J. G., Pandolfi P. P., mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature 541, 228–232 (2017). [DOI] [PubMed] [Google Scholar]
- 79.Williamson C. D., Wong D. S., Bozidis P., Zhang A., Colberg-Poley A. M., Isolation of endoplasmic reticulum, mitochondria, and mitochondria-associated membrane and detergent resistant membrane fractions from transfected cells and from human cytomegalovirus-infected primary fibroblasts. Curr. Protoc. Cell Biol. 68, 3.27.1–3.27.33 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gao W.-W., Xiao R.-Q., Zhang W.-J., Hu Y.-R., Peng B.-L., Li W.-J., He Y.-H., Shen H.-F., Ding J.-C., Huang Q.-X., Ye T.-Y., Li Y., Liu Z.-Y., Ding R., Rosenfeld M. G., Liu W., JMJD6 licenses ERα-Dependent enhancer and coding gene activation by modulating the recruitment of the CARM1/MED12 co-activator complex. Mol. Cell 70, 340–357.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Legends for tables S1 to S3
Tables S1 to S3