Abstract
The las quorum-sensing system of Pseudomonas aeruginosa controls the expression of elastase and rhamnolipid. We report that starvation can select a mutant producing these virulence factors in spite of a lasR deletion. Expression of the autoinducer synthase gene rhlI was increased in this suppressor mutant, suggesting compensation by the rhl system. These data show that P. aeruginosa can restore elastase and rhamnolipid production in the absence of a functional las quorum-sensing system.
Pseudomonas aeruginosa is an opportunistic human pathogen responsible for extensive lung damage in cystic fibrosis patients (8) as well as life-threatening nosocomial pneumonia in intubated (3) and neutropenic cancer patients (1). The pathogenesis of P. aeruginosa infections has been linked to the production of numerous extracellular virulence factors, of which several have been shown to be regulated by quorum sensing (17). A quorum-sensing system is composed of a LuxR-type transcriptional activator protein and a small, presumably diffusible molecule called an autoinducer (AI) that is synthesized by a LuxI-type AI synthase (5). With increasing cell density the concentration of AI reaches a threshold level, at which point it binds to the transcriptional activator. The transcriptional activator-AI complex then induces specific genes in a cell density-dependent manner. In P. aeruginosa two quorum-sensing systems, las (LasR–PAI-1) (16, 19) and rhl (RhlR–PAI-2) (14, 20), have been described. Together they allow the coordinate, cell density-dependent expression of many virulence factors, including elastase (encoded by lasB) (6, 13), LasA protease (lasA) (26), alkaline protease (aprA) (7, 13), exotoxin A (toxA) (7), and rhamnolipid (rhlAB) (2, 14, 21). The las system controls the expression of the rhl system at both transcriptional and posttranslational levels in a hierarchical cascade (12, 22). Mutants with a nonfunctional las system, such as the defined lasR mutant PAO-R1 (Table 1), in which 82% of the lasR open reading frame has been deleted, are unable to produce several virulence factors, including the major protease elastase (6). This strain has also been shown to have substantially reduced virulence in a neonatal mouse pneumonia model (25). Therefore, it has been suggested that the las quorum-sensing system might be a possible target for new therapeutic interventions against P. aeruginosa (18, 22, 27). However, one concern with this approach is the possibility for P. aeruginosa to mutate and restore the production of quorum-sensing controlled virulence factors, such as elastase, in the absence of a functional las quorum-sensing system.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or phenotype | Reference |
|---|---|---|
| P. aeruginosa | ||
| PAO1 | Wild-type, elastolytic prototroph | 9 |
| PAO-R1 | lasR::Tcr; nonelastolytic derivative of strain PAO1 | 6 |
| PR1-E4 | lasR::Tcr; elastolytic mutant of strain PAO-R1 | This study |
| PG201 | Wild-type prototroph | 14 |
| PAO-JP2 | lasI rhlI double mutant derived from PAO1; Hgr Tcr | 21 |
| PAO-JP3 | lasR rhlR double mutant derived from PAO1; Hgr Tcr | 21 |
| Plasmids | ||
| pSW205 | Translational lacZ fusion vector that contains an origin of replication for both E. coli and P. aeruginosa; Apr | 7 |
| pTS400 | pSW205 containing a lasB′-lacZ translational fusion | 16 |
| pECP60 | pSW205 containing an rhlA′-lacZ translational fusion | 22 |
| pLP170 | Transcriptional lacZ fusion vector that contains an origin of replication for both E. coli and P. aeruginosa; Apr | 23 |
| pPCS1002 | pLP170 containing a rhlR′-lacZ transcriptional fusion | 22 |
| pLPRI | rhlI′-lacZ transcriptional fusion; a 904-bp NcoI fragment of strain PG201 chromosomal DNA containing 723 bp upstream of the rhlI translational start site was ligated into the SmaI site of pLP170 | This study |
| pPCS223 | lasI′-lacZ transcriptional fusion; a 407-bp fragment containing 288 bp upstream of the lasI translational start site, with PCR-engineered HindIII and BamHI restriction sites, was ligated into the HindIII-BamHI sites of pLP170 | This study |
In order to address this question, we incubated the non-elastase-producing lasR mutant PAO-R1 in a medium (5 mM KH2PO4, 0.4 mM MgCl2, and 3% casein, pH 7.35) that contains casein as the sole carbon and nitrogen source. In this medium, bacteria unable to produce casein-degrading proteases, such as elastase, starve. Incubation of strain PAO-R1 in the casein medium therefore results in a starvation selection of mutations that would restore protease production. After 10 days of incubation (37°C with shaking), growth became apparent, and isolated colonies were tested for exoproduct secretion. Screening of different clones was performed for proteolytic activity on skim milk plates (50% commercial skim milk incubated for 18 h at 37°C), for elastolytic activity on elastin agar plates (15), and for rhamnolipid production on SW blue plates (14, 24). Several protease-positive but elastase- and rhamnolipid-negative mutants were found. These mutants might have been affected in the expression of LasA protease and/or alkaline protease or another, yet-unknown protease. These other mutants were not further characterized, as we were most interested in mutations affecting the quorum-sensing-controlled protease LasB elastase. Mutant PR1-E4 was selected as the clone with the greatest proteolytic and elastolytic activities and rhamnolipid production. This experiment confirmed that P. aeruginosa can mutate and restore elastase and rhamnolipid production despite a nonfunctional las quorum-sensing system. The same strategy was used in an attempt to isolate elastolytic mutants derived from the double mutants PAO-JP2 (lasI rhlI) (21) and PAO-JP3 (lasR rhlR) (21). We were unable to isolate suppressor mutants of these two strains that were able to grow in casein medium even after incubation times of up to 4 weeks. This result suggested that the restoration of virulence factor production in the absence of the las quorum-sensing system requires an intact rhl system. We were unable to use this approach with a rhlR, lasI, or rhlI mutant, since such mutants still produce enough elastase to grow in the casein medium (21).
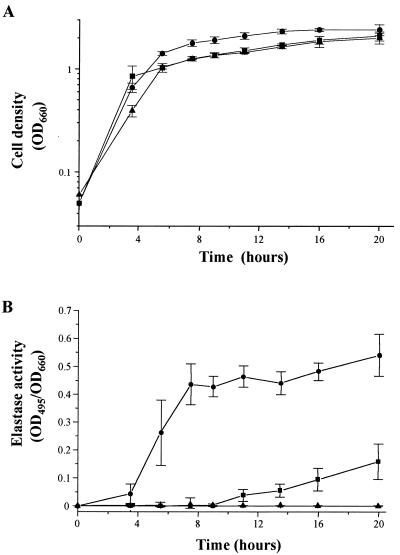
To further characterize the phenotype of mutant PR1-E4, we quantified elastase and rhamnolipid production. Elastin-Congo red assays were performed throughout the growth cycle as described previously (21). Growth rates of strains PAO-R1 and PR1-E4 were identical (Fig. 1A). Slightly higher cell densities were observed for the wild-type strain, PAO1 (Fig. 1A). Therefore, elastase activity was corrected for cell density by dividing by the optical density at 600 nm (OD660). The elastolytic activity of mutant PR1-E4 started to increase linearly in late stationary phase and reached 30% of the production of the wild-type strain, PAO1, at 20 h (Fig. 1B). These kinetics are atypical for a cell density-controlled production and contrast with the exponential increase of elastolytic activity found for the wild-type strain, PAO1, at the end of logarithmic growth (Fig. 1B). As expected, strain PAO-R1 was nonelastolytic throughout the growth cycle (Fig. 1B). Quantification of rhamnolipid production by strains PAO1, PAO-R1, and PR1-E4 was performed by orcinol assays on stationary-phase culture supernatants (OD660 = 3.16 ± 0.4) as previously described (21). Strain PAO-R1 did not produce detectable rhamnolipid in the orcinol assay, confirming results obtained with strain PAO-R1 on SW blue agar plates. Mutant PR1-E4 produced 58% (1.4 mg/ml) of the rhamnolipid level detected in culture supernatants of the wild-type strain, PAO1 (2.4 mg/ml). Therefore, the mutation characterizing strain PR1-E4 substantially restored the production of both elastase and rhamnolipid.
FIG. 1.
Elastin-Congo red assay time course in strains PAO1 (circles), PAO-R1 (triangles), and PR1-E4 (squares). The strains were grown at 37°C with shaking in PTSB medium (starting OD660 = 0.05). Culture filtrate samples were taken at regular intervals, and elastin-Congo red assays were performed as previously described (21). (A) Growth curves; (B) elastin-Congo red assay results. Shown is the average ± standard deviation of three independent experiments performed in triplicate. Elastase activity, measured by OD495, was normalized for bacterial cell density (OD660) in order to express the elastase activity per cell unit.
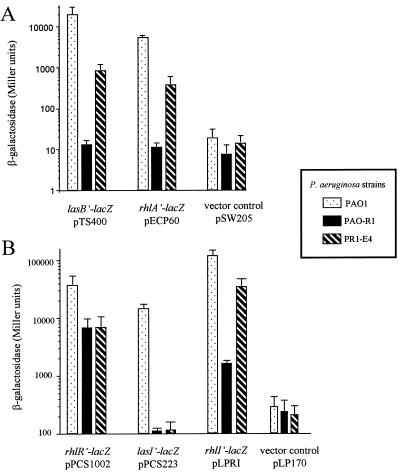
To address whether the elastolytic activity and rhamnolipid production of mutant PR1-E4 was due to a restoration of lasB and rhlA transcription, we determined the expression of lasB and rhlA by using the pTS400 and pECP60 lacZ reporter fusions, respectively. P. aeruginosa cultures were grown in PTSB medium to stationary phase (OD660 = 1.8 ± 0.2), washed twice, and resuspended in A medium prior to duplicate β-galactosidase activity determinations as described previously (22). The expression of both lasB and rhlA was substantially increased in mutant PR1-E4 compared to the background levels detected in the parent strain, PAO-R1 (Fig. 2A). These results confirmed that elastase and rhamnolipid production by mutant PR1-E4 is due to a partial restoration of lasB and rhlA expression. To determine whether the expression of other genes belonging to the las and rhl quorum-sensing systems have been affected by the mutation in strain PR1-E4, we also examined the expression of the transcriptional activator gene rhlR and of the two AI synthase genes, lasI and rhlI, using the pPCS1002, pPCS223, and pLPRI lacZ reporter fusion plasmids, respectively. In both strain PAO-R1 and its derivative mutant PR1-E4, the expression of rhlR was similarly low (18% of the levels detected in the wild-type strain, PAO1), suggesting that the mutation in strain PR1-E4 had not affected the expression of rhlR (Fig. 2B). Both the lasI and rhlI genes were highly expressed in strain PAO1. No lasI transcription could be detected in the absence of LasR in either strain PAO-R1 or mutant PR1-E4. In contrast to lasI, the expression of rhlI differed considerably between strain PAO-R1 and mutant PR1-E4. Indeed, the expression of rhlI, dramatically reduced in strain PAO-R1, was substantially increased in mutant PR1-E4 and reached 30% of the levels observed in strain PAO1 (Fig. 2B). These results suggest that the mutation characterizing strain PR1-E4 significantly affected the transcription of the rhl AI synthase gene.
FIG. 2.
Expression of las- and rhl-controlled genes in strains PAO1, PAO-R1, and PR1-E4. Duplicate β-galactosidase activity determinations were performed as described previously on P. aeruginosa stationary-phase cultures (22). The data are expressed in Miller units as the average of four independent experiments ± standard deviation. (A) Expression of lasB (pTS400) and rhlA (pECP60) translational lacZ fusions. (B) Expression of rhlR (pPCS1002), lasI (pPCS223), and rhlI (pLPRI) transcriptional lacZ fusions.
It is striking that the rhl system controls to some degree the three genes, lasB (2, 14, 21), rhlA (14, 21), and rhlI (12), that show an increased expression in mutant PR1-E4. Moreover, our inability to isolate mutants able to grow in casein medium from strains with deletions in both the las and rhl quorum-sensing systems suggests that an intact rhl system is required for the restoration of proteases such as elastase in the absence of LasR. The expression of lasB in mutant PAO-R1 increased from 15 to 700 Miller units in the presence of 10 μM PAI-2. These levels are similar to those measured in mutant PR1-E4 in the absence of supplemented PAI-2 (915 Miller units). This observation suggests that the mutation characterizing strain PR1-E4 could have primarily affected the transcription of rhlI, leading to a secondary enhanced transcription of lasB and rhlA. It is interesting to note that the partial restoration of virulence gene expression in mutant PR1-E4 could be completely abolished by the overexpression of a P. aeruginosa homologue of the Escherichia coli mutation suppressor gene, dksA (28). The dksA gene was not mutated in strain PR1-E4, and as previously described in E. coli (11), the mutation suppressor effect of dksA was only observed when dksA was expressed from a multicopy plasmid (28). The interference of dksA with quorum sensing, as well as the mechanism responsible for the restoration of elastase and rhamnolipid production in the absence of lasR, is under investigation.
Previous experiments with E. coli have linked starvation selection to mutations in the stationary-phase sigma factor rpoS, which is controlled by homoserine lactone (10, 29). These workers suggested that the synthesis of homoserine-lactone cell signals might be a general signal of starvation (10, 29). Interestingly, P. aeruginosa AIs are acylated homoserine lactones, and the expression of rpoS in P. aeruginosa is regulated by the RhlR–PAI-2 complex (12). Recently an rpoS homologue has been shown to regulate acylhomoserine lactone-dependent autoinduction in the plant pathogen Ralstonia solanacearum (4). The redundancy in the quorum-sensing systems, with overlapping regulation between the las and the rhl systems and rpoS, is intriguing. These systems control each other in a complex circuitry that starts when the bacteria enter stationary phase, where nutrient availability is reduced. At that time quorum sensing-controlled virulence genes are expressed, together with rpoS-regulated genes whose products enhance resistance to various stresses, such as starvation. The combined expression of these genes would then allow the bacteria to survive and to disseminate in order to find new niches with more abundant nutrient supplies. Further work is required to understand the complex circuits between quorum-sensing systems and the stationary sigma factor rpoS.
Our data show, for the first time, that in a P. aeruginosa mutant defective in one of the two quorum-sensing systems (LasR–PAI-1), starvation results in the up-regulation of the second quorum-sensing system (RhlR–PAI-2). This indicates that therapies designed to block only the las quorum-sensing system might result in the rapid appearance of resistant strains with virulence factor production restored. However, the fact that we could not suppress a mutant defective in both quorum-sensing systems suggests that interventions blocking both the las and the rhl quorum-sensing systems might be a promising approach to efficiently reduce virulence factor production by P. aeruginosa.
Acknowledgments
We thank L. Passador and T. DeKievit for discussions, J. Nezezon for pLPRI, P. Seed for pPCS223, and C. D. Cox and I. Kline for preliminary experiments with casein medium.
This work was supported by NIH grant R01A133713-04, NIH predoctoral training grant 5-T32 AI07362-09 (to J.P.P.), Cystic Fibrosis Foundation grant PESCI96FO (to E.C.P.), a Wilmot Foundation grant, and a Swiss Research Foundation grant (to C.V.D.).
REFERENCES
- 1.Bergen G A, Shelhamer J H. Pulmonary infiltrates in the cancer patient. Infect Dis Clin N Am. 1996;10:297–326. doi: 10.1016/s0891-5520(05)70300-7. [DOI] [PubMed] [Google Scholar]
- 2.Brint J M, Ohman D E. Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family. J Bacteriol. 1995;177:7155–7163. doi: 10.1128/jb.177.24.7155-7163.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dunn M, Wunderink R G. Ventilator-associated pneumonia caused by Pseudomonas infection. Clin Chest Med. 1995;16:95–109. [PubMed] [Google Scholar]
- 4.Flavier A B, Schell M A, Denny T P. An RpoS (ςS) homologue regulates acylhomoserine lactone-dependent autoinduction in Ralstonia solanacearum. Mol Microbiol. 1998;28:475–486. doi: 10.1046/j.1365-2958.1998.00804.x. [DOI] [PubMed] [Google Scholar]
- 5.Fuqua C, Winans S C, Greenberg E P. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 6.Gambello M J, Iglewski B H. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J Bacteriol. 1991;173:3000–3009. doi: 10.1128/jb.173.9.3000-3009.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gambello M J, Kaye S, Iglewski B H. LasR of Pseudomonas aeruginosa is a transcriptional activator of the alkaline protease gene (apr) and an enhancer of exotoxin A expression. Infect Immun. 1993;61:1180–1184. doi: 10.1128/iai.61.4.1180-1184.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Govan J R, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkolderia cepacia. Microbiol Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holloway B W, Krishnapillai V, Morgan A F. Chromosomal genetics of Pseudomonas. Microbiol Rev. 1979;43:73–102. doi: 10.1128/mr.43.1.73-102.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huisman G W, Kolter R. Sensing starvation: a homoserine lactone dependent signaling pathway in Escherichia coli. Science. 1994;265:537–539. doi: 10.1126/science.7545940. [DOI] [PubMed] [Google Scholar]
- 11.Kang P J, Craig E A. Identification and characterization of a new Escherichia coli gene that is a dosage-dependent suppressor of a dnaK deletion mutation. J Bacteriol. 1990;172:2055–2064. doi: 10.1128/jb.172.4.2055-2064.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Latifi A, Foglino M, Tanaka K, Williams P, Lazdunski A. A hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhlR (VsmR) to expression of the stationary-phase sigma factor RpoS. Mol Microbiol. 1996;21:1137–1146. doi: 10.1046/j.1365-2958.1996.00063.x. [DOI] [PubMed] [Google Scholar]
- 13.Latifi A, Winson M K, Foglino M, Bycroft B W, Stewart G S, Lazdunski A, Williams P. Multiple homologues of LuxR and LuxI control expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosa PAO1. Mol Microbiol. 1995;17:333–343. doi: 10.1111/j.1365-2958.1995.mmi_17020333.x. [DOI] [PubMed] [Google Scholar]
- 14.Ochsner U A, Reiser J. Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:6424–6428. doi: 10.1073/pnas.92.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohman D E, Cryz S J, Iglewski B H. Isolation and characterization of a Pseudomonas aeruginosa PAO mutant that produces altered elastase. J Bacteriol. 1980;142:836–842. doi: 10.1128/jb.142.3.836-842.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Passador L, Cook J M, Gambello M J, Rust L, Iglewski B H. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science. 1993;260:1127–1130. doi: 10.1126/science.8493556. [DOI] [PubMed] [Google Scholar]
- 17.Passador L, Iglewski B H. Quorum sensing and virulence gene regulation in Pseudomonas aeruginosa. In: Roth J A, editor. Virulence mechanisms of bacterial pathogens. Washington, D.C: American Society for Microbiology; 1995. pp. 65–78. [Google Scholar]
- 18.Passador L, Tucker K D, Guertin K R, Journet M P, Kende A S, Iglewski B H. Functional analysis of the Pseudomonas aeruginosa autoinducer PAI. J Bacteriol. 1996;178:5995–6000. doi: 10.1128/jb.178.20.5995-6000.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pearson J P, Gray K M, Passador L, Tucker K D, Eberhard A, Iglewski B H, Greenberg E P. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc Natl Acad Sci USA. 1994;91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pearson J P, Passador L, Iglewski B H, Greenberg E P. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:1490–1494. doi: 10.1073/pnas.92.5.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pearson J P, Pesci E C, Iglewski B H. Role of Pseudomonas aeruginosa las and rhl quorum-sensing systems in the control of elastase and rhamnolipid biosynthesis genes. J Bacteriol. 1997;179:5756–5767. doi: 10.1128/jb.179.18.5756-5767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pesci E C, Pearson J P, Seed P C, Iglewski B H. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 1997;179:3127–3132. doi: 10.1128/jb.179.10.3127-3132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Preston M J, Seed P C, Toder D S, Iglewski B H, Ohman D E, Gustin J K, Goldberg J B, Pier G B. Contribution of proteases and LasR to the virulence of Pseudomonas aeruginosa during corneal infections. Infect Immun. 1997;65:3086–3090. doi: 10.1128/iai.65.8.3086-3090.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siegmund I, Wagner F. New method for detecting rhamnolipids excreted by Pseudomonas species during growth on mineral plates. Biotechnol Tech. 1991;5:265–268. [Google Scholar]
- 25.Tang H B, DiMango E, Bryan R, Gambello M, Iglewski B H, Goldberg J B, Prince A. Contribution of specific Pseudomonas aeruginosa virulence factors to pathogenesis of pneumonia in a neonatal mouse model of infection. Infect Immun. 1996;64:37–43. doi: 10.1128/iai.64.1.37-43.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toder D S, Gambello M J, Iglewski B H. Pseudomonas aeruginosa LasA: a second elastase under the transcriptional control of lasR. Mol Microbiol. 1991;5:2003–2010. doi: 10.1111/j.1365-2958.1991.tb00822.x. [DOI] [PubMed] [Google Scholar]
- 27.Van Delden, C., and B. H. Iglewski. Cell-signaling and the pathogenesis of Pseudomonas aeruginosa infections. Emerg. Infect. Dis., in press. [DOI] [PMC free article] [PubMed]
- 28.Van Delden, C., J. P. Pearson, E. C. Pesci, and B. H. Iglewski. Submitted for publication.
- 29.Zambrano M M, Siegele D A, Almiron M, Tormo A, Kolter R. Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science. 1993;259:1757–1760. doi: 10.1126/science.7681219. [DOI] [PubMed] [Google Scholar]