Abstract
We produced defined isogenic Helicobacter pylori ureI mutants to investigate the function of UreI, the product of one of the genes of the urease cluster. The insertion of a cat cassette had a strong polar effect on the expression of the downstream urease genes, resulting in very weak urease activity. Urease activity, measured in vitro, was normal in a strain in which ureI was almost completely deleted and replaced with a nonpolar cassette. In contrast to previous reports, we thus found that the product of ureI was not necessary for the synthesis of active urease. Experiments with the mouse-adapted H. pylori SS1 strain carrying the nonpolar ureI deletion showed that UreI is essential for H. pylori survival in vivo and/or colonization of the mouse stomach. The replacement of ureI with the nonpolar cassette strongly reduced H. pylori survival in acidic conditions (1-h incubation in phosphate-buffered saline solution at pH 2.2) in the presence of 10 mM urea. UreI is predicted to be an integral membrane protein and may therefore be involved in a transport process essential for H. pylori survival in vivo.
Helicobacter pylori is a microaerophilic gram-negative bacterium which colonizes the gastric mucosa of humans (9). H. pylori is associated with gastritis and peptic ulcer disease and has been shown to increase the risk of gastric cancers. Urease is a major virulence factor of H. pylori. It is involved in neutralizing the acidic microenvironment of the bacterium and also plays a role in H. pylori metabolism (10, 24).
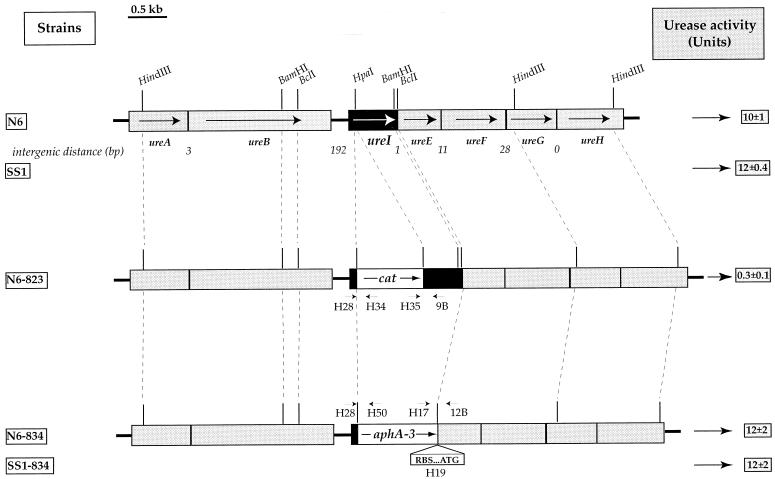
The urease-encoding region of the H. pylori genome is composed of two gene clusters common to all strains (8) (Fig. 1), one comprising the ureAB genes encoding the structural urease subunits and the other containing the ureEFGH genes encoding the accessory proteins required for nickel incorporation into the urease active site. There is a gene of unknown function, ureI, immediately upstream from the latter gene cluster and transcribed in the same direction (Fig. 1). The distances separating ureI from ureE (1 bp) and ureE from ureF (11 bp) suggest that ureI-ureE-ureF constitutes an operon. Cotranscription of ureI and ureE has been demonstrated by Northern blot analysis (1). An H. pylori N6 mutant with the ureI gene disrupted by a MiniTn3-Km transposon was previously obtained (12). This strain (N6-ureI::TnKm-8) presented a urease-negative phenotype, so it was concluded that ureI was an accessory gene required for full urease activity.
FIG. 1.
The urease gene cluster of H. pylori parental strains N6 and SS1 and of the derived mutants deficient in UreI, strains N6-823, N6-834, and SS1-834. The genes are indicated by boxes with arrows showing the direction of their transcription. The distances between the ure genes are given in base pairs. The sites hybridizing to the primers used to confirm correct allelic exchange in strains N6-823, N6-834, and SS1-834 are shown. Blank boxes represent the cassettes containing the genes conferring resistance to CM (cat) or to KM (aphA-3). The urease activities of these strains are given on the right side of the figure. Urease activity was measured at pH7 as the release of ammonia in crude extracts of bacteria grown for 48 h on blood agar plates as described previously (8). One unit corresponds to the amount of enzyme required to hydrolyze 1 μmol of urea/min/mg of total protein. The data are means ± standard deviations calculated from three to five determinations.
The sequence of UreI from H. pylori and those of the AmiS proteins, encoded by the aliphatic amidase operons of Pseudomonas aeruginosa and Rhodococcus sp. strain R312, are similar (4, 25). Aliphatic amidases catalyze the intracellular hydrolysis of short-chain aliphatic amides to produce the corresponding organic acid and ammonia. We have shown that H. pylori also has such an aliphatic amidase, which hydrolyzes acetamide and propionamide in vitro (21).
The sequence similarity between UreI and AmiS together with the very similar structures of the urease and amidase substrates (urea: NH2-CO-NH2; acetamide: CH3-CO-NH2) and the fact that ammonia is produced by both enzymes opened new perspectives for an investigation of the function of the H. pylori UreI protein.
Construction of defined mutations of the H. pylori ureI gene.
H. pylori strains with defined mutations in ureI were generated by allelic exchange to determine whether the UreI protein was necessary for full urease activity. For this purpose, two plasmids (pILL823 and pILL834) with cassettes carrying antibiotic resistance genes inserted in ureI were constructed in Escherichia coli.
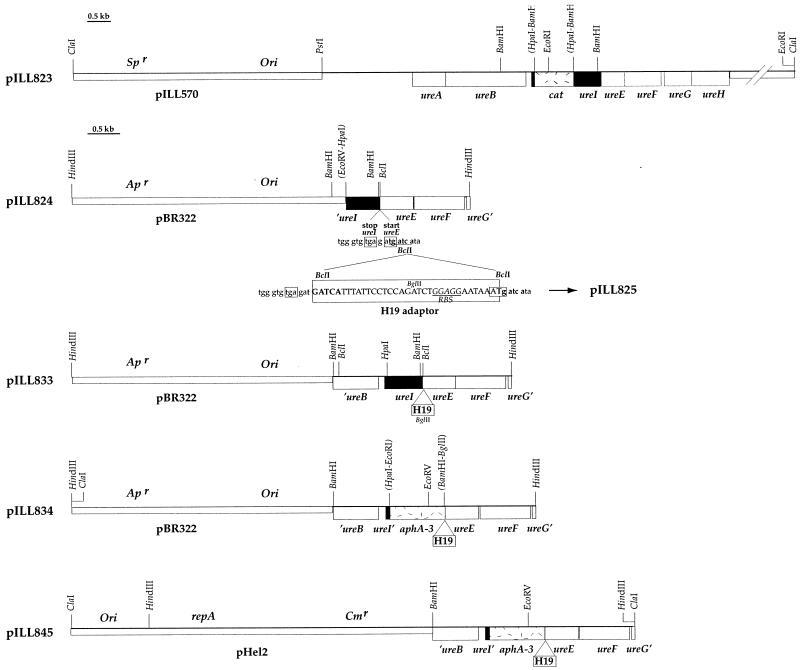
In one plasmid, pILL823 (Fig. 2), the ureI gene was inactivated by the insertion of a promoterless cat gene, conferring resistance to chloramphenicol (CM). A 780-bp blunt-ended BamHI restriction fragment containing the “cat cartridge” from pCM4 (Pharmacia, Uppsala, Sweden) was introduced into a unique HpaI site, between codons 21 and 22 of ureI, in pILL753 (8).
FIG. 2.
Restriction map of pILL823, pILL824, pILL833, pILL834, and pILL845. Small boxes mark the vector of each plasmid; large boxes correspond to genes. Ori indicates the position of the ColE1 origin of replication. repA is the gene coding for the RepA protein, which is necessary for autonomous replication of pHel2 in H. pylori. Sp, Ap, and Cm indicate the genes conferring resistance to spectinomycin, ampicillin, and CM, respectively. The sequence of the DNA region comprising the ureI stop codon and the ureE start codon, including the BclI site where adapter H19 was inserted, is given below pILL824, which was obtained by the insertion of a 1.8-kb HpaI-HindIII fragment from pILL753 (8) into pBR322. Plasmid pILL825 was produced by the insertion of the H19 adapter (carrying an RBS and ATG in frame with ureE) (Table 1) into the BclI site of pILL824; the resulting ureI-ureE intergenic sequence is also shown. The stop codon of ureI and the start codon of ureE are boxed, and the RBS is underlined. In pILL833, the BamHI fragment of pILL825 was replaced by a 1.3-kb blunt-ended PvuII-BamHI fragment from pILL753. Plasmid pILL834 was obtained by replacement of the HpaI-BglII fragment of pILL833 with an 850-bp blunt-ended EcoRI-BamHI fragment of pUC18K2 containing the nonpolar KM cassette (18). Parentheses indicate the position of restriction sites removed by ligation.
The second plasmid, pILL834, carried a ureI gene in which all but the first 21 codons were deleted and replaced with a nonpolar cassette (subcloned from pUC18K2 [18]) composed of the aphA-3 kanamycin (KM) resistance gene (23) with its promoter and terminator regions deleted. In Shigella flexneri (18) and other organisms (such as Yersinia enterocolitica [2]), this cassette has been shown not to affect the transcription of the genes downstream within an operon as long as these distal genes have intact translation signals. There is only 1 bp separating ureI from ureE (Fig. 1), and ureE does not have a ribosome binding site (RBS) of its own; so the expression of ureI and ureE is transcriptionally and translationally coupled. Therefore, the ureI deletion was accompanied by the addition of an RBS immediately upstream from ureE. As shown in Fig. 2, three intermediates, pILL824, pILL825, and pILL833, were constructed in order to produce the final plasmid, pILL834.
Introduction of ureI mutations into H. pylori.
H. pylori ureI mutants were produced by allelic exchange following electroporation with a concentrated preparation of pILL823 and pILL834 (as previously described [21]) of H. pylori N6 (11) and of mouse-adapted H. pylori SS1 (Sydney strain) (16). Bacteria showing chromosomal allelic exchange with pILL823 were selected on CM (4 μg/ml), and those with chromosomal allelic exchange with pILL834 were selected on KM (20 μg/ml). We checked that the desired allelic exchange had taken place in strains N6-823, N6-834, and SS1-834 (Fig. 1) by performing PCR with the appropriate oligonucleotides (Table 1). The PCR products obtained with genomic DNA of these strains were as expected: for strain N6-823, 140 bp with primers H28 and H34, 220 bp with primers H35 and 9B, and 1.2 kilobase pairs (kb) with primers H28 and 9B; for strains N6-834 and SS1-834, 150 bp with primers H28 and H50, 180 bp with primers H17 and 12B, and 1 kb with primers H28 and 12B.
TABLE 1.
Names and nucleotide sequences of oligonucleotides used in this study
| Primera | Oligodeoxynucleotide sequence (5′ to 3′) |
|---|---|
| H17 | TTTGACTTACTGGGGATCAAGCCTG |
| H19 | GATCATTTATTCCTCCAGATCTGGAGGAATAAAT |
| H28 | GAAGATCTCTAGGACTTGTATTGTTATAT |
| H34 | TATCAACGGTGGTATATCCAGTG |
| H35 | GCAGTTATTGGTGCCCTTAAACG |
| H50 | CCGGTGATATTCTCATTTTAGCC |
| 8A | GCGAGTATGTAGGTTCAGTA |
| 9B | GTGATACTTGAGCAATATCTTCAGC |
| 12B | CAAATCCACATAATCCACGCTGAAATC |
H19 was used as the adapter, and the others were used as primers for PCR amplification.
The growth rate of strain N6-834 carrying mutant ureI with a nonpolar cassette was compared to that of the parental strain, N6. No difference in the colony size was observed on blood agar medium plates. Identical doubling times and stationary phase optical densities were measured for both strains grown in brain heart infusion (Oxoid) liquid medium containing 0.2% β-cyclodextrin (Sigma). UreI is thus not essential for H. pylori growth in vitro.
Urease activities of H. pylori ureI mutants.
The urease activities of strains N6-823, N6-834, and SS1-834 were measured in vitro on crude extracts as described previously (8) and compared to the activities of the parental strains, N6 and SS1 (Fig. 1). Urease activity was almost completely abolished in strain N6-823 (0.3 ± 0.1 U). Strains N6-834 and SS1-834, with nonpolar ureI mutations, had wild-type levels of activity (N6-834 and SS1-834: 12 ± 2 U; N6: 10 ± 1 U; SS1: 12 ± 0.4 U).
These results strongly suggested that the urease-negative phenotype of the N6-ureI::TnKm-8 strain (12) and the very weak urease activity of the N6-823 strain were due to a polar effect of the inserted cassettes on the expression of the downstream genes ureE and ureF (Fig. 1). This hypothesis was tested by measuring the urease activity of strain N6-823 complemented in trans with an E. coli/H. pylori shuttle plasmid expressing the ureEF genes. This plasmid, pILL845 (Fig. 2), was obtained by insertion of a 2.8-kb ClaI-BamHI fragment of pILL834 (comprising the 3′ end of ureB, ureI with the nonpolar cassette replacing deleted codons, and intact ureE and ureF genes) into the corresponding sites of the pHel2 shuttle vector (14). Strain N6-823 was electroporated with a DNA preparation of pILL845 (as described in reference 21), and transformants were selected on KM (20 μg/ml) and CM (4 μg/ml). In strain N6-823 harboring pILL845, a high level of urease activity was restored (25 U), confirming that the very low level of urease activity of strain N6-823 was due to a polar effect on the expression of accessory genes ureEF.
Colonization test for the H. pylori SS1-834 mutant in the mouse animal model.
The mouse model for infection by the H. pylori SS1 strain (Sydney strain) (16), validated in our laboratory (6, 13), was used to test the function of UreI in vivo. Mice were infected either with the nonpolar ureI mutant, SS1-834, or with the parental strain, SS1 (which had gone through an equivalent number of in vitro subcultures) as a positive control. This experiment was repeated three times and produced identical results. Two independently constructed SS1-834 mutants were used. The first mutant strain had gone through 30 in vitro subcultures; the second had gone through only 20. R. Ferrero (10a) showed that, under the same experimental conditions, strain SS1 can undergo up to 80 in vitro subcultures without losing its colonization capacity.
In each experiment, aliquots (100 μl) containing 106 H. pylori SS1 or SS1-834 bacteria prepared in peptone broth were administered orogastrically to 10 mice each (6- to 8-week-old Swiss specific-pathogen-free mice) as described by Ferrero et al. (13). Mice were killed 4 weeks after inoculation. We tested for the presence of H. pylori with a direct urease test on biopsies performed on half the stomach (13). The remaining gastric tissues were used for quantitative culture of H. pylori as described by Ferrero et al. (13). In every experiment, the stomachs of the 10 SS1-infected mice all tested positive for urease. The bacterial load was between 5 × 104 and 5 × 105 CFU per g of stomach tissue. None of the stomachs of the mice infected with strain SS1-834 tested positive for urease, and no H. pylori cells were cultured from them. Thus, the UreI protein is essential for H. pylori in vivo survival and/or colonization of the mouse stomach.
Survival of the H. pylori N6-834 mutant in acidic conditions.
Survival under acidic conditions in the presence of 10 mM urea or in the absence of urea was tested with strains N6 and N6-834. The experimental procedures were those described by Clyne et al. (7). Exponentially grown bacteria were harvested and washed in phosphate-buffered saline (PBS; Boehringer, Mannheim, Germany), and 2 × 108 CFU of bacteria per ml were resuspended in PBS at pH 2.2 or 7 in the presence of 10 mM urea or in the absence of urea and incubated for 1 h at 37°C. To evaluate bacterial survival, quantitative cultures (5 days of growth) of the H. pylori strains were performed. At pH 7, in the absence of urea, both strains survived similarly (108 CFU/ml). In agreement with the results of Clyne et al. (7), none of the strains survived at pH 7 in the presence of urea because the final pH rose to 9. As expected (7), both strains were killed at pH 2.2 in the absence of urea, and significant survival at pH 2.2 in the presence of urea was observed with strain N6 (5 × 106 CFU/ml; final pH 6.5). In contrast, when the nonpolar ureI mutant strain N6-834 was incubated at pH 2.2 in the presence of urea, a low level of survival was observed (103 CFU/ml) and the pH was unchanged (pH 2.3) after 1 h of incubation.
Alignment of the UreI and AmiS protein sequences and two-dimensional structure prediction.
A systematic search for UreI homologs in the protein data banks was carried out. We found that H. pylori is not the only ureolytic bacterium with a ureI gene. Two phylogenetically related gram-positive organisms, Streptococcus salivarius, a dental plaque bacterium (5), and Lactobacillus fermentum, a lactic acid bacterium (15), carry genes (the available sequences are only partial) coding for UreI homologs (Fig. 3) located immediately upstream from the urease structural genes. The ureI gene has also been detected in various Helicobacter species; the Helicobacter felis ureI gene has been entirely sequenced (10b) (Fig. 3). PCR experiments have suggested that there is a ureI gene in Helicobacter heilmannii (22) and in Helicobacter mustelae (unpublished data).
FIG. 3.
Alignment of the amino acid sequence of UreI from H. pylori with those of similar proteins and prediction of the two-dimensional structure of members of the protein family comprising UreI and AmiS. Residues identical at one position in at least four sequences are boxed; dashes indicate gaps inserted to optimize alignment. The organisms from which the sequences originated and the degree of identity of each with the H. pylori UreI protein are as follows: UreI-Hp, H. pylori (195 residues; accession no., M84338); UreI-Hf, H. felis; 74% identity over 196 residues (accession no., A41012); UreI-Lacto, L. fermentum; 55% identity over the 46-residue partial sequence (accession no., D10605); UreI-Strepto, S. salivarius; 54% identity over the 129-residue partial sequence (accession no., U35248); AmiS-Myco, M. smegmatis; 39% identity over 172 residues (accession no., X57175); AmiS-Rhod, Rhodococcus sp. strain R312; 37% identity over 172 residues (accession no., Z46523); and AmiS-Pseudo, P. aeruginosa; 37% identity over 171 residues (accession no., X77161). Predicted transmembrane α-helices are shown as shaded boxes. The regions separating these boxes are hydrophilic loops labeled IN when they are predicted to be intracellular and OUT when they are predicted to be extracellular.
Sequence similarities between the UreI protein of H. pylori (21.7 kDa) and the AmiS proteins expressed by the aliphatic amidase operons from P. aeruginosa (25) and Rhodococcus sp. strain R312 (4) have been reported. In Mycobacterium smegmatis, there is an additional AmiS homolog encoded by a gene, open reading frame P3, located immediately upstream from an amidase gene (17).
Alignment of these UreI and AmiS proteins [with the Clustal W(1.60) program] defined strongly conserved stretches of amino acids (Fig. 3). All but one of these conserved blocks are in highly hydrophobic segments. These regions, each 17 to 22 residues long, are probably folded into transmembrane α-helices (Fig. 3). Six transmembrane regions were predicted for the proteins from H. pylori, H. felis, and P. aeruginosa, and seven were predicted for those from Rhodococcus sp. strain R312 and M. smegmatis (these are highly reliable predictions, performed with pHD, a profile-fed neural network system [20]). The orientations of the UreI and AmiS proteins in the membrane were deduced (20) from the charges of the intercalated hydrophilic regions, which are short in these proteins (Fig. 3). These results strongly suggest that the members of the family comprising UreI and AmiS, found in both gram-positive and -negative bacteria, are integral membrane proteins. These proteins have no signal sequence and should therefore be inserted into the cytoplasmic membrane in gram-negative bacteria.
Conclusions.
The urease cluster of H. pylori is unique among the many urease operons of gram-negative bacteria that have been sequenced (19) in that it has an extra gene, ureI. The function of UreI has therefore been the subject of much speculation. It has mostly been assigned the function of an accessory protein required for nickel incorporation at the urease active site or of a nickel transporter. We have demonstrated that UreI is not required for full activation of H. pylori urease during in vitro growth. UreI is thus not a nickel transporter since such a protein, NixA (3), already identified in H. pylori, is necessary for full urease activity. We showed herein that replacing ureI with a nonpolar cassette has no effect on urease activity measured in vitro. This is the first time that a nonpolar cassette (18) has been shown to be functional in H. pylori. This will certainly be a valuable tool for genetic analysis of complex H. pylori operons.
We observed that UreI was essential for survival in vivo and/or for colonization of the mouse stomach. This could be due to the reduced resistance to acidity of the ureI mutant, as suggested by the results of tests of in vitro survival in acidic conditions with 10 mM urea. UreI has a sequence similar to those of the AmiS proteins, proposed to be involved in the transport of short-chain amides (25), molecules structurally similar to urea. The UreI and AmiS proteins have the characteristics of integral membrane proteins, probably of the cytoplasmic membrane. Different roles for UreI can tentatively be proposed. UreI might be involved in (i) transport of urea or short-chain amides, (ii) an uptake system for maintaining appropriate intracellular ammonia concentrations, or (iii) the export of excess intracellular ammonium. An essential role for UreI as an amide transporter seems unlikely because in mouse colonization experiments (performed as described above) an SS1 mutant deficient in aliphatic amidase (carrying the mutation described in reference 21) colonized mice as efficiently as the parental strain, SS1. In addition, amidase activity was not significantly modified by the deletion of ureI in strain N6-834. Our results concerning acidity survival are most compatible with UreI being involved in ammonium export.
Finally, UreI, as a membrane protein essential for the survival of H. pylori in vivo, is an interesting potential target for new antibacterial drugs.
Acknowledgments
S. Skouloubris was supported by OraVax Inc., Boston, Mass. and Pasteur-Mérieux Connaught, Lyon, France.
We thank G. Mendz from the University of New South Wales, Sydney, Australia, A. Vanet, and M.-F. Sagot for helpful discussions; R. Ferrero and P. Jenks from the UPBM for constructive comments on the manuscript; and C. Chevalier for her help in the animal model experiments. We also thank C. Parsot for the gift of pUC18K2 and R. Haas for the gift of shuttle vectors pHel2 and pHel3 prior to publication.
REFERENCES
- 1.Akada J K, Shirai M, Takeuchi H, Tsuda M, Nakazawa T. Transcriptional analysis of urease structural gene and the ureI gene in Helicobacter pylori. Gut. 1997;41:A7. [Google Scholar]
- 2.Allaoui A, Schulte R, Cornelis G R. Mutational analysis of the Yersinia enterocolitica virC operon: characterization of yscE, F, G, H, I, J, K required for Yop secretion and yscH encoding YopR. Mol Microbiol. 1995;18:343–355. doi: 10.1111/j.1365-2958.1995.mmi_18020343.x. [DOI] [PubMed] [Google Scholar]
- 3.Bauerfeind P, Garner R M, Mobley H L T. Allelic exchange mutagenesis of nixA in Helicobacter pylori results in reduced nickel transport and urease activity. Infect Immun. 1996;64:2877–2880. doi: 10.1128/iai.64.7.2877-2880.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chebrou H, Bigey F, Arnaud A, Galzy P. Amide metabolism: a putative ABC transporter in Rhodococcus sp. R312. Gene. 1996;182:215–218. doi: 10.1016/s0378-1119(96)00478-7. [DOI] [PubMed] [Google Scholar]
- 5.Chen Y-Y M, Clancy K A, Burne R A. Streptococcus salivarius urease: genetic and biochemical characterization and expression in a dental plaque streptococcus. Infect Immun. 1996;64:585–592. doi: 10.1128/iai.64.2.585-592.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chevalier, C., J.-M. Thiberge, R. L. Ferrero, and A. Labigne.H. pylori gamma-glutamyltranspeptidase (GGT) is essential for the colonization of the gastric mucosa in a mouse model. Gut 41:A1. [DOI] [PubMed]
- 7.Clyne M, Labigne A, Drumm B. Helicobacter pylori requires an acidic environment to survive in the presence of urea. Infect Immun. 1995;63:1669–1673. doi: 10.1128/iai.63.5.1669-1673.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cussac V, Ferrero R L, Labigne A. Expression of Helicobacter pylori urease genes in Escherichia coli grown under nitrogen-limiting conditions. J Bacteriol. 1992;174:2466–2473. doi: 10.1128/jb.174.8.2466-2473.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunn B E, Cohen H, Blaser M. Helicobacter pylori. Clin Microbiol Rev. 1997;10:720–741. doi: 10.1128/cmr.10.4.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eaton K A, Krakowka S. Effect of gastric pH on urease-dependent colonization of gnotobiotic piglets by Helicobacter pylori. Infect Immun. 1994;62:3604–3607. doi: 10.1128/iai.62.9.3604-3607.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10a.Ferrero, R. Personal communication.
- 10b.Ferrero, R. Unpublished data.
- 11.Ferrero R L, Cussac V, Courcoux P, Labigne A. Construction of isogenic urease-negative mutants of Helicobacter pylori by allelic exchange. J Bacteriol. 1992;174:4212–4217. doi: 10.1128/jb.174.13.4212-4217.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferrero R L, Cussac V, Courcoux P, Labigne A. Construction of isogenic mutants of Helicobacter pylori deficient in urease activity. In: Gasbarrini G, Pretolani S, editors. Basic and clinical aspects of Helicobacter pylori infection: proceedings of the Fourth Workshop of the Helicobacter pylori Study Group held in Bologna, Italy, November 1991. Berlin, Germany: Springer-Verlag; 1994. pp. 179–182. [Google Scholar]
- 13.Ferrero R L, Thiberge J-M, Huerre M, Labigne A. Immune responses of specific-pathogen-free mice to chronic Helicobacter pylori (strain SS1) infection. Infect Immun. 1998;66:1349–1355. doi: 10.1128/iai.66.4.1349-1355.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heuermann D, Haas R. A stable shuttle vector system for efficient genetic complementation of Helicobacter pylori strains by complementation and conjugation. Mol Gen Genet. 1998;257:519–528. doi: 10.1007/s004380050677. [DOI] [PubMed] [Google Scholar]
- 15.Kakimoto S, Sumino Y, Kawahara K, Yamazaki E, Nakatsui I. Purification and characterization of acid urease from Lactobacillus fermentum. Appl Microbiol Biotechnol. 1990;32:538–543. doi: 10.1007/BF00173724. [DOI] [PubMed] [Google Scholar]
- 16.Lee A, O’Rourke J, Corazon De Ungria M, Robertson B, Daskalopoulos G, Dixon M F. A standardized mouse model of Helicobacter pylori infection: introducing the Sydney strain. Gastroenterology. 1997;112:1386–1397. doi: 10.1016/s0016-5085(97)70155-0. [DOI] [PubMed] [Google Scholar]
- 17.Mahenthiralingam E, Draper P, Davis E O, Colston M J. Cloning and sequencing of the gene which encodes the highly inducible acetamidase of Mycobacterium smegmatis. J Gen Microbiol. 1993;139:575–583. doi: 10.1099/00221287-139-3-575. [DOI] [PubMed] [Google Scholar]
- 18.Ménard R, Sansonetti P J, Parsot C. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J Bacteriol. 1993;175:5899–5906. doi: 10.1128/jb.175.18.5899-5906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mobley H L T, Island M D, Hausinger R P. Molecular biology of ureases. Microbiol Rev. 1995;59:451–480. doi: 10.1128/mr.59.3.451-480.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rost B, Casadio R, Fariselli P, Sander C. Prediction of helical transmembrane segments at 95% accuracy. Protein Sci. 1995;4:521–533. doi: 10.1002/pro.5560040318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skouloubris S, Labigne A, De Reuse H. Identification and characterization of an aliphatic amidase in Helicobacter pylori. Mol Microbiol. 1997;25:989–998. doi: 10.1111/j.1365-2958.1997.mmi536.x. [DOI] [PubMed] [Google Scholar]
- 22.Solnick J V, O’Rourke J, Lee A, Tompkins L S. Molecular analysis of urease genes from a newly identified uncultured species of Helicobacter. Infect Immun. 1994;62:1631–1638. doi: 10.1128/iai.62.5.1631-1638.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trieu-Cuot P, Gerbaud G, Lambert T, Courvalin P. In vivo transfer of genetic information between Gram-positive and Gram-negative bacteria. EMBO J. 1985;4:3583–3587. doi: 10.1002/j.1460-2075.1985.tb04120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams C L, Preston T, Hossack M, Slater C, McColl K E L. Helicobacter pylori utilizes urea for amino acid synthesis. FEMS Immunol Med Microbiol. 1996;13:87–94. doi: 10.1111/j.1574-695X.1996.tb00220.x. [DOI] [PubMed] [Google Scholar]
- 25.Wilson S A, Williams R J, Pearl L H, Drew R E. Identification of two new genes in the Pseudomonas aeruginosa amidase operon, encoding an ATPase (AmiB) and a putative integral membrane protein (AmiS) J Biol Chem. 1995;270:18818–18824. doi: 10.1074/jbc.270.32.18818. [DOI] [PubMed] [Google Scholar]