Abstract
The capacity of 20 Mycobacterium avium complex isolates to multiply intracellularly in human monocyte-derived macrophages was assessed and correlated to the clinical relevance of each isolate and its reactivity with several candidate genetic virulence markers. The strongest correlation with a virulence phenotype was found for a conserved coding sequence of the macrophage-induced gene mig identified by a specific mig restriction fragment length polymorphism type.
Mycobacterium avium complex (MAC) is ubiquitous in the environment and can be isolated from a wide range of animate and inanimate samples (5). Clinically significant infection is infrequent, except in individuals infected with human immunodeficiency virus (HIV). There it is associated with low CD4+ T-cell counts in the terminal stage of AIDS and contributes considerably to mortality (10). Restriction fragment length polymorphism (RFLP) analyses of environmental, veterinary, and clinical MAC isolates have indicated that strains causing infection in humans constitute a homogeneous subset of MAC. Pathogenicity for humans was correlated in these studies with the host ranges of a number of mycobacterial insertion sequences (8, 11) and the presence of plasmids (2, 7).
Because monocytes and macrophages are the principal host cells for MAC in mammals, the understanding of the bacterium’s ability to survive and multiply in such a potentially bactericidal environment is of great importance. In this context, we earlier identified a macrophage-induced gene (mig) of M. avium, which is expressed only when the bacilli are growing within macrophages (14). A recombinant mig-expressing strain of Mycobacterium smegmatis showed an advantage for survival inside cultured human monocyte-derived macrophages (HMDM), compared to a mig-negative control strain (13). Here we investigated whether the capacity of 20 environmental and clinical MAC isolates to multiply in HMDM is associated with a number of genetic markers, i.e., a conserved mig coding sequence, the insertion sequence IS1245, the mycobacterial plasmid pLR7, and M. avium- or Mycobacterium intracellulare-specific rRNA sequences.
From a large collection of recently isolated MAC strains, 20 strains were chosen for this study: 7 strains from different water sources (kindly provided by R. Schulze-Röbbecke, Institut für Hygiene, Universität Düsseldorf, Düsseldorf, Germany), 9 strains randomly selected from isolates of patients with confirmed MAC infection, and 4 strains randomly selected from respiratory specimens where the isolate was considered a contaminant based on clinical findings. All clinical strains were primary isolates that had been subcultured only once for differentiation and preparation of inocula. Their exact sources are listed in Table 1.
TABLE 1.
Characteristics of MAC strains and growth rates in the HMDM model
| Strain | Growth rate | Source | RFLP typea | IS1245b | MAC speciesc | pLR7d |
|---|---|---|---|---|---|---|
| SCH 235 | 0.453 | Blood, disseminated infection | A | 25 | M. avium | + |
| SCH 228 | 0.403 | BM,e disseminated infection | A | 18 | M. avium | − |
| 2521 | 0.401 | Water | A | 14 | M. avium | + |
| SCH 193 | 0.387 | Liver, disseminated infection | U | — | X | − |
| SCH 132 | 0.352 | Blood, disseminated infection | A | 19 | M. avium | − |
| SCH 144 | 0.352 | BM, disseminated infection | A | 18 | M. avium | − |
| SCH 273 | 0.350 | BAL, contaminant | B | — | M. intracellulare | − |
| SCH 196 | 0.302 | Blood, disseminated infection | A | 18 | M. avium | − |
| SCH 245 | 0.277 | Blood, disseminated infection | A | 18 | M. avium | − |
| 1816 | 0.220 | Water | A | 10 | M. avium | + |
| SCH 187 | 0.216 | BM, disseminated infection | A | 7 | M. avium | − |
| SCH 180 | 0.210 | BAL, contaminant | A | 5 | M. avium | + |
| SCH 298 | 0.157 | BAL, contaminant | B | — | M. intracellulare | − |
| 3045 | 0.130 | Water | B | — | M. intracellulare | − |
| 2442 | 0.090 | Water | U | 15 | M. avium | + |
| SCH 281 | 0.078 | Sputum, contaminant | B | — | X | − |
| SCH 215 | 0.070 | Cervical lymph node | A | 12 | M. avium | − |
| 1817 | 0.037 | Water | B | — | M. intracellulare | − |
| 2895 | 0.001 | Water | B | — | M. intracellulare | − |
| 2391 | −0.009 | Water | U | — | M. avium | − |
RFLP type A, mig probe-reactive 2.8-kb band after BamHI digestion and 950-bp band after SalI digestion; RFLP type B, mig probe-reactive 6.0-kb band after BamHI digestion and 1,300-bp band after SalI digestion; U, heterogeneous group.
Number of bands showing reactivity with DNA probe. —, no copies of IS1245.
Reactivity with M. avium- or M. intracellulare-specific 16S rRNA probe. X, not reactive with either species-specific probe.
Reactivity with a DNA probe specific for the origin of replication of the mycobacterial plasmid. +, reacts with the probe; −, does not react with the probe.
BM, bone marrow.
The in vitro virulence of each isolate was determined as its capacity to multiply in primary HMDM. Peripheral blood monocytes were isolated by Ficoll (Pharmacia, Uppsala, Sweden) gradient centrifugation (4, 15). Interface mononuclear cells were washed repeatedly, resuspended in warm RPMI 1640 (Life Technologies, Paisley, Scotland) supplemented with 5% autologous serum (AS), and plated into eight-well glass-bottom chamber slides (Nunc, Wiesbaden, Germany). The seeded cultures were incubated overnight at 37°C under a 5% CO2 atmosphere. Nonadherent cells were washed off with warm RPMI 1640, and the remaining monocytes (5 × 104 cells/well) were cultured in RPMI 1640 with 5% AS prior to infection, with daily changes of medium. Inocula of the MAC isolates for infection of the HMDM were prepared from bacterial cultures in 7H9 broth after 2 weeks of incubation at 37°C with moderate agitation. Bacteria were collected by centrifugation and resuspended in 1 ml of fresh AS for opsonization. After 10 min of sonication in a water bath, aliquots of serial 10-fold dilutions in RPMI 1640 were snap frozen at −70°C. The exact number of viable bacilli in each dilution was determined by plating serial dilutions of thawed aliquots and counting CFU.
HMDM cultures were infected with MAC suspensions at a multiplicity of infection (MOI) of 10. After 2 h of incubation at 37°C under a 5% CO2 atmosphere, cell cultures were washed vigorously to remove extracellular bacteria. Macrophages were then incubated in RPMI with 5% AS, with a daily change of medium. Mycobacterial antibiotics were intentionally not included in the culture medium because these may be delivered to the phagosome, confounding the CFU results. Plating of macrophage culture supernatants confirmed negligible growth of the bacteria in the tissue culture medium.
The baseline MOI 2 h after infection and the CFU counts on days 3, 5, and 7 were determined by discarding the supernatants from four replicate wells, lysing the macrophages, and plating serial dilutions for CFU counts as described by Crowle et al. (3). Intracellular growth rates (k) according to the equation y(t) = y(t0) · ekt were determined from the slopes between the means of day 0 (t0) and day 5 by assuming exponential growth between these time points (see Table 2). For genotyping, mycobacterial DNA was purified and Southern blotting was performed as previously described (14). The BamHI mig probe was a 2,845-bp fragment encoding the entire mig and flanking regions (13). The SalI mig probe was a 926-bp fragment covering most of the mig coding sequence and part of the promoter region. Both fragments were purified from plasmid pGPC100 (13) and were labeled with digoxigenin (Boehringer, Mannheim, Germany). The IS1245 probe was generated by PCR using the primers P1 (5′GCCGCCGAAACGATCTAC) and P2 (5′AGGTGGCGTCGAGGAAGAC) as previously described (8). The pLR7 probe was excised from plasmid pMB184 (1) (kindly provided by M. Beggs, John L. McClellan Memorial Veterans Hospital, Little Rock, Ark.) and labeled with digoxigenin as described above. Probes were hybridized to BamHI- or SalI-digested genomic DNA in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–50% formamide–0.2% sodium dodecyl sulfate (SDS)–0.1% N-lauroylsarcosine–2% blocking solution at 42°C for 2 h. This was followed by two washes in 2× SSC–0.1% SDS at 20°C for 5 min and two washes in 0.2× SSC–0.1% SDS at 72°C for 15 min. Any hybridized labeled probe was detected by chemiluminescence according to the procedures recommended by the manufacturer (Boehringer). All strains were analyzed with 16S rRNA probes (Accuprobe; GenProbe Inc., San Diego, Calif.) and showed reactivity with a MAC probe. Thirteen also reacted with the M. avium-specific probe, and five reacted with the M. intracellulare-specific probe. mig RFLP analysis divided our MAC strains into two homogeneous groups and a third, heterogeneous group. Group A was characterized as follows. Digestion of genomic DNA with the restriction endonuclease BamHI followed by Southern blotting and hybridization to a 2,849-bp BamHI mig fragment produced a single band of 2.8 kb. The analogous Southern blot using SalI-digested genomic DNA probed with a 926-bp SalI fragment of mig led to a single 0.9-kb band (Fig. 1, lane 1). These strains carried multiple copies of IS1245, with a mean of 15 and a range of 5 to 25. All IS1245-positive strains had strain-specific RFLP patterns excluding clonality of these strains (data not shown). Eleven of the 13 strains reacting with the M. avium 16S rRNA probe were in group A. Only one of the strains from clinical infections did not fall into this group.
TABLE 2.
Statistical analysis of differences in intracellular growth rates in primary human macrophages between categories of MAC strains by the t test
| Characteristica | Mean k (SD)b for:
|
P | |
|---|---|---|---|
| Positive strainsc | Negative strainsd | ||
| Clinical relevance | 0.3124 (0.1152) | 0.1514 (0.1345) | 0.0109 |
| mig type A | 0.2960 (0.1116) | 0.1357 (0.1431) | 0.0114 |
| IS1245 | 0.2788 (0.1219) | 0.1414 (0.1519) | 0.0378 |
| M. avium species | 0.2567 (0.1414) | 0.1629 (0.1503) | 0.1827 |
| pLR7 | 0.2748 (0.1492) | 0.2069 (0.1486) | 0.3881 |
Clinical relevance, causing clinical infection. mig type A, RFLP type A in a Southern blot using a mig open reading frame-specific DNA probe. IS1245, presence of copies of this insertion element in the bacterial genome. M. avium species, reactivity with the species-specific rRNA probe. pLR7, reactivity with a DNA probe specific for the origin of replication of the mycobacterial plasmid.
Intracellular growth rates (k) were determined from the slopes between the means for day 0 and day 5. Means and standard deviations from four replicate experiments are given.
Strains positive for the given characteristic.
Strains negative for the given characteristic.
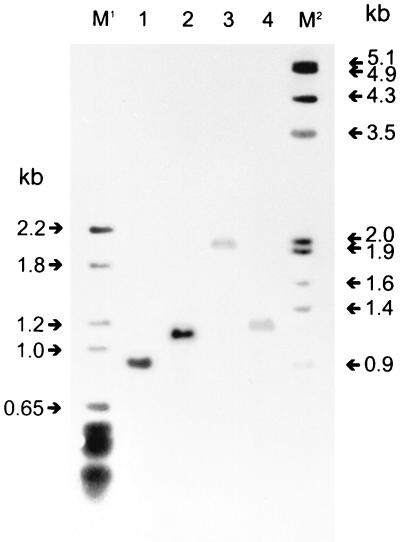
FIG. 1.
mig RFLP of MAC isolates. Genomic DNA was digested with SalI, resolved by agarose gel electrophoresis, blotted to a nylon membrane, and hybridized to a digoxigenin-labeled mig probe. M1, pBR328 DNA digested with BglI and HinfI and used as a size marker. Lane 1, RFLP type A; lane 2, strain 2442; lane 3, strain 2391; lane 4, RFLP type B. M2, λ DNA, digested with HindIII/EcoRI and used as a size marker. RFLP type A shows the conserved SalI mig fragment of the virulent type strain (14).
A second RFLP pattern, common to six strains, could be identified; these strains formed group B. A BamHI Southern blot probed with the 2,849-bp BamHI mig fragment demonstrated a faint 6.0-kb band. Southern analysis with the 900-bp probe after SalI treatment led to a single 1,300-bp band (Fig. 1, lane 4). The insertion element IS1245 was not present. Five of these group B strains reacted with the M. intracellulare 16S rRNA probe. This group included three environmental strains and three clinical isolates of HIV-negative patients without clinical signs of MAC infection.
Three of the 20 strains did not fit into either mig RFLP group. Two were environmental strains, and one was the only AIDS isolate that did not qualify for RFLP type A, as it showed a faint, aberrant 3.5-kb band on the BamHI Southern blot and no bands with the SalI mig probe and IS1245 probe (Table 1). Homologs of the mycobacterial plasmid pLR7 were found in only 5 of the 13 M. avium strains.
Statistical significance for differences in virulence between groups of strains according to the (i) clinical relevance of the isolate, (ii) mig RFLP type, (iii) IS1245 content, (iv) MAC species, and (v) pLR7 plasmid content was analyzed by t test (Table 2). The clinical relevance of the isolate correlated significantly with intracellular growth in the HMDM model, underlining the validity of the macrophage model. A conserved mig coding sequence that is identified by RFLP type A is also significantly associated with faster intracellular growth in macrophages. In contrast, RFLP type B MAC strains were of lower virulence and either were environmental samples or had been considered clinically unimportant when isolated from patients. Analogously, the presence of multiple copy numbers of IS1245 was associated with fast intracellular growth and disseminated infection, whereas this mobile element was not present in avirulent strains. Since one of our previous studies (13) comparing intracellular survival of a mig-expressing M. smegmatis strain with that of a mig-negative control strain already showed strong evidence of mig being a significant virulence factor, the results of this study contribute even more to the importance of mig. Nine of ten clinically relevant MAC isolates carried the virulence phenotype’s mig and possessed significantly higher levels of in vitro virulence than isolates with an aberrant mig. Differences in in vitro virulence and clinical pathogenicity cannot be attributed to species differences alone, since reactivity with the species-specific probes was not statistically significantly correlated with intracellular growth in the HMDM model. The previously observed associations of mycobacterial plasmids, in particular homologs of pLR7, with clinical relevance and virulence are not corroborated by our observations. Only a minority of clinically relevant strains reacted with a pLR7 probe. Several genetic markers seem to indicate the presence of a virulent subtype of MAC that does overlap with the M. avium species, but that is more strictly defined by the presence of a conserved coding sequence for the macrophage-induced gene mig of M. avium. Despite the significant association between virulence, mig, and IS1245, both markers may in fact be only secondary to a repertoire of virulence factors yet to be identified.
One strain not conforming to type A or B seems to support the latter view. Strain SCH 193, in which IS1245 is not present and which does not react with the mig SalI probe, displays a virulence phenotype (Table 1).
Other investigators have found that the majority of AIDS isolates could be characterized as a genetically rather homogenous subset of MAC, which may be considered a virulent epidemic clone (6, 9, 12). This theory is only partly corroborated by our findings. While the highly conserved mig pattern and the presence of IS1245 in AIDS MAC isolates indicate a certain relationship of these strains, the very heterogeneous IS1245 fingerprints exclude the possibility of a clonal infection, a conclusion similar to that of Guerrero et al. (8). Two isolates from tap water and one isolate from bronchoalveolar lavage (BAL), where it was not considered a significant pathogen, seem to indicate that the aquatic environment harbors virulent and nonvirulent types, while only the virulent subset causes disseminated infection in AIDS.
We conclude that MAC strains causing clinical infection in general have a common mig gene that is associated with higher virulence in the HMDM model. This gene may be a selection factor for disseminated MAC infection in AIDS patients, and its use as a genetic marker may be useful to distinguish between possible pathogens and mere colonizers.
Acknowledgments
This work was supported by BMBF-Schwerpunkt Mykobakterielle Infektionen grant 01 KI 9612 from the Bundesministerium für Wissenschaft, Bildung, Forschung, und Technologie of Germany and Köln Fortune Program grant V66/1997 from the Faculty of Medicine, University of Cologne (Cologne, Germany).
We thank Anne Abrams for critical reading of the manuscript.
REFERENCES
- 1.Beggs M L, Crawford J T, Eisenach K D. Isolation and sequencing of the replication region of Mycobacterium avium plasmid pLR7. J Bacteriol. 1995;177:4836–4840. doi: 10.1128/jb.177.17.4836-4840.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crawford J T, Bates J H. Analysis of plasmids in Mycobacterium avium-intracellulare isolates from persons with acquired immunodeficiency syndrome. 1986. [DOI] [PubMed] [Google Scholar]
- 3.Crowle A J, Tsang A Y, Vatter A E, May M H. Comparison of 15 laboratory and patient-derived strains of Mycobacterium avium for ability to infect and multiply in cultured human macrophages. J Clin Microbiol. 1986;24:812–821. doi: 10.1128/jcm.24.5.812-821.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Douvas G S, Looker D L, Vatter A E, Crowle A J. Gamma interferon activates human macrophages to become tumoricidal and leishmanicidal but enhances replication of macrophage-associated mycobacteria. Infect Immun. 1985;50:1–8. doi: 10.1128/iai.50.1.1-8.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Falkinham J O., III Epidemiology of infection by nontuberculous mycobacteria. Clin Microbiol Rev. 1996;9:177–215. doi: 10.1128/cmr.9.2.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frothingham R, Wilson K H. Molecular phylogeny of the Mycobacterium avium complex demonstrates clinically meaningful divisions. J Infect Dis. 1994;169:305–312. doi: 10.1093/infdis/169.2.305. [DOI] [PubMed] [Google Scholar]
- 7.Gangadharam P R, Perumal V K, Crawford J T, Bates J H. Association of plasmids and virulence of Mycobacterium avium complex. Am Rev Respir Dis. 1988;137:212–214. doi: 10.1164/ajrccm/137.1.212. [DOI] [PubMed] [Google Scholar]
- 8.Guerrero C, Bernasconi C, Burki D, Bodmer T, Telenti A. A novel insertion element from Mycobacterium avium, IS1245, is a specific target for analysis of strain relatedness. J Clin Microbiol. 1995;33:304–307. doi: 10.1128/jcm.33.2.304-307.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hampson S J, Portaels F, Thompson J, Green E P, Moss M T, Hermon-Taylor J, McFadden J J. DNA probes demonstrate a single highly conserved strain of Mycobacterium avium infecting AIDS patients. Lancet. 1989;i:65–68. doi: 10.1016/s0140-6736(89)91427-x. [DOI] [PubMed] [Google Scholar]
- 10.Horsburgh C R., Jr Mycobacterium avium complex infection in the acquired immunodeficiency syndrome. N Engl J Med. 1991;324:1332–1338. doi: 10.1056/NEJM199105093241906. [DOI] [PubMed] [Google Scholar]
- 11.Kunze Z M, Wall S, Appelberg R, Silva M T, Portaels F, McFadden J J. IS901, a new member of a widespread class of atypical insertion sequences, is associated with pathogenicity in Mycobacterium avium. Mol Microbiol. 1991;5:2265–2272. doi: 10.1111/j.1365-2958.1991.tb02157.x. [DOI] [PubMed] [Google Scholar]
- 12.McFadden J J, Kunze Z M, Portaels F, Labrousse V, Rastogi N. Epidemiological and genetic markers, virulence factors and intracellular growth of Mycobacterium avium in AIDS. Res Microbiol. 1992;143:423–430. doi: 10.1016/0923-2508(92)90057-u. [DOI] [PubMed] [Google Scholar]
- 13.Plum G, Brenden M, Clark-Curtiss J E, Pulverer G. Cloning, sequencing, and expression of the mig gene of Mycobacterium avium, which codes for a secreted macrophage-induced protein. Infect Immun. 1997;65:4548–4557. doi: 10.1128/iai.65.11.4548-4557.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plum G, Clark Curtiss J E. Induction of Mycobacterium avium gene expression following phagocytosis by human macrophages. Infect Immun. 1994;62:476–483. doi: 10.1128/iai.62.2.476-483.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schnittman S, Lane H C, Witebsky F G, Gosey L L, Hoggan M D, Fauci A S. Host defense against Mycobacterium-avium complex. J Clin Immunol. 1988;8:234–243. doi: 10.1007/BF00916551. [DOI] [PubMed] [Google Scholar]