Abstract
Enterohemorrhagic Escherichia coli (EHEC) strains require intimin to induce attaching and effacing (A/E) lesions in newborn piglets. Infection of newborn calves with intimin-positive or intimin-negative EHEC O157:H7 demonstrated that intimin is needed for colonization, A/E lesions, and disease in cattle. These results suggest that experiments to determine if intimin-based vaccines reduce O157:H7 levels in cattle are warranted.
Enterohemorrhagic Escherichia coli (EHEC) strains of serotype O157:H7 are a major cause of bloody diarrhea in humans in the United States (20). Hemolytic uremic syndrome, a life-threatening complication of EHEC O157:H7 infection, is the primary cause of acute kidney failure in children in the United States and Canada (1). Other serotypes of EHEC have also been associated with outbreaks of bloody diarrhea and hemolytic uremic syndrome (2, 3, 19).
Cattle are important reservoirs of EHEC O157:H7 strains (20, 22). The majority of cases of EHEC disease recognized in the United States are associated with ingestion of undercooked, contaminated hamburger or raw milk. Outbreaks have also been associated with produce contaminated with bovine manure. Therefore, one strategy for reducing the risk of EHEC infections in humans is to reduce the prevalence of EHEC infections in cattle.
All EHEC strains are Shiga toxin-producing E. coli (STEC) strains. They produce cytotoxins, called Shiga toxins (Stx1 and Stx2) or verotoxins, that are considered essential for EHEC virulence in humans. EHEC strains are also characterized by the presence of a ∼90-kb plasmid (12, 14, 21). Many EHEC strains have the capacity to attach intimately to host cell membranes and efface microvilli and cytoplasm in a characteristic pattern referred to as an attaching and effacing (A/E) lesion. EHEC strains cause A/E lesions in selected cell lines in vitro and in the intestines of experimental animals (13, 22).
EHEC-mediated A/E lesions are similar to those produced by enteropathogenic E. coli (EPEC) in humans and animals (13). In EPEC, the eae (for E. coli attaching and effacing; formerly called eaeA) chromosomal locus encodes an outer membrane adhesion protein called intimin (11). The eae gene is necessary, but not sufficient, for EPEC bacteria to cause A/E lesions (6, 7, 10). Some EHEC strains also carry an eae homolog that plays a critical role in the attachment of EHEC O157:H7 to human epithelial cells and the formation of A/E lesions in gnotobiotic pigs (8, 15, 16).
The objectives of the present study were to determine if intimin is required for EHEC-mediated enterocolitis and diarrhea in calves and to extend earlier studies showing its role in A/E lesion formation in neonatal piglets. We compared the pathogenicity of an intimin-negative eae mutant of EHEC O157:H7 (strain 86-24eaeΔ10 [15]) with that of two isogenic intimin-positive (eae+) partners, one a wild-type EHEC isolate (strain 86-24) and the other the eae mutant complemented with the eae gene [strain 86-24eaeΔ10(pEB310) (15)], in neonatal calves and cesarean-derived, colostrum-deprived (CDCD) piglets. We also tested the pathogenicity of an intimin-negative non-O157:H7 wild-type EHEC strain (B2F1) in neonatal calves.
The bacterial strains used in this study are described in Table 1. Each of 23 colostrum-deprived calves (18 holstein, 2 jersey, and 3 mixed breeds; 22 male and 1 female) was inoculated before it was 12 h old via suckling with milk replacer containing 1010 CFU of an EHEC strain that produces intimin (12 calves), an EHEC strain that does not produce intimin (8 calves), or the nonpathogenic control E. coli 123 (3 calves) as previously described (5). Calves were observed every 8 h for signs of disease and euthanatized with sodium pentobarbital at 18 or 42 h postinoculation.
TABLE 1.
E. coli strains used in this study
| Strain | Serotype | eae genea | Stx type | Source | Refer- ence |
|---|---|---|---|---|---|
| 86-24 | O157:H7 | + | Stx2 | Phil Tarr | 15 |
| 86-24eaeΔ10b | O157:H7 | − | Stx2 | 15 | |
| 86-24eaeΔ10 (pEB310)c | O157:H7 | + | Stx2 | 15 | |
| B2F1 | O91:H21 | − | Stx2d | Mohamed Karmali | 9, 17 |
| 123 | O43:H28 | − | 18 |
Presence (+) or absence (−) of eae gene.
In-frame deletion in eae gene in strain 86-24.
eae mutant complemented with the eae gene.
At necropsy, rectal contents and sections from the rectum, colon, cecum, and ileum were collected and frozen at −80°C for bacteriological examination (5). Tissues from the rectum, colon, cecum, and ileum were fixed in formaldehyde, sectioned, and stained with hematoxylin and eosin or immunohistochemically with goat anti-O157:H7 as the primary antibody, biotinylated anti-goat immunoglobulin G (heavy and light chains) as the secondary antibody, and an avidin-biotin-peroxidase conjugate (5). Some sections were removed from formaldehyde and postfixed in glutaraldehyde for transmission electron microscopy (5).
Three of five calves inoculated with wild-type EHEC strain 86-24 developed watery diarrhea by 18 h postinoculation (Table 2). Two of three calves had blood-tinged diarrhea at 42 h postinoculation, and one of these calves died on day 2, about 2 h prior to the scheduled necropsy. Postmortem observations were compatible with enteritis as the cause of death. Similarly, two of seven calves inoculated with the complemented mutant strain 86-24eaeΔ10(pEB310) developed diarrhea by 18 h, and three of four had diarrhea (blood tinged in two calves) by 42 h postinoculation. One such infected calf died on day 2, about 2 h prior to the scheduled necropsy. Again, postmortem observations were compatible with enteritis as the cause of death. In contrast, all calves inoculated with mutant strain 86-24eaeΔ10, with B2F1, or with nonpathogenic control strain 123 remained healthy throughout the experiment.
TABLE 2.
Clinical and histological findings in calves 18 and 42 h after inoculation with an eae+ EHEC strain [86-24 or 86-24eaeΔ10(pEB310)], an eae mutant EHEC strain (86-24eaeΔ10), or a strain lacking the eae gene (B2F1 or nonpathogenic E. coli 123)
| Strain | Time of determination (h postinoculation) | n | No. positive for:
|
|||||
|---|---|---|---|---|---|---|---|---|
| Diarrhea | Death | A/E bacteria ina:
|
||||||
| Rectum | Colon | Cecum | Ileum | |||||
| 86-24 | 18 | 2 | 2 | 0 | 2 | 2 | 2 | 2 |
| 42 | 3 | 3 | 1b | 3 | 3 | 2 | 2 | |
| 86-24eaeΔ10 | 18 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| 42 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 86-24eaeΔ10(pEB310) | 18 | 3 | 1 | 0 | 3 | 3 | 2 | 3 |
| 42 | 4 | 3 | 1b | 3 | 3 | 3 | 3 | |
| B2F1 | 42 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| 123 | 42 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
A/E bacteria stained with E. coli O157:H7 antibody by immunoperoxidase technique.
Calf found dead at ca. 40 h postinoculation; samples were collected for histopathological studies.
Hyperemia, focal petechiae, and fibrinous exudates in the intestines were common postmortem observations in calves inoculated with strain 86-24 or 86-24eaeΔ10(pEB310) but were not noted in any of the calves inoculated with mutant strain 86-24eaeΔ10, strain B2F1, or control strain 123. A/E lesions containing O157:H7+ bacteria were identified by immunostaining in the ileum and large intestines of five of five calves inoculated with strain 86-24 and six of seven calves inoculated with strain 86-24eaeΔ10(pEB310). In addition to A/E lesions, a diffuse mucosal neutrophil infiltration with accompanying hemorrhage, edema, atrophy of ileal villi, and fibrinous to fibrinohemorrhagic exudates in the intestinal lumen was noted in histologic sections from some of these calves. Neutrophil infiltrates also occurred in the one calf that had no A/E lesions. Examination of sections of ileum from two calves [18 and 42 h postinoculation with strain 86-24eaeΔ10(pEB310)] by electron microscopy confirmed the in vivo A/E activity of the complemented mutant (Fig. 1). No A/E lesions or histopathological abnormalities were detected in any calf inoculated with mutant strain 86-24eaeΔ10, strain B2F1, or control strain 123. However, there were patchy layers of O157:H7+ bacteria on the epithelium in the cecum, colon, and ileum of one calf necropsied 18 h after inoculation with mutant strain 86-24eaeΔ10, but these bacteria were not associated with A/E lesions.
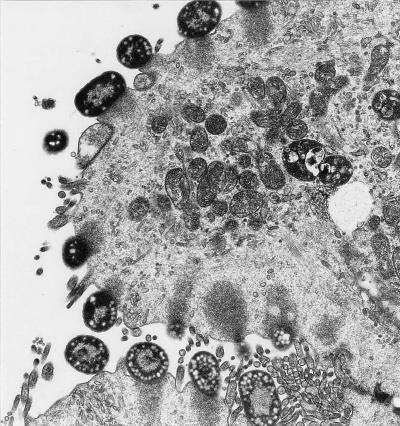
FIG. 1.
Electron micrograph of absorptive cells from the ileum of a calf 18 h after inoculation with EHEC O157:H7 strain 86-24eaeΔ10(pEB310). This strain is an eae mutant which has been complemented with the eae gene. The intestinal lumen is to the left. Bacteria are intimately attached to absorptive-cell membranes with subjacent electron-dense filaments in absorptive-cell cytoplasm. Most of the absorptive-cell microvilli have been effaced. There are pedestals beneath bacteria to the upper left.
The numbers of the inoculated organisms (expressed as CFU per gram) recovered from tissues and feces of calves at 18 or 42 h postinoculation with eae+ or eae strains of E. coli are shown in Fig. 2. Sorbitol-negative EHEC O157:H7 bacteria were quantitated on sorbitol MacConkey agar containing 100 μg of streptomycin per ml (strain 86-24), 100 μg of streptomycin and 20 μg of nalidixic acid per ml (strain 86-24eaeΔ10), or 100 μg of ampicillin and 34 μg of chloramphenicol per ml [strain 86-24eaeΔ10(pEB310)]. Samples from which the inoculated strain were not recovered were recorded as having <103 CFU/g. Selected sorbitol-negative isolates were tested for O157:H7 antigen by a latex agglutination assay (5). Strain B2F1 (O91:H21) and strain 123 (O43:H28) bacteria were quantitated on MacConkey agar containing 100 μg of streptomycin or 20 μg of nalidixic acid per ml, respectively. Colonies were tested for O91 and O43 antigens to identify strains B2F1 and 123, respectively, by filter blot immunoperoxidase assay (4), using anti-O91 and anti-O43 sera (E. coli Reference Center, Pennsylvania State University, University Park) and peroxidase-conjugated anti-rabbit immunoglobulin G (heavy and light chains) (Kirkegaard and Perry Laboratories, Inc., Gaithersburg, Md.).
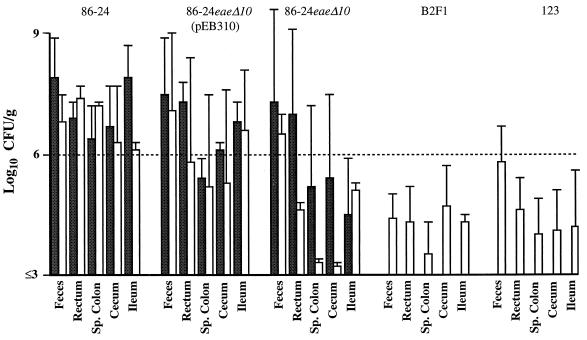
FIG. 2.
CFU of E. coli per gram of tissue or feces in necropsy samples from neonatal calves 18 h (■) or 42 h (□) after inoculation with eae+ EHEC strain 86-24 or 86-24eaeΔ10(pEB310), eae mutant EHEC strain 86-24eaeΔ10, strain B2F1, or nonpathogenic E. coli 123. Data are means ± standard deviations. See Table 2 for the number of calves in each group. Sp., spiral.
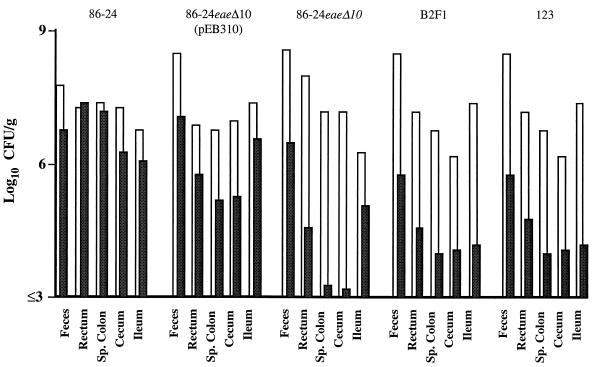
Greater numbers of the inoculated organisms were recovered from the intestines of calves inoculated with strain 86-24 or strain 86-24eaeΔ10(pEB310) than from those of calves inoculated with strain 86-24eaeΔ10, strain B2F1, or control strain 123 at 42 h postinoculation (Fig. 2). Because of the large degree of variation among animals and the small number of animals, there was no significant difference among the numbers of bacteria at the individual tissue level. However, when we took the group average for each tissue and treated the tissues as a block, the mean CFU per gram for all samples obtained at 42 h postinoculation from the group of calves inoculated with eae mutant strain 86-24eaeΔ10 was lower (P < 0.05; analysis of variance and least significant difference test) than the means for calves inoculated with eae+ strain 86-24 or 86-24eaeΔ10(pEB310). The only exception was that the one calf that did not develop clinical signs or have A/E lesions after inoculation with strain 86-24eaeΔ10(pEB310) had bacterial levels comparable to those in calves inoculated with strains that lacked the eae gene. The numbers for strains 86-24eaeΔ10 and B2F1 were similar to those for control strain 123. The number of strain 86-24eaeΔ10 organisms recovered from feces was similar to that of the eae+ strains. The inoculum strain accounted for a larger percentage of the total number of coliforms isolated from calves inoculated with eae+ EHEC than from calves inoculated with eae mutant EHEC, strain B2F1, or the control E. coli strain (Fig. 3). The presence and severity of A/E lesions in tissues from calves inoculated with eae+ strains correlated with the number of inoculated bacteria recovered (data not shown). A/E lesions were only seen in tissues containing ≥106 CFU of eae+ EHEC/g of tissue.
FIG. 3.
Numbers of CFU of inoculated strain (■) and total coliforms (□) per gram of tissue or feces recovered at 42 h postinoculation from calves inoculated with eae+ EHEC strain 86-24 or 86-24eaeΔ10(pEB310), eae mutant EHEC strain 86-24eaeΔ10, strain B2F1, or E. coli control strain 123. Sp., spiral.
In earlier studies we showed that the histopathology of EHEC O157:H7 infection in neonatal calves is similar to that in CDCD piglets, but O157:H7 bacteria do not cause diarrhea in CDCD piglets by 18 h postinoculation (5). In this study, we compared the pathogenicity of isogenic eae+ and eae mutant derivatives of EHEC O157:H7 strain 86-24 in <8-h-old CDCD piglets (8). As shown in Table 3, CDCD piglets developed colonic edema and A/E lesions by 18 h after inoculation with the eae+ strain 86-24 or 86-24eaeΔ10(pEB310) but not with the eae mutant. In contrast to calves, the A/E lesions occurred mainly in the ceca of the piglets. The numbers of inoculated bacteria recovered from the cecum or ileum at 18 h postinoculation were similar in all experimental groups (Table 3), and bacterial counts did not correlate with the presence or absence of A/E lesions. These results indicate that intimin plays a critical role in EHEC O157:H7 pathogenesis in CDCD piglets and extend the findings of earlier studies with these strains in gnotobiotic piglets (15).
TABLE 3.
Findings in CDCD piglets at 18 h after inoculation with an eae+ [86-24 or 86-24eaeΔ10(pEB310)] or an eae mutant (86-24eaeΔ10) EHEC strain or nonpathogenic E. coli 123
| Inoculated strain | No. of piglets tested | No. with colonic edema | No. with A/E bac- teria | Mean log10 CFU ± SD of E. coli O157:H7/g of tissue in:
|
|
|---|---|---|---|---|---|
| Cecum | Ileum | ||||
| 86-24 | 2 | 2 | 2a | 6.9 ± 0.5 | 5.9 ± 0.3 |
| 86-24eaeΔ10 | 3 | 0 | 0 | 7.1 + 0.8 | 5.5 + 0.6 |
| 86-24eaeΔ10 (pEB310) | 3 | 3 | 3b | 5.6 ± 1.4 | 5.5 ± 1.0 |
| 123 | 2 | 0 | 0 | NDc | ND |
A/E bacteria found only in the cecum.
A/E bacteria found in the cecum (three of three) and the ileum (one of three).
ND, not determined.
In this study we have clearly demonstrated that the eae gene locus is required for E. coli O157:H7 strain 86-24 to intensively colonize the intestines and cause diarrhea and A/E lesions in neonatal calves and to cause colonic edema and A/E lesions in CDCD piglets. The eae mutant and B2F1 data indicate that eae-mediated adherence to the intestinal mucosa is critical for EHEC to cause fibrinohemorrhagic enterocolitis and diarrhea in calves. Similarly, the results confirm that eae-mediated colonization is necessary for intestinal lesion formation in CDCD piglets. These results suggest that anti-intimin vaccines might interfere with EHEC infections. Such vaccines could help reduce the levels of EHEC in cattle and thus reduce the number of EHEC infections in humans. The CDCD piglet EHEC infection model will be useful for preliminary experiments to test the efficacy of anti-intimin vaccines.
Acknowledgments
This work was partly supported by grant AI20148-15 from the National Institutes of Health to Alison D. O’Brien.
We thank M. I. Inbody, N. C. Lyon, R. W. Morgan, R. A. Schneider, R. J. Spaete, and J. A. Stasko for technical assistance.
REFERENCES
- 1.Altekruse S F, Cohen M L, Swerdlow D L. Emerging foodborne diseases. Emerg Infect Dis. 1997;3:285–293. doi: 10.3201/eid0303.970304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caprioli A, Tozzi A E, Rizzoni G, Karch H. Non-O157:H7 Shiga toxin-producing Escherichia coli infections in Europe. Emerg Infect Dis. 1997;3:578–579. doi: 10.3201/eid0304.970425. . (Letter.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. Outbreak of acute gastroenteritis attributable to Escherichia coli serotype O104:H21—Helena, Montana 1994. Morbid Mortal Weekly Rep. 1995;44:501–503. [PubMed] [Google Scholar]
- 4.Dean E A, Whipp S C, Moon H W. Age-specific colonization of porcine intestinal epithelium by 987P-piliated enterotoxigenic Escherichia coli. Infect Immun. 1989;57:82–87. doi: 10.1128/iai.57.1.82-87.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dean-Nystrom E A, Bosworth B T, Cray W C, Jr, Moon H W. Pathogenicity of Escherichia coli O157:H7 in the intestines of neonatal calves. Infect Immun. 1997;65:1842–1848. doi: 10.1128/iai.65.5.1842-1848.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donnenberg M S, Kaper J B. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donnenberg M S, Kaper J B. Enteropathogenic Escherichia coli. Infect Immun. 1992;60:3953–3961. doi: 10.1128/iai.60.10.3953-3961.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donnenberg M S, Tzipori S, McKee M L, O’Brien A D, Alroy J, Kaper J B. The role of the eae gene of enterohemorrhagic Escherichia coli in intimate attachment in vitro and in a porcine model. J Clin Invest. 1993;92:1418–1424. doi: 10.1172/JCI116718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ito H, Terai A, Kurazono H, Takeda Y, Nishibuchi M. Cloning and nucleotide sequencing of Vero toxin 2 varian genes from Escherichia coli O91:H21 isolated from a patient with the hemolytic uremic syndrome. Microb Pathog. 1990;8:47–60. doi: 10.1016/0882-4010(90)90007-d. [DOI] [PubMed] [Google Scholar]
- 10.Jerse A E, Kaper J B. The eae gene of enteropathogenic Escherichia coli encodes a 94-kilodalton membrane protein, the expression of which is influenced by the EAF plasmid. Infect Immun. 1991;59:4302–4309. doi: 10.1128/iai.59.12.4302-4309.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jerse A E, Yu J, Tall B D, Kaper J B. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc Natl Acad Sci USA. 1990;87:7839–7843. doi: 10.1073/pnas.87.20.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson W M, Lior H, Bezanson G S. Cytotoxic Escherichia coli O157:H7 associated with haemorrhagic colitis in Canada. Lancet. 1983;i:76. doi: 10.1016/s0140-6736(83)91616-1. [DOI] [PubMed] [Google Scholar]
- 13.Knutton S. Attaching and effacing Escherichia coli. In: Gyles C L, editor. Escherichia coli in domestic animals and humans. Wallingford, United Kingdom: CAB International; 1994. pp. 567–591. [Google Scholar]
- 14.Levine M M, Xu J, Kaper J B, Lior H, Prado V, Tall B, Nataro J, Karch H, Wachsmuth K. A DNA probe to identify enterohemorrhagic Escherichia coli of O157:H7 and other serotypes that cause hemorrhagic colitis and hemolytic uremic syndrome. J Infect Dis. 1987;156:175–182. doi: 10.1093/infdis/156.1.175. [DOI] [PubMed] [Google Scholar]
- 15.McKee M L, Melton-Celsa A R, Moxley R A, Francis D H, O’Brien A D. Enterohemorrhagic Escherichia coli O157:H7 requires intimin to colonize the gnotobiotic pig intestine and to adhere to HEp-2 cells. Infect Immun. 1995;63:3739–3744. doi: 10.1128/iai.63.9.3739-3744.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McKee M L, O’Brien A D. Investigation of enterohemorrhagic Escherichia coli O157:H7 adherence characteristics and invasion reveals a new attachment pattern shared by intestinal E. coli. Infect Immun. 1995;63:2070–2074. doi: 10.1128/iai.63.5.2070-2074.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Melton-Celsa A R, Darnell S C, O’Brien A D. Activation of Shiga-like toxins by mouse and human intestinal mucus correlates with virulence of enterohemorrhagic Escherichia coli O91:H21 isolates in orally infected, streptomycin-treated mice. Infect Immun. 1996;64:1569–1576. doi: 10.1128/iai.64.5.1569-1576.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moon H W, Sorensen D K, Sautter J H. Experimental enteric colibacillosis in piglets. Can J Comp Med. 1968;32:493–497. [PMC free article] [PubMed] [Google Scholar]
- 19.O’Brien A D, Melton A R, Schmitt C K, McKee M L, Batts M L, Griffin D E. Profile of Escherichia coli O157:H7 pathogen responsible for hamburger-borne outbreak of hemorrhagic colitis and hemolytic uremic syndrome in Washington. J Clin Microbiol. 1993;31:2799–2801. doi: 10.1128/jcm.31.10.2799-2801.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su C, Brandt L J. Escherichia coli O157:H7 infection in humans. Ann Intern Med. 1995;123:698–714. doi: 10.7326/0003-4819-123-9-199511010-00009. [DOI] [PubMed] [Google Scholar]
- 21.Wells J G, Davis B R, Wachsmuth I K, Riley L W, Remis R S, Sokolow R, Morris G K. Laboratory investigation of hemorrhagic colitis outbreaks associated with a rare Escherichia coli serotype. J Clin Microbiol. 1983;18:512–520. doi: 10.1128/jcm.18.3.512-520.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whipp S C, Rasmussen M A, Cray W C., Jr Animals as a source of Escherichia coli pathogenic for human beings. J Am Vet Med Assoc. 1994;204:1168–1175. [PubMed] [Google Scholar]