The question of how bacteria are able to overcome species barriers and adapt to new hosts is central to the understanding of both the origin of infectious diseases and the emergence of new pathogens. The analysis of virulence factors used by different Salmonella serotypes can serve as a powerful model for studying mechanisms of host adaptation because these pathogens are physiologically well characterized and lend themselves to genetic analysis. However, they differ greatly with regard to host range and their degree of host adaptation. Salmonella serotypes are closely related as shown by analysis of orthologous genes. Divergence in the nucleotide sequence of orthologous genes ranges between 3.8 and 4.6% and differences in their deduced amino acid sequences range between 0.7 and 1.3% (108). This close DNA relatedness among Salmonella serotypes is evidence for their clonal origin, and based on the degree of sequence divergence, it can be estimated that a common ancestor of the genus existed about 25 to 40 million years ago. Which factors contributed to the clonal divergence of the genus Salmonella from its common ancestor to give rise to serotypes that differ with regard to their host range?
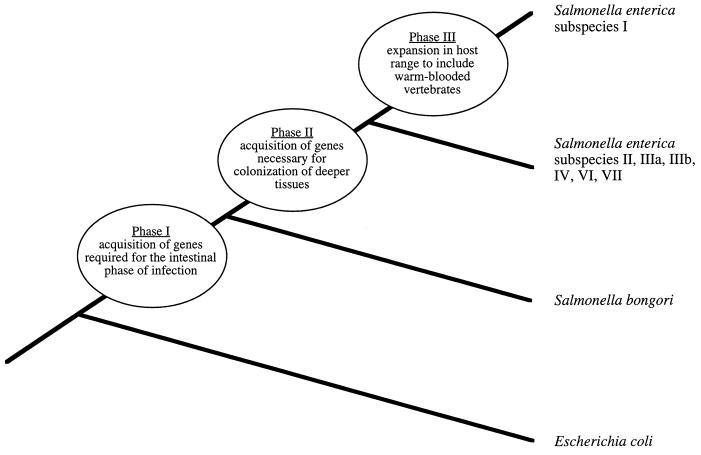
It has been postulated recently that in the genus Salmonella virulence evolved in three phases (11) (Fig. 1). The first phase involved acquisition of Salmonella pathogenicity island 1 (SPI 1) by plasmid- or phage-mediated horizontal gene transfer. SPI 1 was likely obtained by a lineage ancestral to all Salmonella serotypes, since it is present in all phylogenetic lineages of the genus Salmonella but absent from Escherichia coli and other related organisms (70, 79, 85). SPI 1 encodes virulence factors that mediate mechanisms used by Salmonella serotypes during the intestinal phase of infection, including invasion of intestinal epithelial cells (48, 62, 120), induction of neutrophil recruitment (49, 77), and secretion of intestinal fluid (49). Thus, it is likely that these or related virulence mechanisms may have existed early in the evolution of Salmonella serotypes.
FIG. 1.
Model for the evolution of virulence in the genus Salmonella. The three phases in which virulence evolved in the genus Salmonella since its divergence from the E. coli lineage have been proposed previously (11). The phylogenetic tree is not drawn to scale. For explanation see the text.
Multilocus enzyme electrophoresis and comparative sequence analysis of housekeeping and rRNA genes revealed that the genus Salmonella contains two lineages that have diverged considerably from each other during evolution (24, 31, 97). By using genetic distance determined by multilocus enzyme electrophoresis and results of DNA-DNA hybridization studies as criteria, it has been proposed that these lineages represent two distinct species, designated Salmonella enterica and Salmonella bongori (Fig. 1) (69, 97). The formation of these two species could be considered a second phase in the evolution of virulence in the genus Salmonella, since it involved not only divergence of their lineages by point mutation but also acquisition of new virulence determinants by horizontal gene transfer. Serotypes belonging to S. enterica possess a second pathogenicity island, designated SPI 2, that is not present in S. bongori serotypes (55, 84). The virulence genes present on SPI 2 have an average G+C content of 41%, much lower than the overall G+C content of the S. enterica genome, which averages 52% (55). The limited phylogenetic distribution and the atypical G+C content of SPI 2 imply that this pathogenicity island was acquired horizontally after S. enterica branched from the S. bongori lineage (55, 84). A possible mechanism for acquisition of SPI 2 by horizontal transfer is suggested by its insertion into the S. enterica gene encoding tRNAVal, a DNA region which may facilitate integration of newly acquired genetic material because tRNA genes serve as an attachment site for bacteriophage (55, 59). Experiments performed with mice revealed that mutations in SPI 1 attenuate S. enterica serotype Typhimurium between 15- and 50-fold after oral injection but have no attenuating phenotype when the intestinal phase of infection is bypassed by intraperitoneal injection (17, 48). In contrast, mutations in SPI 2 attenuate S. enterica serotype Typhimurium more than 10,000-fold even after intraperitoneal inoculation (86, 109). Although the mechanism by which SPI 2 contributes to pathogenicity has not yet been elucidated, these data imply that virulence genes located on SPI 1 and SPI 2 are required at different stages, namely the intestinal and the systemic phases of infection, respectively.
Finally, the lineage of S. enterica is postulated to have branched into several distinct phylogenetic groups, which by current nomenclature are considered subspecies. The formation of one of these groups, S. enterica subspecies I, involved a dramatic expansion in host range: while S. bongori and S. enterica subspecies II, IIIa, IIIb, IV, VI, and VII are mainly associated with cold-blooded vertebrates, members of S. enterica subspecies I are most frequently isolated from avian and mammalian hosts (2, 93). The host adaptation of S. enterica subspecies I to warm-blooded vertebrates characterized a third phase in the evolution of virulence in the genus Salmonella and is the focus of this review.
LYMPH NODES, A NEW BARRIER ENCOUNTERED IN MAMMALS
Which new barriers did S. enterica subspecies I serotypes encounter in birds and mammals? One obstacle may have been the immune system of higher vertebrates, which is more highly developed and shows a higher level of organization than that of cold-blooded vertebrates. For instance, lymphoid aggregates located in the gut lamina propria of cold-blooded vertebrates appear only as solitary units (82) whereas gut-associated lymphoid nodules of mammals and birds are organized in complex organs, such as Peyer’s patches, tonsils, appendix, or the avian bursa of Fabricius (51, 76). In addition, in birds and mammals B-lymphocyte variants which arise after somatic hypermutation are selected in the germinal centers of lymphoid organs for improving the affinity to their antigen. In contrast, because the lymphoid organs of cold-blooded vertebrates lack germinal centers selection of B cells is impeded, and consequently, antibody affinity does not increase during the immune response (40, 123). Furthermore, the antibody repertoire of lower vertebrates, such as fish, amphibians, and reptiles, is much less diverse than that of mammals (39). B cells selected in germinal centers of higher vertebrates show isotypic switching and are accumulated as memory cells. In cold-blooded vertebrates, on the other hand, immunological memory is poorly developed (39) and repeated immunization with S. enterica induces only the production of immunoglobulin M antibodies in lizards (82), suggesting that isotype switching does not occur during infection with this pathogen.
Finally, the presence of more highly developed peripheral lymphoid filter organs constitutes yet another challenge during infection of warm-blooded hosts. Pathogens that are able to penetrate the intestinal mucosa in fish can spread unchecked to central parts of the body until they are filtered from the blood by phagocytes located in sinusoids of the spleen. During the adaptation to life on land vertebrates developed peripheral lymphoid organs which function as local filtering systems (65). Although less-developed lymph node-like structures have been reported in some reptiles (82), true lymph nodes are present only in mammals and some bird species (41, 42, 51, 123). In mammals, a large number of lymph nodes located at peripheral positions form an highly effective lymph filter system (41, 42). Thus, upon penetration of the intestinal mucosae of mammals, Salmonella serotypes are confronted by an effective barrier to further spread, namely macrophages that line the lymphatic sinuses of regional lymph nodes. In mammals this host defense mechanism can successfully limit bacterial expansion to the intestine, the gut-associated lymphoid tissue, and the mesenteric lymph node. For instance, humans infected with nontyphoidal Salmonella serotypes usually develop an acute gastroenteritis, but in only 1 to 7% of clinical cases do bacteria manage to pass through the mesenteric lymph node and cause bacteremia (19). Therefore, in order to produce systemic disease in warm-blooded hosts, microbial intruders must be able to breach the local defense formed by macrophages of regional lymph nodes.
The capability of S. bongori or S. enterica subspecies II, IIIa, IIIb, IV, VI, and VII serotypes to survive in macrophages of poikilothermic animals has not been studied. Experimental oral exposure of snakes, turtles, or lizards to isolates of S. enterica subspecies I, II, or III resulted in all cases in no overt signs of disease and no colonization of organs other than the intestinal tract (35–37, 111). It has therefore been speculated that Salmonella serotypes evolved in the alimentary tract of reptiles, where they developed from pathogens into commensal organisms (87, 102). In contrast, S. enterica subspecies I serotypes frequently colonize internal organs of warm-blooded animals and the ability to survive and multiply in cells of the reticuloendothelial system correlates with their capability to cause systemic disease in these hosts (8, 45). Since macrophages from distinct homeothermic animal species differ greatly in their abilities to neutralize a particular S. enterica serotype, adaptation to a new host may require adaptation to its mononuclear phagocytes. For instance, the human-adapted S. enterica serotype Typhi is able to survive in vitro in human macrophages but not in murine macrophages, whereas S. enterica serotype Typhimurium, which causes a systemic disease in mice, survives well in vitro in murine macrophages but not in human macrophages (117). Furthermore, these two S. enterica serotypes differ in their mechanism of bacterial internalization and their intracellular trafficking in human and mouse mononuclear phagocytes (3, 60, 61, 91). These data imply that the ability of S. enterica serotypes to cause systemic disease is directly related to the capability to withstand an assault by the macrophages of a given host. Thus it appears that mononuclear phagocytes are an important barrier that restricts the host range of Salmonella serotypes.
COEVOLUTIONARY COURSE TOWARD INCREASED VIRULENCE
Although S. enterica subspecies I contains 1,367 different serotypes (93), only one or a few are associated with the majority of cases of illness in a particular avian or mammalian species (Table 1). These serotypes show different degrees of host adaptation. Pathogens that lack host specificity, such as S. enterica serotype Typhimurium and S. enterica serotype Enteritidis, tend to be more frequently associated with disease in young animals than in adults, suggesting that they are not optimally adapted to cope with a fully mature immune system. Serotypes that are host specific, on the other hand, have acquired the ability to breach defense mechanisms in full-grown animals, as shown by their association, at similar rates, with illness in all age groups (Table 1). Furthermore, host-specific serotypes tend to be more virulent, as illustrated by the fact that they cause higher mortality rates.
TABLE 1.
Diseases caused by Salmonella subspecies I serotypes in humans and higher vertebrates
| Host species | Disease | S. enterica subspecies I serotype(s) most frequently encountered | Most susceptible age groups | Typical symptoms or sign(s) of disease | Reference |
|---|---|---|---|---|---|
| Humans | Salmonella enteritis | Typhimurium, Enteritidis | Children (<4 yr) | Diarrhea, dysentery, fever | 78 |
| Typhoid fever | Typhic | Children and adults | Septicemia, fevera | 78 | |
| Paratyphoid fever | Sendai; Paratyphi A, B, and Cc | Children and adults | Septicemia, fevera | 78 | |
| Cattle | Salmonellosis | Typhimurium | Calves (<8 wk) | Diarrhea, dysentery, septicemia, fever | 104 |
| Dublin | Calves and adult cattle | Diarrhea, dysentery, septicemia, abortion, fever | 99, 104 | ||
| Poultry | Pullorum disease | Pullorumc,d | Newly hatched birds | Diarrhea, septicemia | 27 |
| Fowl typhoid | Gallinarumc,d | Growing stock and adults | Diarrhea, comb discoloration, septicemia | 27 | |
| Avian paratyphoid | Enteritidis, Typhimurium | Newly hatched birds | Diarrhea, septicemia | 27 | |
| Sheep | Salmonellosis | Abortusovisc | Adult sheep | Septicemia, abortion, vaginal discharge | 90 |
| Lambs | Diarrhea, dysentery, septicemia | 90 | |||
| Typhimurium | Lambs | Diarrhea, dysentery, septicemia | 90 | ||
| Pigs | Pig paratyphoid | Choleraesuisc | Weaned and adult pigs | Skin discoloration, septicemia, feverb | 106 |
| Salmonellosis | Typhimurium | Weaned pigs (<4 mo) | Diarrhea | 106 | |
| Chronic paratyphoid | Typhisuis | Intermittent diarrhea | 7 | ||
| Horses | Salmonellosis | Abortusequic | Adult horses | Septicemia, abortion | 110 |
| Foals | Diarrhea, septicemia | 110 | |||
| Typhimurium | Foals | Diarrhea, septicemia | 110 | ||
| Wild rodents | Murine typhoid | Typhimurium, Enteritidis | Septicemia, fever | 28 |
Diarrhea develops only in about one third of typhoid fever patients and usually several days after the onset of fever.
Diarrhea is not a typical sign of pig paratyphoid but may develop by the third or fourth day of disease.
These serotypes have been most frequently associated with illness in the preantibiotic era but are now rare or have been eradicated in most developed countries.
Gallinarum and Pullorum are considered biotypes that belong to the same serotype.
It has been proposed that vertebrate pathogens which cause high mortality rates are new arrivals that are not well adapted to their hosts, because the death of an animal would destroy their habitat, thereby reducing both their transmissibility and fitness. According to this theory, these pathogens will evolve over time into less-virulent forms, which will reflect better adaptation to their hosts (26). However, in S. enterica subspecies I evolution has apparently driven host-pathogen interactions in the opposite direction, since strongly host-adapted serotypes tend to cause higher mortality rates than those with a broad host range. For instance, S. enterica serotype Typhi exhibits host specificity for humans in whom it causes typhoid fever, a disease which results in 12 to 32% mortality (78). In contrast, death occurs in less than 0.5% of cases of human illness caused by zoonotic Salmonella serotypes, such as S. enterica serotype Typhimurium or S. enterica serotype Enteritidis (78). What selective forces were responsible for the increase in virulence that accompanied the adaptation of Salmonella serotypes to humans?
On the basis of a mathematical model, Anderson and May (5) predicted that if transmissibility in a particular host-pathogen association is linked to a high level of virulence, then the coevolutionary course will be toward high virulence. During coevolution of Salmonella serotypes and their human host such a link between transmissibility and high virulence may have been provided by the ability to develop chronic carriage. A chronic carrier has the potential to infect a large number of individuals, thereby increasing transmissibility. Chronic carriage develops more frequently following systemic infection, such as that caused by the host-adapted S. enterica serotype Typhi. For instance, 1 to 4% of patients that recover from typhoid fever but only 0.2 to 0.6% of patients with nontyphoidal salmonellosis become chronic carriers (56, 81). Thus, the link between systemic infection and increased transmissibility resulting from chronic carriage may have exerted evolutionary pressure on the development of a high degree of virulence during the adaptation of typhoidal Salmonella serotypes (causing typhoid or paratyphoid fever in humans) to the human host.
THE VIRULENCE PLASMID IS IMPORTANT FOR SYSTEMIC DISEASE CAUSED BY NONTYPHOIDAL S. ENTERICA SEROTYPES
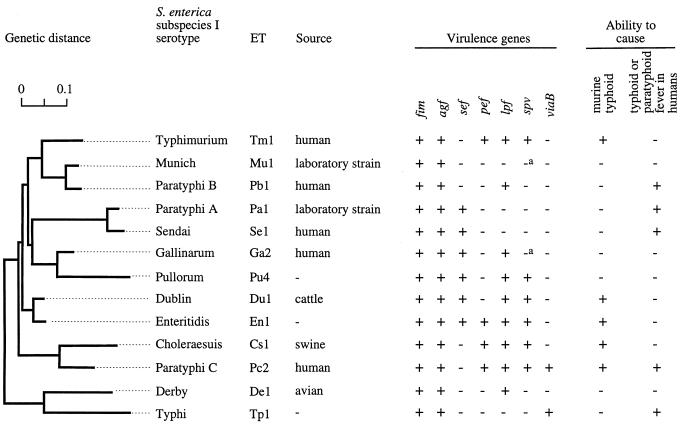
Although nontyphoidal S. enterica serotypes do not cause enteric fever in humans, some do cause similar systemic infections in warm-blooded animals. In S. enterica subspecies I, the spv (Salmonella plasmid virulence [54]) operon is found in only a few serotypes, including S. enterica serotypes Typhimurium, Enteritidis, Choleraesuis, Gallinarum/Pullorum, Abortusovis, Paratyphi C, and Dublin (17, 75, 94, 95, 103, 122) (Fig. 2). Interestingly, these serotypes are also among those most frequently associated with disease in homeothermic vertebrates (Table 1), suggesting a role for spv during infection of these hosts. The spv operon is required for the systemic phase of disease caused by S. enterica serotype Choleraesuis in pigs (34), S. enterica serotype Gallinarum/Pullorum in chickens (9, 10), S. enterica serotype Dublin in cattle (72, 118), and S. enterica serotypes Typhimurium and Enteritidis in mice (53, 63, 83). Epidemiological evidence provides support for the idea that the spv operon is also important for the pathogenesis of extra intestinal infections associated with nontyphoidal Salmonella serotypes in humans (46). Therefore, in a first approximation it appears that the spv operon is required for systemic infections caused by nontyphoidal serotypes in warm-blooded animals, including humans (9, 10, 34, 53, 63, 72, 83, 103, 118). Mechanisms for systemic infection that are spv-dependent may, however, not be restricted to nontyphoidal serotypes since the spv operon is present in S. enterica serotype Paratyphi C (17, 94). Typhoidal serotypes which lack the spv genes, such as S. enterica serotypes Typhi, Paratyphi A, Paratyphi B, and Sendai, produce enteric fever by an spv-independent mechanism. The virulence determinants responsible have not yet been identified (94, 100, 122).
FIG. 2.
Phylogenetic distribution of virulence genes and virulence of different S. enterica subspecies I serotypes in mice and humans. The bacterial isolates shown in this figure have been described previously (23). The left side shows a dendrogram reflecting the phylogenetic relatedness of these strains as reported by Selander and coworkers (23). ET, enzyme type determined by multilocus enzyme electrophoresis (23). The distribution of the spv (Salmonella plasmid virulence), lpf (long polar fimbriae), sef (S. enterica serotype Enteritidis fimbriae), agf (thin aggregative fimbriae), fim (type 1 fimbriae), and pef (plasmid-encoded fimbriae) operons among these isolates has been determined previously (12, 17). The presence in these strains of sequences related to viaB (Vi capsular antigen) is described elsewhere (107). Virulence of these isolates in humans or mice has been reported recently (17, 107). a, although missing in this strain, virulence plasmids are present in most isolates of this serotype.
Although all members of S. enterica subspecies I that are able to produce lethal infection in mice possess the spv operon, its introduction into S. enterica serotype Typhi does not confer mouse virulence to this host-restricted pathogen (103). Furthermore, the spv operon is present in most isolates of S. enterica serotype Gallinarum/Pullorum, which does not cause disease in mice. These data suggest that host-restricted S. enterica serotypes lack additional virulence factors that are encoded on the chromosome of serotypes with a broad host range. Furthermore, in addition to its presence in several serotypes of S. enterica subspecies I, the spv operon has also been detected in some isolates of S. bongori and S. enterica subspecies IIIa and IV (98), and thus the expansion in host range observed for subspecies I cannot be explained merely by the presence of these virulence genes. Therefore, the ability to cause systemic disease in a warm-blooded host is apparently a complex phenotype that cannot be attributed to acquisition of a single virulence determinant. It has been shown recently by subtractive hybridization analysis that 20% of the genome of S. enterica serotype Typhimurium is not present in S. enterica serotype Typhi and vice versa (67). By comparison with the similarly sized E. coli genome, the genome of S. enterica serotype Typhimurium can be estimated to contain approximately 4,400 genes. Serotype-specific DNA may thus encode as many as 880 genes, some of which may contribute to determining the host range. A combination of some of these genes, which are likely to be distributed throughout the chromosome, might determine the ability to infect a particular host. This idea may explain why attempts to extend the host range of host-restricted S. enterica serotypes by transferring small segments of the genome from a broad-host-range serotype have not succeeded.
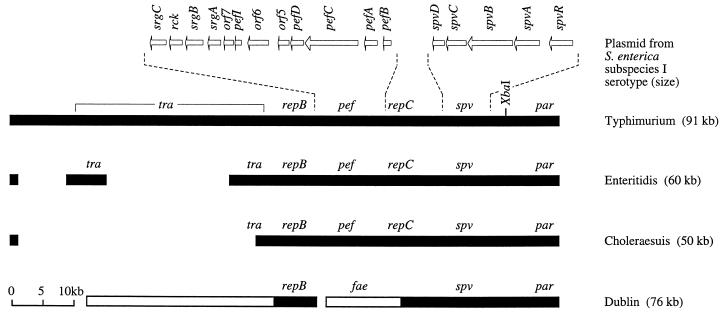
Although the distribution of the spv operon has not been determined for all phylogenetic lineages of the genus Salmonella, its localization on large serotype-specific plasmids in S. enterica subspecies I suggests that this operon was obtained by horizontal gene transfer. Additional regions of virulence plasmids, such as the origin of transfer and the tra genes, hint at conjugation as a possible mechanism by which the spv operon was horizontally transmitted (25, 89, 101). Evidence for the presence of the spv operon on a predecessor plasmid that gave rise to the virulence plasmids found among extant S. enterica serotypes comes from analysis of their patterns of homology. For instance, the virulence plasmids of S. enterica serotypes Enteritidis and Choleraesuis could be seen as variants of the virulence plasmid of S. enterica serotype Typhimurium that were generated by deletion events which occurred during their divergence from a common predecessor (Fig. 3) (25, 80, 101, 120). Similarly, a 25-kb DNA region containing the spv operon is conserved between virulence plasmids of S. enterica serotypes Dublin and Typhimurium, suggesting a common ancestry (18, 68). But where did the virulence plasmids of Salmonella serotypes originate?
FIG. 3.
Linear genetic maps of virulence plasmids of S. enterica subspecies I serotypes. Plasmids were linearized at their conserved par (partitioning) region (29, 113, 114). Regions of homology shared among plasmids are shown as closed bars, which are positioned according to their locations on the S. enterica serotype Typhimurium virulence plasmid. The distribution among virulence plasmids of spv (Salmonella plasmid virulence) (6, 66, 122), repC (replicon C) (113), pef (plasmid encoded fimbriae) (12, 47), repB (replicon B) (113), and the tra (conjugative plasmid transfer) region (66, 101) have been reported recently. Areas of the plasmid of S. enterica serotype Dublin that are not present in S. enterica serotype Typhimurium have been described previously and are shown as open bars (68, 80). The presence on the S. enterica serotype Dublin plasmid of sequences with homology to feaH and feaI, two genes involved in biosynthesis of K88 fimbriae in E. coli, have been reported by Barrow and coworkers (105). The location and size of deletions that may account for the smaller size of plasmids from S. enterica serotypes Choleraesuis and Enteritidis relative to the S. enterica serotype Typhimurium plasmid are reported elsewhere (25, 80, 101). Dashed lines indicate the position of two DNA regions, namely the pef and spv operons, which are shown in more detail above the S. enterica serotype Typhimurium plasmid (1, 47, 52). The positions of genes are indicated by arrows.
The genes finO, traY, and repA are present on the E. coli F plasmid and the virulence plasmids of S. enterica serotypes Enteritidis and Typhimurium (101). Comparison of the nucleotide sequences determined for the finO, traY, and repA genes of several natural isolates of E. coli and S. enterica revealed similar, average, pairwise differences within and between species (21). These data are indicative of one or more recent horizontal transfer events between E. coli and S. enterica and suggest that during formation of S. enterica subspecies I the spv operon was obtained from a F-like plasmid pool (21) shared with ancestral E. coli isolates and possibly other organisms. As proposed for ancestral E. coli strains (87), some of the organisms that contributed to this F-like plasmid pool may have been associated with higher vertebrates long before host adaptations to mammals or birds developed in S. enterica subspecies I. For example, the presence on S. enterica virulence plasmids of tlpA, a gene encoding a regulatory protein that is sensitive to changes from 28°C to physiological temperatures of 37 to 42°C, implies that the donor was already able to sense temperature differences encountered upon entry into warm-blooded vertebrates (58). The identity of this ancestral virulence plasmid donor organism, if extant, remains to be discovered.
ADHESION TO THE MUCOSAL SURFACE AS A MECHANISM FOR HOST ADAPTATION
Salmonella serotypes initiate infection by attaching to the intestinal mucosa of the host. Recent evidence suggests that the recognition of intestinal surfaces by adhesins may contribute to the host adaptation of Salmonella serotypes. A nonfimbrial adhesin which may play a role in host adaptation is encoded by invH, a gene which is present within SPI 1 in S. enterica (22). The idea that invH is involved in adherence rests on the finding that mutational inactivation reduces both attachment to and invasion of cultured epithelial cells by S. enterica serotypes (4). In contrast, mutations in invA and invE, two genes that are part of the type III secretory apparatus encoded on SPI 1, render S. enterica serotypes noninvasive but have no effect on adhesion to cultured epithelial cells (48, 50). The observation that SPI 1-mediated adhesion and invasion are independent events which can be genetically separated suggests that entry of S. enterica into epithelial cells is a two-step process and that InvH may function as an adhesin which mediates an initial attachment step required for invasion (4, 20, 44).
A comparison of the effect on virulence of a mutation in invH with those in genes encoding the invasion-associated type III export apparatus illustrates the importance of adherence during interaction with different hosts. S. enterica serotype Dublin secretes SopB, a virulence factor which is required for eliciting inflammation and fluid secretion in calves, and its translocation into host cells can be abolished by a polar mutation in sipB, a gene located on SPI 1 (49, 121). Similarly, mutations in invA, invB, invC, and sipC render S. enterica serotypes Enteritidis and Typhimurium avirulent in newly hatched chicks (96, 115). Thus, the type III exporter encoded by SPI 1 is apparently required for S. enterica virulence in both chickens and cattle. However, a mutation in the S. enterica serotype Typhimurium invH gene has no effect on oral virulence or colonization of chicks but significantly reduces the severity of enteritis in calves, suggesting a role of the encoded adhesin in adaptation to bovine but not avian hosts (96, 119, 120).
In chicks, the role of InvH in mediating an initial attachment step required for invasion is likely fulfilled by an alternate adhesin. The various fimbrial operons present in S. enterica are possible candidates for encoding such alternate attachment factors involved in host recognition (15, 32). Like binding mediated by InvH, attachment through fimbrial adhesins has been shown to affect entry of S. enterica serotype Typhimurium into some epithelial cell lines (4, 14, 43, 57, 64, 112). In a murine intestinal organ culture model, adhesins encoded by the pef (plasmid-encoded fimbriae) and lpf (long polar fimbriae) operons mediate tissue tropism of S. enterica serotype Typhimurium to villous small intestine and Peyer’s patches, respectively (13, 16). These data suggest that adhesins help to select which epithelial surfaces are colonized by S. enterica in a given animal, a mechanism which is likely to be of importance for determining host range. Analysis of bacterial binding to the mucosa may, in some cases, be complicated by the large number of apparently redundant adhesins. For instance, in the mouse the finding that multiple fimbrial operons are required for full virulence of S. enterica serotype Typhimurium suggests that alternative attachment factors can compensate for the loss of a single adhesin (116).
The repertoire of adhesins expressed by a pathogen is thought to determine which structures are recognized and bound on the surface of intestinal epithelial cells. S. enterica serotype Typhi utilizes the cystic fibrosis transmembrane conductance regulator as a receptor for internalization by intestinal epithelial cells. In contrast, S. enterica serotype Typhimurium appears to use a different epithelial cell receptor for mucosal translocation, because mutations in the cystic fibrosis transmembrane conductance regulator do not reduce the invasiveness of this pathogen for epithelial cell lines or the intestinal wall of transgenic mice (92). This use of different receptors during infection implies the involvement of distinct adhesins which may contribute to differences in host range observed for S. enterica serotypes Typhi and Typhimurium. New combinations of adhesion determinants, generated through horizontal gene transfer and deletion events, may thus have contributed to shifts in host range during evolution of S. enterica serotypes (12).
THE HOST SPECIFICITY GAMBIT
The process of adapting to a host may involve not only acquisition of virulence determinants, such as adhesins or the virulence plasmid, but also loss of gene function. The S. enterica biotypes Gallinarum and Pullorum are both members of the same S. enterica subspecies I serotype (antigen formula 1,9,12:-:-) and show host specificity for poultry and aquatic birds. Multilocus enzyme electrophoresis and comparative sequence analysis revealed that S. enterica biotypes Gallinarum and Pullorum are closely related (71). It has been speculated that their lineage evolved from an S. enterica serotype Enteritidis-like ancestor that had a broad range of hosts, including birds. Since divergence from this ancestor, the ability both to mediate mannose-sensitive hemagglutination (MSHA) and to express flagella (and hence motility) was lost in the S. enterica serotype Gallinarum/Pullorum lineage as a result of point mutation (33, 71, 73, 88). Interestingly, strains of S. enterica serotype Enteritidis and S. enterica serotype Typhimurium that are isolated from cases of avian disease are also frequently nonmotile and lack MSHA (30, 38), suggesting that the niche occupied in birds may select against type 1 fimbriation and flagellation. Although it is arguably an advantage during infection of avian hosts, the simultaneous loss of motility and MSHA results in 100-fold-reduced virulence of S. enterica serotype Typhimurium in mice (74). It is tempting to speculate that the selection for point mutations in type 1 fimbrial and flagellar biosynthesis genes that occurred during the adaptation to avian hosts may account in part for the loss of virulence in mice in the S. enterica serotype Gallinarum/Pullorum lineage (Fig. 2). Thus, in S. enterica subspecies I more-complete adaptation to a particular vertebrate host appears to have evolved, in some cases, at the expense of virulence traits that were important for infection of a wider spectrum of animals, resulting in the loss of virulence for some species. The result of this adaptation was a narrowing of the host range and the development of host specificity.
However, the development of host specificity in S. enterica biotype Gallinarum cannot be explained by these point mutations alone. Other important changes which may have reduced virulence in mice have also been found. For instance, S. enterica serotype Gallinarum/Pullorum appears to be incapable of entering the follicle-associated epithelium of murine Peyer’s patches, is internalized by murine macrophages by a mechanism different from that of S. enterica serotype Typhimurium, and is unable to survive and multiply in cells of the mouse reticuloendothelial system in vivo and in vitro (3, 8, 91). Thus, loss of virulence is a complex phenotype which involved multiple changes that accumulated in the lineage of S. enterica serotype Gallinarum/Pullorum during its evolution toward host specificity.
PERSPECTIVE
Despite the rapid progress in identifying some of the key elements of S. enterica virulence by studying the pathogenesis of murine typhoid, our knowledge about genes and mechanisms involved in the expression of host specificity is still very limited. Studies on the distribution of pathogenicity islands, fimbrial operons, and capsular biosynthesis genes among S. enterica serotypes suggest that during evolution, new combinations of virulence determinants arose through multiple horizontal transfer events (Fig. 2), a process which may have driven the development of host adaptation. In addition, deletion events and sequence divergence by point mutation were likely among the events which contributed to changes in the host ranges of S. enterica serotypes. Thus, adaptation to an animal species is a complex phenotype that doubtless involves a large number of gene products. It is not clear which genetic changes account for the adaptation to a particular mammalian or avian species, and it is likely that many of the responsible virulence mechanisms have not been identified to date. For instance, which virulence factors allowed S. enterica subspecies I to breach the defense formed by regional lymph nodes and to spread to the systemic sites of infection? It is also not known which virulence factors are important during infections that remain localized in the intestine and mesenteric lymph node, such as the gastroenteritis caused by most S. enterica subspecies I serotypes in humans. Future research into the genetic basis of host adaptation will provide answers to these and other open questions regarding the pathogenesis of S. enterica.
ACKNOWLEDGMENTS
We thank Robert A. Kingsley and Stacy M. Townsend for their comments on the manuscript.
Work in A.J.B.’s laboratory is supported by grant AI40124 from the National Institute of Health and a research grant from USDA Formula Animal Health Funds.
REFERENCES
- 1.Ahmer B M M, van Reeuwijk J, Timmers C D, Valentine P J, Heffron F. Salmonella typhimurium encodes an SdiA homolog, a putative quorum sensor of the LuxR family, that regulates genes on the virulence plasmid. J Bacteriol. 1998;180:1185–1193. doi: 10.1128/jb.180.5.1185-1193.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aleksic S, Heinzerling F, Bockemühl J. Human infection caused by salmonellae of subspecies II to VI in Germany, 1977–1992. Zentbl Bakteriol. 1996;283:391–398. doi: 10.1016/s0934-8840(96)80074-0. [DOI] [PubMed] [Google Scholar]
- 3.Alpuche-Aranda C M, Berthiaume E P, Mock B, Swanson J A, Miller S I. Spacious phagosome formation within mouse macrophages correlates with Salmonella serotype pathogenicity and host susceptibility. Infect Immun. 1995;63:4456–4462. doi: 10.1128/iai.63.11.4456-4462.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altmeyer R M, McNern J K, Bossio J C, Rosenshine I, Finlay B B, Galán J E. Cloning and molecular characterization of a gene involved in Salmonella adherence and invasion of cultured epithelial cells. Mol Microbiol. 1993;7:89–98. doi: 10.1111/j.1365-2958.1993.tb01100.x. [DOI] [PubMed] [Google Scholar]
- 5.Anderson R M, May R M. Coevolution of host and parasites. Parasitology. 1982;85:411–426. doi: 10.1017/s0031182000055360. [DOI] [PubMed] [Google Scholar]
- 6.Baird G D, Manning E J, Jones P W. Evidence for related virulence sequences in plasmids of Salmonella dublin and Salmonella typhimurium. J Gen Microbiol. 1985;131:1815–1823. doi: 10.1099/00221287-131-7-1815. [DOI] [PubMed] [Google Scholar]
- 7.Barnes D M, Bergland M E. Salmonella typhisuis infection in Minnesota swine. J Am Vet Med Assoc. 1968;152:1766–1770. [PubMed] [Google Scholar]
- 8.Barrow P A, Huggins M B, Lovell M A. Host specificity of Salmonella infection in chickens and mice is expressed in vivo primarily at the level of the reticuloendothelial system. Infect Immun. 1994;62:4602–4610. doi: 10.1128/iai.62.10.4602-4610.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barrow P A, Lovell M A. The association between a large molecular mass plasmid and virulence in a strain of Salmonella pullorum. J Gen Microbiol. 1988;134:2307–2316. doi: 10.1099/00221287-134-8-2307. [DOI] [PubMed] [Google Scholar]
- 10.Barrow P A, Simpson J M, Lovell M A, Binn M M. Contribution of Salmonella gallinarum large plasmid toward virulence in fowl typhoid. Infect Immun. 1987;55:388–392. doi: 10.1128/iai.55.2.388-392.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bäumler A J. The record of horizontal gene transfer in Salmonella. Trends Microbiol. 1997;5:318–322. doi: 10.1016/S0966-842X(97)01082-2. [DOI] [PubMed] [Google Scholar]
- 12.Bäumler A J, Gilde A J, Tsolis R M, van der Velden A W M, Ahmer B M M, Heffron F. Contribution of horizontal gene transfer and deletion events to the development of distinctive patterns of fimbrial operons during evolution of Salmonella serotypes. J Bacteriol. 1997;179:317–322. doi: 10.1128/jb.179.2.317-322.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bäumler A J, Tsolis R M, Bowe F, Kusters J G, Hoffmann S, Heffron F. The pef fimbrial operon mediates adhesion to murine small intestine and is necessary for fluid accumulation in infant mice. Infect Immun. 1996;64:61–68. doi: 10.1128/iai.64.1.61-68.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bäumler A J, Tsolis R M, Heffron F. Contribution of fimbrial operons to attachment to and invasion of epithelial cell lines by Salmonella typhimurium. Infect Immun. 1996;64:1862–1865. doi: 10.1128/iai.64.5.1862-1865.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bäumler A J, Tsolis R M, Heffron F. Fimbrial adhesins of Salmonella typhimurium—role in bacterial interactions with epithelial cells. In: Paul P S, Francis D H, Benfield D, editors. Mechanisms in the pathogenesis of enteric diseases. Vol. 412. New York, N.Y: Plenum Press; 1997. pp. 149–158. [PubMed] [Google Scholar]
- 16.Bäumler A J, Tsolis R M, Heffron F. The lpf fimbrial operon mediates adhesion to murine Peyer’s patches. Proc Natl Acad Sci USA. 1996;93:279–283. doi: 10.1073/pnas.93.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bäumler A J, Tsolis R M, Valentine P J, Ficht T A, Heffron F. Synergistic effect of mutations in invA and lpfC on the ability of Salmonella typhimurium to cause murine typhoid. Infect Immun. 1997;65:2254–2259. doi: 10.1128/iai.65.6.2254-2259.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beninger P R, Chikami G, Tanabe K, Roudier C, Fierer J, Guiney D G. Physical and genetic mapping of the Salmonella dublin virulence plasmid pSDL2. Relationship to plasmids from other Salmonella strains. J Clin Investig. 1988;81:1341–1347. doi: 10.1172/JCI113461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blaser M J, Feldman R A. Salmonella bacteremia: reports from the centers of disease control and prevention. J Infect Dis. 1981;143:743–746. doi: 10.1093/infdis/143.5.743. [DOI] [PubMed] [Google Scholar]
- 20.Bliska J B, Galán J E, Falkow S. Signal transduction in the mammalian cell during bacterial attachment and entry. Cell. 1993;73:903–920. doi: 10.1016/0092-8674(93)90270-z. [DOI] [PubMed] [Google Scholar]
- 21.Boyd E F, Hartl D L. Recent horizontal transmission of plasmids between natural populations of Escherichia coli and Salmonella enterica. J Bacteriol. 1997;179:1622–1627. doi: 10.1128/jb.179.5.1622-1627.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boyd E F, Li J, Ochman H, Selander R K. Comparative genetics of the inv-spa invasion gene complex of Salmonella enterica. J Bacteriol. 1997;179:1985–1991. doi: 10.1128/jb.179.6.1985-1991.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boyd E F, Wang F-S, Beltran P, Plock S A, Nelson K, Selander R K. Salmonella reference collection B (SARB): strains of 37 serovars of subspecies I. J Gen Microbiol. 1993;139:1125–1132. doi: 10.1099/00221287-139-6-1125. [DOI] [PubMed] [Google Scholar]
- 24.Boyd E F, Wang F-S, Whittam T S, Selander R K. Molecular genetic relationship of the salmonellae. Appl Environ Microbiol. 1996;62:804–808. doi: 10.1128/aem.62.3.804-808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buisán M, Rodrígues-Pena J M, Rotger R. Restriction map of the Salmonella enteritidis virulence plasmid and its homology with the plasmid of Salmonella typhimurium. Microb Pathog. 1994;16:165–169. doi: 10.1006/mpat.1994.1017. [DOI] [PubMed] [Google Scholar]
- 26.Burnet M, White D O. Natural history of infectious disease. Cambridge, United Kingdom: Cambridge University Press; 1972. [Google Scholar]
- 27.Buxton A, Fraser G. Animal microbiology. Vol. 1. London, United Kingdom: Blackwell Scientific Publications; 1977. [Google Scholar]
- 28.Carter P B, Collins F M. The route of enteric infection in normal mice. J Exp Med. 1974;139:1189–1203. doi: 10.1084/jem.139.5.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cerin H, Hackett J. The parVP region of the Salmonella typhimurium virulence plasmid pSLT contains four loci required for incompatibility and partitioning. Plasmid. 1993;30:30–38. doi: 10.1006/plas.1993.1031. [DOI] [PubMed] [Google Scholar]
- 30.Chart H, Conway D, Rowe B. Outer membrane characteristics of Salmonella enteritidis phage type-4 growing in chickens. Epidemiol Infect. 1993;111:449–454. doi: 10.1017/s0950268800057174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Christensen H, Nordentoft S, Olsen J E. Phylogenetic relationships of Salmonella based on rRNA sequences. Int J System Bacteriol. 1998;48:605–610. doi: 10.1099/00207713-48-2-605. [DOI] [PubMed] [Google Scholar]
- 32.Clegg S, Swenson D L. Salmonella fimbriae. In: Klemm P, editor. Fimbriae: adhesion, genetics, biogenesis, and vaccines. Boca Raton, Fla: CRC Press; 1994. pp. 105–113. [Google Scholar]
- 33.Crichton P B, Yakubu D E, Old D C, Clegg S. Immunological and genetical relatedness of type 1 and type 2 fimbriae in salmonellas of serotype Gallinarum, Pullorum and Typhimurium. J Appl Bacteriol. 1989;67:283–291. doi: 10.1111/j.1365-2672.1989.tb02497.x. [DOI] [PubMed] [Google Scholar]
- 34.Danbara H, Moriguchi R, Suzuki S, Tamura Y, Kijima M, Oishi K, Matsui H, Abe A, Nakamura M. Effect of 50 kilobase-plasmid, pKDSC50, of Salmonella choleraesuis RF-1 strain on pig septicemia. J Vet Med Sci. 1992;54:1175–1178. doi: 10.1292/jvms.54.1175. [DOI] [PubMed] [Google Scholar]
- 35.Dimow I. Versuche Landschidkröten der Arten Testudo graeca und Testudo hemanni mit Salmonella-bakterien zu infizieren. Zentbl Bakteriol Parasitenkd Hyg Abt I Orig A. 1966;199:181–184. [PubMed] [Google Scholar]
- 36.Dimow I. Versuche zur künstlichen Infektion von Eidechsen mit Salmonella und Arizona-bakterien. Zentbl Vetmed Reihe B. 1966;13:587–590. [PubMed] [Google Scholar]
- 37.Dimow I, Salwtschew R. Versuche der Experimentalinfizierung von Schlangen Vipera ammodytes mit Salmonella und Arizona-bakterien. Pathol Microbiol. 1967;30:495–497. [PubMed] [Google Scholar]
- 38.Duguid J P, Anderson E S, Alfredsson G A, Barker R, Old D C. A new biotyping scheme for Salmonella typhimurium and its phylogenetic significance. J Med Microbiol. 1975;8:149–166. doi: 10.1099/00222615-8-1-149. [DOI] [PubMed] [Google Scholar]
- 39.Du Pasquier L. Antibody diversity in lower vertebrates—why is it so restricted? Nature. 1982;290:311–313. doi: 10.1038/296311a0. [DOI] [PubMed] [Google Scholar]
- 40.Du Pasquier L. Phylogeny of B-cell development. Curr Opin Immunol. 1993;5:185–193. doi: 10.1016/0952-7915(93)90003-b. [DOI] [PubMed] [Google Scholar]
- 41.Du Pasquire L. Evolution of the immune system. In: Paul W E, editor. Fundamental immunology. 2nd ed. New York, N.Y: Raven Press, Ltd.; 1989. pp. 139–165. [Google Scholar]
- 42.Du Pasquire L. Origin and evolution of the vertebrate immune system. APMIS. 1992;100:383–392. doi: 10.1111/j.1699-0463.1992.tb00888.x. [DOI] [PubMed] [Google Scholar]
- 43.Ernst R K, Dombroski D M, Merrick J M. Anaerobiosis, type 1 fimbriae, and growth phase are factors that affect invasion of HEp-2 cells by Salmonella typhimurium. Infect Immun. 1990;58:2014–2016. doi: 10.1128/iai.58.6.2014-2016.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Falkow S, Isberg R R, Portnoy D A. The interaction of bacteria with mammalian cells. Annu Rev Cell Biol. 1992;8:333–363. doi: 10.1146/annurev.cb.08.110192.002001. [DOI] [PubMed] [Google Scholar]
- 45.Fields P I, Swanson R V, Haidaris C G, Heffron F. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc Natl Acad Sci USA. 1986;83:5189–5193. doi: 10.1073/pnas.83.14.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fierer J, Krause M, Tauxe R, Guiney D. Salmonella typhimurium bacteremia: association with the virulence plasmid. J Infect Dis. 1992;166:639–642. doi: 10.1093/infdis/166.3.639. [DOI] [PubMed] [Google Scholar]
- 47.Friedrich M J, Kinsey N E, Vila J, Kadner R J. Nucleotide sequence of a 13.9 kb segment of the 90 kb virulence plasmid of Salmonella typhimurium: the presence of fimbrial biosynthetic genes. Mol Microbiol. 1993;8:543–558. doi: 10.1111/j.1365-2958.1993.tb01599.x. [DOI] [PubMed] [Google Scholar]
- 48.Galán J E, Curtiss R., III Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc Natl Acad Sci USA. 1989;86:6383–6387. doi: 10.1073/pnas.86.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Galyov E E, Wood M W, Rosqvist R, Mullan P B, Watson P R, Hedges S, Wallis T S. A secreted effector protein of Salmonella dublin is translocated into eukaryotic cells and mediates inflammation and fluid secretion in infected ileal mucosa. Mol Microbiol. 1997;25:903–912. doi: 10.1111/j.1365-2958.1997.mmi525.x. [DOI] [PubMed] [Google Scholar]
- 50.Ginocchio C, Pace J, Galán J E. Identification and characterization of a Salmonella typhimurium gene involved in triggering the internalization of salmonellae into cultured epithelial cells. Proc Natl Acad Sci USA. 1992;89:5976–5980. doi: 10.1073/pnas.89.13.5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Glick B. RES structure and function in aves. In: Cohen N, Sigel M M, editors. The reticuloendothelial system. Vol. 3. New York, N.Y: Plenum Press; 1982. pp. 509–540. [Google Scholar]
- 52.Gulig P A, Caldwell A L, Chiodo V A. Identification, genetic analysis and DNA sequence of a 7.8-kb virulence region of the Salmonella typhimurium virulence plasmid. Mol Microbiol. 1992;6:1395–1411. doi: 10.1111/j.1365-2958.1992.tb00860.x. [DOI] [PubMed] [Google Scholar]
- 53.Gulig P A, Curtiss R. Plasmid-associated virulence of Salmonella typhimurium. Infect Immun. 1987;1987:2891–2901. doi: 10.1128/iai.55.12.2891-2901.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gulig P A, Danbara H, Guiney D G, Lax A J, Norel F, Rhen M. Molecular analysis of spv virulence genes of the Salmonella virulence plasmids. Mol Microbiol. 1993;7:825–830. doi: 10.1111/j.1365-2958.1993.tb01172.x. [DOI] [PubMed] [Google Scholar]
- 55.Hensel M, Shea J E, Bäumler A J, Gleeson C, Blattner F, Holden D W. Analysis of the boundaries of Salmonella pathogenicity island 2 and the corresponding chromosomal region of Escherichia coli K-12. J Bacteriol. 1997;179:1105–1111. doi: 10.1128/jb.179.4.1105-1111.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hoffman T A, Ruiz C J, Counts G W, Sachs J M, Nitzkin J L. Waterborne typhoid fever in Dade County, Florida. Clinical and therapeutic evaluation of 105 bacteremic patients. Am J Med. 1975;59:481–487. doi: 10.1016/0002-9343(75)90255-7. [DOI] [PubMed] [Google Scholar]
- 57.Horiuchi S, Inagaki Y, Okamura N, Nakaya R, Yamamoto N. Type 1 pili enhance the invasion of Salmonella baenderup and Salmonella typhimurium to HeLa cells. Microbiol Immunol. 1992;36:593–602. doi: 10.1111/j.1348-0421.1992.tb02059.x. [DOI] [PubMed] [Google Scholar]
- 58.Hurme R, Berndt K D, Normark S J, Rhen M. A proteinaceous gene regulatory thermometer in salmonella. Cell. 1997;90:55–64. doi: 10.1016/s0092-8674(00)80313-x. [DOI] [PubMed] [Google Scholar]
- 59.Inouye S, Sunshine M G, Six E W, Inouye M. Retrophage ΦR73: an E. coli phage that contains a retroelement and integrates into a t-RNA gene. Science. 1991;252:969–971. doi: 10.1126/science.1709758. [DOI] [PubMed] [Google Scholar]
- 60.Ishibashi Y, Arai T. A possible mechanism for host-specific pathogenesis of Salmonella serovars. Microb Pathog. 1996;21:435–446. doi: 10.1006/mpat.1996.0074. [DOI] [PubMed] [Google Scholar]
- 61.Ishibashi Y, Arai T. Roles of the complement receptor type 1 (CR1) and type 3 (CR3) on phagocytosis and subsequent phagosome-lysosome fusion in Salmonella-infected murine macrophages. FEMS Microbiol Lett. 1990;64:89–96. doi: 10.1111/j.1574-6968.1990.tb03505.x. [DOI] [PubMed] [Google Scholar]
- 62.Jones B D, Ghori N, Falkow S. Salmonella typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of the Peyer’s patches. J Exp Med. 1994;180:15–23. doi: 10.1084/jem.180.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jones G W, Rabert D K, Svinarich M, Whitfield H J. Association of adhesive, invasive, and virulent phenotypes of Salmonella typhimurium with autonomous 60-megadalton plasmids. Infect Immun. 1982;38:476–86. doi: 10.1128/iai.38.2.476-486.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jones G W, Richardson L A. The attachment to and invasion of HeLa cells by Salmonella typhimurium: the contribution of mannose-sensitive and mannose-resistant haemagglutinating activities. J Gen Microbiol. 1981;127:361–370. doi: 10.1099/00221287-127-2-361. [DOI] [PubMed] [Google Scholar]
- 65.Jonsson V. Comparison and definition of spleen and lymph node: a phylogenetic analysis. J Theor Biol. 1985;117:691–699. doi: 10.1016/s0022-5193(85)80247-2. [DOI] [PubMed] [Google Scholar]
- 66.Korpela K, Ranki M, Sukupolvi S, Mäkelä P H, Rehn M. Occurrence of Salmonella typhimurium virulence plasmid-specific sequences in different serovars of Salmonella. FEMS Microbiol Lett. 1989;58:49–54. doi: 10.1016/0378-1097(89)90340-6. [DOI] [PubMed] [Google Scholar]
- 67.Lan R T, Reeves P R. Gene transfer is a major factor in bacterial evolution. Mol Biol Evol. 1996;13:47–55. doi: 10.1093/oxfordjournals.molbev.a025569. [DOI] [PubMed] [Google Scholar]
- 68.Lax A J, Pullinger G D, Baird G D, Williamson C M. The virulence plasmid of Salmonella dublin: detailed restriction map and analysis by transposon mutagenesis. J Gen Microbiol. 1990;136:1117–1123. doi: 10.1099/00221287-136-6-1117. [DOI] [PubMed] [Google Scholar]
- 69.Le Minor L, Popoff M Y. Designation of Salmonella enterica sp. nov., nom. rev., as the type and only species of the genus Salmonella. Int J Syst Bacteriol. 1987;37:465–468. [Google Scholar]
- 70.Li J, Ochman H, Groisman E A, Boyd E F, Solomon F, Nelson K, Selander R K. Relationship between evolutionary rate and cellular location among the Inv/Spa invasion proteins of Salmonella enterica. Proc Natl Acad Sci USA. 1995;92:7252–7256. doi: 10.1073/pnas.92.16.7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li J, Smith N H, Nelson K, Crichton P B, Old D C, Whittam T S, Selander R K. Evolutionary origin and radiation of the avian-adapted non-motile salmonellae. J Med Microbiol. 1993;38:129–139. doi: 10.1099/00222615-38-2-129. [DOI] [PubMed] [Google Scholar]
- 72.Libby S J, Adams L G, Ficht T A, Allen C, Whitford H A, Buchmeier N A, Bossie S, Guiney D G. The spv genes of the Salmonella dublin virulence plasmid are required for severe enteritis and systemic infection in the natural host. Infect Immun. 1997;65:1786–1792. doi: 10.1128/iai.65.5.1786-1792.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lockman H A, Curtiss R., III Isolation and characterization of conditional adherent and non-type 1 fimbriated Salmonella typhimurium mutants. Mol Microbiol. 1992;6:933–945. doi: 10.1111/j.1365-2958.1992.tb01543.x. [DOI] [PubMed] [Google Scholar]
- 74.Lockman H A, Curtiss R., III Virulence of non-type 1-fimbriated and nonfimbriated nonflagellated Salmonella typhimurium mutants in murine typhoid fever. Infect Immun. 1992;60:491–496. doi: 10.1128/iai.60.2.491-496.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mahon J, Lax A J. A quantitative polymerase chain reaction method for the detection in avian faeces of Salmonellas carrying the spvR gene. Epidemiol Infect. 1993;111:455–464. doi: 10.1017/s0950268800057186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Manning M J. Evolution of the vertebrate immune system. J R Soc Med. 1979;72:683–688. doi: 10.1177/014107687907200911. [DOI] [PubMed] [Google Scholar]
- 77.McCormick B A, Miller S I, Carnes D, Madara J L. Transepithelial signaling to neutrophils by salmonellae: a novel virulence mechanism for gastroenteritis. Infect Immun. 1995;63:2302–2309. doi: 10.1128/iai.63.6.2302-2309.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Miller S I, Hohmann E L, Pegues D A. Salmonella (including Salmonella typhi) In: Mandell G L, Bennett J E, Dolin R, editors. Principles and practice of infectious diseases. 4th ed. Vol. 2. New York, N.Y: Churchill Livingstone; 1995. pp. 2013–2033. [Google Scholar]
- 79.Mills D M, Bajaj V, Lee C A. A 40kb chromosomal fragment encoding Salmonella typhimurium invasion genes is absent from the corresponding region of the Escherichia coli K-12 chromosome. Mol Microbiol. 1995;15:749–759. doi: 10.1111/j.1365-2958.1995.tb02382.x. [DOI] [PubMed] [Google Scholar]
- 80.Montenegro M A, Morelli G, Helmuth R. Heteroduplex analysis of Salmonella virulence plasmids and their prevalence in isolates of defined sources. Microb Pathog. 1991;11:391–397. doi: 10.1016/0882-4010(91)90035-9. [DOI] [PubMed] [Google Scholar]
- 81.Musher D M, Rubenstein A D. Permanent carriers of nontyphosal Salmonellae. Public Health Rep. 1973;132:869. [PubMed] [Google Scholar]
- 82.Muthukkaruppan V R, Borysenko M, El Ridi R. RES structure and function in reptilia. In: Cohen N, Sigel M M, editors. The reticuloendothelial system. Vol. 3. New York, N.Y: Plenum Press; 1982. pp. 461–508. [Google Scholar]
- 83.Nakamura M, Sato S, Ohya T, Suzuki S, Ikeda S. Possible relationship of a 36-megadalton Salmonella enteritidis plasmid to virulence in mice. Infect Immun. 1985;47:831–833. doi: 10.1128/iai.47.3.831-833.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ochman H, Groisman E A. Distribution of pathogenicity islands in Salmonella spp. Infect Immun. 1996;64:5410–5412. doi: 10.1128/iai.64.12.5410-5412.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ochman H, Groisman E A. The evolution of invasion by enteric bacteria. Can J Microbiol. 1995;41:555–561. doi: 10.1139/m95-074. [DOI] [PubMed] [Google Scholar]
- 86.Ochman H, Soncini F C, Solomon F, Groisman E A. Identification of a pathogenicity island for Salmonella survival in host cells. Proc Natl Acad Sci USA. 1996;93:7800–7804. doi: 10.1073/pnas.93.15.7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ochman H, Wilson A C. Evolution in bacteria: evidence for a universal substitution rate in cellular genomes. J Mol Evol. 1987;26:74–86. doi: 10.1007/BF02111283. [DOI] [PubMed] [Google Scholar]
- 88.Old D C, Payne S B. Antigens of the type 2 fimbriae of salmonellae: “cross reacting material” (CRM) of type 1 fimbriae. J Med Microbiol. 1971;4:215–225. doi: 10.1099/00222615-4-2-215. [DOI] [PubMed] [Google Scholar]
- 89.Ou J T, Lin M-Y, Chao H-L. Presence of F-like ori-T base-pair sequences on the virulence plasmids of Salmonella serovars Gallinarum, Enteritidis, and Typhimurium, but absent in those of Choleraesuis and Dublin. Microbiol Pathog. 1994;17:13–21. doi: 10.1006/mpat.1994.1048. [DOI] [PubMed] [Google Scholar]
- 90.Pardon P, Sanchis R, Marly J, Lantier F, Papin M, Popoff M. Salmonellose ovine due a Salmonella abortusovis. Ann Rech Vet. 1988;19:221–235. [PubMed] [Google Scholar]
- 91.Pascopella L, Raupach B, Ghori N, Monack D, Falkow S, Small P L. Host restriction phenotypes of Salmonella typhi and Salmonella gallinarum. Infect Immun. 1995;63:4329–4335. doi: 10.1128/iai.63.11.4329-4335.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pier G B, Grout M, Zaidi T, Meluleni G, Mueschenborn S S, Banting G, Ratcliff R, Evans M J, Colledge W H. Salmonella typhi uses CFTR to enter intestinal epithelial cells. Nature. 1998;393:79–82. doi: 10.1038/30006. [DOI] [PubMed] [Google Scholar]
- 93.Popoff M Y, Le Minor L. Antigenic formulas of the Salmonella serovars. 5th ed. Paris, France: WHO Collaborating Center for Reference and Research on Salmonella, Institute Pasteur; 1992. [Google Scholar]
- 94.Popoff M Y, Miras I, Coynault C, Lasselin C, Pardon P. Molecular relationships between virulence plasmids of salmonella serotypes typhimurium and dublin and large plasmids of other Salmonella serotypes. Ann Microbiol. 1984;135A:389–398. doi: 10.1016/s0769-2609(84)80080-0. [DOI] [PubMed] [Google Scholar]
- 95.Poppe C, Curtiss III R, Gulig P A, Gyles C L. Hybridization with a DNA probe derived from the virulence region of the 60 Mdal plasmid of Salmonella typhimurium. Can J Vet Res. 1989;53:378. [PMC free article] [PubMed] [Google Scholar]
- 96.Porter S B, Curtiss R., III Effect of inv mutations on Salmonella virulence and colonization in 1-day-old white leghorn chicks. Avian Dis. 1997;41:45–57. [PubMed] [Google Scholar]
- 97.Reeves M W, Evins G M, Heiba A A, Plikaytis B D, Farmer J J., III Clonal nature of Salmonella typhi and its genetic relatedness to other salmonellae as shown by multilocus enzyme electrophoresis, and proposal of Salmonella bongori comb. nov. J Clin Microbiol. 1989;27:313–320. doi: 10.1128/jcm.27.2.313-320.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rexach L, Dilassier F, Fach P. Polymerase chain reaction for salmonella virulence-associated plasmid genes: a new tool in salmonella epidemiology. Epidemiol Infect. 1994;112:33–43. doi: 10.1017/s0950268800057393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rice D H, Besser T E, Hancock D D. Epidemiology and virulence assessment of Salmonella dublin. Vet Microbiol. 1997;56:111–124. doi: 10.1016/S0378-1135(96)01352-1. [DOI] [PubMed] [Google Scholar]
- 100.Rodriguezpena J M, Alvarez I, Ibanez M, Rotger R. Homologous regions of the salmonella enteritidis virulence plasmid and the chromosome of salmonella typhi encode thiol, disulphide oxidoreductases belonging to the dsba thioredoxin family. Microbiology. 1997;143:1405–1413. doi: 10.1099/00221287-143-4-1405. [DOI] [PubMed] [Google Scholar]
- 101.Rodriguezpena J M, Buisan M, Ibanez M, Rotger R. Genetic map of the virulence plasmid of salmonella enteritidis and nucleotide sequence of its replicons. Gene. 1997;188:53–61. doi: 10.1016/s0378-1119(96)00776-7. [DOI] [PubMed] [Google Scholar]
- 102.Roggendorf M, Muller H E. Enterobacteria from reptiles. Zentbl Bakteriol Parasitenkd Hyg Abt I Orig A. 1976;236:22–35. [PubMed] [Google Scholar]
- 103.Roudier C, Krause M, Fierer J, Guiney D G. Correlation between the presence of sequences homologous to the vir region of Salmonella dublin plasmid pSDL2 and the virulence of twenty-two Salmonella serotypes in mice. Infect Immun. 1990;58:1180–1185. doi: 10.1128/iai.58.5.1180-1185.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Roy J H B. Salmonellosis. In: Roy J H B, editor. The calf. 1 Management of Health. London, United Kingdom: Butterworths; 1990. pp. 118–130. [Google Scholar]
- 105.Rychlik I, Lovell M A, Barrow P A. The presence of genes homologous to the K88 genes faeH and faeI on the virulence plasmid of Salmonella gallinarum. FEMS Microbiol Lett. 1998;159:255–260. doi: 10.1111/j.1574-6968.1998.tb12869.x. [DOI] [PubMed] [Google Scholar]
- 106.Schwartz K J. Salmonellosis in swine. Compend Contin Educ Pract Vet. 1991;13:139–146. [Google Scholar]
- 107.Selander R K, Beltran P, Smith N H, Helmuth R, Rubin F A, Kopecko D J, Ferris K, Tall B D, Cravioto A, Musser J M. Evolutionary genetic relationships of clones of Salmonella serovars that cause human typhoid and other enteric fevers. Infect Immun. 1990;58:2262–2275. doi: 10.1128/iai.58.7.2262-2275.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Selander R K, Li J, Boyd E F, Wang F-S, Nelson K. DNA sequence analysis of the genetic structure of populations of Salmonella enterica and Escherichia coli. In: Priest F G, Ramos-Cormenzana A, Tindall B J, editors. Bacterial diversity and systematics. New York, N.Y: Plenum Press; 1994. pp. 17–49. [Google Scholar]
- 109.Shea J E, Hensel M, Gleeson C, Holden D W. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc Natl Acad Sci USA. 1996;93:2593–2597. doi: 10.1073/pnas.93.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Singh I P, Sharma V K, Kaura Y K. Some aspects of the epidemiology of Salmonella abortus-ovis infection in equines. Br Vet J. 1971;127:378–383. doi: 10.1016/s0007-1935(17)37443-2. [DOI] [PubMed] [Google Scholar]
- 111.Stoll L. Experimentelle Infektionen mit Keimen der Salmonella und Arizona Gruppe bei Schlangen. Nord Vet Med. 1962;14:225–232. [Google Scholar]
- 112.Stone B J, Garcia C M, Badger J L, Hassett T, Smith R I F, Miller V. Identification of novel loci affecting entry of Salmonella enteritidis into eukaryotic cells. J Bacteriol. 1992;174:3945–3952. doi: 10.1128/jb.174.12.3945-3952.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tinge A S, Curtiss R., III Isolation of the replication and partitioning regions of the Salmonella typhimurium virulence plasmid and stabilization of heterologous replicons. J Bacteriol. 1990;172:5266–5277. doi: 10.1128/jb.172.9.5266-5277.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tinge S A, Curtiss R. Conservation of Salmonella typhimurium virulence plasmid maintenance regions among Salmonella serovars as a basis for plasmid curing. Infect Immun. 1990;58:3084–3092. doi: 10.1128/iai.58.9.3084-3092.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Turner A K, Lovell A, Hulme S D, Zhang-Barber L, Barrow P A. Identification of Salmonella typhimurium genes required for colonization of the chicken alimentary tract and for virulence in newly hatched chicks. Infect Immun. 1998;66:2099–2106. doi: 10.1128/iai.66.5.2099-2106.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.van der Velden A W M, Bäumler A J, Tsolis R M, Heffron F. Multiple fimbrial adhesins are required for full virulence of Salmonella typhimurium in mice. Infect Immun. 1998;66:2803–2808. doi: 10.1128/iai.66.6.2803-2808.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Vladoianu I-R, Chang H R, Pechére J-C. Expression of host resistance to Salmonella typhi and salmonella typhimurium: bacterial survival within macrophages of murine and human origin. Microb Pathog. 1990;8:83–90. doi: 10.1016/0882-4010(90)90072-x. [DOI] [PubMed] [Google Scholar]
- 118.Wallis T S, Paulin S M, Plested J S, Watson P R, Jones P W. The Salmonella dublin virulence plasmid mediates systemic but not enteric phases of salmonellosis in cattle. Infect Immun. 1995;63:2755–2761. doi: 10.1128/iai.63.7.2755-2761.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Watson P R, Galyov E E, Paulin S M, Jones P W, Wallis T S. Mutation of invH, but not stn, reduces salmonella-induced enteritis in cattle. Infect Immun. 1998;66:1432–1438. doi: 10.1128/iai.66.4.1432-1438.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Watson P R, Paulin S M, Bland A P, Jones P W, Wallis T S. Characterization of intestinal invasion by Salmonella typhimurium and Salmonella dublin and effect of a mutation in the invH gene. Infect Immun. 1995;63:2743–54. doi: 10.1128/iai.63.7.2743-2754.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wood M W, Rosqvist R, Mullan P B, Edwards M H, Galyov E E. SopE, a secreted protein of Salmonella dublin, is translocated into the target eukaryotic cell via a sip-dependent mechanism and promotes bacterial entry. Mol Microbiol. 1996;22:327–338. doi: 10.1046/j.1365-2958.1996.00116.x. [DOI] [PubMed] [Google Scholar]
- 122.Woodward M J, McLaren I, Wray C. Distribution of virulence plasmids within salmonellae. J Gen Microbiol. 1989;135:503–511. doi: 10.1099/00221287-135-3-503. [DOI] [PubMed] [Google Scholar]
- 123.Zapata A G, Torroba M, Vicente A, Varas A, Sacedon R, Jimenez E. The relevance of cell microenvironments for the appearance of lymphohaemopoietic tissues in primitive vertebrates. Histol Histopathol. 1995;10:761–778. [PubMed] [Google Scholar]