Abstract
The low-Ca2+-response (LCR) plasmid pCD1 of the plague agent Yersinia pestis KIM5 was sequenced and analyzed for its genetic structure. pCD1 (70,509 bp) has an IncFIIA-like replicon and a SopABC-like partition region. We have assigned 60 apparently intact open reading frames (ORFs) that are not contained within transposable elements. Of these, 47 are proven or possible members of the LCR, a major virulence property of human-pathogenic Yersinia spp., that had been identified previously in one or more of Y. pestis or the enteropathogenic yersiniae Yersinia enterocolitica and Yersinia pseudotuberculosis. Of these 47 LCR-related ORFs, 35 constitute a continuous LCR cluster. The other LCR-related ORFs are interspersed among three intact insertion sequence (IS) elements (IS100 and two new IS elements, IS1616 and IS1617) and numerous defective or partial transposable elements. Regional variations in percent GC content and among ORFs encoding effector proteins of the LCR are additional evidence of a complex history for this plasmid. Our analysis suggested the possible addition of a new Syc- and Yop-encoding operon to the LCR-related pCD1 genes and gave no support for the existence of YopL. YadA likely is not expressed, as was the case for Y. pestis EV76, and the gene for the lipoprotein YlpA found in Y. enterocolitica likely is a pseudogene in Y. pestis. The yopM gene is longer than previously thought (by a sequence encoding two leucine-rich repeats), the ORF upstream of ypkA-yopJ is discussed as a potential Syc gene, and a previously undescribed ORF downstream of yopE was identified as being potentially significant. Eight other ORFs not associated with IS elements were identified and deserve future investigation into their functions.
The human-pathogenic yersiniae Yersinia pestis, which causes plague, and Yersinia pseudotuberculosis and Yersinia enterocolitica, which primarily cause gastrointestinal disease, have a ca. 70-kb plasmid that encodes a complex virulence property called the low-Ca2+ response (LCR) (35, 45, 87, 88, 120, 121). The LCR was discovered in Y. pestis growing in vitro, where the bacteria respond to the absence of Ca2+ at 37°C by the strong expression and secretion of a virulence protein called V antigen (now also called LcrV). In certain media, this is accompanied by a growth response termed restriction in which the yersiniae undergo an orderly metabolic shutdown and cease growth (23, 25, 42, 51, 119). It is now known that under these in vitro LCR-inductive conditions, the yersiniae maximally induce the transcription, translation, and secretion of a set of virulence proteins called Yops (Yersinia outer proteins) in addition to LcrV. The operons encoding these proteins and other similarly regulated operons on the LCR plasmid have been referred to as the LCR stimulon (LCRS) (83, 107). Millimolar concentrations of Ca2+ permit a full growth yield at 37°C, weaker expression of LcrV and Yops, and essentially no secretion of these proteins (83, 107). There is only very weak, basal expression of Yops and LcrV at environmental temperatures and no secretion: the LCR is designed to function within a mammal. In addition to the presence or absence of Ca2+, other environmental inputs, such as Mg2+, Cl−, Na+, glutamate, nucleotides, and anaerobiosity, modulate the LCR (23, 42, 60, 118, 119). The molecular basis of these effects has not been determined, but these elements of environmental modulation could be important in adjusting virulence protein expression and secretion in response to the wide range of niches that yersiniae are expected to encounter during an infection (106).
It is believed that the absence of Ca2+ mimics an unidentified signal that yersiniae receive when they are in contact with a mammalian cell (33, 83). The LCR plasmid encodes a type III secretion system called Ysc, for Yop secretion (68), that is dedicated to the secretion of Yops, LcrV, and some regulatory proteins in the LCR; cell contact causes this system to be locally activated at the interface between the bacterium and the eukaryotic cell. Environmentally regulated inner and outer gates of the Ysc (LcrG and LcrE [also called YopN], respectively) then open, permitting the secretion of negative regulatory proteins (a key one being LcrQ, also called YscM). This allows full transcriptional activation of LCRS operons by an AraC-like activator protein, LcrF. Yops are secreted locally, without processing. The secretion mechanism recognizes two signals: one in the first 45 nucleotides of the yop mRNA and one related to a domain that has been found for some Yops to bind a specific Yop chaperone (Syc), also encoded by the LCR plasmid (29, 112). Some Yops (YopB, YopD, and YopK) serve as components of a mechanism for targeting effector Yops directly into the eukaryotic cell. The effector Yops (YopE, YopH, YpkA [Yersinia protein kinase], YopM, and probably YopJ) then act on their intracellular target molecules and derange cellular signaling and cytoskeletal functions. LcrV has a bifunctional role in the LCR: it is a regulatory protein, acting at the levels of Yop secretion and targeting, and it has a role as a potent antihost protein (77, 79, 83). LcrV is the only LCRS protein that is secreted in large amounts into the surrounding medium by yersiniae in contact with eukaryotic cells (79). It is the only LCRS protein that has been shown to have an effect when given by itself to mice (77); all others require delivery by the Ysc machinery from yersiniae in intimate contact with mammalian cells.
The overall effect of the LCR is a profound immunosuppression, resulting from the paralysis of innate defenses at the site of infection and the failure to mobilize an effective cell-mediated immune response. Y. pestis, and also the enteropathogenic yersiniae in immunocompromised individuals, grows unchecked in the lymphoid system in a fulminant disease that has a high mortality if not treated with appropriate antibiotics (26, 83). In contrast, without the LCR plasmid, these bacteria are completely avirulent (33, 83).
It is now apparent that several other important pathogens have virulence systems with many striking similarities to the LCR; however, the LCR is the best characterized of these and remains a prototype for investigations at the forefront of molecular pathogenesis. The fact that the LCR was plasmid borne greatly facilitated its characterization, which began in the early years of the existence of molecular genetic techniques. The early studies delineated a large cluster of genes necessary for the Ca2+ dependence of Y. pestis growth (47, 87, 116, 117). Subsequent studies with the enteropathogenic yersiniae as well as Y. pestis resulted in the following picture for the layout of genes involved in the LCR (83). The Ca2+ dependence region turned out to encode LCR regulatory proteins and the enormously complex Ysc type III secretion mechanism, which is comprised of at least 22 gene products. This LCR cluster now includes the immediately adjacent Yop-targeting and secretion control operon lcrGVH-yopBD and is 25.7 kb in size. Only part of this had been sequenced for Y. pestis (lcrDR, lcrGVH-yopB′, yscN′OPQRS, ysc[A]B-F, and lcrF [7, 37, 49, 53, 76, 82, 84, 89]), but the similarities of LCR-related genes among the three species of human-pathogenic yersiniae have been so high that information from one species has been assumed to apply to the others (83). There are Yops within the LCR cluster (e.g., YopB, YopD, and LcrE [which proved to be the same as YopN] and YscH, which apparently is a Yop [3]), but the effector Yops were found to be scattered outside the cluster (104). The only Yop genes in Y. pestis that had been sequenced were yopM and yopE, with the divergently transcribed SycE-encoding gene (40, 61).
There are two other LCR-associated genes outside the LCR cluster. YlpA is a lipoprotein studied only for Y. enterocolitica; it is a member of the LCRS group, but a knockout of its gene did not affect virulence in an intravenous mouse model (30). Although not a true LCR member, YadA is an adhesin whose gene is activated for transcription by LcrF (99). It is enormously important for virulence of Y. enterocolitica, and in both Y. enterocolitica and Y. pseudotuberculosis, it serves as an adhesin that can promote the productive contact with eukaryotic cells, resulting in Ysc activation and Yop targeting (see, e.g., reference 15). Interestingly, its gene in Y. pestis EV76 has a frameshift mutation that would effectively abolish expression of the protein (98).
The high homology of the LCR cluster and yop genes from the three pathogenic Yersinia species includes a similar genetic organization. In addition, the replication and partitioning functions of the family of LCR plasmids are also highly conserved (6, 12, 32, 68, 83, 87, 88, 108). Sequencing of the replication region of the Y. enterocolitica LCR plasmid pYVe439-80 demonstrated that it possessed all the essential components of an IncFIIA replicon (108). Plasmid pYVe439-80 is maintained at an estimated seven copies per cell (32). Although the replicon region of this plasmid has 68% homology with the IncFIIA replicon of plasmid R100, the two plasmids are compatible. This may be due to differences in the nucleotide sequences in the stem-loop structures of the antisense RNA encoded by copA (64, 108). The observed incompatibility of LCR plasmids with derivatives of the F plasmid appears to be due to a shared partitioning system (6, 12, 40).
Despite all the similarities within the LCR plasmid family, there are also significant differences. Plasmids from Y. pseudotuberculosis and Y. pestis are more closely related than those from Y. enterocolitica. Y. enterocolitica plasmid regions outside the LCRS, replication, and partitioning function regions do not hybridize to analogous regions from Y. pseudotuberculosis or Y. pestis LCR plasmids (88). In addition, the location of the replicon in relation to the LCR cluster and the organization of the yopE-yadA region on Y. enterocolitica plasmids differ from those of LCR plasmids from the other two yersiniae (12, 32, 68, 83). DNA sequencing has revealed two distinct types of lcrV that differ in a hypervariable region between amino acids (aa) 225 and 232 in the translated protein. Serotype O:8 Y. enterocolitica strains synthesize one type, while Y. pestis, Y. pseudotuberculosis, and Y. enterocolitica serotypes O:3, O:9, and O:5,27 produce the other (94). Motin et al. (76) identified a species-specific difference in the lcrV genes from Y. pestis and Y. enterocolitica. These differences likely account for the ability of some anti-LcrV antibodies to protect against infections with Y. pestis and Y. pseudotuberculosis but not against Y. enterocolitica infections (75, 94). Finally, the more stringent growth restriction observed under LCRS-inducing conditions for Y. pestis compared to that for the enteropathogenic yersiniae appears to be due to undetermined differences in the LCR plasmid itself (104). Consequently, it is likely that additional significant differences may reside within the LCR family of plasmids.
To identify new potential virulence factors, discover any further differences between Y. pestis LCRS genes and those previously sequenced for the enteropathogenic yersiniae, and identify other undefined traits of this family of virulence plasmids, we have sequenced one entire LCR plasmid. We chose to sequence the LCR plasmid pCD1 of Y. pestis KIM5 (for Kurdistan Iran man) because it has several advantages over other strains. Derivatives of Y. pestis KIM have been directly demonstrated to retain high-virulence characteristics in both mammals and fleas (8, 22, 52, 83). These derivatives are widely used in research and thus are more genetically characterized than other strains. The vast majority of investigations on the regulation, physiological characteristics, and virulence properties of the Y. pestis LCRS have been performed with derivatives of strain KIM. Finally, nearly all previous DNA sequence information on the Y. pestis LCRS components (∼16 kb) has been derived from pCD1 of strain KIM5 (7, 22, 37, 49, 61, 82–84, 89).
Our analysis of the DNA sequence of pCD1 has revealed a set of LCRS genes very similar to those sequenced for the enteropathogenic yersiniae, a potential new Yop and Yop chaperone, two new insertion sequences (ISs), the IncFIIA replication region, and SopABC partitioning functions. We have also identified IS element remnants scattered throughout the plasmid that suggest that pCD1 has undergone numerous insertional events as well as genetic recombinations and rearrangements during its history.
MATERIALS AND METHODS
Bacterial strains, media, and plasmid isolation.
Y. pestis KIM5 is conditionally avirulent due to deletion of the 102-kb pgm locus; it possesses all three prototypical Y. pestis plasmids, i.e., the 9.5-kb pPCP1, ∼70-kb pCD1, and ∼100-kb pMT1 (83). Plasmid pCD1 was isolated from Y. pestis KIM5 by alkaline lysis followed by precipitation with polyethylene glycol (13, 55). Since pCD1 has no selectable marker, a mixture of pCD1 and pBR322 was transformed into Escherichia coli HB101 (47). Transformants were selected for the ampicillin resistance encoded by pBR322 (5). Five hundred transformants were transferred to nitrocellulose membranes and hybridized against pCD1 radioactively labeled by nick translation. One transformant containing both pCD1 and pBR322 was identified. This isolate was cured of pBR322 by fusaric acid selection (17). pCD1 appears to be stably maintained in E. coli HB101, and this transformant has been stored in buffered glycerol at −70°C. Plasmid DNA from E. coli HB101(pCD1) cells grown in Luria broth was isolated by alkaline lysis (13) followed by further purification with polyethylene glycol (55). This purified pCD1 DNA was used in subsequent sequencing.
DNA sequencing.
Libraries were prepared from nebulized, size-fractionated plasmid DNA (63) in the M13 Janus vector (24). DNA templates were purified from random library clones (81), and sequences were collected by using dye-terminator-labeled fluorescent cycle sequencing Prism reagents and ABI377 automated sequencers (Applied Biosystem Division of Perkin-Elmer). Sequences were assembled into contigs by the SeqMan II program (DNASTAR), and clones were selected for sequencing from the opposite end to fill in coverage, resolve ambiguities, and close gaps (24). Final coverage was about eightfold.
In several instances, the sequence contradicted previously published sequences for the yersiniae or yielded unexpected results. To ensure that this did not result from mutations to pCD1 during carriage in E. coli, we sequenced these regions by using pCD1 isolated from the conditionally virulent Y. pestis KIM5 (83) or pJIT7, a recombinant plasmid containing the IS1616 region adjacent to sopAB (Fig. 1).
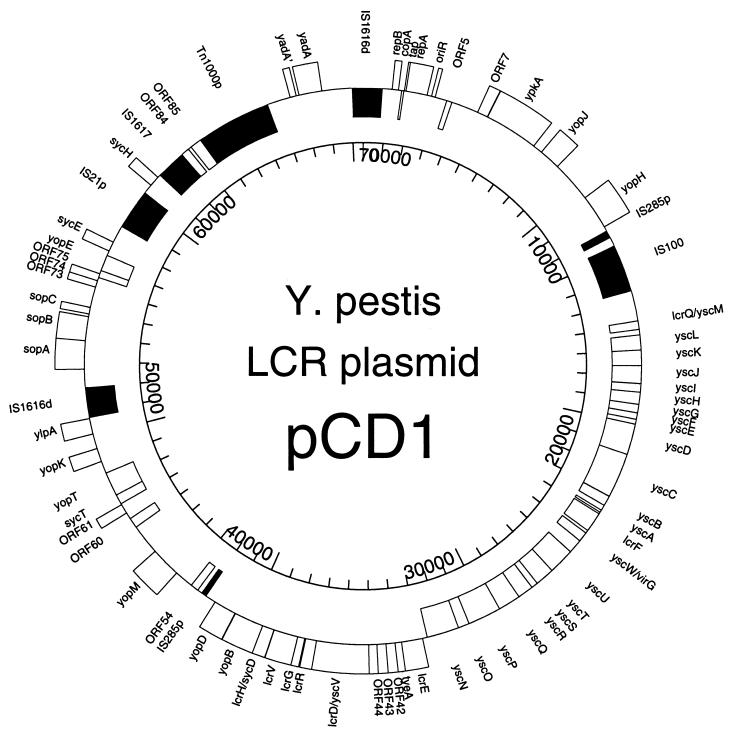
FIG. 1.
Genetic organization of the LCR plasmid, pCD1, of Y. pestis KIM5. The map shows significant ORFs and transposable elements as well as replication and partitioning regions. For genes on the outside of the map, transcription proceeds clockwise. Genes on the inside are transcribed from the complementary strand. The oriR and sopC regions are not transcribed. The placement of transposable elements (solid boxes) does not indicate the direction of transcription. A d indicates a defective element, while a p indicates a partial element. Most IS remnants are not denoted on this figure.
Annotation.
We identified open reading frames (ORFs) encoding at least 50 aa, using Geneplot or GeneQuest (DNASTAR) programs to display start codons (including GUG), stop codons, and codon usage statistics plots for each reading frame. Codon usage analysis helped to predict ORFs. It was assessed in the program by second- and third-order statistical comparisons (20) with a matrix built from all available sequences for Yersinia species. Although this matrix was more useful than one derived from E. coli genes, it was necessarily constructed from a relatively small data set and is no doubt imperfect. Generally, in the absence of experimental data, the start codon farthest upstream was used to annotate the ORF start (14). ORFs with products smaller than 50 aa were included if codon usage statistics showed a high score. ORF amino acid sequences were searched against SWISS-PROT 34 by using the BLOSUM26 matrix, with the DeCypher II System (TimeLogic Inc.). In the first pass, the best hit was automatically saved as an annotation for each ORF, and then known genes and putative functions were assigned for individual ORFs by inspection of the search output.
Subsequent searches of DNA and protein databases for mobile genetic elements, DNA features, and amino acid sequence similarities were performed by using BLAST (4). Analysis and manipulation of DNA sequence data were performed by using programs in the Genetics Computer Group software package version 9.0.
Nucleotide sequence accession number.
The entire sequence of pCD1 from Y. pestis KIM5 has been deposited in the GenBank database and assigned accession number AF074612.
RESULTS AND DISCUSSION
General description.
From our DNA sequencing, we have constructed a genetic map of the Y. pestis KIM5 pCD1 plasmid (Fig. 1), which is 70,509 nucleotides in length. Table 1 lists significant ORFs and their primary characteristics. Of the 61 ORFs in Table 1 (excluding the ylpA and ′yadA pseudogenes), 8 have GTG starts (repA, yscD, yscJ, yscW, Orf7, Orf42, Orf43, and Orf74) and 3 have TTG starts (Orf54, Orf61, and Orf73). We have omitted most IS element remnants and partial ORFs that appear to be nonfunctional due to IS-related events or other deletions and rearrangements.
TABLE 1.
ORFs on pCD1 of Y. pestis KIM5a
| Geneb or ORF | Function of product | Orientation | Beginning of ORF (bp) | End of ORF (bp) | No. of amino acids in product | Isoelectric point of product | Mass of product (kDa) |
|---|---|---|---|---|---|---|---|
| repB (copB) | Negative regulator of repA transcription | + | 1171 | 1425 | 85 | 9.72 | 9.58 |
| tap | Required for translation of repA | + | 1667 | 1741 | 25 | 9.31 | 2.82 |
| repA | Plasmid replication | + | 1734 | 2600 | 289 | 10.96 | 33.55 |
| Orf5 | Unknown | − | 3645 | 3427 | 73 | 9.96 | 8.22 |
| Orf7 | Unknown | + | 4758 | 5186 | 143 | 4.39 | 15.78 |
| ypkA (yopO) | Targeted effector; Ser Thr kinase | + | 5204 | 7402 | 733 | 6.53 | 81.74 |
| yopJ (yopP) | Targeted effector; causes apoptosis in macrophages and interferes with cell signaling | + | 7798 | 8664 | 289 | 7.07 | 32.46 |
| yopH | Targeted effector; protein tyrosine kinase; interferes with cell signaling at focal adhesions | + | 10347 | 11753 | 469 | 8.68 | 50.87 |
| lcrQ (yscM) | Negative regulator of LCR expression | − | 16148 | 15801 | 116 | 6.34 | 12.41 |
| yscL | Type III secretion component | − | 17038 | 16373 | 222 | 4.57 | 24.65 |
| yscK | Type III secretion component | − | 17613 | 16984 | 210 | 6.75 | 23.99 |
| yscJ | Type III secretion component | − | 18347 | 17613 | 245 | 7.43 | 27.04 |
| yscI | Type III secretion component | − | 18701 | 18354 | 116 | 4.47 | 12.67 |
| yscH (yopR) | Secreted; unknown function | − | 19199 | 18702 | 166 | 5.14 | 18.35 |
| yscG | Type III secretion component | − | 19543 | 19196 | 116 | 6.60 | 13.07 |
| yscF | Type III secretion component | − | 19808 | 19545 | 88 | 7.13 | 9.49 |
| yscE | Type III secretion component | − | 20009 | 19809 | 67 | 7.31 | 7.61 |
| yscD | Type III secretion component | − | 21265 | 20006 | 420 | 5.85 | 46.93 |
| yscC | Type III secretion component | − | 23085 | 21262 | 608 | 6.49 | 67.35 |
| yscB | Unknown | − | 23504 | 23091 | 138 | 9.27 | 15.41 |
| yscA | Unknown | − | 23828 | 23730 | 33 | 9.82 | 3.86 |
| lcrF (virF) | Activator of LCR gene expression | − | 24722 | 23907 | 272 | 8.91 | 30.84 |
| yscW (virG) | YscC lipoprotein chaperone | − | 25241 | 24846 | 132 | 10.12 | 14.71 |
| yscU | Type III secretion component | − | 26881 | 25817 | 355 | 8.81 | 40.39 |
| yscT | Type III secretion component | − | 27666 | 26881 | 262 | 5.67 | 28.45 |
| yscS | Type III secretion component | − | 27929 | 27663 | 89 | 6.32 | 9.57 |
| yscR | Type III secretion component | − | 28584 | 27931 | 218 | 4.68 | 24.43 |
| yscQ | Type III secretion component | − | 29504 | 28581 | 308 | 5.08 | 34.42 |
| yscP | Type III secretion component | − | 30868 | 29501 | 456 | 5.44 | 50.42 |
| yscO | Type III secretion component | − | 31332 | 30868 | 155 | 7.84 | 19.00 |
| yscN | Type III secretion component | − | 32648 | 31329 | 440 | 6.48 | 47.81 |
| lcrE (yopN) | Secretion control | + | 32846 | 33727 | 294 | 5.07 | 32.67 |
| tyeA | Secretion and Yop targeting control | + | 33708 | 33986 | 93 | 4.21 | 10.75 |
| Orf42 | Unknown | + | 33973 | 34344 | 124 | 5.54 | 13.61 |
| Orf43 | Unknown | + | 34341 | 34709 | 123 | 6.32 | 13.76 |
| Orf44 | Unknown | + | 34706 | 35050 | 115 | 6.92 | 13.12 |
| lcrD (yscV) | Secretion | + | 35037 | 37151 | 705 | 5.04 | 77.81 |
| lcrR | Unknown | + | 37148 | 37588 | 147 | 10.27 | 16.46 |
| lcrG | Secretion control; efficient Yop targeting | + | 37630 | 37917 | 96 | 8.15 | 11.02 |
| lcrV | Diffusible effector; secretion and targeting control | + | 37919 | 38899 | 327 | 5.66 | 37.24 |
| lcrH (sycD) | YopB and YopD chaperone | + | 38912 | 39418 | 169 | 4.61 | 19.02 |
| yopB | Yop targeting | + | 39396 | 40601 | 402 | 7.09 | 41.83 |
| yopD | Yop targeting; negative regulator | + | 40620 | 41540 | 307 | 6.80 | 33.39 |
| Orf54 | Unknown | − | 42709 | 42386 | 108 | 9.66 | 12.61 |
| yopM | Targeted effector | + | 43481 | 44710 | 410 | 4.23 | 46.21 |
| Orf60 | Unknown | − | 46365 | 45946 | 140 | 7.79 | 15.81 |
| Orf61 | Unknown | + | 46637 | 47026 | 130 | 7.33 | 14.80 |
| sycT | YopT chaperone | − | 47468 | 47070 | 133 | 4.43 | 15.42 |
| yopT | Targeted effector | − | 48436 | 47468 | 323 | 9.13 | 36.31 |
| yopK (yopQ) | Yop targeting | + | 48936 | 49484 | 183 | 4.37 | 21.00 |
| ylpA | Pseudogene | + | 50089 | 50718 | 210 | 5.80 | 22.40 |
| sopA | Plasmid partitioning; negative regulator of sopAB transcription | + | 52730 | 53896 | 389 | 5.82 | 43.41 |
| sopB | Plasmid partitioning; binds to sopC region | + | 53896 | 54858 | 320 | 10.19 | 35.61 |
| Orf73 | Unknown | + | 56087 | 56362 | 92 | 6.16 | 10.10 |
| Orf74 | Unknown | + | 56355 | 56654 | 100 | 5.51 | 11.67 |
| Orf75 | Unknown | − | 56792 | 56496 | 99 | 9.88 | 11.19 |
| yopE | Targeted effector; causes actin depolymerization | − | 57453 | 56794 | 220 | 6.59 | 22.99 |
| sycE (yerA) | YopE chaperone | + | 57647 | 58039 | 131 | 4.49 | 14.65 |
| sycH | YopH chaperone | + | 60796 | 61221 | 142 | 4.81 | 15.76 |
| Orf84 | Unknown | − | 62897 | 62568 | 110 | 8.98 | 13.00 |
| Orf85 | Unknown | − | 63500 | 63036 | 155 | 4.97 | 17.71 |
| yadA′ | Pseudogene | + | 67532 | 67783 | 84 | 5.21 | 8.92 |
| ′yadA | Pseudogene | + | 67900 | 68835 | 312 | 6.84 | 32.47 |
ORFs within transposable elements, as well as disrupted or partial ORFs (except for ylpA, yadA′, and ′yadA), are not included in the table.
Except for copB, yopN, yscV, and yerA, all alternate gene designations (in parentheses) are Y. enterocolitica terminology. copB, plasmid R100 terminology; yopN, Y. enterocolitica and Y. pseudotuberculosis terminology; yscV, proposed terminology change; yerA, Y. pseudotuberculosis terminology.
The LCR-related genes are organized as was proposed in a low-resolution, composite map assembled from numerous studies (83). This consists of a large Ca2+ dependence region (yscM to yopD, LCR cluster) that encodes primarily secretion and regulatory functions, with genes encoding YopK, effector Yops (YpkA, YopJ, YopH, YopM, and YopE), and their chaperones (SycE and SycH) being scattered around the rest of pCD1. The locations and organization of LCR-related genes, as well as the partitioning and replication regions, of pCD1 closely resemble those of the best-characterized LCR plasmid in Y. pseudotuberculosis, pIB1 (12, 95). In contrast, pYVe O:9, from serotype O:9 Y. enterocolitica, has several notable differences. Compared to those of pCD1 and pIB1, the LCR cluster of pYVe O:9 is in the opposite orientation; this is also the case for the sopA-to-sycE region (Fig. 1) (12, 32, 68). In addition, yopM and yopH are located on opposite sides of the LCR cluster in pCD1 compared to pYVe O:9. While yopH and sycH are located adjacent to each other in Y. enterocolitica, they are separated by over 20 kb in pCD1. Finally, ylpA and yopK are located near the partitioning region (sopABC) of pCD1 but near ypkA and the origin of replication in pYVe O:9 (Fig. 1) (12, 32, 68). As noted by others, there is no simple, single mechanism to explain the scrambled locations of these genes among the LCR family of plasmids (12).
We identified a number of intact, defective, and partial IS elements in pCD1. The site of an IS100 insertion, an element with numerous copies in the Y. pestis genome (36, 83), was confirmed and refined. Two new IS elements, which we have named IS1616 and IS1617, were discovered (Fig. 1) and were registered through Esther Lederberg, Plasmid Reference Center, Stanford, Calif. In addition, numerous IS element remnants were identified; these partial ISs cluster primarily in four regions of pCD1 (discussed below).
It is curious that IS100 is near one end of the yscM-to-yopD LCR cluster and that two partial IS285 elements bound this same region (Fig. 1). The type III secretion system and regulatory genes, exemplified by this LCR cluster, are widespread among bacterial pathogens and have been suggested as a possible pathogenicity island (PAI) (67). PAI hallmarks include carriage of virulence genes, a distinct GC content compared to that of the host bacterium, a discrete genetic unit often flanked by direct repeats, association with tRNA genes and/or insertion sequences, the presence of mobility genes (transposase genes, etc.), instability, and absence in less pathogenic strains (48). An additional requirement of a chromosomal location (48) may be somewhat artificial (67) given the large sizes of many virulence plasmids. Although the LCR cluster does have IS elements associated with it, we failed to detect any tRNA genes anywhere on pCD1. In addition, the LCR cluster does not contain genes for effector Yops (except lcrV). Finally, the GC content of this region (44.8%) matches that of the entire plasmid (Table 2) and is similar to the 46 to 47% GC content of the genome of Y. pestis (9, 83).
TABLE 2.
GC compositions of selected genes and regions of pCD1
| Gene or region | Location (bp) | % GC |
|---|---|---|
| Entire pCD1 plasmid | 0–70509 | 44.8 |
| Replication region | 1103–2935 | 48.3 |
| sopABC | 52628–55240 | 43.1 |
| yadA′/′yadA | 67532–68835 | 43.2 |
| IS elements or regions | ||
| IS100 | 12609–14562 | 51.4 |
| IS1616 | 50753–51987 | 50.9 |
| IS1616d | 70056–782 | 51.1 |
| IS1617 | 61336–62549 | 49.9 |
| IS21p | 58749–60418 | 55.1 |
| Tn1000p | 63528–66581 | 50.2 |
| IS285/IS100 region | 11939–15801 | 47.5 |
| IS1617/Tn1000 region | 61231–67532 | 44.8 |
| LCR genes or regions | ||
| yscM-yopD (LCR cluster) | 15801–41540 | 44.8 |
| ypkA | 5204–7402 | 48.1 |
| yopJ | 7798–8664 | 34.8 |
| yopH | 10347–11753 | 47.4 |
| yopM | 43418–44710 | 38.9 |
| yopM upstream region | 41540–43418 | 39.3 |
| yopM downstream region | 44710–47070 | 37.6 |
| yopT | 48436–47468 | 43.1 |
| yopK | 48936–49484 | 33.5 |
| ylpA | 50089–50713 | 47.5 |
| yopE | 57453–56794 | 51.1 |
However, there are intriguing differences in the GC contents of other regions and genes within pCD1 (Table 2). The intact and large partial IS elements all have GC contents significantly higher than that of the Y. pestis genome. In addition, the scattered effector Yop genes have a wide range of GC contents (Table 2) (discussed below). The numerous IS remnants, varied GC content, and scattered yop genes suggest that pCD1 has undergone multiple DNA incursions, rearrangements, and deletions. These alterations may have eliminated or disrupted some of the classic features of a PAI surrounding the LCR cluster.
Replication and partition functions.
Sequencing confirmed that pCD1 has an IncFIIA replication system (Fig. 2). For the IncFIIA resistance plasmid R1 and its close relatives (R100 and R6), a quite detailed analysis of this replication system has been performed. RepA (formerly RepA1) is required for replication at the origin of replication (oriR). Transcription from PrepA is repressed by CopB (RepB in Fig. 2 is its homologue); constitutive transcription from the PcopB promoter (PrepB in Fig. 2) results in a long mRNA encoding RepB, Tap, and RepA that is the source of most of the RepA protein in E. coli (110). Posttranscriptional expression of RepA is controlled by copA, which lies within the leader region of RepA mRNA and encodes an antisense RNA transcribed in the opposite direction compared to RepA mRNA transcription. CopA and its RepA mRNA target site, termed CopT, possess complex secondary stem-loop structures that form a loop-loop RNA-RNA complex. This interaction completely blocks translation of the adjacent gene, tap, which encodes a 24-aa protein. Translation of tap is required for translation of repA (16, 64).
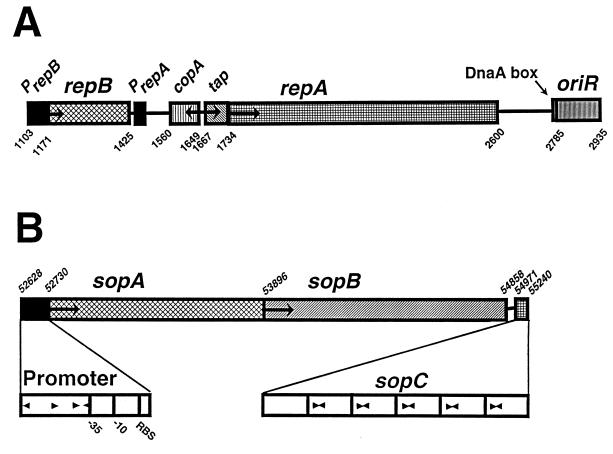
FIG. 2.
Genetic organization of the replication and partitioning regions of pCD1. (A) The replication region contains four genes (repB, copA, tap, and repA) and an origin of replication (oriR). Black boxes indicate the promoter regions for repB and repA; the promoter region for copA is not shown. (B) The par region possesses two putative trans-acting genes (sopA and sopB) and a putative DNA binding site (sopC). Arrowheads within the promoter region show the locations and orientations of short, imperfect repeats. The six 45-bp direct repeats that compose sopC are shown as open boxes, with the locations of five 16-bp inverted repeat structures designated by arrowheads. RBS, ribosome binding site. For both panels, arrows indicate the directions of transcription of individual genes, while locations within pCD1 are indicated by numbers that follow the numbering system in Fig. 1.
The pCD1 replication region showed highest homology to the LCR plasmid pYVe439-80 from Y. enterocolitica (108) and the virulence plasmid from Salmonella enteritidis (93); consequently, we followed the nomenclature used for these two plasmid replicons (Fig. 2). Y. pestis RepB (homologue of CopB) had 100% identity to RepB of Y. enterocolitica and 69.1% identity and 80.3% similarity to Salmonella RepB. Y. pestis RepA was 99.3% identical (100% similar) and 86.5% identical (92.4% similar), respectively, to the Y. enterocolitica and S. enteritidis gene products. For Tap, the homologies were 95.8% identity (100% similarity) with Y. enterocolitica Tap and 68% identity (72% similarity) with S. enteritidis Tap (93, 108). Similar to other IncFIIA replication systems, a DnaA binding site and OriR region lie downstream of repA in pCD1 (Fig. 2).
The copy number and incompatibility of IncFIIA plasmids are determined by the loop-loop interactions between CopA antisense RNA and the CopT mRNA region (16, 64). However, neither the copy number nor the incompatibility characteristics of pCD1 can be empirically determined from the sequence. Sequence changes in the stem and/or loop affect copy number by altering complex formation rates and alter incompatibility specificity (16, 64, 108, 110). However, since the yersinial CopA antisense RNAs are 100% identical (reference 108 and this study), the copy number of seven copies per chromosome determined for the Y. enterocolitica LCR plasmid is probably valid for pCD1. Different CopA stem sequences have been proposed as the reason for compatibility of various IncFIIA plasmids, including pYVe439-80 and R100 (108). Thus, it is likely that pCD1 and pYVE439-80 will also have the same incompatibility characteristics.
As expected from DNA hybridization experiments and plasmid incompatibility testing (6, 12, 40), the partitioning region of pCD1 is a homologue of the sopABC system of the F plasmid (Fig. 2). The pCD1 SopA protein shows 68.8% identity and 83% similarity to F plasmid SopA, while the SopB homologues are 48.9% identical and 64.7% similar. We propose the methionines encoded at bp 52730 and 53896 in the sopA and sopB ORFs of pCD1, respectively, as the initiating methionines. The amino acid sequences after these methionines have strong similarity to the experimentally determined N-terminal amino acid sequences of F plasmid SopA and SopB (73, 74). The sopC regions of both plasmids retain some structural similarities but are less homologous than the other components of this system. The LCR plasmid sopC region has six 45-bp tandem nearly perfect direct repeats, with five of the direct repeats possessing a 16-bp inverted repeat structure (TGGGACCGTGGTCCCA) (Fig. 2). The F plasmid sopC possesses 12 43-bp tandem direct repeats. While the direct repeats are not highly homologous, the inverted repeat structures are identical except for the two central, unpaired nucleotides (reference 73 and this study). Finally, the pCD1 sopABC promoter region has four possible 5- to 6-bp imperfect repeats that show some similarity in sequence and location to imperfect repeats in the F plasmid sopABC promoter region (Fig. 2) (50). These structural and amino acid sequence similarities suggest that the pCD1 partitioning system functions in a manner analogous to that of the F plasmid sopABC system.
In the F plasmid system, SopA binds to the four repeated sequences in the promoter and acts to repress transcription of the operon. SopB appears to enhance, in an unknown manner, binding of SopA to the promoter region. SopB dimers directly bind to the direct repeats of sopC; however, a single sopC direct repeat is sufficient for proper partitioning. In addition to its function as a repressor, SopA may also act, by an unknown mechanism, in the partitioning process. One model has sopC serving a centromere-like function, with DNA wrapping around a core of Sop proteins bound to sopC (10, 11, 50, 73).
Insertion elements.
Several mobile genetic elements have been found in the pathogenic yersiniae, and most of them are present on LCR plasmids as well as on the chromosome (27, 38, 40, 70, 78, 80, 90). ISs known to be associated with the LCR plasmid of Y. pestis include IS100 and IS285 (38, 83, 87). Additional elements are found on the LCR plasmid of Y. enterocolitica but are not present on the Y. pestis plasmid (40, 78). Sequence analysis of pCD1 from Y. pestis KIM5 revealed the presence of three complete insertion elements and numerous partial IS elements. Complete and partial IS elements with >85% identity at the DNA sequence level were considered to be the same as previously described IS elements. For the remaining elements, the element with the highest database match at the amino acid sequence level was considered the closest relative. Only complete IS elements were given new IS number designations.
An intact copy of IS100 is located downstream of yopH in pCD1 (Fig. 1 and Table 2). There are numerous copies of IS100 throughout the genome of Y. pestis KIM strains (36); the IS100 element (bp 12609 to 14562) in pCD1 is 100% identical in size and nucleotide sequence to a copy of IS100 present on the pesticin plasmid of Y. pestis EV76-6 (66). IS100, which appears to have inserted within the relic of another insertion element, is flanked by a 5-bp direct repeat (Fig. 3). Five- and seven-base-pair duplications have been found flanking other IS100 elements in Y. pestis (36, 38, 85).
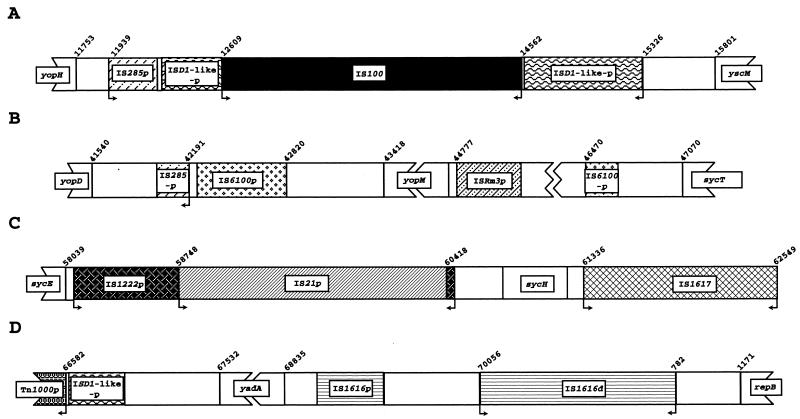
FIG. 3.
Diagrammatic representation of four clusters of intact, defective, and partial IS elements in pCD1. A p or −p indicates that only part of the IS element is present. A d indicates an IS element that is likely defective. Arrows show the presence of inverted repeats within IS elements. Each type of IS element is represented by a unique design that is maintained in each of the four panels. In panel D, the yadA′/′yadA pseudogene is represented as yadA. Numbers correspond to nucleotide positions in Fig. 1.
IS1616 is a new 1,254-bp insertion element located at bp 50753 to 51987, between ylpA and the sopABC partitioning region. The inverted repeats at the ends of IS1616 are 40 bp long and contain nine mismatches. No direct repeats flanking this element were detected. While some elements do not generate a direct repeat upon transposition, the absence of direct repeats could be indicative of changes in the flanking DNA as a result of mutations that have occurred over time. There are three ORFs within IS1616. The first ORF (OrfA, bp 50825 to 51142) is predicted to encode a protein of 105 aa with a pI of 12.6. A second ORF, encoding 186 aa (OrfB, bp 51064 to 51624), overlaps OrfA in the −1 frame. An additional 101 aa (orfC, bp 51625 to 51930), which may have originally been encoded as part of the second ORF, are encoded in the same frame just past the stop codon at bp 51622 for OrfB. A potential ribosome frameshift site, AAAAAG (bp 51078 to 51083), is located within the coding region of OrfA. Translational frameshifting occurs in a number of different bacterial ISs (28). Most frameshift signals consist of a heptanucleotide sequence; however, in some IS elements a tetramer can function as a frameshift site (28, 34). Whether the hexanucleotide in IS1616 is a functional frameshift signal is unknown. IS1616 has the closest amino acid sequence homology to IS1236 from Acinetobacter calcoaceticus. The region of homology includes all of the IS1616 ORFs and spans the stop codon between OrfB and OrfC. IS1236 is a member of the IS3 family and contains two ORFs that could be expressed as a fusion protein as a result of translational frameshifting (46). Remnants of IS1616 are present elsewhere on pCD1 (Table 3 and Fig. 3). A second, presumably defective, copy of IS1616 (IS1616d) is present at bp 70056 to 782 of pCD1. IS1616d contains terminal inverted repeats but is not flanked by direct repeats. An intact OrfA gene is present in IS1616d; however, the downstream ORF has been disrupted by the inversion of a 227-bp fragment corresponding to the complement of nucleotides 51561 to 51787 of IS1616. The inverted segment is flanked by 7-bp inverted repeats in IS1616d. In IS1616, three of the bases in the downstream “inverted repeat” have been changed. When corrected for the inversion, the nucleotide sequence of IS1616d is 99% identical to that of IS1616 and does not possess the stop codon found between OrfB and OrfC of IS1616. While it seems unlikely that either copy of IS1616 is functional, we have designated the element at bp 70056 to 782 as IS1616d in the figures and tables to distinguish it from the element downstream of ylpA. Insufficient sequence information precludes the identification of an IS1616 element downstream of ylpA in Y. enterocolitica or Y. pseudotuberculosis. However, the remnant of IS1616 located downstream of yadA′/′yadA is present in Y. pseudotuberculosis pIB1 as well as in Y. pestis pYV019 but is absent from a similar site in Y. enterocolitica (100).
TABLE 3.
IS element remnants
| IS element homology | Location (bp) | Gene(s) presentb | No. of inverted repeats present |
|---|---|---|---|
| IS1616 | 4187–4519 | ′OrfB | 1 |
| 69062–69456 | ′OrfA, OrfB′ | 1 | |
| ISD1-likea | 12265–12608 | ′OrfB | 0 |
| 14571–15326 | OrfA, OrfB | 1 | |
| 49642–49942* | ′OrfB′ | 0 | |
| 66594–66926* | ′OrfB′ | 0 | |
| IS1617 | 9218–9829 | ′OrfB | 1 |
| IS1222 | 58073–58749 | ′OrfB | 1 |
| 60419–60470 | None | 1 | |
| IS285 | 11939–12249 | None | 1 |
| 41978–42191 | Orf1′ | 1 | |
| IS21 | 58748–60418 | istA, istB′ | 1 |
| Tn1000 | 63528–66582 | tnpR, ΔtnpA | 1 |
| IS6100-like | 42247–42820 | Δtnp | ? |
| 46470–46664 | Δtnp | ? | |
| ISRm3-like | 44777–45191 | Δtransposase | ? |
| IS1327-like | 55656–56005 | ΔtnpA | ? |
Members of this group have amino acid homology. Only those indicated by an asterisk show similarities at the nucleotide sequence level.
OrfA = lcrS; OrfB = lcrT.
IS1617 is a new 1,214-bp element, with inverted repeats of 39 and 40 bp containing 13 mismatches, located downstream of sycH (Fig. 3). The 5 bases flanking each end of IS1617 are identical in four of five positions. Like IS1616, this element belongs to the IS3 family and contains two overlapping ORFs with OrfB in the −1 frame relative to OrfA. OrfA could encode an 88-aa protein (bp 62202 to 62468, complement), while OrfB is open for 289 aa (bp 61369 to 62238, complement). A potential translational frameshift window of AAAAAAG is present in OrfA. IS1617 is more closely related to IS1222 from Enterobacter agglomerans (102) and to ISD1 found in Desulfovibrio vulgaris (43) than to IS1616. A remnant of IS1617 is present downstream of yopJ in pCD1 as well as in Y. pseudotuberculosis pIB1 (Table 3) (44, 59).
In addition to these elements, there are several remnants of other ISs present on pCD1 (Table 3). Portions of IS285 are found at either end of the main LCR gene cluster. The segment upstream of IS100 is 86% identical to the 3′ end of IS285 and includes one copy of the inverted repeat (Fig. 3A). The IS285 remnant located downstream of yopD is 100% identical to the first 214 bp of IS285 and also contains a copy of the inverted repeat (Fig. 3B). However, these two regions together do not make an intact IS. The IS285 segment upstream of the LCR gene cluster is present in the same position in pIB1 from Y. pseudotuberculosis YPIII, but the downstream remnant is absent (19).
Part of an IS21 element (IS21p) is located in the region between sycE and sycH (Fig. 3). This segment, which is 90% identical in nucleotide sequence to IS21 (91), contains one copy of the inverted repeat, an intact istA gene (bp 58849 to 60021), and the 5′ end of istB (bp 60021 to 60418). The Y. pestis IS21p element appears to have disrupted part of an IS1222 element that is 96% identical in nucleotide sequence to IS1222 from E. agglomerans (102). Both IS1222 inverted repeats are present; however, all of OrfA and the 5′ end of OrfB are missing. The IS1222 remnant is not closely related to the new Y. pestis IS elements. Although IS1617 does have amino acid sequence homology to IS1222-encoded products, it is not homologous at the nucleotide level.
Upstream of the yadA′/′yadA pseudogene are the remains of an element that is 85% identical in nucleotide sequence to Tn1000 from E. coli (21). The Y. pestis Tn1000p remnant (Fig. 1 and Table 3) contains one copy of the inverted repeats and an intact tnpR gene (bp 63519 to 64070, complement). The ΔtnpA gene (bp 64234 to 66549) is missing the nucleotides encoding aa 189 to 415. This relic of Tn1000 is only distantly related (69.4%) to a Tn3 homologue found on the LCR plasmid of Y. enterocolitica 29979 (53a). Tn2502, which confers arsenic resistance and also contains a defective tnpA gene, is present downstream of yadA on the LCR plasmid of low-virulence strains of Y. enterocolitica (78). Although Tn2502 and the pCD1 Tn1000 remnant are located in the same general region, they are unrelated.
Several IS remnants are found in the vicinity of yopM (Fig. 3B). Upstream of yopM, there is a portion of IS285 and a region that has amino acid sequence homology to the IS1600 transposase from Mycobacterium fortuitum (65). A similar IS1600-like sequence is present downstream of yopM, as is a segment that is related to the transposase from the Rhizobium meliloti element ISRm3 (Fig. 3 and Table 3) (113).
There are four regions (termed ISD1-like) with homology to ISD1 from D. vulgaris (Fig. 3; Table 3). One segment includes sequences containing genes termed lcrS and lcrT in Y. enterocolitica (92). Other investigators have noted similarity between lcrS and IS ORFs (27). Sequence analysis indicates that the lcrST region is part of an IS element. Consequently we refer to lcrS as OrfA and to lcrT as OrfB. OrfA (bp 14987 to 15253, complement) is 67% identical and 90.9% similar in amino acid sequence to OrfA of ISD1 (43). In Y. pestis, OrfB is longer than lcrT of Y. enterocolitica due to a frameshift mutation and an 11-bp insertion (92). OrfB (bp 14571 to 14951, complement) has the highest homology at the amino acid level to OrfB of ISD1 (43) but is apparently truncated by the insertion of IS100 (Fig. 3A). The homology to ISD1 OrfB is continued in a second ISD1-like segment located upstream of IS100 (Fig. 3A). These two segments may have been part of an ISD1-like element that was disrupted by IS100. The partial ISD1-like element that remains possesses one copy of an inverted repeat which matches the ISD1 repeat in 25 of 44 residues (43). A potential frameshift site (AAAAAAAC, bp 14992 to 14999, complement) is found within OrfA and could yield an OrfAB transframe protein. Two additional regions with amino acid sequence homology to OrfB from ISD1 are located downstream of yopK and Tn1000p, respectively (Table 3). While all of the ISD1-like OrfB remnants are related at the amino acid sequence level, only these last two regions have homologous nucleotide sequences, suggesting that they were derived from an IS element different from that for the other ISD1-like remnants.
A final IS remnant, containing sequences related to IS1327 from Erwinia herbicola (62), is located downstream of the pCD1 sopABC partitioning region. The sequence showing similarity is fairly small, encompassing only 350 bp, and appears to be the only copy of this type on pCD1 (Table 3).
There are several regions that contain clusters of IS elements or remnants (Fig. 3). Two of these IS clusters have already been discussed, the group including IS100 (Fig. 3A) and the remnants around yopM (Fig. 3B). A third collection of IS elements is found in the vicinity of sycH. One of the new IS elements, IS1617, is located downstream of sycH, while the IS21p remnant, contained within an IS1222p remnant, separates sycE from sycH (Fig. 3C). The defective yadA′/′yadA gene also appears to be surrounded by remnants of IS elements (Fig. 3D; Table 3). While there are several interesting correlations with GC content (Table 2), the significance of any of these IS clusters is unknown. They could represent preferred insertion sites for the respective elements. Alternatively, these groupings may simply delineate regions that are nonessential for virulence or plasmid maintenance.
Several of the IS elements appear to have inserted into other mobile genetic elements. Thus, IS100 disrupted an ISD1-like element (Fig. 3A), and IS21p may have inserted into an IS1222p homologue (Fig. 3C). It is difficult to determine if other genes were disrupted by IS transposition. Over time the sequences flanking the ISs or remnants may have diverged so that no apparent ORF remains. However, there are at least two identifiable ORFs whose functions were probably disrupted by IS insertions. These include an ORF encoding a nuclease at positions 782 to 1033 and one encoding a helicase at nucleotides 69568 to 70055. Only portions of these genes remain, to one side of an IS remnant, suggesting that additional genomic rearrangements have occurred since the initial insertion of the IS element.
The nuclease remnant is 94% similar to the carboxy-terminal 82 aa of an endonuclease encoded by the LCR plasmid of Y. enterocolitica 15673 (53a) and 73% similar to a plasmid-encoded endonuclease from Salmonella typhimurium and E. coli. In E. coli and Salmonella this gene is located within a region containing sequences involved in conjugation; however, it is not required for conjugation (86, 114). At this time, the function of this endonuclease is unknown. The portion of helicase remaining in Y. pestis is 66.7% similar to residues 1306 to 1467 of TraI from E. coli. TraI functions during conjugation, not only to unwind but also to nick the DNA at oriT (115).
The presence of these gene remnants in Y. pestis, both of which are associated with conjugation, as well as the genetic linkages of the replication and partitioning functions of pCD1 raise some interesting questions about potential origins of the LCR plasmids. It is possible that at one time the plasmid was capable of conjugation but that the transfer functions were subsequently lost or mutated.
Virulence-related genes.
The LCR genes previously sequenced for Y. enterocolitica and Y. pseudotuberculosis were all present in Y. pestis (with the exception of yscM2, which appears to be unique to Y. enterocolitica [101]). Within previously described operons, the gene order is conserved and the ORFs generally have the same length (three exceptions are discussed below). As anticipated, the homology is high: usually ≥95% and often 98 to 100% identity. Two Ysc components, YscG and YscE, and one effector Yop, YopJ, had 94% identity (95% similarity) to the corresponding proteins of Y. enterocolitica, due to differences scattered within the predicted proteins. Detailed studies will be required to determine if these differences translate into significant functional differences such as arise from the heterogeneity in LcrV (see the introduction). The largest difference between effector Yop proteins of Y. pestis and Y. enterocolitica was in YopM (93% identity [94% similarity] to YopM of Y. enterocolitica), discussed below.
For the genes for the effector Yops and other non-Ysc LCR proteins that are scattered in the regions flanking the central uninterrupted LCR cluster, the base composition varies considerably, from 33.5% GC for yopK to 51.1% for yopE (Table 2), which is suggestive of possible multiple events of Yop gene acquisition by the LCR plasmid. For example, even though ypkA and yopJ lie within an operon in Y. pseudotuberculosis (44), and hence likely also in Y. pestis, they differ significantly in composition. Unlike other LCR operons within the main LCR cluster, which tend to contain tightly packed cistrons, there is a 397-bp spacing between ypkA and yopJ, which itself has 32.2% GC. The DNA upstream of ypkA (including the partial IS sequence) is 45.8% GC, and that downstream from yopJ (including the partial IS sequence) is 46.7% GC. These considerations suggest the possibility that yopJ, together with the intercistronic region, may have been acquired independently of ypkA by the LCR plasmid. Likewise, yopK and the sequence between it and yopT are significantly lower in GC content than are sycT and yopT farther up or than the sequence downstream from yopK, including ylpA. lcrV lies within the cluster of LCR secretion regulation-related genes, but because it also has a virulence role as a secreted protein, it is worth reiterating (89) that it also has a relatively low GC content of 37.7%, as contrasted to yopB and yopD in the same operon, which have 46.7 and 43.1% GC, respectively. These results potentially reflect an interesting evolution for the set of virulence properties on this plasmid.
Previously studied loci.
The YpkA- and YopJ-encoding operon in Y. pestis KIM5, like that in Y. pseudotuberculosis (44), also has a small ORF (Orf7) upstream, but in Y. pestis, this ORF is shorter by 7 residues and is spaced from ypkA by 17, compared to 6 bp. In these two yersiniae these ORFs are identical up to the sequence encoding the C-terminal 10 residues (17 in Y. pseudotuberculosis), where they become very different. An in-frame deletion within this ORF in Y. pseudotuberculosis did not affect expression or secretion of YpkA or YopJ or virulence in mice, leaving it without any obvious role in the LCR or in virulence, and it was speculated that this ORF in fact is not translated (44). The Orf7 product is interesting because it has properties very much like those of the Yop chaperones (Sycs). Its molecular mass of 15,747 Da is similar to the sizes of other described Sycs (111), and like other Sycs, it is predicted to be acidic (pI of 4.39) and to have an amphipathic character. Intriguingly, the database search revealed some homology of this predicted ORF to an ORF upstream of the gene encoding the Avr-like protein HrmA in Pseudomonas syringae pv. syringae (a plant pathogen that secretes virulence proteins by a type III secretion mechanism) (1, 2). These similarities suggest that we should revisit the role of Orf7 to reassess its expression and possible role as a Syc for YopJ, which now is believed to be targeted into eukaryotic cells and has been shown to cause apoptosis of macrophages by Y. pseudotuberculosis and Y. enterocolitica (71, 72). Deletion of this ORF might not have had an effect on virulence, because abolishing YopJ itself did not affect virulence in mice infected orally by Y. pseudotuberculosis (44).
YopM is a leucine-rich repeat (LRR) protein (58) previously noted to be encoded by a gene with a relatively low percent GC content containing a number of exact, directly repeated sequences and inverse complement sequences (61). YopM was reported to be 41,556 Da with a pI of 4.06 (61) but now is seen to be larger, at 46.21 kDa, and to have a predicted pI of 4.23. This new DNA sequence result has been confirmed independently (77a). It is likely that the original analysis was confounded by priming in different, directly repeated regions. YopM is now predicted to have 15 instead of 13 LRRs (77a). Interestingly, the sequence reported for yopM of Y. enterocolitica O:9 (18), which is predicted to encode a protein with 13 LRRs, now differs from the Y. pestis sequence by exactly 2 LRRs, and the difference between the presently predicted Y. pestis YopM and that of Y. enterocolitica comes in a region where the gene contains exact repeats. This kind of genetic structure might be expected to be prone to duplications and deletions, and indeed, yopM in different Yersinia strains varies in size in the LRR-encoding region (77a). Functional studies are needed to determine the significance of this variation.
YopL was designated as the ca. 15-kDa product of a two-cistron operon, yopKL, based on its elimination, along with YopK, from outer membrane fractions of Y. pseudotuberculosis 43 (serotype III) carrying a version of pCD1 of Y. pestis KIM5 that had Mu dI(Ap lac) inserted in yopK; it was the only observed protein species eliminated by a downstream Mu dI(Ap lac) insertion (103). YopL has not been found in Y. pseudotuberculosis or in Y. enterocolitica, although in all three yersiniae there is a spacing of ca. 500 bp between yopK and the downstream monocistronic operon encoding YlpA (30, 54). This spacing is 496 bp in Y. pestis KIM and 482 bp in the sequence from Y. enterocolitica (30); the two sequences of this region are highly similar except for a 14-bp insertion (consisting of a directly duplicated 7-bp sequence) 301 bp after the stop codon for YopK. Although this 496 bp could be large enough to encode YopL, we found that it contains two ORFs that show amino acid homology to the IS element ISD1. Accordingly, the sequence analysis of pCD1 does not support the existence of yopL, and the identity of the second protein eliminated in expression by the Mu d insertion in yopK is not known.
The predicted translation initiation site for YlpA is an uncommon valine codon and lies 40 aa downstream of the initiating methionine predicted for Y. enterocolitica YlpA (30). This surprising finding led us to check further and confirm the sequence in this region by sequencing directly from pCD1 in Y. pestis KIM5. The Y. enterocolitica ylpA 5′ end is indeed present in Y. pestis KIM5, but there is an extra A in a stretch of seven A’s beginning at bp 50070 which is responsible for shifting the reading frame in the pCD1 gene. This 7-A stretch is also present in the pCD1 sequence directly read from Y. pestis KIM5. Accordingly, we believe that YlpA likely is not expressed in Y. pestis, and if it is, it would not be a secreted lipoprotein as it is in Y. enterocolitica: the C residue at the beginning of the mature protein (bp 50020) lies well upstream of the stretch of seven A’s. There are now several instances in which insertions or frameshifts in Y. pestis have abolished expression of genes that are important for virulence in the enteropathogenic yersiniae. These differences are thought to represent adaptations to the vector-borne transmission for Y. pestis and are important for the greater disseminative character of Y. pestis (98). It will be interesting to test the significance of YlpA’s loss in Y. pestis.
The yadA′/′yadA ORF in Y. pestis KIM5 has the same 1-bp deletion as in Y. pestis EV76 (98), showing that this potentially virulence-enhancing loss of a prominent fibrillar adhesin occurred prior to the divergence of the orientalis (strain EV76) and mediaevalis (KIM5) biotypes of Y. pestis.
Potential virulence-related new ORFs.
Fourteen ORFs are not obviously associated with IS elements and either have products with no significant similarity to proteins in the database with known functions or have features suggesting a virulence-related role. These are ORFs that deserve future study as potentially having virulence or virulence-accessory functions.
Orf75 (Table 1; Fig. 1) lies just 1 bp downstream of yopE and lacks an obvious ribosome binding site or upstream promoter. The ORF could encode an 11,192-Da protein with at least one likely transmembrane domain and a noncleavable signal sequence. Its expression conceivably is translationally coupled to that of yopE, suggesting that it could be a member of the LCR. yopE has been called monocistronic, based on its estimated transcript size (750 bases in Y. pseudotuberculosis [41]). The presence of this ORF has not been noted in the literature, even though the beginning of Orf75 is present in the sequences previously submitted for Y. pseudotuberculosis yopE (40, 41), Y. enterocolitica O:9 (69), and Y. pestis EV76 (40). Interestingly, it is intact but is separated from yopE by an insertion element in Y. enterocolitica O:8 strain 8081 (40). At high doses, a Y. pseudotuberculosis mutant containing an insertion in this ORF did not show a loss of virulence in mice infected orally (40). Given that YopE’s importance in virulence was determined with polar insertion mutants (40, 41, 54, 95–97, 104, 105), the significance of this ORF needs to be thoroughly tested.
While preparing this paper, we learned that two new ORFs we found in Y. pestis have been designated yopT and sycT in Y. enterocolitica (56). sycT and yopT are arranged in what appears to be a bicistronic operon 500 bp upstream and on the opposite strand from yopK (Fig. 1). These genes indeed have properties suggesting that they encode a Yop and associated Syc. sycT is predicted to encode an acidic 15.42-kDa peripheral protein (Table 1). The database search brought up weak homology with SycE (with which there is 22% identity). A multiple alignment of SycT with SycE, LcrH (SycD), and SycH shows the greatest similarity toward the C termini of the proteins, as previously demonstrated in a comparison of SycE and LcrH/SycD (111). YopT is predicted to be a peripheral 36.31-kDa basic protein (Table 1). It shows 36.7% identity in residues 98 to 322 with the C terminus (residues 648 to 874) of a surface antigen in Haemophilus somnus that is associated with serum resistance (31). The regulation, mechanism of action, and role in plague of YopT should be investigated.
Orf42 through Orf44, immediately downstream of tyeA (Fig. 1), have been noted to exist in Y. enterocolitica (109). Orf42 has been sequenced for Y. pseudotuberculosis, and a polar insertion near its 3′ end caused a calcium-independent growth phenotype (39), typical of mutations in genes necessary for the functioning of the type III secretion system. As this mutation was complemented by DNA lacking a complete lcrD/yscV gene (downstream of Orf44), this phenotype likely was not caused by disruption of lcrD/yscV. For this reason and because of the location (within the LCR cluster and downstream of tyeA, which is involved in Yop secretion control [57]), we speculate that one or more of Orf42 through Orf44 have a role(s) in secretion or secretion control.
Orf5 (Fig. 1 and Table 1) lies isolated from other virulence-related genes, within a gap between the origin region and an IS1236 remnant. It may not prove to be virulence related.
Orf59, Orf60, and Orf61 (Fig. 1) lie between yopM and sycT. Orf59 is closest to yopM (242 bp away), on the opposite strand, and is predicted to encode a ca. 4-kDa soluble acidic protein (Table 1), significantly smaller than typical Sycs. Orf60 and Orf61 lie 875 bp from Orf59, are separated by 272 bp, and are divergently oriented. Both are predicted to encode membrane-associated proteins with mildly basic pIs that hence do not resemble typical Sycs (acidic, soluble, ca. 16 kDa) or Yops (soluble). Orf60 has an uncommon translation initiation codon (leucine) (Table 1).
Orf73 and Orf74 (Fig. 1) lie in the vicinity of yopE. Their predicted proteins are 10- to 11-kDa soluble acidic proteins that show high similarity to unknown proteins of similar lengths in Mycobacterium tuberculosis; however, neither ORF has a common translation initiation codon (leucine [Orf73] and valine [Orf74]). Both ORFs are predicted to be transcribed in the same direction, with Orf74 overlapping Orf73 by 8 bp (Table 1).
Orf84 and Orf85 (Fig. 1) occupy the region between IS1617 and Tn1000p. They are separated by 139 bp and would be transcribed in the same direction. Both ORFs appear to encode soluble proteins, with the product of Orf84 predicted to be basic and that of Orf85 predicted to be acidic (Table 1).
Summary.
Our analysis of the pCD1 DNA sequence has identified an IncFIIA replicon and Sop-like partitioning system necessary for plasmid maintenance. We noted the insertion sites of IS100, Tn1000p, IS21p, and several partial IS285 elements as well as two new IS elements, IS1616 and IS1617. In addition, there are numerous IS element remnants clustered in four regions of the 70-kb plasmid. We found no evidence for the existence of yopL, and, in Y. pestis, ylpA and yadA are pseudogenes. Although regulatory and secretory components of the LCR constitute a contiguous LCR cluster, elements suggesting that this region is a PAI were not identified. Genes for effector Yops are scattered throughout the plasmid and have widely varying GC contents, indicative of multiple gene acquisition events. This observation coupled with the presence of IS remnants from only distantly related microorganisms suggests that a very complex history of DNA acquisition, insertions, deletions, and rearrangements was required for assembly of pCD1.
We failed to find genes with similarities to putative virulence factors that are not potential members of the LCR. However, we did identify eight ORFs of unknown function (Orfs 5, 59, 60, 61, 73, 74, 84, and 85). The products of Orfs 7, 42, 43, 44, and 75, as well as YopT and its chaperone SycT, are potential new members of the LCR virulence system. Sequence analysis of Orf7 suggests that its product could be a chaperone for YopJ. Clearly, experimental analysis of all of these Orfs is required to determine if they are LCR members or non-LCR virulence determinants.
We corrected the sequence of yopM, showing that its product has two additional LRR repeats that are absent in Y. enterocolitica. While most LCR-related Y. pestis gene products showed 98% identity to their analogous Y. enterocolitica gene products, YopJ, YscG, and YscE were ∼94% identical to Y. enterocolitica products. It will be necessary to determine whether any of the differences in YopM, YopJ, YscG, and YscE and the lack of a functional YlpA gene product are involved in the differing levels of virulence among the pathogenic yersiniae.
ACKNOWLEDGMENTS
We thank G. Plunkett III for help with the codon usage matrix and IS elements, N. T. Perna for performing the initial database searches and organizing their output, and G. F. Mayhew and the technical staff of the Wisconsin Genome Project for DNA sequencing. We thank G. R. Cornelis for informing us of the yopT and sycT designations used in an in-press manuscript from his research group.
This work was supported by Public Health Service (PHS) grant P01 HG01428 to F.R.B. R.D.P. and J.D.F. were supported by PHS grants AI25098 and AI33481. S.C.S. was supported by PHS grants AI21017 and AI41668.
REFERENCES
- 1.Alfano J R, Collmer A. The type III (Hrp) secretion pathway of plant pathogenic bacteria: trafficking harpins, Avr proteins, and death. J Bacteriol. 1997;179:5655–5662. doi: 10.1128/jb.179.18.5655-5662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alfano J R, Kim H-S, Delaney T P, Collmer A. Evidence that the Pseudomonas syringae pv. syringae hrp-linked hrmA gene encodes an Avr-like protein that acts in an hrp-dependent manner within tobacco cells. Mol Plant-Microbe Interact. 1997;10:580–588. doi: 10.1094/MPMI.1997.10.5.580. [DOI] [PubMed] [Google Scholar]
- 3.Allaoui A, Schulte R, Cornelis G R. Mutational analysis of the Yersinia enterocolitica virC operon: characterization of yscE, F, G, I, J, K required for Yop secretion and yscH encoding YopR. Mol Microbiol. 1995;18:343–355. doi: 10.1111/j.1365-2958.1995.mmi_18020343.x. [DOI] [PubMed] [Google Scholar]
- 4.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 5.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1987. [Google Scholar]
- 6.Bakour R, Laroche Y, Cornelis G. Study of the incompatibility and replication of the 70-kb virulence plasmid of Yersinia. Plasmid. 1983;10:279–289. doi: 10.1016/0147-619x(83)90042-2. [DOI] [PubMed] [Google Scholar]
- 7.Barve S S, Straley S C. lcrR, a low-Ca2+-response locus with dual Ca2+-dependent functions in Yersinia pestis. J Bacteriol. 1990;172:4661–4671. doi: 10.1128/jb.172.8.4661-4671.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bearden S W, Fetherston J D, Perry R D. Genetic organization of the yersiniabactin biosynthetic region and construction of avirulent mutants in Yersinia pestis. Infect Immun. 1997;65:1659–1668. doi: 10.1128/iai.65.5.1659-1668.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bercovier H, Mollaret H H, Alonso J M, Brault J, Fanning G R, Steigerwalt A G, Brenner D J. Intra- and interspecies relatedness of Yersinia pestis by DNA hybridization and its relationship to Yersinia pseudotuberculosis. Curr Microbiol. 1980;4:225–229. [Google Scholar]
- 10.Biek D P, Shi J. A single 43-bp sopC repeat of plasmid mini-F is sufficient to allow assembly of a functional nucleoprotein partition complex. Proc Natl Acad Sci USA. 1994;91:8027–8031. doi: 10.1073/pnas.91.17.8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biek D P, Strings J. Partition functions of mini-F affect plasmid DNA topology in Escherichia coli. J Mol Biol. 1995;246:388–400. doi: 10.1006/jmbi.1994.0094. [DOI] [PubMed] [Google Scholar]
- 12.Biot T, Cornelis G R. The replication, partition and yop regulation of the pYV plasmids are highly conserved in Yersinia enterocolitica and Y. pseudotuberculosis. J Gen Microbiol. 1988;134:1525–1534. doi: 10.1099/00221287-134-6-1525. [DOI] [PubMed] [Google Scholar]
- 13.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirpatrick H A, Goeden M A, Rose D J, Mau R, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 15.Bliska J B, Copass M C, Falkow S. The Yersinia pseudotuberculosis adhesin YadA mediates intimate bacterial attachment to and entry into HEp-2 cells. Infect Immun. 1993;61:3914–3921. doi: 10.1128/iai.61.9.3914-3921.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blomberg P, Engdahl H M, Malmgren C, Romby P, Wagner E G H. Replication control of plasmid R1: disruption of an inhibitory RNA structure that sequesters the repA ribosome-binding site permits tap-independent RepA synthesis. Mol Microbiol. 1994;12:49–60. doi: 10.1111/j.1365-2958.1994.tb00994.x. [DOI] [PubMed] [Google Scholar]
- 17.Bochner B R, Huang H-C, Schieven G L, Ames B N. Positive selection for loss of tetracycline resistance. J Bacteriol. 1980;143:926–933. doi: 10.1128/jb.143.2.926-933.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boland A, Sory M-P, Iriarte M, Kerbourch C, Wattiau P, Cornelis G R. Status of YopM and YopN in the Yersinia Yop virulon: YopM of Y. enterocolitica is internalized inside the cytosol of PU5-1.8 macrophages by the YopB, D, N delivery apparatus. EMBO J. 1996;15:5191–5201. [PMC free article] [PubMed] [Google Scholar]
- 19.Bölin I, Wolf-Watz H. The plasmid-encoded Yop2b protein of Yersinia pseudotuberculosis is a virulence determinant regulated by calcium and temperature at the level of transcription. Mol Microbiol. 1988;2:237–245. doi: 10.1111/j.1365-2958.1988.tb00025.x. [DOI] [PubMed] [Google Scholar]
- 20.Borodovsky M, McIninch J. Genmark: parallel gene recognition for both DNA strands. Comput Chem. 1993;17:123–133. [Google Scholar]
- 21.Broom J E, Hill D F, Hughes G, Jones W A, McNaughton J C, Stockwell P A, Petersen G B. Sequence of a transposon identified as TN1000 (γδ) DNA Sequence. 1995;5:185–189. doi: 10.3109/10425179509029361. [DOI] [PubMed] [Google Scholar]
- 22.Brubaker R R. Factors promoting acute and chronic diseases by yersiniae. Clin Microbiol Rev. 1991;4:309–324. doi: 10.1128/cmr.4.3.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brubaker R R, Surgalla M J. The effect of Ca++ and Mg++ on lysis, growth, and production of virulence antigens by Pasteurella pestis. J Infect Dis. 1964;114:13–25. doi: 10.1093/infdis/114.1.13. [DOI] [PubMed] [Google Scholar]
- 24.Burland V, Daniels D L, Plunkett III G, Blattner F R. Genome sequencing on both strands: the Janus strategy. Nucleic Acids Res. 1993;21:3385–3390. doi: 10.1093/nar/21.15.3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burrows T W, Bacon G A. The basis of virulence in Pasteurella pestis: an antigen determining virulence. Br J Exp Pathol. 1956;37:481–493. [PMC free article] [PubMed] [Google Scholar]
- 26.Butler T. Yersinia species (including plague) In: Mandell G L, Bennett J E, Dolin R, editors. Principles and practice of infectious diseases. 4th ed. Vol. 2. New York, N.Y: Churchill Livingstone; 1995. pp. 2070–2078. [Google Scholar]
- 27.Carniel E, Guilvout I, Prentice M. Characterization of a large chromosomal “high-pathogenicity island” in biotype 1B Yersinia enterocolitica. J Bacteriol. 1996;178:6743–6751. doi: 10.1128/jb.178.23.6743-6751.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chandler M, Fayet O. Translational frameshifting in the control of transposition in bacteria. Mol Microbiol. 1993;7:497–503. doi: 10.1111/j.1365-2958.1993.tb01140.x. [DOI] [PubMed] [Google Scholar]
- 29.Cheng L W, Anderson D M, Schneewind O. Two independent type III secretion mechanisms for YopE in Yersinia enterocolitica. Mol Microbiol. 1997;24:757–765. doi: 10.1046/j.1365-2958.1997.3831750.x. [DOI] [PubMed] [Google Scholar]
- 30.China B, Michiels T, Cornelis G R. The pYV plasmid of Yersinia encodes a lipoprotein, YlpA, related to TraT. Mol Microbiol. 1990;4:1585–1593. doi: 10.1111/j.1365-2958.1990.tb02070.x. [DOI] [PubMed] [Google Scholar]
- 31.Cole S P, Guiney D G, Corbeil L B. Molecular analysis of a gene encoding a serum-resistance-associated 76 kDa surface antigen of Haemophilus somnus. J Gen Microbiol. 1993;139:2135–2143. doi: 10.1099/00221287-139-9-2135. [DOI] [PubMed] [Google Scholar]
- 32.Cornelis G R. Yersinia pathogenicity factors. In: Dangl J L, editor. Bacterial pathogenesis of plants and animals. Molecular and cellular mechanisms. New York, N.Y: Springer-Verlag; 1994. pp. 243–263. [Google Scholar]
- 33.Cornelis G R, Wolf-Watz H. The Yersinia Yop virulon: a bacterial system for subverting eukaryotic cells. Mol Microbiol. 1997;23:861–867. doi: 10.1046/j.1365-2958.1997.2731623.x. [DOI] [PubMed] [Google Scholar]
- 34.Farabaugh P J. Programmed translational frameshifting. Annu Rev Genet. 1996;30:507–528. doi: 10.1146/annurev.genet.30.1.507. [DOI] [PubMed] [Google Scholar]
- 35.Ferber D M, Brubaker R R. Plasmids in Yersinia pestis. Infect Immun. 1981;31:839–841. doi: 10.1128/iai.31.2.839-841.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fetherston J D, Perry R D. The pigmentation locus of Yersinia pestis KIM6+ is flanked by an insertion sequence and includes the structural genes for pesticin sensitivity and HMWP2. Mol Microbiol. 1994;13:697–708. doi: 10.1111/j.1365-2958.1994.tb00463.x. [DOI] [PubMed] [Google Scholar]
- 37.Fields K A, Plano G V, Straley S C. A low-Ca2+ response (LCR) secretion (ysc) locus lies within the lcrB region of the LCR plasmid in Yersinia pestis. J Bacteriol. 1994;176:569–579. doi: 10.1128/jb.176.3.569-579.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Filippov A A, Oleinikov P N, Motin V L, Prostsenko O A, Smirnov G B. Sequencing of two Yersinia pestis IS elements, IS285 and IS100. Contrib Microbiol Immunol. 1995;13:306–309. [PubMed] [Google Scholar]
- 39.Forsberg Å, Viitanen A-M, Skrunik M, Wolf-Watz H. The surface-located YopN protein is involved in calcium signal transduction in Yersinia pseudotuberculosis. Mol Microbiol. 1991;5:977–986. doi: 10.1111/j.1365-2958.1991.tb00773.x. [DOI] [PubMed] [Google Scholar]
- 40.Forsberg Å, Wolf-Watz H. Genetic analysis of the yopE region of Yersinia spp.: identification of a novel conserved locus, yerA, regulating yopE expression. J Bacteriol. 1990;172:1547–1555. doi: 10.1128/jb.172.3.1547-1555.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Forsberg Å, Wolf-Watz H. The virulence protein Yop5 of Yersinia pseudotuberculosis is regulated at transcriptional level by plasmid-pIB1-encoded trans-acting elements controlled by temperature and calcium. Mol Microbiol. 1988;2:121–133. doi: 10.1111/j.1365-2958.1988.tb00013.x. [DOI] [PubMed] [Google Scholar]
- 42.Fowler J M, Brubaker R R. Physiological basis of the low calcium response in Yersinia pestis. Infect Immun. 1994;62:5234–5241. doi: 10.1128/iai.62.12.5234-5241.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fu R, Voordouw G. ISD1, an insertion element from the sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough: structure, transposition, and distribution. Appl Environ Microbiol. 1998;64:53–61. doi: 10.1128/aem.64.1.53-61.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Galyov E E, Håkansson S, Wolf-Watz H. Characterization of the operon encoding the YpkA Ser/Thr protein kinase and the YopJ protein of Yersinia pseudotuberculosis. J Bacteriol. 1994;176:4543–4548. doi: 10.1128/jb.176.15.4543-4548.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gemski P, Lazere J R, Casey T, Wohlhieter J A. Presence of a virulence-associated plasmid in Yersinia pseudotuberculosis. Infect Immun. 1980;28:1044–1047. doi: 10.1128/iai.28.3.1044-1047.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gerischer U, D’Argenio D, Ornston L N. IS1236, a newly discovered member of the IS3 family, exhibits varied patterns of insertion into the Acinetobacter calcoaceticus chromosome. Microbiology. 1996;142:1825–1831. doi: 10.1099/13500872-142-7-1825. [DOI] [PubMed] [Google Scholar]
- 47.Goguen J D, Yother J, Straley S C. Genetic analysis of the low calcium response in Yersinia pestis Mu d1 (Ap lac) insertion mutants. J Bacteriol. 1984;160:842–848. doi: 10.1128/jb.160.3.842-848.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hacker J, Blum-Oehler G, Mühldorder I, Tschäpe H. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 49.Haddix P L, Straley S C. Structure and regulation of the Yersinia pestis yscBCDEF operon. J Bacteriol. 1992;174:4820–4828. doi: 10.1128/jb.174.14.4820-4828.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hanai R, Liu R, Benedetti P, Caron P R, Lynch A S, Wang J C. Molecular dissection of a protein SopB essential for Escherichia coli F plasmid partition. J Biol Chem. 1996;271:17469–17475. doi: 10.1074/jbc.271.29.17469. [DOI] [PubMed] [Google Scholar]
- 51.Higuchi K, Kupferberg L L, Smith J L. Studies on the nutrition and physiology of Pasteurella pestis. III. Effects of calcium ions on the growth of virulent and avirulent strains of Pasteurella pestis. J Bacteriol. 1959;77:317–321. doi: 10.1128/jb.77.3.317-321.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hinnebusch B J, Perry R D, Schwan T G. Role of the Yersinia pestis hemin storage (hms) locus in the transmission of plague by fleas. Science. 1996;273:367–370. doi: 10.1126/science.273.5273.367. [DOI] [PubMed] [Google Scholar]
- 53.Hoe N P, Minion F C, Goguen J D. Temperature sensing in Yersinia pestis: regulation of yopE transcription by lcrF. J Bacteriol. 1992;174:4275–4286. doi: 10.1128/jb.174.13.4275-4286.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53a.Hoffmann B, Strauch E, Gewinner C, Nattermann H, Appel B. Characterization of plasmid regions of foodborne Yersinia enterocolitica biogroup 1a strains hybridizing to the Yersinia enterocolitica virulence plasmid. Syst Appl Microbiol. 1998;21:201–211. doi: 10.1016/S0723-2020(98)80024-6. [DOI] [PubMed] [Google Scholar]
- 54.Holmström A, Rosqvist R, Wolf-Watz H, Forsberg Å. Virulence plasmid-encoded YopK is essential for Yersinia pseudotuberculosis to cause systemic infection in mice. Infect Immun. 1995;63:2269–2276. doi: 10.1128/iai.63.6.2269-2276.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Humphreys G O, Willshaw G A, Anderson E S. A simple method for the preparation of large quantities of pure plasmid DNA. Biochim Biophys Acta. 1975;383:457–463. doi: 10.1016/0005-2787(75)90318-4. [DOI] [PubMed] [Google Scholar]
- 56.Iriarte M, Cornelis G R. YopT, a new Yersinia Yop effector protein, affects the cytoskeleton of host cells. Mol Microbiol. 1998;29:915–930. doi: 10.1046/j.1365-2958.1998.00992.x. [DOI] [PubMed] [Google Scholar]
- 57.Iriarte M, Sory M-P, Boland A, Boyd A P, Mills S D, Lambermont I, Cornelis G R. TyeA, a protein involved in control of Yop release and in translocation of Yersinia Yop effectors. EMBO J. 1998;17:1907–1918. doi: 10.1093/emboj/17.7.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kobe B, Deisenhofer J. The leucine-rich repeat: a versatile binding motif. Trends Biochem Sci. 1994;19:415–421. doi: 10.1016/0968-0004(94)90090-6. [DOI] [PubMed] [Google Scholar]
- 59.Krause M, Harwood J, Fierer J, Guiney D. Genetic analysis of homology between the virulence plasmids of Salmonella dulbin and Yersinia pseudotuberculosis. Infect Immun. 1991;59:1860–1863. doi: 10.1128/iai.59.5.1860-1863.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lawton W D, Erdman R L, Surgalla M J. Biosynthesis and purification of V and W antigen in Pasteurella pestis. J Immunol. 1963;91:179–184. doi: 10.21236/ad0299868. [DOI] [PubMed] [Google Scholar]
- 61.Leung K Y, Straley S C. The yopM gene of Yersinia pestis encodes a released protein having homology with the human platelet surface protein GPIbα. J Bacteriol. 1989;171:4623–4632. doi: 10.1128/jb.171.9.4623-4632.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lichter A, Manulis S, Valinsky L, Karniol B, Barash I. IS1327, a new insertion-like element in the pathogenicity-associated plasmid of Erwinia herbicola pv. gypsophilae. Mol Plant-Microbe Interact. 1996;9:98–104. [PubMed] [Google Scholar]
- 63.Mahillon, J., H. A. Kirkpatrick, H. L. Kijenski, C. A. Bloch, C. K. Rode, G. F. Mayhew, D. J. Rose, G. Plunkett III, V. Burland, and F. R. Blattner. Subdivision of Escherichia coli K-12 genome for sequencing: manipulation and DNA sequence of transposable elements introducing unique restriction sites. Gene, in press. [DOI] [PubMed]
- 64.Malmgren C, Wagner E G H, Ehresmann C, Ehresmann B, Romby P. Antisense RNA control of plasmid R1 replication: the dominant product of the antisense RNA-mRNA binding is not a full RNA duplex. J Biol Chem. 1997;272:12508–12512. doi: 10.1074/jbc.272.19.12508. [DOI] [PubMed] [Google Scholar]
- 65.Martin C, Timm J, Rauzier J, Gomez-Lus R, Davies J, Gicquel B. Transposition of an antibiotic resistance element in mycobacteria. Nature. 1990;345:739–743. doi: 10.1038/345739a0. [DOI] [PubMed] [Google Scholar]
- 66.McDonough K A, Hare J M. Homology with a repeated Yersinia pestis DNA sequence IS100 correlates with pesticin sensitivity in Yersinia pseudotuberculosis. J Bacteriol. 1997;179:2081–2085. doi: 10.1128/jb.179.6.2081-2085.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mecsas J, Strauss E J. Molecular mechanisms of bacterial virulence: type III secretion and pathogenicity islands. Emerg Infect Dis. 1996;2:271–285. doi: 10.3201/eid0204.960403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Michiels T, Vanooteghem J-C, Lambert de Rouvroit C, China B, Gustin A, Boudry P, Cornelis G. Analysis of virC, an operon involved in the secretion of Yop proteins by Yersinia enterocolitica. J Bacteriol. 1991;173:4994–5009. doi: 10.1128/jb.173.16.4994-5009.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Michiels T, Wattiau P, Brasseur R, Ruysschaert J-M, Cornelis G. Secretion of Yop proteins by yersiniae. Infect Immun. 1990;58:2840–2849. doi: 10.1128/iai.58.9.2840-2849.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Miller V L, Bliska J B, Falkow S. Nucleotide sequence of the Yersinia enterocolitica ail gene and characterization of the Ail protein product. J Bacteriol. 1990;172:1062–1069. doi: 10.1128/jb.172.2.1062-1069.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mills S D, Boland A, Sory M-P, van der Smissen P, Kerbourch C, Finlay B B, Cornelis G R. Yersinia enterocolitica induces apoptosis in macrophages by a process requiring functional type III secretion and translocation mechanisms and involving YopP, presumably acting as an effector protein. Proc Natl Acad Sci USA. 1997;94:12638–12643. doi: 10.1073/pnas.94.23.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Monack D M, Mecsas J, Ghori N, Falkow S. Yersinia signals macrophages to undergo apoptosis and YopJ is necessary for this cell death. Proc Natl Acad Sci USA. 1997;94:10385–10390. doi: 10.1073/pnas.94.19.10385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mori H, Kondo A, Ohshima A, Ogura T, Hiraga S. Structure and function of the F plasmid genes essential for partitioning. J Mol Biol. 1986;192:1–15. doi: 10.1016/0022-2836(86)90459-6. [DOI] [PubMed] [Google Scholar]
- 74.Mori H, Mori Y, Ichinose C, Niki H, Ogura T, Kato A, Hiraga S. Purification and characterization of SopA and SopB proteins essential for F plasmid partitioning. J Biol Chem. 1989;264:15535–15541. [PubMed] [Google Scholar]
- 75.Motin V L, Nakajima R, Smirnov G B, Brubaker R R. Passive immunity to yersiniae mediated by anti-recombinant V antigen and protein A-V antigen fusion peptide. Infect Immun. 1994;62:4192–4201. doi: 10.1128/iai.62.10.4192-4201.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Motin V L, Pokrovskaya M S, Telepnev M V, Kutyrev V V, Vidyaeva N A, Filippov A A, Smirnov G B. The difference in the lcrV sequences between Y. pestis and Y. pseudotuberculosis and its application for characterization of Y. pseudotuberculosis strains. Microb Pathog. 1992;12:165–175. doi: 10.1016/0882-4010(92)90050-x. [DOI] [PubMed] [Google Scholar]
- 77.Nakajima R, Motin V L, Brubaker R R. Suppression of cytokines in mice by protein A-V antigen fusion peptide and restoration of synthesis by active immunization. Infect Immun. 1995;63:3021–3029. doi: 10.1128/iai.63.8.3021-3029.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77a.Nemeth, J., E. Skrzypek, S. C. Straley, and A. Kajava. Unpublished data.
- 78.Neyt C, Iriarte M, Thi V H, Cornelis G R. Virulence and arsenic resistance in yersiniae. J Bacteriol. 1997;179:612–619. doi: 10.1128/jb.179.3.612-619.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nilles M L, Fields K A, Straley S C. The V antigen of Yersinia pestis regulates Yop vectorial targeting as well as Yop secretion through effects on YopB and LcrG. J Bacteriol. 1998;180:3410–3420. doi: 10.1128/jb.180.13.3410-3420.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Odaert M, Devalckenaere A, Trieu-Cuot P, Simonet M. Molecular characterization of IS1541 insertions in the genome of Yersinia pestis. J Bacteriol. 1998;180:178–181. doi: 10.1128/jb.180.1.178-181.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Olson C H, Blattner F R, Daniels D L. Simultaneous preparation of up to 768 single-stranded DNAs for use as templates in DNA sequencing. Methods. 1991;3:27–32. [Google Scholar]
- 82.Payne P L, Straley S C. YscO of Yersinia pestis is a mobile core component of the Yop secretion system. J Bacteriol. 1998;180:3882–3890. doi: 10.1128/jb.180.15.3882-3890.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Perry R D, Fetherston J D. Yersinia pestis—etiologic agent of plague. Clin Microbiol Rev. 1997;10:35–66. doi: 10.1128/cmr.10.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Plano G V, Barve S S, Straley S C. LcrD, a membrane-bound regulator of the Yersinia pestis low-calcium response. J Bacteriol. 1991;173:7293–7303. doi: 10.1128/jb.173.22.7293-7303.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Podladchikova O N, Dikhanov G G, Rakin A V, Heesemann J. Nucleotide sequence and structural organization of Yersinia pestis insertion sequence IS100. FEMS Microbiol Lett. 1994;121:269–274. doi: 10.1111/j.1574-6968.1994.tb07111.x. [DOI] [PubMed] [Google Scholar]
- 86.Pohlman R F, Liu F, Wang L, Moré M I, Winans S C. Genetic and biochemical analysis of an endonuclease encoded by the IncN plasmid pKM101. Nucleic Acids Res. 1993;21:4867–4872. doi: 10.1093/nar/21.21.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Portnoy D A, Falkow S. Virulence-associated plasmids from Yersinia enterocolitica and Yersinia pestis. J Bacteriol. 1981;148:877–883. doi: 10.1128/jb.148.3.877-883.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Portnoy D A, Wolf-Watz H, Bolin I, Beeder A B, Falkow S. Characterization of common virulence plasmids in Yersinia species and their role in the expression of outer membrane proteins. Infect Immun. 1984;43:108–114. doi: 10.1128/iai.43.1.108-114.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Price S B, Leung K Y, Barve S S, Straley S C. Molecular analysis of lcrGVH, the V antigen operon of Yersinia pestis. J Bacteriol. 1989;171:5646–5653. doi: 10.1128/jb.171.10.5646-5653.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rakin A, Heesemann J. Virulence-associated fyuA/irp2 gene cluster of Yersinia enterocolitica biotype 1B carries a novel insertion sequence IS1328. FEMS Microbiol Lett. 1995;129:287–292. doi: 10.1111/j.1574-6968.1995.tb07594.x. [DOI] [PubMed] [Google Scholar]
- 91.Reimmann C, Moore R, Little S, Savioz A, Willetts N S, Hass D. Genetic structure, function and regulation of the transposable element IS21. Mol Gen Genet. 1989;215:416–424. doi: 10.1007/BF00427038. [DOI] [PubMed] [Google Scholar]
- 92.Rimpiläinen M, Forsberg Å, Wolf-Watz H. A novel protein, LcrQ, involved in the low-calcium response of Yersinia pseudotuberculosis shows extensive homology to YopH. J Bacteriol. 1992;174:3335–3363. doi: 10.1128/jb.174.10.3355-3363.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rodríguez-Peña J M, Buisán M, Ibáñez M, Rotger R. Genetic map of the virulence plasmid of Salmonella enteritidis and nucleotide sequence of its replicons. Gene. 1997;188:53–61. doi: 10.1016/s0378-1119(96)00776-7. [DOI] [PubMed] [Google Scholar]
- 94.Roggenkamp A, Geiger A M, Leitritz L, Kessler A, Heesemann J. Passive immunity to infection with Yersinia spp. mediated by anti-recombinant V antigen is dependent on polymorphism of V antigen. Infect Immun. 1997;65:446–451. doi: 10.1128/iai.65.2.446-451.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rosqvist R, Forsberg Å, Rimpiläinen M, Bergman T, Wolf-Watz H. The cytotoxic protein YopE of Yersinia obstructs the primary host defence. Mol Microbiol. 1990;4:657–667. doi: 10.1111/j.1365-2958.1990.tb00635.x. [DOI] [PubMed] [Google Scholar]
- 96.Rosqvist R, Forsberg Å, Wolf-Watz H. Microinjection of the Yersinia YopE cytotoxin in mammalian cells induces actin microfilament disruption. Biochem Soc Trans. 1991;19:1131–1132. doi: 10.1042/bst0191131. [DOI] [PubMed] [Google Scholar]
- 97.Rosqvist R, Magnusson K-E, Wolf-Watz H. Target cell contact triggers expression and polarized transfer of Yersinia YopE cytotoxin into mammalian cells. EMBO J. 1994;13:964–972. doi: 10.1002/j.1460-2075.1994.tb06341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rosqvist R, Skurnik M, Wolf-Watz H. Increased virulence of Yersinia pseudotuberculosis by two independent mutations. Nature. 1988;334:522–525. doi: 10.1038/334522a0. [DOI] [PubMed] [Google Scholar]
- 99.Skurnik M, Toivanen P. LcrF is the temperature-regulated activator of the yadA gene of Yersinia enterocolitica and Yersinia pseudotuberculosis. J Bacteriol. 1992;174:2047–2051. doi: 10.1128/jb.174.6.2047-2051.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Skurnik M, Wolf-Watz H. Analysis of the yopA gene encoding the Yop1 virulence determinants of Yersinia spp. Mol Microbiol. 1989;3:517–529. doi: 10.1111/j.1365-2958.1989.tb00198.x. [DOI] [PubMed] [Google Scholar]
- 101.Stainier I, Iriarte M, Cornelis G R. YscM1 and YscM2, two Yersinia enterocolitica proteins causing downregulation of yop transcription. Mol Microbiol. 1997;26:833–843. doi: 10.1046/j.1365-2958.1997.6281995.x. [DOI] [PubMed] [Google Scholar]
- 102.Steibl H-D, Lewecke F-M. IS1222: analysis and distribution of a new insertion sequence in Enterobacter agglomerans 339. Gene. 1995;156:37–42. doi: 10.1016/0378-1119(95)00003-o. [DOI] [PubMed] [Google Scholar]
- 103.Straley, S. C. 1988. The plasmid-encoded outer-membrane proteins of Yersinia pestis. Rev. Infect. Dis. 10(Suppl.):S323–S326. [DOI] [PubMed]
- 104.Straley S C, Bowmer W S. Virulence genes regulated at the transcriptional level by Ca2+ in Yersinia pestis include structural genes for outer membrane proteins. Infect Immun. 1986;51:445–454. doi: 10.1128/iai.51.2.445-454.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Straley S C, Cibull M L. Differential clearance and host-pathogen interactions of YopE− and YopK− YopL−Yersinia pestis in BALB/c mice. Infect Immun. 1989;57:1200–1210. doi: 10.1128/iai.57.4.1200-1210.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Straley S C, Perry R D. Environmental modulation of gene expression and pathogenesis in Yersinia. Trends Microbiol. 1995;3:310–317. doi: 10.1016/s0966-842x(00)88960-x. [DOI] [PubMed] [Google Scholar]
- 107.Straley S C, Plano G V, Skrzypek E, Haddix P L, Fields K A. Regulation by Ca2+ in the Yersinia low-Ca2+ response. Mol Microbiol. 1993;8:1005–1010. doi: 10.1111/j.1365-2958.1993.tb01644.x. [DOI] [PubMed] [Google Scholar]
- 108.Vanooteghem J-C, Cornelis G R. Structural and functional similarities between the replication region of the Yersinia virulence plasmid and the RepFIIA replicons. J Bacteriol. 1990;172:3600–3608. doi: 10.1128/jb.172.7.3600-3608.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Viitanen A-M, Toivanen P, Skurnik M. The lcrE gene is part of an operon in the lcr region of Yersinia enterocolitica O:3. J Bacteriol. 1990;172:3152–3162. doi: 10.1128/jb.172.6.3152-3162.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wagner E G H, von Heijne J, Nordström K. Control of replication of plasmid R1: translation of the 7k reading frame in RepA mRNA leader region counteracts the interaction between CopA RNA and CopT RNA. EMBO J. 1987;6:515–522. doi: 10.1002/j.1460-2075.1987.tb04783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wattiau P, Bernier B, Deslée P, Michiels T, Cornelis G R. Individual chaperones required for Yop secretion by Yersinia. Proc Natl Acad Sci USA. 1994;91:10493–10497. doi: 10.1073/pnas.91.22.10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wattiau P, Woestyn S, Cornelis G R. Customized secretion chaperones in pathogenic bacteria. Mol Microbiol. 1996;20:255–262. doi: 10.1111/j.1365-2958.1996.tb02614.x. [DOI] [PubMed] [Google Scholar]
- 113.Wheatcroft R, Laberge S. Identification and nucleotide sequence of Rhizobium melilotii insertion sequence ISRm3: similarity between the putative transposase encoded by ISRm3 and those encoded by Staphylococcus aureus IS256 and Thiobacillus ferrooxidans IST2. J Bacteriol. 1991;173:2530–2538. doi: 10.1128/jb.173.8.2530-2538.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Winans S C, Walker G C. Genetic localization and characterization of a pKM101-coded endonuclease. J Bacteriol. 1983;154:1117–1125. doi: 10.1128/jb.154.3.1117-1125.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yoshioka Y, Fujita Y, Ohtsubo E. Nucleotide sequence of the promoter-distal region of the tra operon of plasmid R100, including traI (DNA helicase I) and traD genes. J Mol Biol. 1990;214:39–53. doi: 10.1016/0022-2836(90)90145-C. [DOI] [PubMed] [Google Scholar]
- 116.Yother J, Chamness T W, Goguen J D. Temperature-controlled plasmid regulon associated with low calcium response in Yersinia pestis. J Bacteriol. 1986;165:443–447. doi: 10.1128/jb.165.2.443-447.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yother J, Goguen J D. Isolation and characterization of Ca2+-blind mutants of Yersinia pestis. J Bacteriol. 1985;164:704–711. doi: 10.1128/jb.164.2.704-711.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zahorchak R J, Brubaker R R. Effect of exogenous nucleotides on Ca2+ dependence and V antigen synthesis in Yersinia pestis. Infect Immun. 1982;38:953–959. doi: 10.1128/iai.38.3.953-959.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zahorchak R J, Charnetzky W T, Little R V, Brubaker R R. Consequences of Ca2+ deficiency on macromolecular synthesis and adenylate energy charge in Yersinia pestis. J Bacteriol. 1979;139:792–799. doi: 10.1128/jb.139.3.792-799.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zink D L, Feeley J C, Wells J G, Vanderzant C, Vickery J C, O’Donovan G A. Possible plasmid-mediated virulence in Yersinia enterocolitica. Trans Gulf Coast Mol Biol Conf. 1978;3:155–163. [Google Scholar]
- 121.Zink D L, Feeley J G, Wells J G, Vanderzant C, Vickery J C, Roof W D, O’Donovan G A. Plasmid-mediated tissue invasiveness in Yersinia enterocolitica. Nature (London) 1980;283:224–226. doi: 10.1038/283224a0. [DOI] [PubMed] [Google Scholar]