Abstract
The oral bacterium Streptococcus gordonii expresses two cell wall-associated polypeptides, designated SspA (1,542 amino acid residues) and SspB (1,462 amino acid residues), that have 70% sequence identity. These polypeptides are members of the antigen I/II family of oral streptococcal adhesins and mediate the binding of streptococci to salivary glycoproteins, collagen, and other oral microorganisms such as Actinomyces naeslundii. To determine if SspA and SspB have differential binding properties, the coding sequences of the sspA and sspB genes were cloned into expression plasmid vector pTREX1-usp45LS to generate pTREX1-sspA and pTREX1-sspB, respectively, and the Ssp polypeptides were displayed on the cell surface of Lactococcus lactis MG1363. Lactococcal cells expressing similar levels of surface SspA or SspB polypeptide were then compared for their abilities to adhere to a range of antigen I/II polypeptide substrates. More than twice as many L. lactis cells expressing SspA bound to immobilized salivary agglutinin glycoprotein (SAG) as did L. lactis cells expressing SspB. In contrast, lactococci expressing SspB adhered twice as well as lactococci producing SspA to collagen type I and to Candida albicans. The binding of A. naeslundii to lactococci was only weakly enhanced by surface expression of Ssp polypeptides. L. lactis(pTREX1-sspB) cells bound in greater numbers to SAG than did Enterococcus faecalis JH2-2 cells expressing SspB from pAM401EB-5. The results suggest that SspA and SspB have markedly different binding affinities for their oral substrates and thus may function to promote site diversity in colonization by S. gordonii.
Streptococci are major primary colonizers of oral hard and soft tissues. The “mutans group” streptococci including Streptococcus mutans and Streptococcus sobrinus are implicated in the development and progression of dental caries. The “mitis group” organisms (24) including Streptococcus gordonii, Streptococcus mitis, Streptococcus oralis, Streptococcus parasanguis, and Streptococcus sanguis are not considered to be highly cariogenic; nevertheless, they play a significant role in the development of the complex microbial accumulations that are associated with caries and other oral diseases such as gingivitis and periodontitis (reviewed in reference 48). Most species of indigenous oral streptococci express on their cell surfaces high-molecular-mass cell wall-associated polypeptides of the antigen I/II family (reviewed in reference 21). These polypeptides are oligospecific adhesins recognizing multiple ligands, and they mediate a wide range of streptococcal adherence properties including binding to salivary glycoproteins (15, 32, 34) (especially parotid salivary agglutinin glycoprotein [SAG] [1, 9]), type I collagen (30, 40), and other oral microbial cells including Actinomyces naeslundii (7, 23), Candida albicans (17), and Porphyromonas gingivalis (4, 27). These antigen I/II polypeptide-binding properties are important not only for initial adhesion of streptococcal cells to saliva-coated surfaces and exposed host tissue matrix but also for interbacterial adhesion and “secondary” colonization by organisms such as P. gingivalis (22).
The antigen I/II family polypeptides have a number of common structural and functional features (21). The polypeptide precursors are between 1500 and 1580 amino acid (aa) residues long, and the primary sequence can be conveniently divided into seven regions. These are, from the N terminus, (i) the leader peptide (approximately 38 aa residues); (ii) an N-terminal charged region (up to 139 aa residues); (iii) alanine-rich helical repeats (approximately 300 aa residues) (A region) with salivary glycoprotein-binding activity; (iv) a V (variable) or D (divergent) region with least sequence similarity among the family members; (v) central proline-rich repeats (P region) comprising about 120 aa residues and highly conserved; (vi) a C-terminal region of approximately 500 aa residues, also highly conserved and carrying sequences implicated in binding SAG, Ca2+, and P. gingivalis; and (vii) a cell wall anchorage region comprising about 85 aa residues. Despite these structural similarities and localized regions of highly conserved sequence among the antigen I/II family polypeptides, evidence suggests that individual members of the protein family have different binding specificities. For example, SAG binding by antigen I/II polypeptide from S. mutans KPSK2 is fucose and lactose sensitive, unlike SAG binding by S. gordonii SspB (10), and S. gordonii SspA and SspB polypeptides both bind P. gingivalis cells whereas S. mutans PAc (antigen I/II) protein does not (4). The molecular basis for these different substrate-binding specificities is not fully understood.
S. gordonii is the only oral Streptococcus species so far identified that expresses two antigen I/II polypeptides. Mature SspA (1,542 aa residues) and SspB (1,462 aa residues) are the products of tandemly arranged, monocistronic chromosomal genes that are independently transcribed (8). Structural and transcriptional start site differences between the sspA and sspB gene promoters indicate that the genes may be differentially regulated in S. gordonii M5. The SspA and SspB polypeptides are 84% identical across their N-terminal regions, 98% identical within the C-terminal regions, but only 27% conserved within the V (or D) region. Insertional inactivation experiments have shown that both sspA and sspB genes are necessary for the binding of S. gordonii cells to SAG, collagen, A. naeslundii, and C. albicans (7, 17, 30). However, these experiments have not enabled the determination of the relative binding properties or affinities of the SspA and SspB polypeptides for their substrates. The SspB protein, when expressed on the surface of Enterococcus faecalis, confers upon the enterococcal cells the ability to bind SAG (6), C. albicans (17), and P. gingivalis (27). Purified SspB binds SAG (9), while recombinant SspA and SspB immobilized onto nitrocellulose are able to support the binding of P. gingivalis cells (4). Thus, the Ssp polypeptides appear to be functionally active when expressed on a heterologous cell surface and retain, to an undefined degree, their binding properties in ex vivo assays.
However, the antigen I/II polypeptides are predicted to form complex tertiary structures (28), and therefore the substrate-binding properties of these proteins may be influenced markedly by conformational folding. To compare the binding functions of the S. gordonii SspA and SspB polypeptides in cell surface conformation, we have expressed the proteins independently as cell surface-anchored molecules in the food-grade organism Lactococcus lactis. Both polypeptides confer upon lactococci a range of antigen I/II-associated adhesion phenotypes. Adhesion assays of lactococcal cells expressing these proteins demonstrate that the SspA and SspB polypeptides have distinct and alternate substrate-binding properties.
MATERIALS AND METHODS
Bacteria and culture conditions.
S. gordonii DL1 (Challis) (36), S. gordonii M5 (a human oral isolate provided by B. Rosan, University of Pennsylvania), E. faecalis JH2-2 (19), and A. naeslundii T14V (3) were cultured at 37°C in brain heart infusion medium (Difco Laboratories, Detroit, Mich.) containing 0.5% (wt/vol) yeast extract (BHY). L. lactis MG1363 (12) and derivatives were cultured at 30°C in M17 medium (Difco) containing 0.5% (wt/vol) glucose. Cultures were inoculated from stock cell suspensions stored at −80°C in BHY medium or M17-glucose medium containing 15% (wt/vol) glycerol and were grown in closed tubes or bottles without shaking. C. albicans ATCC 10261 was grown at 30°C with aeration in a salts-biotin medium containing 1% (wt/vol) glucose (18). Escherichia coli DH5α (16) cells were cultivated in Luria-Bertani medium (39). Antibiotics were incorporated into media where appropriate at the following concentrations: ampicillin, 50 μg/ml; erythromycin, 5 μg/ml (S. gordonii, E. faecalis, and L. lactis) or 50 μg/ml (E. coli); and chloramphenicol, 5 μg/ml (E. faecalis and L. lactis) or 10 μg/ml (E. coli).
Transformation.
E. coli cells were transformed following CaCl2 treatment (39), E. faecalis was electrotransformed following incubation of cells with 4% (wt/vol) glycine essentially as described by Cruz-Rodz and Gilmore (5), and L. lactis was electroporated and transformed with plasmid DNA as previously described (46).
DNA manipulations.
Routine molecular biology techniques were performed by the methods described by Sambrook et al. (39). Chromosomal DNA was isolated from S. gordonii as described previously (20). Plasmid DNA was isolated from E. coli by using Wizard Minipreps (Promega Corp., Madison, Wis.), and from E. faecalis and L. lactis by a modified alkali lysis method (46). DNA restriction and modification enzymes were used under the conditions recommended by the manufacturers.
PCR amplification.
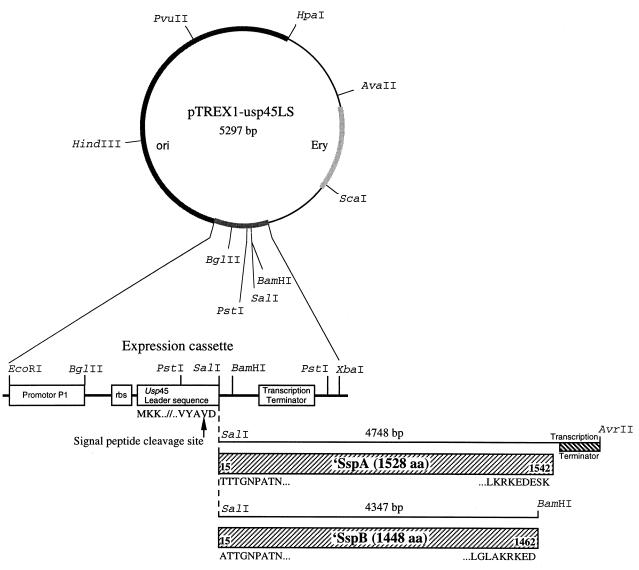
Synthetic oligonucleotides were derived from the nucleotide sequences of sspA and sspB (GenBank accession no. U40025 and U40026, respectively) and were generated to incorporate restriction enzyme sites at their 5′ ends to facilitate cloning. PCR primer pair 1 was designed to amplify DNA from bp 124 to 4871 (U40025) encoding SspA aa residues 15 to 1542 of the mature protein, the TAA stop codon, and 37 bp downstream, within which was a rho-independent transcriptional terminator sequence (stem-loop). The primers were (coding strand) 5′ACGCGTCGACACTACAACTGGGAATCCAGCT3′ (SalI site underlined) and (complementary strand) 5′GCCCTAGGCTTTGAAAACTCAAGAAGCG3′ (AvrII site underlined). PCR primer pair 2 was designed to amplify DNA from bp 669 to 5013 (U40026) encoding SspB aa residues 15 to 1462 and the TAA stop codon. The primers were (coding strand) 5′ACGCGTCGACGCTACTACAGGAAACCCGGCC3′ (SalI site underlined) and (complementary strand) 5′CGGGATCCAATCTTCTTTGCGTTTTGCCAGACC3′ (BamHI site underlined). PCR mixtures contained S. gordonii M5 DNA (50 ng), 3 U of TaqPlus long polymerase mixture (Stratagene, La Jolla, Calif.), and a final Mg2+ concentration of 2 mM. The conditions for DNA amplification were as follows: 40 cycles of a denaturation step (95°C for 30 s), an annealing step (60°C for 60 s), and an extension step (72°C for 4 min; an extra extension (72°C for 10 min) was performed at the end of the final cycle. The PCR products were purified, digested with a combination of SalI and AvrII or SalI and BamHI, and ligated with pTREX1-usp45LS (45) cut with a combination of SalI-XbaI or SalI-BamHI respectively (Fig. 1). In cloning the sspB amplicon, digestion with BamHI generated a fragment with 211 bp missing at the 3′ end owing to the presence of a BamHI site within the sspB gene sequence. Accordingly, the missing 211-bp BamHI fragment was reinserted into the primary clone to generate pTREX1-sspB, and the orientation was confirmed by DNA sequencing. The authenticity of cloning and fusion with the vector usp45 sequence was checked by nucleotide sequencing of recombinant plasmids with custom-synthesized oligonucleotides. The fidelity of PCR amplification was determined across approximately 750 bp of sspA and sspB sequences encompassing the regions encoding the Ad1 and Ad2 adhesion-mediating amino acid sequences that are wholly conserved between the polypeptides (21).
FIG. 1.
Schematic representation of plasmid pTREX1-usp45LS and the sspA and sspB expression plasmids pTREX1-sspA and pTREX1-sspB, respectively. PCR-generated fragments of the S. gordonii genes sspA (encoding aa residues 15 to 1542) and sspB (encoding residues 15 to 1462) were cloned into pTREX1-usp45LS to generate in-frame fusions with the secretion leader peptide of the lactococcal usp45 gene (43). Transcription is driven from a strong lactococcal phage P1 promoter, and the translation initiation region carries a ribosome-binding (Shine-Dalgarno) sequence (rbs) from E. coli bacteriophage T7 gene 10 that has been modified to increase the complementarity to the 16S rRNA of L. lactis (45). Cloning of the sppA sequence carrying the chromosomal transcription terminator replaced the SalI-XbaI fragment within the cassette, while cloning of the sspB sequence replaced the SalI-BamHI fragment, leaving the BamHI-XbaI (152 bp) cassette sequence containing bacteriophage T7 RNA polymerase transcription terminator. The main features of the expression cassette and key restriction sites are indicated. Ery, erythromycin resistance gene.
Analysis of bacterial proteins.
Cell wall polypeptides were released from late-exponential-phase streptococcal cells following generation of spheroplasts by incubating cells with mutanolysin (500 U/ml) (7). Surface proteins were extracted from E. faecalis and L. lactis cells by the same method but with the inclusion of lysozyme (200 μg/ml). Protein concentrations were determined with a protein assay kit (Bio-Rad Laboratories, Richmond, Calif.). Polypeptides were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and stained with silver nitrate or transferred to nitrocellulose by electroblotting as previously described (7, 23). The blots were incubated with rabbit polyclonal antibodies raised to purified P1 (SpaP) polypeptide from S. mutans (provided by N. A. Jacques, Institute of Dental Research, University of Sydney, Sydney, Australia) at a 1:200 dilution, and antibody binding was detected with peroxidase-conjugated swine anti-rabbit immunoglobulins (Dako Corp., Carpinteria, Calif.) (23).
Enzyme-linked immunosorbent assay (ELISA).
Bacterial cells were immobilized onto wells of microtiter plates (Nunc, Roskilde, Denmark) and reacted with SpaP (P1) antiserum, and antibody binding was detected with peroxidase-conjugated secondary antibodies as described elsewhere (17). Antigen concentrations were expressed as absorbance at 492 nm (A492) at a 1:2,000 dilution of serum corrected for A492 values obtained for an irrelevant rabbit antiserum.
Bacterial adherence assays.
Streptococci, enterococci and A. naeslundii cells (BHY medium) and lactococci (M17-glucose medium) were radioactively labeled by growth in medium containing [methyl-3H]thymidine (6 μCi/ml [85 Ci/mmol]; Amersham Corp., Arlington Heights, Ill.) (23) to a specific radioactivity of between 6 × 10−4 and 2 × 10−3 cpm per cell. C. albicans cells were metabolically labeled with [35S]methionine (0.62 MBq, 17 mCi, 103 Ci/mmol) (17). Adherence of cells to immobilized SAG was measured as described previously (23), and adhesion to acid-soluble collagen type I was measured as detailed by Love et al. (30). The binding of radioactively labeled A. naeslundii to immobilized gram-positive cocci was measured by the method of McNab et al. (31), and adhesion of C. albicans to streptococci and lactococci was determined as described previously (17). Student’s t test was used for statistical analysis of the data, and P < 0.05 was considered significant.
RESULTS
Expression of sspB from within pAM401 in E. faecalis and L. lactis.
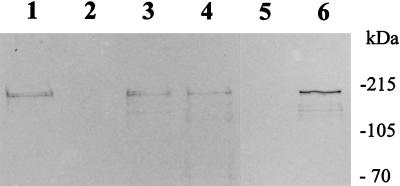
Demuth et al (6) first demonstrated functional expression of an oral streptococcal adhesin (SspB) in a heterologous gram-positive host. The sspB gene and upstream promoter region was cloned into the shuttle plasmid pAM401 (49), and SspB protein was expressed on the E. faecalis S161 cell surface as demonstrated by immunoelectron microscopy (6). By using antibodies raised to P1 (SpaP) protein from S. mutans, which react with both SspA and SspB polypeptides (7), the amount of antibody-reactive SspB protein present on the surface of intact E. faecalis JH2-2(pAM401EB-5) cells was determined by ELISA to be about 45% of the total P1 antibody-reactive protein on the surface of S. gordonii DL1 (Table 1). Western blots of corresponding surface protein extracts reacted with P1 antibodies showed somewhat similar profiles (Fig. 2) and also showed that similar amounts of antigenic material were removed from E. faecalis and S. gordonii cells (Fig. 2).
TABLE 1.
ELISA reactivities and adhesion properties of E. faecalis JH2-2 or L. lactis MG1363 expressing cell surface S. gordonii SspB encoded by plasmid pAM401-EB5
| Strain | ELISA reactivity (mean A492 ± SD [n = 4])a | 106 cells bound to immobilized SAGb (mean ± SD [n = 4])c | 105A. naeslundii T14V cells bound (mean ± SD [n = 4])d |
|---|---|---|---|
| S. gordonii DL1-Challis | 0.79 ± 0.05 | 9.92 ± 0.37 | 64.0 ± 7.10 |
| E. faecalis JH2-2(pAM401) | 0.08 ± 0.00 | 0.52 ± 0.01 | 2.00 ± 0.30 |
| E. faecalis JH2-2(pAM401EB-5) | 0.36 ± 0.01e | 2.58 ± 0.19e | 20.0 ± 2.10e |
| L. lactis MG1363(pAM401) | 0.01 ± 0.00 | <0.01 | 1.50 ± 0.20 |
| L. lactis MG1363(pAM401EB-5) | 0.15 ± 0.01e | 2.07 ± 0.12e | 2.00 ± 0.40 |
Immobilized bacterial cells were incubated with S. mutans SpaP antiserum (diluted 1:2,000) (17), and ELISA values (A492) were corrected for values obtained with an irrelevant rabbit serum (diluted 1:2,000).
A 250-ng portion of SAG was applied to the microtiter plate wells.
Input cell number, 2 × 107.
Input Actinomyces cell number, 1 × 107.
Values significantly different (P < 0.05) from values for the corresponding plasmid control strains.
FIG. 2.
Western blot of surface-extracted polypeptides reacted with polyclonal antibodies to antigen I/II family polypeptide SpaP (P1) from S. mutans. Protein samples were prepared following cell wall hydrolytic enzyme treatment of intact cells (see Materials and Methods), and equal loadings (10 ± 1.0 μg) were electrophoresed in SDS–10% (wt/vol) polyacrylamide gels, electroblotted onto nitrocellulose, and reacted with antiserum diluted 1:200. Lanes: 1, S. gordonii DL1-Challis; 2, L. lactis MG1363(pTREX1); 3, L. lactis MG1363(pTREX1-sspA); 4, L. lactis MG1363(pTREX1-sspB); 5, E. faecalis JH2-2(pAM401); 6, E. faecalis JH2-2(pAM401EB-5). Positions of molecular mass marker proteins are indicated.
Enterococcal cells expressing SspB exhibited 5-fold-increased binding to immobilized SAG (Table 1) and 10-fold-increased binding to A. naeslundii cells (Table 1) compared with E. faecalis JH2-2 wild-type cells. Enterococci expressing SspB also showed enhanced adhesion to C. albicans cells (17). Plasmid pAM401EB-5 carrying sspB was then electroporated into L. lactis MG1363 to determine if the sspB gene was expressed in this organism. L. lactis(pAM401EB-5) cells expressed only about 40% of the level of SspB surface antigen present on enterococcal cells as determined by ELISA (Table 1). Nevertheless, lactococci producing SspB protein showed significantly enhanced binding to SAG (Table 1) over that of control cells. However, L. lactis MG1363(pAM401EB-5) cells did not exhibit significantly increased adherence to A. naeslundii (Table 1). The reactivities of E. faecalis(pAM401) or L. lactis(pAM401) intact cells or protein extracts with antigen I/II antibodies were negligible (Table 1; Fig. 2), and their adhesion properties were identical to those of pAM401 plasmid-free wild-type cells (data not shown). These experiments confirmed that SspB polypeptide expressed heterologously in either E. faecalis or L. lactis confers upon the respective bacterial cells ability to bind SAG.
Controlled expression of sspA and sspB genes in L. lactis.
Having established that SspB polypeptide could be functionally expressed on the cell surface of L. lactis, we designed experiments to compare further the binding properties of SspB and SspA. To achieve equivalent expression levels of these proteins in L. lactis, it was necessary to ensure that transcriptional and translational control signals were identical for both genes. However, the sspA and sspB gene promoters in S. gordonii are somewhat different in sequence and structure (8). Therefore, PCR amplimers of the S. gordonii M5 sspA and sspB genes were each ligated in frame with DNA encoding the leader peptide of the lactococcal secreted protein Usp45 (43) and transcription was initiated from a strong lactococcal P1 promoter present within plasmid pTREX1-usp45LS (Fig. 1). Thus, fusion polypeptides were generated comprising aa residue 15 of the mature SspA or SspB polypeptide bonded with aa residue D29 of the Usp45 leader, with cleavage of the signal peptide for protein secretion occurring within the Usp45 leader between aa residues A27 and V28 (Fig. 1). The only difference between the fusion constructs was that the sspA gene was PCR amplified together with transcriptional terminator sequence whereas the sspB amplimer, lacking its own transcription terminator, was cloned into pTREX1-usp45LS upstream of the transcriptional terminator present within the expression cassette (Fig. 1).
Evidence that equivalent expression levels of SspA and SspB proteins were achieved in recombinant L. lactis MG1363(pTREX1-sspA) and L. lactis MG1363(pTREX1-sspB) was provided by ELISA data. Similar but not identical ELISA reactivities were obtained for intact cells of these strains reacted with polyclonal antigen I/II (SpaP) antibodies (Table 2). The 13% lower ELISA value obtained for L. lactis cells expressing SspB may simply reflect lower overall reactivity of the polyclonal serum antibodies with SspB. Larger amounts of SspB protein were expressed by L. lactis MG1363(pTREX1-sspB) than by L. lactis MG1363(pAM401EB-5) (Table 1). Furthermore, higher levels of ELISA-reactive SspB were present on the L. lactis MG1363(pTREX1-sspB) cell surface than on the E. faecalis JH2-2(pAM401EB-5) cell surface. L. lactis MG1363(pTREX1-sspA) expressed 80% of the ELISA reactivity of S. gordonii cells (Table 2). These results, taken collectively, demonstrate that pTREX1-usp45LS is a highly efficient expression vector. All strains expressed the SspA or SspB polypeptides with an apparent molecular mass of approximately 180 kDa in SDS-PAGE (Fig. 2). These proteins were released from the cell wall following treatment of cells with enzymes hydrolyzing cell wall peptidoglycan and were not extracted by heating intact cells with 1% (wt/vol) SDS.
TABLE 2.
ELISA reactivities and adhesion properties of L. lactis MG1363 strains expressing cell surface S. gordonii SspA or SspB
| Strain | ELISA reactivity (mean A492 ± SD [n = 4])a | 106 cells bound to immobilized SAGb (mean ± SD [n = 4])c | 105 cells bound to immobilized collagend (mean ± SD [n = 4])c | 105A. naeslundii T14V cells bound (mean ± SD [n = 4])e | 104C. albicans ATCC 10261 cells bound (mean ± SD [n = 4])f |
|---|---|---|---|---|---|
| S. gordonii DL1-Challis | 0.79 ± 0.05 | 9.92 ± 0.37 | 46.8 ± 5.70 | 64.0 ± 7.10 | 5.05 ± 0.40 |
| L. lactis MG1363(pTREX1) | 0.01 ± 0.00 | 1.18 ± 0.14 | 6.40 ± 0.40 | 4.00 ± 0.80 | 2.09 ± 0.25 |
| L. lactis MG1363(pTREX1-sspA) | 0.67 ± 0.01g | 13.2 ± 0.93g,h | 10.6 ± 0.47g | 6.80 ± 0.81g | 3.52 ± 0.44g |
| L. lactis MG1363(pTREX1-sspB) | 0.58 ± 0.02g | 5.80 ± 1.10g | 20.0 ± 0.50g,h | 8.80 ± 0.70g | 6.41 ± 0.54g,h |
Immobilized bacterial cells were incubated with S. mutans SpaP antiserum (diluted 1:2,000) (17), and ELISA values (A492) were corrected for values obtained with an irrelevant rabbit serum (diluted 1:2,000).
A 250-ng portion of SAG was applied to the microtiter plate wells.
Input cell number, 2 × 107.
A 5-μg portion of acid-soluble type I collagen was applied to the microtiter plate wells (30).
Input Actinomyces cell number, 1 × 107.
Input Candida cell number, 2 × 105.
Values significantly different (P < 0.05) from those for control strain MG1363(pTREX1).
Values significantly different (P < 0.05) from the alternate ssp-expressing strain.
Plasmids pTREX1, pTREX1-sspA, and pTREX1-sspB were also introduced by electroporation into E. faecalis JH2-2. These plasmids replicated autonomously and conferred erythromycin resistance in E. faecalis transformants; however, Ssp polypeptides were not expressed on the cell surface, as determined by ELISA (results not shown). The most likely explanation for this was that the lactococcal P1 promoter was not recognized efficiently in E. faecalis, although the structural stability of the plasmids was not investigated.
Comparison of SspA and SspB polypeptide-binding properties.
Lactococcal cells expressing cell surface Ssp polypeptides bound in substantially greater numbers to all substrates tested than did the L. lactis MG1363 control strain harboring pTREX1. Background binding levels of L. lactis(pTREX1) were somewhat higher than those of L. lactis(pAM401) (Table 1), which might be related to the different antibiotic selections used. However, the binding properties of lactococci expressing SspA or SspB were quite different. L. lactis expressing SspA adhered in more than twofold-greater numbers to SAG than did L. lactis expressing SspB (Table 2) and in greater numbers than did S. gordonii DL1 itself (Table 2). On the other hand, L. lactis expressing SspB adhered twofold better than did L. lactis expressing SspA to collagen type I (Table 2). Thus, the SspA and SspB proteins appeared to have alternate binding affinities for SAG and collagen substrates. Differences in the binding affinities of the SspA and SspB proteins for A. naeslundii and C. albicans were less pronounced. Strains of lactococci expressing SspA or SspB both showed statistically significant increased binding of A. naeslundii cells over that by controls (Table 2). However, the slightly greater numbers of Actinomyces cells bound to L. lactis expressing SspB were not statistically different from the numbers bound to L. lactis expressing SspA (Table 2). Significantly more (nearly twice as many) C. albicans cells bound to L. lactis expressing SspB than L. lactis expressing SspA (Table 2).
DISCUSSION
Cell wall proteins of gram-positive bacteria are currently the focus of much attention because of their role in a number of processes including adhesion (22) and invasion (2, 29); modulation of immune system function, e.g., by cytokine expression and activation (44); inhibition of phagocytosis (38); and evasion of host immune defense mechanisms (13). These proteins are in the main structurally complex, containing repeat regions of amino acid residues in addition to regions of unique sequence, with both regions being implicated in mediating adhesion. In a number of cases, substrate-binding activities are associated with linear sequences of 100 aa residues or less (22). The S. mutans antigen I/II polypeptide (variously designated AgB, PAc, P1, or SpaP) contains sequences within the alanine-rich (A), variable (V) or divergent (D), and C-terminal regions (see the introduction) that recognize salivary glycoproteins (1, 32, 33). In particular, two linear sequences of 40 aa residues (Ad1) and 20 aa residues (Ad2) are present C-terminal to the central proline-rich (P) region and appear to be major adhesion-mediating sequences (25). Recently, a sequence of approximately 80 aa residues present C-terminal to Ad2 in S. gordonii SspB polypeptide has been shown to be necessary for binding P. gingivalis cells (4) and Ca2+ (11). However, these linear sequences are contained within the complex molecular environments of large polypeptides comprising 1,450 aa residues or more. Consequently, the binding functions are likely to be influenced by the tertiary structure of the protein. Therefore, to characterize further the binding properties of these complex adhesin molecules, it is important to be able to present them in, as far as is possible, their native cell surface conformations.
To begin to address this issue, we have expressed the S. gordonii Ssp adhesins on the cell surface of L. lactis. This organism is nonpathogenic and does not colonize mammals (45), and since there are no reports of high-affinity binding of lactococci to human tissue proteins, epithelial cells, or other bacteria colonizing humans, the organism is ideally suited to studies of streptococcal adhesin function. The SspA and SspB polypeptides were expressed from pTREX1-usp45LS, localized to the bacterial cell surface, and extracted only from cells that had been previously incubated with cell wall hydrolytic enzymes; they could not be extracted from cells with SDS alone. This indicated that the Ssp polypeptides were covalently linked to the lactococcal cell wall, presumably via the gram-positive surface protein anchorage mechanism involving a C-terminal LPXTG motif (35) that is also recognized in lactococci (37, 42). While previous work (6) has demonstrated that E. faecalis also could be used to express functional SspB adhesin, enterococci are less desirable as adhesin display vehicles because they are opportunistic pathogens and may demonstrate a range of phenotypes of adhesion to human tissue components (26, 41). Greater expression levels of SspB polypeptide antigen were obtained in the present study with L. lactis than with E. faecalis, and L. lactis cells expressing SspB bound in larger numbers to SAG than did E. faecalis cells expressing SspB. Interestingly, though, E. faecalis(pAM401EB-5) cells, which produced about 50% less ELISA-reactive cell surface antigen than did L. lactis(pTREX1-sspB) cells, were able to bind at least twice as many A. naeslundii cells than the lactococci did. One possible explanation for this is that the first 15 aa residues of SspA or SspB (which are missing in the constructs expressed in L. lactis) may be critical for antigen I/II polypeptide binding to A. naeslundii. However, this appears to be ruled out by the observation that L. lactis(pAM401EB-5) cells expressing native SspB, while showing an increased ability to bind SAG, did not exhibit an enhanced ability to bind Actinomyces (Table 1). Therefore, it is possible that enterococci express SspB on the cell surface in a form that is better recognized by Actinomyces. Alternatively, SspB-mediated binding of enterococci to A. naeslundii could be promoted through cooperative effects exerted by other surface molecules that are not active on lactococci. Adhesin cooperativity is implicated in the binding reactions of S. gordonii cells to Actinomyces, which involves not only the SspA and SspB polypeptides but also other multiple adhesin-receptor interactions that presumably contribute to strengthening coadhesion (17, 31).
The antigen I/II polypeptides may be responsible, at least in part, for the different binding affinities of streptococcal species for salivary glycoproteins (1). In particular, the Ad1 and Ad2 adhesion-mediating sequences are species specific, and it is speculated that they may determine the specificity of binding to SAG (21). Indeed, the Ad1 and Ad2 sequences within S. gordonii SspA and SspB proteins are identical. However, the SspA and SspB proteins exhibit 70% identical amino acid residues across their lengths (7), while the divergent-region sequences contain only 27% identical amino acid residues. Since lactococci expressing SspA bound SAG more efficiently than did lactococci expressing SspB, this implies that the polypeptides have different affinities for binding salivary glycoprotein ligands. Furthermore, the data strongly support evidence (1, 32, 34) that the binding of antigen I/II polypeptides to salivary glycoproteins involves regions outside of, and in addition to, the Ad1 and Ad2 sequences. Nucleotide sequencing of 750 bp encompassing the Ad1 and Ad2 regions within the pTREX1-sspA and pTREX1-sspB constructs confirmed that the Ad1 and Ad2 sequences in these plasmids were identical to those previously reported (7). This eliminates the possibility that nonfidelity of PCR amplification generated alterations in these adhesion-mediating sequences. Although we cannot exclude the possibility that PCR amplification errors were introduced into other regions, the results argue strongly that sequences outside Ad1 and Ad2 are important for adhesion. Thus, SAG binding by antigen I/II polypeptides may involve an interaction of SAG with more than one binding site, as suggested by monoclonal antibody inhibition studies (1) and synthetic peptide inhibition studies (25, 32). Alternatively, Ad1 and Ad2 sequences may be crucial in forming the SAG-binding pocket, with the affinity of SAG binding being influenced by conformational folding directed by other linearly distal sequences. The ability to express functional antigen I/II polypeptides on the lactococcal cell surface provides a future means of determining more precisely, by genetic and immunological techniques, the sequences and conformations involved in SAG recognition.
Implicit within the interpretation of the comparative binding data for lactococcal cells expressing SspA or SspB proteins is that expression levels of the genes and of the polypeptides at the cell surface were equivalent. The P1 antibody-binding results suggest that similar amounts (within 15%) of SspA or SspB polypeptides were present on the surfaces of the respective lactococcal cells, but the possibility cannot be excluded that the P1 antibodies lack the specificity to detect more significant differences in the surface expression levels of the two polypeptides. Such differences would affect the interpretation of absolute adhesion values and data on the relative affinities of SspA and SspB for various substrates but would not affect the conclusion that the individual SspA and SspB proteins have alternate spectra of substrate recognition. One way to determine more precisely the surface levels of SspA and SspB expressed in heterologous bacteria would be to generate a monoclonal antibody that is reactive with a conserved linear epitope.
The SspA and SspB polypeptides have been shown recently to mediate the adhesion of S. gordonii cells to collagen type I (30). The results reported in this paper extend these data and suggest that SspB polypeptide has a higher affinity than SspA polypeptide for binding to collagen type I. The isolated N-terminal one-third of antigen I/IIf (from S. mutans serotype f) binds collagen, laminin, and fibronectin (40). It is possible, therefore, that these different abilities of SspA and SspB to bind collagen are related to differences in primary sequence within the A regions of the polypeptides that are 84% conserved. Clearly, the ability to functionally express these polypeptides on the lactococcal cell surface should allow future delineation of the collagen-binding regions. It may be envisaged that the alternate binding properties of SspA and SspB for SAG and collagen enable different host environments or surfaces to be colonized by S. gordonii. Hence, the bacteria may be able to bind the salivary pellicle and colonize the tooth surface and may be able to bind collagen present in dentin and invade dentinal tubules (30) without compromising either infection process. Preliminary evidence indicates that expression of the sspA and sspB genes might be differentially regulated (8, 30). This might then impart an additional level of control on the site specificity of adhesion and colonization by S. gordonii.
Interest in heterologous protein expression by L. lactis has centered around the development of these organisms as vaccine delivery vehicles (45–47). Other food-grade bacteria such as Staphylococcus carnosus have also been engineered to display heterologous polypeptides on their surface (14) for biotechnological applications. The work described in this paper demonstrates that L. lactis cells can be used as display vehicles to characterize the binding properties of streptococcal adhesins. Since the displayed polypeptide is covalently linked to an adhesion-inert cell surface, this obviates the requirement for purifying the polypeptide for analysis. Furthermore, the polypeptide is presented in close to native conformation, permitting more detailed investigations of antigen I/II polypeptide structure and function.
ACKNOWLEDGMENTS
We thank N. A. Jacques for providing P1 (SpaP) antiserum, D. B. Clewell and B. Rosan for providing bacterial strains, and R. A. Baker for excellent technical assistance. We also thank R. J. Lamont, D. R. Demuth, and R. McNab for the provision of strains and plasmids and for helpful discussions.
This work was supported by the Health Research Council of New Zealand, and J.M.W. gratefully acknowledges the award of an Advanced Research Fellowship from the Biotechnology and Biological Sciences Research Council, United Kingdom.
REFERENCES
- 1.Brady L J, Piacentini D A, Crowley P J, Oyston P C F, Bleiweis A S. Differentiation of salivary agglutinin-mediated adherence and aggregation of mutans streptococci by use of monoclonal antibodies against the major surface adhesin P1. Infect Immun. 1992;60:1008–1017. doi: 10.1128/iai.60.3.1008-1017.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braun L, Dramsi S, Dehoux P, Bierne H, Lindahl G, Cossart P. InlB: an invasion protein of Listeria monocytogenes with a novel type of surface association. Mol Microbiol. 1997;25:285–294. doi: 10.1046/j.1365-2958.1997.4621825.x. [DOI] [PubMed] [Google Scholar]
- 3.Brecher S M, van Houte J, Hammond B F. Role of colonization in the virulence of Actinomyces viscosus T14-Vi and T14-Av. Infect Immun. 1978;22:603–614. doi: 10.1128/iai.22.2.603-614.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brooks W, Demuth D R, Gil S, Lamont R J. Identification of a Streptococcus gordonii SspB domain that mediates adhesion to Porphyromonas gingivalis. Infect Immun. 1997;65:3753–3758. doi: 10.1128/iai.65.9.3753-3758.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cruz-Rodz A L, Gilmore M S. High efficiency introduction of plasmid DNA into glycine treated Enterococcus faecalis by electroporation. Mol Gen Genet. 1990;224:152–154. doi: 10.1007/BF00259462. [DOI] [PubMed] [Google Scholar]
- 6.Demuth D R, Berthold P, Leboy P S, Golub E E, Davis C A, Malamud D. Saliva-mediated aggregation of Enterococcus faecalis transformed with a Streptococcus sanguis gene encoding the SSP-5 surface antigen. Infect Immun. 1989;57:1470–1475. doi: 10.1128/iai.57.5.1470-1475.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demuth D R, Duan Y, Brooks W, Holmes A R, McNab R, Jenkinson H F. Tandem genes encode cell-surface polypeptides SspA and SspB which mediate adhesion of the oral bacterium Streptococcus gordonii to human and bacterial receptors. Mol Microbiol. 1996;20:403–413. doi: 10.1111/j.1365-2958.1996.tb02627.x. [DOI] [PubMed] [Google Scholar]
- 8.Demuth D R, Duan Y, Jenkinson H F, McNab R, Gil S, Lamont R J. Interruption of the Streptococcus gordonii M5 sspA/sspB intergenic region by an insertion sequence related to IS1167 of Streptococcus pneumoniae. Microbiology. 1997;143:2047–2055. doi: 10.1099/00221287-143-6-2047. [DOI] [PubMed] [Google Scholar]
- 9.Demuth D R, Golub E E, Malamud D. Streptococcal-host interactions. Structural and functional analysis of a Streptococcus sanguis receptor for a human salivary glycoprotein. J Biol Chem. 1990;265:7120–7126. [PubMed] [Google Scholar]
- 10.Demuth D R, Lammey M S, Huck M, Lally E T, Malamud D. Comparison of Streptococcus mutans and Streptococcus sanguis receptors for human salivary agglutinin. Microb Pathog. 1990;9:199–211. doi: 10.1016/0882-4010(90)90022-i. [DOI] [PubMed] [Google Scholar]
- 11.Duan Y, Fisher E, Malamud D, Golub E, Demuth D R. Calcium-binding properties of SSP-5, the Streptococcus gordonii M5 receptor for salivary agglutinin. Infect Immun. 1994;62:5220–5226. doi: 10.1128/iai.62.12.5220-5226.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gasson M J. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983;154:1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gravekamp C, Kasper D L, Michel J L, Kling D E, Carey V, Madoff L C. Immunogenicity and protective efficacy of the alpha C protein of group B streptococci are inversely related to the number of repeats. Infect Immun. 1997;65:5216–5221. doi: 10.1128/iai.65.12.5216-5221.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gunneriusson E, Samuelson P, Uhlen M, Nygren P, Stahl S. Surface display of a functional single-chain Fv antibody on staphylococci. J Bacteriol. 1996;178:1341–1346. doi: 10.1128/jb.178.5.1341-1346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hajishengallis G, Koga T, Russell M W. Affinity and specificity of the interactions between Streptococcus mutans antigen I/II and salivary components. J Dent Res. 1994;73:1493–1502. doi: 10.1177/00220345940730090301. [DOI] [PubMed] [Google Scholar]
- 16.Hanahan D. Techniques for transformation of E. coli. In: Glover D M, editor. DNA cloning. Vol. 1. Oxford, United Kingdom: IRL Press; 1985. pp. 109–135. [Google Scholar]
- 17.Holmes A R, McNab R, Jenkinson H F. Candida albicans binding to the oral bacterium Streptococcus gordonii involves multiple adhesin-receptor interactions. Infect Immun. 1996;64:4680–4685. doi: 10.1128/iai.64.11.4680-4685.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holmes A R, Shepherd M G. Nutritional factors determine germ tube formation of Candida albicans. J Med Vet Mycol. 1988;26:127–131. [PubMed] [Google Scholar]
- 19.Jacob A E, Hobbs S J. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J Bacteriol. 1974;117:360–372. doi: 10.1128/jb.117.2.360-372.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jenkinson H F. Novobiocin-resistant mutants of Streptococcus sanguis with reduced cell hydrophobicity and defective in coaggregation. J Gen Microbiol. 1987;133:1909–1918. doi: 10.1099/00221287-133-7-1909. [DOI] [PubMed] [Google Scholar]
- 21.Jenkinson H F, Demuth D R. Structure, function and immunogenicity of streptococcal antigen I/II polypeptides. Mol Microbiol. 1997;23:183–190. doi: 10.1046/j.1365-2958.1997.2021577.x. [DOI] [PubMed] [Google Scholar]
- 22.Jenkinson H F, Lamont R J. Streptococcal adhesion and colonization. Crit Rev Oral Biol Med. 1997;8:175–200. doi: 10.1177/10454411970080020601. [DOI] [PubMed] [Google Scholar]
- 23.Jenkinson H F, Terry S D, McNab R, Tannock G W. Inactivation of the gene encoding surface protein SspA in Streptococcus gordonii DL1 affects cell interactions with human salivary agglutinin and oral actinomyces. Infect Immun. 1993;61:3199–3208. doi: 10.1128/iai.61.8.3199-3208.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawamura Y, Hou X G, Sultana F, Miura H, Ezaki T. Determination of 16S rRNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of the genus Streptococcus. Int J Syst Bacteriol. 1995;45:406–408. doi: 10.1099/00207713-45-2-406. [DOI] [PubMed] [Google Scholar]
- 25.Kelly C G, Todryk S, Kendal H L, Munro G H, Lehner T. T-cell, adhesion, and B-cell epitopes of the cell surface Streptococcus mutans protein antigen I/II. Infect Immun. 1995;63:3649–3658. doi: 10.1128/iai.63.9.3649-3658.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kreft B, Marre R, Schramm U, Wirth R. Aggregation substance of Enterococcus faecalis mediates adhesion to cultured renal tubular cells. Infect Immun. 1992;60:25–30. doi: 10.1128/iai.60.1.25-30.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lamont R J, Gil S, Demuth D R, Malamud D, Rosan B. Molecules of Streptococcus gordonii that bind Porphyromonas gingivalis. Microbiology. 1994;140:867–872. doi: 10.1099/00221287-140-4-867. [DOI] [PubMed] [Google Scholar]
- 28.LaPolla R J, Haron J A, Kelly C G, Taylor W R, Bohart C, Hendricks M, Pyati J, Graff R T, Ma J K-C, Lehner T. Sequence and structural analysis of surface protein antigen I/II (SpaA) of Streptococcus sobrinus. Infect Immun. 1991;59:2677–2685. doi: 10.1128/iai.59.8.2677-2685.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.LeBrun M, Mengaud J, Ohayon H, Nato F, Cossart P. Internalin must be on the bacterial cell surface to mediate entry of Listeria monocytogenes into epithelial cells. Mol Microbiol. 1996;21:579–592. doi: 10.1111/j.1365-2958.1996.tb02566.x. [DOI] [PubMed] [Google Scholar]
- 30.Love R M, McMillan M D, Jenkinson H F. Invasion of dentinal tubules by oral streptococci is associated with collagen recognition mediated by the antigen I/II family of polypeptides. Infect Immun. 1997;65:5157–5164. doi: 10.1128/iai.65.12.5157-5164.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McNab R, Jenkinson H F, Loach D M, Tannock G W. Cell-surface-associated polypeptides CshA and CshB of high molecular mass are colonization determinants in the oral bacterium Streptococcus gordonii. Mol Microbiol. 1994;14:743–754. doi: 10.1111/j.1365-2958.1994.tb01311.x. [DOI] [PubMed] [Google Scholar]
- 32.Moisset A, Schatz N, Lepoivre Y, Amadio S, Wachsmann D, Scholler M, Klein J-P. Conservation of salivary glycoprotein-interacting and human immunoglobulin G-cross-reactive domains of antigen I/II in oral streptococci. Infect Immun. 1994;62:184–193. doi: 10.1128/iai.62.1.184-193.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Munro G H, Evans P, Todryk S, Buckett P, Kelly C P, Lehner T. A protein fragment of streptococcal cell surface antigen I/II which prevents adhesion of Streptococcus mutans. Infect Immun. 1993;61:4590–4598. doi: 10.1128/iai.61.11.4590-4598.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakai M, Okahashi N, Ohta H, Koga T. Saliva-binding region of Streptococcus mutans surface protein antigen. Infect Immun. 1993;61:4344–4349. doi: 10.1128/iai.61.10.4344-4349.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Navarre W W, Schneewind O. Proteolytic cleavage and cell wall anchoring at the LPXTG motif of surface proteins in gram-positive bacteria. Mol Microbiol. 1994;14:115–121. doi: 10.1111/j.1365-2958.1994.tb01271.x. [DOI] [PubMed] [Google Scholar]
- 36.Pakula R, Walczak W. On the nature of competence of transformable streptococci. J Gen Microbiol. 1963;31:125–133. doi: 10.1099/00221287-31-1-125. [DOI] [PubMed] [Google Scholar]
- 37.Piard J C, Hautefort I, Fischetti V A, Ehrlich S D, Fons M, Gruss A. Cell wall anchoring of the Streptococcus pyogenes M6 protein in various lactic acid bacteria. J Bacteriol. 1997;179:3068–3072. doi: 10.1128/jb.179.9.3068-3072.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Podbielski A, Schnitzler N, Beyhs P, Boyle M D P. M-related protein (Mrp) contributes to group A streptococcal resistance to phagocytosis by human granulocytes. Mol Microbiol. 1996;19:429–441. doi: 10.1046/j.1365-2958.1996.377910.x. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Sciotti M A, Yamodo I, Klein J P, Ogier J A. The N-terminal half part of the oral streptococcal antigen I/IIf contains two distinct binding domains. FEMS Microbiol Lett. 1997;153:439–445. doi: 10.1111/j.1574-6968.1997.tb12608.x. [DOI] [PubMed] [Google Scholar]
- 41.Shorrock P J, Lambert P A. Binding of fibronectin and albumin to Enterococcus (Streptococcus) faecalis. Microb Pathog. 1989;6:61–67. doi: 10.1016/0882-4010(89)90008-9. [DOI] [PubMed] [Google Scholar]
- 42.van Asseldonk M, de Vos W M, Simons G. Functional analysis of the Lactococcus lactis usp45 secretion signal in the secretion of a homologous proteinase and heterologous α-amylase. Mol Gen Genet. 1993;240:428–434. doi: 10.1007/BF00280397. [DOI] [PubMed] [Google Scholar]
- 43.van der Meer J R, Polman J, Beerthuyzen M M, Siezen R J, Kuipers O P, de Vos W M. Characterization of the Lactococcus lactis nisin A operon genes nisP, encoding a subtilisin-like serine protease involved in precursor processing, and nisR, encoding a regulatory protein involved in nisin biosynthesis. J Bacteriol. 1993;175:2578–2588. doi: 10.1128/jb.175.9.2578-2588.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vernier A, Diab M, Soell M, Haan-Archipoff G, Beretz A, Wachsmann D, Klein J-P. Cytokine production by human epithelial and endothelial cells following exposure to oral viridans streptococci involves lectin interactions between bacteria and cell surface receptors. Infect Immun. 1996;64:3016–3022. doi: 10.1128/iai.64.8.3016-3022.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wells J M, Robinson K, Chamberlain L M, Schofield K M, Le Page R W F. Lactic acid bacteria as vaccine delivery vehicles. Antonie Leeuwenhoek. 1996;70:317–330. doi: 10.1007/BF00395939. [DOI] [PubMed] [Google Scholar]
- 46.Wells J M, Wilson P W, Le Page R W F. Improved cloning vectors and transformation procedure for Lactococcus lactis. J Appl Bacteriol. 1993;74:629–636. doi: 10.1111/j.1365-2672.1993.tb05195.x. [DOI] [PubMed] [Google Scholar]
- 47.Wells J M, Wilson P W, Norton P M, Gasson M J, Le Page R W F. Lactococcus lactis: high-level expression of tetanus toxin fragment C and protection against lethal challenge. Mol Microbiol. 1993;8:1115–1162. doi: 10.1111/j.1365-2958.1993.tb01660.x. [DOI] [PubMed] [Google Scholar]
- 48.Whittaker C J, Klier C M, Kolenbrander P E. Mechanisms of adhesion by oral bacteria. Annu Rev Microbiol. 1996;50:513–552. doi: 10.1146/annurev.micro.50.1.513. [DOI] [PubMed] [Google Scholar]
- 49.Wirth R, An F Y, Clewell D B. Highly efficient protoplast transformation system for Streptococcus faecalis and a new Escherichia coli-S. faecalis shuttle vector. J Bacteriol. 1986;165:831–836. doi: 10.1128/jb.165.3.831-836.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]