Abstract
Hypoxia-inducible factor pathway genes are linked to adaptation in both human and nonhuman highland species. EPAS1, a notable target of hypoxia adaptation, is associated with relatively lower hemoglobin concentration in Tibetans. We provide evidence for an association between an adaptive EPAS1 variant (rs570553380) and the same phenotype of relatively low hematocrit in Andean highlanders. This Andean-specific missense variant is present at a modest frequency in Andeans and absent in other human populations and vertebrate species except the coelacanth. CRISPR-base-edited human cells with this variant exhibit shifts in hypoxia-regulated gene expression, while metabolomic analyses reveal both genotype and phenotype associations and validation in a lowland population. Although this genocopy of relatively lower hematocrit in Andean highlanders parallels well-replicated findings in Tibetans, it likely involves distinct pathway responses based on a protein-coding versus noncoding variants, respectively. These findings illuminate how unique variants at EPAS1 contribute to the same phenotype in Tibetans and a subset of Andean highlanders despite distinct evolutionary trajectories.
Variation in hematocrit is mediated by different variants and mechanisms involving the same gene (EPAS1) in Tibetans and Andeans.
INTRODUCTION
High-altitude populations exhibit phenotypes that reflect several hundred generations of genetic adaptations to environmental stress. Many Tibetan and Andean groups have distinct physiological traits that mitigate the decreased availability of oxygen (O2) at high altitude via augmented O2 transport and utilization as well as protection from reproductive and neonatal complications (1, 2). Only a few of these traits have been linked to genetic factors, including relatively lower hemoglobin concentration ([Hb]) (3–5) and altered metabolic function within Tibetans (6, 7) and increased uterine artery diameter, birth weight (8), and exercise capacity within Andeans (9). However, the extent of adaptation involving the same genetic regions across continental highland populations remains an ongoing area of research (10, 11).
Numerous genomic selection studies have implicated a wide range of candidate genes as targets for human high-altitude adaptation (3–5, 9, 12–28). Many of the strongest signals of natural selection emanate from genes within the hypoxia-inducible factor (HIF) pathway, a master transcriptional regulator of cellular responses to low O2 (29). Endothelial PAS Domain Protein 1 (EPAS1), which encodes the alpha subunit of HIF-2 (29), is one of the top selection targets identified in humans (1, 30) and other highland species (31, 32). In addition, variants within EPAS1 have been associated with relatively lower [Hb] in Tibetan groups, which are linked, indirectly or directly, to an adaptive phenotype [e.g., protection from excessive erythrocytosis (EE)] (3, 5). Although Tibetans demonstrate lower average [Hb] than Andeans, Andean highlanders exhibit substantial intrapopulation variation in this phenotype (1), with values ranging from sea level to excessively high levels (33). While many of the same candidate gene regions studied in Tibetans have been identified as potential targets of selection in Andeans (12), including EPAS1 (10, 25), adaptations upon the same genes and associated phenotypes have not been found.
Here, we examine a previously identified EPAS1 missense variant (rs570553380, A>G, p.[His194Arg]) (10, 25) within a cohort of Andean highlanders (Cerro de Pasco, Peru, 4340 m). Through genotype-phenotype analyses and in vitro functional assessments, we show evidence of genotypic-phenotypic, cross-continental convergence between Andean and Tibetan highland groups. We demonstrate that lower hematocrit and altered transcriptional activity is associated with a putatively adaptive, Andean-specific missense EPAS1 variant. In addition, we identify unique metabolomic signatures associated with this EPAS1 allele and hematocrit.
RESULTS
Positive selection of EPAS1 variant rs570553380 in Andean highlanders
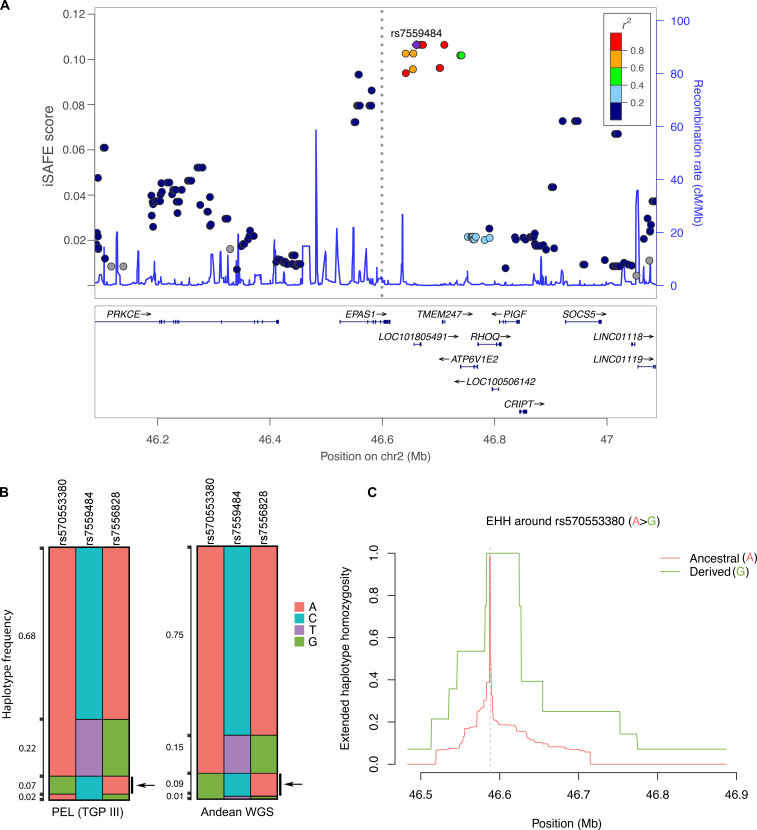
We sequenced the whole genomes of 40 Quechua Andean highlanders (figs. S1 and S2) and examined variants within a single-megabase region surrounding a protein-coding site at rs570553380 using integrated Selection of Allele Favored by Evolution (iSAFE) (34) (Fig. 1A). We observed a signature of selection at this locus as similarly reported in a study of early-stage selection in Colla highlanders (25). The top iSAFE signal localized to a set of intergenic variants (rs7559484, T>C and rs7556828, G>A) approximately 70 kb upstream of rs570553380, which could not be tested directly by iSAFE because of its modest allele frequency. Haplotype analysis of rs7559484, rs7556828, and rs570553380 in the Andean whole genomes (n = 80 haplotypes) and 1000 Genomes Peruvian (PEL) subpopulation (n = 170 haplotypes) showed the putatively adaptive allele of rs570553380 (G) is inherited with derived alleles rs7559484 (C) and rs7556828 (A) (Fig. 1B) (D′ > 0.99). Furthermore, these derived alleles are enriched in Andean highlanders (both 83.8%) compared to the global average (45 and 44%, respectively), suggesting that early-stage selection of rs570553380 may have contributed to an increase in their frequencies. Extended haplotype homozygosity (EHH) (Fig. 1C) and integrated haplotype score (iHS) analyses of rs570553380 in Andeans highlight a signal of natural selection at rs570553380 (G), in which haplotypes with the derived allele (G) were significantly longer than those with the ancestral allele (A) [iHS = −2.43, P < 0.02; |iHS| > 2 indicates strong selection (35)]. In contrast, EHH and iHS analyses of rs7559484 and rs7556828 demonstrated no significant differences in haplotype length (fig. S3), suggesting that selection at the EPAS1 locus in Andeans may be attributed to rs570553380.
Fig. 1. EPAS1 variant under positive selection in Quechua Andean highlanders.
(A) iSAFE results from the 1-Mb window surrounding rs570553380. Regional plots of iSAFE scores (34) generated from the EPAS1 locus in the Andean whole genomes and visualized using Locuszoom. The left y axis shows the iSAFE score ranging from 0 to 1. Markers with iSAFE scores equal to or greater than 0.10 are considered favored by natural selection. The right y axis denotes the recombination rate in hundred crossovers per million base pairs (cM/Mb). The gray dotted line indicates the position of EPAS1 rs570553380 variant (unscored), and the purple diamond denotes the marker in the region with the highest iSAFE score (rs7559484). SNVs in the region are denoted in circles, color-coded based on linkage disequilibrium (LD) with the top iSAFE marker. (B) Haplotype-frequency plot of haplotypes in 1000 Genomes PEL subpopulation (n = 170 haplotypes) and the Andean whole genomes (WGS) (n = 80 haplotypes) containing rs570553380 and top iSAFE markers (rs7559484 and rs7556828). The black arrow indicates the haplotypes carrying the putatively adaptive “G” allele of rs570553380, in which the “C” and “A” alleles of respective iSAFE markers rs7559484 and rs7556828 are nearly always present (D′ > 0.99). (C) EHH plot around rs570553380 (A>G) in the 40 Quechuan Andean whole genomes.
Although the allele frequency of rs570553380 (A>G) is modest among Andean highlanders [Quechua: 10% in this study; Quechua: 9% (10); Colla: 32% (25)], rs570553380 (G) is nearly absent in publicly available genome data with a global frequency of only 0.20% [attributed solely to the Peruvian subpopulation (PEL) = 7.1%] (1000 Genomes, Phase III). The estimated age of rs570553380 is ~9845 years (Genealogical Estimation of Variant Age (GEVA), based on 1000 Genomes data) and ~13,027 years (based off calculations of haplotype heterozygosity in the 40 Andean whole genomes) (table S1). These estimates align approximately with the first human habitation of the Andes (1, 36) and suggest this variant may have originated within early settlers of this region. The age estimates of this variant are consistent with its modest allele frequency in Andeans and early-stage selection at this locus.
Positively selected EPAS1 allele is associated with relatively lower hematocrit in Andeans
Putatively adaptive variants in EPAS1 are associated with significantly lower [Hb] within Tibetan individuals (3, 5). Hematocrit (Hct), the percent volume of red blood cells in whole blood is, on average, higher in Andeans than Tibetans (1) and proportional to [Hb] (Hct ≈ 3*[Hb], [Hb] in grams per deciliter). EE, an overproduction of red blood cells and hallmark of chronic mountain sickness (CMS), is more prevalent in Andean than Tibetan men and associated with pulmonary hypertension, myocardial infarction, and cor pulmonale (33).
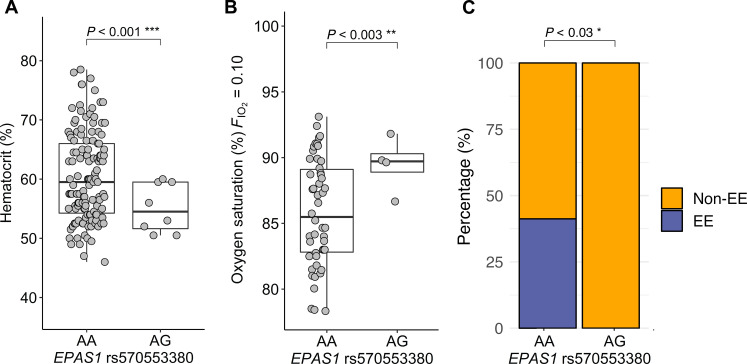
We tested whether rs570553380 (A>G, p.[His194Arg]) was associated with Hct in an expanded cohort of Andean highlanders (n = 139 males, 49 females) (table S2) and validated via a permutation test (fig. S5). Given women have a lower prevalence of EE before menopause (33, 37, 38) and estrogen has a protective effect on EE (39), analyses were stratified by sex. Males with one copy of the putatively adaptive allele “G” (Arg) demonstrated a significantly lower Hct (n = 139) (P < 0.001) (Fig. 2A), higher O2 saturation at 10% inspired O2 ( = 0.10) [n = 55, subset of 139 Andean males (40)] (P < 0.003) (Fig. 2B), and lower incidence of EE (n = 139) (P < 0.03) (Fig. 2C) compared to individuals with the ancestral allele. However, these relationships were not significant in females (fig. S4), most likely due to a smaller sample size of females and well-described differences in Hct and EE with respect to sex and age (before and after menopause) (33, 39, 41) (table S2). Primary and/or secondary effects of this variant on these phenotypes and interplay with other O2 transport components, as well as expanded studies in women, are necessary to understand the underlying physiological mechanisms.
Fig. 2. Positively selected EPAS1 variant is associated with lower hematocrit, higher oxygen saturation, and lower incidence of EE within Andean males.
In Andean males, the EPAS1 missense variant rs570553380 (A>G, p.[His194Arg]) is associated with (A) hematocrit (%) (n = 139), (B) oxygen saturation (%SpO2) at = 0.10 [subset of the 139 Andeans, n = 55; data from (40)], and (C) incidence of EE (n = 139). EE criteria: Hct ≥ 63% in men ([Hb] ≥ 21 g/dl) and ≥ 57% in women ([Hb] ≥ 19 g/dl) (63) (see fig. S4 for females). *P < 0.05, **P < 0.01, and ***P < 0.001. NS, not significant.
EPAS1 site is highly conserved and adaptive variant attenuates expression of HIF target genes
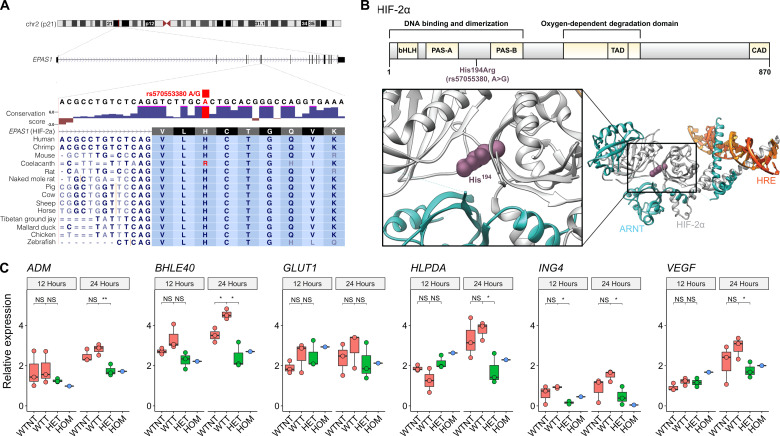
Analysis via phyloP (conservation score) (42) indicates significantly greater conservation of rs570553380 (A>G) than expected due to neutral drift [−log(P) = 6.02] (Fig. 3A). A comparison of rs570553380 (A>G, p.[His194Arg]) across vertebrates at the amino acid level revealed the wild-type histidine residue (His194) as highly conserved across species except the coelacanth, Latimeria chalumnae, a lobe-fined fish found in the depths of the West Indian Ocean. During the past 100 million years, coelacanths have migrated from shallow-brackish to deeper marine waters, during which extreme selective pressures, such as hypoxia, may have acted (43). Although humans and coelacanths diverged more than 400 million years ago (44), this species of lobe-finned fish has the same arginine residue as coded by rs570553380 in Andean highlanders (fig. S6), suggesting potential convergence at this locus.
Fig. 3. EPAS1 variant is highly conserved and results in altered expression of HIF-2α targets under hypoxia.
(A) Conservation of rs570553380 (A>G) in a subset of 100 Species Vertebrate Multiz Alignment & Conservation (fig. S6). Conservation scores (phyloP) of nucleotides across 100 vertebrates showed significant conservation at rs570553380 [−log(P) = 6.02; positive scores indicate slower evolutionary change while negative scores indicate accelerated evolution, greater than expected due to neutral drift (42)]. (B) Location of rs570553380 within the ribbon diagram and crystal structure of HIF-2α (white) as bound to aryl hydrocarbon receptor nuclear translocator (ARNT) (teal) and the hypoxia response element (HRE, orange) (Protein Data Bank ID: 4ZPK). His194 (purple), shown in sphere representation, extends into one of the protein-protein interfaces between HIF-2α and ARNT. (C) Relative expression of HIF-2α target genes Adrenomedullin (ADM), Basic Helix-loop-helix Family Member e40 (BHLE40), Solute Carrier Family 2 Member 1 (GLUT1), Hypoxia Inducible Lipid Droplet Associated (HILPDA), Inhibitor of Growth Family Member 4 (ING4), and Vascular Endothelial Growth Factor A (VEGF) (table S5) in HEK293T cells nontransfected (WTNT, n = 3) and transfected wild-type (WTT, n = 3), heterozygous (HET, n = 3), and homozygous (HOM, n = 1) for rs570553380 (A>G) exposed to hypoxia (1% O2) relative to normoxia (21% O2) for 12 and 24 hours. Gene expression was determined by real-time reverse transcription quantitative polymerase chain reaction (RT-qPCR). Two-way analysis of variance (ANOVA) shows a significant effect of rs570553380 on gene expression of ADM, BHLE40, HILPDA, ING4, and VEGF after 24-hour exposure to hypoxia, as well as a transfection effect on BHLE40 gene expression. The post hoc generalized linear model shows that increased expression of ADM, BHLE40, HILPDA, ING4, and VEGF from 12- to 24-hour exposure to hypoxia is significantly lower in HET compared to WTT. *P < 0.05 and **P < 0.01.
In silico analysis of rs570553380 (A>G, p.[His194Arg]) (Fig. 3B) predicted the Arg residue as destabilizing and deleterious in humans (table S3) (45–47), suggesting the EPAS1 variant may alter HIF-2α structure, stability, and/or transcriptional activity. To assess potential transcriptional effects of rs570553380, we introduced this variant into human endothelial kidney (HEK) 293T cells using CRISPR-Cas9 ABE base editing (48) (fig. S7). Potential off-target mutations from transfection and clonal expansion were assessed via whole-exome sequencing (WES), in which no increase in exome-wide A•T to G•C mutations was observed (fig. S8 and table S4). To evaluate the transcriptional effects of this EPAS1 variant in response to hypoxia, isogenic wild-type, heterozygous, and homozygous clones were cultured under conditions of normoxia (21% O2) and hypoxia (1% O2) for 12 and 24 hours. The selection of these conditions were based on previous literature in which these O2 tensions and time points were implemented to study HIF activity in HEK293T cells (49–52).
Cells with one copy of the putatively adaptive G (Arg) allele demonstrated lower expression of numerous canonical HIF-2 target genes (Fig. 3C) despite showing HIF-2α levels similar to those of wild-type cells under hypoxia (fig. S9). In particular, expression of ADM, BHLE40, HILPDA, ING4, and VEGF (table S5) was significantly decreased in cells heterozygous for the EPAS1 variant relative to wild-type cells from 12- to 24-hour hypoxia exposure relative to normoxia (Fig. 3C). Wild-type cells exhibited lower levels of HIF-2α than cells heterozygous for SNV rs570553380 in normoxia, suggesting potential compensatory changes in response to altered HIF activity (fig. S9). The observed down-regulation of canonical HIF targets in cells with the Andean-specific missense variant parallels the loss-of-function phenotype similarly observed in studies of this variant (10) and Tibetan-specific noncoding variants in EPAS1 (11, 53, 54). While these findings provide insight into altered transcriptional activity as a result of this missense variant in vitro, previous EPAS1 loss-of-function studies in animal models suggest a protective effect against chronic hypoxia, in which heterozygous knockout mice (Epas1+/−) exhibit lower [Hb] and reduced risk of pulmonary hypertension and right ventricular hypertrophy relative to wild-type mice following exposure to sustained hypoxia (54).
EPAS1 variant is associated with serum metabolites in Andeans
To investigate the relationship between circulating metabolites, Hct, and rs570553380, we completed analysis of serum metabolites via liquid chromatography–mass spectrometry (LC-MS) in a subset of the Quechua Andean cohort (n = 115). Of the 5957 metabolites examined from distinctive LC-MS peaks, 298 (21 previously identified, 277 unidentified) were associated with Hct after false discovery rate (FDR) correction (FDR < 0.05). When cross-compared with the FINRISK cohort, a Finnish population survey of chronic, noncommunicable diseases (55) (n = 1170), 12 of these 298 metabolites were associated with higher Hct in Andeans, a finding validated in FINRISK data (table S6).
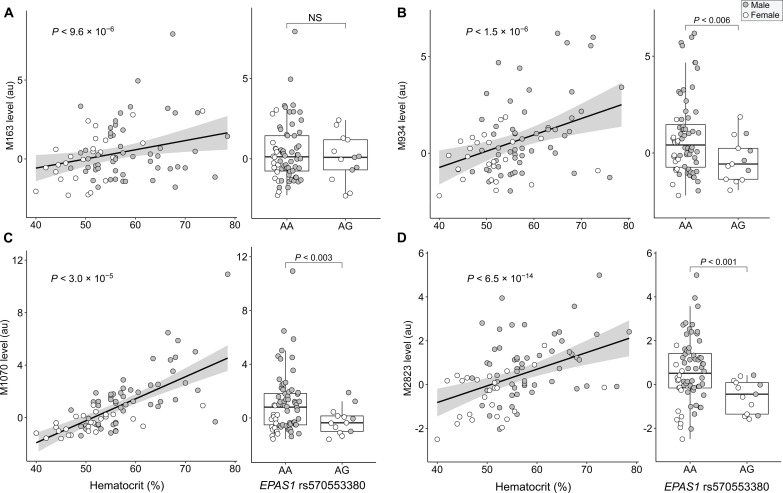
Among these 12 metabolites, endogenous cannabinoid arachidonoyl ethanolamide (M163) was the only previously identified metabolite associated with higher Hct (Fig. 4A). Of the 12 metabolites identified both in Andeans and FINRISK, only three (M934, M1070, and M2832) were further associated with rs570553380 in Andeans (Fig. 4, B to D). All three unidentified metabolites were negatively correlated with increased copies of the putatively adaptive G allele and positively correlated with Hct. Mendelian randomization indicated that Hct is most likely causal to these three metabolites, implicating their role in biological processes downstream of red blood cell production. The only previously identified metabolite associated with Hct in Andeans and FINRISK (M163) was not associated with rs570553380 in the Andean cohort (Fig. 4A).
Fig. 4. Metabolite associations with hematocrit and/or rs570553380 (A>G, p.[His194Arg]).
(A) Hematocrit in the Andean cohort was associated with a previously identified metabolite (ID: M163) classified as endogenous cannabinoid arachidonoyl ethanolamide. Metabolites with putative IDs M934 (B), M1070 (C), and M2823 (D) were associated both with hematocrit and the EPAS1 rs570553380 variant in the Andean cohort. These metabolites were found in a large-cohort population study (FINRISK) and are yet to be classified. Male and female data points are indicated with closed and open circles, respectively.
DISCUSSION
Associations between adaptive EPAS1 variants and [Hb] in Tibetan groups at high altitude have been well replicated (3, 5). Although overlapping signals of selection have been observed across Tibetans and Andeans (12, 25), convergent adaptation involving the same genes and phenotypes has not been shown. Here, we demonstrate evidence for shared genotypic-phenotypic signals between Tibetan and Andean highlanders. Although the average [Hb] ([Hb] ∝ Hct) is notably lower in Tibetans compared to Andeans (56), Andeans with the positively selected EPAS1 variant (rs570553380, A>G, p.[His194Arg]) demonstrate relatively lower Hct and protection from EE. Furthermore, this Andean-specific, protein-coding EPAS1 variant attenuates activity of certain canonical HIF-2α targets under hypoxia, a molecular phenotype similarly associated with noncoding Tibetan-specific EPAS1 variants (11, 53, 54).
In contrast to EPAS1 variants that are found at high frequency and nearly fixed in many Tibetan groups (3), the Andean-specific variant presented here is observed at low frequency. Considering the notably young age (~9845 to ~13,027 years) and modest frequency of the Andean-specific variant, rs570553380(G) is likely under early-stage selection in highlander populations of the Andes, as identified and noted in Colla Andean highlanders (25). In contrast, the adaptive EPAS1 haplotype identified in Tibetan groups was inherited from Denisovans more than 48,000 years ago (26, 57, 58). Furthermore, ancestors of populations in present-day Tibet resided at high altitudes much longer than populations in the Andes (1, 56), allowing more time for natural selection at this locus to occur. Significant iSAFE, as well as EHH and iHS results provide strong evidence for selection in this Andean cohort, demonstrating that haplotypes containing the derived rs570553380 allele (G) are significantly longer than those with the ancestral allele (A). The modest frequency of this adaptive variant could explain the inability of prior selection scans to identify rs570553380 as an outlier at the genome-wide level (10, 12, 18–20, 24, 27, 28), as only methods designed to detect recent positive selection in Andeans have identified rs570553380 (25).
The in vitro results suggest the protein-coding variant in EPAS1 has broad effects beyond correlations with Hct, O2 saturation, and EE. Presence of the His194Arg variant is associated with decreased expression of ADM (adrenomedullin) and VEGF (vascular endothelial growth factor A), suggesting that rs570553380 may confer protection from pulmonary hypertension and right ventricular hypertrophy (59, 60), as recently demonstrated in a CRISPR mouse model (10). The Andean EPAS1 variant is also associated with decreased expression of BHLE40 (basic helix-loop-helix family member E40), which prevents myogenic repair following ischemic injury (61), and HILPDA (hypoxia inducible lipid droplet associated), a mediator of intracellular fatty acid metabolism (62). Whether rs570553380 directly or indirectly affects these traits and biomarkers of functional relevance will require investigation from multiple angles. Such associations may or may not be immediately apparent; e.g., recent studies in Andean groups at high altitude show that variation in levels of erythropoietin, which stimulates red blood cell production, does not correlate with Hct (31). It is plausible that changes in gene expression involving this HIF-2α variant lead to pleiotropic effects and alterations at various steps of the O2 transport cascade with concomitant or compensatory effects that remain to be determined.
The association between rs570553380 and Hct in Andean highlanders parallels similar findings observed in Tibetans (3, 5) and suggests that this variant may improve evolutionary fitness at high altitude. As noted in Tibetans, elevated [Hb] (or EE) is a hallmark of CMS (63, 64), a debilitating disease more prevalent in males than females (37, 38, 64). Andean males with the adaptive EPAS1 allele demonstrate significantly lower Hct, higher O2 saturations under conditions of hypoxia, and lower incidence of EE. The lack of significance in females could be explained by the notably reduced sample size compared to males (n = 139 males, 49 females) and inability to control for relevant covariates, such as estrogen levels, menstruation cycle, and menopause status (33, 39). Considering that Hct and [Hb] levels are generally lower in females than males (65), it is also plausible that the effects of rs570553380 become more pronounced with increasing severity of EE or CMS. Molecularly, this could manifest as protection from chronic hyperactivation of the HIF pathway due to prolonged exposure to high-altitude hypoxia (30) and worsening pulmonary arterial hypertension (64). This disease severity–dependent relationship could also explain the lack of significance between [Hb] and rs570553380 in a recent study of Andean highlanders (10) with a younger average participant age (24 years, versus 41 years in this study). It is well documented that EE and CMS severity worsen notably with age (64), suggesting that the protective effects of rs570553380 may be more pronounced in older individuals.
Although the Andean-specific and Tibetan-specific alleles converge upon a similar phenotype, they likely manifest via distinct molecular and physiological changes. The adaptive EPAS1 variants characterized in Tibetans are located within cis-regulatory elements that alter gene expression of EPAS1 (11, 53, 54) and affect protein levels of HIF-2α, likely in a temporal- and cell-specific manner. In contrast, the missense EPAS1 variant described here in Andeans is protein coding, altering the structure and function of HIF-2α itself (10). Despite these differences, both population-specific alleles appear to attenuate HIF-2 activity in some form. Recent work in murine models demonstrates that Tibetan-specific EPAS1 variants are likely pleiotropic in nature (11). The observed associations between rs570553380, Hct, and unknown metabolites in Andeans suggest that this missense EPAS1 variant may also alter additional phenotypes beyond Hct (or [Hb]) that may be more direct targets of selection. Although not correlated with rs570553380, endogenous cannabinoid arachidonoyl ethanolamide (anandamide) was positively associated with Hct in both Andeans and FINRISK. Anandamide has been observed to alter sleep (66), neuroprotection, and immunosuppression via transient receptor potential vanilloid type-1 receptor and nuclear peroxisome proliferator–activated receptors (67), suggesting the endocannabinoid system may play a role in mediating the effects of EE. However, the relationship between Hct (or [Hb]), downstream metabolites, EE, and red blood cell turnover in Tibetans has not been studied (68). Further cross-population studies between Hct and metabolites could help elucidate the similarities and differences in the production and destruction of erythrocytes in Andeans and Tibetans.
While we provide cross-population evidence for genotype-phenotype adaptation in high-altitude humans involving different variants within the same gene, natural selection at various EPAS1 sites in other highland species has been observed (31, 32). The presence of the same EPAS1 amino acid substitution (H>R) in coelacanths poses questions of potential convergent evolution at this residue. Substantial adaptations in lung function and morphology since the Cretaceous period (69) between the extant coelacanth, Latimeria, and its extinct predecessor, Axelrodichthys, further suggest potential impacts of selective forces, in which this H>R substitution may have arisen independently and been advantageous in the coelacanth genetic background.
Although these findings provide a step forward in the understanding of hypoxia adaptations, there are various limitations of this work. The use of an in vitro cell culture model, with set tensions and time courses, fails to capture the dynamic chemical gradients that exist within a vascularized organ and the nuances of chronic hypoxia in highlanders. Furthermore, the use of HEK293T cells to model the transcriptional effects of rs570553380 on HIF-2 activity is not fully representative of the diverse spectrum of cell types within the human body. Despite these drawbacks, this study adds to our understanding of human evolution and has important implications for health and disease. Integrative genomic and physiological assessments with gene editing and multiomics analyses will continue to provide crucial insights into functional adaptive variation at high altitude (70), and natural selection within our species, with biological and clinical implications for hypoxia-related diseases and personalized medicine (68).
MATERIALS AND METHODS
Participant recruitment and criteria
The study was approved by the University of California, San Diego Human Research Protection Program (UC San Diego Project, nos. 140235 and 171772) and the Institutional Ethics Committee for Humans of Universidad Peruana Cayetano Heredia (CIEH-UPCH Project, no. 59285). All participants received and signed a detailed consent form in Spanish that explained the study. Males and nonpregnant females ages 19 to 65 of Quechua Andean ancestry living in Cerro de Pasco, Peru (4340 m), were recruited (n = 224), including individuals with and without CMS and EE [Hct ≥ 63% in males ([Hb] ≥ 21 g/dl) and ≥ 57% in females ([Hb] ≥ 19 g/dl)]. Ancestry was self-reported, and individuals self-identified as Quechua with both maternal and paternal grandparents resident at high altitude. Anyone with a history of cardiovascular/pulmonary disease besides CMS or EE was excluded from the study. Participants were also excluded if they had traveled to lower elevation (<3000 m) for more than 7 days in the 6 months before the study or had undergone phlebotomies and/or blood transfusions within 6 months before study.
Whole-genome sequencing
We performed high-coverage whole-genome sequencing (WGS) analysis of DNA from 40 unrelated Quechua Andean men and women selected at random from the 224 recruited Andeans in this study on the Illumina HiSeqX sequencing platform (average and minimum call rate, 92.4 and 91.7%, respectively). An average of 2.86 Gb of sequencing data was aligned for each genome. We obtained an average of 30× coverage (76.0% of the genome) with 99.5% exonic coverage. Approximately 3.76 million of the total 12.51 million single-nucleotide variants (SNVs) were identified across each genome with 14.1% previously unidentified SNVs. For each sample, we identified an average of 188,852 SNVs across the whole exome, of which 0.29% were novel. As determined by Annotate Variation (ANNOVAR), there were 41,236 synonymous and 48,057 nonsynonymous SNVs (10,600 missense, 82 stopgain, and 10 stoploss on average).
Principal components analysis and admixture
We used the program ADMIXTURE to determine the distribution of genetic ancestry among Andean participants from Cerro de Pasco (71). A single population was determined through ADMIXTURE analysis, determined by fivefold cross validation, for the 40 Andean genomes. We combined these data with the 1000 Genomes Yoruba in Ibadan, Nigeria (YRI), Japanese in Tokyo (JPT), Iberian population in Spain (IBS), Colombian (CLM), Mexican (MXL), and Peruvian (PEL) genomes and ran an unsupervised analysis to estimate the best fit among populations and the distribution of genetic ancestry in Andeans (72). According to the best fit (K = 4) mode, the Andean cohort’s ancestry reflects a small fraction resembling the component identified in an Iberian population from Spain (IBS) and a large non-IBS, likely Native American, component (fig. S1). This is further illustrated by a projection of the Andean samples onto a PC space created using data from several populations in the 1000 Genomes dataset: Northern Europeans from Utah (CEU), Iberian (IBS), PEL, CLM, MXL, CHB, JPT, and YRI (fig. S2). We also ran Flare (73) to determine local ancestry at the EPAS1 gene region (chr2:46,524,546-46,613,836) (GRCh37/hg19 by NCBI Gene) plus 100 kb up- and down-stream of the EPAS1 gene using the Thousand Genome Project phase 3 as the reference population. No significant admixture was noted at this locus.
Selection analyses
We used the iSAFE to further examine a 1-Mb region surrounding the EPAS1 SNV rs570553380 in the Andean whole genomes using default parameters (34). Results were visualized using Locuszoom (74). The Andean whole genomes were polarized against the ENSEMBL Homo sapiens GRCh37/hg19 ancestral allele file. EHH and iHS analyses were conducted on rs570553380, rs7559484, and rs7556828 in the Andean whole genomes via rehh (75) using default parameters.
Dating of variants
The ages of rs570553380, rs7559484, and rs7556828 were determined using the joint clock configuration on Genealogical Estimation of Variant Age (GEVA), which incorporates 1000 Genomes phase 3 into their software (76). The age of rs570553380 was additionally estimated within the 40 Andean whole genomes by setting the EHH threshold (probability of homozygote = 0.05) and calculating the distance to either side from rs570553380 (77). All analyses assumed a generation length of 25 years.
Genotyping
Venous blood samples (~10 ml) were collected in EDTA tubes (BD Vacutainer, BD, Franklin Lakes, NJ, USA). Buffy coat was isolated from whole blood via centrifugation and processed for storage in a cell lysis solution (Qiagen, Germantown, MD, USA) in Cerro de Pasco, Peru, and all samples were shipped to the University of California, San Diego. Genomic DNA was obtained from cell lysis and purified using a Gentra Puregene Blood Kit (Qiagen, Germantown, MD, USA). Genomic DNA samples were stored at 4°C for less than 1 year before genotyping. DNA samples were diluted to working stock concentrations of 5 ng/ul. DNA quality was verified with NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA USA, 260/280 absorbance ratios of 1.8 to 2.0 and 260/230 absorbance ratios of 2.0 to 2.2) and gel electrophoresis to ensure samples were not degraded. After whole-genome sequence data were analyzed, targeted SNV genotyping at the rs570553380 locus was completed using Applied Biosystems TaqMan SNV Genotyping Assays (Thermo Fisher Scientific, Waltham, MA, USA). Quantitative real-time polymerase chain reactions (PCRs) were conducted on a QuantStudio 3 (Applied Biosystems, Foster City, CA, USA) with a 0.1-ml of thermal block.
Genotyping calls were determined with the Applied Biosystems Genotyping Analysis Module. The presence of each allele was determined by the multicomponent amplification patterns of fluorescent VIC and FAM dyes in relation to a ROX reference dye. The real-time experimental analysis method was used with heterozygote-permitted autocalling. Each automatic call was verified visually by the experimenter.
Phenotype analyses
Phenotypes of the recruited participants were obtained in Cerro de Pasco, Peru, in August 2015, December 2015, December 2016, December 2017, and March 2019. Methods and equipment for phenotype collection were consistent across trips. Height, weight, resting heart rate, and resting O2 saturation were collected during the initial screening session as participants were seated with their right arm elevated to heart level. O2 saturation (SpO2) under conditions of inspired O2 at 10% ( = 0.10) was obtained in a subset of individuals as previously described (40). All O2 saturation and heart rate measurements were collected via pulse oximeter (Nellcor model N-560, Medtronic, MN, USA). Hematocrit (Hct), the proportion of red blood cells in whole blood, was measured from venous blood using capillary tubes. Individuals with a Hct ≥ 63% in men ([Hb] ≥ 21 g/dl) and ≥ 57% in women ([Hb] ≥ 19 g/dl) were considered to have EE (63).
Genotype-phenotype associations
A multivariate generalized linear model was used to assess associations between EPAS1 genotypes and phenotypic measurements of unrelated individuals. We collected self-reported information regarding familial relationships and excluded all but one of any related individuals. Our WGS analyses in 40 individuals confirmed self-reported nonrelatedness and ancestry. The regression model accounts for the various plausible modes of inheritance of potential genetic variants and can be written as:
where Phenotype is a continuous variable stratified by sex, Genotype is a factor with n levels (n ∈ {2,3}) dependent on the zygosity of the variant, Age and Study are covariates whose fixed and random effects were controlled. Age and study batch effects of measurements collected in m studies were no longer considered by the model if their corresponding coefficients were not significantly different from zero. A robust SW estimator was used to account for heteroskedasticity of the phenotypic data (78).
Constructs and molecular cloning
Guide RNA (gRNA) expression plasmids with the spacers GGTCTTGCACTGCACGGGCC (for EPAS1 H194R editing) and CAGTCAGCCGGTGGTGCAGA (for deadEGFP editing) were generated via site-directed mutagenesis cloning using pFYF1230 (Addgene plasmid no. 47511) as a template and Phusion High-Fidelity DNA Polymerase (Thermo Fisher Scientific, no. F534L) according to the manufacturer’s instructions. All DNA vector amplification was carried out using NEB 10-β competent cells (NEB, no. C3019H). All plasmids were purified using the ZymoPURE II Plasmid Midiprep Kit (Zymo Research, no. 11-550B).
HEK293T cell culture and transfections for base editing experiments
HEK293T cells (ATCC CRL-3216) were cultured and maintained in high-glucose Dulbecco’s modified Eagle’s medium (DMEM) supplemented with GlutaMAX (Thermo Fisher Scientific, no. 10566024), 10% (v/v) fetal bovine serum (Life Technologies, no. 10437-028), and penicillin-streptomycin (P/S) (100 U/ml; Gibco, no. 15070063), at 37°C with 5% CO2. Transfection of plasmid DNA into HEK293T cells for the purpose of generating isogenic cell lines has been previously described (79). Briefly, cells were plated at a density of 50,000 cells per well in 250 μl of P/S-free medium per well in 48-well plates and transfected 16 hours after plating. For each transfection, 200 ng of gRNA plasmid (described above) and 800 ng of base editor plasmid (pABEmax-NG-P2A-EGFP, Addgene, no. 140005) were combined with 1.5 μl of Lipofectamine 2000 (Life Technologies, no. 11668-019) and Opti-MEM (Life Technologies, no. 31985070) according to the manufacturer’s instructions. For the generation of the EPAS1 cell lines in this study, cells were sorted to enrich for high base-editing activity using a mCherry-P2A-deadEGFP plasmid. In these transfections, 300 ng of the mCherry-P2A-deadEGFP plasmid and 100 ng of its corresponding gRNA expressing plasmid were included in the lipofectamine mixture. A similar technique has been previously described (80). Briefly, a successful A•T to G•C edit within a nonfluorescent EGFP results in GFP fluorescence, allowing for fluorescence-activated cell sorting (FACS) enrichment.
Sequencing to assess base editing efficiencies
Cells were cultured for 3 days following transfection. Genomic DNA was extracted after rinsing with 150 μl of phosphate-buffered saline (PBS, Gibco Life, no. 10010-023) with 100 μl of freshly prepared cell lysis solution [10 mM tris-HCl (pH 7.5), 0.05% SDS, and proteinase K (25 μg/ml; NEB, no. P8107S)] for each well. The cell lysate was transferred to 0.2-ml PCR tubes and incubated at 37°C for 1 hour and then heat-inactivated at 80°C for 30 min. This cell lysate (0.5 μl; gDNA) was used as a PCR template with the forward primer ATCGTGAGGCTGCTGGACGAGT and the reverse primer TCTCCACTCAGGAAGCTCCGGC. The PCR amplicon was sequenced with Sanger sequencing using the forward primer. After confirmation of >10% A•T to G•C editing efficiency at H194 of the EPAS1 genomic locus using EditR (81), the transfection step was repeated. However, instead of harvesting the cells after 3 days, they were sorted using FACS and allowed to clonally expand as described below.
Utilization of FACS to isolate base-edited single-cell EPAS1 H194R clones
Three days after transfection, the old medium was aspirated off and the cells were rinsed with 150 μl of PBS. The cells were then detached from the plate with 300 μl of Accumax (Innovative-Cell Technology, no. AM-105) and resuspended with the addition of 500 μl of PBS. The cell suspension was transferred to a 15-ml conical tube, which was centrifuged for 5 min at 200 rcf at room temperature. After aspirating off the supernatant, the cells were resuspended in FACS buffer [1% fetal bovine serum (FBS), 50 μM EDTA (pH 8.0), and propidium iodide (PI) (2 μg/ml; Sigma-Aldrich, no. P3566)] and filtered through a cell strainer. Using a FACSAria II system, nonviable cells and doublets were eliminated via gating parameters and under sterile conditions, single cells fluorescent in both mCherry and EGFP were sorted into individual wells of 96-well plates containing 100 μl of DMEM supplemented with 50% FBS and 1% Pen/Strep. The 96-well plate was immediately placed into a tissue culture incubator after sorting. Subculturing and genotyping of single cells after clonal expansion was conducted as previously described (79). Genotyping of 26 clones confirmed the presence of three clones containing a heterozygous genotype and one clone containing a homozygous genotype (fig. S7). Heterozygotes were defined as containing at least one mutant and one wild-type allele. These, along with two unedited clones, were cryopreserved and subjected to further experiments.
WES analysis of edited cell lines
Whole-exome sequencing (WES) data for a subset of the isogenic cell lines ran in duplicate were generated on the Illumina NovaSeq 6000 (DNA Link Precision Genomics, Seoul Korea). Raw WES data were aligned to the reference human genome (hg19) using the Burrows-Wheeler aligner (82), and variants of the aligned genome were called using Freebayes (83). Variants with call quality scores ≤50 and/or inconsistent calls across runs were excluded. Variant call files were then annotated using Qiagen Clinical Insight (Qiagen, Germantown, MD, USA) (table S4). To assess relative enrichment of off-target mutations across cell lines (fig. S8A), the mutation rates of all observed SNVs were compared to those of the nontransfected control (fig. S8B).
In vitro hypoxia experiments
For all hypoxia experiments, low passage, isogenic clones wild-type (WT), heterozygous (HET), and homozygous (HOM) for the EPAS1 variant (rs570553380, A>G, p.[His194Arg]) were used within several passages of defrosting from cryopreservation. Data from the single HOM clone are shown but not included in statistical analyses. Both wild-type HEK293T cells that had [wild-type transfected (WTT)] and had not [wild-type nontransfected (WTNT)] undergone the transfection and clonal expansion process were used as controls. Cells were grown within a 37°C humidified incubator (Heracell VIOS 160i Tri-Gas CO2, Thermo Fisher Scientific) under hypoxia (1% O2) and normoxia (21% O2) for 0, 12, and 24 hours. The use of a cell culture model system with O2 exposure ranging from room air to as low as 0.5% provides a powerful tool to study cellular mechanisms in hypoxia. The selection of 1% O2 was based off prior studies in which O2 tension induced HIF-2 stabilization in HEK293T cells (49–52). O2 and CO2 levels were calibrated before experimentation against external sensors. Pericellular dissolved O2 and pH levels were measured every 5 min using the sbi ID Developer’s Kit and Data Hub (Scientific Bioprocessing, Pittsburgh, PA, USA). For each time point (normoxia, 12 hours, 24 hours), total RNA and protein were immediately extracted for downstream applications.
RNA extraction and quantitative real-time PCR
Following a 2-ml PBS wash to remove dead cells and waste, total RNA was immediately extracted for all time points (normoxia, 12 hours, 24 hours) using the RNeasy Mini kit with DNase digestion according to manufacturer’s instructions (Qiagen, Germantown, MD, USA). After extraction, RNA concentration and quality were measured via NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA USA). Total RNA (1 μg) was then converted into cDNA using the High-Capacity cDNA Reverse Transcriptase kit (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions.
Quantitative real-time PCR was carried out in duplicate for all biological samples on the QuantStudio 3 (Applied Biosystems, Foster City, CA, USA) with a 0.1-ml thermal block using the TaqMan Fast Advanced Master Mix (Thermo Fisher Scientific, Waltham, MA, USA). TaqMan assays (Thermo Fisher Scientific) for canonical HIF-2α target genes (table S5) were normalized to β-actin (ACTNB) as a housekeeping gene in all edited cell lines. The ΔΔCt method was conducted to calculate differential gene expression of each isogenic cell line at all three time points normalized by their normoxia counterparts, where ΔCt = Ct target − Ctβ-actin. Relative expression was then quantified by ΔΔCt = ΔCt hypoxia − ΔCt normoxia and then compared between nontransfected and transfected wild-type cell lines as well as between genotypes using a two-way analysis of variance (ANOVA) test. The genes used for quantitative real-time PCR (table S5) were assayed in prior studies assessing HIF-2 activity (84–87).
Protein extraction and Western blot analysis
HEK293 cells cultured in T25 flasks were exposed to normoxia (21% O2) or 24-hour hypoxia (1% O2). At time of collection, flasks were rapidly taken out from the incubator, cell media was discarded, and cells were washed with 1.6 ml of chilled PBS. Cells were retrieved with a cell scraper in 600 μl of radioimmunoprecipitation assay buffer (no. 9806S, Cell Signaling Technology, MA, USA) with 0.1% of Protease Inhibitor Cocktail (no. P8340, Sigma-Aldrich, USA) and collected in Eppendorf tubes. For lysis, the cells were incubated on ice for 15 min with successive vortex agitation cycles and then stored at - 80°C. Protein content was determined using DC Protein Assay (BioRad, CA, USA). Proteins were separated in 10% Bis Tris-Plus premade gels (Mini Gel Tank using Bolt buffers, Thermo Fisher Scientific, Carlsbad, CA), and transferred to polyvinyldene difluoride membranes (iBlot, Invitrogen, Carlsbad, CA). Membranes were blocked for 1 hour with 5% bovine serum albumin (no. A-7030, Sigma-Aldrich, USA), diluted in PBS with 0.1% Tween 20 at room temperature, and then incubated with primary antibodies against HIF-2α (Novus, no. NB-100-122, 1/250) and β-actin (Cell Signaling Technology, no. 4970S, 1/1000) overnight at 4°C, washed, and incubated with fluorescent secondary antibodies for imaging detection (Odyssey Infrared Imager, LiCOR). Protein band quantification was performed with ImageJ software. Data are presented as the ratio of HIF-2α to β-actin band intensities.
Metabolomics and association validation
To understand the upstream and downstream metabolomic effects on high-altitude adaptation, we analyzed 116 plasma samples from the subset of the full Andean cohort, which consisted of 59 men and 57 women. LC-MS was used to identify unique peaks suggesting the existence/abundance of previously identified and unidentified metabolites (88). Metabolite signals were standardized by dividing the peak-median intensity difference by the median absolute deviation. To test for metabolite-phenotype associations, we used a generalized linear model defined as
where M is the standardized signal of metabolite level, Phe refers to the measurements of hematocrit (%), and sex, age, and body mass index (BMI) were considered as covariates. We controlled for sex in these analyses for direct comparison to publicly available metabolomics data from FINRISK. FDR correction was conducted based off the number of distinct LC-MS peaks detected. Furthermore, associations of the top metabolites revealed in the Andean cohort were validated using the FINRISK cohort (n = 1170) with the same model. Mendelian randomization was additionally performed (89) on the significant hits to infer statistical causality.
Molecular graphics representations
The ribbon diagram of HIF-2α was generated on IBS software (90), while the structure of HIF-2α [Protein Data Bank (PDB) ID: 4ZPK] was annotated on UCSF Chimera (91).
Structural analysis and prediction of rs570553380
Analysis of rs570553380 on the structure and stability of HIF-2α (PDB ID: 4ZP4) was conducted using PROVEAN (45), MutPred2 (46), and DynaMut2 (47).
Acknowledgments
We are grateful to individuals from the community of Cerro de Pasco, Peru, for their involvement in this study. We thank L. B. Jorde, A. R. Rogers, and P. D. Wagner for review of the manuscript and S. Fassardi for assistance with molecular assays.
Funding: This work was supported by National Institutes of Health, National Heart, Lung, Blood Institute R01HL145470, National Geographic Society Explorer Award, and John B West Endowment in Respiratory Physiology (T.S.S.); Wellcome Trust Award 107544/Z/15/Z (F.C.V.); European Union’s Horizon 2020 Research and Innovation Program under a Marie Skłodowska-Curie grant agreement No 890768 (K.A.O.); National Institutes of Health T32 grant T32HL134632 (J.E.H.); Howard Hughes Medical Institute Gilliam Fellowship and the National Academies of Sciences, Engineering, and Medicine Ford Foundation Fellowship (C.A.V.); National Science Foundation Graduate Research Fellowship Program under Grant No DGE-2038238 (P.T.B.); Research Corporation for Science Advancement through Cottrell Scholar Award 27502 (A.C.K.); Investigators Programme grant from Science Foundation Ireland 12/IP/1727 (G.L.C.); Finnish Foundation for Cardiovascular Research and Juho Vainio Foundation (V.S.); and Academy of Finland (A.S.H.).
Author contributions: Conceptualization: E.S.L., W.G., J.J.Y., K.A.O., C.A.V., J.E.H., E.A.M., J.J.R., R.M.S., G.V.-G., K.P.F., F.L.P., M.J., A.C.K., C.D.H., F.C.V., and T.S.S. Data curation: E.S.L., W.G., C.A.-R., C.A.V., K.M., M.A., J.-L.M., R.F.-M., N.C., K.P.F., V.S., M.J., A.C.K., C.D.H., F.C.V., and T.S.S. Formal analysis: E.S.L., W.G., R.J.B., A.M.C., J.J.Y., H.H., C.A.V., K.M., M.A., T.L., J.E.H., E.A.M., J.J.R., M.A.B., R.M.S., K.P.F., M.J., C.D.H., and T.S.S. Funding acquisition: W.G., K.A.O., V.S., A.M., M.J., A.C.K., G.L.C., C.D.H., F.C.V., and T.S.S. Investigation: E.S.L., W.G., C.A.-R., A.M.C., J.J.Y., E.C.H., K.A.O., C.A.V., J.E.H., E.A.M., M.A.B., J.J.R., M.C.K., R.M.S., G.V.-G., J.-L.M., R.F.-M., D.B., N.C., E.G., K.P.F., V.S., A.S.H., A.C.K., G.L.C., C.D.H., F.C.V., and T.S.S. Methodology: E.S.L., W.G., R.J.B., C.A.-R., J.J.Y., K.A.O., C.A.V., Q.T.C., T.L., J.E.H., E.A.M., R.M.S., K.P.F., M.J., A.C.K., C.D.H., F.C.V., and T.S.S. Project administration: E.S.L., W.G., C.A.-R., J.J.Y., E.C.H., J.E.H., J.J.R., K.P.F., A.M., M.J., A.C.K., C.D.H., F.C.V., and T.S.S. Resources: E.S.L., W.G., C.A.V., Q.T.C., M.A., J.E.H., E.A.M., M.A.B., V.S., R.M.S., G.V.-G., A.S.H., M.J., A.C.K., G.L.C., C.D.H., F.C.V., T.S.S. Software: E.S.L., W.G., R.J.B., K.P.F., and C.D.H. Supervision: E.S.L., W.G., E.C.H., P.T.B., J.E.H., R.M.S., K.P.F., A.J.M., A.M., F.L.P., A.C.K., C.D.H., F.C.V., and T.S.S. Validation: E.S.L., W.G., J.J.Y., K.A.O., C.A.V., T.L., J.E.H., E.A.M., J.J.R., M.C.K., K.P.F., R.M.S., A.C.K., C.D.H., F.C.V., and T.S.S. Visualization: E.S.L., W.G., R.J.B., J.E.H., E.A.M., D.B., K.P.F., M.J., A.C.K., and T.S.S. Writing—original draft: E.S.L., W.G., J.E.H., E.A.M., J.J.R., K.P.F., F.L.P., C.D.H., F.C.V., and T.S.S. Writing—review and editing: E.S.L., W.G., R.J.B., C.A.-R., A.M.C., J.J.Y., E.C.H., K.A.O., P.T.B., M.A., J.E.H., E.A.M., J.J.R., M.C.K., R.M.S., G.V.-G., J.-L.M., R.F.-M., D.B., N.C., K.P.F., V.S., A.S.H., F.L.P., M.J., A.C.K., G.L.C., C.D.H., F.C.V., and T.S.S.
Competing interests: A.C.K. is a member of the SAB of Pairwise Plants, is an equity holder for Pairwise Plants and Beam Therapeutics, and receives royalties from Pairwise Plants, Beam Therapeutics, and Editas Medicine via patents licensed from Harvard University. A.C.K.’s interests have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies. M.J. and T.L. are affiliated with Sapient Bioanalytics, LLC. V.S. has received an honorarium from Sanofi for consulting and also has ongoing research collaboration with Bayer Ltd. (all unrelated to the present study). All other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Data including selection scan results, whole-genome sequencing, genotyping arrays, phenotypic measurements, as well as CRISPR gene expression data and scripts used to analyze the datasets are available on GitHub (https://github.com/SimonsonLabBioinfomatics/epas1_data) and Zonodo (10.5281/zenodo.10574122). The FINRISK data for the present study are available with a written application to the THL Biobank as instructed on the website of the Biobank (https://thl.fi/en/research-and-development/thl-biobank/for-researchers). A separate permission is needed from FINDATA (www.findata.fi/en/) for use of the EHR data.
Supplementary Materials
This PDF file includes:
Figs. S1 to S9
Tables S1 to S3 and S5
Legends for tables S4 and S6
Other Supplementary Material for this manuscript includes the following:
Tables S4 and S6
REFERENCES AND NOTES
- 1.Simonson T. S., Altitude Adaptation: A Glimpse Through Various Lenses. High Alt. Med. Biol. 16, 125–137 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moore L. G., Measuring high-altitude adaptation. J. Appl. Physiol. 123, 1371–1385 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beall C. M., Cavalleri G. L., Deng L., Elston R. C., Gao Y., Knight J., Li C., Li J. C., Liang Y., McCormack M., Montgomery H. E., Pan H., Robbins P. A., Shianna K. V., Tam S. C., Tsering N., Veeramah K. R., Wang W., Wangdui P., Weale M. E., Xu Y., Xu Z., Yang L., Zaman M. J., Zeng C., Zhang L., Zhang X., Zhaxi P., Zheng Y. T., Natural selection on EPAS1 (HIF2alpha) associated with low hemoglobin concentration in Tibetan highlanders. Proc. Natl. Acad. Sci. U.S.A. 107, 11459–11464 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simonson T. S., Yang Y., Huff C. D., Yun H., Qin G., Witherspoon D. J., Bai Z., Lorenzo F. R., Xing J., Jorde L. B., Prchal J. T., Ge R., Genetic evidence for high-altitude adaptation in Tibet. Science 329, 72–75 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Yi X., Liang Y., Huerta-Sanchez E., Jin X., Cuo Z. X. P., Pool J. E., Xu X., Jiang H., Vinckenbosch N., Korneliussen T. S., Zheng H., Liu T., He W., Li K., Luo R., Nie X., Wu H., Zhao M., Cao H., Zou J., Shan Y., Li S., Yang Q., Asan, Ni P., Tian G., Xu J., Liu X., Jiang T., Wu R., Zhou G., Tang M., Qin J., Wang T., Feng S., Li G., Huasang, Luosang J., Wang W., Chen F., Wang Y., Zheng X., Li Z., Bianba Z., Yang G., Wang X., Tang S., Gao G., Chen Y., Luo Z., Gusang L., Cao Z., Zhang Q., Ouyang W., Ren X., Liang H., Zheng H., Huang Y., Li J., Bolund L., Kristiansen K., Li Y., Zhang Y., Zhang X., Li R., Li S., Yang H., Nielsen R., Wang J., Wang J., Sequencing of 50 human exomes reveals adaptation to high altitude. Science 329, 75–78 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ge R.-L., Simonson T. S., Cooksey R. C., Tanna U., Qin G., Huff C. D., Witherspoon D. J., Xing J., Zhengzhong B., Prchal J. T., Jorde L. B., McClain D. A., Metabolic insight into mechanisms of high-altitude adaptation in Tibetans. Mol. Genet. Metab. 106, 244–247 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horscroft J. A., Kotwica A. O., Laner V., West J. A., Hennis P. J., Levett D. Z. H., Howard D. J., Fernandez B. O., Burgess S. L., Ament Z., Gilbert-Kawai E. T., Vercueil A., Landis B. D., Mitchell K., Mythen M. G., Branco C., Johnson R. S., Feelisch M., Montgomery H. E., Griffin J. L., Grocott M. P. W., Gnaiger E., Martin D. S., Murray A. J., Metabolic basis to Sherpa altitude adaptation. Proc. Natl. Acad. Sci. U.S.A. 114, 6382–6387 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bigham A. W., Julian C. G., Wilson M. J., Vargas E., Browne V. A., Shriver M. D., Moore L. G., Maternal PRKAA1 and EDNRA genotypes are associated with birth weight, and PRKAA1 with uterine artery diameter and metabolic homeostasis at high altitude. Physiol. Genomics 46, 687–697 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brutsaert T. D., Kiyamu M., Elias Revollendo G., Isherwood J. L., Lee F. S., Rivera-Ch M., Leon-Velarde F., Ghosh S., Bigham A. W., Association of EGLN1 gene with high aerobic capacity of Peruvian Quechua at high altitude. Proc. Natl. Acad. Sci. U.S.A. 116, 24006–24011 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jorgensen K., Song D., Weinstein J., Garcia O. A., Pearson L. N., Inclán M., Rivera-Chira M., León-Velarde F., Kiyamu M., Brutsaert T. D., Bigham A. W., Lee F. S., High-altitude Andean H194R HIF2A allele is a hypomorphic allele. Mol. Biol. Evol. 40, msad162 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gray O. A., Yoo J., Sobreira D. R., Jousma J., Witonsky D., Sakabe N. J., Peng Y.-J., Prabhakar N. R., Fang Y., Nobréga M. A., Di Rienzo A., A pleiotropic hypoxia-sensitive EPAS1 enhancer is disrupted by adaptive alleles in Tibetans. Sci. Adv. 8, eade1942 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bigham A., Bauchet M., Pinto D., Mao X., Akey J. M., Mei R., Scherer S. W., Julian C. G., Wilson M. J., López Herráez D., Brutsaert T., Parra E. J., Moore L. G., Shriver M. D., Identifying signatures of natural selection in Tibetan and Andean populations using dense genome scan data. PLOS Genet. 6, e1001116 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peng Y., Yang Z., Zhang H., Cui C., Qi X., Luo X., Tao X., Wu T., Ouzhuluobu B., Ciwangsangbu D., Chen H., Shi H., Su B., Genetic variations in Tibetan populations and high-altitude adaptation at the Himalayas. Mol. Biol. Evol. 28, 1075–1081 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Wang B., Zhang Y.-B., Zhang F., Lin H., Wang X., Wan N., Ye Z., Weng H., Zhang L., Li X., Yan J., Wang P., Wu T., Cheng L., Wang J., Wang D.-M., Ma X., Yu J., On the origin of Tibetans and their genetic basis in adapting high-altitude environments. PLOS One 6, e17002 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu S., Li S., Yang Y., Tan J., Lou H., Jin W., Yang L., Pan X., Wang J., Shen Y., Wu B., Wang H., Jin L., A genome-wide search for signals of high-altitude adaptation in Tibetans. Mol. Biol. Evol. 28, 1003–1011 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Alkorta-Aranburu G., Beall C. M., Witonsky D. B., Gebremedhin A., Pritchard J. K., Di Rienzo A., The genetic architecture of adaptations to high altitude in Ethiopia. PLOS Genet. 8, e1003110 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huerta-Sánchez E., Degiorgio M., Pagani L., Tarekegn A., Ekong R., Antao T., Cardona A., Montgomery H. E., Cavalleri G. L., Robbins P. A., Weale M. E., Bradman N., Bekele E., Kivisild T., Tyler-Smith C., Nielsen R., Genetic signatures reveal high-altitude adaptation in a set of Ethiopian populations. Mol. Biol. Evol. 30, 1877–1888 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou D., Udpa N., Ronen R., Stobdan T., Liang J., Appenzeller O., Zhao H. W., Yin Y., Du Y., Guo L., Cao R., Wang Y., Jin X., Huang C., Jia W., Cao D., Guo G., Gamboa J. L., Villafuerte F., Callacondo D., Xue J., Liu S., Frazer K. A., Li Y., Bafna V., Haddad G. G., Whole-genome sequencing uncovers the genetic basis of chronic mountain sickness in Andean highlanders. Am. J. Hum. Genet. 93, 452–462 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eichstaedt C. A., Antão T., Cardona A., Pagani L., Kivisild T., Mormina M., Genetic and phenotypic differentiation of an Andean intermediate altitude population. Physiol. Rep. 3, e12376 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foll M., Gaggiotti O. E., Daub J. T., Vatsiou A., Excoffier L., Widespread signals of convergent adaptation to high altitude in Asia and America. Am. J. Hum. Genet. 95, 394–407 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeong C., Alkorta-Aranburu G., Basnyat B., Neupane M., Witonsky D. B., Pritchard J. K., Beall C. M., Di Rienzo A., Admixture facilitates genetic adaptations to high altitude in Tibet. Nat. Commun. 5, 3281 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Udpa N., Ronen R., Zhou D., Liang J., Stobdan T., Appenzeller O., Yin Y., Du Y., Guo L., Cao R., Wang Y., Jin X., Huang C., Jia W., Cao D., Guo G., Claydon V. E., Hainsworth R., Gamboa J. L., Zibenigus M., Zenebe G., Xue J., Liu S., Frazer K. A., Li Y., Bafna V., Haddad G. G., Whole genome sequencing of Ethiopian highlanders reveals conserved hypoxia tolerance genes. Genome Biol. 15, R36 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wuren T., Simonson T. S., Qin G., Xing J., Huff C. D., Witherspoon D. J., Jorde L. B., Ge R.-L., Shared and unique signals of high-altitude adaptation in geographically distinct Tibetan populations. PLOS One 9, e88252 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crawford J. E., Amaru R., Song J., Julian C. G., Racimo F., Cheng J. Y., Guo X., Yao J., Ambale-Venkatesh B., Lima J. A., Rotter J. I., Stehlik J., Moore L. G., Prchal J. T., Nielsen R., Natural selection on genes related to cardiovascular health in high-altitude adapted Andeans. Am. J. Hum. Genet. 101, 752–767 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eichstaedt C. A., Pagani L., Antao T., Inchley C. E., Cardona A., Mörseburg A., Clemente F. J., Sluckin T. J., Metspalu E., Mitt M., Mägi R., Hudjashov G., Metspalu M., Mormina M., Jacobs G. S., Kivisild T., Evidence of Early-Stage Selection on EPAS1 and GPR126 genes in Andean high altitude populations. Sci. Rep. 7, 13042 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu H., Petousi N., Glusman G., Yu Y., Bohlender R., Tashi T., Downie J. M., Roach J. C., Cole A. M., Lorenzo F. R., Rogers A. R., Brunkow M. E., Cavalleri G., Hood L., Alpatty S. M., Prchal J. T., Jorde L. B., Robbins P. A., Simonson T. S., Huff C. D., Evolutionary history of Tibetans inferred from whole-genome sequencing. PLOS Genet. 13, e1006675 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stobdan T., Akbari A., Azad P., Zhou D., Poulsen O., Appenzeller O., Gonzales G. F., Telenti A., Wong E. H. M., Saini S., Kirkness E. F., Venter J. C., Bafna V., Haddad G. G., New insights into the genetic basis of Monge’s disease and adaptation to high-altitude. Mol. Biol. Evol. 34, 3154–3168 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacovas V. C., Couto-Silva C. M., Nunes K., Lemes R. B., de Oliveira M. Z., Salzano F. M., Bortolini M. C., Hünemeier T., Selection scan reveals three new loci related to high altitude adaptation in Native Andeans. Sci. Rep. 8, 12733 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Semenza G. L., The genomics and genetics of oxygen homeostasis. Annu. Rev. Genomics Hum. Genet. 21, 183–204 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Bigham A. W., Lee F. S., Human high-altitude adaptation: forward genetics meets the HIF pathway. Genes Dev. 28, 2189–2204 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pamenter M. E., Hall J. E., Tanabe Y., Simonson T. S., Cross-species insights into genomic adaptations to hypoxia. Front. Genet. 11, 743 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Storz J. F., Cheviron Z. A., Physiological genomics of adaptation to high-altitude hypoxia. Annu Rev Anim Biosci. 9, 149–171 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Villafuerte F. C., Simonson T. S., Bermudez D., León-Velarde F., High-altitude erythrocytosis: mechanisms of adaptive and mal-adaptive responses. Physiology 37, 0 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Akbari A., Vitti J. J., Iranmehr A., Bakhtiari M., Sabeti P. C., Mirarab S., Bafna V., Identifying the favored mutation in a positive selective sweep. Nat. Methods 15, 279–282 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson K. E., Voight B. F., Patterns of shared signatures of recent positive selection across human populations. Nat. Ecol. Evol. 2, 713–720 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rademaker K., Hodgins G., Moore K., Zarrillo S., Miller C., Bromley G. R. M., Leach P., Reid D. A., Álvarez W. Y., Sandweiss D. H., Paleoindian settlement of the high-altitude Peruvian Andes. Science 346, 466–469 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Leon-Velarde F., Ramos M. A., Hernandez J. A., De Idiaquez D., Munoz L. S., Gaffo A., Cordova S., Durand D., Monge C., The role of menopause in the development of chronic mountain sickness. Am. J. Physiol. Regul. Integr. Comp. Physiol. 272, R90–R94 (1997). [DOI] [PubMed] [Google Scholar]
- 38.León-Velarde F., Rivera-Chira M., Tapia R., Huicho L., Monge-C C., Relationship of ovarian hormones to hypoxemia in women residents of 4,300 m. Am. J. Physiol. Regul. Integr. Comp. Physiol. 280, R488–R493 (2001). [DOI] [PubMed] [Google Scholar]
- 39.Azad P., Villafuerte F. C., Bermudez D., Patel G., Haddad G. G., Protective role of estrogen against excessive erythrocytosis in Monge’s disease. Exp. Mol. Med. 53, 125–135 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heinrich E. C., Orr J. E., Gilbertson D., Anza-Ramirez C., DeYoung P. N., Djokic M. A., Corante N., Vizcardo-Galindo G., Macarlupu J. L., Gaio E., Powell F. L., Malhotra A., Villafuerte F. C., Simonson T. S., Relationships between chemoreflex responses, sleep quality, and hematocrit in Andean men and women. Front. Physiol. 11, 437 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hancco I., Bailly S., Baillieul S., Doutreleau S., Germain M., Pépin J.-L., Verges S., Excessive erythrocytosis and chronic mountain sickness in dwellers of the highest city in the world. Front. Physiol. 11, 773 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pollard K. S., Hubisz M. J., Rosenbloom K. R., Siepel A., Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 20, 110–121 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cupello C., Clément G., Meunier F. J., Herbin M., Yabumot Y., Brito P. M., The long-time adaptation of coelacanths to moderate deep water: reviewing the evidences. Bulletin of the Kitakyushu Museum of Natural History and Human History, Series A (Natural History) 17, 29–35 (2019). [Google Scholar]
- 44.Amemiya C. T., Alföldi J., Lee A. P., Fan S., Philippe H., MacCallum I., Braasch I., Manousaki T., Schneider I., Rohner N., Organ C., Chalopin D., Smith J. J., Robinson M., Dorrington R. A., Gerdol M., Aken B., Biscotti M. A., Barucca M., Baurain D., Berlin A. M., Blatch G. L., Buonocore F., Burmester T., Campbell M. S., Canapa A., Cannon J. P., Christoffels A., De Moro G., Edkins A. L., Fan L., Fausto A. M., Feiner N., Forconi M., Gamieldien J., Gnerre S., Gnirke A., Goldstone J. V., Haerty W., Hahn M. E., Hesse U., Hoffmann S., Johnson J., Karchner S. I., Kuraku S., Lara M., Levin J. Z., Litman G. W., Mauceli E., Miyake T., Mueller M. G., Nelson D. R., Nitsche A., Olmo E., Ota T., Pallavicini A., Panji S., Picone B., Ponting C. P., Prohaska S. J., Przybylski D., Saha N. R., Ravi V., Ribeiro F. J., Sauka-Spengler T., Scapigliati G., Searle S. M. J., Sharpe T., Simakov O., Stadler P. F., Stegeman J. J., Sumiyama K., Tabbaa D., Tafer H., Turner-Maier J., van Heusden P., White S., Williams L., Yandell M., Brinkmann H., Volff J.-N., Tabin C. J., Shubin N., Schartl M., Jaffe D. B., Postlethwait J. H., Venkatesh B., Di Palma F., Lander E. S., Meyer A., Lindblad-Toh K., The African coelacanth genome provides insights into tetrapod evolution. Nature 496, 311–316 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choi Y., Chan A. P., PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 31, 2745–2747 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pejaver V., Urresti J., Lugo-Martinez J., Pagel K. A., Lin G. N., Nam H.-J., Mort M., Cooper D. N., Sebat J., Iakoucheva L. M., Mooney S. D., Radivojac P., Inferring the molecular and phenotypic impact of amino acid variants with MutPred2. Nat. Commun. 11, 5918 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodrigues C. H. M., Pires D. E. V., Ascher D. B., DynaMut2: Assessing changes in stability and flexibility upon single and multiple point missense mutations. Protein Sci. 30, 60–69 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Komor A. C., Kim Y. B., Packer M. S., Zuris J. A., Liu D. R., Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muñiz-García A., Romero M., Falcόn-Perez J. M., Murray P., Zorzano A., Mora S., Hypoxia-induced HIF1α activation regulates small extracellular vesicle release in human embryonic kidney cells. Sci. Rep. 12, 1443 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang V., Davis D. A., Haque M., Huang L. E., Yarchoan R., Differential gene up-regulation by hypoxia-inducible factor-1alpha and hypoxia-inducible factor-2alpha in HEK293T cells. Cancer Res. 65, 3299–3306 (2005). [DOI] [PubMed] [Google Scholar]
- 51.Liu X., Chen Z., Xu C., Leng X., Cao H., Ouyang G., Xiao W., Repression of hypoxia-inducible factor α signaling by Set7-mediated methylation. Nucleic Acids Res. 43, 5081–5098 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bracken C. P., Fedele A. O., Linke S., Balrak W., Lisy K., Whitelaw M. L., Peet D. J., Cell-specific regulation of hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha stabilization and transactivation in a graded oxygen environment. J. Biol. Chem. 281, 22575–22585 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Xin J., Zhang H., He Y., Duren Z., Bai C., Chen L., Luo X., Yan D.-S., Zhang C., Zhu X., Yuan Q., Feng Z., Cui C., Qi X., Ouzhuluobu W. H., Wong Y., Wang B. S., Chromatin accessibility landscape and regulatory network of high-altitude hypoxia adaptation. Nat. Commun. 11, 4928 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peng Y., Cui C., He Y., Ouzhuluobu H., Zhang D., Yang Q. Z., Bianbazhuoma L., Yang Y., He K., Xiang X., Zhang S., Bhandari P. S., Yangla D., Baimakangzhuo D., Pan Y., Cirenyangji B., Gonggalanzi C. B., Bianba B., Ciwangsangbu S., Xu H., Chen S., Liu T., Wu X., Qi B. S., Down-regulation of EPAS1 transcription and genetic adaptation of Tibetans to high-altitude hypoxia. Mol. Biol. Evol. 34, 818–830 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Borodulin K., Tolonen H., Jousilahti P., Jula A., Juolevi A., Koskinen S., Kuulasmaa K., Laatikainen T., Männistö S., Peltonen M., Perola M., Puska P., Salomaa V., Sundvall J., Virtanen S. M., Vartiainen E., Cohort Profile: The national FINRISK study. Int. J. Epidemiol. 47, 696–696i (2018). [DOI] [PubMed] [Google Scholar]
- 56.Beall C. M., Two routes to functional adaptation: Tibetan and Andean high-altitude natives. Proc. Natl. Acad. Sci. U.S.A. 104, 8655–8660 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang X., Witt K. E., Bañuelos M. M., Ko A., Yuan K., Xu S., Nielsen R., Huerta-Sanchez E., The history and evolution of the Denisovan-EPAS1 haplotype in Tibetans. Proc. Natl. Acad. Sci. U.S.A. 118, e2020803118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huerta-Sánchez E., Jin X., Asan Z., Bianba B. M., Peter N., Vinckenbosch Y., Liang X., Yi M., He M., Somel P., Ni B., Wang X. O., Huasang J., Luosang Z. X. P., Cuo K., Li G., Gao Y., Yin W., Wang X., Zhang X., Xu H., Yang Y., Li J., Wang J., Wang R. N., Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature 512, 194–197 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou W., Liu K., Zeng L., He J., Gao X., Gu X., Chen X., Jing Li J., Wang M., Wu D., Cai Z., Claesson-Welsh L., Ju R., Wang J., Zhang F., Chen Y., Targeting VEGF-A/VEGFR2 Y949 signaling-mediated vascular permeability alleviates hypoxic pulmonary hypertension. Circulation 146, 1855–1881 (2022). [DOI] [PubMed] [Google Scholar]
- 60.Bouzina H., Rådegran G., Plasma adrenomedullin peptides and precursor levels in pulmonary arterial hypertension disease severity and risk stratification. Pulm. Circ. 10, 1–9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang C., Liu W., Liu Z., Chen L., Liu X., Kuang S., Hypoxia inhibits myogenic differentiation through p53 protein-dependent induction of Bhlhe40 protein. J. Biol. Chem. 290, 29707–29716 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.de la Rosa Rodriguez M. A., Kersten S., Regulation of lipid droplet homeostasis by hypoxia inducible lipid droplet associated HILPDA. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 1865, 158738 (2020). [DOI] [PubMed] [Google Scholar]
- 63.Oberholzer L., Lundby C., Stauffer E., Ulliel-Roche M., Hancco I., Pichon A., Lundby A.-K. M., Villafuerte F. C., Verges S., Robach P., Reevaluation of excessive erythrocytosis in diagnosing chronic mountain sickness in men from the world’s highest city. Blood 136, 1884–1888 (2020). [DOI] [PubMed] [Google Scholar]
- 64.Villafuerte F. C., Corante N., Chronic Mountain Sickness: Clinical aspects, etiology, management, and treatment. High Alt. Med. Biol. 17, 61–69 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Murphy W. G., The sex difference in haemoglobin levels in adults–mechanisms, causes, and consequences. Blood Rev. 28, 41–47 (2014). [DOI] [PubMed] [Google Scholar]
- 66.Murillo-Rodriguez E., Blanco-Centurion C., Sanchez C., Piomelli D., Shiromani P. J., Anandamide enhances extracellular levels of adenosine and induces sleep: an in vivo microdialysis study. Sleep 26, 943–947 (2003). [DOI] [PubMed] [Google Scholar]
- 67.Snider N. T., Walker V. J., Hollenberg P. F., Oxidation of the endogenous cannabinoid arachidonoyl ethanolamide by the cytochrome P450 monooxygenases: physiological and pharmacological implications. Pharmacol. Rev. 62, 136–154 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu J. J., Non A. L., Heinrich E. C., Gu W., Alcock J., Moya E. A., Lawrence E. S., Tift M. S., O’Brien K. A., Storz J. F., Signore A. V., Khudyakov J. I., Milsom W. K., Wilson S. M., Beall C. M., Villafuerte F. C., Stobdan T., Julian C. G., Moore L. G., Fuster M. M., Stokes J. A., Milner R., West J. B., Zhang J., Shyy J. Y., Childebayeva A., Vázquez-Medina J. P., Pham L. V., Mesarwi O. A., Hall J. E., Cheviron Z. A., Sieker J., Blood A. B., Yuan J. X., Scott G. R., Rana B. K., Ponganis P. J., Malhotra A., Powell F. L., Simonson T. S., Time domains of hypoxia responses and -omics insights. Front. Physiol. 13, 885295 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cupello C., Brito P. M., Herbin M., Meunier F. J., Janvier P., Dutel H., Clément G., Allometric growth in the extant coelacanth lung during ontogenetic development. Nat. Commun. 6, 8222 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hall J. E., Lawrence E. S., Simonson T. S., Fox K., Seq-ing higher ground: Functional investigation of adaptive variation associated with high-altitude adaptation. Front. Genet. 11, 471 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Alexander D. H., Novembre J., Lange K., Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.1000 Genomes Project Consortium, Auton A., Brooks L. D., Durbin R. M., Garrison E. P., Kang H. M., Korbel J. O., Marchini J. L., McCarthy S., McVean G. A., Abecasis G. R., A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Browning S. R., Waples R. K., Browning B. L., Fast, accurate local ancestry inference with FLARE. Am. J. Hum. Genet. 110, 326–335 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Boughton A. P., Welch R. P., Flickinger M., VandeHaar P., Taliun D., Abecasis G. R., Boehnke M., LocusZoom.js: Interactive and embeddable visualization of genetic association study results. Bioinformatics 37, 3017–3018 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gautier M., Vitalis R., rehh: an R package to detect footprints of selection in genome-wide SNP data from haplotype structure. Bioinformatics 28, 1176–1177 (2012). [DOI] [PubMed] [Google Scholar]
- 76.Albers P. K., McVean G., Dating genomic variants and shared ancestry in population-scale sequencing data. PLoS Biol. 18, e3000586 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Voight B. F., Kudaravalli S., Wen X., Pritchard J. K., A map of recent positive selection in the human genome. PLoS Biol. 4, e72 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.MacKinnon J. G., White H., Some heteroskedasticity-consistent covariance matrix estimators with improved finite sample properties. J. Econom. 29, 305–325 (1985). [Google Scholar]
- 79.Vasquez C. A., Cowan Q. T., Komor A. C., Base editing in human cells to produce single-nucleotide-variant clonal cell lines. Curr. Protoc. Mol. Biol. 133, e129 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee C., Hyun Jo D., Hwang G.-H., Yu J., Kim J. H., Park S.-E., Kim J.-S., Kim J. H., Bae S., CRISPR-Pass: Gene Rescue of Nonsense Mutations Using Adenine Base Editors. Mol. Ther. 27, 1364–1371 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kluesner M. G., Nedveck D. A., Lahr W. S., Garbe J. R., Abrahante J. E., Webber B. R., Moriarity B. S., EditR: A method to quantify base editing from sanger sequencing. CRISPR J. 1, 239–250 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.E. Garrison, G. Marth, Haplotype-based variant detection from short-read sequencing. arXiv:1207.3907 [q-bio.GN] (17 July 2012).
- 84.Percy M. J., Furlow P. W., Lucas G. S., Li X., Lappin T. R. J., McMullin M. F., Lee F. S., A gain-of-function mutation in the HIF2A gene in familial erythrocytosis. N. Engl. J. Med. 358, 162–168 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lin Q., Cong X., Yun Z., Differential hypoxic regulation of hypoxia-inducible factors 1alpha and 2alpha. Mol. Cancer Res. 9, 757–765 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gameiro P. A., Yang J., Metelo A. M., Pérez-Carro R., Baker R., Wang Z., Arreola A., Rathmell W. K., Olumi A., López-Larrubia P., Stephanopoulos G., Iliopoulos O., In vivo HIF-mediated reductive carboxylation is regulated by citrate levels and sensitizes VHL-deficient cells to glutamine deprivation. Cell Metab. 17, 372–385 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bartoszewski R., Moszyńska A., Serocki M., Cabaj A., Polten A., Ochocka R., Dell’Italia L., Bartoszewska S., Króliczewski J., Dąbrowski M., Collawn J. F., Primary endothelial cell-specific regulation of hypoxia-inducible factor (HIF)-1 and HIF-2 and their target gene expression profiles during hypoxia. FASEB J. 33, 7929–7941 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Watrous J. D., Niiranen T. J., Lagerborg K. A., Henglin M., Xu Y.-J., Rong J., Sharma S., Vasan R. S., Larson M. G., Armando A., Mora S., Quehenberger O., Dennis E. A., Cheng S., Jain M., Directed Non-targeted Mass Spectrometry and chemical networking for discovery of eicosanoids and related oxylipins. Cell Chem Biol. 26, 433–442.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhu Z., Zheng Z., Zhang F., Wu Y., Trzaskowski M., Maier R., Robinson M. R., McGrath J. J., Visscher P. M., Wray N. R., Yang J., Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat. Commun. 9, 224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu W., Xie Y., Ma J., Luo X., Nie P., Zuo Z., Lahrmann U., Zhao Q., Zheng Y., Zhao Y., Xue Y., Ren J., IBS: an illustrator for the presentation and visualization of biological sequences. Bioinformatics 31, 3359–3361 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E., UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S9
Tables S1 to S3 and S5
Legends for tables S4 and S6
Tables S4 and S6