Abstract
PURPOSE
Matching patients to clinical trials is cumbersome and costly. Attempts have been made to automate the matching process; however, most have used a trial-centric approach, which focuses on a single trial. In this study, we developed a patient-centric matching tool that matches patient-specific demographic and clinical information with free-text clinical trial inclusion and exclusion criteria extracted using natural language processing to return a list of relevant clinical trials ordered by the patient's likelihood of eligibility.
MATERIALS AND METHODS
Records from pediatric leukemia clinical trials were downloaded from ClinicalTrials.gov. Regular expressions were used to discretize and extract individual trial criteria. A multilabel support vector machine (SVM) was trained to classify sentence embeddings of criteria into relevant clinical categories. Labeled criteria were parsed using regular expressions to extract numbers, comparators, and relationships. In the validation phase, a patient-trial match score was generated for each trial and returned in the form of a ranked list for each patient.
RESULTS
In total, 5,251 discretized criteria were extracted from 216 protocols. The most frequent criterion was previous chemotherapy/biologics (17%). The multilabel SVM demonstrated a pooled accuracy of 75%. The text processing pipeline was able to automatically extract 68% of eligibility criteria rules, as compared with 80% in a manual version of the tool. Automated matching was accomplished in approximately 4 seconds, as compared with several hours using manual derivation.
CONCLUSION
To our knowledge, this project represents the first open-source attempt to generate a patient-centric clinical trial matching tool. The tool demonstrated acceptable performance when compared with a manual version, and it has potential to save time and money when matching patients to trials.
AI improves patient-centric clinical trials matching.
INTRODUCTION
Clinical trials are a critical step in translating scientific discoveries from bench to bedside. Many drugs that show promise in preclinical studies are ultimately not brought to market because of lack of efficacy. Yet, even for effective drugs, the clinical trials process poses many challenges, including inadequate enrollment. Roughly 86% of trials do not meet their enrollment timeline, and one third of phase III trials are terminated because of inadequate participation.1-3 As a result, the process of drug development—which is estimated to take 10-15 years and costs up to $2 billion US dollars4—often fails. Given that the clinical trial phase of development is often the most expensive,5 there is a pressing need to optimize patient recruitment.
CONTEXT
Key Objective
To develop a tool for processing free-text clinical trial inclusion and exclusion criteria and matching patients to relevant clinical trials.
Knowledge Generated
The automated tool for criteria extraction and patient-trial matching demonstrated acceptable performance and significantly reduced manual workload. However, the tool failed to identify key criteria extracted by human abstractors.
Relevance
Automated clinical trial inclusion and exclusion criteria extraction can be combined with manual subject matter review to optimize clinical trials matching performance while reducing human workload.
Since clinical trial eligibility criteria are nonstandardized and semantically complex,6 screening patients for eligibility is generally conducted manually. This process is cumbersome and costly, requiring a significant amount of time from clinicians or administrative staff who have domain-specific expertise.5 To streamline this process, there is significant interest in developing technology to automate the process of patient-trial matching. However, the lack of eligibility criteria in readily computable form is a major bottleneck.
Previous studies have sought to match patients to clinical trials by computing a match score between free-text criteria and free-text notes from the electronic health record (EHR) although these studies have demonstrated wide-ranging positive predictive values from 13% to 63%.5,7 Studies have sought to validate the use of IBM Watson to match patients to lung and breast cancer trials, reporting positive predictive values as high as 76.5%.8,9 However, these studies' methodologies are largely opaque because of the technology's proprietary nature. Other private companies have emerged with similar aims, including Deep Lens,10 Deep 6 AI,11 Antidote,12 Mendel.ai,13 and Massive Bio.14 To date, none of these have published information on their performance outside of small studies or abstracts with restricted cohorts.9,15
CancerLinQ, which develops structured data sets from a combination of automatically and manually curated data from a patient's record, is developing capabilities to allow sites to match patients to clinical trials.16 Genomic analysis companies, including Tempus17 and Foundation Medicine,18 have developed genomic-based clinical trials matching mechanisms to identify patients with rare molecular alterations and match them to clinical trials to increase enrollment for trials studying rare subtypes. For example, Foundation Medicine has partnered with the National Cancer Institute to notify physicians when a patient's genomic testing includes an alteration that may make a patient eligible for participation on the Molecular Analysis for Therapy Choice (MATCH) study, which is assessing efficacy of targeted therapies.18
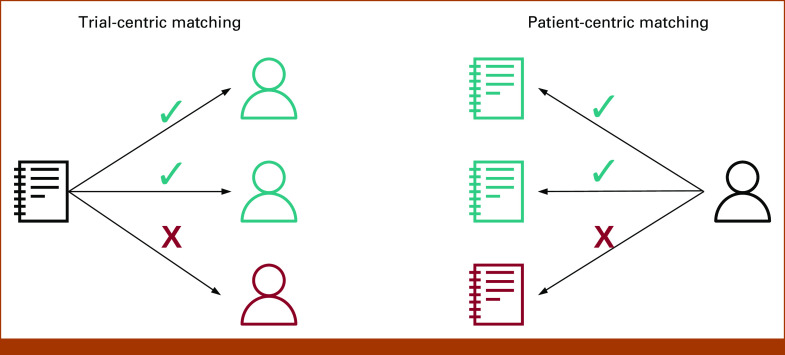
Despite progress in clinical trials matching, nearly all efforts rely on the cumbersome process of manual data extraction from patient records.19 The dominant methodology used in previous work is known as trial-centric cohort identification,5 in which patients from a large cohort are screened for eligibility for a specific trial. An alternative approach is patient-centric trial recommendation, in which physicians enter data regarding an individual patient into prespecified fields, after which a recommendation engine generates probabilistic matches to available trials (Fig 1). This approach gives patients more freedom to choose clinical trials that fit their goals and preferences, even if those trials are not enrolling at a local center. While some patients' values and preferences may lead them to only enroll on clinical trials available at a local center, others may choose to travel to a larger referral center for clinical trials participation—something which trial-centric cohort identification is not well-suited to support.
FIG 1.
Comparison of trial-centric and patient-centric matching approaches. Trial-centric matching attempts to identify one or more patients who are eligible to participate in a clinical trial. By contrast, patient-centric matching attempts to identify one or more clinical trials a patient may be eligible for. The directional one-to-many asymmetry in patient-centric matching approaches may lend to greater patient choice.
This study aims to address methodological gaps in patient-centric clinical trial recommendation via creation of an open-source, patient-centric trial recommendation tool, validated for the specific use case of pediatric acute leukemia.
MATERIALS AND METHODS
Protocol Data Set
A total of 216 trial protocols were used as training data for the matching pipeline. Selection criteria included all phase I, II, and III clinical trials listed on ClinicalTrials.gov20 that enrolled children and adolescents with leukemia between 1987 and 2018. Both completed and active clinical trials were included. Criteria formats (in order of increasing complexity) observed included the following: inclusion/exclusion criteria in a bulleted list (14%), inclusion/exclusion criteria in a bulleted list with nested sub-bullets (16%), category headers and subheaders (27%, eg, disease characteristics, biologic therapy) in a bulleted list, and category headers/subheaders in a bulleted list with nested sub-bullets (43%). Less complex criteria structures (eg, simple bulleted list) were observed with increasing frequency in more contemporary trials.
Text Extraction
To assemble a training data set, trial information from 216 protocols was downloaded as extensible markup language (XML) files. XML files are structured data with syntax that can readily be consumed by computing platforms (Fig 2). XML files were parsed to extract a free-text block of eligibility criteria for each trial. Individual criteria were discretized from each text block on the basis of recognition of common criterion separation patterns (eg, bulleted list, numbered outline) via the application of flexible regular expressions. Text preprocessing involved lowercasing text (except for medical abbreviations), removal of special characters (except semicolons, which are used to specify genetic mutations), removal of single-letter words, removal of stop words, and lemmatization (the grouping together of inflected forms).
FIG 2.

Eligibility criteria excerpt from extensible markup language file from ClinicalTrials.gov identifier: NCT02883049.
Sentence Embedding and Labeling
After processing, individual text criteria were represented numerically as sentence embeddings using FastText,21 an open-source algorithm developed by Facebook. Embeddings were constructed using a continuous bag-of-words model22 and a vector size of 256, with otherwise default hyperparameters. Word embeddings were transformed into sentence embeddings by taking the average of all word embedding vectors of a sentence (known as mean/average-pooling or centroid method).23,24 Each criterion was manually assigned a label corresponding to a clinically relevant category (eg, required renal function) by a study team member with input from clinical subject matter experts. Thirteen label categories (12 labels plus other category) were derived from previous semantic categories defined in the literature25 and variable categories prominent in the leukemia data dictionaries of the Pediatric Cancer Data Commons, the flagship project of Data for the Common Good.26,27 Remaining criteria were labeled as unclassified.
Training of Multilabel Support Vector Machine Classifier
The sentence embeddings with labels were used to train a multilabel support vector machine (SVM). Modeled criteria were selected on the basis of their frequency (ie, higher likelihood of being encountered in other trials) and syntactic regularity (ie, higher likelihood of being matched to patient characteristics without multiple comparators and deep nested logic). Criteria represented in <2% of trials were not included in the model. A one-versus-all approach was used to select the label with the highest probability of match. Platt scaling28,29 was used to transform decision values into probabilities. Grid search30 with five-fold cross-validation was performed for the cost (C) hyperparameter, whose values were inversely weighted to the outcome rate (ie, class frequency) to adjust for class imbalance. In addition, to flag text data that did not fit a prespecified category, a variety of confidence thresholds were tested at which to leave a sentence unclassified.
Patient-Trial Matching
To compute a similarity score for each trial, available structured fields (ie, age and diagnosis) were extracted from input XML files and converted into if-then logic to flag a potentially matched criterion. For machine-classified free text with a relatively predictable structure in the data set (eg, required renal function), logic filters were constructed using flexible regular expressions to extract numbers, comparators, and relationships. After extraction, comparison with an input patient field (eg, creatinine) could be made by computing the proportion of matches to potential matches. Individual criteria match scores were summed and divided by the total number of potential matches to produce a normalized composite match score for each trial. A penalty was added to each composite score on the basis of a trial's percentage of unclassified criteria. Composite scores could then be ranked in order of highest potential likelihood of eligibility.
Pipeline Validation
For validation, a cohort of five currently enrolling trials being used in a nonautomated version of this matching tool (GEARBOx31) was placed into the extraction, classification, and scoring pipeline described above. To assess how the model would perform with input formats other than XML, three of the five trials were extracted as full protocols from Microsoft Word documents—a file format commonly used for protocols that were not represented in the training data. Text classification performance was reported as accuracy, precision, and recall. The proportion of automatically extracted information as compared with that manually extracted by the tool's current version was reported as a percentage. A synthetic cohort of 20 patients was generated using Python by defining all eligibility criteria used by the trial matching tool and randomly populating patient characteristics within realistic bounds. The synthetic cohort was used to assess the tool's top-3 accuracy in patient matching as compared with the manual tool. The synthetic cohort sample size was limited to 20 patients to balance the time required to validate against the nonautomated tool as this portion of the validation required manual entry of all patient characteristics. Finally, a time analysis estimate was conducted to compare manual versus automatic extraction.
Software and Tools
Text extraction and processing, data analysis, and visualization were conducted using the Python programming language32 in the Jupyter Notebook environment.33 Machine learning and natural language processing (NLP) libraries used included scikit-learn,30 Natural Language ToolKit,34 and gensim.35
RESULTS
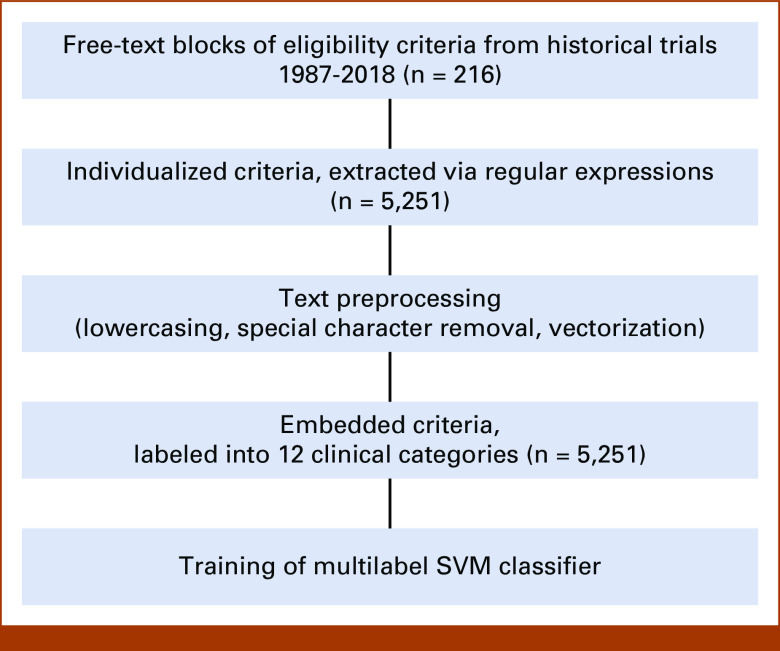
A total of 5,251 discretized criteria were extracted from the protocols (Fig 3).
FIG 3.
Process diagram with descriptive results of text processing. SVM, support vector machine.
Eligibility criteria label types included previous chemotherapy/biologics (17%), patient pregnancy and/or contraception use (7%), renal function (6%), diagnosis (5%), disease status (5%, eg, relapsed/refractory to therapy), active infection (5%), hepatic function (5%), performance status (4%), cardiovascular function (4%), previous radiotherapy (3%), age (3%), and central nervous system involvement (2%). Approximately 35% of criteria did not fall into a prespecified category and were categorized as others.
Classifier Performance
The held-out validation data set of five trials comprised a total of 216 discretized eligibility criteria. The one- versus all-text classifier demonstrated 75% pooled accuracy, 76% precision, and 75% recall. Individual accuracy as compared with a human abstractor was highest for renal function (83%), performance status (83%), and active infection (73%). Lowest accuracy was observed for stem-cell donor availability (59%) and concurrent medications (59%).
Comparative Performance With a Nonautomated Patient-Matching Tool
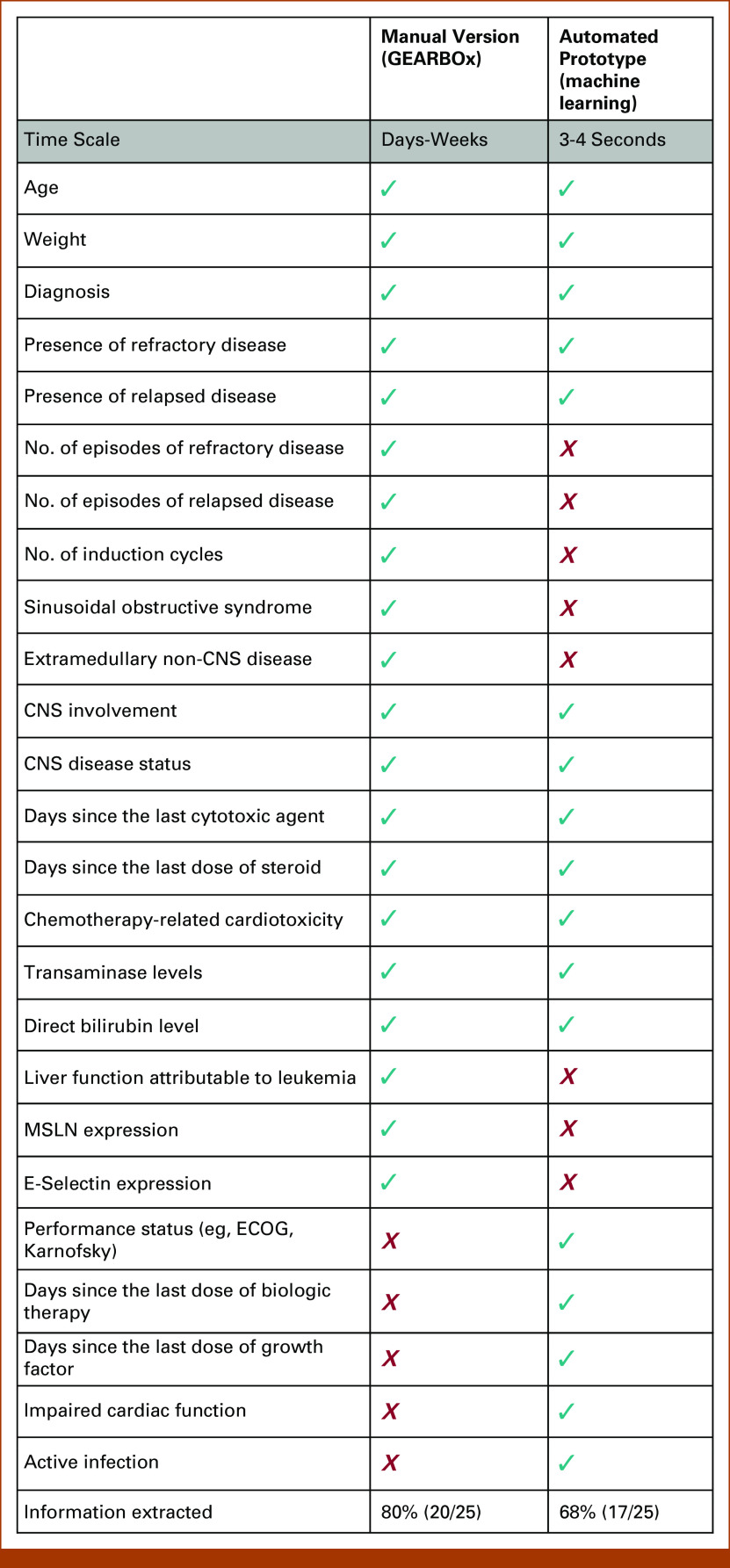
The nonautomated version of this tool represents approximately 20 patient-criteria match fields that were manually chosen as relevant inclusion criteria, extracted from free text and hand-coded into if-then logic. The automated prototype of this study was able to recognize and extract 12 of 20 (60%) represented fields automatically and extract eligibility rules for five additional criteria fields not represented in the manual version. These additional five fields that were leveraged by the automated tool were not included in the manual version as they were not deemed to be as relevant as other criteria when the manual version was generated. This represents an information extraction of 68% for the automated version as compared with 80% for the manually derived tool (Fig 4).
FIG 4.
Comparison of feature extraction between automated and manual tools. ECOG, Eastern Cooperative Oncology Group.
When compared with the manually derived version across a cohort of 20 synthetically generated patients, the automated matching tool generated ranked lists with a top-3 trial accuracy of 100%. In other words, the top trial identified by the manual version was present in the top 3 identified as most likely from the automated version 100% of the time. The top 1 accuracy was 15%.
The automated matching algorithm took approximately 3-4 seconds to derive eligibility criteria for multiple input trials and return a ranked list of trials in order of most likely eligibility for an individual patient. On the other hand, manual derivation eligibility criteria rules may take hours to hand-code if-then logic for every new trial. An estimate provided by the technical team that developed GEARBOx is that the manual process takes approximately 8 hours to extract eligibility criteria and code into if-then logic for each trial.
DISCUSSION
We successfully developed a patient-centric, automated text extraction and classification pipeline for matching children with leukemia to clinical trials. The prototype demonstrated acceptable performance when compared with a manually derived version of the same tool. The automation process led to a modest 12% drop in information extraction compared with manual methods, which can reduce the workload for human abstractors. Classification of discrete clinical trial criteria via a multilabel SVM achieved a pooled accuracy of 75%, which is similar in performance to other studies examining semantically complex biomedical text.36 Further validation across a larger set of clinical trials and real-world patients will provide additional insights into performance. Eligibility criteria categories for which individual accuracy was the highest were typically those with the most repeated and predictable syntax, such as organ function laboratory value requirements. Automated recognition of more variable text categories, such as contraindicated medications, proved to be more difficult as pharmaceutical-related text can have very high degrees of freedom and medications may be referred to by category (eg, corticosteroid) or drug name. Composite matching scores generated from the natural language processing pipeline were able to achieve a 100% top-3 accuracy in recommending patients from a synthetic cohort to currently enrolling trials. Furthermore, the prototype was able to achieve these results with a processing time of approximately 4 seconds, as compared with an estimated manual extraction time of 8 hours. This demonstrates potential to save time via automated classification of well-recognized trial criteria, leaving only poorly processed sentences for manual review. Although the most ideal future state would include authoring of clinical trials in a tool that includes structured data elements from the start, we envision an intermediate future state where an automated machine learning model performs initial extraction and programming of if-then logic on the basis of inclusion criteria in an effort to leverage large data sets to identify pertinent inclusion criteria, which are then reviewed by a subject matter expert for completeness, accuracy, and relative importance. This process combines the efficiency of an automated process that uses a large data set with the strengths of human abstractors to increase the ease with which new trials can be added to automated clinical trials matching tools while also optimizing precision and recall for clinical trials matching.
Strengths of this work include transparent methods and assessment of algorithm performance. TrialJectory is a similar tool that takes a patient-driven approach, encouraging patients to answer questions about their diagnosis and personal preferences and attempts to match patients to clinical trials on the basis of artificial intelligence–based extraction of eligibility criteria from ClinicalTrials.gov.37 However, we were unable to compare our performance with that of TrialJectory as the details of their proprietary method and its performance on the same data set were not available.
This study has multiple limitations. First, the training data set used came exclusively via ClinicalTrials.gov. Although this is a readily available public resource, it may not contain all eligibility criteria included in a trial's full protocol (often buried within a PDF or Microsoft Word document). However, typically, the most pertinent information is included in ClinicalTrials.gov, and given that this tool applies a probabilistic recommendation approach to screen for potential eligibility, the entirety of the eligibility criteria may not be necessary. Second, although comparative performance metrics were reported, the manually derived version of the tool is not directly comparable with the automated prototype. This is because the manual version uses a more deterministic approach to trial matching (ie, yes/no) on the basis of pertinent eligibility criteria deemed most relevant and extracted by human abstractors, whereas the automated version assigns a probability on the basis of criteria that were identified through an automated process. Because of this difference, trials assigned as a poor match with the manual version could demonstrate a relatively high composite probability when using automation. Moreover, at the time of validation, there was no immediately available cohort of real-world patients with leukemia for comparison of the tool's performance. Although the synthetic cohort was generated to represent characteristics of realistic patients with leukemia, these results may not be generalizable to real-world cohorts as they were randomly generated. The synthetic cohort therefore may introduce bias. Future validation work could include applying the algorithm to a deidentified set of real-world patients that includes a larger sample size than the synthetic cohort and development of automated methods to enter data into the nonautomated matching tool for larger-scale validation. Because the training data set represented pediatric leukemia trials conducted in the United States, the model may not perform well with trials conducted on other diseases (which may focus on different categories of eligibility criteria) or trials performed outside of the United States, which may format or phrase eligibility criteria differently. The automated tool was validated against a relatively small number of clinical trials; validation against a larger set of clinical trials would improve our understanding of the tool's performance across a broader set of clinical trials. As the tool was not tested in a real-world clinical setting, we were unable to assess the burden of false-positive matches on the patient and clinical team. When implemented in real-world practice, the optimal tradeoff between sensitivity and specificity may need to be calibrated to optimize the matching process while also minimizing the burden of false-positive matches. Finally, one key limitation of the automated model was the ability to extract named pharmaceutical agents. We expect improvements in model performance by exposing the model to a larger corpus of drug names (eg, RxNorm).
In conclusion, to our knowledge, this represents the first open-source and publicly documented attempt to generate a patient-centric trial matching tool for pediatric leukemia, in which providers anywhere can enter information regarding their patient and find a probabilistic match to currently enrolling trials. The majority of previous studies in patient-trial matching do not offer this capability as cohort-centric matching tools often focus on a single trial. Moreover, existing clinical trials matching systems are proprietary and lack transparency about how trial lists are generated. Nonetheless, the performance of the system we describe could be compared with that of proprietary systems head-to-head by entering patient information and assessing—through subject matter expert review—accuracy of top clinical trials matches, assuming that both systems include the same clinical trials against which to match patients. While trial-centric approaches are designed to maximize enrollment on a particular trial, patient-centric approaches are designed to maximize patients' choice for clinical trials enrollment.
By automating the process for clinical trials matching, barriers to access for clinical trials enrollment can be lowered. Future work will seek to take advantage of more sophisticated classification techniques, such as bidirectional encoder representations from transformers (BERT38), and to validate probabilistic patient-trial matching scores on real-world cohorts across multiple disease groups. Although the approach we describe automates clinical trials eligibility criteria extraction, the extraction and entry of patient characteristics remain a manual process. Data standards, including HL7 Fast Healthcare Interoperability Resources, have the potential to automate data extraction from the EHR, bringing us one step closer to the holy grail of end-to-end automation for clinical trials matching.
Although NLP capabilities have significantly advanced over the past decade, NLP remains imperfect and manual human review remains the gold standard for determining patient eligibility for clinical trials. Researchers, pharmaceutical companies, research cooperative groups, clinicians, and patients would all benefit from the use of data standards, with common data elements to represent clinical trials eligibility criteria. This ideal state would allow for eligibility criteria to be unambiguously coded in machine-interpretable form and compared with discrete data elements within the EHR (eg, laboratory tests, patient demographics, diagnosis codes). Thus far, a lack of incentive alignment has precluded widespread adoption of such an ideal state, requiring the type of third-party, downstream tool and platform development as we have described. As the community increases adoption of trial authoring platforms that produce structured standardized data, there will be an increase in fidelity and availability of trial matching tools for clinicians.
Brian Furner
Stock and Other Ownership Interests: United Therapeutics
Samuel L. Volchenboum
Stock and Other Ownership Interests: Litmus Health
Consulting or Advisory Role: Accordant, Westat
Travel, Accommodations, Expenses: Sanford Health
No other potential conflicts of interest were reported.
DISCLAIMER
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
SUPPORT
Supported by The Fund for Innovation in Cancer Informatics (ICI Foundation) and the National Cancer Institute of the National Institutes of Health under award number 1U01CA269420-01. The Center for Research Informatics was funded by the Biological Sciences Division at the University of Chicago with additional funding provided by the Institute for Translational Medicine, CTSA grant number UL1 TR000430 from the National Institutes of Health.
DATA SHARING STATEMENT
The data sets used to train the model, synthetic cohort, and trained model are publicly available at https://github.com/chicagopcdc/Automated-Matching-of-Patients-to-Clinical-Trials.
AUTHOR CONTRIBUTIONS
Conception and design: Samuel Kaskovich, Tomasz Oliwa, Luca Graglia, Brian Furner, Anoop Mayampurath, Samuel L. Volchenboum
Financial support: Samuel L. Volchenboum
Administrative support: Samuel L. Volchenboum
Collection and assembly of data: Samuel Kaskovich, Brian Furner, Samuel L. Volchenboum
Data analysis and interpretation: Samuel Kaskovich, Kirk D. Wyatt, Tomasz Oliwa, Luca Graglia, Brian Furner, Jooho Lee, Samuel L. Volchenboum
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/cci/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Brian Furner
Stock and Other Ownership Interests: United Therapeutics
Samuel L. Volchenboum
Stock and Other Ownership Interests: Litmus Health
Consulting or Advisory Role: Accordant, Westat
Travel, Accommodations, Expenses: Sanford Health
No other potential conflicts of interest were reported.
REFERENCES
- 1.Huang GD, Bull J, Johnston McKee K, et al. : Clinical trials recruitment planning: A proposed framework from the Clinical Trials Transformation Initiative. Contemp Clin Trials 66:74-79, 2018 [DOI] [PubMed] [Google Scholar]
- 2.Sacks LV, Shamsuddin HH, Yasinskaya YI, et al. : Scientific and regulatory reasons for delay and denial of FDA approval of initial applications for new drugs, 2000-2012. JAMA 311:378-384, 2014 [DOI] [PubMed] [Google Scholar]
- 3.Ross JS, Dzara K, Downing NS: Efficacy and safety concerns are important reasons why the FDA requires multiple reviews before approval of new drugs. Health Aff 34:681-688, 2015 [DOI] [PubMed] [Google Scholar]
- 4.Harrer S, Shah P, Antony B, et al. : Artificial intelligence for clinical trial design. Trends Pharmacol Sci 40:577-591, 2019 [DOI] [PubMed] [Google Scholar]
- 5.Ni Y, Wright J, Perentesis J, et al. : Increasing the efficiency of trial-patient matching: Automated clinical trial eligibility pre-screening for pediatric oncology patients. BMC Med Inform Decis Mak 15:28, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ross J, Tu S, Carini S, et al. : Analysis of eligibility criteria complexity in clinical trials. Summit Transl Bioinform 2010:46-50, 2010 [PMC free article] [PubMed] [Google Scholar]
- 7.Ni Y, Kennebeck S, Dexheimer JW, et al. : Automated clinical trial eligibility prescreening: Increasing the efficiency of patient identification for clinical trials in the emergency department. J Am Med Inform Assoc 22:166-178, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Helgeson J, Rammage M, Urman A, et al. : Clinical performance pilot using cognitive computing for clinical trial matching at Mayo Clinic. J Clin Orthod 36, 2018. (15 suppl; abstr e18598) [Google Scholar]
- 9.Alexander M, Solomon B, Ball DL, et al. : Evaluation of an artificial intelligence clinical trial matching system in Australian lung cancer patients. JAMIA Open 3:209-215, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deep Lens. https://www.deeplens.ai
- 11.Deep6 : Deep6.ai. 2020. https://deep6.ai/
- 12.Antidote : Clinical trial patient recruitment. https://www.antidote.me
- 13.Mendel AI—Unlock the power of your unstructured clinical data. https://www.mendel.ai/
- 14.Massive Bio : Massive Bio. 2021. https://massivebio.com/
- 15.Calaprice-Whitty D, Galil K, Salloum W, et al. : Improving clinical trial participant prescreening with artificial intelligence (AI): A comparison of the results of AI-assisted vs standard methods in 3 oncology trials. Ther Innov Regul Sci 54:69-74, 2020 [DOI] [PubMed] [Google Scholar]
- 16.Supporting each cancer patient at every step of their journey: CancerLinQ in 2022 and beyond. https://www.cancerlinq.org/supporting-each-cancer-patient-every-step-their-journey-cancerlinq-2022-and-beyond
- 17.Clinical Trial Matching. Tempus. 2020. https://www.tempus.com/oncology/clinical-trial-matching/ [Google Scholar]
- 18.Foundation Medicine to Identify Patients Eligible for National Cancer Institute's NCI-MATCH (Molecular Analysis for Therapy Choice) Study. Foundation Medicine. https://www.foundationmedicine.com/press-releases/6c196a9b-ce54-4005-a5e3-2c8f9c10bcce [Google Scholar]
- 19.Tate C: US hospital EMR market share 2020. 2020. https://klasresearch.com/report/us-hospital-emr-market-share-2020/1616
- 20.Home—ClinicalTrials.gov. https://clinicaltrials.gov
- 21.fastText. https://fasttext.cc/index.html
- 22.Mikolov T, Chen K, Corrado G, et al. : Efficient estimation of word representations in vector space. arXiv 10.48550/ARXIV.1301.3781 [epub ahead of print on January 16, 2013] [DOI] [Google Scholar]
- 23.Rudkowsky E, Haselmayer M, Wastian M, et al. : More than bags of words: Sentiment analysis with word embeddings. Commun Methods Meas 12:140-157, 2018 [Google Scholar]
- 24.Pennington J, Socher R, Manning C: GloVe: Global vectors for word representation, in Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP). Doha, Qatar, Association for Computational Linguistics, 2014, pp 1532-1543 [Google Scholar]
- 25.Bhattacharya S, Cantor MN: Analysis of eligibility criteria representation in industry-standard clinical trial protocols. J Biomed Inform 46:805-813, 2013 [DOI] [PubMed] [Google Scholar]
- 26.Pediatric Cancer Data Commons : Pediatric cancer data commons. 2019. https://commons.cri.uchicago.edu/
- 27.Plana A, Furner B, Palese M, et al. : Pediatric cancer data commons: Federating and democratizing data for childhood cancer research. JCO Clin Cancer Inform 5:1034-1043, 2021 [DOI] [PubMed] [Google Scholar]
- 28.Platt JC: Probabilities for SV machines, in Smola AJ, Bartlett PL, Schölkopf B, Schuurmans D (eds): Advances in Large Margin Classifiers. Cambridge, MA, MIT Press, 2000, pp 61-74. [Google Scholar]
- 29.Platt JC: Probabilistic outputs for support vector machines and comparisons to regularized likelihood methods. 1999. https://home.cs.colorado.edu/∼mozer/Teaching/syllabi/6622/papers/Platt1999.pdf
- 30.Pedregosa F, Varoquaux G, Gramfort A, et al. : Scikit-learn: Machine learning in Python. J Mach Learn Res 12:2825-2830, 2011 [Google Scholar]
- 31.GEARBOx. GEARBOx.pedscommons.org
- 32.Python : Python.org. https://www.python.org/
- 33.Project Jupyter. https://www.jupyter.org
- 34.NLTK : Natural Language Toolkit. https://www.nltk.org/
- 35.Řehůřek R, Sojka P: Software framework for topic modelling with large corpora. Presented at the LREC 2010 Workshop New Challenges for NLP Frameworks, Valletta, Malta, May 22, 2010
- 36.Joachims T: Text categorization with support vector machines: Learning with many relevant features, in Machine Learning: ECML-98. Lecture Notes in Computer Science. Springer Berlin Heidelberg, 1998, pp 137-142. [Google Scholar]
- 37.Geva N: TrialJectory for Oncologists. TrialJectory. 2020. https://www.trialjectory.com/for-oncologists/ [Google Scholar]
- 38.Li Y, Rao S, Solares JRA, et al. : BEHRT: Transformer for electronic health records. Sci Rep 10:7155, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets used to train the model, synthetic cohort, and trained model are publicly available at https://github.com/chicagopcdc/Automated-Matching-of-Patients-to-Clinical-Trials.