Abstract
Introduction
A set of genetic mutations to classify hepatocellular carcinoma (HCC) useful to clinical studies is an unmet need. Hepatitis B virus-related HCC (HBV-HCC) harbors a unique genetic mutation, namely, the HBV integration, among other somatic endogenous gene mutations. We explored a combination of HBV DNA integrations and common somatic mutations to classify HBV-HCC by using a capture-sequencing platform.
Methods
A total of 153 HBV-HCCs after surgical resection were subjected to capture sequencing to identify HBV integrations and three common somatic mutations in genomes. Three mutually exclusive mutations, HBV DNA integration into the TERT promoter, HBV DNA integration into MLL4, or TERT promoter point mutation, were identified in HBV-HCC.
Results
They were used to classify HBV-HCCs into four groups: G1 with HBV-TERT integration (25.5%); G2 with HBV-MLL4 integration (10.5%); G3 with TERT promoter mutation (30.1%); and G4 without these three mutations (34.0%). Clinically, G3 has the highest male-to-female ratio, cirrhosis rate, and associated with higher early recurrence and mortality after resection, but G4 has the best outcome. Transcriptomic analysis revealed a grouping different from the published ones and G2 with an active immune profile related to immune checkpoint inhibitor response. Analysis of integrated HBV DNA provided clues for HBV genotype and variants in carcinogenesis of different HCC subgroup. This new classification was also validated in another independent cohort.
Conclusion
A simple and robust genetic classification was developed to aid in understanding HBV-HCC and in harmonizing clinical studies.
Keywords: Virus DNA integration, Classification, TERT, Liver cancer
Introduction
Hepatocellular carcinoma (HCC) ranks 5th among common human cancers and usually has a very poor prognosis [1]. In the wake of molecular targeting and immunotherapy, major progress with significant improvement in survival for HCC patients has recently been achieved [2]. Management of HCC depends on the Barcelona Clinic Liver Cancer (BCLC) staging system, which is based upon tumor load and liver function. However, this system does not yet include the nature of individual HCCs, namely, their classification.
Traditionally, HCC is classified by the degree of differentiation according to tumor histopathology, but such classification does not correlate well with prognosis or treatment outcome. As human cancer has been considered a disease of genetic mutations, mutations are already essential in classifying leukemia or lung cancers into distinct subgroups for predicting survival and guiding therapeutic options. Many common somatic mutations of HCC have been discovered by genomic studies [3–6]. RNA sequencing of HCC has also generated different expression profiles, suggesting heterogeneity in this cancer. Despite these efforts to classify HCC, the currently proposed genetic classifications of HCC need to be improved because the genetic mutation criteria do not separate the subgroups clearly. Therefore, a simple and useful genetic classification of human HCC is still an unresolved challenge.
The majority of HCC cases worldwide are due to chronic hepatitis B virus (HBV) or hepatitis C virus infection, accounting for ∼70% of liver cancers [7]. HBV-related HCC (HBV-HCC) is the single most important factor and is present in 50% of all HCC cases; it shares many somatic mutations with HCC of other etiologies. However, HBV-HCC harbors a unique somatic mutation, namely, insertional mutagenesis by HBV DNA integration into the chromosomes of infected hepatocytes. HBV integration occurs early in infection, inserting viral DNA into chromosomes in a random manner at the rate of 0.1% of infected hepatocyte [8]. However, approximately 90% of HBV-HCC involves HBV DNA integration in chromosomes [4, 9–13]. In addition, a few integration hotspots in HBV-HCCs have been identified in gene TERT, MLL4 (KMT2B), and CCNE1[10, 11, 14]. Both a dramatic increase in the HBV DNA integration rate from infected hepatocytes to HCC and the presence of integration hotspots in HCC support insertional carcinogenesis via HBV DNA. Therefore, exploring whether HBV integration is useful for the classification of HBV-HCC is warranted.
In our previous study on the identification of HBV DNA integration in HBV-HCCs, we noted HBV integration in the TERT promoter or in MLL4 and somatic mutations in the TERT promoter, G(-124)A or G(-146)A from ATG, to be quite common, in 28%, 14%, and 20% of HBV-HCCs, respectively. More importantly, these three mutations appeared to occur in a mutually exclusive manner [15]. This raises the hypothesis that the three somatic mutations are independent and might be useful in defining subgroups of HBV-HCCs. To address this, we expanded our HBV integration analysis in a larger HCC series. Here, we report data supporting that HBV-TERT, HBV-MLL4 integration, and TERT promoter mutation are truly independent and can be used to classify HBV-HCC into four distinct subgroups. The four subgroups show unique somatic mutations, RNA expression profiles, and different clinical outcomes, with indications of response to molecular or immunotherapy. Analysis of integrated HBV DNA further suggests HBV genetic variations influencing the clinical outcome of subgroup. This new classification may provide a simple and reproducible platform for investigators to classify HBV-HCC in a common format to improve clinical management.
Materials and Methods
Sample and Clinical Data Collection
For the main cohort, 122 HBV-HCC tumor tissues after surgical resection were collected at National Taiwan University Hospital (NTUH), and 31 HBV-HCC tumor gDNA were applied from Taiwan Liver Center Network (TLCN). Another 112 HBV-HCC tumor cases were included in validation cohort from National Taiwan University, Taipei Veterans General Hospital, Chang Gung Memorial Hospital, and Tri-Service General Hospital. Clinical information including gender, age, tumor size, tumor grade, cirrhosis, microvascular invasion, AFP, recurrence, and survival was collected if available. The Institutional Review Boards approved the use of these samples and clinical information.
Whole-Genome Sequence and HBV Capture-Next Generation Sequence
Genomic DNA was extracted from tumor tissues by the standard phenol-chloroform method for library preparation using TruSeq DNA Sample Preparation Kit (Illumina, CA, USA) and sequenced with the NovaSeq system (Illumina) for whole-genome sequence (WGS). Clonality of WGS-sequenced junctions was defined as the ratio of detected junction sequencing depth to the average sequencing depth of the entire sequenced genome. Details of the capture-next generation sequence (capture-NGS) and analysis were described in our previous study [15]. In brief, the HBV-containing DNA fragments and targeted endogenous gene DNA fragments in tumor genomic DNA libraries were enriched by biotin-labeled DNA probes for NGS with Illumina MiSeq or NextSeq system (Illumina). The probes target HBV genome and endogenous genes including TP53 (coding sequence), CTNNB1 (exon3), TERT promoter, and GAPDH. Clonality of capture-sequenced junctions was defined as the ratio of detected junction sequencing depth to the sequencing depth of GAPDH- or TP53-targeted regions. Integrated HBV sequences were assembled with in-house-developed BBAP [16]. For the analysis of integrated HBV, only full HBV reads (reads with >90% read length aligned to HBV reference with >90% identity) were included in the assemblies. The HBV genotype and the HBV basal core promoter (BCP) mutation and pre-core (PC) frequencies were identified according to the assembly results.
RNA-Seq and Data Analysis
Sequencing libraries were prepared using a TruSeq Stranded mRNA Preparation Kit (Illumina) according to the manufacturer’s instructions, with sequencing using the Illumina NovaSeq 6000 system (performed by Welgene Biotech, Taiwan). Read quality was evaluated by FastQC (v0.11.9), and adaptor sequences were trimmed using cutadapt (v3.0). Qualified reads were aligned to human reference genome GRCh38 using STAR (v.2.7.2) [17], and read counts for individual genes annotated based on GENCODE (v.35) were subsequently determined using featureCounts [18]. Read counts of all genes across all samples were normalized using the TMM (trimmed mean of M-values) normalization method and converted into log-2-transformed transcripts per million (log2TPM) for downstream analyses. Differential expression analysis was performed using limma [19]. For preranked gene-set enrichment analysis (GSEA), a ranking metric was calculated for each gene as R = sign (log2FC)*−log10 (p value). Preranked GSEA was performed using a curated collection of gene sets from MSigDB (v7.4). The gene expression signature and pathway activation of each sample were assessed using the R package GSVA.
To identify subgroup-specific genes, ANOVA was performed to determine differentially expressed genes (DEGs) among subgroups. The R function Tukey honest significant difference (HSD) was used to identify which subgroup pairs showed significant differences. Genes were considered subgroup-specific if (1) Benjamini-Hochberg-adjusted F test p value <0.05 and (2) any cluster pair showed a significant difference in the HSD test (absolute difference of the cluster pair = 3 and BH-adjusted p value of Tukey’s HSD method <0.05). Overrepresentative analysis of group-specific genes was performed by clusterProfiler using gene sets from MSigDB (v7.4).
HCC transcriptome-based molecular subtypes were predicted using the R package MS.liverK with default parameters [20]. Two independent methods were employed to estimate the immune cell composition from bulk RNA-seq data. The first method was to score each immune cell by averaging the gene expression of the corresponding markers [21]. The second method was to characterize the cell composition by the CIBERSORT algorithm with the absolute mode and LM22 signature matrix [22]. Sia’s immune scores [23] and treatment benefit score (TBS) for regorafenib and sorafenib [24, 25] were calculated using GSVA. In brief, TBS for regorafenib in each HCC sample was calculated using GSVA algorithm on 11 genes (HIF1A, CTSK, SKC2A1, KLH1L12, CDKN1A, CA12, WDR1, CD53, CBR4, NUFK-AS1, and RAB30-DT). TBS for sorafenib was based on the following formula: (0.118910*mTOR) + (0.138561*VEGFR2) + (0.258877*c-KIT) + (0.147012 * c-RAF). T-cell function and inflammation scores followed a previously described method [26].
Immunohistochemical Staining
The paraffin-embedded HCC tissue sections (5 μm thick) were prepared for immunohistochemical (IHC) staining, with first Abs for CD45 (760-2505, Ventana Medical Systems, AZ, USA), CD8 (790-4460, Ventana Medical Systems), and PD-1 (780-4895, Ventana Medical Systems) and corresponding secondary Abs, using the Ventana Benchmark XT autostainer (Ventana Medical Systems) by following the manufacture’s instruction. The expression of each marker was independently assessed by two pathologists, graded as negative, weak, moderate, and strong, respectively. Based on the IHC staining results, tumors were divided into two groups, low expression group (negative and weak) and high expression group (moderate and strong), for further comparison analysis. Images were acquired with AxioImager.M1 (Zeiss, Germany) at ×20 magnification.
Telomere Length Measurement
Telomere length was estimated by the ratio of telomere repeat (T) to single copy gene (S), i.e., (T/S). Telomere repeats were quantified by telomere PCR (F: CGG TTT GTT TGG GTT TGG GTT TGG GTT TGG GTT TGG GTT, R: GGC TTG CCT TAC CCT TAC CCT TAC CCT TAC CCT TAC CCT) using POLG2 (F: GAGCTGTTGACGGAAAGGAG, R: CAGAAGAGAATCCCGGCTAAG) as the single copy gene control [27, 28]. Twenty ng of gDNA was used for each PCR reaction and triplicate was performed for each sample. PCR efficiencies were corrected before calculation and the quantification results were relative to the reference samples.
Statistical Analysis
RNA-seq data and bioinformatics analyses were carried out in R language (v4.1.0). Comparisons of mutation rates and features between groups were calculated by Fisher’s exact test and χ2 test. The Kaplan-Meier estimate was applied to determine a significant difference between the classification and recurrence-free survival (RFS) or overall survival (OS). The hazard ratio (HR) and 95% confidence intervals of HCC recurrence or mortality were estimated by the Cox proportional-hazards regression model using classification results and clinical characteristics, including age, sex, tumor size, tumor grade (Edmondson-Steiner), microvascular invasion, cirrhosis, and AFP levels, as variables in the Cox proportional-hazards regression model analysis. The same variables were included in the logistic regression model for the estimation of odds ratios and 95% confidence intervals of HCC recurrence within 2 years. Regression analysis was conducted using Stata (StataCorp LLC, TX, USA). The level of significance was defined as p < 0.05 by two-tailed tests.
Results
HBV-HCC Can Be Classified into Four Groups by Two HBV Integration Hotspots and TERT Promoter Mutation through a Capture-NGS Platform
To demonstrate the authenticity of our capture-NGS platform in identifying HBV-human chimeric junction sequences and targeted somatic mutations, we performed WGS (60 G) and capture-NGS (1 G) in parallel using 12 HBV-HCC tumor tissues. The distribution of identified HBV-human junctions of HBV integration sites in host chromosomes is illustrated in online supplementary Figure 1 (see online suppl. material at https://doi.org/doi/10.1159/000530699). WGS identified 75 (16.1%) clonal junctions from 467 HBV-human chimeric junctions. In comparison, capture-NGS identified 68 (42.5%) clonal junctions from 160 HBV-human chimeric junctions. Using the detection results of WGS as reference, capture-NGS has the detection sensitivity and specificity of 93.7% and 90.7% for the detection of clonal junctions. All the somatic mutations in the targeted region of CTNNB1 and TP53 identified by WGS were also detected by capture-NGS. The results indicate that capture-NGS is an accurate and efficient platform for identifying HBV-human integration and selected tumor-associated mutations for HBV-HCC.
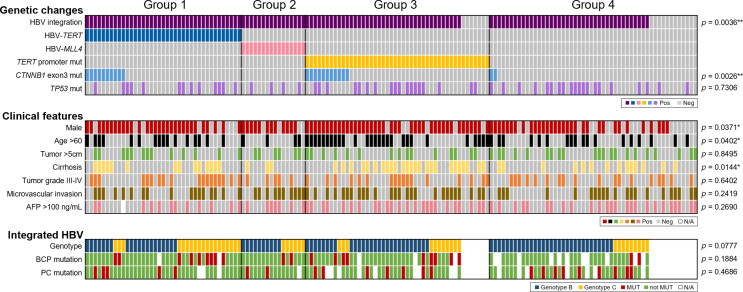
The detection results for HBV hotspot integrations and selected mutations of 153 HBV-HCC tumors by the capture-NGS platform are shown in Figure 1. Clonal HBV integration was detected in 135 (88.2%) cases. Consistent with 35 previous studies identifying HBV integration sites that are integrated in the Viral Integration Site Database [29] (online suppl. Fig. 2), integrations at the TERT and MLL4 (KMT2B) genes are the two major integration hotspots identified in the current study. There were 39 cases (25.5%) with HBV integration into the TERT gene, 16 (10.5%) with HBV integration into the MLL4 gene and 46 (30.1%) with TERT promoter point mutations.
Fig. 1.
Features of HBV-HCCs divided by hotspot HBV integrations and TERT promoter mutation (N = 153). Features were categorized into panels including major genetic changes in host genome, clinical features, and integrated HBV-related features. The p value is the results of comparison for each parameter between the four groups.
Interestingly, the three mutations occurred in a mutually exclusive manner (Fig. 1, genetic changes). The results confirmed our previous findings [15] and suggested HBV-HCC can be divided into four nonoverlapping subgroups: group 1 (G1), HBV-TERT integration; group 2 (G2), HBV-MLL4 integration; group 3 (G3), TERT promoter point mutation; and group 4 (G4), without the above three genetic changes. For the somatic mutations, CTNNB1 exon3 mutation rates were 25.6%, 0%, 23.9%, and 3.8% for G1–G4, respectively (p = 0.0026**, Fig. 1; Table 1, genetic changes), which is higher in the TERT-related subgroups (G1 + G3 vs. G2 + G4, 24.7% vs. 2.9%, p = 0.0002***), suggesting possible synergy between CTNNB1 and TERT in carcinogenesis, whereas TP53 (p = 0.7306, Fig. 1; Table 1, genetic changes) and other common mutations (online suppl. Fig. 3) have similar mutation rate in different HCC groups.
Table 1.
Characteristics and HBV-related features of HBV-HCC subgroups (N = 153)
| Group | All | G1 HBV-TERT | G2 HBV-MLL4 | G3 TERTp mut | G4 others | Statistics | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N | 153 | (100%) | 39 | (25%) | 16 | (10%) | 46 | (30%) | 52 | (34%) | |
| Genetic changes | |||||||||||
| HBV integration | 134 | (88%) | 39 | (100%) | 16 | (100%) | 39 | (85%) | 40 | (77%) | G1 versus non-G1, p = 0.0038** G4 versus non-G4, p = 0.0041** G1 versus G4, p = 0.0009a |
| CTNNB1 exon3 mutation | 23 | (15%) | 10 | (26%) | 0 | (0%) | 11 | (24%) | 2 | (4%) | G1 versus non-G1, p = 0.0318* G3 versus non-G3, p = 0.0439* G4 versus non-G4, p = 0.0042** G1 versus G4, p = 0.0036# G3 versus G4, p = 0.0056a |
| TP53 mutation | 55 | (36%) | 13 | (33%) | 4 | (25%) | 18 | (39%) | 20 | (38%) | n.s. |
| Clinical features | |||||||||||
| Gender | |||||||||||
| Male | 114 | (75%) | 30 | (77%) | 12 | (75%) | 40 | (87%) | 32 | (62%) | G3 versus non-G3, p = 0.0205* G4 versus non-G4, p = 0.0082** G3 versus G4, p = 0.0045a |
| Female | 39 | (25%) | 9 | (23%) | 4 | (25%) | 6 | (13%) | 20 | (38%) | |
| Age | |||||||||||
| >60 years | 65 | (42%) | 16 | (41%) | 4 | (25%) | 27 | (59%) | 18 | (35%) | G3 versus non-G3, p = 0.0078** |
| ≤60 years | 88 | (58%) | 23 | (59%) | 12 | (75%) | 19 | (41%) | 34 | (65%) | |
| Tumor size | |||||||||||
| >5 cm | 55 | (36%) | 13 | (33%) | 6 | (38%) | 15 | (33%) | 21 | (40%) | n.s. |
| ≤5 cm | 98 | (64%) | 26 | (67%) | 10 | (63%) | 31 | (67%) | 31 | (60%) | |
| Cirrhosis | |||||||||||
| Positive | 56 | (37%) | 14 | (36%) | 3 | (19%) | 25 | (54%) | 14 | (27%) | G3 versus non-G3, p = 0.0028** G3 versus G4, p = 0.0056a |
| Negative | 97 | (63%) | 25 | (64%) | 13 | (81%) | 21 | (46%) | 38 | (73%) | |
| Tumor grade (Edmondson) | |||||||||||
| G1-G2 | 95 | (62%) | 21 | (54%) | 11 | (69%) | 29 | (63%) | 34 | (65%) | n.s. |
| G3-G4 | 58 | (38%) | 18 | (46%) | 5 | (31%) | 17 | (37%) | 18 | (35%) | |
| Microvascular invasion | |||||||||||
| Yes | 74 | (48%) | 17 | (44%) | 10 | (63%) | 26 | (57%) | 21 | (40%) | n.s. |
| No | 79 | (52%) | 22 | (56%) | 6 | (38%) | 20 | (43%) | 31 | (60%) | |
| AFP | |||||||||||
| >100 ng/mL | 68 | (45%) | 12 | (32%) | 7 | (44%) | 24 | (52%) | 25 | (48%) | n.s. |
| ≤100 ng/mL | 84 | (55%) | 26 | (68%) | 9 | (56%) | 22 | (48%) | 27 | (52%) | |
| Integrated HBV-related features | |||||||||||
| Number of clonal junctions | |||||||||||
| Range | 1–18 | 1–17 | 1–12 | 1–9 | 1–18 | G1 versus non-G1, p = 0.0114b G3 versus non-G3, p = 0.0118b G1 versus G3, p = 0.0160c |
|||||
| Average ± SD | 4.9±3.7 | 6.0±4.0 | 5.0±3.1 | 3.6±2.0 | 5.2±4.5 | ||||||
| HBV genotype | |||||||||||
| B | 89 | (66%) | 20 | (51%) | 10 | (63%) | 28 | (72%) | 31 | (78%) | G1 versus non-G1, p = 0.0175* |
| C | 45 | (34%) | 19 | (49%) | 6 | (38%) | 11 | (28%) | 9 | (23%) | |
| BCP mutation (A1762T/G1764A) | |||||||||||
| Not MUT | 96 | (81%) | 28 | (76%) | 14 | (88%) | 27 | (75%) | 27 | (93%) | n.s. |
| Mutant | 22 | (19%) | 9 | (24%) | 2 | (13%) | 9 | (25%) | 2 | (7%) | |
| PC mutation (G1896A) | |||||||||||
| Not MUT | 91 | (80%) | 31 | (86%) | 13 | (87%) | 23 | (77%) | 24 | (73%) | n.s. |
| Mutant | 23 | (20%) | 5 | (14%) | 2 | (13%) | 7 | (23%) | 9 | (27%) | |
n.s., not significant.
*p < 0.05.
**p < 0.01.
***p < 0.001.
a p < 0.0083 in pairwise comparison by χ2 test or Fisher’s exact test.
b p < 0.05 in Mann-Whitney U Test.
cAdjusted p < 0.05 in pairwise comparison after Kruskal-Wallis Test.
Distinct Clinical Characteristics of Specific Subgroups: G4 Has the Best Survival after Tumor Resection and G3 Has the Highest 2-Year Recurrence Rate
To find HCC subgroup-specific features, we compared common clinical characteristics, including sex, age, tumor size, cirrhosis, tumor grade, microvascular invasion, and AFP, among the different groups of HCC patients (Fig. 1; Table 1, clinical features). The G3 group had the highest male-to-female ratio (G3 vs. non-G3, 6.7:1 vs. 2.2:1, p = 0.0205*), more patient with age >60 years (G3 vs. non-G3, 58.7% vs. 35.5%, p = 0.0078**), and higher cirrhosis rate (G3 vs. non-G3, 54.3% vs. 29.0%, p = 0.0028**). In contrast, G4 had the lowest male-to-female ratio (G4 vs. non-G4, 1.6:1 vs. 4.3:1, p = 0.0082**). No difference in tumor size, tumor grade, microvascular invasion, and AFP was found by pairwise comparison of different groups.
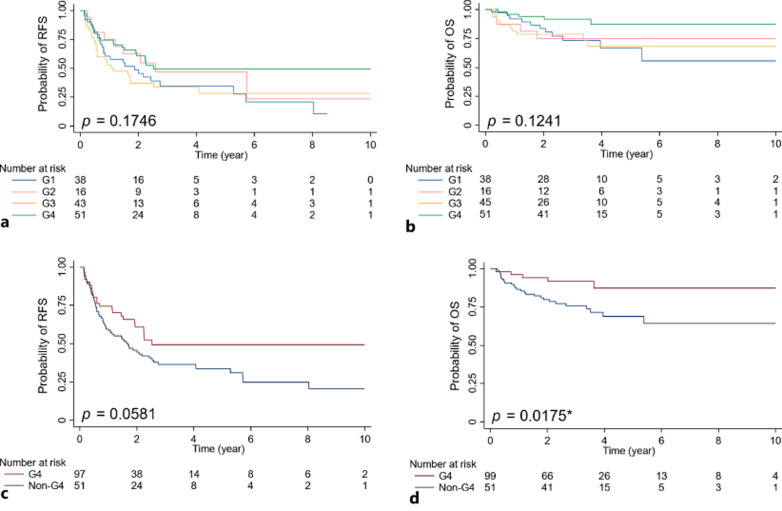
In terms of outcome, we compared RFS and OS among the G1–G4 groups with the Kaplan-Meier estimate, which revealed no significant difference (Fig. 2a, b). The univariate Cox regression (Table 2) indicates G4 has the lowest HR for both recurrence (HR = 0.63, p = 0.060) and death (HR = 0.33, p = 0.024*). Indeed, when comparing G4 versus non-G4 groups, G4 showed marginal better RFS and significant better OS than the non-G4 groups (Fig. 2c, d, G4 vs. non-G4: p = 0.0581 for RFS, p = 0.0175* for OS). The multivariable Cox regression model (Table 2) suggests Group 4 as an independent variable for mortality (HR = 0.26, p = 0.015*).
Fig. 2.
Recurrence-free survival (RFS) and overall survival (OS) of different HBV-HCC subgroups by Kaplan-Meier estimate. a RFS analysis of the four groups. b OS analysis of the four groups. c RFS analysis of G4 and non-G4 group. d OS analysis of G4 and non-G4 group.
Table 2.
Univariate and multivariable Cox proportional-hazards regression models for various variables in predicting the recurrence and mortality of HBV-HCC
| End point and variables | Univariate Cox regression | Multivariate Cox regression | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| group 1 | group 2 | group 3 | group 4 | |||||||||||||||||
| HR | (95% CI) | p value | HR | (95% CI) | p value | HR | (95% CI) | p value | HR | (95% CI) | p value | HR | (95% CI) | p value | ||||||
| Recurrence | ||||||||||||||||||||
| G1 versus non-G1 | 1.26 | 0.80–2.00 | 0.323 | 1.08 | 0.66–1.76 | 0.760 | ||||||||||||||
| G2 versus non-G2 | 0.83 | 0.42–1.65 | 0.593 | 0.85 | 0.42–1.74 | 0.658 | ||||||||||||||
| G3 versus non-G3 | 1.41 | 0.90–2.22 | 0.134 | 1.50 | 0.91–2.47 | 0.114 | ||||||||||||||
| G4 versus non-G4 | 0.63 | 0.39–1.02 | 0.060 | 0.69 | 0.42–1.15 | 0.156 | ||||||||||||||
| Age >60 versus ≤60 | 1.15 | 0.75–1.76 | 0.536 | 1.16 | 0.74–1.80 | 0.522 | 1.14 | 0.73–1.78 | 0.556 | 1.06 | 0.68–1.68 | 0.789 | 1.12 | 0.72–1.74 | 0.628 | |||||
| Male versus female | 1.38 | 0.82–2.33 | 0.223 | 1.47 | 0.86–2.53 | 0.157 | 1.50 | 0.87–2.57 | 0.145 | 1.40 | 0.82–2.42 | 0.222 | 1.37 | 0.79–2.38 | 0.256 | |||||
| Tumor size >5 cm versus ≤5 cm | 2.53 | 1.66–3.88 | <0.001 | *** | 2.75 | 1.69–4.47 | <0.001 | *** | 2.75 | 1.70–4.46 | <0.001 | *** | 3.03 | 1.85–4.96 | <0.001 | *** | 2.89 | 1.79–4.66 | <0.001 | *** |
| Microinvasion Y versus N | 1.93 | 1.25–2.98 | 0.003 | ** | 1.21 | 0.73–2.01 | 0.465 | 1.22 | 0.73–2.05 | 0.440 | 1.17 | 0.71–1.95 | 0.539 | 1.16 | 0.70–1.93 | 0.564 | ||||
| Cirrhosis Y versus N | 1.58 | 1.03–2.43 | 0.037 | * | 1.78 | 1.11–2.84 | 0.016 | * | 1.77 | 1.10–2.83 | 0.018 | * | 1.71 | 1.06–2.74 | 0.027 | * | 1.72 | 1.08–2.74 | 0.023 | * |
| Tumor grade 34 versus 12 | 1.84 | 1.20–2.81 | 0.005 | ** | 1.76 | 1.11–2.81 | 0.017 | * | 1.78 | 1.13–0.280 | 0.013 | * | 1.81 | 1.15–2.85 | 0.011 | * | 1.70 | 1.06–2.71 | 0.026 | * |
| AFP >100 ng/mL versus ≤100 ng/mL | 1.39 | 0.91–2.14 | 0.128 | 0.86 | 0.52–1.43 | 0.561 | 0.85 | 0.52–1.40 | 0.525 | 0.82 | 0.50–1.35 | 0.447 | 0.88 | 0.53–1.45 | 0.617 | |||||
| Mortality | ||||||||||||||||||||
| G1 versus non-G1 | 1.57 | 0.75–3.28 | 0.230 | 1.49 | 0.66–3.37 | 0.339 | ||||||||||||||
| G2 versus non-G2 | 1.18 | 0.41–3.39 | 0.753 | 1.20 | 0.40–3.59 | 0.745 | ||||||||||||||
| G3 versus non-G3 | 1.55 | 0.74–3.23 | 0.247 | 1.94 | 0.84–4.46 | 0.120 | ||||||||||||||
| G4 versus non-G4 | 0.33 | 0.13–0.86 | 0.024 | * | 0.26 | 0.09–0.77 | 0.015 | * | ||||||||||||
| Age >60 versus ≤60 | 0.58 | 0.27–1.26 | 0.167 | 0.55 | 0.24–1.26 | 0.156 | 0.55 | 0.24–1.28 | 0.165 | 0.50 | 0.22–1.16 | 0.108 | 0.50 | 0.22–1.15 | 0.103 | |||||
| Male versus female | 1.39 | 0.57–3.40 | 0.466 | 1.68 | 0.66–4.25 | 0.277 | 1.61 | 0.63–4.11 | 0.324 | 1.46 | 0.57–3.78 | 0.430 | 1.22 | 0.47–3.20 | 0.681 | |||||
| Tumor size >5 cm versus ≤5 cm | 3.99 | 1.91–8.33 | <0.001 | *** | 3.09 | 1.36–7.00 | 0.007 | ** | 3.35 | 1.47–7.61 | 0.004 | ** | 3.89 | 1.66–9.10 | 0.002 | ** | 3.99 | 1.76–9.06 | 0.001 | ** |
| Microinvasion Y versus N | 3.28 | 1.47–7.33 | 0.004 | ** | 1.71 | 0.70–4.17 | 0.237 | 1.64 | 0.68–3.99 | 0.274 | 1.50 | 0.61–3.67 | 0.375 | 1.48 | 0.61–3.62 | 0.390 | ||||
| Cirrhosis Y versus N | 1.45 | 0.72–2.95 | 0.300 | 1.92 | 0.86–4.27 | 0.110 | 2.02 | 0.89–4.58 | 0.092 | 1.84 | 0.82–4.16 | 0.141 | 1.84 | 0.83–4.10 | 0.134 | |||||
| Tumor grade 34 versus 12 | 1.89 | 0.93–3.83 | 0.077 | 1.42 | 0.63–3.19 | 0.400 | 1.56 | 0.72–3.39 | 0.257 | 1.53 | 0.71–3.30 | 0.280 | 1.14 | 0.49–2.66 | 0.770 | |||||
| AFP >100 ng/mL versus ≤100 ng/mL | 2.39 | 1.14–5.02 | 0.022 | * | 1.57 | 0.64–3.80 | 0.323 | 1.39 | 0.60–3.24 | 0.447 | 1.35 | 0.59–3.12 | 0.477 | 1.80 | 0.75–4.31 | 0.189 | ||||
*p < 0.05.
**p < 0.01.
***p < 0.001.
In addition to long-term outcome, we also explored the association between early recurrence (<2 years) with HCC subgroups by logistic regression (online suppl. Table 1). The OR for G3 was 2.12 (p = 0.042*) in univariate and 2.35 (p = 0.058) in multivariable analysis, suggesting that G3 may be associated with early recurrence of HCC.
Analysis of Integrated HBV DNA Revealing Timing of HBV Integration and Role of HBV Genotypes and Basal Core/PC Variants in Carcinogenesis of Different Subgroups
HBV genotype and HBV variants, the BCP A1762T/G1764A and PC G1896A from circulating HBV, are known risk factors associated with HCC development [30]. The large amount of HBV sequences detected by capture-NGS allowed us to investigate the possible role of the genotype and variants in the integrated HBV in HCC (Fig. 1; Table 1, integrated HBV-related features). Genotype B and C HBV accounts for 66% and 34% of the HBV integration in HBV-HCC. Interestingly, higher portion of genotype C is observed in G1 (G1 vs. non-G1, 48.7% vs. 27.4%, p = 0.0175*), implying that integrated genotype C HBV sequence may provide positive selection of hepatocytes with HBV-TERT integration.
HBV integration can occur multiple times during chronic HBV infection; however, the timing of tumor-promoting integration events is unexplored. Since BCP and PC mutation occurs around HBeAg seroconversion, the mutations may serve as the marker for the integration occurs then. Therefore, for HCC with integrated HBV DNA with more than 90% viral sequences with BCP and PC variations, such events most likely occurred nearby or after HBeAg seroconversion. Overall, such HBV-HCC with predominantly BCP or PC variation is 19% and 20%, respectively (Fig. 1; Table 1, integrated HBV-related features) in HBV-HCC. In sum, BCP or PC variation was detected in 31.3% of integration-positive HBV-HCC, without significant difference between subgroups of HBV-HCC. In other words, the results also indicate that the majority of HBV DNA integration in HCC occurs at the stage before HBeAg seroconversion.
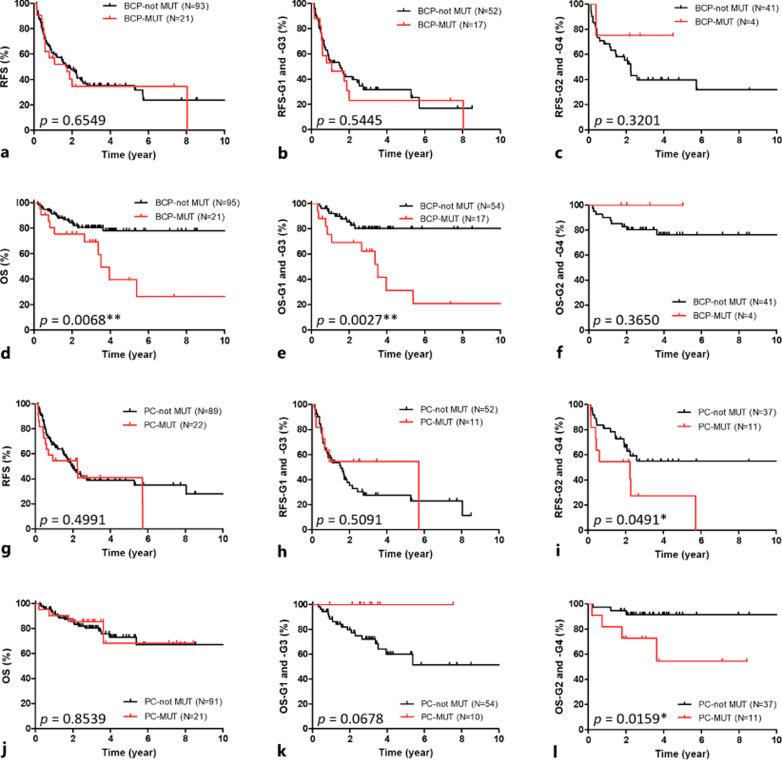
To investigate if the genotype or variants of integrated HBV has impact on clinical outcomes, we stratified the RFS and OS by BCP, PC mutation, and genotype in the integrated HBV (Fig. 3; online suppl. Fig. 4). There is no difference in RFS or OS between HCC with genotype B or C integrated HBV (online suppl. Fig. 4). However, shorter OS was found in HCC with integrated HBV DNA containing BCP mutation (Fig. 3d, p = 0.0068**), especially in G1 and G3 (Fig.3e, p = 0.0027**). Both shorter RFS and OS were observed in G2 and G4 tumor with PC mutation (Fig. 3i, p = 0.0491*; Fig. 3l, p = 0.0159*). The results suggest that combination of BCP/PC mutation in integrated HBV with HCC subgroup may help further identify the HBV-HCC patients with worse prognosis.
Fig. 3.
Recurrence-free survival (RFS) and overall survival (OS) stratified by BCP or PC mutation in integrated HBV between different HBV-HCC subgroups. a–c RFS analysis stratified by BCP in all HBV-HCCs (a), G1 and G3 tumor (b), and G2 and G4 tumor (c). d–f OS analysis stratified by BCP in all HBV-HCCs (d), G1 and G3 tumor (e), and G2 and G4 tumor (f). g–i RFS analysis stratified by PC in all HBV-HCCs (g), G1 and G3 tumor (h), and G2 and G4 tumor (i). j–l OS analysis stratified by PC in all HBV-HCCs (j), G1 and G3 tumor (k), and G2 and G4 tumor (l).
Transcriptome of HBV-HCC Subgroups Corresponding to Genetic Classification by Capture-NGS
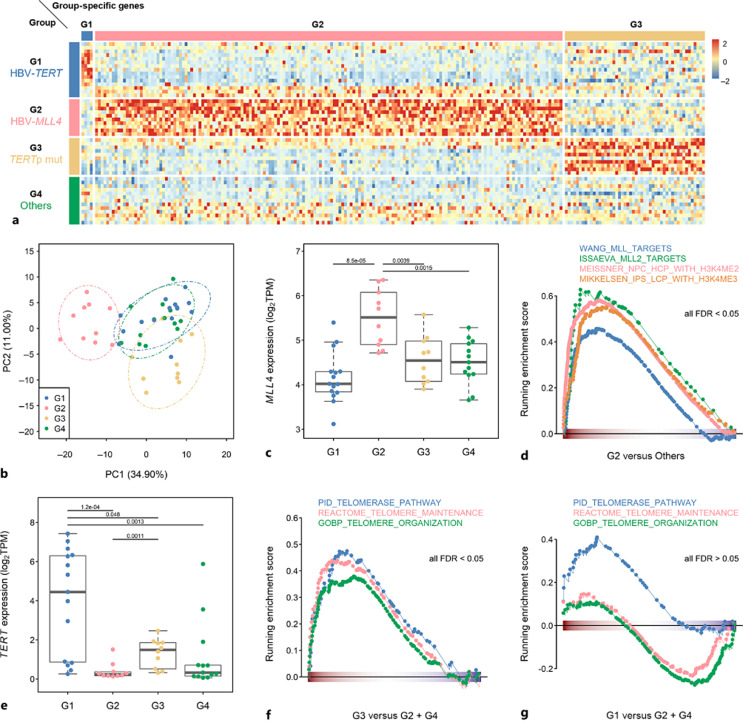
To investigate the intrinsic molecular mechanisms of distinct HBV-related subgroups, we performed transcriptomic analysis of 48 HCCs, including 15, 10, 10, and 13 samples in G1–G4, respectively. Hierarchical clustering and principal component analyses of all samples, based on expression profiles of the entire set of genes analyzed, revealed a modest distinction of HBV-related subgroups due to case heterogeneity (online suppl. Fig. 5a, b). Therefore, ANOVA was performed to identify DEGs among subgroups (adjusted p < 0.05), after which the significantly upregulated DEGs in a given subgroup were used to define subgroups. As depicted in Figure 4a, 4 G1-, 167 G2-, and 50 G3-specific genes were identified. The principal component analysis using subgroup-specific genes showed a clear distinction between G2, G3, and G1 + G4 (Fig. 4b). Due to the limited number of G1-specific genes and no G4-specific genes were identified, G1 and G4 were clustered together in the PC dimension.
Fig. 4.
TERT and MLL4 expression patterns consistent with classification results in RNA-seq profiling of HBV-HCCs (N = 48). a Heatmap representation of group-specific genes. b PCA using subgroup-specific genes. c Expression of MLL4 (log2TPM) in G1–G4. d Gene-set enrichment analysis (GSEA) of gene sets of MLL-regulated genes and H3K4 methylation-related genes in G2 relative to other groups. e Expression of TERT (log2TPM) in G1–G4. f, g GSEA of gene sets for telomere-related genes in G3 relative to G2 and G4 (f), and in G1 relative to G2 and G4 (g). FDR, false discovery rate.
With HBV-MLL4 integration, MLL4 was significantly highly expressed in G2, and expression of gene signatures for MLL and H3K4me2/3 responses was increased in G2 relative to other subgroups (Fig. 4c, d). Overrepresentative analysis also revealed genes associated with the H3K27me2/3 response, myogenesis, and apoptosis, and G3-specific genes were enriched in pathways related to JAK2, NRG1, and GSK3 (online suppl. Fig. 5c). These results suggest distinct carcinogenesis programs and intrinsic molecular properties in G2 and G3.
TERT was highly expressed in both G1 and G3 (Fig. 4e), and the expression level in G1 was even higher than that in G3. The activation status in terms of TERT enzyme activity in these four groups of HCC was estimated by the EXTEND (EXpression-based Telomerase ENzymatic activity Detection) algorithm [31]. The EXTEND analysis showed that G1 (HBV-TERT) and G3 (TERTp mut) HCC had relatively higher telomerase activity than G2 and G4 HCC (online suppl. Fig. 6a), correlating well with TERT mRNA levels measured by qRT-PCR (online suppl. Fig. 6b).
Despite this, genes involved in telomere organization, the telomerase pathway, and the TERT signature were significantly enriched only in G3 (Fig. 4f, g; online suppl. Fig. 7a). Moreover, both the telomerase pathway activity indicated by GSVA and the telomere length were correlated strongly with TERT expression in G3 but not in G1 HCC (online suppl. Fig. 7b–d). In addition, the GSEA identified the hallmark gene sets involved in the regulation of cell cycle and proliferation (e.g., E2F and c-Myc) were highly expressed in G3 HCC. However, diverse biological processes were enriched in the G1 group, such as angiogenesis, EMT, glycolysis, and INF response (online suppl. Fig. 8). Therefore, downstream carcinogenic mechanisms may differ between G1 and G3 although the TERT expression level was elevated in both subgroups of HCC.
Transcriptome Analysis of HBV-HCC Subgroups Uncovers Distinct Mechanisms of Tumorigenesis
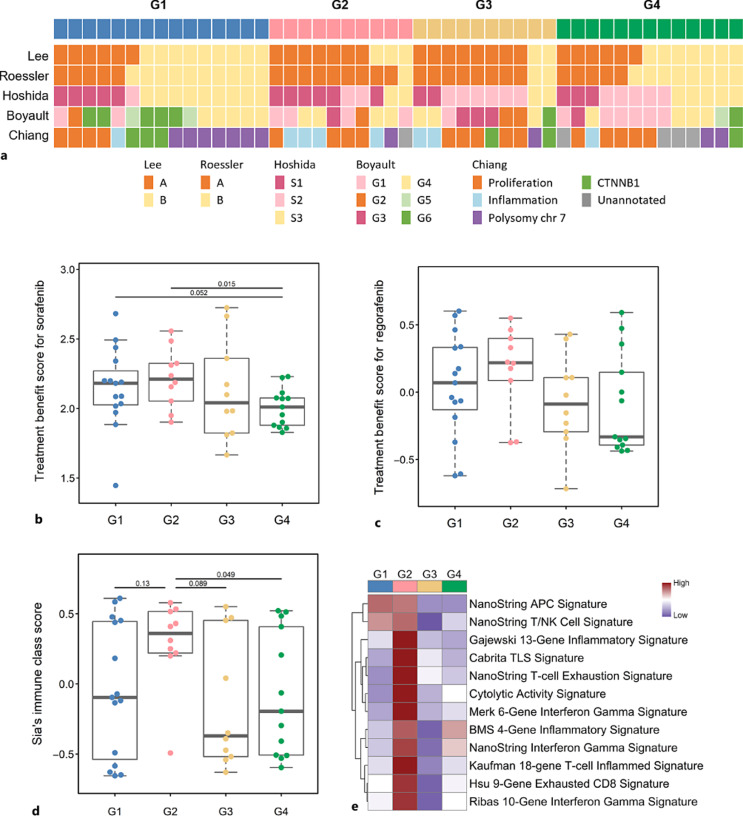
During the past two decades, several groups have proposed transcriptome-based molecular classifications for HCC [32–36]. Thus, we compared our HBV-related subgroups to these previously defined HCC classifications (Fig. 5a). Our HBV-related classification was significantly associated with Roessler classification (p = 0.0084**), where most G2 and G3 samples were also classified into subgroup A of Roessler’s classification. G1 was significantly linked to S3 in Hoshida’s classification (p = 0.0330*), G6 in Boyault’s classification (p = 0.0240*), and polysomy chr 7 in Chiang’s classification (p = 0.0130*).
Fig. 5.
Association of HBV-based classification with other HCC molecular classifications and therapeutic signatures. a HBV-HCC classified by other reported classification systems. b Sorafenib treatment score of the four groups. c Regorafenib treatment score of the four groups. d Immune class score of the four groups. e ICI treatment signature analysis of the four groups.
To explore the clinical significance of our new classification in transcription analysis, we examined signaling pathways among HBV-related subgroups and explored which subgroup may benefit from current HCC molecular or immune-oncology treatments. Transcriptome-based pathway activities were assessed using the GSVA algorithm (online suppl. Fig. 9). Genes responsive to VEGFR, a major target of multikinase inhibitors, were highly expressed in G2. MET and PDGFR have higher activities in TERT-related subgroups (G1 and G3). There was no pathway specifically highly expressed in G4. We also examined TBSs for sorafenib [24] and regorafenib [25] proposed in previous publications. Patients in G4 had a lower rate of sorafenib and regorafenib response, consistent with the results of pathway activity analyses (Fig. 5b, c).
The tumor immune microenvironment plays a critical role in tumors and is related to the efficacy of immune checkpoint inhibitor (ICI) therapy. We used two independent computational methods to dissect immune cell compositions in tumors [21, 22]. There was no obvious difference in immune cell compositions among the HBV-HCC groups (online suppl. Fig. 10). Several works have demonstrated that inflammatory and T-cell expression signatures are associated with sensitivity to ICI therapy [26, 37, 38]. Tumors of G2 have higher immune scores, as defined by Sia et al. [23]. Concordant with the immune score (Fig. 5d), G2 showed higher expression scores of inflammatory and T-cell signatures (Fig. 5e), suggesting that G2 has a higher probability of response to ICI therapy.
In a validation analysis, we have performed IHC staining for CD45 (for assessing infiltrating immune cells), CD8 and PD-1 (for assessing potential ICI response) on HCC tissues subjected to RNA-seq analysis. The results showed that immune scores determined by immune cell infiltration were higher in G2 versus non-G2 HCC, 56% versus 23% (p = 0.0976). CD8 and PD-1-positive immune cells were also higher in G2 versus non-G2 HCC, 78% versus 26% (p = 0.0066**) and 44% versus 17% (p = 0.1748), respectively (online suppl. Fig. 11a–e). Compared to non-G2 HCC, G2 HCC showed a higher proportion of immune cells with high CD8 and PD-1 signals, suggesting that G2 HCC may be more responsive to ICI treatment (online suppl. Fig. 11f), consistent with higher “predictive signatures for response to ICI therapy” in G2 HCC (Fig. 5d, e).
Validation of the New Classification System
To validate our results, we classified another HCC cohort containing 112 HBV-HCCs by HBV-TERT, HBV-MLL4, and TERT promoter mutation (online suppl. Fig. 12; Table 2). HBV-HCCs in validation cohort can be classified into four groups without overlapping. As noted, the CTNNB1 mutation rate was much higher in the TERT-related subgroups (G1 + G3 vs. G2 + G4, 29.0% vs. 11.6%, p = 0.0319*). Interestingly, no CTNNB1 mutation was identified in G2, the same as in the main cohort. The high male ratio in G3 (G3 vs. non-G3, 91.4% vs. 74.0%, p = 0.0433*) and high female ratio in G4 (G4 vs. non-G4, 48.3% vs. 10.8%, p < 0.0001***), more >60-year patients in G3 (G3 vs. non-G3, 68.6% vs. 39.0%, p = 0.0037**) were also observed in the validation cohort. In terms of integrated HBV, the percentage of HBV genotype C integration is highest in G1 than other groups (47.1% vs. 28.8%, p = 0.0640), and the BCP/PC mutation pattern was similar as that of the main cohort.
Discussion
In the current study, we explored a new classification system based on two HBV integration hotspots, namely, HBV-TERT and HBV-MLL4, and TERT promoter point mutation. HBV-HCCs can be classified unambiguously into four groups by these three genetic mutations, suggesting that the carcinogenic mechanisms are independent among groups. Our study confirmed the importance of TERT and MLL4 in HBV-HCC carcinogenesis, and the two genes may become new therapeutic targets relevant for HCC. However, new and reliable animal models are required to understand the carcinogenic mechanisms and to explore therapeutic agents.
As shown by WGS, HBV integrations in nontumorigenic liver are randomly distributed on all chromosomes, yet in HCC there are hotspot integrations at the TERT and MLL4 genes. This suggests a positive selection for hepatocytes containing hotspot gene integrations during carcinogenesis. The mechanisms of oncogenesis initiated by these two driver genes are expected to be different because of distinct activity of the telomerase versus histone methyltransferase, respectively. The progress from HBV integrations to carcinogenesis takes decades and likely involves distinct mechanisms and accompanying mutations; for example, the β-catenin mutations are frequently found in G1 HCC but not in G2 HCC. Admittedly, more studies are needed to elucidate the mechanism for different groups of HCC.
TERT is silent in most hepatocytes; thus, telomeres are progressively shortened during hepatocyte replication in the aging liver, which eventually leads to replicative senescence [39, 40]. However, telomere attrition is reversed in some HCCs, which is more likely due to reactivation of TERT by genetic mutations, like hepatitis C virus-related HCC; the rate of TERT promoter mutation reaches approximately 50–70% [41, 42]. In HBV-HCC, 25% HBV-TERT integration and 30% TERT promoter mutation rates were found in the current study, consistent with other studies [3, 43, 44], contributing to TERT reactivation. Interestingly, despite increased expression of TERT in G1 and G3, the telomerase pathway and telomere maintenance signatures correlated only with the TERT expression level in G3. This suggests that carcinogenesis may be promoted in G1 via pathways other than increasing telomerase enzymatic activity. TERT promoter mutation has been identified in cirrhotic nodules with dysplasia or early HCC but not in normal-appearing cirrhotic nodules [45, 46], indicating that TERT promoter mutation is selected out in response to telomere attrition during chronic inflammation and repetitive regeneration. In contrast, HBV-TERT integration occurs during infection, a stage when the telomere is not sufficiently shortened to induce replicative senescence. Therefore, the carcinogenesis promoted by HBV-TERT may be through telomere length-independent manner, supported by the different transcription profiles in G1 and G3.
The mutations of MLL4 (MMT2B), a ubiquitous histone methyltransferase, were documented in several tumors, such as endometrial, skin, and peritoneal tumors. Most are loss-of-function mutations due to missense, nonsense, or frameshift mutations affecting the SET or PHD domain [47]. However, in HCC, HBV integration results in N’-terminal truncated MLL4 without affecting its enzymatic domain or creating new MLL4 isoforms by alternative splicing [48]. In addition to hypermethylation caused by increased MLL4 levels, remodeling of the immune microenvironment is one possible function, as revealed by our RNA-seq analysis. A new function introduced by N’-terminal truncation or virus fusion may be dominant as the other wild-type allele MLL4 is not affected by HBV integration.
Without common, specific driver mutations, G4 is likely a heterogeneous group of HBV-HCC, which require other markers for a better classification. For example, we found three cases with TERT gene duplication in G4, which might also lead to TERT reactivation. At present, clinical features such as female dominance and best prognosis are associated with this group. The exceptionally high percentage of female patients in G4 may imply the difference in HBV-HCC carcinogenesis and clinical outcome between male and female patients.
In addition to the dysregulation of integrated host genes, the analysis of HBV in tumor indicates that integrated HBV may also help to shape the carcinogenic process or disease outcome, which is worthy to be included for HCC subgrouping analysis. From our studies, about 70% of the HBV integrations in HCC likely occurs before HBeAg seroconversion, and certain HCC subgroups with BCP or PC variant integrated DNA suffered worse prognosis. The results suggested that combination of BCP/PC mutation in integrated HBV with HCC subgroups may help to further identify the HBV-HCC patients with poor prognosis. This may be an impelling cause to consider the anti-viral treatment to be applied as soon as possible after infection to minimize carcinogenesis by the insertional mutagenesis.
The current classification system may also be applied as a surrogate marker for choosing ICI treatment or molecular targets. Transcriptome analysis showed that G2 had the highest immune class score and ICI-related signatures, which has been validated by the histological data in terms of immune cell infiltration status and ICI markers. It will be interesting to assess whether G2 HCC responds to ICI therapy. Meanwhile, there are many ongoing clinical trials of adjuvant MTT or ICI therapies for HCCs after curative treatment. The efficacy of these adjuvant therapies is affected by the heterogeneity of HCCs between control and experimental arms and compromises the likelihood of success. If these post-curative HCC patients can be further classified by the current system, these patient heterogeneities can be better balanced, and the outcomes can be compared. This classification may also benefit systemic therapies or trials for advanced HBV-HCCs.
In conclusion, we put forth a new HBV-HCC classification system that is simple and robust. As the capture-sequencing platform used for this new classification is modified from the platform well adopted for detecting mutations in lung cancer or leukemia to guide targeted therapy, it is expected to be widely applicable in different institutions in the future. Adopting the classification can help with information sharing and data harmonizing among studies, eventually improving HCC patient management.
Acknowledgments
We thank the Taiwan Liver Cancer Network (TLCN) for providing samples and related data (all anonymous) for our research. We thank the staff of the Sequencing and Biochemistry Core, Department of Medical Research, National Taiwan University Hospital, for technical support and also National Center for High-performance Computing (NCHC) of National Applied Research Laboratories (NARLabs) of Taiwan for providing computational resources and storage resources. The assistance in IHC provided by the Department of Pathology, National Taiwan University Hospital, is greatly appreciated.
Statement of Ethics
This study protocol was reviewed and approved by Institutional Review Board of the National Taiwan University Hospital and National Health Research Institutes (Number: 1053700400, 1083703901, and 1083706055). Signed written informed consents were obtained from participants during recruitment or initial enrollment in Taiwan Liver Cancer Network.
Conflict of Interest Statement
P.J.C. is the consultant and received a grant from TCM Biotech. S.T.T., C.F.H., and Y.C.W. are employees of TCM Biotech.
Funding Sources
This study was supported by grants from the Ministry of Science and Technology, Taiwan (MOST 111-2326-B-002-016-, 111-2634-F-002-017-, and 111-2326-B-002-017-), and the “Center of Precision Medicine” from the Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan (NTU-CC-112L901401) and Academia Sinica, Taiwan (AS-KPQ-110-NGT and AS-KPQ-111-KNT-18-1).
Author Contributions
S.H.Y. and P.J.C. contributed to study concept and design, provided critical revision of the manuscript for important intellectual content, and obtained funding. C.L.L., C.L.H., T.C.H., Y.F.L., and S.F.T. contributed to acquisition of data. C.L.L., C.L.H., Y.Y.L., S.H.Y., and P.J.C. contributed to the analysis and interpretation of data. C.L.L., C.L.H., and Y.Y.L. contributed to the drafting of the manuscript. C.L.L., C.L.H., and C.L.C. contributed to statistical analysis. M.C.H. and R.H.H. contributed to patient recruitments. S.T.T., C.F.H., and Y.C.W. contributed to technical support. P.J.C. contributed to study supervision. All authors approved the final version to be published.
Funding Statement
This study was supported by grants from the Ministry of Science and Technology, Taiwan (MOST 111-2326-B-002-016-, 111-2634-F-002-017-, and 111-2326-B-002-017-), and the “Center of Precision Medicine” from the Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan (NTU-CC-112L901401) and Academia Sinica, Taiwan (AS-KPQ-110-NGT and AS-KPQ-111-KNT-18-1).
Data Availability Statement
All data generated or analyzed during this study are included in this article and its online supplementary material. RNA-sequencing data can be accessed on GEO using the accession number GSE198946. Further inquiries can be directed to the corresponding author.
Supplementary Material
References
- 1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49. 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2. Finn RS, Qin S, Ikeda M, Galle PR, Ducreux M, Kim T-Y, et al. Atezolizumab plus bevacizumab in unresectable hepatocellular carcinoma. N Engl J Med. 2020;382(20):1894–905. 10.1056/NEJMoa1915745. [DOI] [PubMed] [Google Scholar]
- 3. Totoki Y, Tatsuno K, Covington KR, Ueda H, Creighton CJ, Kato M, et al. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat Genet. 2014;46(12):1267–73. 10.1038/ng.3126. [DOI] [PubMed] [Google Scholar]
- 4. Fujimoto A, Furuta M, Totoki Y, Tsunoda T, Kato M, Shiraishi Y, et al. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat Genet. 2016;48(5):500–9. 10.1038/ng.3547. [DOI] [PubMed] [Google Scholar]
- 5. Schulze K, Imbeaud S, Letouze E, Alexandrov LB, Calderaro J, Rebouissou S, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet. 2015;47(5):505–11. 10.1038/ng.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kan Z, Zheng H, Liu X, Li S, Barber TD, Gong Z, et al. Whole-genome sequencing identifies recurrent mutations in hepatocellular carcinoma. Genome Res. 2013;23(9):1422–33. 10.1101/gr.154492.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. GBD 2019 Diseases and Injuries Collaborators . Global burden of 369 diseases and injuries in 204 countries and territories, 1990-2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet. 2020;396(10258):1204–22. 10.1016/S0140-6736(20)30925-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tu T, Budzinska MA, Vondran FWR, Shackel NA, Urban S. Hepatitis B virus DNA integration occurs early in the viral life cycle in an in vitro infection model via sodium taurocholate cotransporting polypeptide-dependent uptake of enveloped virus particles. J Virol. 2018;92(11):e02007–17. 10.1128/JVI.02007-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Imazeki F, Omata M, Yokosuka O, Okuda K. Integration of hepatitis B virus DNA in hepatocellular carcinoma. Cancer. 1986;58(5):1055–60. [DOI] [PubMed] [Google Scholar]
- 10. Sung WK, Zheng H, Li S, Chen R, Liu X, Li Y, et al. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat Genet. 2012;44(7):765–9. 10.1038/ng.2295. [DOI] [PubMed] [Google Scholar]
- 11. Kawai-Kitahata F, Asahina Y, Tanaka S, Kakinuma S, Murakawa M, Nitta S, et al. Comprehensive analyses of mutations and hepatitis B virus integration in hepatocellular carcinoma with clinicopathological features. J Gastroenterol. 2016;51(5):473–86. 10.1007/s00535-015-1126-4. [DOI] [PubMed] [Google Scholar]
- 12. Peneau C, Imbeaud S, La Bella T, Hirsch TZ, Caruso S, Calderaro J, et al. Hepatitis B virus integrations promote local and distant oncogenic driver alterations in hepatocellular carcinoma. Gut. 2022;71(3):616–26. 10.1136/gutjnl-2020-323153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhao LH, Liu X, Yan HX, Li WY, Zeng X, Yang Y, et al. Genomic and oncogenic preference of HBV integration in hepatocellular carcinoma. Nat Commun. 2016;7:12992. 10.1038/ncomms12992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jiang S, Yang Z, Li W, Li X, Wang Y, Zhang J, et al. Re-evaluation of the carcinogenic significance of hepatitis B virus integration in hepatocarcinogenesis. PLoS One. 2012;7(9):e40363. 10.1371/journal.pone.0040363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li CL, Li CY, Lin YY, Ho MC, Chen DS, Chen PJ, et al. Androgen receptor enhances hepatic telomerase reverse transcriptase gene transcription after hepatitis B virus integration or point mutation in promoter region. Hepatology. 2019;69(2):498–512. 10.1002/hep.30201. [DOI] [PubMed] [Google Scholar]
- 16. Lin YY, Hsieh CH, Chen JH, Lu X, Kao JH, Chen PJ, et al. De novo assembly of highly polymorphic metagenomic data using in situ generated reference sequences and a novel BLAST-based assembly pipeline. BMC Bioinformatics. 2017;18(1):223. 10.1186/s12859-017-1630-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liao Y, Smyth GK, Shi W. FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–30. 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 19. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Petitprez F, Meunier L, Letouzé E, Hoshida Y, Villanueva A, Llovet J, et al. MS.liverK: an R package for transcriptome-based computation of molecular subtypes and functional signatures in liver cancer. BioRxiv. 2019. [Google Scholar]
- 21. Danaher P, Warren S, Dennis L, D’Amico L, White A, Disis ML, et al. Gene expression markers of tumor infiltrating leukocytes. J Immunother Cancer. 2017;5:18. 10.1186/s40425-017-0215-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–7. 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sia D, Jiao Y, Martinez-Quetglas I, Kuchuk O, Villacorta-Martin C, Castro de Moura M, et al. Identification of an immune-specific class of hepatocellular carcinoma, based on molecular features. Gastroenterology. 2017;153(3):812–26. 10.1053/j.gastro.2017.06.007. [DOI] [PubMed] [Google Scholar]
- 24. Santangelo A, Rossato M, Lombardi G, Benfatto S, Lavezzari D, De Salvo GL, et al. A molecular signature associated with prolonged survival in glioblastoma patients treated with regorafenib. Neuro Oncol. 2021;23(2):264–76. 10.1093/neuonc/noaa156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim CM, Hwang S, Keam B, Yu YS, Kim JH, Kim DS, et al. Gene signature for sorafenib susceptibility in hepatocellular carcinoma: different approach with a predictive biomarker. Liver Cancer. 2020;9(2):182–92. 10.1159/000504548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hsu CL, Ou DL, Bai LY, Chen CW, Lin L, Huang SF, et al. Exploring markers of exhausted CD8 T cells to predict response to immune checkpoint inhibitor therapy for hepatocellular carcinoma. Liver Cancer. 2021;10(4):346–59. 10.1159/000515305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cawthon RM. Telomere measurement by quantitative PCR. Nucleic Acids Res. 2002;30(10):e47. 10.1093/nar/30.10.e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cote HC, Soudeyns H, Thorne A, Alimenti A, Lamarre V, Maan EJ, et al. Leukocyte telomere length in HIV-infected and HIV-exposed uninfected children: shorter telomeres with uncontrolled HIV viremia. PLoS One. 2012;7(7):e39266. 10.1371/journal.pone.0039266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tang D, Li B, Xu T, Hu R, Tan D, Song X, et al. VISDB: a manually curated database of viral integration sites in the human genome. Nucleic Acids Res. 2020;48(D1):D633–41. 10.1093/nar/gkz867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang HI, Yeh SH, Chen PJ, Iloeje UH, Jen CL, Su J, et al. Associations between hepatitis B virus genotype and mutants and the risk of hepatocellular carcinoma. J Natl Cancer Inst. 2008;100(16):1134–43. 10.1093/jnci/djn243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Noureen N, Wu S, Lv Y, Yang J, Alfred Yung WK, Gelfond J, et al. Integrated analysis of telomerase enzymatic activity unravels an association with cancer stemness and proliferation. Nat Commun. 2021;12(1):139. 10.1038/s41467-020-20474-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee JS, Chu IS, Heo J, Calvisi DF, Sun Z, Roskams T, et al. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40(3):667–76. 10.1002/hep.20375. [DOI] [PubMed] [Google Scholar]
- 33. Roessler S, Jia HL, Budhu A, Forgues M, Ye QH, Lee JS, et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 2010;70(24):10202–12. 10.1158/0008-5472.CAN-10-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hoshida Y, Nijman SM, Kobayashi M, Chan JA, Brunet JP, Chiang DY, et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009;69(18):7385–92. 10.1158/0008-5472.CAN-09-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Boyault S, Rickman DS, de Reynies A, Balabaud C, Rebouissou S, Jeannot E, et al. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology. 2007;45(1):42–52. 10.1002/hep.21467. [DOI] [PubMed] [Google Scholar]
- 36. Chiang DY, Villanueva A, Hoshida Y, Peix J, Newell P, Minguez B, et al. Focal gains of VEGFA and molecular classification of hepatocellular carcinoma. Cancer Res. 2008;68(16):6779–88. 10.1158/0008-5472.CAN-08-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sangro B, Melero I, Wadhawan S, Finn RS, Abou-Alfa GK, Cheng AL, et al. Association of inflammatory biomarkers with clinical outcomes in nivolumab-treated patients with advanced hepatocellular carcinoma. J Hepatol. 2020;73(6):1460–9. 10.1016/j.jhep.2020.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Litchfield K, Reading JL, Puttick C, Thakkar K, Abbosh C, Bentham R, et al. Meta-analysis of tumor- and T cell-intrinsic mechanisms of sensitization to checkpoint inhibition. Cell. 2021;184(3):596–614 e14. 10.1016/j.cell.2021.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Takubo K, Nakamura K-I, Izumiyama N, Furugori E, Sawabe M, Arai T, et al. Telomere shortening with aging in human liver. J Gerontol A Biol Sci Med Sci. 2000;55(11):B533–6. 10.1093/gerona/55.11.b533. [DOI] [PubMed] [Google Scholar]
- 40. Paradis V, Youssef N, Dargère D, Bâ N, Bonvoust F, Deschatrette J, et al. Replicative senescence in normal liver, chronic hepatitis C, and hepatocellular carcinomas. Hum Pathol. 2001;32(3):327–32. 10.1053/hupa.2001.22747. [DOI] [PubMed] [Google Scholar]
- 41. Nault JC, Mallet M, Pilati C, Calderaro J, Bioulac-Sage P, Laurent C, et al. High frequency of telomerase reverse-transcriptase promoter somatic mutations in hepatocellular carcinoma and preneoplastic lesions. Nat Commun. 2013;4(1):2218–7. 10.1038/ncomms3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen Y-L, Jeng Y-M, Chang C-N, Lee H-J, Hsu H-C, Lai P-L, et al. TERT promoter mutation in resectable hepatocellular carcinomas: a strong association with hepatitis C infection and absence of hepatitis B infection. Int J Surg. 2014;12(7):659–65. 10.1016/j.ijsu.2014.05.066. [DOI] [PubMed] [Google Scholar]
- 43. Fujimoto A, Totoki Y, Abe T, Boroevich KA, Hosoda F, Nguyen HH, et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet. 2012;44(7):760–4. 10.1038/ng.2291. [DOI] [PubMed] [Google Scholar]
- 44. Ding D, Lou X, Hua D, Yu W, Li L, Wang J, et al. Recurrent targeted genes of hepatitis B virus in the liver cancer genomes identified by a next-generation sequencing–based approach. PLoS Genet. 2012;8(12):e1003065. 10.1371/journal.pgen.1003065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nault JC, Calderaro J, Di Tommaso L, Balabaud C, Zafrani ES, Bioulac-Sage P, et al. Telomerase reverse transcriptase promoter mutation is an early somatic genetic alteration in the transformation of premalignant nodules in hepatocellular carcinoma on cirrhosis. Hepatology. 2014;60(6):1983–92. 10.1002/hep.27372. [DOI] [PubMed] [Google Scholar]
- 46. Brunner SF, Roberts ND, Wylie LA, Moore L, Aitken SJ, Davies SE, et al. Somatic mutations and clonal dynamics in healthy and cirrhotic human liver. Nature. 2019;574(7779):538–42. 10.1038/s41586-019-1670-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019;47(D1):D941–7. 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dong H, Zhang L, Qian Z, Zhu X, Zhu G, Chen Y, et al. Identification of HBV-MLL4 integration and its molecular basis in Chinese hepatocellular carcinoma. PLoS One. 2015;10(4):e0123175. 10.1371/journal.pone.0123175. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this article and its online supplementary material. RNA-sequencing data can be accessed on GEO using the accession number GSE198946. Further inquiries can be directed to the corresponding author.