Abstract
Mycoplasma hyopneumoniae colonizes the swine respiratory tract at the level of ciliated cells by attaching specifically to the cilium membrane. This interaction involves an adhesin called P97; the cilium binding activity of this protein was localized to the carboxy terminus, which included two repeat regions, R1 and R2 (T. Hsu, S. Artiushin, and F. C. Minion, J. Bacteriol. 179:1317–1323, 1997). To further delineate the molecular mechanisms of M. hyopneumoniae interactions with ciliated epithelium, we used a bank of transposon inserts in the cloned P97 gene to identify the site for cilium binding by testing the truncated gene products in an in vitro microtiter plate adherence assay. These studies showed that the cilium binding site was located in the AAKPV(E) repeat sequence of P97, referred to as the R1 repeat. For functional binding, at least seven AAKPV(E) repeats were required. The adherence-blocking monoclonal antibody F1B6 also recognized this region but required fewer AAKPV(E) repeats for recognition. We then constructed R1 region-lacZ gene fusions and used the resulting R1 repeat–β-galactosidase fusion proteins in an in vitro assay to confirm the role of R1 in cilium binding. A comparison of the R1 regions of M. hyopneumoniae strains displaying variation in cilium adherence failed to identify changes that could account for the differences in adherence shown by the strains. Thus, we concluded that other proteins, in addition to P97, must be involved in cilium adherence, possibly in combination with P97.
Mycoplasma hyopneumoniae constitutes a significant threat to swine health and is responsible for estimated losses to the swine industry of more than $200 million per year. Alone it causes a prevalent and persistent disease of swine called enzootic pneumonia. In combination with other respiratory pathogens, i.e., porcine reproductive and respiratory syndrome virus or swine influenza virus, it produces pneumonia significantly more severe than that after infection with either agent alone (12). Vaccination against M. hyopneumoniae alone does not prevent colonization or protect sufficiently against disease, nor does it obviate the enhancing role of M. hyopneumoniae in dual infection with other infectious pathogens. In the absence of more effective intervention strategies to reduce disease, vaccines must be improved if we are to reduce economic losses due to enzootic pneumonia. This is only possible if we have a full understanding of the mechanisms employed by M. hyopneumoniae to cause disease so that effective therapeutic approaches to circumvent those strategies can be developed. Therefore, it is essential that the virulence mechanisms of M. hyopneumoniae be addressed.
The initial event in M. hyopneumoniae colonization of swine is its adherence to the cilia of the respiratory tract epithelial cells (8). This is followed by an extensive loss of cilia from the epithelial cells of the trachea, bronchi, and bronchioles (8). The molecular basis for the cilium binding specificity is unknown, but a 97-kDa protein, designated P97, was shown to be involved. Recent studies by Zhang et al. (18–20) were instrumental in establishing a swine cilium-specific adherence assay and in identifying potential receptors and ligands involved in the adherence process. A monoclonal antibody (MAb), F1B6, was identified as being able to block M. hyopneumoniae adherence to porcine cilia in the in vitro adherence assay (20). In addition, the gene coding for the ciliary adhesin has been cloned and sequenced (6, 7). It codes for a 125-kDa protein which undergoes a posttranslational cleavage event to produce a final protein product of approximately 102.3 kDa. The product migrates as a 97-kDa protein by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (20) and has been designated P97. The study reported here focused on the identification and analysis of the cilium binding site of P97.
Study of the molecular basis of mycoplasmal adherence has led to the identification of putative binding sites for the P1 adhesin of M. pneumoniae (2, 3) and the P1-like MgPa adhesin of Mycoplasma genitalium (11), but these studies lacked a functional assay to test well-defined mutants. Our studies used the defined adherence assay described by Zhang et al. (19) with purified swine cilia and MAb F1B6 to detect binding of P97. By analysis of a series of Tn1000 insertions in the P97 sequence that resulted in progressive truncation of the recombinant protein, we were able to demonstrate both the location of the MAb epitope and the cilium binding site on the P97 protein. Both activities reside within an AAKPV(E) repeating motif in the carboxy terminus of the protein. A minimal number of repeats are required for functional activity for both the antigenic epitope and cilium binding. The presence of an adequate number of repeats is not sufficient to confer the ability to adhere, however, suggesting that additional factors or proteins are required for adherence of M. hyopneumoniae to swine cilia.
MATERIALS AND METHODS
Bacteria.
Escherichia coli strains included the general cloning host LE392 (5), the opal suppressor host ISM612 (6), and the conjugal F+ donor DPWC (14). BW26 is a kanamycin-resistant conjugal recipient (Gold Biotechnology, Inc., St. Louis, Mo.). CSH50 is ara Δ(lac-pro) strA thi F−. All E. coli strains except ISM612 were started from stock cultures maintained at −70°C and were grown in Luria-Bertani (LB) broth or on LB agar medium. Strain ISM612 was grown in superbroth medium (32 g of tryptone, 20 g of yeast extract, and 5 g of NaCl per liter). Antibiotics were used at the following concentrations: ampicillin, 100 μg per ml; kanamycin, 50 μg per ml; and chloramphenicol, 10 μg per ml.
All M. hyopneumoniae strains were obtained from Richard F. Ross (Iowa State University) and grown in Friis medium as described previously (6). Strain 232A is a virulent, swine cilium-adherent strain. Individual clonal isolates of 232A with different cilium binding characteristics were obtained by picking single colonies and testing for adherence activity; 232A.H (232A 91-3) had high adherence activity, 232A.M (232A 20-10) had moderate adherence activity (63% of that of strain 232A.H), and 232A.L (232A 61-3) had low adherence activity (17% of that of strain 232A.H) (17). Strain J (ATCC 25934), the M. hyopneumoniae type strain, is avirulent and has low swine cilium adherence activity (20). Strain 144L is a virulent, cilium-adherent field strain (20).
Tn1000 mutagenesis.
Tn1000 mutagenesis was performed by conjugal mating of DPWC(pISM2159) with kanamycin-resistant recipient E. coli BW26 (4). Plasmid pISM2159 contained the P97 gene on a 5.1-kb Tsp509I fragment as described previously (6). The locations of the Tn1000 inserts were determined by digestion with restriction enzymes SalI and EcoRV and by DNA sequencing.
DNA sequencing and PCR.
DNA sequencing was performed by the Iowa State University DNA facility, using cycle sequencing protocols and an automated DNA sequencer (model 373 or 377; Applied Biosystems, The Perkin-Elmer Corporation, Norwalk, Conn.). DNA sequence analysis was performed with MacVector software (Eastman Kodak Company, Rochester, N.Y.).
All PCR amplifications were performed with a TwinBlock system (model EZ cycler; Ericomp Inc., San Diego, Calif.). The annealing temperature was determined by Oligo primer analysis software (National Biosciences, Inc., Plymouth, Minn.). For analysis of mycoplasmal DNA, the basic PCR mixture contained 2 mM MgCl2, 25 pmol of each primer, 1 to 50 ng of template DNA, and 0.25 U of Taq DNA polymerase in 50 μl of 1× manufacturer’s reaction buffer. The PCR conditions were as follows: denaturation of the DNA at 94°C for 5 min, followed by 35 cycles (94°C denaturation for 1 min, 55°C annealing for 1.5 min, and 72°C extension for 1 min) and a final 5-min 72°C extension step.
Primer pairs TH120 (AAGGTAAAAGAGAAGAAGTAG)-TH121 (TTGTAAGTGAAAAGCCAGTAT) and TH122 (AGCGAGTATGAAGAACAAGAA)-TH123 (TTTTTACCTAAGTCAGGAAGG) were used to amplify the two repeat regions of P97, R1 and R2, respectively. For most reactions, chromosomal DNAs were used as templates. PCR products were analyzed by agarose electrophoresis and by DNA sequencing. For construction of pISM1244, phosphorylated primers TH120 and TH121 were used with Pfu polymerase (Stratagene). The template DNA for that reaction was pISM2159, a plasmid containing the P97 sequence on a 5.1-kb fragment (6). For plasmids pISM1257 and pISM1258, chromosomal DNAs of M. hyopneumoniae J and 144L, respectively, were used as templates.
Plasmid constructions.
Plasmid pMLB1107, obtained from Greg Phillips (Iowa State University), was originally constructed by Michael Berman (1). The plasmid is a pBR322 derivative with the tetracycline resistance marker replaced by the lacI and lacZ genes; lacZ has the original Plac promoter upstream controlling its expression. The lacZ gene has the pUC8 polylinker inserted at its 5′ end within the coding sequence, resulting in unique SmaI, BamHI, SalI, and HindIII cloning sites at that location. Cloning into any of these sites results in a lacZ gene fusion that can be induced with 1 mM isopropyl-β-d-thiogalactopyranoside. After induction, the fusion protein can be studied by using standard assays (9). pISM1244, pISM1257, and pISM1258 were constructed by cloning R1 region PCR products from strains 232A, J, and 144L, respectively, into the SmaI site of pMLB1107 and transforming the ligation mixture into Lac-deficient CSH50. The orientation and reading frame of each fragment was confirmed by DNA sequencing using primer TH121 as described above.
Immunoblot analysis.
Immunoblotting was performed as described by Towbin et al. (15). Proteins resolved by SDS-PAGE were transferred to nitrocellulose membranes. After electroblotting, membranes were blocked with TS-Tween buffer (0.01 M Tris, 0.140 M NaCl, 0.01% [vol/vol] Tween 20) plus 5% powdered milk for 1 h and washed three times with TS-Tween for 15 min each time. The blots were developed with MAb F1B6, which recognized P97 on immunoblots and inhibited binding of intact M. hyopneumoniae to purified cilia (20). It was also used previously to identify clones containing the P97 coding sequence (6).
Adherence assay.
To prepare lysates of recombinant E. coli, 30- to 40-ml aliquots of overnight ISM612 superbroth cultures were induced with 2.5 mM isopropyl-β-d-thiogalactopyranoside for 5 to 6 h in order to enhance suppression of opal UGA codons within the P97 gene sequence (10, 13). The E. coli pellets were resuspended in 5 ml of TES-2 buffer (50 mM Tris-HCl, 10 mM EDTA, 10% sucrose [pH 8.0]) and centrifuged again. The final E. coli pellets were suspended in 2 ml of TES-2 buffer. Cells were lysed by sonication, and lysates were collected after centrifugation at 12,000 × g for 20 min. Protein concentration was determined by Bio-Rad protein assay (Bio-Rad Laboratories, Richmond, Calif.), using bovine serum albumin as the protein standard. Swine cilia were prepared, and the adherence assay was performed with MAb F1B6 as described in detail elsewhere (19). Negative controls included wells with negative control antigens or wells with blocking buffer. All experiments were performed in triplicate with freshly prepared antigens, and experiments were repeated at least twice. For experiments with the R1 repeat–β-galactosidase fusion proteins, the antibodies were omitted, and the plate was developed with standard β-galactosidase assay reagents in place of alkaline phosphate substrate (9).
RESULTS AND DISCUSSION
Mapping of the P97 MAb F1B6 antigenic and ciliary binding epitopes.
Studies were performed to map both the MAb F1B6 binding epitope and the cilium binding site on the gene sequence. Both analyses were begun by Tn1000 insertional mutagenesis of plasmid pISM2159, which contained the P97 gene sequence, followed by analysis of the resulting recombinant P97 gene product in E. coli. Mutagenized pISM2159 plasmids were isolated, and the Tn1000 insertions were mapped by restriction digests. The location of the Tn1000 insertion was also determined by DNA sequencing using Tn1000 end-specific primers (Fig. 1). Selected plasmids were transformed into E. coli ISM612, the transformants were induced, and the lysates were examined for MAb reactivity and for ciliary binding activity.
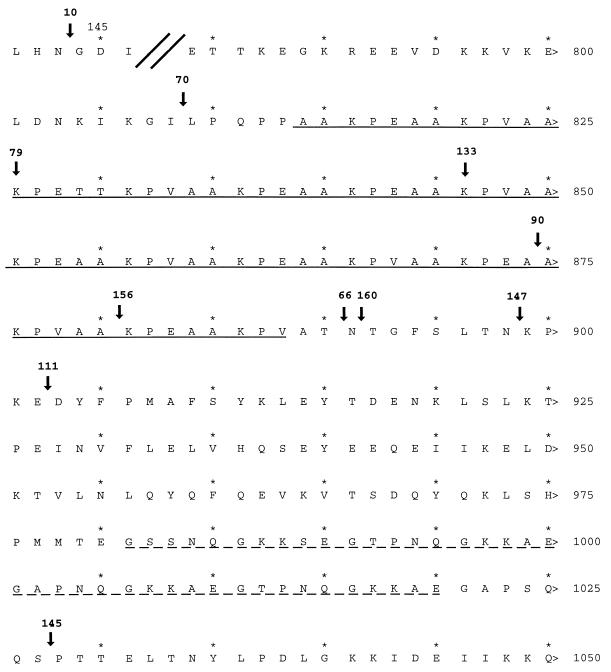
FIG. 1.
Locations of Tn1000 insertions used to identify the MAb F1B6 binding epitope and the cilium binding site of P97. Shown is the translated amino acid sequence (accession no. U50901); amino acid positions are indicated by numbers at the right. Arrows indicate transposon insertion locations. The transposon insert number is given above each arrow. Insertion 10 is located upstream between amino acid positions 143 and 144. The solid underline indicates R1; the broken underline indicates R2.
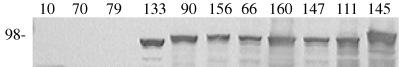
MAb reactivity was determined by immunoblotting as shown in Fig. 2. Immunoblot analysis with lysates from a series of Tn1000 insertions in pISM2159 showed that transposon insertions at or upstream of pISM2159::Tn1000.79 (bp 2476 of P97) failed to express MAb F1B6-reactive antigens (Fig. 2). Plasmids with Tn1000 inserts at or downstream of pISM2159::Tn1000.133 retained the ability to produce MAb F1B6-reactive proteins. Inserts 79 and 133 are separated by 62 bp, corresponding to amino acids 830 to 850 in the protein. This portion of the protein contains the amino acid R1 repeat sequence AAKPV(E) (Fig. 1). It was concluded that the MAb epitope requires at least 2.5 repeating units of the R1 repeat.
FIG. 2.
Mapping the P97 F1B6 MAb binding epitope by immunoblotting using Tn1000 insertions in pISM2159. Whole-cell antigens were prepared from recombinant E. coli as described in Materials and Methods. The proteins were resolved on an SDS–10% polyacrylamide gel (3 μg per lane), and the resulting blot was developed with MAb F1B6. Each lane number indicates the number of the Tn1000 insertion (Fig. 1). The molecular mass given on the left in kilodaltons was determined from molecular weight markers (Bio-Rad). P97 does not undergo posttranslational cleavage in E. coli and would normally migrate as a 125-kDa protein, but the Tn1000 insertion results in truncation of the protein at the point of insertion.
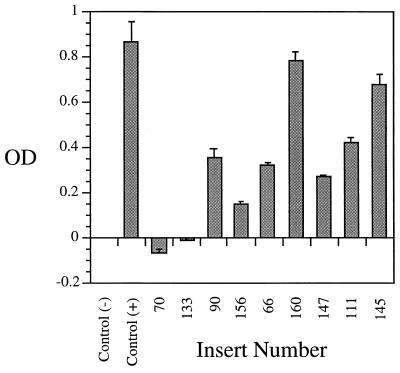
To locate the P97 ciliary binding epitope, microtiter plate adherence assays were performed with E. coli lysates from a series of strains carrying Tn1000 insertions in pISM2159. Lysates prepared from pISM2159::Tn1000.70 and pISM2159::Tn1000.133 failed to bind to cilia, whereas lysates prepared from pISM2159::Tn1000.90 and those downstream of pISM2159::Tn1000.90 showed ciliary binding activity (Fig. 3). The amount of binding varied, but this variation could be explained by the variation in the amount of truncated P97 product made in E. coli as illustrated in Fig. 2. A standard amount of total lysate protein was analyzed in both the immunoblot and cilium binding assays, because it was not possible to accurately quantitate the amount of the truncated P97 protein in each preparation. The strains with the highest binding, 160 and 145, had substantially more P97 protein than those with less binding, 90, 156, 66, 147, and 111 (Fig. 3). Strain 133 had no binding, although sufficient protein was available for binding (Fig. 2). The two inserts pISM2159::Tn1000.133 (7th repeat of R1) and pISM2159::Tn1000.90 (12th repeat of R1) are separated by 75 bp. Thus, our data identify R1 as the probable binding domain and show that an active cilium binding site requires a minimum of seven repeats of the AAKPV(E) sequence (Fig. 1).
FIG. 3.
Microtiter plate adherence assay analysis of Tn1000 insertions in P97. Lysates were prepared from ISM612 (pISM2159::Tn1000) strains and tested in the microtiter plate adherence assay as described in Materials and Methods. The data are presented as mean optical density (OD) ± standard error of triplicate wells. Numbers refer to positions of Tn1000 insertions in the P97 DNA sequence and correspond to lane numbers in Fig. 2. The positive control contains nonmutated pISM2159, and the negative control is lysate from ISM612 containing vector only.
Analysis of size variation in the P97 repeat regions by PCR.
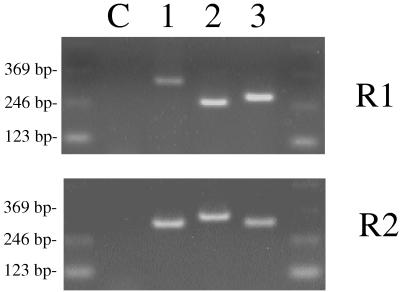
Since our data are consistent with the idea that the R1 region of P97 is involved in cilium binding, it was of interest to determine if cilium binding activities of different M. hyopneumoniae strains which vary in adherence could be correlated with changes in this region of P97. Also, the presence of both R1 and R2 at the carboxy-terminal end of P97 suggested that there might be a relationship between the size of these repeat regions in different strains and the size of the P97 product as has been shown for other mycoplasmal surface proteins (16). To study these two possibilities, we examined the sizes of the R1 and R2 regions of M. hyopneumoniae strains with different cilium adherence activities by PCR analysis using the two primer pairs TH120-TH121 for amplification of R1 and TH122-TH123 for amplification of R2, respectively. In addition to the virulent strain 232A, strains 144L (adherent) and J (nonadherent, avirulent) were sequenced. Also, single-colony isolates of 232A with different adherence activities were identified and sequenced. Figure 4 shows the results of the PCR amplification of R1 and R2 from chromosomal DNA of M. hyopneumoniae 232A, 144L, and J. M. hyopneumoniae 232A gave rise to a product of the R1 region 57 bp larger than that of strain 144L, which in turn was 21 bp larger than that of strain J. In contrast, amplification of R2 from strain J produced a fragment 24 bp larger than that from strain 144L which was 6 bp larger than that from strain 232A (Fig. 4). There was no size variation in either repeat region among 232A single-colony isolates even though adherence activity varied (data not shown). Thus, we were unable to make a direct correlation between the sizes of the two repeat regions and either the size of P97 or the binding activity of the strain in which it was found.
FIG. 4.
Analysis of size variation in the P97 repeat regions by PCR analysis. The bands represent PCR products produced from template DNA from different M. hyopneumoniae strains. Lane C, no template DNA control; lane 1, strain 232A (adherent); lane 2, strain J (nonadherent); lane 3, strain 144L (adherent). R1 and R2 were amplified with primer sets TH120-TH121 and TH122-TH123, respectively.
DNA sequence analysis of the P97 repeat sequence R1 in different strains.
Zhang et al. (20) had previously shown that M. hyopneumoniae exhibited variable ciliary binding activity among different strains. Analysis of the P97 sequence for the cilium binding epitope by transposon mutagenesis indicated that it was located within the central portion of the R1 region. One hypothesis for the loss of cilium binding activity in different M. hyopneumoniae strains is a modification of the R1 region of P97. None of the strains analyzed above by PCR lacked R1, and all of the strains seemed to have regions large enough to code for the epitope. Another possibility for loss of binding is that the repeat sequence has been modified to create a missense or loss-of-function mutation. This possibility was addressed by sequencing PCR-amplified R1 regions of different strains, using chromosomal DNAs as templates. Primers TH120 (upper primer for R1) and TH123 (lower primer for R2) were used for these studies. Following gel purification, sequences of the PCR products were determined by using primer TH120 and then analyzed. The R1 sequences exhibited a high degree of sequence homology among the six strains analyzed, two of which (J and 232 A.L) had low cilium binding activity (data not shown). All six strains retained the AAKPV(E) R1 repeat sequences. There was no sequence variation in the R1 repeat between the 232A colony isolates, although their binding activity varied significantly (data not shown). The R1 regions in strains J and 144L had only 10 and 11 repeats, respectively.
Functional analysis of the R1 region.
Our data indicated that the R1 region encoded the amino acids forming the cilium binding site. These studies, however, did not rule out the possibility that other regions of the protein contributed to the binding in some important way, i.e., as part of a three-dimensional structure forming a binding cleft or pocket. To study this in more detail, we sought a way to isolate the repeat region from other P97 sequences and measure its ability to bind to swine cilia. This was accomplished by using plasmid pMLB1107 to construct R1 region-lacZ gene fusions and inducing expression of the β-galactosidase fusion protein in a Lac-deficient background. By exposing E. coli lysates to cilia in a microtiter plate adherence assay followed by an assay for bound β-galactosidase activity, it was possible to measure the contribution of R1 independent of other P97 sequences. Using this same approach, we were also able to determine if the R1 regions were functional in M. hyopneumoniae strains J (nonadherent) and 144L (adherent).
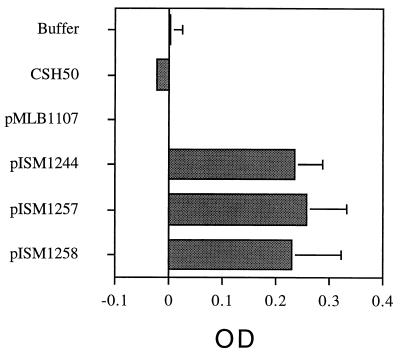
Figure 5 shows the results of this study. E. coli lysates were normalized with respect to amount of total protein (20 μg) added per well. The total amount of β-galactosidase activity added to each well was not the same in every case, but the variation was less than 20% (data not shown). As expected, wild-type β-galactosidase (pMLB1107) failed to bind to swine cilia. All three of the R1 repeat–β-galactosidase fusions bound equally to cilia, however, proving that R1 could function independently as a cilium binding domain. In addition, each β-galactosidase fusion protein reacted with MAb F1B6 by immunoblotting (data not shown).
FIG. 5.
Cilium binding activity of β-galactosidase fusion proteins containing R1 regions from different M. hyopneumoniae strains. The assay was performed as described previously (6) except that antibodies were omitted and o-nitrophenyl galactopyranoside was substituted as the substrate (9). Twenty micrograms of protein containing 28 U (pISM1244), 23 U (pISM1257), or 26 U (pISM1258) of β-galactosidase activity from E. coli lysates was added to each well, and the plates were incubated at 37°C for 90 min to allow binding of the fusion proteins to cilia. The control with pMLB1107 contained 44 U of activity. The data represent the mean optical density (OD) and standard deviation of six independent assays.
Interestingly, the fusion containing the R1 repeat from strain J also bound to cilia even though strain J has low binding activity (20). It is still not clear why strain J fails to bind to swine cilia since P97 is produced and the repeat region is fully competent in binding. DNA sequence analysis of low-, medium-, and high-adherence colony isolates of strain 232A gave similar results; neither the DNA sequence of the R1 region nor P97 expression seemed to vary among the strains in spite of the loss of binding activity. Our conclusion is that other proteins or factors independent of P97 contribute to cilium binding in critical ways. While it is clear from our studies that P97 alone is fully competent for cilium binding, these factors could participate indirectly in the intact cell by participating in the posttranslational modification of P97, its translocation across the membrane, or its placement in the mycoplasma membrane. The protein is positioned at a distance from the cell membrane, connected through an electron-translucent component (20). It is also possible that other elements interact with P97 to enhance binding in the intact cell, or they may act independently through mechanisms not yet identified.
ACKNOWLEDGMENTS
We thank Richard F. Ross and Theresa Young for the M. hyopneumoniae strains. We also thank Qijing Zhang for work in isolating and characterizing the high-, medium-, and low-adherence clonal isolates of 232A. We are indebted to Kendall King for Mhp1 (P97) deletion mutants, which provided preliminary information on the importance of the carboxy terminus of P97 in cilium binding.
REFERENCES
- 1. Berman, M. Unpublished data.
- 2.Dallo S F, Su C J, Horton J R, Baseman J B. Identification of P1 gene domain containing epitope(s) mediating Mycoplasma pneumoniae cytadherence. J Exp Med. 1988;167:718–723. doi: 10.1084/jem.167.2.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerstenecker B, Jacobs E. Topological mapping of the P1-adhesin of Mycoplasma pneumoniae with adherence-inhibiting monoclonal antibodies. J Gen Microbiol. 1990;136:471–476. doi: 10.1099/00221287-136-3-471. [DOI] [PubMed] [Google Scholar]
- 4.Guyer M S. Uses of transposon γδ in the analysis of cloned genes. Methods Enzymol. 1983;101:362–369. doi: 10.1016/0076-6879(83)01027-7. [DOI] [PubMed] [Google Scholar]
- 5.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 6.Hsu T, Artiushin S, Minion F C. Cloning and analysis of P97, a respiratory cilium adhesin gene of Mycoplasma hyopneumoniae. J Bacteriol. 1997;179:1317–1323. doi: 10.1128/jb.179.4.1317-1323.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.King K W, Faulds D H, Rosey E L, Yancey R J. Characterization of the gene encoding Mhp1 from Mycoplasma hyopneumoniae and examination of Mhp1’s vaccine potential. Vaccine. 1996;15:25–35. doi: 10.1016/s0264-410x(96)00121-1. [DOI] [PubMed] [Google Scholar]
- 8.Mebus C A, Underdahl N R. Scanning electron microscopy of trachea and bronchi from gnotobiotic pigs inoculated with Mycoplasma hyopneumoniae. Am J Vet Res. 1977;43:1249–1254. [PubMed] [Google Scholar]
- 9.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 10.Minion F C, VanDyk C, Smiley B K. Use of an Escherichia coli enhanced opal suppressor strain to screen a Mycoplasma hyopneumoniae library. FEMS Microbiol Lett. 1995;131:81–85. doi: 10.1016/0378-1097(95)00239-2. [DOI] [PubMed] [Google Scholar]
- 11.Opitz O, Jacobs E. Adherence epitopes of Mycoplasma genitalium adhesin. J Gen Microbiol. 1992;138:1785–1790. doi: 10.1099/00221287-138-9-1785. [DOI] [PubMed] [Google Scholar]
- 12.Ross R F. Mycoplasmal disease. In: Leman A D, Straw B E, Mengeling W L, D’Allaire S, Taylor D J, editors. Diseases of swine. Ames, Iowa: Iowa State University Press; 1992. pp. 537–551. [Google Scholar]
- 13.Smiley B K, Minion F C. Enhanced readthrough of opal (UGA) codons and production of Mycoplasma pneumoniae P1 epitopes in Escherichia coli. Gene. 1993;134:33–40. doi: 10.1016/0378-1119(93)90171-x. [DOI] [PubMed] [Google Scholar]
- 14.Strathmann M, Hamilton B A, Mayeda C A, Simon M I, Meyerowitz E M, Palazzolo M J. Transposon-facilitated DNA sequencing. Proc Natl Acad Sci USA. 1991;88:1247–1250. doi: 10.1073/pnas.88.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wise K S, Yogev D, Rosengarten R. Antigenic variation. In: Maniloff J, McElhaney R N, Finch L R, Baseman J B, editors. Mycoplasmas: molecular biology and pathogenesis. Washington, D.C: American Society for Microbiology; 1992. pp. 473–489. [Google Scholar]
- 17.Young T F, Zhang Q, Erickson B Z, Ross R F. Abstracts of the 10th International Congress of the International Organization for Mycoplasmology. Bordeaux, France: International Organization for Mycoplasmology; 1994. Isolation and characterization of high and low adherent clones of Mycoplasma hyopneumoniae; pp. 684–685. [Google Scholar]
- 18.Zhang Q, Young T F, Ross R F. Glycolipid receptors for attachment of Mycoplasma hyopneumoniae to porcine respiratory ciliated cells. Infect Immun. 1994;62:4367–4373. doi: 10.1128/iai.62.10.4367-4373.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Q, Young T F, Ross R F. Microtiter plate adherence assay and receptor analogs for Mycoplasma hyopneumoniae. Infect Immun. 1994;62:1616–1622. doi: 10.1128/iai.62.5.1616-1622.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Q, Young T F, Ross R F. Identification and characterization of a Mycoplasma hyopneumoniae adhesin. Infect Immun. 1995;63:1013–1019. doi: 10.1128/iai.63.3.1013-1019.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]