Abstract
Background
Urinary metabolomics has demonstrated considerable potential to assess kidney function and its metabolic corollaries in health and disease. However, applications in epidemiology remain sparse due to technical challenges.
Methods
We added 17 metabolites to an open-access urinary nuclear magnetic resonance metabolomics platform, extending the panel to 61 metabolites (n = 994). We also introduced automated quantification for 11 metabolites, extending the panel to 12 metabolites (+creatinine). Epidemiological associations between these 12 metabolites and 49 clinical measures were studied in three independent cohorts (up to 5989 participants). Detailed regression analyses with various confounding factors are presented for body mass index (BMI) and smoking.
Results
Sex-specific population reference concentrations and distributions are provided for 61 urinary metabolites (419 men and 575 women), together with methodological intra-assay metabolite variations as well as the biological intra-individual and epidemiological population variations. For the 12 metabolites, 362 associations were found. These are mostly novel and reflect potential molecular proxies to estimate kidney function, as the associations cannot be simply explained by estimated glomerular filtration rate. Unspecific renal excretion results in leakage of amino acids (and glucose) to urine in all individuals. Seven urinary metabolites associated with smoking, providing questionnaire-independent proxy measures of smoking status in epidemiological studies. Common confounders did not affect metabolite associations with smoking, but insulin had a clear effect on most associations with BMI, including strong effects on 2-hydroxyisobutyrate, valine, alanine, trigonelline and hippurate.
Conclusions
Urinary metabolomics provides new insight on kidney function and related biomarkers on the renal-cardiometabolic system, supporting large-scale applications in epidemiology.
Keywords: Metabolomics, urine, biomarkers, kidney function, metabolism
Key Messages.
This work appears as the first comprehensive quantitative urine metabolomics study at an epidemiological scale, and presents 362 associations (most of them novel) between 12 urinary metabolites and 49 clinical and biochemical measures, with replication in three independent population cohorts of up to 5989 participants.
All individuals have amino acids in the urine. Thus, despite the high efficiency of the amino acid transporters, the large volume of plasma filtered would result in unspecific leakage of amino acids into the urine, similarly to the situation with glucose.
Seven urinary metabolites associated with smoking, providing questionnaire-independent proxy measures of smoking status in epidemiological studies. Common confounders did not affect these associations.
Of the 12 urinary metabolites, only dimethylamine and urea did not associate with body mass index (BMI). Insulin had a clear effect on most associations with BMI, including strong effects on 2-hydroxyisobutyrate, valine, alanine, trigonelline and hippurate.
The novel extensive quantitative urinary metabolomics data and the plethora of associations with clinically relevant measures and outcomes support large-scale epidemiological studies for new insight on kidney function and related disease biomarkers.
Introduction
A limited number of urinary biomarkers are widely used as diagnostic aids in kidney disease (creatinine and albumin) and diabetes (glucose). The measurement rationale for these is mainly to pinpoint high values that cross pre-set diagnostic limits, for example with the standard urinary glucose test strips with detection limits as high as 5.6 mmol/L.1 Quantitative metabolic approaches with large enough numbers of individuals for appropriate epidemiological studies, targeting improved understanding of urinary metabolites in health and as potential biomarkers of disease risk, are almost non-existent.2–8 This is at an immense contrast to the situation with various metabolomics and lipidomics approaches already in widespread use in genetics and epidemiology for large-scale quantitative studies of systemic blood biomarkers.9–15
Nevertheless, the potential of urinary metabolites in epidemiology and translational medicine has been recognized for quite some time.2–5,7,16 The molecular content of urine is physiologically connected to the glomerular filtration and molecular reabsorption processes in the kidneys, and reflects multiple key biochemical pathways in relation to cardiometabolic conditions, gut microbial metabolic activities and dietary characteristics. Detailed quantitative data on urinary metabolites may thus provide direct molecular probes to assess kidney function and its corollaries in various metabolic conditions. Towards this far-reaching aim, we have recently developed a basis for an open-access methodology for quantitative high-throughput urinary nuclear magnetic resonance (NMR) metabolomics.1,7,17
The focus of this work is to extend the population-level quantitative data to 61 urinary metabolites, and to provide their sex-specific reference concentrations and distributions in a population sample of 994 individuals. The first coherent set of automated quantification models for 12 urinary metabolites (+ creatinine) is also presented, together with large-scale assessment of these metabolite concentrations in morning spot urine samples and their associations with wide-ranging clinical data, in three independent population cohorts of up to 5989 participants. This work extends the epidemiological scale of urine metabolomics to a new level, incorporates independent replication and provides a plethora of novel metabolic findings in relation to kidney function, with potential translational relevance.
Material and methods
General aspects and data
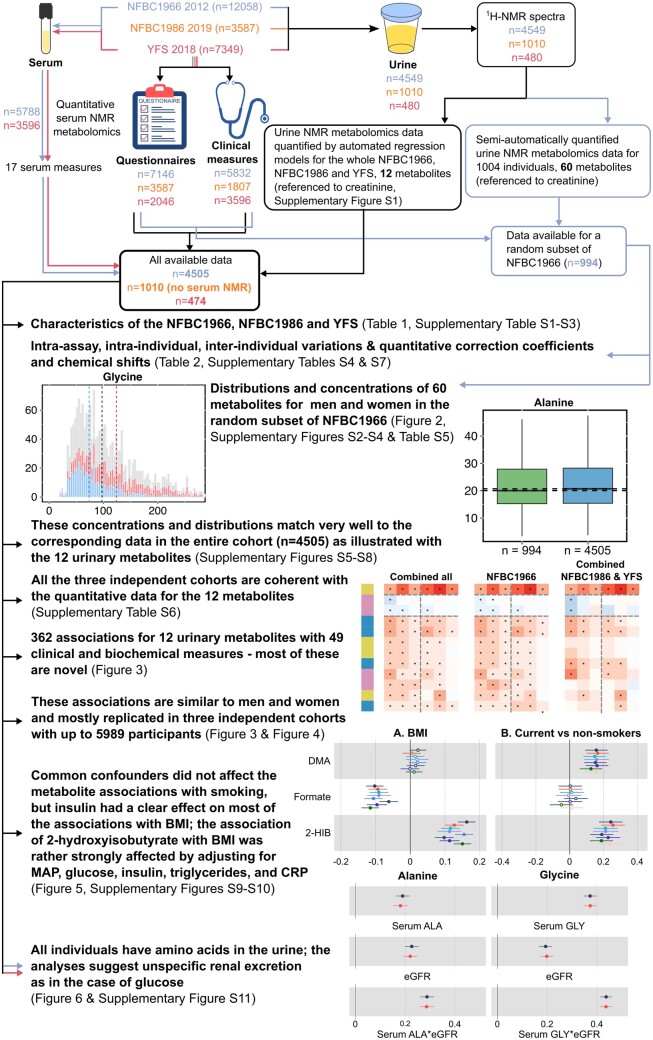
This work is based on an open-access proton NMR spectroscopy methodology we have recently introduced.7 A study outline and a summary of various analyses performed are shown in a schematic form in Figure 1. The characteristics of the three independent population cohorts are given in Table 1. The cohorts are described in more detail in the online Supplementary data together with the detailed list of the 49 clinical and biochemical measures. The sex-specific characteristics for each cohort are given in Supplementary Table S1, available as Supplementary data at IJE online (Northern Finland Birth Cohort 1966; NFBC1966, n = 4505), Supplementary Table S2, available as Supplementary data at IJE online (Northern Finland Birth Cohort 1986; NFBC1986, n = 1010)18 and Supplementary Table S3, available as Supplementary data at IJE online (Cardiovascular Risk in Young Finns Study; YFS, n = 474).
Figure 1.
A flowchart illustrating the study design, statistical analyses and key findings. The data from NFBC1966, NFBC1986 and YFS are indicated by colour-coded arrows: blue, orange and red, respectively. The black arrows represent analyses for all the cohorts. NFBC, Northern Finland Birth Cohort; YFS, Cardiovascular Risk in Young Finns Study; NMR, nuclear magnetic resonance; 1H-NMR, proton nuclear magnetic resonance; BMI, body mass index; MAP, mean arterial pressure; CRP, C-reactive protein; eGFR, estimated glomerular filtration rate
Table 1.
Characteristics of the three independent study populationsa
| Characteristic | NFBC1966 | NFBC1986 | YFS |
|---|---|---|---|
| Number | 4505 | 1010 | 474 |
| Age (years) | 46.7 (46.2–47.1) | 33.7 (33.4–34.1) | 50.1 (31.3–68.5) |
| BMI (kg/m2) | 26 (23–29) | 25 (23–28) | 27 (24–30) |
| Waist-to-hip ratio | 0.91 (0.85–0.97) | 0.91 (0.87–0.97) | |
| Body fat (%) | 28 (22–35) | 26 (20–34) | |
| Visceral fat area (cm2) | 99 (76–127) | 81 (59–121) | |
| Total body water (L) | 39 (34–47) | 39 (33–47) | |
| Systolic blood pressure (mmHg) | 124 (114–135) | 111 (103–120) | 128 (117–142) |
| Diastolic blood pressure (mmHg) | 84 (77–92) | 74 (68–80) | 78 (71–85) |
| Pulse (beats/min) | 69 (62–77) | 71 (64–79) | 69 (62–77) |
| Fitness score | 74 (69–79) | 75 (70–80) | |
| Basal metabolic rate (calories) | 1509 (1358–1756) | 1520 (1353–1753) | |
| Grip strength average (kg) | 32 (26–45) | 35 (30–47) | |
| Smoking prevalence (%)b | 17.5 | 14 | 18.7 |
| Leucine (mmol/L) | 0.08 (0.07–0.10) | 0.12 (0.10–0.14) | |
| Isoleucine (mmol/L) | 0.05 (0.05–0.07) | 0.06 (0.05–0.07) | |
| Valine (mmol/L) | 0.20 (0.18–0.23) | 0.23 (0.21–0.26) | |
| Alanine (mmol/L) | 0.45 (0.41–0.5) | 0.37 (0.32–0.43) | |
| Glutamine (mmol/L) | 0.57 (0.53–0.61) | 0.75 (0.70–0.80) | |
| Glycine (mmol/L) | 0.29 (0.26–0.33) | 0.26 (0.23–0.32) | |
| Phenylalanine (mmol/L) | 0.08 (0.07–0.08) | 0.06 (0.05–0.07) | |
| Tyrosine (mmol/L) | 0.06 (0.05–0.06) | 0.06 (0.06–0.07) | |
| Glycated haemoglobin (%) | 5.5 (5.2–5.7) | 5.2 (5.0–5.4) | 5.5 (5.3–5.8) |
| Fasting insulin (IU/L) | 7.9 (5.4–11.7) | 9.2 (5.4–13.4) | |
| Fasting glucose (mmol/L) | 5.4 (5.1–5.8) | 4.9 (4.7–5.2) | 5.4 (5.1–5.8) |
| Lactate (mmol/L) | 1.4 (1.2–1.7) | 2.0 (1.7–2.4) | |
| Pyruvate (mmol/L) | 0.09 (0.08–0.12) | 0.06 (0.05–0.08) | |
| Citrate (mmol/L) | 0.12 (0.11–0.13) | 0.04 (0.04–0.05) | |
| Glycerol (mmol/L) | 0.07 (0.06–0.09) | 0.12 (0.09–0.15) | |
| Apolipoprotein B (g/L) | 1.02 (0.88–1.2) | 0.93 (0.79–1.11) | |
| Total triglycerides (mmol/L) | 1.03 (0.76–1.47) | 0.78 (0.57–1.09) | 1.13 (0.85–1.55) |
| Apolipoprotein A-I (g/L) | 1.7 (1.6–1.9) | 1.5 (1.4–1.7) | |
| HDL cholesterol (mmol/L) | 1.5 (1.3–1.8) | 1.4 (1.2–1.7) | 1.3 (1.1–1.6) |
| Acetoacetate (mmol/L) | 0.04 (0.03–0.05) | 0.02 (0.01–0.04) | |
| Beta-hydroxybutyrate (mmol/L) | 0.12 (0.10–0.16) | 0.05 (0.02–0.11) | |
| C-reactive protein (mg/L) | 0.82 (0.45–1.65) | 0.71 (0.36–1.55) | 1.03 (0.53–2.37) |
| GlycA (mmol/L) | 1.4 (1.3–1.5) | 0.89 (0.81–0.96) | |
| Haemoglobin (g/L) | 141 (132–150) | 136 (128–146) | 144 (137–152) |
| Leukocytes (× 109 cells/L) | 5.4 (4.5–6.4) | 6.1 (5.2–7.1) | |
| Platelets (× 109 cells/L) | 247 (215–286) | 239 (210–274) | 254 (219–292) |
| Erythrocytes (× 1012 cells/L) | 4.7 (4.4–4.9) | 4.6 (4.3–4.9) | 4.8 (4.5–5.1) |
| Bilirubin (µmol/L) | 11 (9–15) | 12 (9–16) | |
| Alkaline phosphatase (U/L) | 61 (51–73) | 55 (45–66) | |
| Alanine aminotransferase (U/L) | 25 (18–36) | 21 (16–31) | 22 (15–31) |
| Gamma-glutamyl transferase (U/L) | 23 (15–38) | 15 (11–24) | 24 (17–38) |
| Uric acid (µmol/L) | 297 (249–353) | 303 (252–356) | |
| Creatinine (µmol/L) | 67 (59–75) | 65 (58–74) | 76 (67–87) |
| eGFR (mL/min/1.73m2) | 104 (95–107) | 115 (107–118) | 87 (75–100) |
| FINRISK | 0.57 (0.20–1.66) | 0.10 (0.04–0.32) | 1.19 (0.2–6.62) |
| CKD Nelson risk | 1.4 (1.0–2.4) | 0.29 (0.23–0.47) | 6.6 (1.2–31.7) |
| CKD O’Seaghdha risk | 0.76 (0.76–1.61) | 0.24 (0.23–0.25) | 3.65 (0.50–20.89) |
| CKD Chien risk | 6.1 (4.7–8.4) | 1.8 (1.4–2.3) | 10.2 (2.7–40) |
BMI, body mass index; CKD, chronic kidney disease; eGFR, estimated glomerular filtration rate; FINRISK, a large Finnish population survey of risk factors for chronic, noncommunicable diseases; GlycA, glycoprotein acetyls; HDL, high-density lipoprotein; NFBC, Northern Finland Birth Cohort; YFS, Cardiovascular Risk in Young Finns Study.
Values are median (interquartile range).
The number of current smokers/the total number of cohort participants.
In addition to the novel data on clinical and biochemical associations, we extend the methodology here from the previously quantified 43 urinary metabolites (+creatinine) to 60 metabolites (+creatinine), and present the methodological intra-assay metabolite coefficients of variation (CV %s) as well as 30-day consecutive intra-individual and inter-individual population variation for the added 17 metabolites (Table 2). The corresponding previously published information7 for the 43 metabolites is given for the convenience of the readers in Supplementary Table S4, available as Supplementary data at IJE online (together with the data for the added 17 metabolites). These analyses were done in a random subset of 994 participants in the NFBC1966, and the distributions for all these metabolites are illustrated in Figure 2.
Table 2.
Intra-assay, intra-individual and inter-individual variation of 17 quantified urine metabolites (introduced in this work)f
| Metabolite | Intra-assay | Intra-individual | Inter-individual |
|---|---|---|---|
| CV (%) a , b | CV (%) a , c | CV (%) a , d | |
| Amino acids | |||
| Leucine | 6.34 | 26.48 | 58.25 |
| Metabolism of amino acids | |||
| Betaine | 3.23 | 43.96 | 176.13 |
| Phenylacetylglutamine | 3.12 | 31.90 | 54.00 |
| Pyroglutamate | 4.40 | 17.64 | 27.64 |
| Carbohydrate metabolism | |||
| Fumarate | —e | 176.23 | 218.72 |
| Succinate | 13.36 | 32.08 | 179.91 |
| Mannitol | —e | 164.85 | 222.25 |
| Caffeine metabolism | |||
| 3-methylxanthine | 5.02 | 174.27 | 84.08 |
| Microbial metabolism | |||
| Methanol | 1.91 | 60.33 | 114.21 |
| Dietary metabolites | |||
| 1-methylhistidine | 2.04 | 21.15 | 31.26 |
| Levoglucosan | 1.52 | 304.04 | 190.10 |
| Proline-betaine | 2.71 | 132.03 | 139.93 |
| Quinate | 3.51 | 262.56 | 81.67 |
| Scyllitol | 1.19 | 22.05 | 57.91 |
| Trans-ferulate | 4.71 | 31.12 | 101.66 |
| Miscellaneous | |||
| 4-deoxyerythronate | 1.59 | 18.15 | 38.46 |
| 4-deoxythreonate | 1.67 | 30.42 | 38.60 |
Identical data for 43 metabolites from a previous publication are shown for the convenience of readers in Supplementary Table S4 (available as Supplementary data at IJE online).
CV, coefficient of variation.
Concentrations are scaled to the concentration of creatinine; CV (%) = (standard deviation/average) * 100%.
One urine sample prepared and analysed as 10 replicates; reflects the entire quantitative process, i.e. including all the sample preparation steps, nuclear magnetic resonance experimentation and mathematical quantification protocols.
A 30-day consecutive urine collection, averaged over three different volunteers.
In 1003 different individuals from the Northern Finland Birth Cohort 1966.
Concentration of the metabolite below the detection limit in this urine sample.
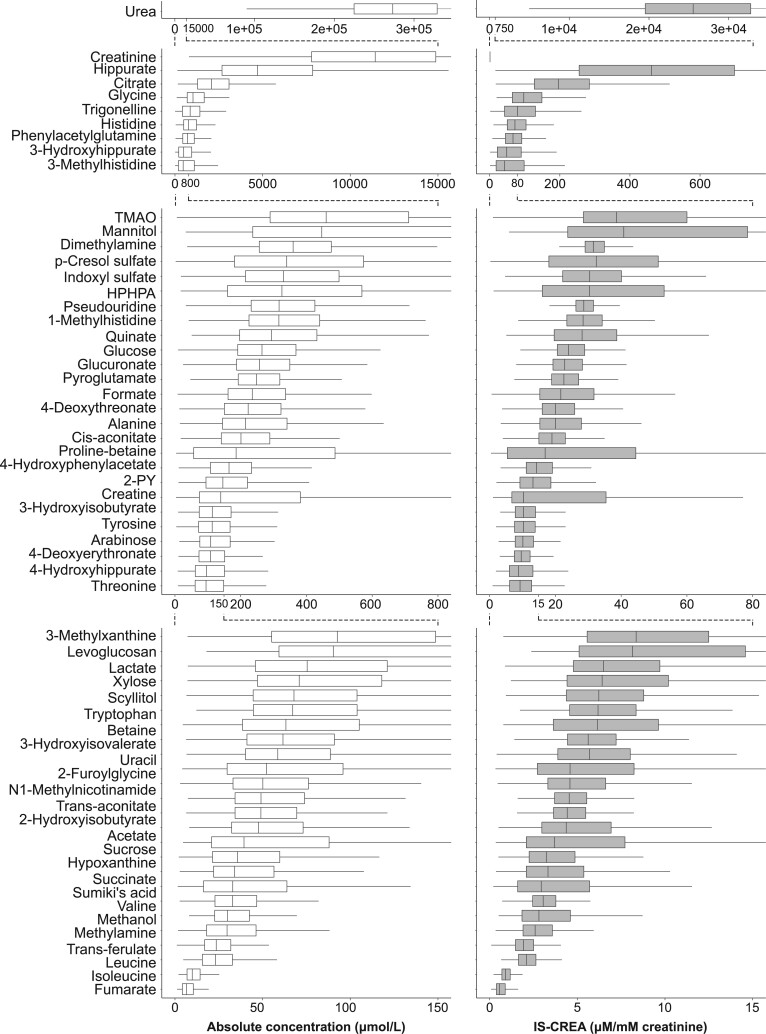
Figure 2.
Absolute (left) and creatinine-referenced (right) concentrations of 61 quantified urinary metabolites in a random subset (n = 994) of morning spot urine samples in the Northern Finland Birth Cohort 1966. The metabolites are presented in the descending order of median absolute concentrations. Several different scales are used for the x-axes to provide a clear visualization for the large concentration ranges. TMAO, trimethylamine N-oxide; HPHPA, 3-(3-hydroxyphenyl)-3-hydroxypropanoate; 2-PY, N1-methyl-2-pyridone-5-carboxamide; IS-CREA, use the creatinine concentration as the internal standard
In this work we present automated quantification models for 12 urinary metabolites (+ creatinine) (Supplementary Figure S1, available as Supplementary data at IJE online). These models enabled coherent analyses of urinary metabolite concentrations in three independent population cohorts (up to 5989 participants), and replicated association analyses with wide-ranging clinical data. A representative set of 49 clinical and biochemical measures, 17 of which are based on serum NMR metabolomics,7,10,19,20 were chosen for the association analyses. For the details of the urine sample preparation and the NMR spectroscopy experimentation, we refer to earlier open-access publications.7,17
Metabolite quantification and analytical issues
The 61 urinary metabolites (Figure 2) were quantified from the NMR spectra with a semi-automated methodology using sophisticated constrained total line shape (CTLS) fitting analysis.7,21,22 These analyses are tedious and time consuming, and are not feasible in large-scale epidemiology applications. This is the very reason why we started to develop an automated regression analysis approach for urine NMR metabolomics,7 as this approach has proved superior in the case of quantitative serum NMR metabolomics, with current data available for a plethora of various epidemiological and genetic applications and spanning to 1.5 million samples and counting.10,13,14,23,24
Additional boxplots (Supplementary Figure S2, available as Supplementary data at IJE online) and histograms (Supplementary Figures S3 and S4, available as Supplementary data at IJE online) are available in the online Supplementary data for the 61 urinary metabolites. Importantly, the concentration and distribution data for the random subset match very well to the corresponding data in the entire cohort of 4505 participants (Supplementary Figures S5–S8, available as Supplementary data at IJE online). The numerical data for the urinary metabolite concentrations in men and women for the NFBC1966 subset (60 metabolites + creatinine) can be found in Supplementary Table S5 (available as Supplementary data at IJE online), and for all the three cohorts (12 metabolites + creatinine) in Supplementary Table S6 (available as Supplementary data at IJE online).
Results from only two automated regression models, glucose and creatinine, have been published previously.1,7 In this work we add 11 automated quantification models, namely 2-hydroxyisobutyrate, valine, alanine, pseudouridine, dimethylamine, glycine, citrate, urea, formate, trigonelline and hippurate. Assessments of the automated regression models for these metabolites are available in Supplementary Figure S1 (available as Supplementary data at IJE online) together with the population distributions for ∼4500 individuals in NFBC1966.
In this study we also determined the correction coefficients for each quantified metabolite, to lead to the true absolute metabolite concentrations (Supplementary Table S7, available as Supplementary data at IJE online).
Statistical analyses
Urinary metabolite concentrations normalized to urinary creatinine concentration were used in all analyses, but absolute concentrations are also presented (Figure 2). Referencing to creatinine has been a long-term standard choice in urine NMR metabolomics, but we have also recently comprehensively studied the effects of various normalization methods, also supporting the use of creatinine referencing in epidemiological studies.17
Partial rank correlations (adjusted for sex in all the cohorts and in addition for age in YFS) were used to illustrate the associations between the 12 automatically quantified urinary metabolites and the 49 clinical and biochemical measures. The results are shown in colour-coded heat maps in Figure 3 for the biggest individual cohort, NFBC1966, with separate maps for the entire cohort (n = 4505) as well as for men (n = 1950) and women (n = 2555). The two-dimensional hierarchical clustering is based on the results for the entire cohort, and the resulting ordering is preserved in all the following heat maps. The chemical taxonomy of the metabolites can be seen in Table 2.17 Replication of the associations is illustrated in Figure 4. In all, 56 principal components explained over 99% of variation in the 60 creatinine-referenced urinary metabolite concentrations and the 49 clinical and biochemical measures in NFBC1966. Therefore, we used a multiple comparison corrected P-value threshold of 0.0009 to suggest evidence in favour of an association (0.05/56 via the Bonferroni method; P <0.0009 is denoted in the figures with an asterisk).
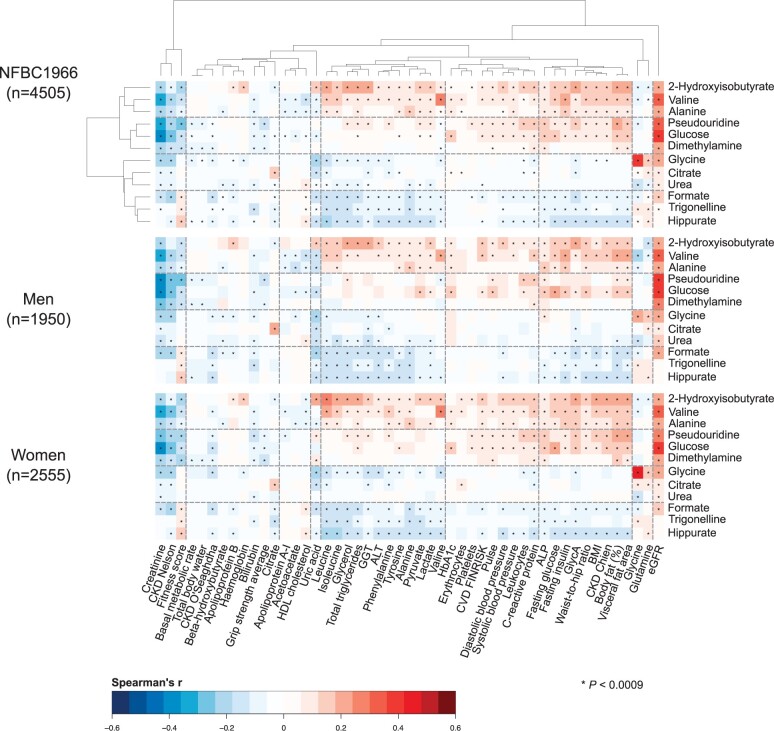
Figure 3.
The associations between the 12 automatically quantified urinary metabolites (referenced to urinary creatinine) and 49 customary clinical and biochemical measures as indicated by Spearman’s rank correlations (adjusted for sex) for the entire Northern Finland Birth Cohort 1966 (n = 4505) as well as for men (n = 1950) and women (n = 2555). The two-dimensional hierarchical clustering is based on the results for the entire cohort, and the resulting ordering is preserved in all the following heat maps. Four three-metabolite clusters were rendered that reflect the clinical and biochemical associations of the urinary metabolites. P-value <0.0009 is marked with an asterisk in the map to indicate a multiple testing corrected association. ALP, alkaline phosphatase; ALT, alanine aminotransferase; GGT, gamma-glutamyl transferase; eGFR, estimated glomerular filtration rate; CKD, chronic kidney disease; HbA1c, glycated haemoglobin; BMI, body mass index; GlycA, glycoprotein acetyls; FINRISK, a large Finnish population survey of risk factors for chronic, noncommunicable diseases
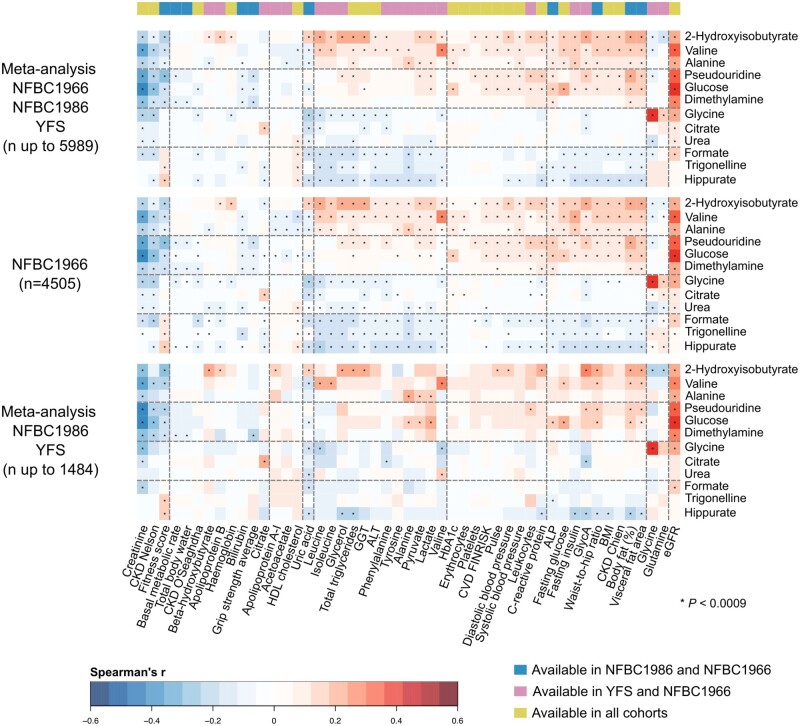
Figure 4.
Meta-analyses of the associations (Spearman’s rank correlations adjusted for sex) between the 12 automatically quantified urinary metabolites (referenced to urinary creatinine) and 49 customary clinical and biochemical measures, to illustrate the replication of the findings in all the three independent population cohorts. The uppermost heat map shows the full meta-analyses for all the available data (n up to 5989). The heat map in the middle is for the entire NFBC1966 (the same heat map as in Figure 3, to facilitate visual comparison). The lowermost heat map shows the meta-analysis for NFBC1986 and YFS (n up to 1484). The heat maps are presented in the same order of metabolites and clusters as in Figure 3. The colour key on the top of the figure represents the availability of clinical and biochemical measures in the three cohorts. There were 20 measures available in all three cohorts (green), 19 measures available only in NFBC1966 and YFS (pink) and 10 measures available only in NFBC1966 and NFBC1986 (blue). P-value <0.0009 is marked with an asterisk in the map to indicate a multiple testing corrected association. ALP, alkaline phosphatase; ALT, alanine aminotransferase; GGT, gamma-glutamyl transferase; eGFR, estimated glomerular filtration rate; CKD, chronic kidney disease; HbA1c, glycated haemoglobin; BMI, body mass index; GlycA, glycoprotein acetyls; FINRISK, a large Finnish population survey of risk factors for chronic, noncommunicable diseases; NFBC, Northern Finland Birth Cohort; YFS, Cardiovascular Risk in Young Finns Study
Associations between the urinary metabolites and body mass index (BMI), as well as smoking history (current smokers vs non-smokers), were analysed via linear regression analyses adjusted for sex in all the cohorts and in addition for age in YFS. Extreme metabolite levels (metabolites >third quartile + 8 * interquartile range) were truncated to the values of the upper bound, and the metabolite concentrations were log-transformed. The truncation was done as a precaution, since the extreme values are rare (Supplementary Table S8, available as Supplementary data at IJE online), represent real metabolite concentrations (not artefacts) and did not have strong effects on the associations. All measures were scaled to standard deviation (SD) units (by subtracting the mean and dividing by the standard deviation). Association magnitudes are reported in SD units to ease the comparison across multiple measures with different initial units and scales. All models were further individually adjusted for mean arterial pressure, fasting glucose, fasting insulin, smoking history (BMI analysis only), total triglycerides, C-reactive protein (CRP), estimated glomerular filtration rate (eGFR) and BMI (smoking analysis only). Individual adjustments for the confounders were performed to understand the relations and potential mediation of the urinary metabolite associations with the clinical and biochemical measures and outcomes. All analyses were done separately in the individual cohorts (Supplementary Figure S9, available as Supplementary data at IJE online, for BMI and Supplementary Figure S10, available as Supplementary data at IJE online, for smoking) and then meta-analysed (Figure 5A for BMI and Figure 5B for smoking).
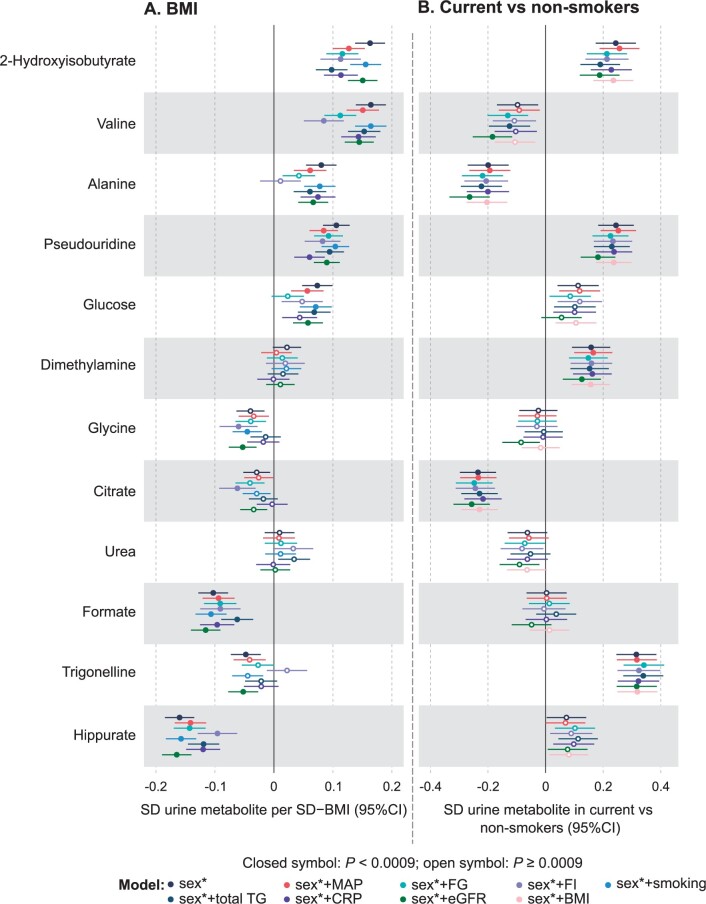
Figure 5.
Meta-analyses of the regression models for body mass index (A) and smoking (B) with the 12 automatically quantified urinary metabolites (referenced to creatinine). The effects of sex (black), sex + MAP (red), sex + fasting glucose (cyan), sex + fasting insulin (lila), sex + smoking (light blue, applied to the BMI models only), sex + total triglycerides (blue), sex + CRP (violet), sex + eGFR (green) and sex + BMI (blush, applied to the smoking models only) were examined; asterisk indicates that age was also adjusted for YFS. The smoking data for the cohorts are: NFBC1966, 750 current and 3544 non-smokers; NFBC1986, 115 current and 706 non-smokers; and YFS, 85 current and 370 non-smokers. MAP, mean arterial pressure; CRP, C-reactive protein; eGFR, estimated glomerular filtration rate; YFS, Cardiovascular Risk in Young Finns Study; NFBC, Northern Finland Birth Cohort
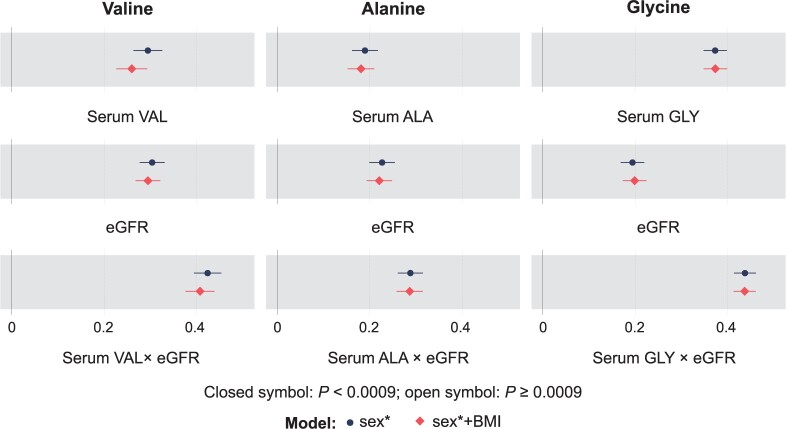
Three regression analyses for each of the three urinary amino acid concentrations (valine, alanine and glycine) were also performed (as in our previous work for glucose1), namely with their corresponding serum concentrations, eGFR and the multiplication of serum concentration and eGFR. The models were further adjusted for BMI. These data were available for NFBC1966 and YFS, so the analyses were first done separately in these two cohorts (Supplementary Figure S11, available as Supplementary data at IJE online) and then meta-analysed (Figure 6).
Figure 6.
Meta-analyses of the regression models for the three automatically quantified urinary amino acids (valine, alanine and glycine) concentrations (referenced to creatinine) and their corresponding serum concentrations, eGFR, and the multiplication of the serum concentration and eGFR in Northern Finland Birth Cohort 1966 (n = 4505) and Cardiovascular Risk in Young Finns Study (n = 474). The effects of sex (black circle) and sex + BMI (red diamond) were examined; asterisk indicates that age was also adjusted for in YFS. eGFR, estimated glomerular filtration rate; YFS, Cardiovascular Risk in Young Finns Study
Results
Metabolite distributions, abundance and sex differences
Figure 2 illustrates the absolute and urine creatinine-referenced concentrations of 61 quantified urinary metabolites in 994 morning spot urine samples. The set of 61 metabolites presented here represents all the most abundant signals in the urine NMR spectra. Urea is by far the most abundant metabolite, with a median absolute concentration greater than 200 mM. Creatinine is also an abundant metabolite, with a median absolute concentration greater than 10 mM. Hippurate and citrate are present in median absolute concentrations greater than 1 mM. The population variation of the metabolites is substantial (Figure 2, Table 2 and Supplementary Table S4, available as Supplementary data at IJE online) and many of the metabolite distributions are positively skewed (Supplementary Figures S2–S4, available as Supplementary data at IJE online). In creatinine-referenced data, many metabolites are slightly more abundant in women than in men, but the concentration differences are small (Supplementary Figures S2–S4 and Supplementary Table S5, available as Supplementary data at IJE online).
The methodological intra-assay metabolite CV%s for the new 17 metabolites are similar to those for the earlier 43 metabolites, i.e. mostly less than 5%, indicating high consistency and accuracy of urine NMR spectroscopy per se. Also their 30-day consecutive intra-individual and inter-individual population variations follow the same overall pattern as reported earlier,7 with rather large intra-individual and typically even larger inter-individual variation (Table 2 and Supplementary Table S4, available as Supplementary data at IJE online). Figure 2, together with the detailed information in Supplementary Table S5 (available as Supplementary data at IJE online), provide valuable reference concentrations for key urinary metabolites at a population level.
Association clusters
Figure 3 illustrates a colour-coded heat map of associations between 12 urinary metabolites (referenced to urinary creatinine) and 49 customary clinical and biochemical measures (detailed descriptions are available in online Supplementary data). The associations depicted are mostly novel, since quantitative data on urinary metabolites at an epidemiological scale are scarce. Even though the associations are overall rather weak, 362 associations were detected which fulfilled the statistical multiple comparison corrected P-value threshold of 0.0009. To facilitate the metabolic interpretation of the results, a two-dimensional hierarchical clustering of the heat map was done based on the sex-adjusted associations in the entire NFBC1966 cohort (n = 4505). Four three-metabolite clusters were rendered which reflect the clinical and biochemical associations of the 12 urinary metabolites.
The strongest association appears between urinary and serum glycine, and the association between urinary valine and serum valine is rather strong. The estimated glomerular filtration rate (eGFR) associates positively with all the urinary metabolite clusters, the strongest associations being with glucose, pseudouridine and valine. These associations are, as expected, mirrored by negative associations with serum creatinine (which is used in the estimation of glomerular filtration rate). The two uppermost metabolite clusters in Figure 3 (the first one consisting of 2-hydroxyisobutyrate, valine and alanine and the second one of pseudouridine, glucose and dimethylamine) behave generally similarly regarding their associations. They are overall positive with over 60% of the clinical and biochemical measures, including eGFR as noted above, multiple serum amino acids, serum triglycerides, glycaemic traits, lactate, pyruvate, inflammation (CRP and GlycA), liver function markers [alkaline phosphatase (ALP), alanine aminotransferase (ALT) and gamma-glutamyl transferase (GGT)], various obesity measures, blood pressure and the FINRISK cardiovascular disease (CVD) and the Chien chronic kidney disease (CKD) risk scores.
The associations of the two downmost metabolite clusters in Figure 3 (the first one consisting of glycine, citrate and urea and the second one of formate, trigonelline and hippurate) also behave generally similarly in their associations, though the former of these has the lowest number of associations with the clinical and biochemical measures. Their associations are, in general, negative for most of the above-mentioned positive associations of the two uppermost metabolite clusters. However, the associations are somewhat reversed for serum glycine and glutamine, with weak negative associations with the two uppermost metabolite clusters and mixed negative and positive associations with the two downmost clusters. In addition to serum creatinine, serum citrate, bilirubin and the CKD Nelson, as well as the O'Seaghdha risk scores, tend to associate negatively with all the urinary metabolite clusters and metabolites. Apolipoprotein B associates positively with the uppermost urinary metabolite cluster and negatively with the downmost cluster. The associations for high-density lipoprotein (HDL) cholesterol are opposite to those of apolipoprotein B. The fitness score associates negatively with the two uppermost metabolite clusters and positively with the lowermost cluster.
The associations of the urinary metabolite clusters and of the individual urinary metabolites with the clinical and biochemical measures are similar for men and women (Figure 3). In addition, the associations appear coherent between three independent cohorts, as illustrated in Figure 4. The NFBC1986 (n = 1010) and YFS (n = 474) had fewer urine samples available than the NFBC1966 (n = 4505), and thus only the most prominent associations reach the multiple comparison corrected P-value threshold of 0.0009. Nevertheless, the entire association pattern matches excellently with the one for NFBC1966.
Metabolite associations with BMI
The associations of the 12 urinary metabolites, referenced to creatinine, with BMI are shown in Figure 5A, meta-analysed for the three independent cohorts (n up to 5989). The results for the individual cohorts are shown in Supplementary Figure S9 (available as Supplementary data at IJE online). Only urea and dimethylamine did not associate with BMI. Fasting insulin had a strong effect on the associations of several urinary metabolites with BMI. Apart from the effects of fasting insulin, the other adjustments in the regression models had overall very little, if any, effects on the associations. However, adjusting for fasting glucose had a similar but less pronounced effect on valine and alanine as fasting insulin. Adjusting for fasting glucose also had (an expected) strong effect on diluting the association of urinary glucose with BMI. The association of 2-hydroxyisobutyrate was rather strongly affected by adjusting for mean arterial pressure, fasting glucose, fasting insulin, total triglycerides and CRP.
Metabolite associations with smoking
The associations of the 12 urinary metabolites referenced to creatinine with smoking are shown in Figure 5B, meta-analysed for the three independent cohorts (n up to 5989). The results for the individual cohorts are shown in Supplementary Figure S10 (available as Supplementary data at IJE online). Seven metabolites associated with smoking at the multiple testing corrected P-value threshold <0.0009, namely 2-hydroxyisobutyrate, valine, alanine, pseudouridine, dimethylamine, citrate and trigonelline. The various adjustments had very little, if any, effect on the urinary metabolite associations with smoking.
Unspecific renal excretion of amino acids
The 12 quantified urinary metabolites include three amino acids, valine, alanine and glycine. Figure 6 illustrates how these urinary amino acid concentrations associate with corresponding serum concentrations and eGFR, and that these associations are strengthened for the multiplication of the serum concentration and eGFR. The results shown are meta-analysed for the NFBC1966 and YFS, for which the serum amino acid data were available. Results for the individual cohorts are given in Supplementary Figure S11 (available as Supplementary data at IJE online).
Discussion
Novel quantitative data are presented here for 60 urinary metabolites (+ creatinine) in a 994-individual subset of the NFBC1966 cohort. This random subset is representative of the entire cohort of 4505 participants (Supplementary Figures S5–S8, available as Supplementary data at IJE online) and thus the quantitative data provide valuable reference concentrations for key urinary metabolites at a population level for men and women (Figure 2, Supplementary Figures S2–S4 and Supplementary Table S5, available as Supplementary data at IJE online). The concentration differences between males and females are small, but the creatinine-referenced values tend to be slightly higher for women. On average, women have lower muscle mass, leading to lower concentrations of circulating creatinine and thus lower amounts of excreted creatinine into the urine. Whereas in random urine samples absolute metabolite concentrations would not be relevant, in the case of morning spot urine samples, as here and as reflected by Figure 2, the biological variability is to some extent reduced due to the corresponding times and conditions in which the urine has been accumulating in all the participants, i.e. overnight in mostly a fasting physiological state.25
The first coherent set of automated quantification models for 12 urinary metabolites (+ creatinine) is also presented in this work (Supplementary Figure S1, available as Supplementary data at IJE online). Application of these models made it feasible to analyse these metabolite concentrations in urine samples for almost 6000 people in three independent population cohorts, and to study their associations with a comprehensive set of 49 clinical and biochemical measures. Hierarchical clustering of these results revealed four three-metabolite clusters that comprehensively summarized their association patterns (Figure 3 and Figure 4). Most of the detected associations are novel, since no epidemiological studies have been carried out combining a comprehensive quantitative metabolomics approach in urine samples with an extensive set of attached clinical and biochemical data. Since urinary metabolites overall correlate weakly with systemic metabolic measures,7 urine samples are a potential source of unique metabolic information. In addition, quantitative data on specific urinary metabolites provide a direct individual measurement of kidney function and can potentially alleviate generalized approximations in estimated glomerular filtration rate, a known marker of ageing and cardiometabolic diseases.26,27
We noted recently that glucose in the urine is a normal phenomenon (though some renal physiology textbooks still claim otherwise) and that typical absolute glucose concentrations in urine are between 0.1 and 0.5 mmol/L.1 This phenomenon is likely a reflection of unspecific renal excretion of glucose also at low concentration ranges of circulating glucose.1,28 Amino acids—as well as glucose—are indispensable in human metabolism, and thus basically all amino acids filtered by the kidneys are also reabsorbed into the circulation (or used in the kidneys) via a set of specific amino acid transporters.29,30 The population distributions for all the nine quantified amino acids in the urine samples (Figure 2; and Supplementary Figures S2–S4 and Supplementary Table S5, available as Supplementary data at IJE online) are similar and resemble those of glucose.1 In fact, similarly to glucose, all individuals have amino acids in the urine. Thus, despite the high efficiency of the amino acid transporters, the large volume of plasma filtered would result in some unspecific leakage of amino acids into the urine.28,29 As in the case of glucose, the unspecific leakage is suggested for the amino acids by the rather strong correlations between their serum and urine concentrations (Figure 3 and Figure 4) as well as by the additional contribution of the eGFR to the urinary amino acid concentration (Figure 6). Since quantitative metabolic studies of urine samples at the population level are scarce, we know very little about these types of unspecific molecular processes in the kidneys and their potential role as population-level biomarkers for kidney function and/or disease risk.1
The multiple associations between urinary metabolites and various clinical and biochemical measures suggest that urine metabolites may well have general value as population-level health and disease biomarkers. For example, the urinary branched-chain amino acid (BCAA) valine associates positively to various obesity markers (e.g. BMI and waist-to-hip ratio, as well as body and visceral fat), clinical diabetes indicators and risk factors [glycated haemoglobin (HbA1c), fasting glucose, fasting insulin, and serum BCAAs valine, leucine and isoleucine), systemic inflammation (CRP and GlycA), serum triglycerides, blood pressure, liver function (ALP, ALT and GGT), and CKD as well as CVD risk (Figure 3). The direction of association is reversed to overall fitness and HDL-related measures. These findings are in accordance with findings related to circulating valine, and in general with serum BCAAs, concentrations. Apart from adding confirmatory data to systemic metabolic findings, a key aspiration in the case of urinary metabolites would be that, if added into the systemic metabolic risk assessment, they might be able to bring in additional information, directly reflecting kidney function. This is supported by our previous findings that urine and serum metabolites generally correlate weakly.7 Furthermore, in the regression models of smoking with the urinary metabolite concentrations, adjusting for various key systemic measures (e.g. CRP, blood pressure, BMI, fasting insulin) had very minor or no effects on the associated metabolites 2-hydroxyisobutyrate, valine, alanine, pseudouridine, dimethylamine, citrate and trigonelline (Figure 5B; and Supplementary Figure S10, available as Supplementary data at IJE online). This conclusion also applies to adjustments with eGFR, suggesting that the associations of urinary metabolites are typically such that they cannot be simply explained by a standard clinical estimate of kidney function. A similar conclusion regarding eGFR is valid for the urinary metabolite associations with BMI. However, contrary to the overall minor effects due to the adjustments, insulin had a clear effect on most of the associations with BMI (Figure 5A). These findings are in line with recent longitudinal finding on systemic metabolic ageing trends and obesity.31,32
The positive associations of urinary 2-hydroxyisobutyrate with BMI appear most broadly affected by the adjustments, including those for mean arterial pressure, fasting glucose, fasting insulin, total triglycerides and CRP; 2-hydroxyisobutyrate associated positively also with smoking, but none of the adjustments had a clear effect on the association. In general, the associations of 2-hydroxyisobutyrate are very similar to those of valine (Figure 3 and Figure 4), except that urinary valine associates negatively to smoking (Figure 5B). Potentially originating from gut microbial valine degradation, 2-hydroxyisobutyrate is a tertiary alcohol.4,33,34 Epidemiological data on this metabolite are almost completely lacking,35 except its urinary concentration has also previously been associated with BMI.4 In addition, a recent study in individuals with type 1 diabetes found urinary 2-hydroxyisobutyrate positively associated with the progression of diabetic nephropathy.8 All these finding suggest urinary 2-hydroxyisobutyrate concentrations being linked with insulin resistance.36,37 Some other associations have also been reported, e.g. 2-hydroxyisobutyrate being part of a ‘peculiar obese urinary metabotype’38 and associating with various issues of pregnancy.39,40 However, all these studies have applied orthogonal projections to latent structures discriminant analysis (OPLS-DA) supervised analyses that are well-known to lead to spurious findings, particularly when a lot of spectral data points are used as the basis for the classifications in very small datasets.9,41–46
The results discussed above in relation to 2-hydroxyisobutyrate demonstrate how urinary metabolomics can provide substantial scientific novelty. First, because comprehensive quantitative data on urinary metabolites from large-scale epidemiological studies are scarce, and second, because urine as a waste biofluid—tightly connected to the kidney function—provides a metabolic view that is interdependent with and complementing systemic metabolism. In addition, quantitative metabolite data are indispensable to avoid the common limitations of multivariate metabolomics applications (typically the use of OPLS-DA) that result in spurious findings, due to overtraining of classification models with high numbers of variables (usually spectral data points) with very small numbers of individuals.9,20,41,45–47 Quantitative metabolite data (identical to data from standard clinical chemistry analyses) also provide easy means for confounding adjustments as applied in this work48 and replication of the findings, in this case done in up to three independent cohorts. These are essential elements allowing triangulation49 and leading to scientific reliability.50
Even though urinary metabolites intrinsically disclose what we eat and drink, the search for discriminatory molecular signals for individual nutrients or even dietary patterns has proved to be futile, 41 though some systemic metabolomics studies have been able to link multi-metabolic profiles of individuals with their habitual diets, for example, a preponderance of ‘fruits and vegetables’ or ‘junk food’.51 These types of questionnaire-independent ways of assessing true food consumption would be valuable in epidemiological studies. This applies also for explicit gauging of smoking. We showed here that seven metabolites (Figure 5B; and Supplementary Figure S10, available as Supplementary data at IJE online) associated with smoking (current vs non-smokers). These findings might prove valuable in large epidemiological studies as a questionnaire-independent assessment of smoking status.
Conclusion
It is fundamental to keep in mind with these observational epidemiological results that they cannot apprise of any mechanisms. In addition, we did not have direct measures of organ function available, which limits our ability to assess the potential clinical utility of the new metabolites compared with established biomarkers. However, this work gives one of the first demonstrations of the rich and diverse association pattern of urinary metabolites, with multiple descriptors of the renal-cardiometabolic system. Many associations of urinary metabolites with clinical outcomes appear independent of key systemic metabolic regulators, thus suggesting that quantitative urinary metabolomics may inherently convey rather specific information on tubular filtration and reabsorption, as well as on kidney-specific molecular interactions. Keeping in mind that urine is a waste product, a coherent metabolic association pattern in three independent population cohorts is an intriguing result per se. These large-scale results also point towards a very high analytical reliability of the new quantitative methodology, as we noted earlier, based on biologically relevant genetic associations with only a few hundred individuals.7 The presented extensive data and results give a dependable rationale to extend quantitative urinary metabolomics to large-scale epidemiological studies for new insight on kidney function and related metabolic disease biomarkers. All the three population cohorts studied in this work are from Finland. Hence, replication of the findings in other ethnicities and geographical locations would be valuable and also of high scientific interest, due to potential societal and environmental effects on urinary metabolite profiles and their clinical associations.
Ethics approval
The Northern Finland Birth Cohort 1966 and 1986 were approved by the Northern Ostrobothnia Hospital District, Finland. The Cardiovascular Risk in Young Finns Study was approved by the five universities with medical schools in Finland that were involved in the study (Turku, Helsinki, Tampere, Kuopio and Oulu). All participants gave written informed consent.
Supplementary Material
Contributor Information
Tianqi Li, Systems Epidemiology, Faculty of Medicine, University of Oulu, Oulu, Finland; Research Unit of Population Health, Faculty of Medicine, University of Oulu, Oulu, Finland; Biocenter Oulu, University of Oulu, Oulu, Finland.
Andrei Ihanus, Systems Epidemiology, Faculty of Medicine, University of Oulu, Oulu, Finland; Research Unit of Population Health, Faculty of Medicine, University of Oulu, Oulu, Finland; Biocenter Oulu, University of Oulu, Oulu, Finland; NMR Metabolomics Laboratory, School of Pharmacy, University of Eastern Finland, Kuopio, Finland.
Pauli Ohukainen, Systems Epidemiology, Faculty of Medicine, University of Oulu, Oulu, Finland; Research Unit of Population Health, Faculty of Medicine, University of Oulu, Oulu, Finland; Biocenter Oulu, University of Oulu, Oulu, Finland.
Marjo-Riitta Järvelin, Research Unit of Population Health, Faculty of Medicine, University of Oulu, Oulu, Finland; Unit of Primary Health Care, Oulu University Hospital, OYS, Oulu, Finland; Department of Epidemiology and Biostatistics, MRC-PHE Centre for Environment and Health, Imperial College London, London, UK; Department of Life Sciences, College of Health and Life Sciences, Brunel University London, London, UK.
Mika Kähönen, Department of Clinical Physiology, Tampere University Hospital, and Finnish Cardiovascular Research Center Tampere, Tampere University, Tampere, Finland.
Johannes Kettunen, Systems Epidemiology, Faculty of Medicine, University of Oulu, Oulu, Finland; Research Unit of Population Health, Faculty of Medicine, University of Oulu, Oulu, Finland; Biocenter Oulu, University of Oulu, Oulu, Finland; Department of Public Health and Welfare, Finnish Institute for Health and Welfare, Helsinki, Finland.
Olli T Raitakari, Research Centre of Applied and Preventive Cardiovascular Medicine, University of Turku, Turku, Finland; Centre for Population Health Research, University of Turku and Turku University Hospital, Turku, Finland; Department of Clinical Physiology and Nuclear Medicine, Turku University Hospital, Turku, Finland.
Terho Lehtimäki, Department of Clinical Chemistry, Fimlab Laboratories, and Finnish Cardiovascular Research Center Tampere, Tampere University, Tampere, Finland.
Ville-Petteri Mäkinen, Systems Epidemiology, Faculty of Medicine, University of Oulu, Oulu, Finland; Research Unit of Population Health, Faculty of Medicine, University of Oulu, Oulu, Finland; Biocenter Oulu, University of Oulu, Oulu, Finland.
Tuulia Tynkkynen, Systems Epidemiology, Faculty of Medicine, University of Oulu, Oulu, Finland; Research Unit of Population Health, Faculty of Medicine, University of Oulu, Oulu, Finland; Biocenter Oulu, University of Oulu, Oulu, Finland; NMR Metabolomics Laboratory, School of Pharmacy, University of Eastern Finland, Kuopio, Finland.
Mika Ala-Korpela, Systems Epidemiology, Faculty of Medicine, University of Oulu, Oulu, Finland; Research Unit of Population Health, Faculty of Medicine, University of Oulu, Oulu, Finland; Biocenter Oulu, University of Oulu, Oulu, Finland; NMR Metabolomics Laboratory, School of Pharmacy, University of Eastern Finland, Kuopio, Finland.
Data availability
The datasets used in the current study are available from the cohorts through the application process for researchers who meet the criteria for access to confidential data: [https://www.oulu.fi/nfbc/] and [http://youngfinnsstudy.utu.fi]. Regarding the YFS data, the Ethics Committee has concluded that under applicable law, the data from this study cannot be stored in public repositories or otherwise made publicly available. The data controller may permit access on case-by-case basis for scientific research, not however to individual participant-level data but to aggregated statistical data, which cannot be traced back to the individual participants' data.
Supplementary data
Supplementary data are available at IJE online.
Author contributions
T.L. performed the epidemiological statistical analyses, interpreted the results and wrote parts of the manuscript. T.T. performed the NMR experiments and the quantitative spectral CTLS fitting analyses, interpreted the results and wrote parts of the manuscript. A.I. designed and performed the quantitative regression modelling of the urinary metabolite concentrations. M-R.J., M.K., O.T.R., T.L. and M.A-K. provided samples and the phenotype data for the epidemiological cohorts. V.-P.M. interpreted the results and contributed to the statistical analyses of the data. M.A-K. conceived of and designed the study, interpreted the results and wrote the first version of the manuscript. All authors discussed the results, commented, contributed to the manuscript and approved the final version. M.A-K. supervised the study with help from T.T., P.O., J.K. and V-P.M. M.A-K. is the guarantor of the work.
Funding
Study funders: EU, Academy of Finland, Sigrid Juselius Foundation, Finnish Foundation for Cardiovascular Research, Emil Aaltonen Foundation, PREcisE, Turku University Foundation, Tampere University Hospital Supporting Foundation, Diabetes Research Foundation of Finnish Diabetes Association, Tampere Tuberculosis Foundation, the Finnish Cultural Foundation, Joint Programming Initiative a Healthy Diet for a Healthy Life (no. 655), UK Medical Research Council, Biotechnology and Biological Sciences Research Council (MR/S03658X/1), and European Regional Development Fund, (grant no. 539/2010 A31592) EU Horizon 2020 (grant 755320 for TAXINOMISIS) and European Research Council (grant 742927 for MULTIEPIGEN project). The study funders were not involved in the design of the study; the collection, analysis, and interpretation of data ;or writing the report; and did not impose any restrictions regarding the publication of the report.
Conflict of interest
None declared.
References
- 1. Li T, Ihanus A, Ohukainen P. et al. There is always glucose in normal urine: unspecific excretion associated with serum glucose and glomerular filtration rate. Int J Epidemiol. 2022;51:2022–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Holmes E, Loo RL, Stamler J. et al. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature 2008;453:396–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nicholson G, Rantalainen M, Maher AD. et al. Human metabolic profiles are stably controlled by genetic and environmental variation. Mol Syst Biol 2011;7:525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Elliott P, Posma JM, Chan Q. et al. Urinary metabolic signatures of human adiposity. Sci Transl Med 2015;7:285ra62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Emwas A-H, Roy R, McKay RT. et al. Recommendations and standardization of biomarker quantification using NMR-based metabolomics with particular focus on urinary analysis. J Proteome Res 2016;15:360–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. van Duynhoven JPM, Jacobs DM.. Assessment of dietary exposure and effect in humans: the role of NMR. Prog Nucl Magn Reson Spectrosc 2016;96:58–72. [DOI] [PubMed] [Google Scholar]
- 7. Tynkkynen T, Wang Q, Ekholm J. et al. Proof of concept for quantitative urine NMR metabolomics pipeline for large-scale epidemiology and genetics. Int J Epidemiol 2019;48:978–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mutter S, Valo E, Aittomäki V. et al. Urinary metabolite profiling and risk of progression of diabetic nephropathy in 2670 individuals with type 1 diabetes. Diabetologia 2022;65:140–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ala-Korpela M, Davey Smith G.. Metabolic profiling–multitude of technologies with great research potential, but (when) will translation emerge? Int J Epidemiol 2016;45:1311–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Würtz P, Kangas AJ, Soininen P, Lawlor DA, Davey Smith G, Ala-Korpela M.. Quantitative serum nuclear magnetic resonance metabolomics in large-scale epidemiology: a primer on -omic technologies. Am J Epidemiol 2017;186:1084–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huynh K, Barlow CK, Jayawardana KS. et al. High-throughput plasma lipidomics: detailed mapping of the associations with cardiometabolic risk factors. Cell Chem Biol 2019;26:71–84.e4. [DOI] [PubMed] [Google Scholar]
- 12. Deelen J, Kettunen J, Fischer K. et al. A metabolic profile of all-cause mortality risk identified in an observational study of 44,168 individuals. Nat Commun 2019;10:3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Locke AE, Steinberg KM, Chiang CWK. et al. ; FinnGen Project. Exome sequencing of Finnish isolates enhances rare-variant association power. Nature 2019;572:323–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ala-Korpela M, Zhao S, Järvelin M-R, Mäkinen V-P, Ohukainen P.. Apt interpretation of comprehensive lipoprotein data in large-scale epidemiology: disclosure of fundamental structural and metabolic relationships. Int J Epidemiol 2022;51:996–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ekholm J, Ohukainen P, Kangas AJ. et al. EpiMetal: an open-source graphical web browser tool for easy statistical analyses in epidemiology and metabolomics. Int J Epidemiol 2020;49:1075–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bouatra S, Aziat F, Mandal R. et al. The human urine metabolome. PLoS One 2013;8:e73076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li T, Tynkkynen T, Ihanus A, Zhao S, Mäkinen V-P, Ala-Korpela M.. Characteristics of normalization methods in quantitative urinary metabolomics—implications for epidemiological applications and interpretations. Biomolecules 2022;12:903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nordström T, Miettunen J, Auvinen J. et al. Cohort Profile: 46 years of follow-up of the Northern Finland Birth Cohort 1966 (NFBC1966). Int J Epidemiol 2022;50:1786–87j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Soininen P, Kangas AJ, Würtz P. et al. High-throughput serum NMR metabonomics for cost-effective holistic studies on systemic metabolism. Analyst 2009;134:1781–85. [DOI] [PubMed] [Google Scholar]
- 20. Soininen P, Kangas AJ, Würtz P, Suna T, Ala-Korpela M.. Quantitative serum nuclear magnetic resonance metabolomics in cardiovascular epidemiology and genetics. Circ Cardiovasc Genet 2015;8:192–206. [DOI] [PubMed] [Google Scholar]
- 21. Mierisová S, Ala-Korpela M.. MR spectroscopy quantitation: a review of frequency domain methods. NMR Biomed 2001;14:247–59. [DOI] [PubMed] [Google Scholar]
- 22. Soininen P, Haarala J, Vepsäläinen J, Niemitz M, Laatikainen R.. Strategies for organic impurity quantification by 1H NMR spectroscopy: constrained total-line-shape fitting. Anal Chim Acta 2005;542:178–85. [Google Scholar]
- 23. Julkunen H, Cichońska A, Slagboom PE, Würtz P; Nightingale Health UK Biobank Initiative. Metabolic biomarker profiling for identification of susceptibility to severe pneumonia and COVID-19 in the general population. Elife 2021;10:e63033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Julkunen H, Cichońska A, Tiainen M. et al. Atlas of plasma NMR biomarkers for health and disease in 118,461 individuals from the UK Biobank. Nat Commun 2023;14:604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vart P, Grams ME.. Measuring and assessing kidney function. Semin Nephrol 2016;36:262–72. [DOI] [PubMed] [Google Scholar]
- 26. Manjunath G, Tighiouart H, Ibrahim H. et al. Level of kidney function as a risk factor for atherosclerotic cardiovascular outcomes in the community. J Am Coll Cardiol 2003;41:47–55. [DOI] [PubMed] [Google Scholar]
- 27. Sarnak MJ, Amann K, Bangalore S. et al. ; Conference Participants. Chronic kidney disease and coronary artery disease: JACC state-of-the-art review. J Am Coll Cardiol 2019;74:1823–38. [DOI] [PubMed] [Google Scholar]
- 28. Vallon V. Glucose transporters in the kidney in health and disease. Pflugers Arch 2020;472:1345–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li X, Zheng S, Wu G.. Amino acid metabolism in the kidneys: nutritional and physiological significance. Adv Exp Med Biol 2020;1265:71–95. [DOI] [PubMed] [Google Scholar]
- 30. Verrey F, Singer D, Ramadan T, Vuille-dit-Bille RN, Mariotta L, Camargo SMR.. Kidney amino acid transport. Pflugers Arch 2009;458:53–60. [DOI] [PubMed] [Google Scholar]
- 31. Mäkinen V-P, Karsikas M, Kettunen J. et al. Longitudinal profiling of metabolic ageing trends in two population cohorts of young adults. Int J Epidemiol 2022;51:1970–83. [DOI] [PubMed] [Google Scholar]
- 32. Mäkinen V-P, Kettunen J, Lehtimäki T. et al. Longitudinal metabolomics of increasing body-mass index and waist-hip ratio reveals two dynamic patterns of obesity pandemic. Int J Obes (Lond) 2023;47:453–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Miccheli A, Capuani G, Marini F. et al. Urinary (1)H-NMR-based metabolic profiling of children with NAFLD undergoing VSL#3 treatment. Int J Obes (Lond) 2015;39:1118–25. [DOI] [PubMed] [Google Scholar]
- 34. Schifano E, Conta G, Preziosi A. et al. 2-hydroxyisobutyric acid (2-HIBA) modulates ageing and fat deposition in Caenorhabditis elegans. Front Mol Biosci 2022;9:986022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wishart DS, Feunang YD, Marcu A. et al. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res 2018;46:D608–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lotta LA, Gulati P, Day FR. et al. ; Cambridge FPLD1 Consortium. Integrative genomic analysis implicates limited peripheral adipose storage capacity in the pathogenesis of human insulin resistance. Nat Genet 2017;49:17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang Q, Holmes MV, Davey Smith G, Ala-Korpela M.. Genetic support for a causal role of insulin resistance on circulating branched-chain amino acids and inflammation. Diabetes Care 2017;40:1779–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Calvani R, Miccheli A, Capuani G. et al. Gut microbiome-derived metabolites characterize a peculiar obese urinary metabotype. Int J Obes (Lond) 2010;34:1095–98. [DOI] [PubMed] [Google Scholar]
- 39. Diaz SO, Pinto J, Graça G. et al. Metabolic biomarkers of prenatal disorders: an exploratory NMR metabonomics study of second trimester maternal urine and blood plasma. J Proteome Res 2011;10:3732–42. [DOI] [PubMed] [Google Scholar]
- 40. Gil AM, Duarte D, Pinto J, Barros AS.. Assessing exposome effects on pregnancy through urine metabolomics of a Portuguese (Estarreja) cohort. J Proteome Res 2018;17:1278–89. [DOI] [PubMed] [Google Scholar]
- 41. Ala-Korpela M. Objective metabolomics research. Clin Chem 2018;64:30–33. [DOI] [PubMed] [Google Scholar]
- 42. Ala-Korpela M. Critical evaluation of 1H NMR metabonomics of serum as a methodology for disease risk assessment and diagnostics. Clin Chem Lab Med 2008;46:27–42. [DOI] [PubMed] [Google Scholar]
- 43. Ala-Korpela M. Metabolomics in cardiovascular medicine: not personalised, not diagnostic. Eur J Prev Cardiol 2016;23:1821–22. [DOI] [PubMed] [Google Scholar]
- 44. Madsen R, Lundstedt T, Trygg J.. Chemometrics in metabolomics—a review in human disease diagnosis. Anal Chim Acta 2010;659:23–33. [DOI] [PubMed] [Google Scholar]
- 45. Kjeldahl K, Bro R.. Some common misunderstandings in chemometrics. J Chemom 2010;24:558–64. [Google Scholar]
- 46. Bevilacqua M, Bro R.. Can we trust score plots? Metabolites 2020;10:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ala-Korpela M, Kangas AJ, Soininen P.. Quantitative high-throughput metabolomics: a new era in epidemiology and genetics. Genome Med 2012;4:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Davey Smith G, Lawlor DA, Harbord R, Timpson N, Day I, Ebrahim S.. Clustered environments and randomized genes: a fundamental distinction between conventional and genetic epidemiology. PLoS Med 2007;4:e352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lawlor DA, Tilling K, Davey Smith G.. Triangulation in aetiological epidemiology. Int J Epidemiol 2016;45:1866–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ioannidis JPA. How to make more published research true. PLoS Med 2014;11:e1001747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bogl LH, Pietiläinen KH, Rissanen A. et al. Association between habitual dietary intake and lipoprotein subclass profile in healthy young adults. Nutr Metab Cardiovasc Dis 2013;23:1071–78. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used in the current study are available from the cohorts through the application process for researchers who meet the criteria for access to confidential data: [https://www.oulu.fi/nfbc/] and [http://youngfinnsstudy.utu.fi]. Regarding the YFS data, the Ethics Committee has concluded that under applicable law, the data from this study cannot be stored in public repositories or otherwise made publicly available. The data controller may permit access on case-by-case basis for scientific research, not however to individual participant-level data but to aggregated statistical data, which cannot be traced back to the individual participants' data.