Abstract
Streptococcus mutans glucosyltransferases (GTFs; GtfB, -C, and -D) synthesize water-soluble and -insoluble glucan polymers from sucrose. We have identified previously a conserved region of 19 amino acids (aa) (Gtf-P1; aa 409 to 427 of GtfB and aa 435 to 453 of GtfC) which is functionally important for both enzymatic activity and bacterial adherence. Monoclonal antibodies directed against Gtf-P1 selectively inhibited insoluble glucan synthesis by GtfB and -C but had no effect on soluble glucan synthesis by GtfD, suggesting that despite an apparent near identity of sequence, corresponding residues may function differently in these enzymes. To test this hypothesis, we used different strategies of mutagenesis to analyze amino acid residues of GtfB and GtfC in Gtf-P1. In-frame insertion of 6 amino acids preceding, or deletion of 14 amino acids within, this conserved region abolished the enzymatic activities of both GtfB and GtfC. Substitution of several residues in combination by random mutagenesis resulted in GtfB, but not GtfC, enzymes exhibiting decreased glucan synthesis and reduced rates of sucrose hydrolysis. Amino acid substitutions of Asp residues in GtfB or GtfC were found to be more critical for enzymatic activity than at other positions of this region. Interestingly, single mutation at Asp411 or Asp413 of GtfB resulted in enzymes retaining about 20% of wild-type activity, whereas mutagenesis of the corresponding Asp at position 437 or 439 in GtfC resulted in complete loss of enzymatic activity. Furthermore, single amino acid substitution of a Val residue between the two Asp residues enhanced the sucrase- and glucan-synthesizing activities of GtfB and GtfC. These results confirmed the report from another laboratory that Asp residues in the Gtf-P1 region are essential for enzymatic catalysis and provide new evidence that identical residues may function differently in closely related Gtf enzymes.
Glucosyltransferases (GTFs; EC 2.4.1.5) of mutans streptococci are enzymes responsible for the synthesis of water-soluble and -insoluble glucose polymers (glucans) from sucrose. These polysaccharides enhance the colonization of cariogenic bacteria and promote the formation of dental plaque on tooth surfaces (11). Genetic and biological analyses have identified several GTFs with distinct characteristics in Streptococcus mutans and Streptococcus sobrinus, the two mutans streptococci most frequently isolated from the human oral cavity (20). Three GTFs of S. mutans have been described (2, 12–14, 28, 33): two enzymes, GtfB (GTF-I; 162 kDa) and GtfC (GTF-SI; 149 kDa), that synthesize primarily insoluble glucan, and another, GtfD (GTF-S; 155 kDa), that synthesizes exclusively water-soluble glucan. GtfB and GtfC share a high degree of nucleotide and amino acid sequence similarity. GtfD is dependent on the acceptor for glucan synthesis; GtfB and -C are independent of the exogenous glucan acceptor, whereas their enzymatic activity is enhanced in the presence of dextran. Assays of adherence to glass surfaces in studies using an in vitro model suggest that GtfB and GtfC are more important than GtfD for bacterial attachment (7, 8). However, recent experiments in vivo indicated that all three enzymes are required for maximal cariogenesis in animal model systems (34).
The enzymatic activities of GTFs include sucrose hydrolysis and glucan synthesis, and they also bind glucan. Studies of the structure and functional relationships of the GTFs have identified several important domains. A carboxyl-terminal region composed of multiple, homologous direct repeat segments constitutes the glucan-binding domain (GBD) (16, 19). A deletion study using the gtfD gene has demonstrated that the GBD encompasses the C-terminal one-third, approximately 510 residues (19). A similar study of the gtf-I gene from S. sobrinus, or domain shuffling between GtfB and GtfD, showed that GBD was essential for glucan synthesis but not for sucrase activity (1, 24). The sucrose-binding domain, capable of binding and hydrolysis of sucrose, has been attributed to the N-terminal two-thirds of the GTFs. Mooser et al. (23) identified an active site of 9 amino acids (aa) in the N-terminal one-third of S. sobrinus GTFs by sequencing a peptide from the stabilized glucosyl-enzyme complex. This peptide is conserved in the GTFs from mutans streptococci and contains a putative catalytic aspartic acid which is common to a broad array of α-glucosidases and related transferases (23). Site-directed mutagenesis of the corresponding Asp residue in GtfB completely inactivated the enzyme (17). Additional analyses by mutagenesis of further N-terminal residues, which are conserved in GtfB and GtfD, have shown that substitution of a single amino acid can affect the structure of the glucan product synthesized by either enzyme (27). Taken together, results from these investigations suggested that the N-terminal one-third of the GTFs may play a central role in the enzymatic activities of sucrose splitting and glucan synthesis. However, the subdomains and amino acid residues directly involved in catalysis or regulating the incorporation of glucose residues into glucan polymers remain to be determined.
Using genetic and immunological approaches, we identified another N-terminal conserved region of 19 aa the sequence of which is almost identical among the GTFs of several mutans streptococci and of Streptococcus salivarius (1, 10, 14, 28, 29, 33). This region, corresponding to residues 409 to 427 of GtfB and 435 to 453 of GtfC, is located 19 residues upstream of the active site described by Mooser et al. (23) (GtfB residues 446 to 454; GtfC residues 472 to 480). Previous studies in our laboratory have shown this region is important for both enzymatic activity and sucrose-dependent adherence of S. mutans in vitro (4). More recently, we named the 19-aa region Gtf-P1 and found that it contains one of the major B-cell epitopes recognized in the natural human antibody response to S. mutans infection (6). A recent mutagenesis analysis of GtfB has found that Asp413 in the GtfB-P1 region is essential for the enzymatic catalysis (32). Moreover, the 19-aa Gtf-P1 region share complete sequence identity with another active-site peptide in the dextransucrase from Leuconostoc mesenteroides (22). However, aside from the nine-residue region proposed by Mooser et al. (23), Gtf-P1 shares no homology with other α-glucosidases or transferases such as α-amylase. Thus far, Gtf-P1 and homologous domains seem to be found specifically in the GTFs of prokaryotic organisms such as mutans streptococci, S. salivarius, and Leuconostoc species.
Previously we found that monoclonal antibodies (MAbs) directed against Gtf-P1 would inhibit the enzymatic activities of GtfB and GtfC but not that of GtfD, even though GtfD has an almost identical 19-aa sequence (4). We hypothesized that despite an apparent near identity of sequence, corresponding residues may function differently in these closely related GTFs. This report presents evidence to support this hypothesis and shows that two Asp residues in the Gtf-P1 region are essential for enzymatic catalysis of both GtfB and GtfC, but not to the same degree. Moreover, we also found that a single amino acid substitution in this region could enhance the enzymatic activities of both enzymes. These findings are novel and may shed light on the structure and function of the GTFs.
MATERIALS AND METHODS
Bacteria and plasmids.
Escherichia coli JM109 (35) and ES1301 mutS (Alter Sites II kit; Promega, Madison, Wis.) were used as plasmid hosts and for site-directed mutagenesis. Cultures were grown in Luria-Bertani (LB) medium (25) supplemented with ampicillin (100 μg/ml) or tetracycline (40 μg/ml) and/or agar (2%) as required. Plasmid pAlter-1 (Alter Sites II kit; Promega) was used for single- and multiple-site mutagenesis. Plasmids pYNB13, expressing GtfB, and pNH3, expressing GtfC, were kindly provided by H. K. Kuramitsu, State University of New York, Buffalo. Plasmid pNH3 contained the intact gtfC gene under control of its own promoter (13). Plasmid pYNB13 contained the gtfB gene under control of the lac repressor. All other plasmids were constructed for the present study; each contained the desired mutation (Table 1).
TABLE 1.
Plasmids used
| Gtf | Relevant characteristicsa |
|---|---|
| GtfB | pBluescript KS derivative containing intact gtfB (pYNB13)b |
| GtfC | pUC18 derivative containing intact gtfC (pNH3)c |
| pAlter1 derivative containing EcoRI-BamHI fragment of gtfC | |
| GtfB-In1 | In-frame insertion of 6 aa, FVDSRQ, between E405 and F406 of GtfB |
| GtfB-In2 | In-frame insertion of 6 aa, LSRVDR, between E405 and F406 of GtfB |
| GtfB-Dm1 | In-frame deletion of 14 aa from D414 to F427 of GtfB |
| GtfB-Dm2 | GtfB-Dm1 with additional aa substitution, L408S |
| GtfB-ms2 | GtfB with 2 aa substitutions, W426F and L427D |
| GtfB-ms3 | GtfB with 3 aa substitutions, L408S, W426F, and L427D |
| GtfB-ms4 | GtfB with 4 aa substitutions, L408S, D413E, W426F, and L427D |
| GtfB-ms41 | GtfB with 4 aa substitutions, L408S, A421L, W426F, and L427D |
| GtfB-ms5 | GtfB with 5 aa substitutions, L408S, D413E, A421L, W426F, and L427D |
| GtfB-D411N | GtfB with single aa conversion, D411N |
| GtfB-D413N | GtfB with single aa conversion, D413N |
| GtfB-D411/413N | GtfB with 2 aa conversions, D411N and D413N |
| GtfB-V412I | GtfB with single aa conversion, V412I |
| GtfB-E422Q | GtfB with single aa conversion, E422Q |
| GtfC-In1 | In-frame insertion of 6 aa, FVDSRQ, between E431 and F432 of GtfC |
| GtfC-In2 | In-frame insertion of 6 aa, LSRVDR, between E431 and F432 of GtfC |
| GtfC-Dm1 | In-frame deletion of 14 aa from D440 to F453 of GtfC |
| GtfC-Dm2 | In-frame deletion of 14 aa from D440 to F453 of GtfC with additional aa substitution, L434S |
| GtfC-ms3 | GtfC with 3 aa substitutions, L434S, W426F, and L427D |
| GtfC-ms4 | GtfC with 4 aa substitutions, L434S, D413E, W426F, and L427D |
| GtfC-ms41 | GtfC with 4 aa substitutions, L434S, A421L, W426F, and L427D |
| GtfC-ms5 | GtfC with 5 aa substitutions, L434S, D413E, A421L, W426F, and L427D |
| GtfC-D437N | GtfC with single aa conversion, D437N |
| GtfC-D439N | GtfC with single aa conversion, D439N |
| GtfC-V438I | GtfC with single aa conversion, V438I |
| GtfC-E448Q | GtfC with single aa conversion, E448Q |
All plasmids carried ampicillin resistance markers and are derivatives of pYNB13 or pNH3. E. coli JM109 was the host for expression of wild-type or mutated GtfB and -C.
pYNB13 contains gtfB under control of the lac promoter.
pNH3 contains the gtfC endogenous promoter which is recognized by E. coli (13). Unlisted constructs are GtfB-ms2R, -ms3R, -ms4R, -ms41R, and -ms5R and GtfC-ms3R, -ms4-R, -ms41R, and -ms5R (see text).
Oligonucleotide primers and mutagenesis.
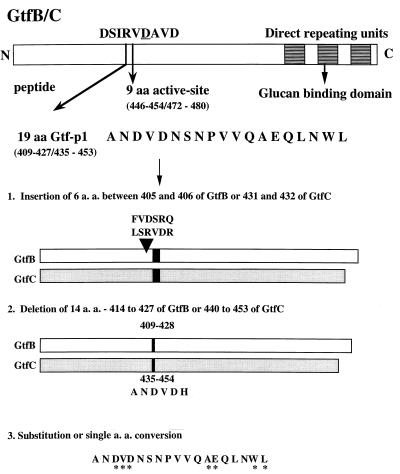
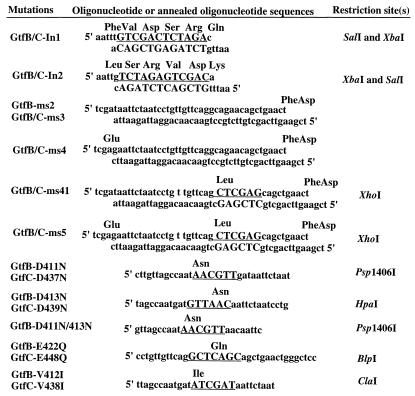
The amino acid sequence analyzed is shown in Fig. 1, and oligonucleotide primers designed for mutagenesis experiments are described in Fig. 2. Another primer (17 nucleotides), selected from a homologous region in both gtfB (1825 to 1841) and gtfC (1393 to 1409), was used as an internal sequencing primer. All primers were synthesized and purified by Clontech (Palo Alto, Calif.). The insertion mutants were constructed by ligation of an annealed oligonucleotide encoding 6 aa (three codons N terminal of the 19-aa region; chosen by unique restriction sites [Fig. 2] into the unique EcoRI site in gtfB and gtfC). Four GtfB and -C mutants were generated: GtfB-In1 and GtfB-In2, derived from GtfB; and GtfC-In1 and GtfC-In2, derived from GtfC. PCR with overlap extension (15) was used to generate deletion mutations GtfB-Dm1, GtfB-Dm2, and GtfC-Dm1. Substitution of amino acid residues between aa 414 to 427 of GtfB and aa 440 to 453 of GtfC was performed by ligation of the DNA fragments into the deletion mutants by using the SalI site (generated by PCR). Four DNA fragments containing the mutated sequences were derived by annealing the synthesized oligonucleotides with appropriate restriction sites (Fig. 2). Because the synthesized DNA fragments could also be ligated in either orientation, we generated additional mutated GtfBs (GtfB-ms2R to -ms5R) and GtfCs (GtfC-ms2R to -ms5R), with reversed amino acid sequences in addition to the substitutions. Site-directed mutagenesis was carried out with the Altered Sites II in vitro mutagenesis system as instructed by the manufacturer (Promega). The EcoRI-BamHI fragment of the gtfC gene was cloned into the pAlter-1 vector for single or additional rounds of mutagenesis using synthetic oligonucleotides. After nucleotide sequence analysis to confirm the desired mutations, the EcoRI-BamHI fragment was purified and cloned back into the gtfB and gtfC genes.
FIG. 1.
Amino acid sequence and positions analyzed by mutagenesis in GtfB and GtfC from S. mutans GS-5. The 19-aa Gtf-P1 (dark region) was identical in GtfB and GtfC. C-terminus direct repeats (shaded) contain 3.5 segments of 65 aa each in GtfB and 2 segments of 49 aa each in GtfC. The numbers refer to the positions of amino acids before and after mutagenesis. Asterisks mark amino acid residues substituted by random or site-directed mutagenesis.
FIG. 2.
Oligonucleotides used for mutagenesis. Substituted nucleotides and restriction sites are underlined. Altered amino acid residues are shown above the sequences of oligonucleotides, and changes in individual mutants are described in Table 1.
DNA sequence analysis.
The plasmid DNAs of PCR-derived deletion mutations, insertion mutations, and mutated gtfB and -C fragments were purified (Minipreps; Promega), denatured, and sequenced by the dideoxy-chain termination method (26), using Sequenase version 2 as instructed by the manufacturer (U.S. Biochemical, Cleveland, Ohio).
Detection of proteins.
E. coli JM109 transformants carrying wild-type or mutant gtfB or gtfC were cultured in LB broth with or without isopropyl-β-d-thiogalactopyranoside. Cells in the stationary phase of growth (optical density at 600 nm of 0.8) were harvested, washed, and suspended in extraction buffer (20 mM Tris hydrochloride buffer [pH 8.3] containing 1.0 mM phenylmethylsulfonyl fluoride and 2.5 mM EDTA). After disruption by sonication and centrifugation at 12,000 × g for 30 min, the supernatant was collected, dialyzed against sodium phosphate buffer (50 mM, pH 6.5), and used as the crude enzyme preparation. Proteins were analyzed by sodium dodecyl sulfate–7.5% polyacrylamide gel electrophoresis and then stained with Coomassie brilliant blue or subjected to Western immunoblotting as described previously, using anti-GtfB/C rabbit serum; proteins were detected by using alkaline phosphatase-labeled goat anti-rabbit immunoglobulin G, followed by a p-nitrophenyphosphate substrate (4, 5). Protein concentrations were determined by using a modification of the method of Lowry et al. (21), with bicinchoninic acid as the colorimetric detection reagent (BCA protein assay reagent; Pierce Chemical Co., Rockford, Ill.).
Enzyme assay.
Sucrase activity was determined by the Somogyi-Nelson procedure as described previously (31). To determine the rate of sucrose hydrolysis, fractions of reaction mixture, taken at different time intervals, were analyzed by high-pressure liquid chromatography (HPLC) combined with a pulsed ampherometric detection system (Dionex, Sunnyvale, Calif.) for estimation of the number of glucose and fructose molecules released. Standard glucose, fructose, fucose, and sucrose solutions were purchased from the manufacturers. The system detected the carbohydrates at a sensitivity as low as 1 pmol/ml.
Glucan-synthesizing activity was determined by the [14C]glucose-sucrose (NEN; New England Nuclear Corp., Boston, Mass.) incorporation assay as described previously (18). Briefly, the reaction mixture consisted of enzyme, 2.9 mM labeled (0.017 mCi/mmol) sucrose (final sucrose concentration, labeled plus unlabeled, was 20 mM), without or with dextran T10, and 0.10 M potassium phosphate buffer (pH 6.0) in a total volume of 0.5 ml. The reaction mixtures were incubated at 37°C for 1 h, and synthesis was terminated by the addition of 5 ml of methanol (for total glucan synthetic activity) or by heating at 100°C for 5 min (for insoluble glucan synthesis). The methanol-precipitated samples were filtered through a 2.4-cm-diameter glass fiber filter (Whatman, Maidstone, England) and washed three times with methanol. The heated samples were pipetted onto the filters and washed with 0.9% sodium chloride–methanol. Radioactivity was measured with a liquid scintillation counter (Beckman). One unit of enzyme activity is defined as the amount of enzyme required to incorporate 1.0 mmol of glucose from sucrose into glucan per minute under standard assay conditions. Crude enzymes of either GtfB, GtfC, or mutants with residual activities exhibited equivalent estimated specific activities (milliunits per milligram of protein) when different concentrations of crude preparations were assayed. For comparison, crude enzymes of individual wild-type and mutant were adjusted to 200 μg per assay, and relative amounts of the GTFs were similar, as judged from the intensities and molecular sizes of the bands on Western blots. The band intensities were quantified by scanning the blots (duplicate) and subsequently analyzed with the program NIH Image 1.6. For comparison between blots, intensities of prestained molecular size markers (Bio-Rad, Hercules, Calif.) and background intensities were used as internal controls for efficiency of transfer and development conditions in individual blots.
To determine the Km of GtfB or -C for sucrose, the enzyme (2 mU) was incubated in reaction mixture containing 1 to 30 mM sucrose plus [14C]glucose-sucrose in 0.1 M sodium phosphate buffer at 37°C for 1 h. The glucan synthesized was determined as described above. The substrate saturation kinetics were determined by using the Lineweaver-Burk double-reciprocal plot method in three replicate examinations.
RESULTS
Mutagenesis of gtfB and gtfC genes.
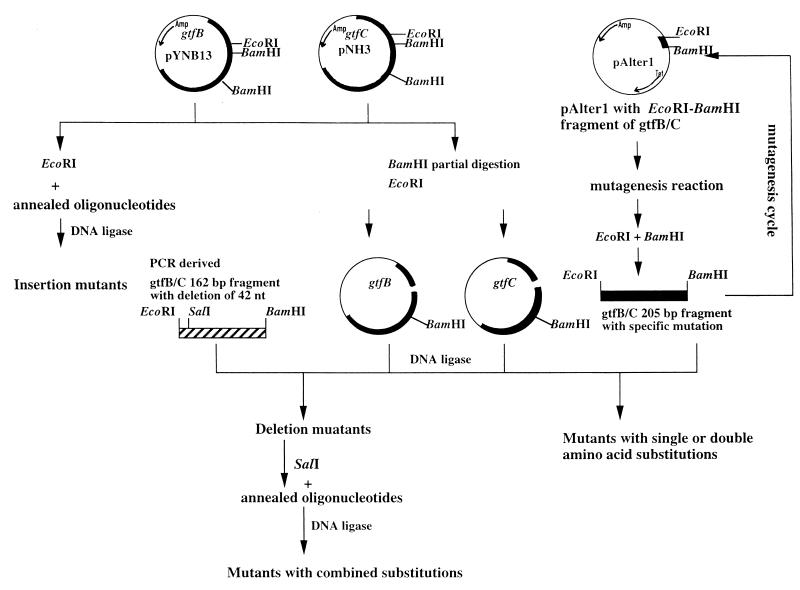
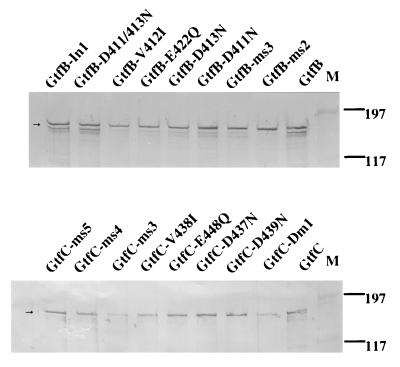
To investigate the specific role of the 19-aa Gtf-P1 region in the N-terminal third of the GTFs, we used a genetic approach involving mutagenesis of the gtfB and gtfC genes, including deletions, insertions, and substitutions (Fig. 1). Strategies for mutagenesis and the resultant constructs expressing the mutated GtfB or GtfC are summarized in Fig. 3. All GtfB and -C mutations were confirmed by DNA sequencing and Western blot analyses. Western blot analysis revealed no difference in the expression level or stability of the mutated enzymes expressed in E. coli (Fig. 4). The insertion mutations were constructed by ligation of an annealed oligonucleotide of 6 aa (Table 1; Fig. 2) into the unique EcoRI site in gtfB or gtfC. Because the DNA fragment could be ligated in both orientations, four mutants (GtfB-In1 and -In2, from GtfB, and GtfC-In1 and -In2, from GtfC) were generated. The deletion mutations GtfB-Dm1 and GtfC-Dm1 were obtained by cloning of the PCR-generated EcoRI-BamHI fragment with deleted nucleotide sequence and the generation of a unique SalI site without affecting the amino acid residues at positions 412 and 413 immediately adjacent to the deleted peptide region (Fig. 3). After DNA sequencing analysis, one of the clones contained a PCR-generated spontaneous mutation that resulted in conversion of Leu to Ser at residues 408 of GtfB and 434 of GtfC. Therefore, GtfB-Dm2 and GtfC-Dm2 contained a single amino acid substitution in addition to the deletion.
FIG. 3.
Strategy for mutagenesis and construction of plasmids expressing GtfB and -C with insertion, deletion, and single amino acid mutations. Black bars represent gtfB and gtfC genes. pYNB13-D6 and -D201 were generated by cloning of the PCR-derived fragment containing in-frame deletion of 14 aa. pNH3-D1 was generated by a similar procedure. pAlter1-EB was generated by cloning of EcoRI-BamHI fragment from gtfC into identical multiple cloning sites in plasmid pAlter1, which carries two antibiotic resistance markers, ampicillin and tetracycline. nt, nucleotides.
FIG. 4.
Western blot analysis of GtfB and GtfC. The amount of total protein loaded in each lane was the same (20 μg), and the amounts of GtfB and -C were estimated from the band intensities, which were quantified by scanning the blots (duplicate) and subsequently analyzed with the program NIH Image 1.6. Band intensities were comparable between wild-type and individual mutants (except GtfC-Dm1 and GtfC-ms3), and bands of degraded forms could be detected in both wild-type and mutant enzymes. The decreased intensity found for GtfC-Dm1 and GtfC-ms3 was due to the greater tendency of mutated protein to form insoluble fractions in crude lysate. Positions of prestained molecular mass markers (lanes M) are given in kilodaltons. Arrows indicate the predicted molecular weights of the proteins.
A total of 18 mutant enzymes carrying amino acid substitutions between aa 414 and 427 of GtfB and aa 440 and 453 of GtfC were subsequently generated (GtfB/C-ms and -msR [Table 1]) by replacement of the DNA fragments into the deletion mutants by using the SalI site (Fig. 3). Preliminary functional assays from these mutants suggested that amino acid substitution of Asp411 in GtfB was more critical for activity than at other positions in the Gtf-P1 region. Therefore, the Asp residues at 411 and 413 of GtfB and at residues 437 and 439 of GtfC were converted to Asn, individually or in combination. Because previous results from analysis of the dextransucrase of L. mesenteroides suggested that catalysis involved a carboxyl group in a sequence homologous to Gtf-P1 in GtfB/C (9), the other residues containing a carboxyl group, Glu at aa 422 of GtfB and aa 448 of GtfC, was converted to Gln. The Val residues at 412 of GtfB and 438 of GtfC were converted to Ile, which is present in GtfD. This Ile (instead of Val) is the only discrepancy between the amino acid sequences of GtfB/C and GtfD in the Gtf-P1 region.
Enzyme activities of GtfB and GtfC mutants.
The insertion or deletion mutations in either GtfB or GtfC abolished enzymatic activities completely (data not shown). Random substitution of amino acid residues at both ends of the 19-aa region resulted in two GtfB constructs, GtfB-ms2 and GtfB-ms3, with reduced activities. Furthermore, in contrast to the counterpart GtfB mutants, the sucrase- and glucan-synthesizing activities of the GtfC -constructs GtfC-ms2 and GtfC-ms3 were completely inactivated (data not shown). In the case of GtfB, the sucrase activity was preserved unless the Asp residue at 411 was converted to Glu (GtfB-ms4), in combination with three other substitutions. This result suggested that Asp 411 of GtfB might be essential for the sucrase activity of this enzyme. Notably, mutants with single substitutions, GtfB-D411N and GtfB-D413N, exhibited sucrase- and glucan-synthesizing activities, although glucan synthesis was greatly reduced (Table 2). Further analysis found that sucrase activity was still detectable unless both Asp residues at aa 411 and 413 in GtfB were simultaneously converted to Asn, as shown by GtfB-D411/413N. In contrast to GtfB, single amino acid conversion of Asp to Asn, at either aa 437 or aa 439, resulted in complete inactivation of GtfC enzyme activities. No sucrase- or glucan-synthesizing activity was detected in the single-substitution mutants GtfC-D437N and GtfC-D439N. GtfB-E422Q and GtfC-E448Q were still capable of hydrolyzing sucrose, but the synthesized levels of glucans lower than those produced by the wild-type enzymes (Table 2). Therefore, two Asp residues in the Gtf-P1 region of GtfB and GtfC are important for catalysis, but not to the same degree. In addition, identical residues in Gtf-P1 may play different roles structurally and/or functionally in GtfB and GtfC.
TABLE 2.
Soluble and insoluble glucan-synthesizing activities of Gtfs
| Gtf | Without dextran T10
|
With dextran T10
|
Stimulation by dextranb
|
|||||
|---|---|---|---|---|---|---|---|---|
| Glucan-synthesizing activitya (mU/mg of protein)
|
SG/IG ratio | Glucan-synthesizing activity (mU/mg of protein)
|
SG/IG ratio | |||||
| SG | IG | SG | IG | SG | IG | |||
| GtfB | 0.33 | 2.98 | 0.11 | 0.67 | 3.60 | 0.19 | 2.01 | 1.21 |
| GtfC | 0.26 | 1.81 | 0.14 | 0.69 | 2.44 | 0.28 | 2.69 | 1.35 |
| GtfB-ms2 | 0.03 | 1.98* (−34) | 0.02 | 0.33 | 2.36* (−34) | 0.14 | 10.53# | 1.19 |
| GtfB-ms3 | 0.29 | 0.59* (−80) | 0.50 | 0.39 | 1.06* (−71) | 0.37 | 1.34 | 1.79 |
| GtfB-D411N | 0.16 | 0.56* (−81) | 0.28 | 0.17 | 0.65* (−82) | 0.27 | 1.09 | 1.16 |
| GtfB-D413N | 0.08 | 0.64* (−78) | 0.13 | 0.10 | 0.63* (−83) | 0.16 | 1.22 | 0.97 |
| GtfB-V412I | 0.01 | 3.33* (+12) | 0.003 | 0.56 | 3.97* (+10) | 0.14 | 59.12# | 1.19 |
| GtfC-V438I | 0.02 | 2.17* (+19) | 0.01 | 0.40 | 2.66* (+8.8) | 0.15 | 22.63# | 1.12 |
| GtfB-E422Q | 0.08 | 0.64* (−62) | 0.13 | 0.24 | 1.45* (−59) | 0.17 | 3.06 | 1.08 |
| GtfC-E448Q | 0.24 | 0.58* (−68) | 0.41 | 0.42 | 1.03* (−58) | 0.40 | 1.73 | 1.76 |
| GtfB-D411/413N | 0 | 0 | 0 | 0 | ||||
Expressed as specific activity for SG and as IG synthesized from [U-14C]sucrose. Values in parentheses represent percentages of reduction or increase of IG synthesized by the mutants, calculated as 100-% IG for the original GtfB or GtfC, which was normalized to 100%. Average values from quadruplicate assays are given, and the standard deviation of individual set of assays is within 12% of the average value. ∗, significant difference between mutant and wild-type proteins (P < 0.01 by Student’s t test; t = 3.707).
Expressed as the ratio of SG or IG in the presence and absence of dextran T10. #, significant increase in soluble glucan synthesis stimulated by dextran (P < 0.001).
Among the mutants listed in Table 1, only single amino acid substitutions at two positions (Glu and Val) in the Gtf-P1 region resulted in similar changes in enzymatic characteristics. Surprisingly, substitution of the Val residue (residue 412 for GtfB and residue 438 for GtfC) between the Asp residues resulted in an increase in glucan-synthesizing activities (Table 2). As shown in Table 2, insoluble glucan synthesis by GtfB-V412I and GtfC-V438I was enhanced about 10 to 20%, whereas soluble glucan synthesis by these enzymes was significantly lower than for the wild-type enzymes or other mutants in the absence of dextran. In the presence of dextran, both soluble glucan synthesis and insoluble glucan synthesis were enhanced significantly, with stimulation increased to a greater extent in the former than in the latter (59% versus 10% in GtfB-V412I and 23% versus 8.8% in GtfC-V438I). A significant increase of soluble glucan synthesis stimulated by dextran was also found in GtfB-ms2 compared to wild-type GtfB, while all other GtfB/C mutant and wild-type enzymes showed similar increases in soluble glucan synthesis stimulated by dextran. The significance of this differential increase in activity relative to the acceptor dextran due to single amino acid substitutions in Gtf-P1 of both enzymes awaits further investigation.
Effects of mutagenesis on enzyme kinetics.
To investigate in greater detail sucrose hydrolysis and its relation to altered glucan synthesis in several mutated GtfB or GtfC enzymes, the products glucose and fructose were assessed directly by HPLC followed by identification of the individual sugars. Consistent with the findings obtained by the colorimetric method, individual substitutions of residues other than the Asp resulted in mutated GtfB or GtfC enzymes exhibiting sucrase activity, but the rate of the hydrolysis was reduced in these mutants. The decreased rate of hydrolysis of sucrose could easily be detected by comparing the concentrations of fructose released over different time intervals. GtfB-D411N and GtfB-D413N, which showed sucrase activity comparable to that the wild-type enzyme in the colorimetric assay to detect reducing sugars, were found to act at rates far below the wild-type GtfB rate. The concentrations of fructose released by wild-type GtfB after 5- and 30-min reactions were 0.40 and 1.94 nmol/ml, respectively; the corresponding concentrations released by GtfB-411N and GtfB-D413N were 0.15 and 0.68 nmol/ml 0.19 and 0.74 nmol/ml. The reduction in the rate of sucrose hydrolysis by these two GtfB mutants relative to the wild-type GtfB was about 78%, which is consistent with the extent of reduction detected by measuring glucan synthesis. Analogous results were found for other GtfB and GtfC mutants exhibiting residual enzymatic activities except for GtfB-E422Q and GtfC-E448Q, which exhibited significant higher reductions in sucrolytic than in glucan synthesis activity (85% versus 60%). Therefore, the reduced glucan detected earlier in both GtfB and GtfC mutants was coupled to a decrease in the rate of sucrose hydrolysis. Consistent with the enhanced glucan synthesis activity detected for GtfB-V412I and GtfC-V438I, significant increases in the rate of sucrose hydrolysis were found in both mutants. For GtfB, the increase in the amount of fructose released in 5- and 30-min reactions were 20 and 5%, respectively.
To determine whether the reduced rate of sucrose hydrolysis was due to the changes in substrate binding affinity which resulted from the conformational changes induced by mutations, we determined the Kms for the wild-type and mutant GtfB and GtfC enzymes exhibiting residual glucan-synthesizing activity. The Kms of GtfB and GtfC expressed in E. coli were 12 ± 2.2 and 20 ± 3.7 mM, respectively, in the absence of dextran T10. The Kms of all but one of the GtfB mutants listed in Table 1 were similar to those of wild-type GtfB; the exception was GtfB-ms3, which exhibited a two- to threefold increase in its Km, 30 ± 2.4 mM. Kms of GtfC, GtfC-V438I, and GtfC-448Q were similar. These results suggested that the abilities of GtfB and GtfC to bind to sucrose were not altered significantly, except in the case of GtfB-ms3, by single amino acid substitutions in the 19-aa Gtf-P1 domain. Except for the soluble glucan synthesis activities of GtfB-ms2, GtfB-V412I, and GtfC-V438I as described above, the activities of both soluble and insoluble glucan synthesis for altered GtfB and GtfC enzymes were stimulated by addition of dextran T10 to a degree similar to that for the wild-type proteins (Table 2).
DISCUSSION
The conserved 19-residue N-terminal Gtf-P1 region, extending from aa 407 to 427 in GtfB (or aa 435 to 453 of GtfC), initially was identified due to an EcoRI polymorphism in the gtfB and -C genes of clinical isolates of S. mutans (3). This region is highly conserved among the GTFs of several streptococci, and DNA sequence variation did not affect the amino acid sequence of S. mutans GtfB and -C (5); this conservation suggests some biological importance of the domain. We have investigated this possibility and demonstrated previously that MAbs which reacted with Gtf-P1 were able to inhibit the synthesis of insoluble glucans by GtfC and the attachment of S. mutans to glass surfaces (4). The results of the present study provide further evidence that the 19-aa Gtf-P1 region is essential for both the sucrose hydrolysis and glucan synthesis activities of the GtfB and GtfC enzymes. In Gtf-P1, substitution of Asp residues, singly or in combination, appeared to be more critical for sucrase activity than substitutions at other positions in this region. Without crystallography data, we cannot exclude the possibility that substitution of Asp residues induces conformational changes, although the substitutions of the intervening valine residues (GtfB-V412I and GtfC-V438I) did not reduce either enzymatic activity or Km, suggesting that the Asp mutations resulted in a functional, rather than structural, change. The finding that substitutions of amino acids adjacent to the Asp residues resulted in GtfB mutants (GtfB-ms2 and GtfB-ms3) exhibiting reduced but detectable sucrase activity with unaltered Kms supported this hypothesis.
Consistent with our findings, mutagenesis of Asp413 to Thr in GtfB reported by another laboratory also resulted in a significant reduction of enzymatic activities, and this mutated GtfB exhibited a Km similar to that of wild-type GtfB (32). Nevertheless, in that study, conversion of Asp411 to Thr did not inhibit GtfB’s activity, whereas we found that the activity of GtfB-D411N was approximately 20% of wild-type enzyme activity. Further investigation is needed to determine whether substitution of Asp411 with other charged or uncharged residues may have similar effect on enzymatic activities. Functional analysis conducted by Funnae and coworkers (9) suggested that both Asp and Glu residues in the Gtf-P1 region are directly involved in enzymatic catalysis by dextransucrase of L. mesenteroides. Nevertheless, our data suggested that the Asp residues may be more important than Glu in catalysis by the GTFs, particularly in the case of GtfC. Specifically, substitution of Glu but not the Asp residues (GtfC-E448Q) did not result in loss of sucrase activity, whereas substitution of the Asp residues (GtfC-D437N and GtfC-D439N) resulted in complete loss of activity. Moreover, in the case of GtfB, the results of substitution analysis suggested that the functional role of Asp may not be replaced by Glu: GtfB-ms3, a GtfB mutant which contains three amino acid substitutions (L408S, W426F, and L427D), had detectable sucrase activity which was abolished when Asp 411 was additionally converted to Glu (in GtfB-ms4). Both GtfB and GtfC synthesize primarily insoluble glucan in a primer-independent manner, but both soluble and insoluble glucan synthesis could be enhanced in the presence of dextran (8). Significant increases in soluble glucan synthesis by GtfB were found earlier when corresponding residues were converted to those in GtfD, either singly or in multiple combinations (27). However, enhanced synthesis of both soluble and insoluble glucan as we found in GtfB-V412I and GtfC-V438I due to single amino acid substitution was an unexpected finding. Moreover, the results of two different assays confirmed that these two mutant also exhibited enhanced sucrase activity.
Another interesting finding was the discrepancy between GtfB and GtfC induced by substitutions of identical residues in Gtf-P1. This was observed initially when sucrase activity was assessed with a colorimetric method and subsequently confirmed by the use of a more sensitive method. Our results indicated that two Asp residues are indispensable for the activity of GtfC, whereas residual activity of GtfB remains when only one or the other Asp is converted to Asn. Both GtfB-D411N and GtfB-D413N can hydrolyze sucrose and synthesize glucan, although the rate of hydrolysis was about one-third of the wild-type GtfB rate at various substrate concentrations (79% reduction for GtfB-D411N and 81% reduction for GtfB-V413I). Differences between GtfB and GtfC also were observed for mutations with substitutions other than of Asp residues. For example, GtfB-ms2 and GtfB-ms3 both exhibited detectable sucrase and glucan synthesis activities, whereas identical substitution of corresponding residues in GtfC resulted in inactivation of the enzymes. These results confirmed that although they are closely related, GtfB and GtfC are distinct entities structurally and/or functionally. The inconsistency found between the GtfB and GtfC mutants also indicated that identical amino acid residues may play distinct roles, structurally or functionally, in these closely related enzymes. However, the results of mutagenesis alone are not sufficient to determine whether differences also exist in the catalytic mechanisms for the same sucrase reaction. Currently, we are also investigating the effects of mutagenesis on the enzymatic activities of GtfD to determine the characteristics of this third GTF of S. mutans. We have shown previously that although MAbs directed to this 19-aa peptide reacted with the three GTFs on when tested by enzyme-linked immunosorbent assay and Western blotting, unlike the activities of GtfB and GtfC, the enzymatic activity of GtfD was not inhibited even though the sequences of this region are almost identical. It will also be interesting to investigate the effect of mutations on other GTFs, such as those of S. sobrinus and S. salivarius. The 19-aa Gtf-P1 is well conserved in all GTFs, and they all contain two Asp residues at corresponding positions.
The GTFs are considered to be significant virulence factors for bacterial adherence and initiation of dental caries on smooth surfaces (20). Details of the enzyme structure and function are important not only in evaluating the nature of virulence but also for the development of a vaccine against dental caries. Several lines of evidence supported the view that protection induced by immunization with GTFs may involve antibody-mediated inhibition of their catalytic and/or the glucan-binding activities. We demonstrated previously that polyclonal or monoclonal antibodies which inhibited the enzymatic activities of the GTFs also could inhibit adherence in vitro. However, polyclonal antibodies which failed to inhibit the enzymatic activities of the GTFs could not interfere with the adherence of S. mutans (5). More recently, we found that this 19-aa Gtf-P1 peptide is one of the major B-cell epitopes in the human humoral immune response and that the anti-peptide immunoglobulin A antibody level in saliva correlated with disease activity (6). Furthermore, Gtf-P1 has recently been demonstrated in an animal model to be able to induce protective immunity against dental caries (30). Results of the present mutagenesis study confirmed the functional role of this peptide region in the GTFs. Together, our results suggest that peptides derived from functionally important regions in the GTFs, such as Gtf-P1, may serve as candidate subunit vaccines against S. mutans infection.
ACKNOWLEDGMENTS
We thank H. K. Kuramitsu for providing plasmids. We thank Tim J. Harrison, Reader in Molecular Virology, University Department of Medicine, Royal Free Hospital School of Medicine, for his kind review and help in preparation of the manuscript.
The pulsed ampherometric detection system for sugar analysis was supported by funding from the Instrument Center of the National Science Council. This work was supported by grants NSC-842331-B002-159, 852331-B002-024, and 862314-B002-113 and National Health Research Institute grant DOH88-HR-814.
REFERENCES
- 1.Abo H, Matsumura T, Kodama T, Ohta H, Fukui K, Kato K, Kagawa H. Peptide sequences for sucrose splitting and glucan binding within Streptococcus sobrinus glucosyltransferase (water-insoluble glucan synthesis) J Bacteriol. 1991;173:989–996. doi: 10.1128/jb.173.3.989-996.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aoki H, Shiroza T, Hayakawa M, Sato S, Kuramitsu H K. Cloning of a Streptococcus mutans glucosyltransferase gene coding for insoluble glucan synthesis. Infect Immun. 1986;53:587–594. doi: 10.1128/iai.53.3.587-594.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chia J S, Hsu T Y, Teng L J, Chen J Y, Hahn L J, Yang C S. Glucosyltransferase gene polymorphism among Streptococcus mutans strains. Infect Immun. 1991;59:1656–1660. doi: 10.1128/iai.59.5.1656-1660.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chia J S, Lin R H, Lin S W, Chen J Y, Yang C S. Inhibition of glucosyltransferase activities of Streptococcus mutans by a monoclonal antibody to a subsequence peptide. Infect Immun. 1993;61:1656–1660. doi: 10.1128/iai.61.11.4689-4695.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chia J S, Lin S W, Hsu T Y, Chen J Y, Kwan H W, Yang C S. Analysis of a DNA polymorphic region in the gtfB and gtfC genes of Streptococcus mutans strains. Infect Immun. 1993;61:1563–1566. doi: 10.1128/iai.61.4.1563-1566.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chia J S, Lin S W, Yang C S, Chen J Y. Antigenicity of a synthetic peptide from glucosyltransferases of Streptococcus mutans in humans. Infect Immun. 1997;65:1126–1130. doi: 10.1128/iai.65.3.1126-1130.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujiwara T, Tamesada M, Bian Z, Kawabata S, Kimura S, Hamada S. Deletion and reintroduction of glucosyltransferase genes of Streptococcus mutans and role of their gene products in sucrose dependent cellular adherence. Microb Pathol. 1996;20:225–233. doi: 10.1006/mpat.1996.0021. [DOI] [PubMed] [Google Scholar]
- 8.Fukushima K, Ikeda T, Kuramitsu H K. Expression of Streptococcus sobrinus gtf genes in Streptococcus milleri. Infect Immun. 1992;60:2815–2822. doi: 10.1128/iai.60.7.2815-2822.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Funane K, Shiraiwa M, Hashimoto K, Ichishima E, Kobayashi M. An active-site peptide containing the second essential carboxyl group of dextransucrase from Leuconostoc mesenteroides by chemical modification. Biochemistry. 1993;32:13696–13702. doi: 10.1021/bi00212a039. [DOI] [PubMed] [Google Scholar]
- 10.Giffard P M, Simpson C L, Milward C P, Jacques N A. Molecular characterization of a cluster of at least two glucosyltransferase genes in Streptococcus salivarius ATCC 25975. J Gen Microbiol. 1991;137:2577–2593. doi: 10.1099/00221287-137-11-2577. [DOI] [PubMed] [Google Scholar]
- 11.Hamada S, Slade H D. Biology, immunology, and cariogenicity of Streptococcus mutans. Microbiol Rev. 1980;44:331–384. doi: 10.1128/mr.44.2.331-384.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanada N, Kuramitsu H K. Isolation and characterization of the Streptococcus mutans gtfD gene, coding for primer-dependent soluble glucan synthesis. Infect Immun. 1989;57:2079–2085. doi: 10.1128/iai.57.7.2079-2085.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanada N, Kuramitsu H K. Isolation and characterization of the Streptococcus mutans gtfC gene, coding for synthesis of both soluble and insoluble glucans. Infect Immun. 1988;56:1999–2005. doi: 10.1128/iai.56.8.1999-2005.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Honda O, Kato C, Kuramitsu H K. Nucleotide sequence of the Streptococcus mutans gtfD gene encoding the glucosyltransferase-S enzyme. J Gen Microbiol. 1990;136:2099–2105. doi: 10.1099/00221287-136-10-2099. [DOI] [PubMed] [Google Scholar]
- 15.Horton R M, Hunt H D, Ho S N, Pullen J K, Pease L R. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 16.Kato C, Kuramitsu H K. Carboxyl-terminal deletion analysis of the Streptococcus mutans glucosyltransferase-I enzymes. FEMS Microbiol Lett. 1990;72:299–302. doi: 10.1016/0378-1097(90)90321-g. [DOI] [PubMed] [Google Scholar]
- 17.Kato C, Nakano Y, Lis M, Kuramitsu H K. Molecular genetic analysis of the catalytic site of Streptococcus mutans glucosyltransferase. Biochem Biophys Res Commun. 1992;189:1184–1188. doi: 10.1016/0006-291x(92)92329-v. [DOI] [PubMed] [Google Scholar]
- 18.Kuramitsu H K. Characterization of extracellular glucosyltransferase activity of Streptococcus mutans. Infect Immun. 1975;12:738–749. doi: 10.1128/iai.12.4.738-749.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lis M, Shiroza T, Kuramitsu H K. Role of the C-terminal direct repeating units of the Streptococcus mutans glucosyltransferase-S in glucan binding. Applied Environ Microbiol. 1995;61:2040–2042. doi: 10.1128/aem.61.5.2040-2042.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loesche W J. Role of Streptococcus mutans in human dental decay. Microbiol Rev. 1986;50:353–380. doi: 10.1128/mr.50.4.353-380.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 22.Monchois V, Willemot R M, Remaud-Simeon M, Croux C, Monsan P. Cloning and sequencing of a gene coding for a novel dextransucrase from Leuconostoc mesenteroides NRRL B-1299 synthesizing only α(1-6) and α(1-3) linkages. Gene. 1996;182:23–32. doi: 10.1016/s0378-1119(96)00443-x. [DOI] [PubMed] [Google Scholar]
- 23.Mooser G, Hefta S A, Paxton R J, Shively J E, Lee T D. Isolation and sequence of an active-site peptide containing a catalytic aspartic acid from two Streptococcus sobrinus glucosyltransferase. J Biol Chem. 1991;266:8916–8922. [PubMed] [Google Scholar]
- 24.Nakano Y J, Kuramitsu H K. Mechanism of Streptococcus sobrinus glucosyltransferases: hybrid-enzyme analysis. J Bacteriol. 1992;174:5639–5646. doi: 10.1128/jb.174.17.5639-5646.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 26.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimamura A, Nakano Y J, Mukasa H, Kuramitsu H K. Identification of amino acid residues in Streptococcus mutans glucosyltransferases influencing the structure of the glucan product. J Bacteriol. 1994;176:4845–4850. doi: 10.1128/jb.176.16.4845-4850.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shiroza T, Ueda S, Kuramitsu H K. Sequence analysis of the gtfB gene from Streptococcus mutans. J Bacteriol. 1987;169:4263–4270. doi: 10.1128/jb.169.9.4263-4270.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simpson C L, Giffard P M, Jacques N A. Streptococcus salivarius ATCC 25975 possesses at least two genes coding for primer-independent glucsyltransferases. Infect Immun. 1995;63:609–621. doi: 10.1128/iai.63.2.609-621.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith D J, Shoushtari B, Heschel R L, King W F, Taubman M A. Immunogenecity and protective immunity induced by syntehtic peptides associated with a catalytic subdomain of mutans group streptooccal glucosyltransferase. Infect Immun. 1997;65:4424–4430. doi: 10.1128/iai.65.11.4424-4430.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Somogyi M. A new reagent for the determination of sugars. J Biol Chem. 1945;160:61–68. [Google Scholar]
- 32.Tsumori H, Minami T, Kuramitsu H K. Identification of essential amino acids in the Streptococcus mutans glucosyltransferases. J Bacteriol. 1997;11:3391–3396. doi: 10.1128/jb.179.11.3391-3396.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ueda S, Shiroza T, Kuramitsu H K. Sequence analysis of the gtfC gene from Streptococcus mutans GS-5. Gene. 1988;69:101–109. doi: 10.1016/0378-1119(88)90382-4. [DOI] [PubMed] [Google Scholar]
- 34.Yamashita Y, Bowen W H, Burne R A, Kuramitsu H K. Role of the Streptococcus mutans gtf genes in caries induction in the specific-pathogen-free rat model. Infect Immun. 1993;61:3811–3817. doi: 10.1128/iai.61.9.3811-3817.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]