Highlights
-
•
Utility of ctDNA analysis by Next Generation Sequencing in patients with NSCLC.
-
•
Driver druggable variants were detected in patients at diagnosis and at progression.
-
•
Liquid biopsy allows detection of tumour heterogeneity in NSCLC patients.
Keywords: Liquid biopsy, NGS, NSCLC, Predictive biomarkers, Resistance to treatment
Abstract
Background
The present study evaluates the utility of NGS analysis of circulating free DNA (cfDNA), which incorporates small amounts of tumor DNA (ctDNA), at diagnosis or at disease progression (PD) in NSCLC patients.
Methods
Comprehensive genomic profiling on cfDNA by NGS were performed in NSCLC patients at diagnosis (if tissue was unavailable/insufficient) or at PD to investigate potential druggable molecular aberrations. Blood samples were collected as routinary diagnostic procedures, DNA was extracted, and the NextSeq 550 Illumina platform was used to run the Roche Avenio ctDNA Expanded Kit for molecular analyses. Gene variants were classified accordingly to the ESCAT score.
Results
A total of 106 patients were included in this study; 44 % of cases were requested because of tissue unavailability at the diagnosis and 56 % were requested at the PD. At least one driver alteration was observed in 62 % of cases at diagnosis. Driver druggable variants classified as ESCAT level I were detected in 34 % of patients, including ALK-EML4, ROS1-CD74, EGFR, BRAF, KRAS p.G12C, PI3KCA. In the PD group, most patients were EGFR-positive, progressing to a first line-therapy. Sixty-three percent of patients had at least one driver alteration detected in blood and 17 % of patients had a known biological mechanism of resistance allowing further therapeutic decisions.
Conclusions
The present study confirms the potential of liquid biopsy to detect tumour molecular heterogeneity in NSCLC patients at the diagnosis and at PD, demonstrating that a significant number of druggable mutations and mechanisms of resistance can be detected by NGS analysis on ctDNA.
List of abbreviations
- NGS
next-generation sequencing.
- ctDNA
circulating tumour DNA.
- NSCLC
non-small cell lung cancer.
- SCLC
small cell lung cancer.
Introduction
Non Small Cell Lung Cancer (NSCLC) accounts for about the 85 % of all lung cancers, and counts more than 200 different actionable mutations [1]. The identification of clinically actionable alterations expanded patients’ treatment options improving their management, survival, and quality of life [2]. Up to date, more than 20 targeted therapies are approved for NSCLC in Europe, including first or later lines of targeted agents for mutant EGFR, ALK, ROS1, RET, MET, BRAF, KRAS p.G12C, HER2 exon 20 insertion, and NTRK [3]. The most common alteration found in NSCLC is represented by an aberrant activation of the EGFR gene (up to 50 % in Asians), followed by MET (20 %), HER2 (15–30 %), KRAS p.G12C (13 %), ALK and BRAF (3–5 %), RET and ROS1 (1–2 %), NTRK (0.2 %) [1,4,5].
The ESCAT score is a classification developed by ESMO which describes the evidence level for genomic alterations as biomarkers for targeted therapies. In detail, level I corresponds to actionable alterations with available targeted treatments. Level II identifies molecular alterations that could benefit from a specific drug, but further evaluation is required. Level III belongs to molecular alterations for which a drug is available with evidence in other tumour types. Level IV, includes mutations for which there is pre-clinical evidence of activity. Finally, level V and level X correspond respectively to mutations to which a drug has given an objective response but without obvious clinical benefit, and molecular alterations for which there is no evidence of actionability [6]. The ESCAT scale implements and harmonises the information about the actionability of detected variants, allowing a better interpretation of cancer genomics, and facilitating the communication between professionals.
Nowadays, the genomic profile of NSCLC obtained from tissue profiling remains the gold standard for guiding treatment choice in patients with advanced NSCLC. However, tissue biopsy presents several limitations, including screening failures due to limited tissue availability and the inability to capture intratumor spatial and temporal heterogeneity [7]. On the contrary, the analysis of cell-free DNA circulating in plasma (cfDNA), also called liquid biopsy, which contains a small fraction of tumour DNA (ctDNA) allows the genomic profiling of a neoplasm with many advantages over tissue biopsy, being repeatable, non-invasive, and capable of providing information on the entire molecular profile of tumours, catching their complex molecular heterogeneity [8,9]. Therefore, ctDNA analysis represents an important alternative to define the genomic profile of the tumour, both at diagnosis and at the progression of the disease (PD). Nonetheless, the potential of advanced technologies such as Next Generation Sequencing (NGS), which allows a comprehensive genomic profiling of the tumour, has yet to be fully exploited in clinical practice.
The present study aims to evaluate the usefulness of ctDNA analysis by using NGS for molecular assessment at the diagnosis or to investigate mechanisms of resistance at PD in patients affected by NSCLC.
Patients and methods
The present study collected data routinely obtained by genomic profiling of ctDNA of NSCLC patients at diagnosis, when tissue biopsy was unavailable or insufficient for molecular profiling, or at the disease progression. The analysis was carried out on samples collected between March 2021 and February 2023. All patients provided written informed consent in sharing and using non-identifiable personal data, according to the institutional policies and Ethics Committee of referring hospitals.
Blood sampling and circulating free DNA isolation
Six ml of blood were collected from patients in tubes containing EDTA, centrifuged for 10 min at 3000 rpm to obtain plasma, and stored at −20 °C until the analysis. Samples were centrifuged for 5 min at 4000 rpm; the extraction of circulating free DNA (cfDNA) was performed using the QIAmp circulating nucleic acid kit from Qiagen® (Qiagen®, Valencia, CA). cfDNA amounts were assessed using a Qubit fluorometer and Qubit dsDNA High Sensitivity Assay Kit (Invitrogen, Carlsbad, CA, USA). Quality of cfDNA samples was determined with the 2100 Bioanalyzer using an Agilent High Sensitivity DNA Kit (Agilent Technologies, Santa Clara, CA, USA).
Next-generation sequencing
Genomic analysis was performed from 10 to 50 ng of cfDNA using the AVENIO ctDNA Expanded kit from Roche® according to the manufacturer's instructions (Roche Diagnostics, Basilea, CHE). The panel covered 77 genes, including hot spots (single nucleotide variants [SNVs] and short indels), copy number variations (CNVs) and gene fusions. Library preparation started with an adapter ligation and 10 μl of unique sample adapter were added to each sample with overnight hybridization for 16–18 h. Libraries were cleaned up using AVENIO cleanup beads and enrichment of target genes was performed using streptavidin-conjugated magnetic beads (Dynabeads M-270 Streptavidin, ThermoFisher Scientific, Waltham, MA, USA) at 47 °C. Target-enriched libraries were size-selected for an average fragment size of 300 bp. Library size was verified by Agilent High Sensitivity DNA Kit (Agilent Technologies, Santa Clara, CA, USA) on the 2100 Agilent Bioanalyzer and quantified using Qubit dsDNA High Sensitivity Assay Kit (Invitrogen, Carlsbad, CA, USA) on Qubit fluorometer. The pooled libraries were diluted to 4 nM, spiked with 15 % PhiX control library, and sequenced on the Illumina NextSeq 550 platform (Illumina, San Diego, CA) using the 300-cycle NextSeq 500/550 Mid Output v2 kit in paired-end mode (2 × 151 cycles).
Data analysis
Alignment and gene variant calling were performed using the AVENIO ctDNA analysis software (Roche Diagnostics, Basilea, CHE), with default parameter settings for the expanded panel. The software's bioinformatic algorithms and checkpoints have been optimized to enable accurate variant calls across all mutation classes. The AVENIO Oncology Analysis Software integrates a digital error suppression strategy combining molecular barcodes with in silico error suppression techniques, allowing the detection of low frequency alleles down to 0.1 % with high sensitivity and specificity. The software integrates 5 leading oncology databases, a curated loci of interest list and a customizable annotation database, all integrated into the AVENIO Oncology Analysis Software (Data on file with Roche; [10]). Three default reports are automatically generated by the analysis software: a sample metrics report, an initial variant report (unfiltered or all variants), and a second variant report (Roche default filter). Sample metric report provides an overview of all the quality field and run metrics, including the number of read pairs in the lane passing Illumina filter, the percentage of all read bases with a quality score of at least 30, the percentage of reads aligned to Illumina Phix sequencing control, the mean sequencing depth and the mean depth of coverage with unique reads.
The percentage of aligned reads to the human genome that is within the targeted region (unique depth) according to the manufacturer's instructions should be >40 %. Similarly, the expected median unique depth across bases in the targeted region should be at least 2500 ×, given 50 ng input cfDNA.
To investigate pathogenicity value, the target variants were submitted to the disease-associated databases COSMIC, VARSOME, and OncoKB, and only variants annotated as pathogenic or likely pathogenic were taken into account. Only somatic alterations with variant allele fraction (VAF) ≥ 0.5 % were considered for the following analysis. In order to exclude potential clonal hematopoiesis variants (CHIP) the peripheral blood sample was stored and analyzed in case of suspected CHIP using a dPCR (Qiagen®, Valencia, CA).
Actionable alterations were classified according to the ESCAT classification [11].
Results
Study population
Data from a total of 106 tests were included in this study, including 47 (44 %) samples at the diagnosis and 59 (56 %) at PD. Table 1 reports the clinical characteristics of the 2 groups of patients included in the study. Turnaround time was 24 days; timelines between testing and treatment initiation were in accordance to hospital procedures.
Table 1.
Clinical characteristics of patients.
| Diagnosis n = 47 | Progression of disease n = 59 | |
|---|---|---|
| Age average (range), years | 69 (32–89) | 68 (48–90) |
| Gender, n (%) | ||
| Males | 21 (45 %) | 23 (39 %) |
| Females | 26 (55 %) | 36 (61 %) |
| Smokinghistory, n (%) | ||
| Current | 1 (2 %) | 4 (7 %) |
| Former | 3 (6 %) | 6 (10 %) |
| Never | 1 (2 %) | 9 (15 %) |
| Unknown | 42 (90 %) | 40 (68 %) |
| Stage, n (%) | ||
| IIIA | – | 1 (2 %) |
| IIIB | 1 (2 %) | 1 (2 %) |
| IV | 46 (98 %) | 57 (96 %) |
| Tumor histology | ||
| Adenocarcinoma | 43 (91 %) | 52 (88 %) |
| Squamous cell carcinoma | 4 (9 %) | 7 (12 %) |
| ECOG PS, n (%) | ||
| 0–1 | 5 (11 %) | 20 (34 %) |
| ≥ 2 | 2 (4 %) | 3 (5 %) |
| Unknown | 40 (85 %) | 36 (61 %) |
| Line of treatment, n (%) | ||
| 1 | – | 43 (73 %) |
| 2 | – | 7 (12 %) |
| 3 | – | 2 (3 %) |
| 4 | – | 7 (12 %) |
| Therapy, n (%) | ||
| Gefitinib/erlotinib/afatinib | – | 9 (15 %) |
| Osimertinib | – | 33 (56 %) |
| Alectinib/lorlatinib | – | 6 (10 %) |
| Sotorasib | – | 1 (2 %) |
| Poziotinib | – | 1 (2 %) |
| Chemotherapy-based treatments | – | 9 (15 %) |
Sequencing performance
On average, 52 million reads were obtained from each sample (range, 40 to 62 million). The mean sequencing depth across all the analyses was 9354, and the mean depth of coverage with unique reads was 3606,3.
Gene variant detection
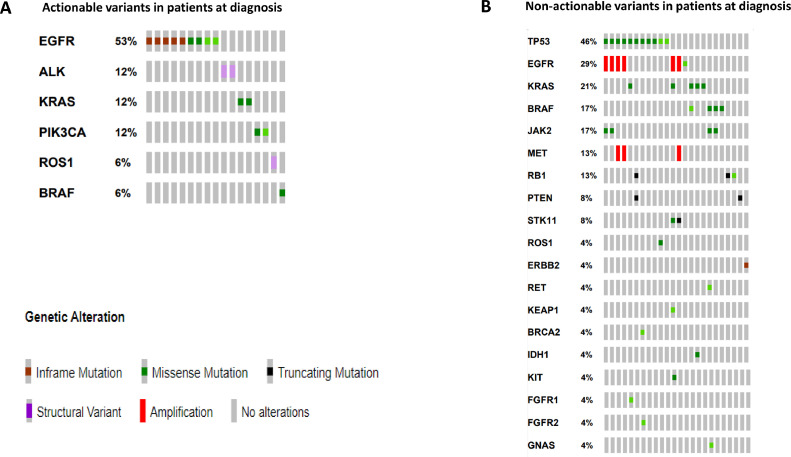
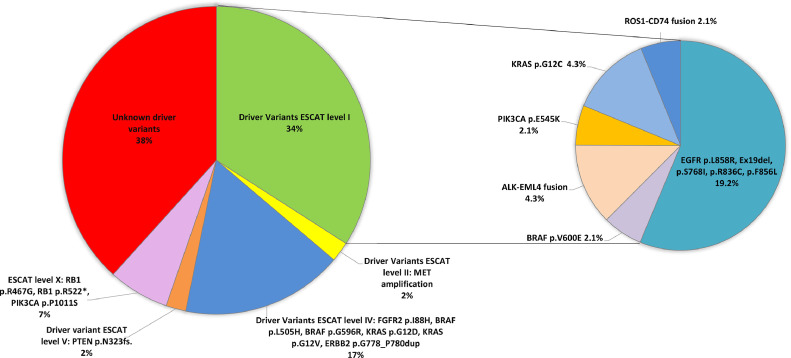
In the group of patients for which cfDNA was analyzed at the diagnosis, the presence of at least one driver alteration in ctDNA was found in 62 % of cases (Fig. 1). Driver druggable variants classified as ESCAT level I were detected in 34 % of patients, including ALK-EML4 fusion, ROS1-CD74 fusion, activating EGFR mutations, BRAF p.V600E, KRAS p.G12C, and PI3KCA (Fig. 2). Amplification of MET alone, classified as ESCAT level II, was also identified at diagnosis. The median allelic frequency (AF) of variants found at the diagnosis was 1 % (min 0,02 % – max 33,76 %). In 28 % of cases, only undruggable driver alterations were found. In detail, among ESCAT level IV alterations, KRAS p.G12D, p.G12V, BRAF p.L505H, p.G596R, FGFR2 p.I88H, and ERBB2 p.G778_P780 duplication were found. Among ESCAT level V mutations, frameshift PTEN p.N323 was found. Finally, among ESCAT level X mutations the RB1 p.R467G, p.R552*, and PIK3CA p.P1011S variants were detected (Fig. 2). The concomitant presence of a druggable driver variant and other driver events was found in 4 cases, including ALK-EML4 fusion together with MET and EGFR amplification, EGFR and MET amplification, ROS1-CD74 fusion in association with ROS1 non-synonymus variants (p.G2101A, p.G2095A, p.G2032R, p.G2026R, p.F2004C, p.F1998C, p.L1946V, p.L1941V), BRAF p.V600E in combination with KRAS p.A146P and IDH1 p.R132C.
Fig. 1.
Concomitant alterations in NSCLC patients at diagnosis. (A) Actionable variants detected in NSCLC patients. (B) Non-actionable variants detected in NSCLC patients.
Fig. 2.
ESCAT classification of driver alterations detected in NSCLC patients at diagnosis.
In addition, variants with potential prognostic significance and potential predictive power were also detected, such as STK11 p.D194Y and p.Q305*, KEAP1 p.H311L and different TP53 alterations.
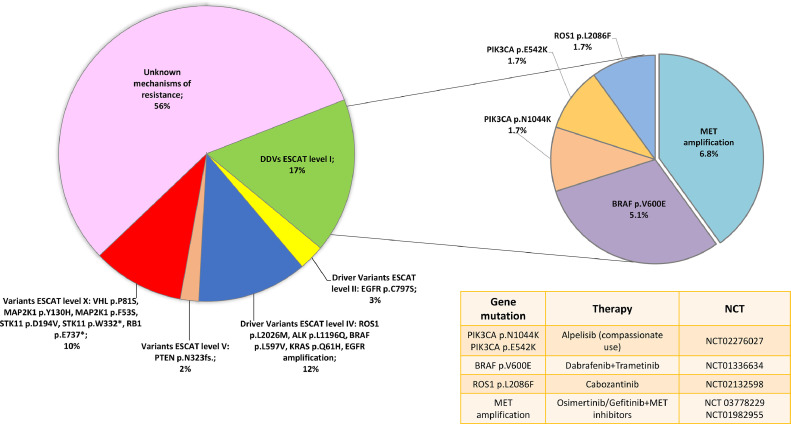
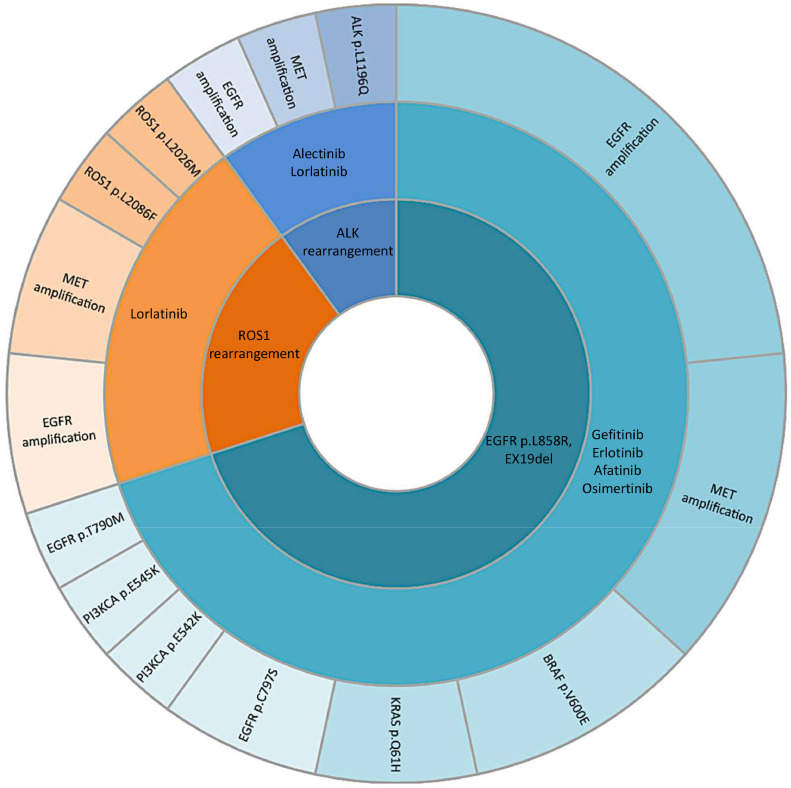
Fifty-nine samples were analysed to investigate the mechanisms of resistance to treatment at disease progression. ctDNA analysis was requested after treatment with first/second-generation EGFR-TKIs in 15 % of cases, in the 56 % after a third-generation EGFR-TKIs, in 10 % after an ALK-TKI treatment, in 15 % after first-line chemo-based treatments and after poziotinib and sotorasib therapy in 2 % of cases, respectively (Table 1). At least one driver alteration was detected in 63 % of patients of which 10 % were amenable of an approved targeted therapy (Fig. 3, Fig. 4). Based on the actionable effect and preclinical/clinical evidence to benefit from targeted therapy, 7 % of patients who progressed to previous standard treatments may have a potential treatment sequence strategy based on available clinical trials (data source: clinicaltrials.gov; last updated Feb. 2023) (Fig. 4 and Table 1S). Among ESCAT level II, the EGFR p.C797S ESCAT level II mutation was found, associated with EGFR exon 19 deletion in one case and with EGFR p.L858R mutation together with EGFR p.T790M in the other one. Instead, among ESCAT level IV mutations, variant ROS1 p.L2026M, ALK p.L1196Q, BRAF p.L597V, KRAS p.Q61H, and EGFR amplification were identified. PTEN p.N323fs variant was the only ESCAT level V present. Finally, among the ESCAT level X mutations, MAP2K1 p.Y130H, p.F53S, STK11 p.D194V, p.W332*, VHL p.P81S and RB1 p.E737* variants were detected (Fig. 4). In 19 % of cases, only the primary mutation was found without any known mechanisms of resistance. In this regard, in the oncogene-addicted population, the primary mutation was confirmed in 21/42 EGFR, 2/5 ALK-EML4, and 1/1 ROS1-CD74 of cases by liquid biopsy. In the EGFR mutant population treated with 1st-2nd generation TKIs, the most frequent mechanisms of resistance were EGFR p.T790M mutation and EGFR amplification. In EGFR mutant patients treated with the 3rd generation TKI osimertinib, detected mechanisms of resistance included: EGFR and MET amplifications, EGFR p.C797S, BRAF p.V600E, PI3KCA p.E542K and p.E545K, and KRAS p.Q61H (Figs. 5 and 1S). In the ALK-EML4 and ROS1-CD74 positive patients who progressed to ALK-TKIs, secondary point mutations in ALK and ROS1 were found as mechanisms of resistance, respectively (Figs. 5 and 1S). A detailed description of the therapy administered, and the mutations detected among all the patients evaluated at PD is reported in Table 2S.
Fig. 3.
Concomitant alterations in NSCLC patients at disease progression. (A) Actionable variants detected in NSCLC patients. (B) Non-actionable variants detected in NSCLC patients.
Fig. 4.
ESCAT classification of alterations detected in NSCLC patients at disease progression.
Fig. 5.
Representation of resistance mechanisms identified in patients at disease progression, depending on previously administered therapy and driver alterations.
Finally, a JAK2 p.V617F mutation classified as clonal haematopoiesis of indeterminate potential (CHIP) was detected in 7 patients with a median AF of 1.77 % (min 0.16 % – max 28.21 %). Fig. 2S summarizes the study design and the results.
Discussion
The present study demonstrates the utility of performing NGS analysis of cfDNA in a routine practice setting for patients affected by NSCLC, resulting in 34 % of patients with a druggable driver alteration at the diagnosis and 17 % of patients with druggable alterations when progressing to previous treatments. cfDNA demonstrated its value to investigate and identify tumour molecular heterogeneity across different tumours and stages. Up to date, the request of a cfDNA test is recommended by national and international scientific societies for patients affected by lung, breast, prostate, colorectal cancers and cholangiocarcinoma, to identify predictive biomarkers of response or resistance to treatment [12]. The NGS has the advantage of detecting multiple types of mutations in different genes in a single analysis [13] and is becoming increasingly important as the number of biomarkers to be tested in NSCLC is continuously increasing [13,14]. In the present study, the most frequent gene alterations found in blood were the EGFR activating mutations, particularly the p.L858R and exon 19 deletions, in agreement with previous studies [15,16]. The presence of druggable driver variants at the diagnosis was in many cases associated with other undruggable variants, such as EGFR amplification or TP53 mutations, usually correlated with worse prognosis [17]. Considering the most recent biomarkers introduced as druggable [18,19], the KRAS p.G12C and the BRAF p.V600E mutations were found in 2 cases at diagnosis, respectively. Activating PIK3CA variants, for which alpelisib is approved [20], were found in 2.1 % at diagnosis. Interestingly, both at the diagnosis and at the PD, the analysis of ctDNA was able to identify fusions in ALK and ROS1 genes, confirming also its potential in fusion detection. Interestingly, in one of the cases requested for the molecular assessment at diagnosis, the ROS1-CD74 fusion had 8 ROS1 concomitant variants, of which 3 are known to be mechanisms of resistance to ALK TKIs. In detail, the ROS1 p.F2004C is resistant to entrectinib, crizotinib, and cabozantinib [21], ROS1 p.G2101A is resistant to crizotinib but sensitive to lorlatinib [22], and ROS1 p.G2032A is resistant to lorlatinib [23]. Although this is a clear example of how complex is the molecular heterogeneity of NSCLC, posing challenges in terms of treatment strategy, it also highlights the advantages of using the NGS testing on cfDNA, allowing to detect multiple variants, which helps us to predict the response to treatment [24]. Unfortunately, most of the driver alterations found in NSCLC are still waiting for the development of effective targeted treatment and, accordingly, 62 % of cases at diagnosis, included in the present study, showed undruggable driver alterations, such as KRAS variants other than p.G12C and ERBB2 variant.
Among patients who progressed to treatment, 17 % had at least a druggable driver variant. The majority of patients were treated with first-line osimertinib, and known mechanisms of resistance were detected, including amplifications of MET and EGFR, BRAF p.V600E, PIK3CA p.E542K and p.E545K and EGFR p.C797S [25], [26], [27], allowing patients treatment with a second targeted-therapy such as the EGFR/MET combination of osimertinib and savolitinib, or the trametinib-dabrafenib combination for patients with a BRAF V600E mutation [28]. In case of acquired resistance due to EGFR p.C797S in patients treated with second-line osimertinib harbouring the EGFR activating and the p.T790M variants, according to published studies, if the p.C797S and p.T790M are in trans, they seem to be sensitive to the combination of osimertinib with a first-generation EGFR TKIs [29,30]. Instead, if the 2 alterations are in cis they seem to respond to brigatinib with an anti-EGFR antibody [31,32] or fourth-generation EGFR TKIs [33]. To overcome this type of resistance to therapy, a phase 1/2 study of the use of a novel EGFR inhibitor (BLU-945) in combination with osimertinib is also under investigation (NCT04862780). In ALK-rearranged tumors, mechanisms of resistance including MET amplification or the ALK variant p.L1196Q were found in patients treated with alectinib. In these cases, a subsequent treatment with crizotinib or brigatinib may be effective, respectively [34,35]. Finally, a ROS1-CD74 positive tumor treated with lorlatinib showed a ROS1 p.L2086F resistance variant, which turned out to respond to cabozantinib [23]. Importantly, ctDNA analysis may also suggest if a patient needs a re-biopsy. These are the cases where the concomitant presence of RB1 loss and TP53 alterations are found, suggesting a potential SCLC transformation, a known mechanism of resistance to treatment in NSCLC [36]. In patients in whom NSCLC has progressed, liquid biopsy is crucial for monitoring changes in the molecular profile of disease and guiding treatment choices, as re-biopsy is not possible in most cases. Even if the evidence of clinical utility of cfDNA analysis is continuously increasing, it is highly needed the development of robust assays or software tools able to differentiate ‘true’ from possibly ‘false’ positive (i.e. CHIP) or negative results. The analysis of ctDNA through liquid biopsy has the advantage of catching the intra-tumor spatial and temporal molecular heterogeneity, overcoming tissue biopsy limitations and allowing a maximisation of the concept of “tailored treatment”.
We acknowledge that the present study has some major limitations including the small number of patients and the lack of comparison of ctDNA with tissue re-biopsy in patients at the PD [37]. However, the unavailability of tissue biopsy sufficient for molecular analysis (at diagnosis or at PD) led to the request, as per clinical practice, of a cfDNA test in our population. Moreover, a critical aspect of our study is the lack of information about the clinical outcomes and follow-up data of our patient population, which should be addressed in further studies enrolling a larger patient population, to better evaluate the clinical significance of incorporating the NGS analysis of cfDNA in clinical routine.
Conclusions
Our study confirms the ability of NGS to detect targetable alterations and mechanisms of resistance, guiding therapeutic choices in patients affected by NSCLC both at diagnosis and disease progression, also emphasizing the usefulness of ctDNA in monitoring the evolution of the molecular pattern of the tumour without the invasiveness of tissue biopsy.
Ethics approval and consent to participate
This study was conducted in accordance with the Declaration of Helsinki and approved by the local Ethics Committee of Area Vasta Nord-Ovest, Tuscany Region, Italy (reference code 2014–363).
Consent for publication
All patients gave their signed informed consent before blood collection and data analysis.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Funding
This study was supported by institutional fundings to Prof. Romano Danesi and Dr. Marzia Del Re (grants no. 2017NRW5K-PRIN 2017, 20209KY3Y7-PRIN 2020, ECS00000017-PNRR) from Ministero dell’Università e della Ricerca (MUR), Italy.
CRediT authorship contribution statement
Marzia Del Re: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Writing – original draft, Writing – review & editing. Giovanna Irene Luculli: Conceptualization, Data curation, Investigation, Methodology, Writing – original draft, Writing – review & editing. Iacopo Petrini: Resources, Writing – review & editing, Writing – original draft. Andrea Sbrana: Resources, Writing – review & editing, Writing – original draft. Vieri Scotti: Resources, Writing – review & editing, Writing – original draft. Diego de Miguel Perez: Data curation, Investigation, Writing – review & editing, Writing – original draft. Lorenzo Livi: Resources, Writing – review & editing, Writing – original draft. Stefania Crucitta: Data curation, Investigation, Writing – original draft, Writing – review & editing. Mauro Iannopollo: Resources, Writing – review & editing, Writing – original draft. Francesca Mazzoni: Resources, Writing – review & editing, Writing – original draft. Martina Ruglioni: Data curation, Investigation, Writing – original draft, Writing – review & editing. Carmelo Tibaldi: Resources, Writing – review & editing, Writing – original draft. Emanuela Olmetto: Resources, Writing – review & editing, Writing – original draft. Irene Stasi: Resources, Writing – review & editing, Writing – original draft. Editta Baldini: Resources, Writing – review & editing, Writing – original draft. Giacomo Allegrini: Resources, Writing – review & editing, Writing – original draft. Lorenzo Antonuzzo: Resources, Writing – review & editing, Writing – original draft. Franco Morelli: Resources, Writing – review & editing, Writing – original draft. Andrea Pierini: Software, Writing – original draft. Nicola Panzeri: Software, Writing – original draft. Stefano Fogli: Data curation, Writing – review & editing, Writing – original draft. Antonio Chella: Resources, Writing – review & editing, Writing – original draft. Christian Rolfo: Data curation, Investigation, Supervision, Writing – review & editing, Writing – original draft. Romano Danesi: Conceptualization, Funding acquisition, Supervision, Data curation, Writing – review & editing, Writing – original draft.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
None.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2023.101869.
Appendix. Supplementary materials
References
- 1.Hendriks L.E., Kerr K.M., Menis J., Mok T.S., Nestle U., Passaro A., et al. Oncogene-addicted metastatic non-small-cell lung cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann. Oncol. 2023;34(4):339–357. doi: 10.1016/j.annonc.2022.12.009. [DOI] [PubMed] [Google Scholar]
- 2.Wang Z., Xing Y., Li B., Li X., Liu B., Wang Y. Molecular pathways, resistance mechanisms and targeted interventions in non-small-cell lung cancer. Mol. Biomed. 2022;3(1) doi: 10.1186/s43556-022-00107-x. [cited 2023 Apr 3]; Available from: /pmc/articles/PMC9743956/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tan A.C., Tan D.S.W. Targeted therapies for lung cancer patients with oncogenic driver molecular alterations. J. Clin. Oncol. 2022;40(6):611–625. doi: 10.1200/JCO.21.01626. [DOI] [PubMed] [Google Scholar]
- 4.Midha A., Dearden S., McCormack R. EGFR mutation incidence in non-small-cell lung cancer of adenocarcinoma histology: a systematic review and global map by ethnicity (mutMapII) Am J. Cancer Res. 2015;5(9):2892–2911. [PMC free article] [PubMed] [Google Scholar]
- 5.Hong D.S., Fakih M.G., Strickler J.H., Desai J., Durm G.A., Shapiro G.I., et al. KRASG12C inhibition with sotorasib in advanced solid tumors. N. Engl. J. Med. 2020;383(13):1207–1217. doi: 10.1056/NEJMoa1917239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mateo J., Chakravarty D., Dienstmann R., Jezdic S., Gonzalez-Perez A., Lopez-Bigas N., et al. A framework to rank genomic alterations as targets for cancer precision medicine: the ESMO Scale for Clinical Actionability of molecular Targets (ESCAT) Ann. Oncol. 2018;29(9):1895–1902. doi: 10.1093/annonc/mdy263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esagian S.M., Grigoriadou G., Nikas I.P., Boikou V., Sadow P.M., Won J.K., et al. Comparison of liquid-based to tissue-based biopsy analysis by targeted Next Generation Sequencing in advanced non-small cell lung cancer: a comprehensive systematic review. J. Cancer Res. Clin. Oncol. 2020;146(8):2051–2066. doi: 10.1007/s00432-020-03267-x. https://pubmed.ncbi.nlm.nih.gov/32462295/ [cited 2023 May 3]; Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sonmezler O., Boga I., Bisgin A. Integration of liquid biopsies into clinical laboratory applications via NGS in cancer diagnostics. Clin. Lab. 2020;66(5):763–769. doi: 10.7754/Clin.Lab.2019.190836. https://pubmed.ncbi.nlm.nih.gov/32390404/ [cited 2023 May 3]; Available from: [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y.C., Zhou Q., Wu Y.L. The emerging roles of NGS-based liquid biopsy in non-small cell lung cancer. J. Hematol. Oncol. 2017;10(1) doi: 10.1186/s13045-017-0536-6. https://pubmed.ncbi.nlm.nih.gov/29061113/ [cited 2023 May 3]; Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Newman A.M., Lovejoy A.F., Klass D.M., Kurtz D.M., Chabon J.J., Scherer F., et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat. Biotechnol. 2016;34(5):547–555. doi: 10.1038/nbt.3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mateo J., Chakravarty D., Dienstmann R., Jezdic S., Gonzalez-Perez A., Lopez-Bigas N., et al. A framework to rank genomic alterations as targets for cancer precision medicine: the ESMO Scale for Clinical Actionability of molecular Targets (ESCAT) Ann. Oncol. 2018;29(9):1895–1902. doi: 10.1093/annonc/mdy263. https://pubmed.ncbi.nlm.nih.gov/30137196/ [cited 2023 May 3]; Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pascual J., Attard G., Bidard F.C., Curigliano G., De Mattos-Arruda L., Diehn M., et al. ESMO recommendations on the use of circulating tumour DNA assays for patients with cancer: a report from the ESMO Precision Medicine Working Group. Ann. Oncol. 2022;33(8):750–768. doi: 10.1016/j.annonc.2022.05.520. [DOI] [PubMed] [Google Scholar]
- 13.Miller T.E., Yang M., Bajor D., Friedman J.D., Chang R.Y.C., Dowlati A., et al. Clinical utility of reflex testing using focused next-generation sequencing for management of patients with advanced lung adenocarcinoma. J. Clin. Pathol. 2018;71(12):1108–1115. doi: 10.1136/jclinpath-2018-205396. https://pubmed.ncbi.nlm.nih.gov/30228211/ [cited 2023 May 3]; Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waterhouse D.M., Tseng W.Y., Espirito J.L., Robert N.J. Understanding contemporary molecular biomarker testing rates and trends for metastatic NSCLC among community oncologists. Clin. Lung Cancer. 2021;22(6):e901–e910. doi: 10.1016/j.cllc.2021.05.006. https://pubmed.ncbi.nlm.nih.gov/34187757/ [cited 2023 May 3]; Available from: [DOI] [PubMed] [Google Scholar]
- 15.Zhao S., Cong X., Liu Z. Mutation profile assessed by next-generation sequencing (NGS) of circulating tumor DNA (ctDNA) in Chinese lung adenocarcinoma patients: analysis of real-world data. Biomed. Res. Int. 2021 doi: 10.1155/2021/8817898. https://pubmed.ncbi.nlm.nih.gov/33997043/ [cited 2023 May 3]; Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.D'Angelo S.P., Pietanza M.C., Johnson M.L., Riely G.J., Miller V.A., Sima C.S., et al. Incidence of EGFR exon 19 deletions and L858R in tumor specimens from men and cigarette smokers with lung adenocarcinomas. J. Clin. Oncol. 2011;29(15):2066–2070. doi: 10.1200/JCO.2010.32.6181. https://pubmed.ncbi.nlm.nih.gov/21482987/ [cited 2023 May 3]; Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao J., Han Y., Li J., Chai R., Bai C. Prognostic value of KRAS/TP53/PIK3CA in non-small cell lung cancer. Oncol. Lett. 2019;17(3) doi: 10.3892/ol.2019.10012. https://pubmed.ncbi.nlm.nih.gov/30867754/ [cited 2023 May 3]; Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh N., Temin S., Baker S., Blanchard E., Brahmer J.R., Celano P., et al. Therapy for stage IV non-small-cell lung cancer with driver alterations: ASCO living guideline. J. Clin. Oncol. 2022;40(28):3310–3322. doi: 10.1200/JCO.22.00824. https://pubmed.ncbi.nlm.nih.gov/35816666/ [cited 2023 May 3]; Available from: [DOI] [PubMed] [Google Scholar]
- 19.Khunger A., Khunger M., Velcheti V. Dabrafenib in combination with trametinib in the treatment of patients with BRAF V600-positive advanced or metastatic non-small cell lung cancer: clinical evidence and experience. Ther. Adv. Respir. Dis. 2018;12 doi: 10.1177/1753466618767611. https://pubmed.ncbi.nlm.nih.gov/29595366/ [cited 2023 May 3]; Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farmaci a Uso Compassionevole, Agenzia Italiana del Farmaco [Internet]. [cited 2023 May 3]. Available from: https://www.aifa.gov.it/web/guest/farmaci-a-uso-compassionevole.
- 21.Keddy C., Shinde P., Jones K., Kaech S., Somwar R., Shinde U., et al. Resistance profile and structural modeling of next-generation ROS1 tyrosine kinase inhibitors. Mol. Cancer Ther. 2022;21(2):336–346. doi: 10.1158/1535-7163.MCT-21-0395. https://pubmed.ncbi.nlm.nih.gov/34907086/ [cited 2023 May 3]; Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Begum P., Cui W., Popat S. Crizotinib-resistant ROS1 G2101A mutation associated with sensitivity to lorlatinib in ROS1-rearranged NSCLC: case report. JTO Clin. Res. Rep. 2022;3(9) doi: 10.1016/j.jtocrr.2022.100376. https://pubmed.ncbi.nlm.nih.gov/35966191/ [cited 2023 May 3]; Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin J.J., Choudhury N.J., Yoda S., Zhu V.W., Johnson T.W., Sakhtemani R., et al. Spectrum of mechanisms of resistance to Crizotinib and Lorlatinib in ROS1 fusion-positive lung cancer. Clin. Cancer Res. 2021;27(10):2899–2909. doi: 10.1158/1078-0432.CCR-21-0032. https://pubmed.ncbi.nlm.nih.gov/33685866/ [cited 2023 May 3]; Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crucitta S., Cucchiara F., Mathijssen R., Mateo J., Jager A., Joosse A., et al. Treatment-driven tumour heterogeneity and drug resistance: lessons from solid tumours. Cancer Treat. Rev. 2022;104 doi: 10.1016/j.ctrv.2022.102340. https://pubmed.ncbi.nlm.nih.gov/35151155/ [cited 2023 May 3]; Available from: [DOI] [PubMed] [Google Scholar]
- 25.Lee K., Kim D., Yoon S., Lee D.H., Kim S.W. Exploring the resistance mechanisms of second-line osimertinib and their prognostic implications using next-generation sequencing in patients with non-small-cell lung cancer. Eur. J. Cancer. 2021;148:202–210. doi: 10.1016/j.ejca.2021.01.052. https://pubmed.ncbi.nlm.nih.gov/33744716/ [cited 2023 Jan 25]; Available from: [DOI] [PubMed] [Google Scholar]
- 26.Yang Z., Yang N., Ou Q., Xiang Y., Jiang T., Wu X., et al. Investigating novel resistance mechanisms to third-generation EGFR tyrosine kinase inhibitor osimertinib in non-small cell lung cancer patients. Clin. Cancer Res. 2018;24(13):3097–3107. doi: 10.1158/1078-0432.CCR-17-2310. https://pubmed.ncbi.nlm.nih.gov/29506987/ [cited 2023 May 3]; Available from: [DOI] [PubMed] [Google Scholar]
- 27.Ho C.C., Liao W.Y., Lin C.A., Shih J.Y., Yu C.J., Chih-Hsin Yang J. Acquired BRAF V600E mutation as resistant mechanism after treatment with osimertinib. J. Thorac. Oncol. 2017;12(3):567–572. doi: 10.1016/j.jtho.2016.11.2231. https://pubmed.ncbi.nlm.nih.gov/27923714/ [cited 2023 May 3]; Available from: [DOI] [PubMed] [Google Scholar]
- 28.Planchard D., Smit E.F., Groen H.J.M., Mazieres J., Besse B., Helland Å., et al. Dabrafenib plus trametinib in patients with previously untreated BRAFV600E-mutant metastatic non-small-cell lung cancer: an open-label, phase 2 trial. Lancet Oncol. 2017;18(10):1307–1316. doi: 10.1016/S1470-2045(17)30679-4. https://pubmed.ncbi.nlm.nih.gov/28919011/ [cited 2023 May 3]; Available from: [DOI] [PubMed] [Google Scholar]
- 29.Wang Z., Yang J.J., Huang J., Ye J.Y., Zhang X.C., Tu H.Y., et al. Lung adenocarcinoma harboring EGFR T790M and in trans C797S responds to combination therapy of first- and third-generation EGFR TKIs and shifts allelic configuration at resistance. J. Thorac. Oncol. 2017;12(11):1723–1727. doi: 10.1016/j.jtho.2017.06.017. https://pubmed.ncbi.nlm.nih.gov/28662863/ [cited 2023 May 3]; Available from: [DOI] [PubMed] [Google Scholar]
- 30.Niederst M.J., Hu H., Mulvey H.E., Lockerman E.L., Garcia A.R., Piotrowska Z., et al. The allelic context of the C797S mutation acquired upon treatment with third-generation EGFR inhibitors impacts sensitivity to subsequent treatment strategies. Clin. Cancer Res. 2015;21(17):3924–3933. doi: 10.1158/1078-0432.CCR-15-0560. https://pubmed.ncbi.nlm.nih.gov/25964297/ [cited 2023 May 3]; Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uchibori K., Inase N., Araki M., Kamada M., Sato S., Okuno Y., et al. Brigatinib combined with anti-EGFR antibody overcomes osimertinib resistance in EGFR-mutated non-small-cell lung cancer. Nat. Commun. 2017;8 doi: 10.1038/ncomms14768. https://pubmed.ncbi.nlm.nih.gov/28287083/ [cited 2023 May 3]; Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X., Zhou L., Yin J.C., Wu X., Shao Y.W., Gao B. Lung adenocarcinoma harboring EGFR 19del/C797S/T790M triple mutations responds to brigatinib and anti-EGFR antibody combination therapy. J. Thorac. Oncol. 2019;14(5):e85–e88. doi: 10.1016/j.jtho.2019.01.015. https://pubmed.ncbi.nlm.nih.gov/30711650/ [cited 2023 May 3]; Available from: [DOI] [PubMed] [Google Scholar]
- 33.Kashima K., Kawauchi H., Tanimura H., Tachibana Y., Chiba T., Torizawa T., et al. CH7233163 overcomes osimertinib-resistant EGFR-Del19/T790M/C797S mutation. Mol. Cancer Ther. 2020;19(11):2288–2297. doi: 10.1158/1535-7163.MCT-20-0229. https://pubmed.ncbi.nlm.nih.gov/32943545/ [cited 2023 May 3]; Available from: [DOI] [PubMed] [Google Scholar]
- 34.Latif H., Liu S.V. Novel ALK mutation with durable response to brigatinib-a case report. Transl. Lung Cancer Res. 2020;9(5):2145–2148. doi: 10.21037/tlcr-20-145. https://pubmed.ncbi.nlm.nih.gov/33209633/ [cited 2023 May 3]; Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Camidge D.R., Otterson G.A., Clark J.W., Ignatius Ou S.H., Weiss J., Ades S., et al. Crizotinib in patients with MET-Amplified NSCLC. J. Thorac. Oncol. 2021;16(6):1017–1029. doi: 10.1016/j.jtho.2021.02.010. https://pubmed.ncbi.nlm.nih.gov/33676017/ [cited 2023 May 3]; Available from: [DOI] [PubMed] [Google Scholar]
- 36.Offin M., Chan J.M., Tenet M., Rizvi H.A., Shen R., Riely G.J., et al. Concurrent RB1 and TP53 alterations define a subset of EGFR-mutant lung cancers at risk for histologic transformation and inferior clinical outcomes. J. Thorac. Oncol. 2019;14(10):1784–1793. doi: 10.1016/j.jtho.2019.06.002. https://pubmed.ncbi.nlm.nih.gov/31228622/ [cited 2023 May 3]; Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gerratana L., Movarek M., Wehbe F., Katam N., Mahalingam D., Donahue J., et al. Genomic landscape of advanced solid tumors in circulating tumor DNA and correlation with tissue sequencing: a single institution's experience. JCO Precis. Oncol. 2022;6(6) doi: 10.1200/PO.21.00289. https://pubmed.ncbi.nlm.nih.gov/35772051/ [cited 2023 May 3]; Available from: [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.