Abstract
The probiotic bacterium Escherichia coli Nissle 1917 (EcN) holds significant promise for use in clinical and biological industries. However, the reliance on antibiotics to maintain plasmid-borne genes has overshadowed its benefits. In this study, we addressed this issue by engineering the endogenous cryptic plasmids pMUT1 and pMUT2. The non-essential elements were removed to create more stable derivatives pMUT1NR△ and pMUT2HBC△. Synthetic promoters by integrating binding motifs on sigma factors were further constructed and applied for expression of Bacteroides thetaiotaomicron heparinase III and the biosynthesis of ectoine. Compared to traditional antibiotic-dependent expression systems, our newly constructed antibiotic-free expression systems offer considerable advantages for clinical and synthetic biology applications.
Keywords: Cryptic plasmids, Antibiotic-free, Gene expression, Plasmid stability, E. coli Nissle 1917
1. Introduction
The Gram-negative model microorganism Escherichia coli has been widely used in areas of metabolic engineering and synthetic biology [1] because of the available diverse toolboxes for genome engineering [2,3], gene expression and regulation [[4], [5], [6]]. In particular, E. coli Nissle 1917 (EcN), a probiotic strain that isolated by Dr. Alfred Nissle from the feces of a World War I soldier who remarkably unaffected by epidemic shigella diarrhea [7], has traditionally been used in the prevention and treatment of various inflammatory disorders and enteric infections [[7], [8], [9], [10], [11]]. Because of its probiotic attributes, EcN has attracted intensive attention for potential applications in industrial production and gene therapy [12,13]. Many valuable products such as heparosan [14,15], 5-aminolevulinic acid [16], 3-hydroxybutyrate [17], Butyric acid [18], β-alanine [19] and γ-aminobutyric acid [20] have been biosynthesized by utilizing EcN chassis. With the development of CRISPR-Cas and CRISPRi regulatory systems [21,22], many EcN strains have been engineered to precisely deliver drugs for the treatment of diverse gastrointestinal and immunological diseases [23,24], cancers [16,25], Parkinson's disease [26], hyperuricemia [27] and diabetes [28].
For industrial production and in vivo therapy, the utilization of antibiotics is always subjected to stringent limitations [29]. To attain antibiotic-free expression, heterologous genes are commonly integrated into the genome for expression [30,31]. However, genome integration poses challenges as its time-consuming operation and low dose gene expression [32]. To circumvent the need for antibiotic addition, auxotrophy complementation systems such as purine, pyrimidine, and amino acid auxotrophies have been devised [32]. However, the presence of amino acids and other substances is generally inevitable in high-density culture, which often poses challenges for the stable retention of the recombinant plasmid maintained by the auxotrophic complementation system [33]. Recently, it has been demonstrated that the toxin-antitoxin (TA) systems that function with the expression of a long-lived toxin and a shorter-lived antitoxin outperform the commonly used hok/sok system [34]. In this regard, the type II TA systems have been engineered to construct antibiotic-free expression systems [35,36].
In EcN, two stably replicating cryptic plasmids pMUT1 (Accession No. MW240712) and pMUT2 (Accession No. CP023342) have been characterized [33] and applied for the expression of ospA/ospG antigens [37] and cystatin [38]. Recently, Zainuddin and colleagues demonstrated the nonexistence of metabolic burden from cryptic plasmids and the stable existence of the engineered pMUT2 containing an antibiotic resistance reporter without selection [39], suggesting the plasticity and potential applications of the cryptic plasmids. To simplify the development of therapeutic EcN bacteria, Kan and colleagues systematically engineered pMUT1 and pMUT2 by inserting selection markers (ampicillin and tetracycline), fluorescent markers, temperature responding elements, and secretion signal peptide sequences [29]. More recently, Dong et al. conducted similar studies and found that different insertion sites in the vectors generated significantly different expression level of proteins of interest [40]. In parallel, the commonly used inducible promoters Plac, Ptrc, Ptac, PBAD, and PT7 have been introduced into EcN for gene expressions [41]. Despite the increasing applications of cryptic plasmids, the essential elements for maintaining the plasmid stability are still unclear. Moreover, even antibiotics are not used during cell cultivation, the presence of antibiotic resistance encoding genes in cell still raises concerns [32] Additionally, the shortage of strong constitutive promoters for EcN would also hinder the applications of EcN for industrial production.
In this study, we employed the CRISPR/Cas9 system to delete pMUT plasmids and constructed EcN strains cured of one or both cryptic plasmids. Then, all elements in pMUT1 and pMUT2 were analyzed and deleted, and the effects on stability and cell growth were examined. After combinatorial truncation, minimized plasmids with higher stability were constructed. To expand the application of the engineered cryptic plasmids, an artificial synthetic promoter library was further engineered by interlocking the binding motifs on sigma factors σ70 and σ38 [42,43]. Eventually, the resistance genes that assisting screening were eliminated by introducing a FLP recombinase encoding helper plasmid which is easily curable. The developed antibiotic-free expression systems present a platform for the rapid development of robust industrial strains and therapeutic EcN strains.
2. Material and methods
2.1. Strains and media
All bacterial strains utilized in this study are documented in Table S2. The E. coli Top10 strain was employed for plasmid amplification and construction purposes. EcN served as the parental strain for breeding all genetically modified EcN strains. Antibiotics (100 mg/L ampicillin, 50 mg/L kanamycin, and 15 mg/L chloramphenicol) were added when necessary. For cloning operations and specific protein expression, Luria-Bertani medium (containing 10 g/L tryptone, 5 g/L yeast extract, and 10 g/L NaCl) was used. Ectoine production employed a glucose-defined medium consisting of 20 g/L glucose, 0.1 g/L thiamine, 13.5 g/L KH2PO4, 4.0 g/L (NH4)2HPO4, 1.4 g/L MgSO4·H2O, 1.7 g/L citric acid, and 10.0 mL trace metal solution per liter (composed of 10.0 g/L FeSO4·7H2O, 2.0 g/L CaCl2, 2.2 g/L ZnSO4·7H2O, 0.5 g/L MnSO4·4H2O, 1.0 g/L CuSO4·5H2O, 0.1 g/L (NH4)6Mo7O24·4H2O, and 0.02 g/L Na2B4O7·10H2O).
2.2. Plasmid and DNA manipulation
The plasmids used in this study are listed in Table S3, while the primers employed are listed in Table S4. The elements derived from cryptic plasmids are presented in Table 1. To knockout the native pMUT1 and pMUT2 using the CRISPR/Cas9 system, gRNA was ligated into amplified pTarget using the designated primers anti-T1-F/R and anti-T2-F/R, resulting in the generation of plasmids pTargetF-1 and pTargetF-2, respectively. For the subsequent expression of green fluorescence protein coding gene (gfp), ampicillin resistance gene (ampR) and kanamycin resistance gene (kanR), the linearized gfp gene, antibiotic resistance gene and the pMUT1 and pMUT2 vectors were amplified using the primers gfp-F/R, kan-F/R, amp-F/R and gfp-zaiti-F/R, respectively. The gfp genes were then ligated into the amplified pMUT1 and pMUT2 vectors using Gibson assembly, resulting in the formation of pMUT1-gfp and pMUT2-gfp plasmids. To delete specific elements from pMUT1, pMUT1-gfp was amplified using the designated primers del-NikA-F/R, del-Hyp1-F/R, del-HTH-F/R, del-RNAI-F/R, and del-Rop-F/R, leading to the generation of plasmids pMUT1N-gfp, pMUT1R-gfp, pMUT1Ht-gfp, pMUT1H-gfp and pMUT1NR-gfp. Similarly, to delete specific elements from pMUT2, pMUT2-gfp was amplified using the designated primers del-mobB-F/R, del-mobC-F/R, del-mobD-F/R, del-Hyp-F/R, del-DUF-F/R, del-Toxin-F/R, and del-Antitoxin-F/R, generating plasmids pMUT2B-gfp, pMUT2C-gfp, pMUT2D-gfp, pMUT2H-gfp, pMUT2Du-gfp, pMUT2T-gfp, pMUT2HD-gfp, pMUT2BC-gfp, pMUT2CD-gfp, pMUT2HB-gfp, pMUT2HC-gfp, pMUT2HD-gfp, pMUT2HBC-gfp, and pMUT2HCD-gfp.To remove the antibiotic resistance genes from the engineered cryptic plasmids, we introduced FRT (flippase recognition target) sites on both the left and right flanks of the resistance genes. Following the successful selection of positive transformants, the pCP20 plasmid was transferred into the host, allowing for the removal of the antibiotic resistance gene through the action of flippase recombination enzyme (FLP) [44].
Table 1.
Predicted elements of cryptic plasmids.
| Plasmids | Functional elements | Predicted function |
|---|---|---|
| pMUT1 | Rep | Replicase |
| Hyp1 | Hypothestical protein | |
| Hyp2 | Hypothestical protein | |
| HTH | Helix-turn-helix domain-containing protein | |
| Cer | Cer region | |
| NikA | Relaxase accessory protein of the conjugative plasmid | |
| RNAI | Inihibite the processing of precursor primer RNAII | |
| Rop | Rop family plasmid primer RNA-binding protein | |
| pMUT2 | Rep | Replicase |
| Hyp | Hypothetical protein | |
| DUF | DUF4868 domain-containing protein | |
| MobC | MobC family plasmid mobilization relaxosome | |
| MobA | Relaxase | |
| MobB | Mobilization protein | |
| MobD | MbeD family mobilization protein | |
| Toxin | Type II toxin-antitoxin system RelE/ParE family toxin | |
| Antitoxin | Type II toxin-antitoxin system RelB/DinJ family antitoxin |
To construct the library of artificial promoters, the BhepIII encoding gene (hepC) was amplified using primers BhepIII-F/R and inserted into the linearized plasmids pMUT1NR-P119-gfp and pMUT1NR-Ptac-gfp. This resulted in the generation of pMUT1NR-BhepIII-gfp and pMUT1NR-Ptac-BhepIII-gfp. Subsequently, the pMUT1NR-P119-BhepIII-gfp plasmid was amplified using sigma-F/R primers to construct the mutation library pMUT1NR-Psigma-BhepIII-gfp.
For the application of the engineered cryptic plasmids, amplification of pMUT1NR△, pMUT2HBC△, pCOLADuet, pRSFDuet-1, and pACYCDuet-1 was performed using the designated primers listed in Table S4. Additionally, genes including ectABC (encoding L-2,4-diaminobutyrate transaminase, 2,4-diaminobuty-rate acetyltransferase, and ectoine synthase from Halomonas elongata ATCC 3317), asd (aspartate-semialdehyde dehydrogenase from H. elongata ATCC 3317), lysCG1A, C932T (aspartate kinase with G1A and C932T mutations, Corynebacterium glutamicum ATCC 13032), and the BhepIII encoding gene (heparinase III from Bacteroides thetaiotaomicron VPI-5482) were amplified using primers listed in Table S4. The amplified genes, ectABC, asd, lysC, and hepC, were then ligated into the linearized pMUT1NR△, pMUT2HBC△, pCOLADuet, pRSFDuet-1, and pACYCDuet-1 plasmids, generating pMUT1NR△-BhepIII, pMUT1NR△-EctABC, pMUT2HBC△-LA, pMUT2HBC△-BhepIII, pCOLA-BhepIII, pRSF-EctABC, and pACYC-LA, respectively. T1-P1-F/R, T2-P1-F/R, pCO-P1-F/R, pRSF-P1-F/R, and pAC-P1-F/R primers were employed to replace the original promoter in pMUT1NR△-BhepIII, pMUT1NR△-EctABC, pMUT2HBC△-LA, pMUT2HBC△-BhepIII, pCOLAD-BhepIII, pRSF-EctABC, and pACYC-LA with the P1 promoter.
For verifying Hyp1-Hyp2 as a toxin-antitoxin complex, amplification of Hyp1, Hyp2 and Hyp1-Hyp2 was performed using the designated primers listed in Table S4. The amplified genes were then ligated into pBAD33, generating pBAD33-Hyp1, pBAD33-Hyp2 and pBAD33-Hyp12, respectively.
2.3. Analysis of engineered plasmids segregation stability in EcN
To evaluate the retention rate of plasmids in EcN, the EcN△1 or EcN△2 strains carrying plasmids derived from pMUT1 or pMUT2 were cultivated in 25 mL LB media with and without antibiotics at 37 °C for 12 h. Subsequently, every 12 h, the cells were re-inoculated into fresh LB media with and without antibiotics. During each 12-h interval, cell dilutions were collected and measured for green fluorescence intensity (excitation: 490 nm; emission: 530 nm) and cell densities (absorbance: 600 nm) using an Infinite 200 PRO plate reader. Concurrently, serial dilutions of the cells were plated onto LB agar plates with and without antibiotics. The plasmid-containing cells were determined based on the ratio of fluorescence to OD600 without antibiotics versus fluorescence to OD600 with antibiotics, approximated as the ratio of counted colonies without antibiotics to counted colonies with antibiotics. This defined the proportion of plasmid-containing cells.
2.4. Plasmid copy number determination by quantitative PCR
Templates for qPCR were obtained following the method described by Skulj, M. et al. [45]. The cells were subjected to incubation at 98 °C for 10 min, followed by centrifugation to collect the supernatant, which served as the template for qPCR. The qPCR primers were designed to target specific regions of both the EcN chromosome and the cryptic plasmid. Additionally, an AT-rich sequence (AATAAATCATAA) was added to the 5′ end of the primers, enhancing the qPCR fluorescence signal and improving the amplification efficiency [29,46]. We selected the recA and ldh genes on the EcN chromosome as reference genes, and their corresponding primers were recA-F/R and ldh-F/R, respectively. For targeting pMUT1 and pMUT2, we used T1-Rep-F/R and T2-Rep-F/R primers, respectively. The qPCR reactions were performed in a 50 μL mixture containing 25 μL of 2x SYBR Green®RPCR Master Mix (Vazyme), 200 nM of each primer (final concentration), and 4 μL of the sample. All qPCR reactions, including positive controls and negative controls (non-template control), were run on a QuantStudio3 instrument (Applied Biosystems). The universal cycling conditions for all reactions were as follows: an initial denaturation step at 95 °C for 30 s, followed by 40 cycles of denaturation at 95 °C for 10 s, annealing at 95 °C for 15 s, extension at 60 °C for 60 s, and a final extension at 95 °C for 15 s. The efficiency and specificity of the primers were evaluated by analyzing the standard curve and melting curve. The plasmid copy number was calculated using the comparative CT method (ΔΔC) based on the CT values obtained for the plasmid amplicons and the chromosome amplicons.
2.5. Construction and screening artificial of artificial promoters
The artificial promoter libraries constructed by interlocking the binding site of sigma factor σ70 and σ38 were performed as described previously [42]. To conduct the screening of efficient artificial promoters, the centrifuged cells were subjected to two washes with PBS (containing 8 g/L NaCl, 0.2 g/L KCl, 1.44 g/L Na2HPO4, and 8 g/L KH2PO4). The cells were then diluted to an OD600 of 0.3 and run at a rate of 0.5 μL/s. After gating, the cells were collected for analysis. Fluorescence characterization analysis was performed using FV10-ASW Viewer software and flow cytometry. The sorted plasmids were subsequently transformed into EcN to evaluate the efficiency of the promoters. The recombinant EcN strains were inoculated into 96-well plates for fluorescence measurement (following the method described above). Simultaneously, the recombinant EcN strains were also inoculated in 25 mL LB medium for enzyme activity measurement.
2.6. Heparinase III enzyme activity assay
The BhepIII enzyme activity towards heparin was assessed by measuring the A232 nm of unsaturated uronic acid using a UV-2450 spectrophotometer, with a molar extinction coefficient of 3800 L/(mole·cm). For the assay, a mixture consisting of 0.96 mL of 20 mM Tris-HCl buffer (pH 7.4), 20 g/L heparin, and 40 μL of the enzyme solution was prepared. The reaction was allowed to proceed for 1 min at 50 °C. The amount of protein required to produce 1 μmol of unsaturated uronic acid per minute at 50 °C was defined as one unit (U)
2.7. Quantification the titer of ecotine
Cell growth was monitored by measuring the A600nm using an Uvmini-1280 spectrometer. The extracellular and intracellular ectoine levels was quantified by HPLC (Agilent 1100) using an Thermo Scientific Hypersil GOLD aQ C18 column with an acetonitrile/water mixture (2:98 v/v) as the mobile phase (flow rate 0.6 mL/min). Ectoine was detected by a UV detector at a wavelength of 210 nm [47].
3. Results
3.1. Characterization and minimization of pMUT1
Although it has been documented the stable existence of native plasmids pMUT1 (Fig. 1A) and pMUT2 (Fig. 1B) in EcN [33], the presence of native pMUT plasmids impedes the transformation of subsequent recombinant pMUT plasmids. Therefore, we employed the CRISPR/Cas9 system and replicon incompatibility to eliminate the native pMUT1 and pMUT2 plasmids (Fig. 1C) to generate strains EcN△1 (with cure of pMUT1), EcN△2 (with cure of pMUT2), and EcN△12 (with cure of both pMUT1 and pMUT2), respectively. Given the ambiguous functions of the elements on cryptic plasmids, the sequences of pMUT1 (Fig. 1A) and pMUT2 (Fig. 1B) were annotated with plannotate [48], Uniprot [49], and NCBI Conserved Domain Database [50]. As shown in Table 1, pMUT1 consists of 8 coding sequences: Rep (ColEl-type replicon), RNAI (RNA for regulating plasmid replication) [51], Cer (recombinant site derived from ColE1, aiding in maintaining the plasmid as a monomer), HTH (protein containing a helical-turn-helix domain), Rop (a protein helps RNAI function) [52,53], NikA (relaxase accessory protein of the conjugative plasmid), Hyp1 (unknown functional protein) and Hyp2 (unknown functional protein). In consideration of the probably critical roles of elements Rep, RNAI and Cer in controlling plasmid replication, we selected HTH, Rop, NikA, Hyp1 and Hyp2 for the next knockout analyses.
Fig. 1.
Plasmid curing of pMUT1 and pMUT2. A. The map of the cryptic plasmid pMUT1 with a red frame, which designates the fragment to be verified using PCR. B. The map of the cryptic plasmid pMUT2 with a blue frame, which designates the fragment to be verified using PCR. C. The agarose gel images display the results of the removal of pMUT1 and pMUT2 (M: Marker, 1: EcN, 2: EcN with pMUT1 plasmid removed, 3: EcN with pMUT2 plasmid removed 4: EcN with pMUT1 and pMUT2 plasmids removed).
To assist the necessity of each segment on plasmid stability, the green fluorescence protein encoding gene (as a reporter) and the ampicillin resistance gene ampR (facilitating the screening of transformants) expression cassette that flanked by FRT sites were inserted into pMUT1 between Cer and HTH coding regions [29] to generate pMUT1-gfp (Fig. 2A, pMUT1 containing FRT site, gene gfp and ampicillin resistance encoding gene). Specifically, the plasmid loss rate was determined by calculating the ratio of fluorescent protein intensity with the absence and presence of antibiotics, which was also verified consistency with spread plate method by determining stability of pCOLAD-gfp (Fig S3, pCOLADuet-1 containing gene gfp). In general, a plasmid was considered lost when the rate of plasmid-containing cells was consistently below 95 % for two consecutive passages. As shown in Fig. 2A, pMUT1-gfp amplified stably in EcN for 6.5 days without the presence of ampicillin. In contrast, without the addition of kanamycin, the plasmid pCOLAD-gfp started to lose after only 12 h. After 6 days, the positive cells harboring pCOLAD-gfp dropped to less 20 % (Fig. 2B). Then, the putative coding sequences HTH, Rop, NikA, Hyp1 and Hyp2 were individually deleted and the impacts on plasmid stability were examined. In comparison, it could be found that deletion of HTH and Hyp1 coding sequences shorten the plasmid stability periods of pMUT1Ht-gfp (Fig. 2C, pMUT1-gfp with HTH coding sequence removed) and pMUT1H-gfp (Fig. 2D, pMUT1-gfp with Hyp1 coding sequence removed) to 2 days and 3 days, respectively. On the contrary, when deleting NikA and Rop coding sequences, the stability periods of pMUT1N-gfp (Fig. 2E, pMUT1-gfp with NikA coding sequence removed) and pMUT1R-gfp (Fig. 2F, pMUT1-gfp with Rop coding sequence removed) were extended to 9 days and 7 days, respectively, suggesting deletion of NikA and Rop is favorable to plasmid stability. Interestingly, we always failed to delete Hyp2 (no colony was observed on plate after transformation), suggesting its possible important role in interacting with other elements. Accordingly, the minimized plasmid pMUT1NR-gfp (Fig. 2G, pMUT1-gfp with NikA and Rop coding sequences removed) with deletion of both NikA and Rop was constructed which showed similar stability with pMUT1N-gfp (Fig. 2E). In addition, EcN cells harboring pMUT1NR-gfp exhibited better growth (Fig. 2H).
Fig. 2.
Investigating the impact of deleting elements from pMUT1 on segregation stability and growth via green fluorescence method. Segregation stability test of A. pMUT1-gfp, B. pCOLAD-gfp C. pMUT1Ht-gfp, D. pMUT1H-gfp, E. pMUT1N-gfp, F. pMUT1R-gfp and G. pMUT1NR-gfp. H. The growth of EcNP-gfp (EcN carrying pCOLAD-gfp), EcN1-gfp (EcN△1 carrying pMUT1-gfp), EcN1Ht-gfp (EcN△1 carrying pMUT1Ht-gfp), EcN1H-gfp (EcN△1 carrying pMUT1H-gfp), EcN1N-gfp (EcN△1 carrying pMUT1N-gfp), EcN1R-gfp (EcN△1 carrying pMUT1R-gfp), EcN1NR-gfp (EcN△1 carrying pMUT1NR-gfp).
3.2. Characterization and minimization of pMUT2
The pMUT2 plasmid was predicted to comprise the following components coding sequences: Rep (replicase), Hyp3 (protein-coding region with an unknown function), DUF (protein containing the DUF4868 domain), MobC (mobC family plasmid mobilization relaxosome), MobA (relaxase), MobB (mobilization protein), MobD (mbeD family mobilization protein), RelE (type II toxin-antitoxin system RelE/ParE family toxin), and RelB (type II toxin-antitoxin system RelB/DinJ family antitoxin) (Fig. 1B). In view of the critical roles of Rep and RelB in plasmid replication and neutralization of toxin, respectively, we skipped the deletion of these two genes.
In order to establish a dual plasmid expression system, the plasmid pMUT2-gfp was constructed by inserting the gene gfp and the kanamycin resistance gene between the DUF and MobC coding region [29]. Obviously, compared with pMUT1-gfp (Fig. 2A), pMUT2-gfp (Fig. 3A) showed better stability (8.5 days). With single deletion of MobB, MobC, MobD and Hyp3 coding sequences from pMUT2-gfp, the stability was extended to 13 days (Figs. 3B), 12 days (Figs. 3C), 12 days (Figs. 3D), and 13 days (Fig. 3E), respectively. Meanwhile, no obvious adverse effect on cell growth was observed (Fig. 3G). On the contrary, deletion of RelB or DUF decreased the plasmid stability (Fig. 3F, Fig. S1) while deletion of DUF also impacted cell growth (Fig. 3G). Additionally, similar with Hyp2 in pMUT1, the element MobA was always failed to remove. The results underscored the critical role of MobA, DUF and RelB elements in maintaining the separational stability and function of pMUT2 in EcN.
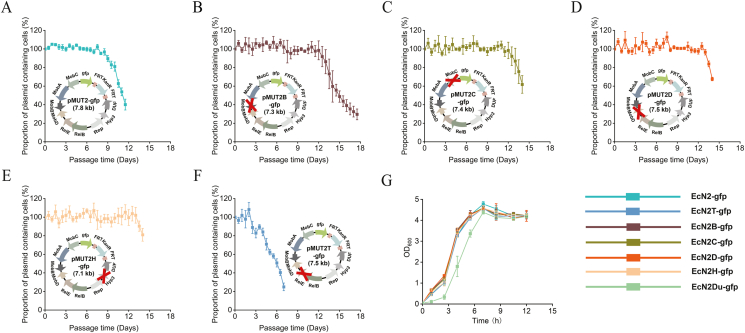
Fig. 3.
Examining the impact of individually deleting elements from pMUT2 on segregation stability and growth via green fluorescence method. Segregational stability test of plasmids A. pMUT2-gfp, B. pMUT2B-gfp, C. pMUT2C-gfp, D. pMUT2D-gfp, E. pMUT2H-gfp and F. pMUT2T-gfp. G. The growth of EcN2-gfp (EcN△2 carrying pMUT2-gfp), EcN2T-gfp (EcN△2 carrying pMUT2T-gfp), EcN2B-gfp (EcN△2 carrying pMUT2B-gfp), EcN2C-gfp (EcN△2 carrying pMUT2C-gfp), EcN2D-gfp (EcN△2 carrying pMUT2D-gfp), EcN2TA-gfp (EcN△2 carrying pMUT2TA-gfp), EcN2H-gfp (EcN△2 carrying pMUT2H-gfp), EcN2Du-gfp (EcN△2 carrying pMUT2Du-gfp).
Subsequently, double deletion of Hyp3, MobB, MobC, and MobD coding regions was further carried out. The stability periods of pMUT2HB-gfp (pMUT2-gfp with Hyp3 and MobB coding sequences removed), pMUT2HC-gfp(pMUT2-gfp with Hyp3 and MobC coding sequences removed), pMUT2HD-gfp (pMUT2-gfp with Hyp3 and MobD coding sequences removed), pMUT2BC-gfp (pMUT2-gfp with MobB and MobC coding sequences removed), and pMUT2CD-gfp (pMUT2-gfp with MobC and MobD coding sequences removed) with separate deletion of Hyp3-MobB, Hyp3-MobC, Hyp3-MobD, MobB-MobC, and MobC-MobD were 8 days (Fig. 4A), 11.5 days (Figs. 4B), 8.5 days (Figs. 4C), 10 days (Figs. 4D), and 13 days (Fig. 4E), respectively. In view of the failure for simultaneous deletion of MobB-MobD, triple deletions of Hyp3-MobB-MobC and Hyp3-MobC-MobD were performed to generate the plasmids pMUT2HBC-gfp (Fig. 4F, pMUT2-gfp with Hyp3, MobB and MobC coding sequences removed) and pMUT2HCD-gfp (Fig. 4G, pMUT2-gfp with Hyp3, MobC and MobD coding sequences removed), whose stability periods were 13 days and 12 days, respectively. Furthermore, the triple deletions did not affect cell growth (Fig. 4H). Taken together, the engineered plasmids pMUT1NR and pMUT2HBC were selected for next experiments.
Fig. 4.
Examining the impact of multiple element deletion from pMUT2 on segregation stability and growth via green fluorescence method. Segregational stability test of plasmids A. pMUT2HB-gfp, B. pMUT2HC-gfp, C. pMUT2HD-gfp, D. pMUT2BC-gfp, E. pMUT2CD-gfp, F. pMUT2HBC-gfp and G. pMUT2HCD-gfp. (H) The growth of EcN2HB-gfp (EcN△2 carrying pMUT2HB-gfp), EcN2HC-gfp (EcN△2 carrying pMUT2HC-gfp), EcN2HD-gfp (EcN△2 carrying pMUT2HD-gfp), EcN2BC-gfp (EcN△2 carrying pMUT2BC-gfp), EcN2CD-gfp (EcN△2 carrying pMUT2CD-gfp), EcN2HBC-gfp (EcN△2 carrying pMUT2HBC-gfp), EcN2HCD-gfp (EcN△1 carrying pMUT2HCD-gfp).
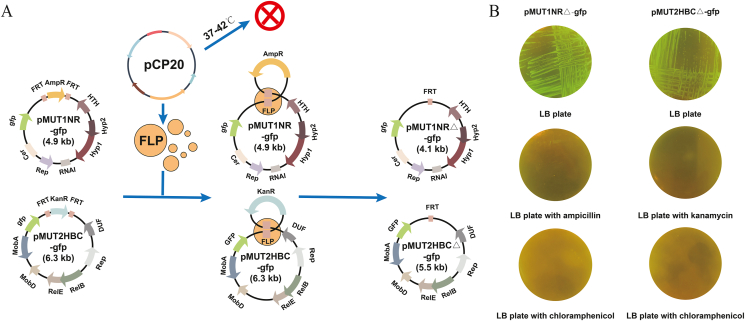
3.3. Elimination of the antibiotic resistance gene after selection
After selection, the presence of antibiotic resistance genes in host cells generally raised concerns about the spread of antibiotic resistance genes in the environment. In many cases, it is forbidden to use recombinant strains that containing antibiotic resistance genes even no antibiotic was added during cultivation [29]. Thus, the resistance genes in the engineered plasmids pMUT1NR-gfp (ampicillin) and pMUT2HBC-gfp (kanamycin) were eliminated by co-transforming the helper plasmid pCP20 (Fig. 5A) [44]. After thermal induction at 42 °C, the expressed FLP recombinase selectively acts on the repeated FRT sites flanking the resistance gene. At the same time, pCP20 was cured because of the temperature-sensitive replication. Eventually, the minimized cryptic plasmids pMUT1NR△ (pMUT1NR with ampicillin resistance gene removed) or pMUT2HBC△ (pMUT2HBC with kanamycin resistance encoding gene removed) with good separational stability were used for expressing genes of interest, for instance gene gfp here (Fig. 5B). Also, the copy numbers of plasmids pMUT1 (20 copies per cell), pMUT2 (12 copies per cell), pMUT1NR△ (30 copies per cell) and pMUT2HBC△ (15 copies per cell) were determined using qPCR [45] while no significant changes were detected (Table S1).
Fig. 5.
Removal of antibiotic resistance gene. A. The process of eliminating the antibiotic resistance gene through FLP/FRT. B. Host cells harboring plasmid pMUT1NR△ or pMUT2HBC△ were inoculated onto LB plates supplemented with ampicillin or kanamycin to confirm the elimination of the respective resistance genes. Additionally, these cells were also inoculated onto LB plates supplemented with chloramphenicol to verify the removal of plasmid pCP20.
3.4. Construction of an artificial promoter library with gradient strength
To broaden the potential applications of the engineered EcN expression systems, we next developed a synthetic promoter library by adopting the strategy of interlocking the binding motifs of different sigma factors [42,54,55]. In view of the leadership of σ70 and σ38 for regulating most of genes in E. coli [56,57], we devised an artificial promoter sequence consisting of four segments: the −35 region recognition sequence of σ70 (TTGACA) and the −10 region recognition sequence of σ70 (TATAAT), as well as the −35 region recognition sequence of σ38 (TCTTGT) and the −10 region recognition sequence of σ38 (GCTATACT). These recognition sequences are interspersed with random sequences, while an AT-rich sequence is positioned immediately upstream of the recognition sequence. As shown in Fig. 6A, to screen promoters supporting the expression of BhepIII, a heparinase from B. thetaiotaomicron, the BhepIII encoding sequence was designed at N-terminal and fused by gene gfp with the commonly used GS linker (GGTGGTGGTTCTGGTGGTGGTTCTGGTGGTGGTTCCGGTGGTGGTTCT).
Fig. 6.
Construction of screening artificial promoters. A. The arrangement of the artificial promoters involved assembling interlocking sigma factor σ70 and σ38 binding motifs. The consensus sequences −10 and −35, recognized by sigma factors σ70 and σ38, respectively, were highlighted in red and blue. The letters N and W represent the generated bases A, T, C, or G. The reporter for the artificial promoter was the fusion of BhepIII encoding gene with gfp using the GS sequence. B. Flow cytometry analysis was conducted on strain Top10-sigma using the ScrABBLE device. The plasmids located in the “4 region” of the cells were transformed into EcN. C. A total of 15 colonies were selected and inoculated into shaking flasks for measuring fluorescence and BhepIII activity. The promoters PJ23119 and Ptac were used as controls.
The artificial promoter library was screened using flow cytometry. Recombinant E. coli Top10 cells exhibiting fluorescence intensities (4 region in Fig. 6B) were collected and a series of promoters with varying intensity gradients were screened (Fig. 6C). Following the sorting process, plasmids were extracted and transformed into EcN△12 with cure of both pMUT1 and pMUT2. The fluorescence values of the recombinant EcN strains were determined with 96-well plates. The enzymatic activities of BhepIII in strains exhibiting higher fluorescence values of were measured in parallel. Notably, a list of synthetic promoters with a broad range of regulation strength were constructed (Table 2). Compared with the artificial strong promoter PJ23119, the strongest promoter P1 (Fig. 6C and Table 2) resulted in higher BhepIII activity and green fluorescence intensity values (Fig. 6C).
Table 2.
Sequence of artificial promoters (The red region denotes the binding sequence of σ70, while the blue region signifies the binding sequence of σ38).
| Name | Sequence (5′–3′) |
|---|---|
| P1 | TATTTTATTGACAATCTTGTACATAGTCTTTATAATGCTATACT |
| P2 | TATTTATTGACATTGTTGAATACGAAATTTATAATGATATACT |
| P3 | TAAATTTTGACAATCTTGTTCGCAGCTCGTATAATGCTATACT |
| P4 | TAATATTTGACACCTCTTGTCGACGTACTATATAATGCTATACT |
| P5 | TAAAATTTGACAAATCTTGTTCTTCCATCGTATAATGCTATACT |
| P6 | TATAATTTGACAATTCTTGTTCCTTCTTCCTATAATGCTATACT |
| P7 | TTATATTTGACATTTCTTGTTTCTTTCTTTTATAATGCTATACT |
| P8 | TATTTTTTGACACCTCTTGTCGTGACTGCTTATAATGCTATACT |
| P9 | AATTTTTTGACACGTCTTGTATACCCTGTGTATAATGCTATACT |
| P10 | ATAAAATTGACACCTCTTGTACTAGACCGTTATAATGCTATAC |
| P11 | AATAATTTGACACATCTTGTAAGTTATAACTATAATGCTATACT |
| P12 | AAAAAATTGACACCTCTTGTACTGCTCCCGTATAATGCTATACT |
| P13 | TTAATTTTGACACATCTTGTTTCCTAGCCGTATAATGCTATACT |
| P14 | TTATAATTGACAACTCTTGTCCTTCCGCTTTATAATGCTATACT |
| P15 | TTTAAATTGACAATTCTTGTTTATAACAAATATAATGCTATACT |
3.5. Application of the antibiotic-free systems for protein expression and metabolic engineering
To illustrate the practicality of the engineered cryptic plasmids embedded with synthetic promoters, the enzyme BhepIII was expressed in EcN△12 by utilizing the optimized cryptic plasmids pMUT1NR△ and pMUT2HBC△. In parallel, we also comparatively investigated the performance of the original cryptic plasmids pMUT1 and pMUT2, as well as the traditional plasmid pCOLADuet-1. Specifically, all the BhepIII expression cassettes were driven by the engineered promoter PP1 (Fig. 7A). As shown in Fig. 7B, both pMUT1NR△ and pMUT2HBC△ generated much higher activities comparing to pCOLADuet-1, pMUT1 and pMUT2. The enzymatic activity of BhepIII reached 5400 U/L when applying the minimized pMUT1NR△, which was 210 % of that from pCOLADuet-1. In parallel, we also evaluate the capacity of pMUT1NR△ and pMUT2HBC△ for metabolic pathway engineering of ectoine in EcN. Accordingly, three enzymes (EctA, EctB and EctC) encoding genes and two enzymes LysC G1A, C932T and Asd encoding genes (Fig. 7D) [47] were designed as two expression cassettes and assembled with pMUT1NR△ and pMUT2HBC△ to generate pMUT1NR△-ectABC and pMUT2HBC△-lysC G1A, C932T-asd, respectively (Fig. 7C). As a control, the two expression cassettes were also inserted into pRSFDuet-1 and pACYCDuet-1 to produce pRSFDuet-ectABC and pACYCDuet-lysC G1A, C932T-asd, respectively (Fig. 7C). After transformation, the recombinant EcN strains with two different systems were cultivated and the titer of ectoine was measured. As shown in Fig. 7E, compared with the EcNPP-EctABC-LA (EcN△12 carrying pRSF-P1-EctABC and pACYC-P1-LA) cultured with antibiotic, the EcN12-EctABC-LA (EcN△12 carrying pMUT1NR△-P1-EctABC and pMUT2HBC△-gfp-P1-LA) cultured without antibiotic displayed higher titer of ectoine, suggesting the superiority of the antibiotic-free systems for constructing robust microbial cell factories.
Fig. 7.
Application of the artificial promoters and engineered cryptic plasmids system to ectoine and BhepIII overproduction. A. Schematic diagram of BhepIII synthesis via the cryptic plasmids and traditional plasmids. B. Activity profiles of BhepIII carried by pCOLADuet-1, pMUT1NR△ and pMUT2HBC△. C. Schematic diagram of ectoine synthesis via the antibiotic-dependent plasmids and cryptic plasmids. D. Ectoine is synthesized by the ectoine synthase (encoded by gene ectC) enzyme using N-acetyl-2,4-diamino-butyrate as a precursor. Enzymes that are overexpressed are highlighted with colorful arrows. 2,4-diaminobutyrate acetyltransferase (encoded by gene ectA), L-2,4-diaminobutyrate transaminase (encoded by gene ectB); aspartate-semialdehyde dehydrogenase (encoded by gene asd); aspartate kinase with G1A and C932T mutation (encoded by gene lysCG1A, C932T). E. Growth and metabolism of strains EcN12-EctABC-LA (EcN△12 carrying pMUT1NR△-P1-EctABC and pMUT2HBC△-P1-LA) cultured with antibiotic and EcNPP-EctABC-LA (EcN△12 carrying pRSF-P1-EctABC and pACYC-P1-LA) cultured without antibiotic.
4. Discussion
In this study, we eliminated the non-essentail elements of two cryptic plasmids pMUT1 and pMUT2 and investigated their contributions to plasmid separational stability. Eventually, pMUT1NR△ and pMUT2HBC△ with minimized size and better stability were constructed and applied for enzyme expression and metabolic pathway engineering.
For pMUT1 minimization, we found that deletion of the element HTH or Hyp1 exhibited significantly adverse effects on plasmid stability (Fig. 2CD) while Hyp2 always failed to eliminate. The helix-turn-helix (HTH) motif, predominantly present in prokaryotic transcription factors, performs essential functions such as DNA repair, transcription, plasmid replication, and RNA metabolism [58]. This may explain the observed alterations in plasmid segregation stability following the knockout of HTH (Fig. 2D). In pMUT2, the toxin (RelE)-antitoxin (RelB) cassette has been annotated. In contrast, no corresponding toxin-antitoxin partners have been characterized in pMUT1. Recently, more and more studies have demonstrated the wide distribution of the toxin-antitoxin fragment and its plasmid-stabilizing capabilities [34,59,60]. Thus, in view of the negative effect of Hyp1 deletion on plasmid stability, the unerasability of Hyp2, and the natural location of Hyp1 and Hyp2 in one gene cluster, we speculated that Hyp1 and Hyp2 form a toxin-antitoxin system to uphold the segregation stability of pMUT1 (Fig. 1A). Further investigations with the arabinose-induced rigorous plasmid pBAD33 confirmed that Hyp1 and Hyp2 form a toxin-antitoxin system while Hyp1 acts as a toxin (Fig. S2). On the contrary, the good performance of pMUT1N-gfp with deletion of NikA might be ascribed to the decreased interaction between NikA (relaxase accessory protein) and Rep [61]. Furthermore, Rop has the ability to enhance the inhibitory activity of RNAI on RNAII [52,53]. As a result, it was reasonable that the absence of Rop resulted in increased segregation stability (Fig. 2F).
In pMUT2, the Mob proteins MobA, MobB, MobC and MobD are involved in facilitating the mobility of mobilizable ColE1 plasmids between cells [62,63]. Herein, our results showed that single deletion of MobA or double deletion of MobB and MobD is lethal, suggesting that the balanced expression of these Mob proteins for cell normal growth. The segment DUF with unknown function is also found within other E. coli plasmids. The decreased stability revealed that DUF might be is associated with plasmid replication (Fig. 3G). Furthermore, it is noteworthy that Hyp3 exhibits 99 % amino acid sequence similarity with KwaA, which is prokaryotic antiviral defense protein in a given genomic sequence [64].
In conclusion, two compatible minimized cryptic plasmids, namely pMUT1NR△ and pMUT2HBC△ from pMUT1 and pMUT2 were constructed, respectively. By combining the engineered promoters with different strengths, two antibiotic-free expression systems were used for heparinase III expression and ectoine biosynthesis in EcN. Comparing with the traditional antibiotic-dependent expression systems, the antibiotic-free expression systems would boost the synthetic biology studies and clinical and industrial applications for EcN.
CRediT authorship contribution statement
Siyan Zhou: Investigation, Formal analysis, Software, Writing – original draft. Linlin Zhao: Investigation, Formal analysis, Software, Writing – original draft. Wenjie Zuo: Investigation. Yilin Zheng: Investigation. Ping Zhang: Investigation. Yanan Sun: Investigation. Yang Wang: Reviewing. Guocheng Du: Reviewing. Zhen Kang: Conceptualization, Funding acquisition, Supervision, Writing – review & editing.
Declaration of competing interest
The authors declare there is no conflict of interest.
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (32370066, 32000058), the Fundamental Research Funds for the Central Universities (JUSRP622003), National First-class Discipline Program of Light Industry Technology and Engineering (QGJC20230202) and the Postgraduate Research & Practice Innovation Program of Jiangsu Province (KYCX23_2487).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.synbio.2024.01.006.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
References
- 1.Pontrelli S., Chiu T.Y., Lan E.I., Chen F.Y., Chang P., Liao J.C. Escherichia coli as a host for metabolic engineering. Metab Eng. 2018;50:16–46. doi: 10.1016/j.ymben.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 2.Li Y., Lin Z., Huang C., Zhang Y., Wang Z., Tang Y.-j., Chen T., Zhao X. Metabolic engineering of Escherichia coli using CRISPR–Cas9 meditated genome editing. Metab Eng. 2015;31:13–21. doi: 10.1016/j.ymben.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 3.Jiang Y., Chen B., Duan C., Sun B., Yang J., Yang S. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl Environ Microbiol. 2015;81:2506–2514. doi: 10.1128/aem.04023-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu C.C., Qi L., Lucks J.B., Segall-Shapiro T.H., Wang D., Mutalik V.K., Arkin A.P. An adaptor from translational to transcriptional control enables predictable assembly of complex regulation. Nat Methods. 2012;9:1088–1094. doi: 10.1038/nmeth.2184. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y., Yin G., Weng H., Zhang L., Du G., Chen J., Kang Z. Gene knockdown by structure defined single-stem loop small non-coding RNAs with programmable regulatory activities. Synth Syst Biotechnol. 2023;8:86–96. doi: 10.1016/j.synbio.2022.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu Y., Li Y., Xu Y., Zhang J., Ma L., Qi Q., Wang Q. Development of bifunctional biosensors for sensing and dynamic control of glycolysis flux in metabolic engineering. Metab Eng. 2021;68:142–151. doi: 10.1016/j.ymben.2021.09.011. [DOI] [PubMed] [Google Scholar]
- 7.Sonnenborn U. Escherichia coli strain Nissle 1917-from bench to bedside and back: history of a special Escherichia coli strain with probiotic properties. FEMS Microbiol Lett. 2016;363 doi: 10.1093/femsle/fnw212. [DOI] [PubMed] [Google Scholar]
- 8.Geldart K.G., Kommineni S., Forbes M., Hayward M., Dunny G.M., Salzman N.H., Kaznessis Y.N. Engineered E. coli Nissle 1917 for the reduction of vancomycin-resistant Enterococcus in the intestinal tract. Bioeng Transl Med. 2018;3:197–208. doi: 10.1002/btm2.10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palmer J.D., Piattelli E., McCormick B.A., Silby M.W., Brigham C.J., Bucci V. Engineered probiotic for the inhibition of Salmonella via tetrathionate-induced production of microcin H47. ACS Infect Dis. 2018;4:39–45. doi: 10.1021/acsinfecdis.7b00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sarate P.J., Heinl S., Poiret S., Drinić M., Zwicker C., Schabussova I., Daniel C., Wiedermann U., coli Nissle E. Is a safe mucosal delivery vector for a birch-grass pollen chimera to prevent allergic poly-sensitization. Mucosal Immunol. 1917;12:132–144. doi: 10.1038/s41385-018-0084-6. 2019. [DOI] [PubMed] [Google Scholar]
- 11.Yu X., Lin C., Yu J., Qi Q., Wang Q. Bioengineered Escherichia coli Nissle 1917 for tumour-targeting therapy. Microb Biotechnol. 2020;13:629–636. doi: 10.1111/1751-7915.13523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu M., Hu S., Tang B., Yang H., Sun D. Engineering Escherichia coli Nissle 1917 as a microbial chassis for therapeutic and industrial applications. Biotechnol Adv. 2023;67 doi: 10.1016/j.biotechadv.2023.108202. [DOI] [PubMed] [Google Scholar]
- 13.Lynch J.P., Goers L., Lesser C.F. Emerging strategies for engineering Escherichia coli Nissle 1917-based therapeutics. Trends Pharmacol Sci. 2022;43:772–786. doi: 10.1016/j.tips.2022.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Datta P., Fu L., Brodfuerer P., Dordick J.S., Linhardt R.J. High density fermentation of probiotic E. coli Nissle 1917 towards heparosan production, characterization, and modification. Appl Microbiol Biotechnol. 2021;105:1051–1062. doi: 10.1007/s00253-020-11079-9. [DOI] [PubMed] [Google Scholar]
- 15.Hu S., Zhao L., Hu L., Xi X., Zhang Y., Wang Y., Chen J., Chen J., Kang Z. Engineering the probiotic bacterium Escherichia coli Nissle 1917 as an efficient cell factory for heparosan biosynthesis. Enzym Microb Technol. 2022;158 doi: 10.1016/j.enzmictec.2022.110038. [DOI] [PubMed] [Google Scholar]
- 16.Chen J., Li X., Liu Y., Su T., Lin C., Shao L., Li L., Li W., Niu G., Yu J., Liu L., Li M., Yu X., Wang Q. Engineering a probiotic strain of Escherichia coli to induce the regression of colorectal cancer through production of 5-aminolevulinic acid. Microb Biotechnol. 2021;14:2130–2139. doi: 10.1111/1751-7915.13894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan X., Liu X.Y., Zhang D., Zhang Y.D., Li Z.H., Liu X., Wu F., Chen G.Q. Construction of a sustainable 3-hydroxybutyrate-producing probiotic Escherichia coli for treatment of colitis. Cell Mol Immunol. 2021;18:2344–2357. doi: 10.1038/s41423-021-00760-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park Y.T., Kim T., Ham J., Choi J., Lee H.S., Yeon Y.J., Choi S.I., Kim N., Kim Y.R., Seok Y.J. Physiological activity of E. coli engineered to produce butyric acid. Microb Biotechnol. 2022;15:832–843. doi: 10.1111/1751-7915.13795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu S., Fei M., Fu B., Yu M., Yuan P., Tang B., Yang H., Sun D. Development of probiotic E. coli Nissle 1917 for β-alanine production by using protein and metabolic engineering. Appl Microbiol Biotechnol. 2023;107:2277–2288. doi: 10.1007/s00253-023-12477-5. [DOI] [PubMed] [Google Scholar]
- 20.Lan Y.-J., Tan S.-I., Cheng S.-Y., Ting W.-W., Xue C., Lin T.-H., Cai M.-Z., Chen P.-T., Ng I.S. Development of Escherichia coli Nissle 1917 derivative by CRISPR/Cas9 and application for gamma-aminobutyric acid (GABA) production in antibiotic-free system. Biochem Eng J. 2021;168 doi: 10.1016/j.bej.2021.107952. [DOI] [Google Scholar]
- 21.Xu J., Xia K., Li P., Qian C., Li Y., Liang X. Functional investigation of the chromosomal ccdAB and hipAB operon in Escherichia coli Nissle 1917. Appl Microbiol Biotechnol. 2020;104:6731–6747. doi: 10.1007/s00253-020-10733-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Azam M.W., Khan A.U. CRISPRi-mediated suppression of E. coli Nissle 1917 virulence factors: a strategy for creating an engineered probiotic using csgD gene suppression. Front Nutr. 2022;9 doi: 10.3389/fnut.2022.938989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodríguez-Nogales A., Algieri F., Garrido-Mesa J., Vezza T., Utrilla M.P., Chueca N., Fernández-Caballero J.A., García F., Rodríguez-Cabezas M.E., Gálvez J. The administration of Escherichia coli Nissle 1917 ameliorates development of DSS-induced colitis in mice. Front Pharmacol. 2018;9:468. doi: 10.3389/fphar.2018.00468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sarate P.J., Srutkova D., Geissler N., Schwarzer M., Schabussova I., Inic-Kanada A., Kozakova H., Wiedermann U. Pre- and neonatal imprinting on immunological homeostasis and epithelial barrier integrity by Escherichia coli Nissle 1917 prevents allergic poly-sensitization in mice. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.612775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harimoto T., Hahn J., Chen Y.Y., Im J., Zhang J., Hou N., Li F., Coker C., Gray K., Harr N., Chowdhury S., Pu K., Nimura C., Arpaia N., Leong K.W., Danino T. A programmable encapsulation system improves delivery of therapeutic bacteria in mice. Nat Biotechnol. 2022;40:1259–1269. doi: 10.1038/s41587-022-01244-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X., Pang G., Sun T., Liu X., Pan H., Zhang Y., Liu J., Chang J., Wang H., Liu D. A red light-controlled probiotic bio-system for in-situ gut-brain axis regulation. Biomaterials. 2023;294 doi: 10.1016/j.biomaterials.2023.122005. [DOI] [PubMed] [Google Scholar]
- 27.Zhao R., Li Z., Sun Y., Ge W., Wang M., Liu H., Xun L., Xia Y. Engineered Escherichia coli Nissle 1917 with urate oxidase and an oxygen-recycling system for hyperuricemia treatment. Gut Microb. 2022;14 doi: 10.1080/19490976.2022.2070391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yue J., Gou X., Li Y., Wicksteed B., Wu X. Engineered epidermal progenitor cells can correct diet-induced obesity and diabetes. Cell Stem Cell. 2017;21:256–263. doi: 10.1016/j.stem.2017.06.016. e4. 10.1016/j.stem.2017.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kan A., Gelfat I., Emani S., Praveschotinunt P., Joshi N.S. Plasmid vectors for in vivo selection-free use with the probiotic E. coli Nissle 1917. ACS Synth Biol. 2021;10:94–106. doi: 10.1021/acssynbio.0c00466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu M., Kim J., Ahn J.H., Moon Y. Nononcogenic restoration of the intestinal barrier by E. coli-delivered human EGF. JCI Insight. 2019;4 doi: 10.1172/jci.insight.125166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kurtz C.B., Millet Y.A., Puurunen M.K., Perreault M., Charbonneau M.R., Isabella V.M., Kotula J.W., Antipov E., Dagon Y., Denney W.S., Wagner D.A., West K.A., Degar A.J., Brennan A.M., Miller P.F. An engineered E. coli Nissle improves hyperammonemia and survival in mice and shows dose-dependent exposure in healthy humans. Sci Transl Med. 2019;11 doi: 10.1126/scitranslmed.aau7975. [DOI] [PubMed] [Google Scholar]
- 32.LE Yi-Lin S.Y., Wang Hong-Cheng, Wei-Lan S.H.A.O. Advances in selection markers and their bio-safety in applications of transformed microorganisms. Microbiology China. 2016;43(8):1814–1821. [Google Scholar]
- 33.Blum-Oehler G., Oswald S., Eiteljörge K., Sonnenborn U., Schulze J., Kruis W., Hacker J. Development of strain-specific PCR reactions for the detection of the probiotic Escherichia coli strain Nissle 1917 in fecal samples. Res Microbiol. 2003;154:59–66. doi: 10.1016/s0923-2508(02)00007-4. [DOI] [PubMed] [Google Scholar]
- 34.Fedorec A.J.H., Ozdemir T., Doshi A., Ho Y.K., Rosa L., Rutter J., Velazquez O., Pinheiro V.B., Danino T., Barnes C.P. Two new plasmid post-segregational killing mechanisms for the implementation of synthetic gene networks in Escherichia coli. iScience. 2019;14:323–334. doi: 10.1016/j.isci.2019.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang S., Kang Z., Cao W., Du G., Chen J. Construction of a novel, stable, food-grade expression system by engineering the endogenous toxin-antitoxin system in Bacillus subtilis. J Biotechnol. 2016;219:40–47. doi: 10.1016/j.jbiotec.2015.12.029. [DOI] [PubMed] [Google Scholar]
- 36.Chen Z., Yao J., Zhang P., Wang P., Ni S., Liu T., Zhao Y., Tang K., Sun Y., Qian Q., Wang X. Minimized antibiotic-free plasmid vector for gene therapy utilizing a new toxin-antitoxin system. Metab Eng. 2023;79:86–96. doi: 10.1016/j.ymben.2023.07.003. [DOI] [PubMed] [Google Scholar]
- 37.Buddenborg C., Daudel D., Liebrecht S., Greune L., Humberg V., Schmidt M.A. Development of a tripartite vector system for live oral immunization using a gram-negative probiotic carrier. Int J Med Microbiol. 2008;298:105–114. doi: 10.1016/j.ijmm.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 38.Whelan R.A., Rausch S., Ebner F., Günzel D., Richter J.F., Hering N.A., Schulzke J.D., Kühl A.A., Keles A., Janczyk P., Nöckler K., Wieler L.H., Hartmann S. A transgenic probiotic secreting a parasite immunomodulator for site-directed treatment of gut inflammation. Mol Ther. 2014;22:1730–1740. doi: 10.1038/mt.2014.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zainuddin H.S., Bai Y., Mansell T.J. CRISPR-based curing and analysis of metabolic burden of cryptic plasmids in Escherichia coli Nissle 1917. Eng Life Sci. 2019;19:478–485. doi: 10.1002/elsc.201900003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dong M.M., Song L., Xu J.Q., Zhu L., Xiong L.B., Wei D.Z., Wang F.Q. Improved cryptic plasmids in probiotic Escherichia coli Nissle 1917 for antibiotic-free pathway engineering. Appl Microbiol Biotechnol. 2023;107:5257–5267. doi: 10.1007/s00253-023-12662-6. [DOI] [PubMed] [Google Scholar]
- 41.Fiege K., Frankenberg-Dinkel N. Construction of a new T7 promoter compatible Escherichia coli Nissle 1917 strain for recombinant production of heme-dependent proteins. Microb Cell Factories. 2020;19:190. doi: 10.1186/s12934-020-01447-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y., Liu Q., Weng H., Shi Y., Chen J., Du G., Kang Z. Construction of synthetic promoters by assembling the sigma factor binding -35 and -10 boxes. Biotechnol J. 2019;14 doi: 10.1002/biot.201800298. [DOI] [PubMed] [Google Scholar]
- 43.Wang Y., Shi Y., Hu L., Du G., Chen J., Kang Z. Engineering strong and stress-responsive promoters in Bacillus subtilis by interlocking sigma factor binding motifs. Synth Syst Biotechnol. 2019;4:197–203. doi: 10.1016/j.synbio.2019.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Datsenko K.A., Wanner B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Skulj M., Okrslar V., Jalen S., Jevsevar S., Slanc P., Strukelj B., Menart V. Improved determination of plasmid copy number using quantitative real-time PCR for monitoring fermentation processes. Microb Cell Factories. 2008;7:6. doi: 10.1186/1475-2859-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Afonina I., Ankoudinova I., Mills A., Lokhov S., Huynh P., Mahoney W. Primers with 5' flaps improve real-time PCR. Biotechniques. 2007;43:770–772. doi: 10.2144/000112631. 774. [DOI] [PubMed] [Google Scholar]
- 47.Wang D., Chen J., Wang Y., Du G., Kang Z. GreenChE; 2021. Engineering Escherichia coli for high-yield production of ectoine. [DOI] [Google Scholar]
- 48.McGuffie M.J., Barrick J.E. pLannotate: engineered plasmid annotation. Nucleic Acids Res. 2021;49:W516–w522. doi: 10.1093/nar/gkab374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blum M., Chang H.Y., Chuguransky S., Grego T., Kandasaamy S., Mitchell A., Nuka G., Paysan-Lafosse T., Qureshi M., Raj S., Richardson L., Salazar G.A., Williams L., Bork P., Bridge A., Gough J., Haft D.H., Letunic I., Marchler-Bauer A., Mi H., Natale D.A., Necci M., Orengo C.A., Pandurangan A.P., Rivoire C., Sigrist C.J.A., Sillitoe I., Thanki N., Thomas P.D., Tosatto S.C.E., Wu C.H., Bateman A., Finn R.D. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 2021;49:D344–d354. doi: 10.1093/nar/gkaa977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marchler-Bauer A., Derbyshire M.K., Gonzales N.R., Lu S., Chitsaz F., Geer L.Y., Geer R.C., He J., Gwadz M., Hurwitz D.I., Lanczycki C.J., Lu F., Marchler G.H., Song J.S., Thanki N., Wang Z., Yamashita R.A., Zhang D., Zheng C., Bryant S.H. CDD: NCBI's conserved domain database. Nucleic Acids Res. 2015;43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cesareni G., Helmer-Citterich M., Castagnoli L. Control of ColE1 plasmid replication by antisense RNA. Trends Genet. 1991;7:230–235. doi: 10.1016/0168-9525(91)90370-6. [DOI] [PubMed] [Google Scholar]
- 52.Chaillou S., Stamou P.E., Torres L.L., Riesco A.B., Hazelton W., Pinheiro V.B. Directed evolution of colE1 plasmid replication compatibility: a fast tractable tunable model for investigating biological orthogonality. Nucleic Acids Res. 2022;50:9568–9579. doi: 10.1093/nar/gkac682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cesareni G., Muesing M.A., Polisky B. Control of ColE1 DNA replication: the rop gene product negatively affects transcription from the replication primer promoter. Proc Natl Acad Sci U S A. 1982;79:6313–6317. doi: 10.1073/pnas.79.20.6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Y., Shi Y., Hu L., Du G., Chen J., Kang Z. Engineering strong and stress-responsive promoters in Bacillus subtilis by interlocking sigma factor binding motifs. Synth. Syst. Biotechnol. 2019;4:197–203. doi: 10.1016/j.synbio.2019.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mejía-Almonte C., Busby S.J.W., Wade J.T., van Helden J., Arkin A.P., Stormo G.D., Eilbeck K., Palsson B.O., Galagan J.E., Collado-Vides J. Redefining fundamental concepts of transcription initiation in bacteria. Nat Rev Genet. 2020;21:699–714. doi: 10.1038/s41576-020-0254-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cho B.K., Kim D., Knight E.M., Zengler K., Palsson B.O. Genome-scale reconstruction of the sigma factor network in Escherichia coli: topology and functional states. BMC Biol. 2014;12:4. doi: 10.1186/1741-7007-12-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kang Z., Wang Q., Zhang H., Qi Q. Construction of a stress-induced system in Escherichia coli for efficient polyhydroxyalkanoates production. Appl Microbiol Biotechnol. 2008;79:203–208. doi: 10.1007/s00253-008-1428-z. [DOI] [PubMed] [Google Scholar]
- 58.Aravind L., Anantharaman V., Balaji S., Babu M.M., Iyer L.M. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol Rev. 2005;29:231–262. doi: 10.1016/j.femsre.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 59.Ni S., Li B., Tang K., Yao J., Wood T.K., Wang P., Wang X. Conjugative plasmid-encoded toxin-antitoxin system PrpT/PrpA directly controls plasmid copy number. Proc Natl Acad Sci U S A. 2021;118 doi: 10.1073/pnas.2011577118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jurėnas D., Fraikin N., Goormaghtigh F., Van Melderen L. Biology and evolution of bacterial toxin-antitoxin systems. Nat Rev Microbiol. 2022;20:335–350. doi: 10.1038/s41579-021-00661-1. [DOI] [PubMed] [Google Scholar]
- 61.Moran R.A., Hall R.M. Analysis of pCERC7, a small antibiotic resistance plasmid from a commensal ST131 Escherichia coli, defines a diverse group of plasmids that include various segments adjacent to a multimer resolution site and encode the same NikA relaxase accessory protein enabling mobilisation. Plasmid. 2017;89:42–48. doi: 10.1016/j.plasmid.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 62.Varsaki A., Lamb H.K., Eleftheriadou O., Vandera E., Thompson P., Moncalián G., de la Cruz F., Hawkins A.R., Drainas C. Interaction between relaxase MbeA and accessory protein MbeC of the conjugally mobilizable plasmid ColE1. FEBS Lett. 2012;586:675–679. doi: 10.1016/j.febslet.2012.01.060. [DOI] [PubMed] [Google Scholar]
- 63.Meyer R. Functional organization of MobB, a small protein required for efficient conjugal transfer of plasmid R1162. J Bacteriol. 2011;193:3904–3911. doi: 10.1128/jb.05084-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tesson F., Hervé A., Mordret E., Touchon M., d'Humières C., Cury J., Bernheim A. Systematic and quantitative view of the antiviral arsenal of prokaryotes. Nat Commun. 2022;13:2561. doi: 10.1038/s41467-022-30269-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.