Abstract
Esophageal cancer is an upper gastrointestinal malignancy with a bleak prognosis. It is still being explored in depth due to its complex molecular mechanisms of occurrence and development. Lipids play a crucial role in cells by participating in energy supply, biofilm formation, and signal transduction processes, and lipid metabolic reprogramming also constitutes a significant characteristic of malignant tumors. More and more studies have found esophageal cancer has obvious lipid metabolism abnormalities throughout its beginning, progress, and treatment resistance. The inhibition of tumor growth and the enhancement of antitumor therapy efficacy can be achieved through the regulation of lipid metabolism. Therefore, we reviewed and analyzed the research results and latest findings for lipid metabolism and associated analysis techniques in esophageal cancer, and comprehensively proved the value of lipid metabolic reprogramming in the evolution and treatment resistance of esophageal cancer, as well as its significance in exploring potential therapeutic targets and biomarkers.
Keywords: Lipid metabolism, Esophageal cancer, Progression, Treatment resistance, New therapeutic targets
Graphical abstract
Highlights
-
•
The molecular characteristics of esophageal cancer through lipid metabolism is explored.
-
•
Potential therapeutic targets or biomarkers related to lipid metabolism is analyzed.
-
•
The analysis technology of lipid metabolism in esophageal cancer is summarized.
1. Introduction
The latest statistics showed that in 2020, approximately 604,100 new cases and 544,100 deaths were reported worldwide due to esophageal cancer [1]. One out of every eighteen cancer-related mortalities might be caused by esophageal cancer [2], which showed that esophageal cancer has high morbidity and mortality and seriously threatens human health. Common histological subtypes include esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC), which account for 85% and 14% of esophageal cancers, respectively [1]. As the symptoms of esophageal cancer in the early stage are not obvious, more than half of patients are in middle and advanced stages when they are initially diagnosed [3]. In operable locally advanced esophageal cancer, drug and radiation therapy are usually performed before surgery to reduce the volume of the tumor and increase the probability of complete resection of the lesion [4]. Definitive chemoradiotherapy is the standard treatment for patients with unresectable, locally advanced esophageal cancer [5]. The most commonly used drugs to treat esophageal cancer include paclitaxel, fluorouracil, platinum-based drugs, capecitabine, S-1, immune checkpoint inhibitors (mainly programmed cell death protein 1 and programmed cell death ligand 1 (PD-1/PD-L1) inhibitors), etc.. Noteworthy, immune checkpoint inhibitors combined with chemotherapy have become the first-line treatment for advanced esophageal cancer [6,7]. In recent years, strategies for managing esophageal cancer have shown advancements in surgery, radiotherapy, and drug therapy. However, further improvement in therapeutic efficacy is still needed.
The esophageal mucosa is repeatedly subjected to physical or chemical injury, such as hot beverages, alcohol, tobacco smoke, stomach acid, etc., resulting in DNA damage and mutation of epithelial cells and eventually leading to malignant changes in normal cells. Among them, esophageal cancer-associated abnormal pathways are enriched in cell cycle regulators, tyrosine kinase receptors, chromatin remodeling, and embryonic pathways [8]. Given the complexity of its pathogenesis, active research on esophageal cancer is still ongoing. Aberrant lipid metabolism and metabolic reprogramming constitute significant biological hallmarks of malignant tumors and exhibit a strong association with the onset and advancement of esophageal cancer by promoting cancer cells uncontrolled proliferation, survival, invasion, and resistance to antineoplastic therapy [9,10]. Lipids are a class of small-molecular metabolites with molecular weights of less than 1,500 Da, and play important roles in the course of the life activity of the organism, such as energy supply, biological membrane formation, and signal transduction. Therefore, normal regulation of lipid metabolism is essential to maintaining cellular homeostasis [11].
With an in-depth understanding of the biological functions of lipids and the development of mass spectrometry (MS) technology, “lipidomics” has gradually formed based on “metabolomics” [12]. Lipidomics is devoted to the study of the pathways and networks of lipid metabolism and the variations of whole-lipidome profiling in various biological phenomena by analyzing the composition, structure, and quantification of lipids from single cells to whole-organisms [13]. Compared to genes, proteins, and peptides, lipid metabolites are structurally unstable and insoluble in water, which poses challenges for lipidomics analysis, particularly in acquiring high lipid throughput, coverage, and accuracy [14]. As the main tool for lipid analysis, MS has greatly improved the coverage and accuracy of lipid detection by using single MS techniques or integrated MS-based multiple omics techniques. Traditional MS-based lipid analysis requires homogenized tissues for lipid analysis, losing the spatial location of lipid molecules in the tissues or organs. However, this problem has been further solved by the emergence and development of MS imaging (MSI) technology, which visualizes the spatial distribution of lipids and promises to enable multiple characterizations of molecular information in cancer analysis [15]. Above all, MS and MSI-based lipidomics had made considerable progress, providing qualitative, quantitative, and spatial distribution variation of lipids to explore the complex link between lipid metabolism and tumors [12,[16], [17], [18]]. Analyzing the characteristics of lipid metabolism in esophageal cancer is helpful to reveal its malignant performance and resistance mechanisms and identify potential new drug targets. Therefore, we summarized and analyzed the relevant findings of lipid metabolic reprogramming in esophageal cancer, most of which are studies in the past five years, hoping to comprehensively show the potential value of lipid metabolism and explore possible therapeutic targets or biomarkers, hoping to further cement management of esophageal cancer and related diseases.
2. Lipid composition and metabolism
2.1. Composition
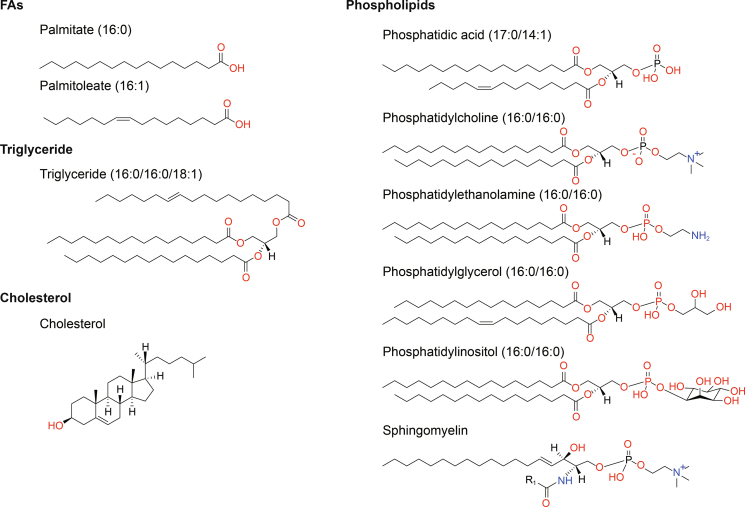
Lipids are structurally rich and diverse, and nearly 50,000 distinct lipid structures can be acquired in the LIPID MAPS Structure Database as of March 2023 (https://www.lipidmaps.org/databases/lmsd), which can be divided into eight main categories, including fatty acyls, glycerolipids, glycerophospholipids (GP), sphingolipids, sterol lipids, prenol lipids, saccharolipids, and polyketides [19,20]. Among them, fatty acids (FAs) are the main components of fatty acyls and the basic building blocks of most lipids. FAs can be further categorized in line with carbon chain length and the number and position of C=C bonds, such as palmitate (16:0) and palmitoleate (16:1) (Fig. 1). Triglycerides are the most common glycerolipids and the major energy-storing form in living organisms (Fig. 1). Phospholipids are the main components of biological membranes, including GP and sphingomyelin (SM) (the major sphingolipids in mammals). GP can be further divided into phosphatidic acid (PA), phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylglycerol (PG), phosphatidylinositol (PI), and phosphatidylserine (PS) according to changes in its polar head. The nonpolar tail of GP comprises one or two long-chain FAs, often containing unsaturated bonds, and an even carbon skeleton (Fig. 1). Under inflammatory stimulation, cytosolic phospholipase A2 can catalyze phospholipids in the cell membrane to release arachidonic acid. Subsequently, arachidonic acid can be further metabolized to bioactive lipids (such as prostaglandins, leukotrienes, hydroxyeicosatetraenoic acids, and epoxyeicosatrienoic acids) to participate in and promote the inflammatory response, which may be closely related to the occurrence and development of tumors [21,22]. Sphingomyelin, usually synthesized from PC and ceramide (Cer), is not only an important membrane phospholipid but also a reservoir of Cer. Cholesterol is one of the more broadly investigated sterol lipids and, together with phospholipid, participates in biofilm formation (Fig. 1). Because FA, triglyceride, phospholipid, and cholesterol are lipids of greater concern in esophageal cancer, this review mainly introduces their metabolism in detail.
Fig. 1.
Representative structures of various classes of lipids. The structures of the above lipid molecules were obtained from LIPID MAPS (https://lipidmaps.org/databases/lmsd/overview). R1 indicates the aliphatic chain of fatty acid (FA) residue.
2.2. Metabolism
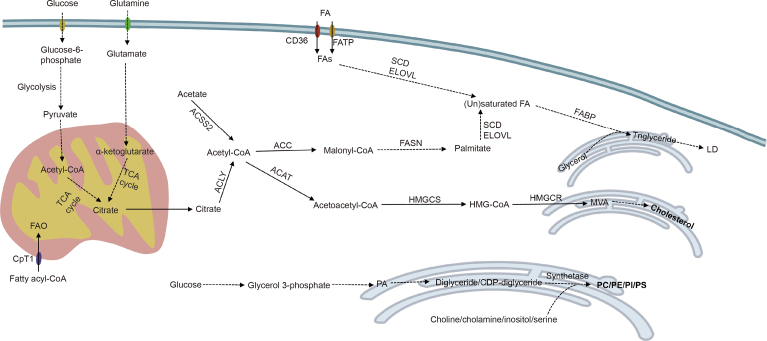
Lipid metabolism is a complex and vital biochemical process in disease-related biological systems. With the assistance of various enzymes, the absorption, synthesis, and breakdown of lipids can ensure the normal operation of cell physiology, which is of important significance for life activities. In the organism, both glucose and glutamine can be converted into FAs, which are de novo synthesis of FAs (Fig. 2). De novo FA synthesis is carried out in the cytosol using acetyl-CoA as the direct feedstock. Citrate, derived from the tricarboxylic acid cycle (TCA cycle) or glutamine metabolism in the mitochondria, enters the cytosol and produces acetyl-CoA under ATP-citrate lyase (ACLY). Besides, acetyl-CoA can also be synthesized by acetyl-CoA synthetase 2 (ACSS2), which catalyzes acetate in the cytoplasm [23]. Subsequently, acetyl-CoA is irreversibly translated to malonyl-CoA by acetyl-CoA carboxylase (ACC), which is a rate-limiting step [24]. Under the catalysis of FA synthetase (FASN), acetyl-CoA continuously reacts with malonyl-CoA until palmitate (16:0) is formed [25]. Longer-chain FAs can be synthesized by increasing the length of the palmitate carbon chain under the elongation of very-long-chain FAs protein enzymes [26]. The FAs synthesized as described above can be catalyzed by stearoyl-CoA desaturase (SCD) to produce monounsaturated FAs, such as palmitoleate (C16:1) and oleate (C18:1) [27]. The transcription factor sterol regulatory element-binding protein 1 (SREBP1) controls the activation of various lipogenic enzyme genes, including ACC, FASN, and SCD, which is central to the regulation of FA synthesis [28].
Fig. 2.
Simplified diagram of major lipid metabolic pathways. TCA cycle: tricarboxylic acid cycle; FAO: fatty acid oxidation; CPT1: carnitine palmitoyl transferase; FA: fatty acid; CD36: cluster of differentiation 36; FATP: fatty acid transport protein; FA: fatty acid; SCD: stearoyl-CoA desaturase; ELOVL: elongation of very-long-chain fatty acids protein; FABP: fatty-acid-binding protein; LD: lipid droplet; ACSS2: acetyl-CoA synthetase 2; ACLY: ATP-citrate lyase; ACC: acetyl-CoA carboxylase; FASN: fatty acid synthetase; ACAT: acetyl-CoA acetyltransferase; HMGCS: 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) synthetase; HMGCR: HMG-CoA reductase; MVA: mevalonate; PA: phosphatidic acid; PC: phosphatidylcholine; PE: phosphatidylethanolamine; PI: phosphatidylinositol; PS: phosphatidylinositol.
In addition, extracellular FAs, especially the essential polyunsaturated FAs, can also be directly taken up by cells via FA translocase (including cluster of differentiation 36 (CD36)), FA transport protein (FATP, also known as SLC27A), and FA-binding protein (FABP) in the plasma membrane [[29], [30], [31]] (Fig. 2). Intracellularly, FAs are transported to different organelles for storage and utilization via FABP. Typically, storage is carried out in the form of lipid droplets (LDs), organelles consisting of a monolayer of phospholipid membranes that store neutral lipids. FAs react with glycerol in the endoplasmic reticulum (ER) to produce triglycerides, and the aggregation of triglycerides leads to the formation of LDs protruding from the ER membrane. The energy provided by FAs primarily occurs through FA oxidation (FAO) in mitochondria (Fig. 2). After long-chain FAs are catalyzed to fatty acyl-CoA by acyl-CoA synthetase, carnitine palmitoyl transferase 1 (CPT1), a pivotal regulatory enzyme in FAO, mediates the uptake of fatty acyl-CoA into mitochondria, where they participate in the oxidation reaction and TCA cycle to release energy.
Phospholipids are mainly synthesized through the Kennedy pathway, which requires CTP to activate choline, ethanolamine, and diglycerides to eventually generate the corresponding phospholipids [32]. Among them, PC is the most abundant phospholipid in biological membranes, and lysophosphatidylcholine acyltransferase (LPCAT) can promote the transformation of newborn PC into mature membrane PC and participate in PC remodeling [33]. Besides, through the hydrolysis of phospholipases (phospholipase C (PLC), phospholipase D (PLD), and phospholipase A (PLA)), phospholipids can be degraded into many bioactive lipid mediators such as diacylglycerol, PA, lysophosphatidic acid (lysoPA), and arachidonic acid, which are important in intracellular and intercellular signaling [34]. This process can modulate various cellular variations, including proliferation, survival, and inflammation, during disease progression [35].
Enzymes for cholesterol synthesis are found in the cytoplasm and smooth ER. Acetyl-CoA in the cytoplasm is utilized as a substrate and catalyzed by acetyl-CoA acetyltransferase (ACAT) and 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) synthetase to form HMG-CoA. As a key enzyme in cholesterol synthesis, HMG-CoA reductase catalyzes HMG-CoA to mevalonate (MVA), which is subsequently modified to produce various cholesterols with biological functions (Fig. 2).
2.3. Reprogramming
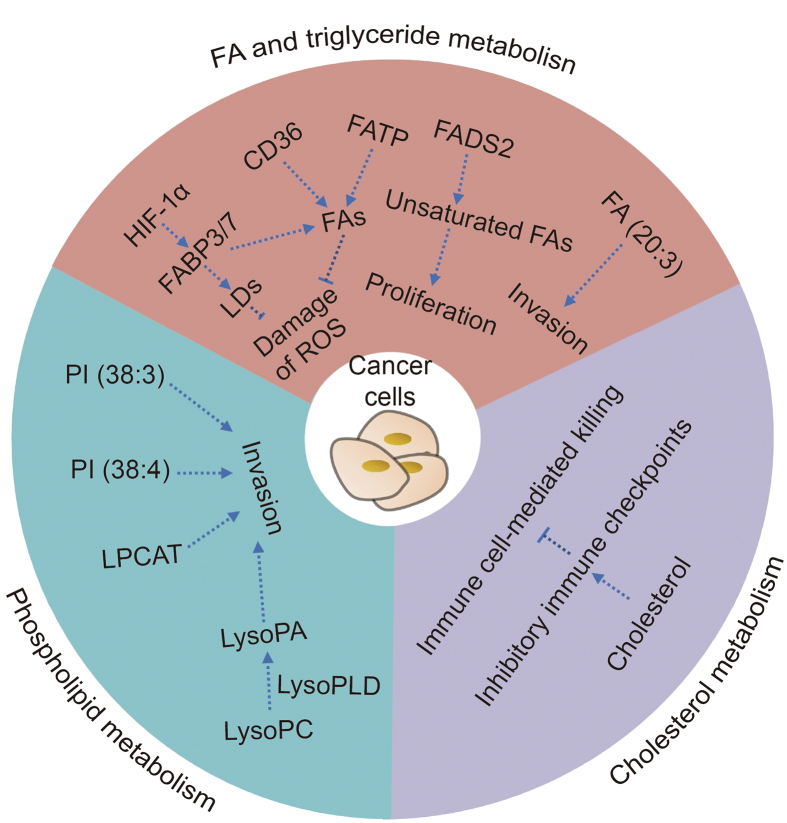
To fulfill the need for continuous proliferation and rapid growth, tumor cells show different characteristics in lipid metabolism compared to normal cells. This phenomenon, known as lipid metabolic reprogramming, simultaneously enhances the multiplication and invasion capacities of tumor cells, thereby increasing their malignancy [36,37] (Fig. 3). For example, tumor cells have an enhanced ability to synthesize FAs de novo compared to normal cells, which is one of the metabolic characteristics of tumor cells [27]. With the uncontrolled growth of tumor cells, the vascular system that transports oxygen and nutrients in tumor tissues is disordered and hypoplastic, resulting in an abnormal hypoxic tumor microenvironment. In hypoxic tumor cells, de novo FA synthesis is reduced, and the increased expression of hypoxia-inducible factor 1α further induces upregulation of FABP3/7 expression to promote increased FA input and intracellular LDs accumulation, which can reduce the damage caused by reactive oxygen species (ROS) to tumor cells and increase the survival of hypoxia-reoxygenated cells [38]. In addition, the expression of genes and proteins for CD36 and FATP in tumor cells also increased significantly to facilitate the uptake of extracellular FA and maintain tumor cell proliferation [39]. Inhibiting CD36 expression can reduce the accumulation of cholesterol and LDs induced by adipocytes in tumor cells and reduce the content of intracellular ROS, preventing the development of adipocyte-induced malignant tumor phenotypes [40]. Additionally, SREBP can also regulate FA and cholesterol metabolism, and many proto-oncogenic signaling pathways can regulate the expression of SREBP, motivating the reprogramming of lipid metabolism in tumors [16].
Fig. 3.
Reprogrammed fatty acid (FA) and triglyceride, phospholipid, and cholesterol metabolism in cancer cells. HIF-1α: hypoxia-inducible factor-1α; FABP3/7: fatty-acid-binding protein 3/7; LDs: lipid droplets; ROS: reactive oxygen species; CD36: cluster of differentiation 36; FATP: fatty acid transport protein; FADS2: fatty acid desaturase 2; LysoPC: lysophosphatidylcholine; LysoPLD: lysophospholipase D; LysoPA: lysophosphatidic acid; LPCAT: lysophosphatidylcholine acyltransferase; PI: phosphatidylinositol.
Unsaturated FAs, as the raw materials for biofilm synthesis, are crucial to cells, so alternative FA desaturation pathways other than the SCD pathway exist to meet their demand for unsaturated FAs [41]. For instance, FA desaturase 2 can convert palmitate (C16:0) into sapienate (C16:1). When SCD is inhibited, tumor cells undergo metabolic adjustments to continue producing unsaturated FAs through the FASD2 pathway to meet the needs of biofilm formation and tumor cell proliferation [42]. While FA synthesis and utilization are increased in tumor cells, it is possible that FA catabolism may also be altered. Compared to healthy persons, the peripheral blood of ESCC patients has significantly lower medium- and long-chain acylcarnitines (the main substrates for energy produced by FAO in mitochondria), which may imply altered activity of β-oxidation and the TCA cycle in ESCC [43].
Zang et al. [44] examined and compared the metabolic properties of an in vitro model of multicellular tumor spherical (MCTS) esophageal cancer with clinical ESCC samples by matrix-assisted laser desorption/ionization mass spectrometry imaging (MALDI-MSI). Interestingly, they discovered in vitro model of esophageal cancer MCTS and clinical ESCC tissues exhibited similar spatial and temporal metabolic features; for example, compared with normal esophageal tissues, the expression of FA (20:3), PI (38:3), and PI (38:4) was significantly increased in all cell and human samples of esophageal cancer [44]. Additionally, in esophageal cancer MCTS, overexpressions of FA (20:3), PI (38:3), and PI (38:4) were located in the outer region (proliferation zone), which also supported the perspective that metabolic signatures of FA and phospholipid accumulation may be associated with tumor cell aggressiveness [38,45]. Moreover, the reprogramming of lipid metabolism in esophageal cancer cells is bound up with abnormal expression of metabolic enzymes [46]. Both esophageal cancer MCTS and clinical ESCC tissues indeed exhibited abnormally increased expression of FASN compared to normal tissues [44]. As a key enzyme in de novo FA synthesis, FASN is overexpressed in most cancers, which may represent a common phenotypic alteration in response to adverse microenvironmental changes [25]. Besides, lysophospholipase D (lysoPLD) is highly expressed in various tumors and catalyzes the conversion of lysophosphatidylcholine (lysoPC) into lysoPA, which may contribute to the occurrence, progression, and metastasis of cancer [47]. The composition of phospholipids and the expression of LPCAT are also altered in various tumor tissues, including esophageal cancer, and have been proven to contribute to tumor development [48].
Recent studies have revealed the crucial role of mevalonate pathway enzymes in the survival of cancer cells, underscoring the importance of MVA pathway metabolites for cancer cell survival [49]. For example, high cholesterol levels in the tumor microenvironment can promote immune cells to express inhibitory immune checkpoints to protect tumor cells from immunologic attack, which implies that cancer patients with hypercholesterolemia may have a better response to immunotherapy [50].
However, with the occurrence, development, and treatment resistance of tumors, lipid metabolism in tumor cells changes dynamically at different stages. Thus, it is necessary to conduct specific analysis at each stage to fully understand these alterations. Fortunately, significant progress has been made in the lipidomics analysis technique, which further meets the needs of researchers to explore the characteristics of lipid metabolism in tumors.
3. Lipidomics analysis techniques
3.1. MS-based lipidomics
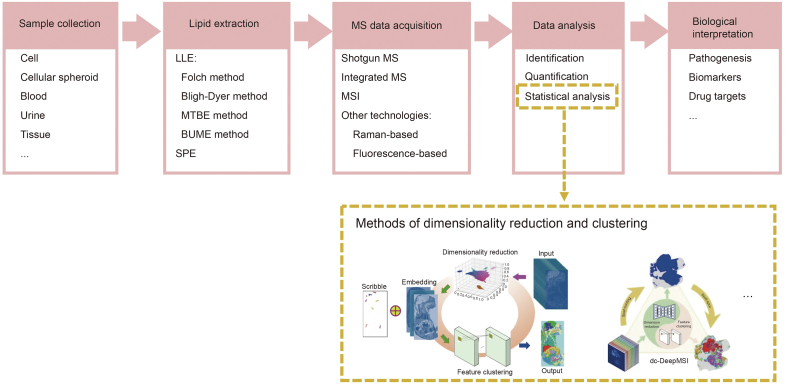
The MS platform has been widely used as a fundamental technique in lipidomics research. Fig. 4 [51,52] shows the experimental procedures for MS-based lipidomics studies, including data analysis. MS-based lipidomics can be split into untargeted and targeted lipidomics. The purpose of untargeted metabolomics is to conduct a systematic and comprehensive analysis of endogenous metabolites in a sample, obtain a large amount of data on metabolites, and best reflect the total metabolite traits of the sample. The purpose of targeted metabolomics is to verify whether the target metabolite exists in the sample and to obtain the absolute content of the target metabolite in different samples. The techniques of MS-based lipidomics analysis can be mainly divided into three types: shotgun MS analysis using direct injection detection, integrated MS methods including separation techniques (such as liquid chromatography (LC) and gas chromatography (GC)), and MSI techniques acquiring the spatial distribution of lipids from single cells to whole organisms [53]. Among them, most analytical methods require enrichment and extraction of lipids from samples before MS or MSI, which is an important prerequisite for successful lipid analysis.
Fig. 4.
The operating procedures in lipidomics research. Typically, lipidomics research includes five parts: sample collection, lipid extraction, mass spectrometry (MS) data acquisition, data analysis, and biological interpretation. LLE: liquid-liquid extraction; MTBE: methyl tert-butyl ether; BUME: butanol and methanol; SPE: solid-phase extraction; MSI: mass spectrometry imaging. Reprinted from Refs. [51,52] with permission.
The commonly used lipid extraction approaches involve liquid-liquid extraction (LLE) and solid-phase extraction (SPE). LLE can extract many species of lipids by using water and water-immiscible organic solvents, which is suitable for untargeted lipidomics analysis [54]. As the earliest LLE method for lipid extraction, the Folch method consisted of chloroform, methanol, and water (8:4:3, V/V/V), which can make the extraction rate of lipids greater than 90%, including glyceride/PC/PE/sphingolipids [55]. Based on the Folch method, Bligh and Dyer adjusted the volume ratio of chloroform/methanol/water to 1:2:0.8 and added acid additives to improve the extraction efficiency of FAs and acidic phospholipids [56]. However, the chloroform phase is located below the aqueous phase, which may be disturbed by water-soluble substances during operation. Due to the harmfulness of chloroform, chloroform-free methods, such as methyl tert-butyl ether (MTBE) and butanol and methanol (BUME), are developed for lipid extraction. The MTBE method consists of MTBE, methanol, and water (5:1.5:1.45, V/V/V), and the BUME method requires butanol/methanol (3:1, V/V), heptane/ethyl acetate (3:1, V/V), and 1% acetic acid [57,58]. Their extraction efficiency on lipids is comparable to or higher than that of the gold-standard Folch method. For example, the MTBE method can extract polar and negatively charged phospholipids more efficiently [57]. In addition, SPE can be used alone or in combination with LLE and requires the analyte to be adsorbed onto the stationary phase, and then the solution with different elution capacities is used for stepwise elution to achieve lipid separation and enrichment. In general, SPE is often used to extract one or several classes of lipids that need to be studied in depth, especially low-abundance lipids from targeted lipidomics analysis [59].
Shotgun MS is a commonly used method for untargeted lipidomics analysis, in which MS techniques such as high-resolution MS, tandem MS, and ion mobility MS can be utilized to improve resolution and accuracy [60,61]. The common MS for untargeted lipidomics includes quadruple time-of-flight MS, orbitrap MS, and Fourier transform ion cyclotron resonance MS. For targeted lipidomics, triple quadrupole MS and quadrupole linear-ion trap MS are the main modes for qualitative and quantitative analysis of lipids. In addition, the combination of chromatography technique and MS represents the integration of high resolution and high sensitivity, which plays an important role in improving detection accuracy, identifying lipid isomers, and detecting low-abundance lipids. The most frequently used separation techniques include GC, LC, and supercritical fluid chromatography (SFC). GC takes the gas phase as the mobile phase and achieves the effective separation of lipids by gasifying the sample and transferring it to the chromatographic column. GC is appropriate for the extraction of volatile lipids, and GC-MS is the main method for FA and cholesterol analysis and quantification [62,63]. Generally, acid derivatization methods and basic derivatization methods are often utilized to further improve the separation efficiency of GC-MS, which also facilitates the separation of non-volatile lipids. By contrast, LC is the most broadly used chromatographic method due to its high sensitivity, easy integration with MS, and suitability for concentrating volatile, non-volatile, and heat-labile lipids. The mobile phase of LC-MS is the liquid phase. Impurities such as small particles in the mobile phase need to be removed to avoid clogging the chromatographic column and affecting its lifetime. Bubbles from dissolution or mixing in the mobile phase should also be removed to avoid affecting the detection results. SFC is a novel separation method that can separate a variety of lipids from polar to non-polar with high efficiency, high resolution, and high throughput [64].
Usually, lipid classification is identified by comparing and integrating the information of carbon atoms and double bond numbers, chemical formula, and monoisotopic mass matched to lipid-related databases, which is very important because accurate identification and quantification are the basis of research and analysis [65]. Therefore, it may be necessary to understand the characteristic fragmentation patterns of various lipids [66]. Available online databases include LIPID MAPS (http://www.lipidmaps.org/), LipidBank (https://lipidbank.jp/), and LipidBlast (https://fiehnlab.ucdavis.edu/), etc.. The output of lipidomics data produces a complex and high-throughput dataset that requires dimensionality reduction and clustering to discover lipid characteristics between samples. Dimensionality reduction and clustering methods include but are not limited to principal component analysis [67], linear discriminant analysis [68], isometric mapping [69], t-distributed stochastic neighbor embedding [18], uniform manifold approximation and projection [70], orthogonal partial least squares-discriminant analysis [67], partial least squares regression [71], and k-means [72]. Post-processing of lipidomics data, such as statistical analysis and visualization, is an important step to finding the biological significance of lipid features [73]. Therefore, with the development of lipid detection platforms, the technology of lipid analysis has also progressed. Currently, commonly used tools for statistical analysis and visualization in lipidomic analysis include lipidr [74], LipidSuite [75], MetaboAnalyst [76], etc..
When analyzing biological samples, researchers frequently employ targeted and non-targeted lipidomics techniques in conjunction with multivariate data analysis to investigate the lipid profiles of samples and identify distinctions between tumor and normal samples [65,77]. In addition, metabolic pathway matching analysis can be performed on differential lipids obtained from the analysis of the lipidomic profiles of various samples to further identify potential tumor-related metabolic enzymes. Moreover, traditional biological experimental methods such as Western blot, immunohistochemistry, and quantitative real-time polymerase chain reaction (PCR) can be employed to detect the expression of these metabolic enzymes and validate them in conjunction with the results obtained from lipid analysis.
3.2. MSI-based lipidomics
Usually, MS-based lipidomics analysis can be performed on homogenized conditions for various biological samples, including cells, body fluids (blood, urine, and cerebrospinal fluid), and tissues, lacking representative spatial information to reflect the sample heterogeneity. The development of MSI has effectively solved this problem [15].
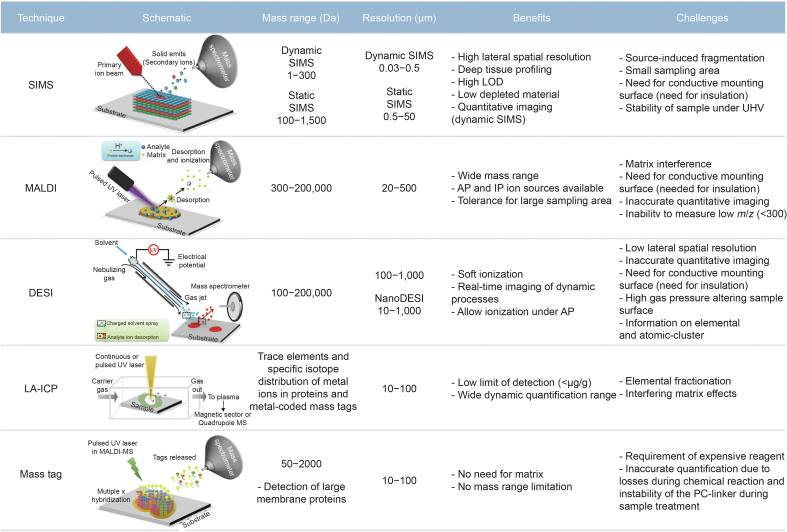
Based on the integrated MS and advanced imaging technique, MSI can obtain the spatial distribution of hundreds to thousands of molecules at the section level with a label-free analysis. Combined with histopathological information, MSI can accurately reflect the metabolic heterogeneity among organelles, cells, and tissues [78]. Previously, we described in detail the different sample preparation requirements from the cellular to tissue level in MSI-based lipidomics [79]. Suitable sample sections are a prerequisite for getting the accurate location and abundance of lipids and sustaining adequate spatial resolution and MS signals [79]. There is a high requirement for sample integrity: all sections must not be deformed, folded, ripped, or cracked during sample preparation, preservation, cryosectioning, and sticking [79]. In addition, the emergence of more optimized ion sources is also necessary to promote the application of MSI in lipidomic analysis. The main function of an ion source is to provide energy for sample ionization to form ion beams with different mass-to-charge ratios and finally transfer them to a mass analyzer for mass-to-charge ratio analysis, which is the cornerstone of subsequent data analysis and obtaining real lipid profiles. Therefore, reducing or avoiding ion source-generated artifacts can further improve the sensitivity and accuracy of MSI and promote the development of precise lipid analysis [80]. Depending on the ionization method, the main MSI used for lipidomics analysis includes three technologies: secondary ion mass spectrometry MSI (SIMS-MSI), MALDI-MSI, and desorption electrospray ionization MSI (DESI-MSI) [15]. SIMS-MSI is the earliest MSI method with the highest spatial resolution (up to tens of nanometers). SIMS-MSI utilizes a primary ion beam to strafe the sample surface, and the mass-to-charge ratio of the sputtering secondary ions with charge is detected by a mass spectrometer. Due to the high energy of the primary ion beam using SIMS-MSI, a large number of fragmented ions can disturb the lipid identification. MALDI-MSI is a broadly used MSI method and has high spatial resolution, but it requires the application of matrix on the sample surface to promote compound ionization. Therefore, the correct selection and application of the MALDI matrix is crucial for MALDI-MSI, which directly affects the ionization efficiency of analytes and the spatial resolution of imaging. DESI-IMS uses a charged solvent spray to bind to the sample surface to desorb and ionize lipids. Compared with the first two techniques, DESI-MSI possesses the least destructive ionization method and allows the ionization of molecules at atmospheric pressure without matrix spray with a lower resolution. By introducing high-rate airflow into the ion source of DESI-MSI, Abliz and co-workers [81] developed air flow-assisted desorption electrospray ionization mass spectrometry imaging (AFADESI-MSI), which improves the detection of metabolites and lipids, especially low-abundance molecules, with wide coverage, high sensitivity, and fast analysis. The comparison of MALDI-MSI, DESI-MSI, and SIMS-MSI in terms of their main principles, benefits, and challenges is shown in Fig. 5 [82].
Fig. 5.
Main principles, benefits, and challenges of several mass spectrometry imaging (MSI) methods. SIMS: secondary ion mass spectrometry; LOD: limit of detection; UHV: ultra-high vacuum; MALDI: matrix-assisted laser desorption/ionization; UV: ultroviolet; AP: atmosphere pressure; IP: intermediate pressure; DESI: desorption electrospray ionization; LA-ICP: laser ablation inductively coupled plasma; MS: mass spectrometry; PC: photocleavable. Reprinted from Ref. [82] with permission.
Compared to the analysis process of MS-based lipid data, the analysis of MSI-based lipid data is more difficult because the latter recovers the complex spatial information of lipids, which multiplies the amount of research data. And MSI segmentation is one of the key and most difficult problems in data processing. Cai and co-workers have also explored this aspect and invented an interactive strategy to improve spatial segmentation of MSI, called iSegMSI, and an unsupervised segmentation mode based on deep learning, namely divide-and-conquer (dc)-DeepMSI (Fig. 4) [51,52]. iSegMSI can fine-tune the segmentation results to integrate or separate regions of interest based on the scribble-regulated prior information to improve MSI segmentation [51]. dc-DeepMSI can identify regions of interest and perform metabolic heterogeneity analysis without prior knowledge in spat-contig and spat-spor modes [52]. Although there is software for MSI data analysis, such as SCiLS Lab, Image Quest, MSiReader, etc., in order to deeply mine the characteristics of the lipid metabolism of tumors and potential targets for intervention, more effective MSI data analysis algorithms may still need to be developed.
3.3. Other analytical techniques for lipids
In addition to MS and MSI, there are other methods available for lipid detection, including Raman-based technologies and fluorescence-based technologies. These methods are commonly used for lipid analysis in cells and subcellular compartments. Raman-based technologies utilize the Raman scattering of laser-illuminated samples, which is a non-elastic scattering phenomenon of light. When combined with microscopy, Raman-based technologies allow for the non-destructive identification and imaging of specific lipid molecules present in the sample by analyzing the Raman scattering patterns [83,84]. However, Raman-based imaging techniques face challenges in identifying molecules with similar vibrational modes and detecting low concentrations of lipids [85]. Compared to Raman-based techniques, fluorescence microscopy offers higher sensitivity for measuring extremely low concentrations of cellular lipids, although it does not possess the same level of specificity as MS [85]. Using fluorescent probes, fluorescence microscopy is primarily employed for imaging plasma membranes, organelle membranes, and lipid droplets [86]. However, the labeling process of fluorescent probes may alter lipid behavior in studies. Additionally, achieving absolute specificity in binding to specific lipids with fluorescent probes also presents a challenge.
4. Lipid metabolism-associated occurrence, progression, and metastasis in esophageal cancer
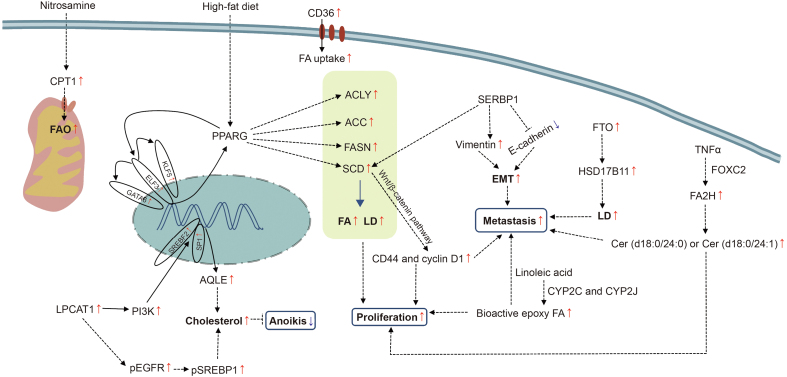
Tumor development is accompanied by lipid metabolic reprogramming in tumor cells. Abnormal lipid metabolism not only enables tumor cells to produce more energy to adapt to the nutrient-poor microenvironment but also generates signaling molecules to activate tumor-related signaling pathways and promote tumor occurrence, progression, and metastasis [87]. Concomitantly, it makes scholars aware of the extraordinary significance of lipid metabolism in tumors, which is an important part of tumors and one of the fields that must be understood for effective tumor control. Given the complexity of lipid metabolism and the heterogeneity of lipidomics profiles across different tumors, we enumerated the specific regulatory effects of several important lipids and their interactions with other molecules in esophageal cancer (Fig. 6).
Fig. 6.
Major molecular mechanisms of tumorigenesis and development associated with lipid metabolic networks in esophageal cancer. CPT1: carnitine palmitoyl transferase 1; FAO: fatty acid oxidation; CD36: cluster of differentiation 36; PPARG: peroxisome proliferator-activated receptor gamma; ELF3/KLF5/GATA6: transcription factors; ACLY: ATP-citrate lyase; ACC: acetyl-CoA carboxylase; FASN: fatty acid synthetase; SCD: stearoyl-CoA desaturase; FA: fatty acid; LD: lipid droplet; SREBP1: sterol regulatory element-binding protein 1; EMT: epithelial-mesenchymal transition; FTO: fat-mass and obesity-associated protein; HSD17B11: hydroxysteroid 17-beta dehydrogenase 11; TNFα: tumor necrosis factor α; FOXC2: forkhead box protein C2; FA2H: fatty acid 2-hydroxylase; Cer: ceramide; CYP2C and CYP2J: cytochrome P450 superfamily members; LPCAT1: lysophosphatidylcholine acyltransferase; PI3K: phosphatidylinositol 3-kinases; SP1/SREBF2: transcription factors; SQLE: squalene epoxidase; pEGFR: phosphorylated epidermal growth factor receptor; pSREBP1: phosphorylated sterol regulatory element-binding protein 1.
4.1. FA metabolism
Because of the importance of FA to cell structure and survival, changes in FA metabolism are involved in the initiation and progression of esophageal cancer. For example, obesity is relevant to the increased risk of EAC [88]. Ma et al. [89] identified a transcriptional feedback loop to explain the molecular mechanisms associated with obesity, FA metabolism, and EAC. Master regulator transcription factors (MRTFs), including ELF3, KLF5, and GATA6 in EAC, can promote the transcription and overexpression of nuclear receptor peroxisome proliferator-activated receptor gamma (PPARG) by directly regulating the promoter and enhancer regions of PPARG, which can promote the upregulation of downstream genes (ACLY, ACC, FASN, and SCD), motivate de novo synthesis of FAs, increase the abundance of intracellular LDs, and enhance the proliferation ability of tumor cells [89]. In addition, high-fat diets, such as stearic acid (C18:0), oleic acid (C18:1), and arachidonic acid (C20:4), in turn activate PPARG, which further activates the ELF3 super-enhancer to promote ELF3 expression, and subsequently upregulates the expression of other MRTFs in EAC cells via an interconnected regulatory circuitry [89]. Besides, PPARG and MRTF GATA6 and KLF5 are also upregulated in Barrett's esophagus, suggesting that MRTF/PPARG/FA synthesis may facilitate esophageal squamous cell metaplasia and contribute to the initiation and progression of EAC [[89], [90], [91]].
Abnormal FA metabolism has also been observed in the early stages of nitrosamine-induced esophageal cancer. It has been reported that nitrosamine exposure led to a decrease in specific FAs (palmitic acid, linoleic acid, y-linoleic acid, arachidonic acid, docosahexaenoic acid, and eicosapentaenoic acid) and an increase in inflammatory mediators in the peripheral blood of mice, which may be due to the upregulation of FA utilization, as polyunsaturated FAs can be metabolized into proinflammatory mediators [[92], [93], [94]]. Besides, nitrosamine exposure also promoted the expression of CPT1 to facilitate FAs entry into mitochondria for FAO [92]. However, Molendijk et al. [95] discovered that during the progression from normal esophageal epithelium to EAC, the proportion of polyunsaturated and long-chain lipids in the esophageal epithelium significantly increased to promote DNA damage in tumor cells due to bile acids. These findings may reflect the heterogeneity in the process of esophageal carcinogenesis under different causes and highlight the differences between various tissue types of esophageal cancer.
The imbalance of FA-regulated enzymes and metabolic molecules also plays an active role in the progression of esophageal cancer. High expression of CD36 in tumor cells has been associated with increased proliferation and invasion capabilities, thereby promoting tumor progression [96]. Inhibition of CD36 leads to a shift in energy sources from FAs to essential amino acids, resulting in attenuated cell proliferation and invasion, suggesting the importance of FA metabolism-derived energy in ESCC and the critical regulatory role of CD36 [96]. Moreover, overexpression of SREBP1 in ESCC can promote the expression of vimentin, inhibit the expression of E-cadherin, and trigger epithelial-mesenchymal transition (EMT) in tumors [97]. SREBP1 can also promote SCD1 expression, activate the Wnt/β-catenin signaling pathway, and upregulate the expression of downstream proteins (CD44 and cyclin D1), thereby enhancing the proliferation and stemness of tumor cells and promoting tumor growth and metastasis [97]. Recently, Zhang et al. [98] demonstrated that pre-messegner RNA (mRNA) processing factor 19 could interfere with FA metabolism by enhancing the stability of SREBP1 mRNA, leading to increased expression of SREBP1 and the proliferation and progression of ESCC. In addition, epigenetic regulation is also involved in lipid metabolic reprogramming and tumor progression [99,100]. For instance, fat-mass and obesity-associated protein (FTO), an m6A demethylase enzyme highly expressed in esophageal cancer, can upregulate the expression of hydroxysteroid 17-beta dehydrogenase 11 of the short-chain dehydrogenase/reductase family, promote LD formation in tumor cells, and enhance the aggressiveness and stemness of esophageal cancer cells [101].
4.2. Metabolism of phospholipid and cholesterol
Phospholipids, such as GP and SM, as well as cholesterol, are also important components of cellular structures, and their metabolic processes are closely associated with the proliferation and invasion of esophageal cancer cells, involving various enzymes and metabolites in metabolic regulation. FA 2-hydroxylase (FA2H) is an enzyme that catalyzes the formation of 2-hydroxy FA from FA, in which 2-hydroxy FA can be further metabolized to sphingolipid [102]. As an important raw material for sphingolipid synthesis and the main product of catabolism, Cer is indispensable for sphingolipid metabolism, and can also participate in the regulation of mitochondrial respiration and glycolysis in cells [103]. Recently, Zhou et al. [104] indicated that the expression of FA2H in tumor tissues of ESCC was higher than that in adjacent normal tissues, with the highest expression observed in advanced ESCC. The content of Cer (d18:0/24:0) and Cer (d18:0/24:1) in FA2H-silenced tumor cells was significantly increased, and treatment of metastatic ESCC cells with Cer (d18:0/24:0) or Cer (d18:0/24:1) supplementation significantly reduced the oxygen consumption rate and extracellular acidification rate of tumor cells and decreased the proliferation, movement, and metastatic abilities of tumor cells, indicating that FA2H promoted tumor metastasis by reducing the abundance of Cer (d18:0/24:0) and Cer (d18:0/24:1) in tumor cells [104]. And tumor necrosis factor α can promote the binding of the transcription factor forkhead box protein C2 to the FA2H gene, thereby promoting the expression of FA2H and tumor cell metastasis [104].
Besides, cholesterol metabolism in tumor cells is also altered in order to obtain more cholesterol for cell survival. LPCAT1 is not only associated with phospholipid metabolism but also promotes cholesterol synthesis, thereby promoting anoikis resistance of tumor cells, inhibiting tumor cell apoptosis, and promoting the progression of ESCC [105]. In ESCC, LPCAT1 promotes cholesterol synthesis via two major pathways. On one hand, LPCAT1 can promote the phosphorylation and activation of PI 3-kinases (PI3K), promote the transcription factors specificity protein 1 and sterol regulatory element binding transcription factor 2 into the nucleus, upregulate the expression of squalene epoxidase, and promote cholesterol synthesis [105]. On the other hand, LPCAT1 can also stimulate the phosphorylation and activation of epidermal growth factor receptor, inhibit the expression of INSIG-1 in the ER, which is tightly bound to SREBP1, and promote the phosphorylation of SREBP1 and its entry into the nucleus to promote cholesterol synthesis [105].
4.3. Clinical research-based lipid metabolic features and biomarkers to indicate esophageal cancer development
With the in-depth analysis of lipidomics in precious clinical specimens, more lipid metabolic signatures of esophageal cancer have been revealed, which gives us a clearer and more specific understanding of the disease. Among them, compared with healthy people, the levels of plasma lysoPCs (lysoPC (14:0), lysoPC (16:0), lysoPC (16:1), lysoPC (18:0), lysoPC (20:3), and lysoPC (20:5)) in ESCC patients were significantly reduced, which may be caused by the overexpression of lysoPLD catalyzing the conversion of lysoPC to lysoPA to promote tumor progression [106,107]. Consistently, Li et al. [108] also discovered that the levels of lysoPC (16:0) in tumor lesions and peripheral blood of esophageal cancer patients were distinctly lower than those of healthy people, and the levels of PC (16:0/18:1) in tumor lesions and peripheral blood were observably higher in esophageal cancer patients than those in healthy people. By analyzing and comparing esophageal biopsy samples from healthy people and patients with Barrett's esophagus and EAC, Molendijk et al. [95] indicated that PC, PE, and Cer gradually increased during the development of esophageal cancer, while triglycerides and dihydroceramide gradually decreased, and these lipid changes were mainly concentrated in GP and sphingolipid synthesis. Wang et al. [43] considered that lipid metabolism dysregulation related to the diagnosis and prognosis of ESCC patients mainly focused on the linoleic acid metabolic pathway. Linoleic acid can be oxidized by cytochromes P450 (CYP2C and CYP2J) to generate bioactive epoxy FA and induce angiogenesis, cell proliferation, and migration to accelerate tumor growth and metastasis, suggesting that cytochrome P450 may be an underlying therapeutic target for ESCC [43,109].
5. Clinical application of lipid metabolism
5.1. Early diagnosis
The intricate process of tumorigenesis involves the systemic disorder of metabolic pathways. Multiple biomarkers can provide more accurate information for clinical practice and further promote the early diagnosis and treatment of tumors. By comparing the metabolomic information in the peripheral blood of ESCC patients and healthy individuals, Xu et al. [106] found a metabolomic biomarker panel that can significantly discern ESCC patients from healthy individuals, and the area under the curve (AUC) for the generated receiver operating characteristic (ROC) was 0.96, with sensitivity of 90.2% and specificity of 96.0% at the optimal cut-off. This panel included decanoylcarnitine, octanoylcarnitine, lysoPC (16:0), lysoPC (16:1), lysoPC (18:0), linoleic acid, and uric acid [106]. The abundance of LPCAT1 in ESCC was significantly higher than that in adjacent normal esophageal tissues. The level of LPCAT1 in the peripheral blood of ESCC patients was also significantly higher than that of healthy individuals, with an AUC value of about 0.80 for distinguishing ESCC [105]. It was demonstrated that LPCAT1 was considered as a potential biomarker for ESCC diagnosis [105]. Yang et al. [110] also found significant differences in GP metabolism between ESCC and normal esophageal tissues, among which LPCAT1 and PS synthase 1 were highly expressed in ESCC, and the AUCs for identifying ESCC were 0.914 and 0.980, respectively.
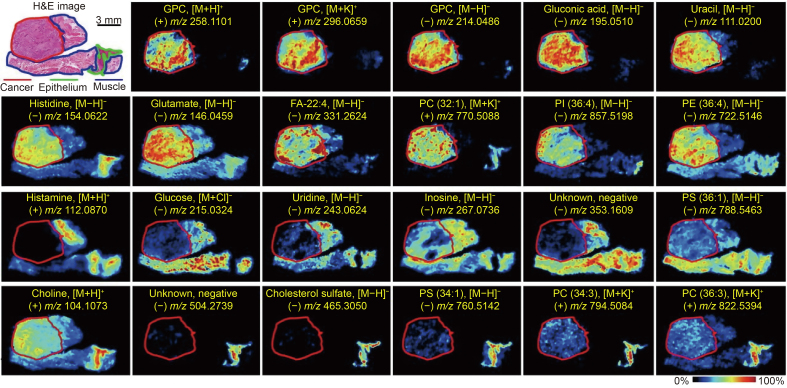
In addition, Abbassi-Ghadi et al. [111] further revealed the functions of the GP profile in the early diagnosis of primary EAC by DESI-MSI. Compared with normal esophageal epithelium, PG and polyunsaturated acyls were significantly increased in EAC, which also had longer acyl chains [111]. The reduction of de novo lipogenesis could reverse the abnormal GP acyl phenotype to normal [111]. By analyzing the phospholipidome, EAC was objectively identified from other non-malignant tissues, including normal tissues and Barrett's dysplasia. Previously, this research group, Abbassi-Ghadi et al. [112], found that primary lesions of EAC had similar lipidomic characteristics to metastatic lymph nodes, which differed from those of non-metastatic lymph nodes and metastatic lymph nodes that respond to chemotherapy. Therefore, it may be possible to determine lymph node metastasis by analyzing the lipidomic profile during esophagectomy, which would be a more rapid and objective method. These studies demonstrate the feasibility of lipid metabolomics as a potential early diagnostic tool for esophageal cancer. Besides, Fig. 7 [46] shows the different metabolic maps between tumor tissue and normal esophageal epithelium in ESCC based on MSI.
Fig. 7.
Mass spectrometry imaging (MSI) images of the spatial distribution of metabolites in esophageal squamous cell carcinoma (ESCC) tissue sections. GPC: glycerophosphorylcholine; GPE: glycerophosphorylethanolamine; FA: fatty acid; PC: phosphatidylcholine; PI: phosphatidylinositol; PE: phosphatidylethanolamine; PS: phosphatidylserine. Reproduced from Ref. [46] with permission.
5.2. Prediction of efficacy and prognosis
The metabolic characteristics of tumors may provide a more accurate reflection of the response of esophageal cancer cells to treatment at the molecular level. Recently, Buck et al. [113] explored the value of metabolomics in appraising the response to neoadjuvant therapy in EAC through MALDI Fourier-transform ion cyclotron resonance MSI. They retrospectively collected postoperative paraffin specimens and clinicopathological characteristics from 144 patients with EAC who received neoadjuvant therapy. Then, they analyzed the metabolomics identities of neoplasms and stromal regions in various specimens. Tumor regression grading is the most widely used clinical system for evaluating tumor response to neoadjuvant therapy. In this study, the accuracy of tumor regression grading in predicting patient prognosis was 70.5%, and the accuracy of prediction based on tumor metabolic characteristics was 89.7%, which indicated that the latter may be more suitable as an indicator for patient prognosis stratification [113]. Metabolic profiling of tumors holds potential value as an evaluation criterion for neoadjuvant therapy and a prognostic stratification criterion in esophageal cancer. Esophageal cancer patients with different effects of chemoradiotherapy not only have different lipid metabolism characteristics in tumor tissues, but also show different lipid metabolism characteristics in peripheral blood. In ESCC patients who achieved a complete response after chemoradiotherapy, the levels of decanoylcarnitine, lysoPC (16:1), and octanoylcarnitine in the peripheral blood gradually increased and returned to healthy human levels [106]. Conversely, this alteration was not observed in non-complete responders. These findings imply that these metabolites may serve as potential predictive indicators for assessing the efficacy of chemoradiotherapy in esophageal cancer.
Given the important of lipid metabolic reprogramming in tumor cells, scholars have not only analyzed the role of lipid metabolism in the prediction of efficacy in esophageal cancer but also explored the relationship between lipid metabolism, related enzymes, and patient prognosis by integrating clinical survival information (Table 1) [96,101,104,105,[114], [115], [116], [117], [118]]. Among them, upregulation of certain regulatory enzymes involved in lipid metabolism, such as ACSS2, CD36, SREBP1, SCD, FTO, FA2H, and LPCAT1, may be linked to a poor prognosis for esophageal cancer patients. Exosomes are extracellular vesicles secreted by almost all living cells that contain rich biological information and are involved in the initiation and progression of cancer [119]. By comparing the metabolism of exosomes in the peripheral blood of ESCC patients with and without recurrence, Zhu et al. [118] identified a potential metabolomic feature, including palmitoleic acid, palmitaldehyde, isobutyl decanoate, and 3′-UMP, for predicting ESCC recurrence with an AUC of up to 98%. Although it has been found that these lipid metabolism-related biomarkers have an impact on the prognosis of esophageal cancer, the initiation and progression of esophageal cancer involve a complex interplay of multiple factors, which is a process with highly sophisticated molecular mechanisms and may still need to be comprehensively analyzed.
Table 1.
Lipid metabolism-related markers for the prognosis of esophageal cancer.
| Biomarkers | The associated lipids | Histologic subtypes of cancer | Source of specimens | Number of cases | Clinical prognostic significance | Refs. |
|---|---|---|---|---|---|---|
| CD36 | FA | ESCC | Tumor tissues | 160 | High CD36 expression was associated with shorter overall survival (P = 0.023), disease-specific survival (P = 0.046) and recurrence-free survival (P = 0.048) | [96] |
| FTO | LD | Esophageal cancer | Tumor tissues | 106 | Patients with low expression of FTO had a better prognosis (P < 0.001) | [101] |
| FA2H | Cer (d18:0/24:0) and Cer (d18:0/24:1) | ESCC | Tumor tissues | 306 | Patients with high FA2H expression had poor disease-free survival (P = 0.0209) and overall survival (P = 0.0125) | [104] |
| LPCAT1 | Cholesterol | ESCC | Tumor tissues | 154 | Low mRNA (P < 0.001) and protein expression (P = 0.021) of LPCAT1 was associated with a better prognosis. | [105] |
| ACSS2 | Acetyl-CoA | ESCC | Tumor tissues | 28 | Patients with low expression of ACSS2 achieved longer disease-free survival (P = 0.0311) | [114] |
| SCD | Oleic aid | ESCC | Tumor tissues | 236 | Patients with high SCD expression had a shorter disease-free survival (P = 0.025) | [115] |
| SREBP1 | FA | ESCC | Tumor tissues | 200 | Patients with low SREBP1 expression had longer overall survival (P = 0.001) | [116] |
| OTUB2 | PS | ESCC | Tumor tissues | 103, 95, and 114 | Patients with high expression of OTUB2 received longer overall survival (P = 0.0058, P < 0.0001, and P = 0.0011) | [117] |
| Palmitoleic acid, palmitaldehyde, isobutyl decanoate, and 3′-UMP | Palmitoleic acid | ESCC | Blood samples | 71 | A marker set of metabolomic signatures used to predict ESCC recurrence had an AUC of 98% | [118] |
CD36: cluster of differentiation 36; FA: fatty acid; ESCC: esophageal squamous cell carcinoma; FTO: fat-mass and obesity-associated protein; LD: lipid droplet; FA2H: fatty acid 2-hydroxylase; Cer: ceramide; LPCAT1: lysophosphatidylcholine acyltransferase 1; mRNA: messenger RNA; ACSS2: acetyl-CoA synthetase 2; SCD: stearoyl-CoA desaturase; SREBP1: sterol regulatory element-binding protein 1; OTUB2: otubain 2; PS: phosphatidylserine; AUC: area under the curve.
5.3. Treatment resistance and drug discovery
Considering the consequence of lipid metabolism in the initiation and progression of esophageal cancer, scholars are exploring the causes of clinical treatment resistance from the perspective of lipid metabolism, looking for new therapeutic targets to interfere with tumor lipid metabolism, and exploring new treatment options.
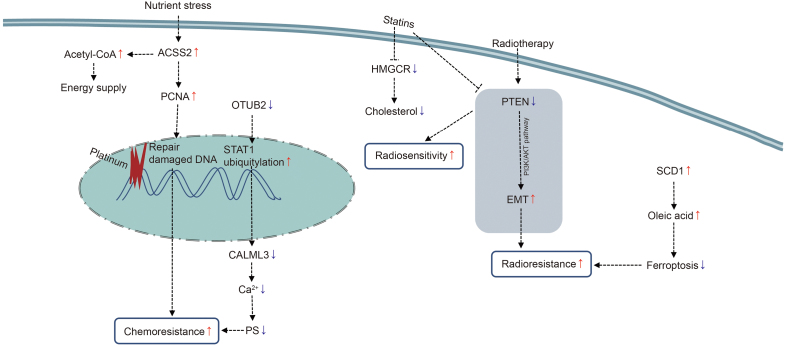
Platinum-based double-agent chemotherapy is a common clinical treatment for esophageal cancer, and chemoresistance is also a difficult problem for doctors. Platinum-based drugs, such as cisplatin, carboplatin, and oxaliplatin, can enter cells through transmembrane transport, undergo chloro-ligand(s) replacement to become activated, and finally couple with intra-strands or between strands of double-stranded DNA to cause DNA damage and cancer cell death [120]. However, due to the uncontrolled proliferation of tumor cells and the disorder of vascular distribution, the nutritional status in the tumor microenvironment is very poor. Under this condition, tumor cells can promote the production of acetyl-CoA by overexpressing ASCC2 to ensure the energy supply of the cells. Simultaneously, ACSS2 can also stabilize the expression of proliferating cell nuclear antigen, promote the rapid repair of DNA damage induced by cisplatin, and lead to the chemoresistance of tumor cells [114] (Fig. 8). At present, the developed ACSS2 inhibitors include ACSS2-IN-2, VY-3-135, and MTB-9655, but there is still no relevant study on ACSS2 inhibitors combined with chemotherapy in the treatment of esophageal cancer. In the future, researchers are expected to continue to explore the combination of the two, which may be a potential way to improve the treatment effects of cancer patients. In addition, metabolic heterogeneity within and between tumors may also lead to differences in the efficacy of drugs. Otubain 2 (OTUB2), a deubiquitinating enzyme, can promote the occurrence and progression of lung and breast cancers [121,122]. But Chang et al. [117] found that OTUB2 was generally lost in ESCC due to DNA hypermethylation, while it was widely expressed in normal esophageal tissues. In ESCC, OTUB2 can deubiquitinate signal transducer and activator of transcription 1 (STAT1), motivate the phosphorylation and activation of STAT1, and activate the transcription and expression of calmodulin-like protein 3 to improve intracellular calcium level and the capacity of oxidative phosphorylation and PS synthesis, thereby inhibiting tumor cell growth and enhancing the sensitivity of tumor cells to chemotherapy [117] (Fig. 8). And oral administration of PS can restore the chemosensitivity in the OTUB2-silenced ESCC xenograft mice [117].
Fig. 8.
Lipid metabolism molecules associated with treatment tolerance in esophageal cancer. ACSS2: acetyl-CoA synthetase 2; PCNA: proliferating cell nuclear antigen; OTUB2: otubain 2; STAT1: signal transducer and activator of transcription 1; CALML3: calmodulin-like protein 3; PS: phosphatidylserine; HMGCR: 3-hydroxy-3-methylglutaryl-coenzyme A reductase; PTEN: phosphatase and tensin homolog; PI3K/AKT pathway: phosphatidylinositol 3-kinases/AKT pathway; EMT: epithelial-mesenchymal transition; SCD1: stearoyl-CoA desaturase 1.
When tumor cells have an excess of iron ions, a large amount of ROS can be produced by reacting with hydrogen peroxide, and the accumulated ROS can trigger extensive peroxidation of unsaturated phospholipids in the cell membrane, resulting in membrane rupture and cell death. This kind of cell death caused by oxygen-free radicals produced by iron ions is called ferroptosis, which is a regulated form of non-apoptotic cell death [123]. Because ferroptosis is closely related to lipids, the reprogramming of lipid metabolism may affect the occurrence of ferroptosis in tumor cells, leading to tumor cell resistance to treatment [124]. As one of the indispensable means for the treatment of esophageal cancer, radiation therapy can directly destroy the DNA double-strand of tumor cells to induce cell death and also induce the ionization and decomposition of intracellular water molecules, producing ROS to cause cell damage and indirectly lead to cell death, including ferroptosis [125]. Nevertheless, tumor cells can undergo lipid metabolism reprogramming under such stress conditions to resist ferroptosis, leading to radiation resistance and tumor cell survival [126]. Luo et al. [115] observed that upregulated expression of SCD1 in ESCC can promote the production of oleic acid (C18:1), alleviate lipid peroxidation, and attenuate radiation-induced cell ferroptosis, thereby inducing radiation resistance in tumor cells. The use of SCD1 inhibitors (MF-438) can enhance the radiosensitivity of ESCC tumor cells by increasing ferroptosis and immunogenic cell death (Fig. 8). As found in melanoma cells, oleic acid can protect tumor cells from ferroptosis through acyl-CoA synthetase long-chain family member 3 [127]. In addition, existing drugs regulating lipid metabolism may also enhance the anti-tumor efficacy of radiotherapy in esophageal cancer. For instance, statins can competitively inhibit endogenous HMGCR, resulting in decreased intracellular cholesterol synthesis [128]. Jin et al. [129] found that simvastatin could reverse the low expression of phosphatase and tensin homologs and the activation of the PI3K/AKT pathway induced by radiotherapy, inhibit EMT of tumor cells, and improve the radiosensitivity of esophageal cancer (Fig. 8). Simvastatin can also induce cell death by increasing lipid peroxidation in tumor cells [130].
Researchers can also investigate drug development for new targets by intervening in the regulation of lipid metabolism directly based on the metabolic characteristics of esophageal cancer tumor cells. For example, tumor cell survival can be affected by intervening in key targets of FA metabolism such as FASN, SREBP, and ACC. TVB-3166, an oral FASN inhibitor, can effectively inhibit FASN, leading to tumor cell death, inhibit the growth of xenograft tumors in vivo, and have no significant effect on normal cells [131]. ACC inhibitors, including ND-646, TOFA, and CP-640186, can induce apoptosis in various types of tumor cells and inhibit tumor development to a certain extent [132]. Besides, Huang et al. [133] found that fatostatin, an inhibitor of SREBP1, reduced the percentage of CD133 cells, decreased the expression of SREBP1 and ZEB1 (a transcription factor that promotes EMT), and increased the sensitivity of ESCC to drug therapy. According to the high expression of LPCAT1 in ESCC, Jun et al. [134] developed nanoparticles of doxorubicin combined with siLPCAT1. The results of animal experiments revealed that the expression of LPCAT1 was strikingly reduced and the tumor volume was notably shrunk after the nanomedicine treatment compared with doxorubicin alone, without obvious toxicity to the heart, lung, liver, or kidney, which demonstrated the potential feasibility of targeting LPCAT1 [134]. It is hoped that more nanomaterials targeting the regulation of lipid metabolism can be developed and used in esophageal cancer research.
Although research on intervening in lipid metabolism for the treatment of esophageal cancer is still in the preclinical stage, it has shown obvious antitumor effects and potential for enhancing the effectiveness of traditional antitumor therapy. However, due to the complexity and universality of lipid metabolism, further research is still needed to advance its clinical application.
6. Conclusions and perspectives
For the moment, the treatment of esophageal cancer has entered a bottleneck period, and it is urgent to develop new therapeutic targets to ameliorate the prognosis of esophageal cancer patients. More and more studies have demonstrated significantly abnormal lipid metabolism in esophageal cancer compared with normal esophageal tissue, which is intimately correlated with the initiation and development of esophageal cancer and clinical treatment tolerance. Consequently, there is a growing interest among researchers in targeting and regulating lipid metabolism as a potential therapeutic approach, which will provide exciting new therapeutic targets for the treatment of esophageal cancer, improve the treatment status of esophageal cancer, and bring hope to esophageal cancer patients. However, there are still several challenges to be overcome in the technology of lipid detection and analysis. For example, the accuracy and coverage of lipid detection by MSI-based lipidomics need to be improved to achieve more accurate qualitative, quantitative, and spatial positioning. In addition, there is a lack of lipid databases, which are still in the stage of continuous improvement. It is necessary to continue to build a complete database that can be utilized by researchers using different instrument platforms, different experimental methods, and from different countries. Finally, multi-omics combined analysis can more systematically and comprehensively understand the initiation and progression of tumors. Therefore, the combination of lipidomics and genomics, transcriptomics, proteomics, radiomics, and other omics technologies may be the direction we need to continue exploring in the future.
CRediT author statement
Ruidi Jiao: Methodology, Formal analysis, Investigation, Data curation, Writing - Original draft preparation, Reviewing and Editing, Visualization; Wei Jiang: Data Curation, Supervision, Funding acquisition; Kunpeng Xu: Methodology, Supervision; Qian Luo: Writing - Reviewing and Editing; Luhua Wang: Conceptualization, Writing - Reviewing and Editing, Supervision, Funding acquisition; Chao Zhao: Conceptualization, Writing - Reviewing and Editing, Supervision, Project administration, Funding acquisition.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgments
The work was supported by the National Natural Science Foundation of China (Grant Nos.: 22176195 and 82127801), National Key R&D Program of China (Grant No.: 2022YFF0705003), the Shenzhen Key Laboratory of Precision Diagnosis and Treatment of Depression (Grant No.: ZDSYS20220606100606014), the Guangdong Province Zhu Jiang Talents Plan, China (Grant No.: 2021QN02Y028), the Natural Science Foundation of Guangdong Province, China (Grant No.: 2021A1515010171), the Key Program of Fundamental Research in Shenzhen, China (Grant No.: JCYJ20210324115811031), the Sustainable Development Program of Shenzhen, China (Grant No.: KCXFZ202002011008124), and the National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital & Shenzhen Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Shenzhen (Grant Nos.: SZ2020ZD002 and SZ2020QN005).
Footnotes
Peer review under responsibility of Xi’an Jiaotong University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpha.2023.08.019.
Contributor Information
Luhua Wang, Email: wangluhua@csco.org.cn.
Chao Zhao, Email: chao.zhao@siat.ac.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Morgan E., Soerjomataram I., Rumgay H., et al. The global landscape of esophageal squamous cell carcinoma and esophageal adenocarcinoma incidence and mortality in 2020 and projections to 2040: New estimates from GLOBOCAN 2020. Gastroenterology. 2022;163:649–658.e2. doi: 10.1053/j.gastro.2022.05.054. [DOI] [PubMed] [Google Scholar]
- 2.Sung H., Ferlay J., Siegel R.L., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 3.Zeng H., Ran X., An L., et al. Disparities in stage at diagnosis for five common cancers in China: A multicentre, hospital-based, observational study. Lancet Public Health. 2021;6:e877–e887. doi: 10.1016/S2468-2667(21)00157-2. [DOI] [PubMed] [Google Scholar]
- 4.Eyck B.M., van Lanschot J.J.B., Hulshof M.C.C.M., et al. Ten-year outcome of neoadjuvant chemoradiotherapy plus surgery for esophageal cancer: The randomized controlled CROSS trial. J. Clin. Oncol. 2021;39:1995–2004. doi: 10.1200/JCO.20.03614. [DOI] [PubMed] [Google Scholar]
- 5.Shah M.A., Kennedy E.B., Catenacci D.V., et al. Treatment of locally advanced esophageal carcinoma: ASCO guideline. J. Clin. Oncol. 2020;38:2677–2694. doi: 10.1200/JCO.20.00866. [DOI] [PubMed] [Google Scholar]
- 6.Sun J.M., Shen L., Shah M.A., et al. Pembrolizumab plus chemotherapy versus chemotherapy alone for first-line treatment of advanced oesophageal cancer (keynote-590): A randomised, placebo-controlled, phase 3 study. Lancet. 2021;398:759–771. doi: 10.1016/S0140-6736(21)01234-4. [DOI] [PubMed] [Google Scholar]
- 7.Janjigian Y.Y., Shitara K., Moehler M., et al. First-line nivolumab plus chemotherapy versus chemotherapy alone for advanced gastric, gastro-oesophageal junction, and oesophageal adenocarcinoma (CheckMate 649): A randomised, open-label, phase 3 trial. Lancet. 2021;398:27–40. doi: 10.1016/S0140-6736(21)00797-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smyth E.C., Lagergren J., Fitzgerald R.C., et al. Oesophageal cancer. Nat. Rev. Dis. Primers. 2017;3 doi: 10.1038/nrdp.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Broadfield L.A., Pane A.A., Talebi A., et al. Lipid metabolism in cancer: New perspectives and emerging mechanisms. Dev. Cell. 2021;56:1363–1393. doi: 10.1016/j.devcel.2021.04.013. [DOI] [PubMed] [Google Scholar]
- 10.Petan T. Lipid droplets in cancer. Rev. Physiol. Biochem. Pharmacol. 2023;185:53–86. doi: 10.1007/112_2020_51. [DOI] [PubMed] [Google Scholar]
- 11.Yoon H., Shaw J.L., Haigis M.C., et al. Lipid metabolism in sickness and in health: Emerging regulators of lipotoxicity. Mol. Cell. 2021;81:3708–3730. doi: 10.1016/j.molcel.2021.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han X. Lipidomics for studying metabolism. Nat. Rev. Endocrinol. 2016;12:668–679. doi: 10.1038/nrendo.2016.98. [DOI] [PubMed] [Google Scholar]
- 13.Alves M.A., Lamichhane S., Dickens A., et al. Systems biology approaches to study lipidomes in health and disease. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2021;1866 doi: 10.1016/j.bbalip.2020.158857. [DOI] [PubMed] [Google Scholar]
- 14.Ko C.W., Qu J., Black D.D., et al. Regulation of intestinal lipid metabolism: Current concepts and relevance to disease. Nat. Rev. Gastroenterol. Hepatol. 2020;17:169–183. doi: 10.1038/s41575-019-0250-7. [DOI] [PubMed] [Google Scholar]
- 15.Mellinger A.L., Muddiman D.C., Gamcsik M.P. Highlighting functional mass spectrometry imaging methods in bioanalysis. J. Proteome Res. 2022;21:1800–1807. doi: 10.1021/acs.jproteome.2c00220. [DOI] [PubMed] [Google Scholar]
- 16.Bian X., Liu R., Meng Y., et al. Lipid metabolism and cancer. J. Exp. Med. 2021;218 doi: 10.1084/jem.20201606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lam S.M., Tian H., Shui G. Lipidomics, en route to accurate quantitation. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2017;1862:752–761. doi: 10.1016/j.bbalip.2017.02.008. [DOI] [PubMed] [Google Scholar]
- 18.Guo X., Cao W., Fan X., et al. Tandem mass spectrometry imaging enables high definition for mapping lipids in tissues. Angew. Chem. Int. Ed. Engl. 2023;62 doi: 10.1002/anie.202214804. [DOI] [PubMed] [Google Scholar]
- 19.Fahy E., Subramaniam S., Murphy R.C., et al. Update of the LIPID MAPS comprehensive classification system for lipids. J. Lipid Res. 2009;50:S9–S14. doi: 10.1194/jlr.R800095-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liebisch G., Fahy E., Aoki J., et al. Update on LIPID MAPS classification, nomenclature, and shorthand notation for ms-derived lipid structures. J. Lipid Res. 2020;61:1539–1555. doi: 10.1194/jlr.S120001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masoodi M., Eiden M., Koulman A., et al. Comprehensive lipidomics analysis of bioactive lipids in complex regulatory networks. Anal. Chem. 2010;82:8176–8185. doi: 10.1021/ac1015563. [DOI] [PubMed] [Google Scholar]
- 22.Greten F.R., Grivennikov S.I. Inflammation and cancer: Triggers, mechanisms, and consequences. Immunity. 2019;51:27–41. doi: 10.1016/j.immuni.2019.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pietrocola F., Galluzzi L., Bravo-San Pedro J.M., et al. Acetyl coenzyme A: A central metabolite and second messenger. Cell Metab. 2015;21:805–821. doi: 10.1016/j.cmet.2015.05.014. [DOI] [PubMed] [Google Scholar]
- 24.Cronan J.E., Jr, Waldrop G.L. Multi-subunit acetyl-CoA carboxylases. Prog. Lipid Res. 2002;41:407–435. doi: 10.1016/s0163-7827(02)00007-3. [DOI] [PubMed] [Google Scholar]
- 25.Menendez J.A., Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat. Rev. Cancer. 2007;7:763–777. doi: 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- 26.Jakobsson A., Westerberg R., Jacobsson A. Fatty acid elongases in mammals: Their regulation and roles in metabolism. Prog. Lipid Res. 2006;45:237–249. doi: 10.1016/j.plipres.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 27.Currie E., Schulze A., Zechner R., et al. Cellular fatty acid metabolism and cancer. Cell Metab. 2013;18:153–161. doi: 10.1016/j.cmet.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shao W., Espenshade P.J. Expanding roles for SREBP in metabolism. Cell Metab. 2012;16:414–419. doi: 10.1016/j.cmet.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Febbraio M., Hajjar D.P., Silverstein R.L. CD36: A class B scavenger receptor involved in angiogenesis, atherosclerosis, inflammation, and lipid metabolism. J. Clin. Invest. 2001;108:785–791. doi: 10.1172/JCI14006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kazantzis M., Stahl A. Fatty acid transport proteins, implications in physiology and disease. Biochim. Biophys. Acta. 2012;1821:852–857. doi: 10.1016/j.bbalip.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Furuhashi M., Hotamisligil G.S. Fatty acid-binding proteins: Role in metabolic diseases and potential as drug targets. Nat. Rev. Drug Discov. 2008;7:489–503. doi: 10.1038/nrd2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kennedy E.P., Weiss S.B. The function of cytidine coenzymes in the biosynthesis of phospholipides. J. Biol. Chem. 1956;222:193–214. [PubMed] [Google Scholar]
- 33.Law S.H., Chan M.L., Marathe G.K., et al. An updated review of lysophosphatidylcholine metabolism in human diseases. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20051149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spaulding S.C., Bollag W.B. The role of lipid second messengers in aldosterone synthesis and secretion. J. Lipid Res. 2022;63 doi: 10.1016/j.jlr.2022.100191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anand P.K. Lipids, inflammasomes, metabolism, and disease. Immunol. Rev. 2020;297:108–122. doi: 10.1111/imr.12891. [DOI] [PubMed] [Google Scholar]
- 36.Cheng C., Geng F., Cheng X., et al. Lipid metabolism reprogramming and its potential targets in cancer. Cancer Commun. (Lond) 2018;38 doi: 10.1186/s40880-018-0301-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He W., Li Q., Li X. Acetyl-CoA regulates lipid metabolism and histone acetylation modification in cancer. Biochim. Biophys. Acta Rev. Cancer. 2023;1878 doi: 10.1016/j.bbcan.2022.188837. [DOI] [PubMed] [Google Scholar]
- 38.Bensaad K., Favaro E., Lewis C.A., et al. Fatty acid uptake and lipid storage induced by HIF-1α contribute to cell growth and survival after hypoxia-reoxygenation. Cell Rep. 2014;9:349–365. doi: 10.1016/j.celrep.2014.08.056. [DOI] [PubMed] [Google Scholar]
- 39.Koundouros N., Poulogiannis G. Reprogramming of fatty acid metabolism in cancer. Br. J. Cancer. 2020;122:4–22. doi: 10.1038/s41416-019-0650-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ladanyi A., Mukherjee A., Kenny H.A., et al. Adipocyte-induced CD36 expression drives ovarian cancer progression and metastasis. Oncogene. 2018;37:2285–2301. doi: 10.1038/s41388-017-0093-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Z., Park H.G., Wang D.H., et al. Fatty acid desaturase 2 (FADS2) but not FADS1 desaturates branched chain and odd chain saturated fatty acids. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2020;1865 doi: 10.1016/j.bbalip.2019.158572. [DOI] [PubMed] [Google Scholar]
- 42.Vriens K., Christen S., Parik S., et al. Evidence for an alternative fatty acid desaturation pathway increasing cancer plasticity. Nature. 2019;566:403–406. doi: 10.1038/s41586-019-0904-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang P.P., Song X., Zhao X.K., et al. Serum metabolomic profiling reveals biomarkers for early detection and prognosis of esophageal squamous cell carcinoma. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.790933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zang Q., Sun C., Chu X., et al. Spatially resolved metabolomics combined with multicellular tumor spheroids to discover cancer tissue relevant metabolic signatures. Anal. Chim. Acta. 2021;1155 doi: 10.1016/j.aca.2021.338342. [DOI] [PubMed] [Google Scholar]
- 45.Ackerman D., Simon M.C. Hypoxia, lipids, and cancer: Surviving the harsh tumor microenvironment. Trends Cell Biol. 2014;24:472–478. doi: 10.1016/j.tcb.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun C., Li T., Song X., et al. Spatially resolved metabolomics to discover tumor-associated metabolic alterations. Proc. Natl. Acad. Sci. U S A. 2019;116:52–57. doi: 10.1073/pnas.1808950116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barbayianni E., Kaffe E., Aidinis V., et al. Autotaxin, a secreted lysophospholipase D, as a promising therapeutic target in chronic inflammation and cancer. Prog. Lipid Res. 2015;58:76–96. doi: 10.1016/j.plipres.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 48.Wang B., Tontonoz P. Phospholipid remodeling in physiology and disease. Annu. Rev. Physiol. 2019;81:165–188. doi: 10.1146/annurev-physiol-020518-114444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bathaie S.Z., Ashrafi M., Azizian M., et al. Mevalonate pathway and human cancers. Curr. Mol. Pharmacol. 2017;10:77–85. doi: 10.2174/1874467209666160112123205. [DOI] [PubMed] [Google Scholar]
- 50.Perrone F., Minari R., Bersanelli M., et al. The prognostic role of high blood cholesterol in advanced cancer patients treated with immune checkpoint inhibitors. J. Immunother. 2020;43:196–203. doi: 10.1097/CJI.0000000000000321. [DOI] [PubMed] [Google Scholar]
- 51.Guo L., Liu X., Zhao C., et al. iSegMSI: An interactive strategy to improve spatial segmentation of mass spectrometry imaging data. Anal. Chem. 2022;94:14522–14529. doi: 10.1021/acs.analchem.2c01456. [DOI] [PubMed] [Google Scholar]
- 52.Guo L., Dong J., Xu X., et al. Divide and conquer: A flexible deep learning strategy for exploring metabolic heterogeneity from mass spectrometry imaging data. Anal. Chem. 2023;95:1924–1932. doi: 10.1021/acs.analchem.2c04045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu T., Zhang J.L. Mass-spectrometry-based lipidomics. J. Sep. Sci. 2018;41:351–372. doi: 10.1002/jssc.201700709. [DOI] [PubMed] [Google Scholar]
- 54.Aldana J., Romero-Otero A., Cala M.P. Exploring the lipidome: Current lipid extraction techniques for mass spectrometry analysis. Metabolites. 2020;10 doi: 10.3390/metabo10060231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Folch J., Lees M., Sloane Stanley G.H. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 56.Zhang T., Chen S., Syed I., et al. A LC-MS-based workflow for measurement of branched fatty acid esters of hydroxy fatty acids. Nat. Protoc. 2016;11:747–763. doi: 10.1038/nprot.2016.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matyash V., Liebisch G., Kurzchalia T.V., et al. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J. Lipid Res. 2008;49:1137–1146. doi: 10.1194/jlr.D700041-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Löfgren L., Forsberg G.B., Ståhlman M. The BUME method: A new rapid and simple chloroform-free method for total lipid extraction of animal tissue. Sci. Rep. 2016;6 doi: 10.1038/srep27688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Silva M.S., Tavares A.P.M., de Faria H.D., et al. Molecularly imprinted solid phase extraction aiding the analysis of disease biomarkers. Crit. Rev. Anal. Chem. 2022;52:933–948. doi: 10.1080/10408347.2020.1843131. [DOI] [PubMed] [Google Scholar]
- 60.Rustam Y.H., Reid G.E. Analytical challenges and recent advances in mass spectrometry based lipidomics. Anal. Chem. 2018;90:374–397. doi: 10.1021/acs.analchem.7b04836. [DOI] [PubMed] [Google Scholar]
- 61.Hinz C., Liggi S., Griffin J.L. The potential of ion mobility mass spectrometry for high-throughput and high-resolution lipidomics. Curr. Opin. Chem. Biol. 2018;42:42–50. doi: 10.1016/j.cbpa.2017.10.018. [DOI] [PubMed] [Google Scholar]
- 62.Chiu H.H., Kuo C.H. Gas chromatography-mass spectrometry-based analytical strategies for fatty acid analysis in biological samples. J. Food Drug Anal. 2020;28:60–73. doi: 10.1016/j.jfda.2019.10.003. [DOI] [PubMed] [Google Scholar]
- 63.Son H.H., Moon J.Y., Seo H.S., et al. High-temperature GC-MS-based serum cholesterol signatures may reveal sex differences in vasospastic angina. J. Lipid Res. 2014;55:155–162. doi: 10.1194/jlr.D040790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang Y., Liang Y., Yang J., et al. Advances of supercritical fluid chromatography in lipid profiling. J. Pharm. Anal. 2019;9:1–8. doi: 10.1016/j.jpha.2018.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Avela H.F., Sirén H. Advances in lipidomics. Clin. Chim. Acta. 2020;510:123–141. doi: 10.1016/j.cca.2020.06.049. [DOI] [PubMed] [Google Scholar]
- 66.Yang K., Cheng H., Gross R.W., et al. Automated lipid identification and quantification by multidimensional mass spectrometry-based shotgun lipidomics. Anal. Chem. 2009;81:4356–4368. doi: 10.1021/ac900241u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yan P., Wei Y., Wang M., et al. Network pharmacology combined with metabolomics and lipidomics to reveal the hypolipidemic mechanism of Alismatis rhizoma in hyperlipidemic mice. Food Funct. 2022;13:4714–4733. doi: 10.1039/d1fo04386b. [DOI] [PubMed] [Google Scholar]
- 68.Matthiesen R., Lauber C., Sampaio J.L., et al. Shotgun mass spectrometry-based lipid profiling identifies and distinguishes between chronic inflammatory diseases. EBioMedicine. 2021;70 doi: 10.1016/j.ebiom.2021.103504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu L., Chen Y., Feng A., et al. Study on detection method of microplastics in farmland soil based on hyperspectral imaging technology. Environ. Res. 2023;232 doi: 10.1016/j.envres.2023.116389. [DOI] [PubMed] [Google Scholar]
- 70.Hoji A., Kumar R., Gern J.E., et al. Cord blood sphingolipids are associated with atopic dermatitis and wheeze in the first year of life. J. Allergy Clin. Immunol. Glob. 2022;1:162–171. doi: 10.1016/j.jacig.2022.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sharma N., Yadav M., Tripathi G., et al. Bile multi-omics analysis classifies lipid species and microbial peptides predictive of carcinoma of gallbladder. Hepatology. 2022;76:920–935. doi: 10.1002/hep.32496. [DOI] [PubMed] [Google Scholar]
- 72.Sun Y., Liu B., Chen Y., et al. Multi-omics prognostic signatures based on lipid metabolism for colorectal cancer. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.811957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hoffmann N., Mayer G., Has C., et al. A current encyclopedia of bioinformatics tools, data formats and resources for mass spectrometry lipidomics. Metabolites. 2022;12 doi: 10.3390/metabo12070584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mohamed A., Molendijk J., Hill M.M. Lipidr: A software tool for data mining and analysis of lipidomics datasets. J. Proteome Res. 2020;19:2890–2897. doi: 10.1021/acs.jproteome.0c00082. [DOI] [PubMed] [Google Scholar]
- 75.Mohamed A., Hill M.M. LipidSuite: Interactive web server for lipidomics differential and enrichment analysis. Nucleic Acids Res. 2021;49:W346–W351. doi: 10.1093/nar/gkab327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pang Z., Chong J., Zhou G., et al. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021;49:W388–W396. doi: 10.1093/nar/gkab382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wolrab D., Jirásko R., Cífková E., et al. Lipidomic profiling of human serum enables detection of pancreatic cancer. Nat. Commun. 2022;13 doi: 10.1038/s41467-021-27765-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhao C., Guo L., Dong J., et al. Mass spectrometry imaging-based multi-modal technique: Next-generation of biochemical analysis strategy. Innovation (Camb) 2021;2 doi: 10.1016/j.xinn.2021.100151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhao C., Cai Z. Three-dimensional quantitative mass spectrometry imaging in complex system: From subcellular to whole organism. Mass Spectrom. Rev. 2022;41:469–487. doi: 10.1002/mas.21674. [DOI] [PubMed] [Google Scholar]
- 80.Hu C., Luo W., Xu J., et al. Recognition and avoidance of ion source-generated artifacts in lipidomics analysis. Mass Spectrom. Rev. 2022;41:15–31. doi: 10.1002/mas.21659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.He J., Sun C., Li T., et al. A sensitive and wide coverage ambient mass spectrometry imaging method for functional metabolites based molecular histology. Adv. Sci. (Weinh) 2018;5 doi: 10.1002/advs.201800250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Han J.Y., Permentier H., Bischoff R., et al. Imaging of protein distribution in tissues using mass spectrometry: An interdisciplinary challenge. Trends Anal. Chem. 2019;112:13–28. [Google Scholar]
- 83.Eberhardt K., Stiebing C., Matthäus C., et al. Advantages and limitations of Raman spectroscopy for molecular diagnostics: An update. Expert Rev. Mol. Diagn. 2015;15:773–787. doi: 10.1586/14737159.2015.1036744. [DOI] [PubMed] [Google Scholar]
- 84.Egoshi S., Dodo K., Sodeoka M. Deuterium Raman imaging for lipid analysis. Curr. Opin. Chem. Biol. 2022;70 doi: 10.1016/j.cbpa.2022.102181. [DOI] [PubMed] [Google Scholar]
- 85.Yao M., Vaithiyanathan M., Allbritton N.L. Analytical techniques for single-cell biochemical assays of lipids. Annu. Rev. Biomed. Eng. 2023;25:281–309. doi: 10.1146/annurev-bioeng-110220-034007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Klymchenko A.S. Fluorescent probes for lipid membranes: From the cell surface to organelles. Acc. Chem. Res. 2023;56:1–12. doi: 10.1021/acs.accounts.2c00586. [DOI] [PubMed] [Google Scholar]
- 87.Snaebjornsson M.T., Janaki-Raman S., Schulze A. Greasing the wheels of the cancer machine: The role of lipid metabolism in cancer. Cell Metab. 2020;31:62–76. doi: 10.1016/j.cmet.2019.11.010. [DOI] [PubMed] [Google Scholar]
- 88.Turati F., Tramacere I., La Vecchia C., et al. A meta-analysis of body mass index and esophageal and gastric cardia adenocarcinoma. Ann. Oncol. 2013;24:609–617. doi: 10.1093/annonc/mds244. [DOI] [PubMed] [Google Scholar]
- 89.Ma S., Zhou B., Yang Q., et al. A transcriptional regulatory loop of master regulator transcription factors, PPARG, and fatty acid synthesis promotes esophageal adenocarcinoma. Cancer Res. 2021;81:1216–1229. doi: 10.1158/0008-5472.CAN-20-0652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rogerson C., Britton E., Withey S., et al. Identification of a primitive intestinal transcription factor network shared between esophageal adenocarcinoma and its precancerous precursor state. Genome Res. 2019;29:723–736. doi: 10.1101/gr.243345.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rogerson C., Ogden S., Britton E., et al. Repurposing of KLF5 activates a cell cycle signature during the progression from a precursor state to oesophageal adenocarcinoma. Elife. 2020;9 doi: 10.7554/eLife.57189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang H., Zhao C., Liu Q., et al. Dysregulation of fatty acid metabolism associated with esophageal inflammation of ICR mice induced by nitrosamines exposure. Environ. Pollut. 2022;297 doi: 10.1016/j.envpol.2021.118680. [DOI] [PubMed] [Google Scholar]
- 93.Serhan C.N. Pro-resolving lipid mediators are leads for resolution physiology. Nature. 2014;510:92–101. doi: 10.1038/nature13479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang H., Zhao C., Zhang Y., et al. Multi-omics analysis revealed NMBA induced esophageal carcinoma tumorigenesis via regulating PPARα signaling pathway. Environ. Pollut. 2023;324 doi: 10.1016/j.envpol.2023.121369. [DOI] [PubMed] [Google Scholar]
- 95.Molendijk J., Kolka C.M., Cairns H., et al. Elevation of fatty acid desaturase 2 in esophageal adenocarcinoma increases polyunsaturated lipids and may exacerbate bile acid-induced DNA damage. Clin. Transl. Med. 2022;12 doi: 10.1002/ctm2.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yoshida T., Yokobori T., Saito H., et al. CD36 expression is associated with cancer aggressiveness and energy source in esophageal squamous cell carcinoma. Ann. Surg. Oncol. 2021;28:1217–1227. doi: 10.1245/s10434-020-08711-3. [DOI] [PubMed] [Google Scholar]
- 97.Wang J., Ling R., Zhou Y., et al. SREBP1 silencing inhibits the proliferation and motility of human esophageal squamous carcinoma cells via the Wnt/β-catenin signaling pathway. Oncol. Lett. 2020;20:2855–2869. doi: 10.3892/ol.2020.11853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang G.C., Yu X.N., Guo H.Y., et al. PRP19 enhances esophageal squamous cell carcinoma progression by reprogramming SREBF1-dependent fatty acid metabolism. Cancer Res. 2023;83:521–537. doi: 10.1158/0008-5472.CAN-22-2156. [DOI] [PubMed] [Google Scholar]
- 99.Song T., Yang Y., Wei H., et al. Zfp217 mediates m6A mRNA methylation to orchestrate transcriptional and post-transcriptional regulation to promote adipogenic differentiation. Nucleic Acids Res. 2019;47:6130–6144. doi: 10.1093/nar/gkz312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Deng X., Su R., Weng H., et al. RNA N6-methyladenosine modification in cancers: Current status and perspectives. Cell Res. 2018;28:507–517. doi: 10.1038/s41422-018-0034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Duan X., Yang L., Wang L., et al. m6A demethylase FTO promotes tumor progression via regulation of lipid metabolism in esophageal cancer. Cell Biosci. 2022;12 doi: 10.1186/s13578-022-00798-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Alderson N.L., Rembiesa B.M., Walla M.D., et al. The human FA2H gene encodes a fatty acid 2-hydroxylase. J. Biol. Chem. 2004;279:48562–48568. doi: 10.1074/jbc.M406649200. [DOI] [PubMed] [Google Scholar]
- 103.Liu R., Cao K., Tang Y., et al. C16:0 ceramide effect on melanoma malignant behavior and glycolysis depends on its intracellular or exogenous location. Am. J. Transl. Res. 2020;12:1123–1135. [PMC free article] [PubMed] [Google Scholar]
- 104.Zhou X., Huang F., Ma G., et al. Dysregulated ceramides metabolism by fatty acid 2-hydroxylase exposes a metabolic vulnerability to target cancer metastasis. Signal Transduct. Target. Ther. 2022;7 doi: 10.1038/s41392-022-01199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tao M., Luo J., Gu T., et al. LPCAT1 reprogramming cholesterol metabolism promotes the progression of esophageal squamous cell carcinoma. Cell Death Dis. 2021;12 doi: 10.1038/s41419-021-04132-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Xu J., Chen Y., Zhang R., et al. Global and targeted metabolomics of esophageal squamous cell carcinoma discovers potential diagnostic and therapeutic biomarkers. Mol. Cell. Proteom. 2013;12:1306–1318. doi: 10.1074/mcp.M112.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhu Z.J., Qi Z., Zhang J., et al. Untargeted metabolomics analysis of esophageal squamous cell carcinoma discovers dysregulated metabolic pathways and potential diagnostic biomarkers. J. Cancer. 2020;11:3944–3954. doi: 10.7150/jca.41733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Li H., Zhang C., Zhang M., et al. Angustoline inhibited esophageal tumors through regulating LKB1/AMPK/ELAVL1/LPACT2 pathway and phospholipid remodeling. Front. Oncol. 2020;10 doi: 10.3389/fonc.2020.01094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Edin M.L., Duval C., Zhang G., et al. Role of linoleic acid-derived oxylipins in cancer. Cancer Metastasis Rev. 2020;39:581–582. doi: 10.1007/s10555-020-09904-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yang T., Hui R., Nouws J., et al. Untargeted metabolomics analysis of esophageal squamous cell cancer progression. J. Transl. Med. 2022;20 doi: 10.1186/s12967-022-03311-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Abbassi-Ghadi N., Antonowicz S.S., McKenzie J.S., et al. De novo lipogenesis alters the phospholipidome of esophageal adenocarcinoma. Cancer Res. 2020;80:2764–2774. doi: 10.1158/0008-5472.CAN-19-4035. [DOI] [PubMed] [Google Scholar]
- 112.Abbassi-Ghadi N., Golf O., Kumar S., et al. Imaging of esophageal lymph node metastases by desorption electrospray ionization mass spectrometry. Cancer Res. 2016;76:5647–5656. doi: 10.1158/0008-5472.CAN-16-0699. [DOI] [PubMed] [Google Scholar]
- 113.Buck A., Prade V.M., Kunzke T., et al. Metabolic tumor constitution is superior to tumor regression grading for evaluating response to neoadjuvant therapy of esophageal adenocarcinoma patients. J. Pathol. 2022;256:202–213. doi: 10.1002/path.5828. [DOI] [PubMed] [Google Scholar]
- 114.Mi L., Zhou Y., Wu D., et al. ACSS2/AMPK/PCNA pathway-driven proliferation and chemoresistance of esophageal squamous carcinoma cells under nutrient stress. Mol. Med. Rep. 2019;20:5286–5296. doi: 10.3892/mmr.2019.10735. [DOI] [PubMed] [Google Scholar]
- 115.Luo H., Wang X., Song S., et al. Targeting stearoyl-coa desaturase enhances radiation induced ferroptosis and immunogenic cell death in esophageal squamous cell carcinoma. Oncoimmunology. 2022;11 doi: 10.1080/2162402X.2022.2101769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shao F., Bian X., Jiang H., et al. Association of phosphoenolpyruvate carboxykinase 1 protein kinase activity-dependent sterol regulatory element-binding protein 1 activation with prognosis of oesophageal carcinoma. Eur. J. Cancer. 2021;142:123–131. doi: 10.1016/j.ejca.2020.09.040. [DOI] [PubMed] [Google Scholar]
- 117.Chang W., Luo Q., Wu X., et al. OTUB2 exerts tumor-suppressive roles via STAT1-mediated CALML3 activation and increased phosphatidylserine synthesis. Cell Rep. 2022;41 doi: 10.1016/j.celrep.2022.111561. [DOI] [PubMed] [Google Scholar]
- 118.Zhu Q., Huang L., Yang Q., et al. Metabolomic analysis of exosomal-markers in esophageal squamous cell carcinoma. Nanoscale. 2021;13:16457–16464. doi: 10.1039/d1nr04015d. [DOI] [PubMed] [Google Scholar]
- 119.Xu R., Rai A., Chen M., et al. Extracellular vesicles in cancer – implications for future improvements in cancer care. Nat. Rev. Clin. Oncol. 2018;15:617–638. doi: 10.1038/s41571-018-0036-9. [DOI] [PubMed] [Google Scholar]
- 120.Johnstone T.C., Suntharalingam K., Lippard S.J. The next generation of platinum drugs: Targeted Pt(II) agents, nanoparticle delivery, and Pt(IV) prodrugs. Chem. Rev. 2016;116:3436–3486. doi: 10.1021/acs.chemrev.5b00597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Li J., Cheng D., Zhu M., et al. OTUB2 stabilizes U2AF2 to promote the Warburg effect and tumorigenesis via the AKT/mTOR signaling pathway in non-small cell lung cancer. Theranostics. 2019;9:179–195. doi: 10.7150/thno.29545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhang Z., Du J., Wang S., et al. OTUB2 promotes cancer metastasis via hippo-independent activation of YAP and TAZ. Mol. Cell. 2019;73:7–21.e7. doi: 10.1016/j.molcel.2018.10.030. [DOI] [PubMed] [Google Scholar]
- 123.Dixon S.J., Lemberg K.M., Lamprecht M.R., et al. Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jiang X., Stockwell B.R., Conrad M. Ferroptosis: Mechanisms, biology and role in disease. Nat. Rev. Mol. Cell Biol. 2021;22:266–282. doi: 10.1038/s41580-020-00324-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lei G., Zhang Y., Koppula P., et al. The role of ferroptosis in ionizing radiation-induced cell death and tumor suppression. Cell Res. 2020;30:146–162. doi: 10.1038/s41422-019-0263-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Li D., Li Y. The interaction between ferroptosis and lipid metabolism in cancer. Signal Transduct. Target. Ther. 2020;5 doi: 10.1038/s41392-020-00216-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ubellacker J.M., Tasdogan A., Ramesh V., et al. Lymph protects metastasizing melanoma cells from ferroptosis. Nature. 2020;585:113–118. doi: 10.1038/s41586-020-2623-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jiang W., Hu J.W., He X.R., et al. Statins: A repurposed drug to fight cancer. J. Exp. Clin. Cancer Res. 2021;40 doi: 10.1186/s13046-021-02041-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Jin Y., Xu K., Chen Q., et al. Simvastatin inhibits the development of radioresistant esophageal cancer cells by increasing the radiosensitivity and reversing EMT process via the PTEN-PI3K/AKT pathway. Exp. Cell Res. 2018;362:362–369. doi: 10.1016/j.yexcr.2017.11.037. [DOI] [PubMed] [Google Scholar]
- 130.Chen Y., Li L.B., Zhang J., et al. Simvastatin, but not pravastatin, inhibits the proliferation of esophageal adenocarcinoma and squamous cell carcinoma cells: A cell-molecular study. Lipids Health Dis. 2018;17 doi: 10.1186/s12944-018-0946-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ventura R., Mordec K., Waszczuk J., et al. Inhibition of de novo palmitate synthesis by fatty acid synthase induces apoptosis in tumor cells by remodeling cell membranes, inhibiting signaling pathways, and reprogramming gene expression. EBioMedicine. 2015;2:808–824. doi: 10.1016/j.ebiom.2015.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wang Y., Yu W., Li S., et al. Acetyl-CoA carboxylases and diseases. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.836058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Huang C.M., Huang C.S., Hsu T.N., et al. Disruption of cancer metabolic SREBP1/miR-142-5p suppresses epithelial-mesenchymal transition and stemness in esophageal carcinoma. Cells. 2019;9 doi: 10.3390/cells9010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Jun Y., Tang Z., Luo C., et al. Leukocyte-mediated combined targeted chemo and gene therapy for esophageal cancer. ACS Appl. Mater. Interfaces. 2020;12:47330–47341. doi: 10.1021/acsami.0c15419. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.