Abstract
During recent decades, the emergence of pathogenic fungi has posed an increasing public health threat, particularly given the limited number of antifungal drugs available to treat invasive infections. In this Review, we discuss the global emergence and spread of three antifungal-resistant emerging fungi: Candida auris, driven by global healthcare transmission and possibly facilitated by climate change; azole-resistant Aspergillus fumigatus, driven by the selection facilitated by azole fungicide use in agricultural and other settings; and Trichophyton indotineae, driven by the underregulated use of over-the-counter high potency corticosteroid-containing antifungal creams. The diversity of the fungi themselves and the drivers of their emergence make it clear that we cannot predict what might emerge next. Therefore, vigilance is critical to monitoring for fungal emergence, as well as the rise in overall antifungal resistance.
Graphical Abstract
In this Review, Lockhart, Chowdhary and Gold discuss the global emergence and spread of three antifungal-resistant emerging fungi: Candida auris, azole-resistant Aspergillus fumigatus and Trichophyton indotineae, with the common thread that all three are currently emerging across the globe and have a high rate of acquired resistance.
Introduction
Fungi are among the most abundant and diverse organisms in the world. They are found in and on most humans as a part of the human microbiome and we breathe in hundreds of fungal spores each day (supplementary Box 1). Out of several million species of fungi, only several hundred are known to cause human illness. Most fungal infections are superficial, such as onychomycosis of the nails, dermatophyte infections of the skin such as athlete’s foot and mucosal infections (for example, vulvovaginal candidiasis). However, for some people, especially those with frequent healthcare interactions or weakened immune systems, encounters with fungi poses a serious health threat.
The emergence of pathogenic fungi is not a rare event. In recent decades, Cryphonectria parasitica nearly destroyed all chestnut trees in the United States and Fusarium oxysporum var. cubense nearly decimated banana monoculture; Batrachochytrium dendrobatidis is destroying amphibian populations, and Pseudogymnoascus destructans is killing entire colonies of bats 1–4. Fungal diseases of plants threaten food security worldwide by causing epiphytotics in staple crops and by contaminating food supplies with toxins. Rice-blast disease, caused by Magnaporthe oryzae, destroys 12–30% of annual rice harvests, and a new, virulent strain of Puccinia graminis that causes stem rust disease on wheat jeopardizes harvests in Europe, Asia and the Americas5,6.
Emerging fungi that directly affect human health have been less common. During the early HIV epidemic, Candida dubliniensis emerged alongside Candida albicans as a cause of azole-resistant mucosal candidiasis, and both Cryptococcus neoformans and Pneumocystis jirovecii exploited a new population of immunosuppressed hosts to become major causes of death in patients with advanced HIV disease7–9. With the HIV epidemic, cancer and immunomodulating medicine, fungal infections in humans have become more common, but until now, the emergence of new human pathogenic fungi has been rare. We are now witnessing both the emergence of new species of fungi, such as Candida auris and Trichophyon indotineae, as well the emergence and spread of drug-resistant lineages of well-known common fungi such as azole-resistant Aspergillus fumigatus 10–12 (Figure 1).
Figure 1. Global disbursement of the three emerging fungi Candida auris, azole-resistant Aspergillus fumigatus and Trichophyton indotineae.
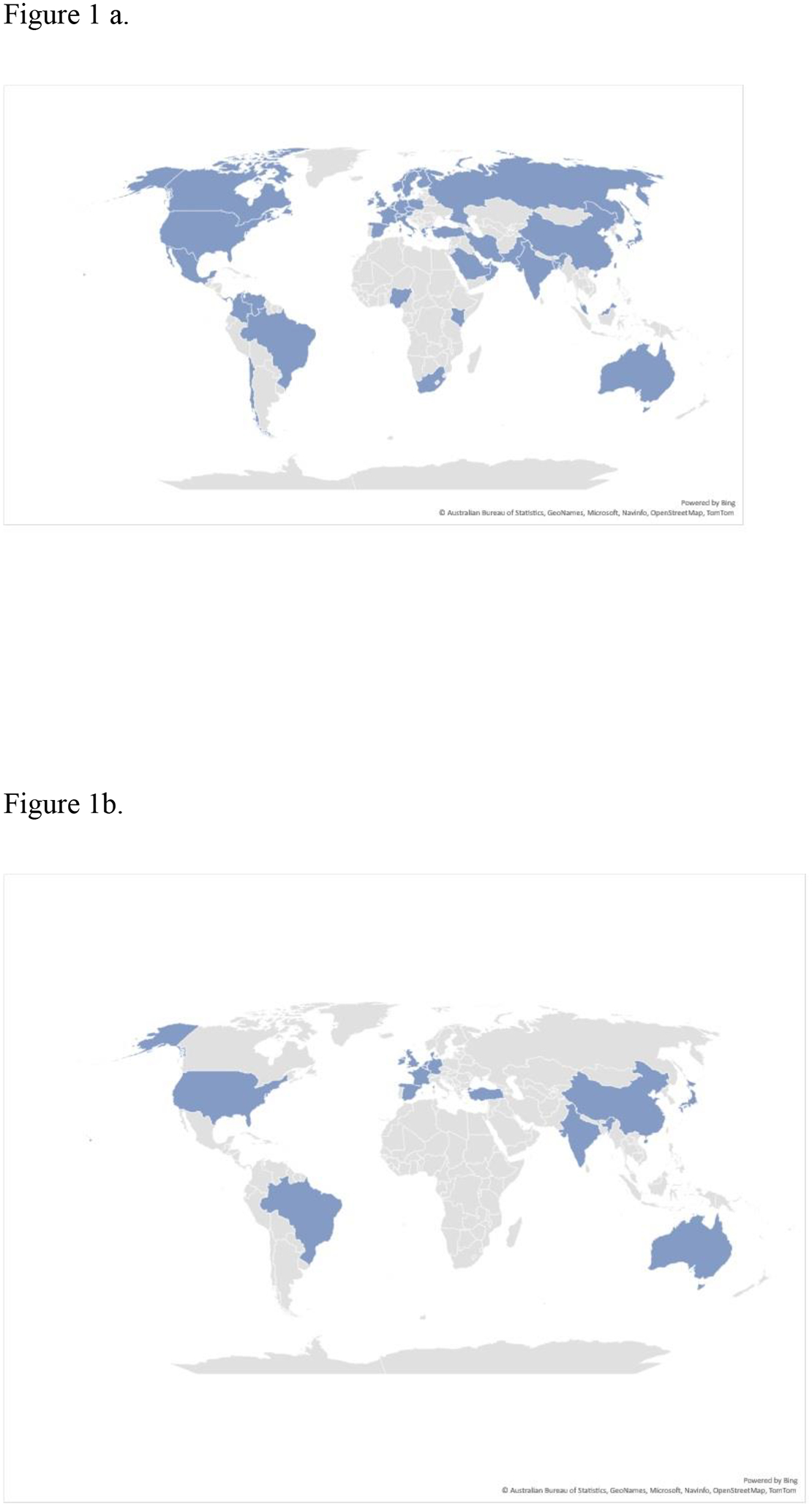
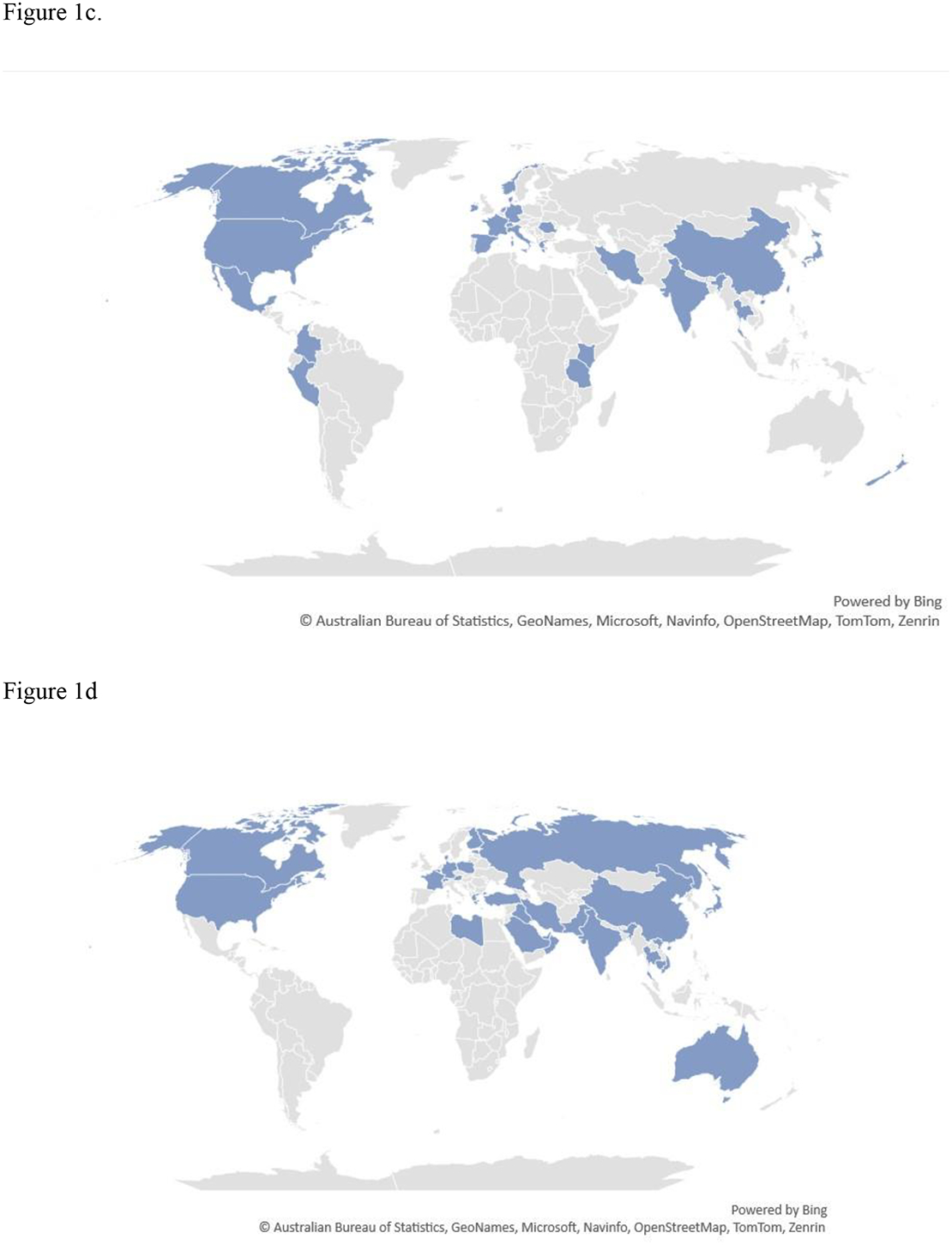
The presence of any of the organisms in a country does not mean they are ubiquitous there, and in some cases may be represented by a single isolate. a). Map of countries that have reported cases of Candida auris. b). Map of countries that have reported azole-resistant Aspergillus fumigatus containing the TR34/L98H or TR46/Y121F/T289A mutation in patients. c) Map of countries that have reported azole-resistant A. fumigatus containing the TR34/L98H or TR46/Y121F/T289A mutation in the environment. D) Map of countries where Trichophyton indotineae has been identified from patients or the country where the infection was most likely acquired.
Two important factors are common among C. auris, T. indotineae and azole-resistant A. fumigatus. First, these pathogens are spreading across the globe as essentially clonal lineages, C. auris as four clonal clades, T. indotineae as a single clonal clade and A. fumigatus as a clone (the clonal mutations, TR34, and TR46 to be discussed later with recombination. Although this has occurred before with fungi that infect amphibians (Batrachochytrium dendrobatidis3) and plants (Puccinia graminis6), this is the first time that human-pathogenic fungi have spread intercontinentally in such a short time frame (a decade for T. indotineae and C. auris, and several decades for azole-resistant A. fumigatus). The mechanisms for this spread may be different among the fungi, but the rapid rate is similar and concerning. What is surprising is that the emergence and spread of these three disparate species has occurred almost simultaneously.
The second commonality among these three fungi is that they share a very high rate of acquired antimicrobial resistance (Box 1). The mechanisms of acquired resistance and the antifungals to which they are resistant are different for each of the fungi, but it is nonetheless remarkable to have three fungi with high levels of acquired resistance emerge simultaneously, particularly because antifungal resistance in fungi is rare. In the cases of C. auris and T. indotineae, resistance to more than one class of antifungal can be present, putting these pathogens in the rare category of multidrug-resistant fungi. Although the cause of the sudden emergence and spread of C. auris, azole-resistant A. fumigatus and T. indotineae is unknown, researchers have proposed several potential contributing factors. It has been proposed that climate change may be contributing to the emergence of fungal species, including C. auris, but the data confirming this hypothesis for most fungi is lacking13–16. As global temperatures rise, selective pressure will favor fungi that survive at increased temperatures such as the temperatures found in the human body. A second proposed factor is the widespread use of antifungal compounds in human and animal medicine, as well for other applications, including for crop agriculture and the preservation of wood products, plastics and paints 17. In addition to selecting for resistant organisms, this widespread usage of antifungals may also have a secondary harmful role: increasing antimicrobial usage could potentially be changing microbiomes of not only of human hosts but also natural microbiomes 18–20. Changes to the established microbiome might promote the emergence of never-before-seen colonization or infections by microorganisms not previously detected in microbiomes. These microorganisms might emerge in the setting of the alterations of the natural microbiome that has developed over tens of thousands of years on the surfaces of our mucosa, gastrointestinal tracts and skin. C. auris colonization is correlated with changes in the skin microbiome21,22. T. indotineae infection is associated with the use of combination antifungal, antibiotic and topical steroid products, and research has shown that antibiotics alter the skin microbiome, but a direct correlation with T. indotineae infection and changes to the skin microbiome has not yet been shown23. Whether the spread of azole-resistant A. fumigatus is linked to changes in the environmental microbiome is unclear, but the presence of resistance-causing azoles in soil has been shown to change the soil microbiome24–27.
Box 1. Antifungal resistance.
Two categories of antifungal resistance exist: intrinsic and acquired resistance. Intrinsic resistance means that the wild type of a fungal species naturally exhibits resistance to antifungal compounds. By contrast, acquired resistance means that a fungal species is normally susceptible to antifungal compounds, but certain isolates have acquired mutations or epigenetic changes that confer resistance 186. In general, when researchers and clinicians discuss antifungal resistance, they are referring to acquired resistance.
Intrinsic antifungal resistance is a common phenomenon. For example, Basidiomycetes, Mucormycetes, and Fusarium species all exhibit resistance to echinocandins, a class of antifungal drugs that targets one of the proteins responsible for synthesis of β-1,3 glucan in fungal cell walls 187,188. Because β-1,3 glucan constitutes only a minor component of Basidiomycete, Mucormycete, and Fusarium species, these fungi are intrinsically resistant to this drug class. Another example is the intrinsic resistance of Candida krusei to fluconazole, an antifungal drug that targets the enzyme lanosterol deacetylase, a protein in the ergosterol synthesis pathway. Fluconazole is ineffective against C. krusei because this Candida species has an amino acid polymorphism in its lanosterol deacetylase that prevents fluconazole from binding to the protein and inhibiting ergosterol synthesis 189,190.
Acquired resistance is uncommon in fungi. For Candida species, among those with established breakpoints, the overall resistance rate to antifungals is 7% (Ref. 191). Acquired resistance in molds is thought to be even less common, but because antifungal resistance testing of molds is infrequently performed, the true extent of acquired resistance in molds is unknown.
Fungi acquire antifungal resistance through numerous mechanisms. The most common forms of acquired resistance involve mutations to target protein binding-sites, which prevent the antifungal from binding 192. This mechanism is common among azole-resistant strains of Candida species such as C. albicans, C. tropicalis and C. parapsilosis, as well as in azole-resistant Aspergillus species 192. Certain yeast species acquire resistance through mutations in transcription factors that lead to increased expression of the antifungal drug’s target, diluting the antifungal’s effect. In C. glabrata, the most common mechanism of fluconazole resistance involves mutations in transcription factors which lead to increased expression of efflux pumps that remove the antifungal from the cytoplasm before it can bind the target protein 193. Overexpression of the target protein through polyploidy and chromosomal duplication is another way that fungi overexpress target proteins and dilute the effect of antifungals 194,195. Although the mechanisms have not been well studied, some fungi may acquire resistance to azoles and amphotericin B by changing the composition of the fungal cell wall or cell membrane, thus compensating for the loss of the product of the target protein 192.
Fungal cells have evolved over hundreds of millions of years to become as efficient as possible. Mutation or overexpression of housekeeping genes can lead to a fitness cost for the cells. With the advent of RNAseq and whole-genome sequencing, data increasingly suggest that most cells that acquire antifungal resistance undergo multiple other compensatory changes 196. Some of these changes are epigenetic changes in expression; others are mutations in other proteins. These secondary mutations are generally compensatory, and their role is most likely to alleviate fitness costs that may be associated with the acquired resistance 197–200.
The emergence of antifungal resistance has been a concern for clinicians, as the armamentarium for treating invasive fungal infections is essentially limited to three drug classes: azoles, echinocandins, and polyenes. For invasive mold infections, treatment options are further limited to certain azoles and the polyenes. When a mold becomes azole resistant, the choices remaining are to use combination treatment with an echinocandin or a less effective antifungal like terbinafine, or to use the polyene amphotericin B 131. Although amphotericin B has excellent efficacy against most fungal infections, it can cause severe side effects.
In this Review, we discuss C. auris, azole-resistant A. fumigatus and T. indotineae, with the common thread that all three are currently emerging across the globe and have a high rate of acquired resistance, two characteristics that are rare among fungi. Although the precise details of their emergence might remain elusive, we outline and discuss our current understanding of the emergence, epidemiology, evolution, pathogenicity, clinical manifestations, resistance mechanisms and treatment options for these pathogens (Table 1).
Table 1.
Features of Candida auris, azole-resistant Aspergillus fumigatus and Trichophyton indotineae.
| Feature | Candida auris | Azole-resistant Aspergillus fumigatus | Trichophyton indotineae |
|---|---|---|---|
| Antifungal resistance | Nearly all isolates are fluconazole resistant; amphotericin B, echinocandin and flucytosine resistance has been recorded | TR34 – itraconazole; TR46 – voriconazole; through additional mechanisms, some isolates may become pan-azole resistant | Frequently resistant to terbinafine, sometimes resistant to azole antifungals (for example, itraconazole) |
| Country or region where first cases have been identified | East Asia, Indian subcontinent, South Africa and Venezuela | Europe | Southern and Southeast Asia |
| Current treatment options | For azole- and amphotericin B resistant isolates the treatment of choice is an echinocandin; for pan-resistant isolates there are ongoing clinical trials for new the antifungals ibrexafungerp and fosmanogepix | Amphotericin B; there are ongoing clinical trials for the new antifungals ibrexafungerp and olorofim | Terbinafine remains a treatment option, but for resistant isolates itraconazole or other triazoles are an option |
| Current distribution | Worldwide distribution, predominantly from the clades that originate from the Indian subcontinent, South Africa and Venezuela | Predominant in Europe but has been identified in Africa, North America, South America, Asia and Australia | Predominantly in India; cases have been reported throughout Asia, Europe, Africa, and North America; cases in Europe have generally been associated with travel or migration |
| Clinical presentation | Can be present as both a skin or urinary tract commensal with no symptoms or a pathogen in the bloodstream with typical symptoms of candidemia | Typical presentation for aspergillosis but may be refractory to initial therapy | Inflammatory dermatophyte infection, most commonly affecting the face and body, occasionally affecting nails |
Candida auris
Emergence and epidemiology.
The emergence of C. auris has been rapid and overwhelming. The first recognized clinical isolate was recovered from the ear of a patient in Japan in 2006 10 (Table 1). By 2014, C. auris had been identified in South Korea, India and South Africa; by 2016, four separate emerging clades had been described on three different continents 28–31(Figure 1a). In just over a decade after its initial discovery, researchers have identified C. auris in over 40 countries and on all continents except Antarctica 32 (Figure 1a). A retrospective review of culture collections across the globe identified some older isolates of C. auris, with the oldest isolates now identified from a collection in South Korea from 1996, a collection from Japan in 1997, a collection in France from 2007, and an isolate from Pakistan from 2008 (Refs. 28,31,33–35). However, these are one-off isolates, which suggests that C. auris was extremely rare before the early 2010s, and that the emergence and spread across three continents has been sudden. Molecular clock analysis using whole-genome sequences indicates that the time to the most recent common ancestor of three of the four clades happened as recently as the 1980’s, whereas the oldest clade is likely to have only diverged from a common ancestor around 400 years ago 36. Whole-genome sequencing of isolates from around the world also reveals that the global spread occurred solely through the previously identified four clades and not through emergence of new clades, although a fifth clade has been recently identified exclusively in Iran 36,37.
The rapid emergence of C. auris is tied to its epidemiology. Three factors differentiate C. auris from other species of Candida: C. auris is primarily associated with the skin rather than the gut or mucosal surfaces; it spreads rapidly in healthcare environments leading to frequent outbreaks; and most isolates have acquired antifungal resistance, with some isolates demonstrating resistance to all three classes of systemic antifungals 38. Most Candida species are considered commensals of mucosal surfaces and the gastrointestinal tract, although some of the same species found on mucosal surfaces can also be found on the skin 39,40. By contrast, C. auris is primarily associated with the skin, although it is occasionally found in the respiratory and gastrointestinal tracts 41. C. auris colonizes the skin of patients in healthcare settings and has been associated with a disruption to the normal skin flora 21,22. Extensive screening of persons outside of a healthcare setting has not been performed, but two studies that looked for colonization of healthcare workers failed to detect colonization, which implies that an intact skin microbiome may be a barrier to C. auris colonization, although it has been shown to be transient on the hands of healthcare workers 41–43. Because C. auris can persist on the skin, it can spread widely in healthcare environments, including both among patients and on environmental surfaces. The degree of skin colonization has been shown to be proportional to the spread within the patient environment 22,44. Further, skin colonization with C. auris is a risk factor for bloodstream infections; approximately 5–10% of patients who are colonized with C. auris develop bloodstream infections 45. Because C. auris is a nationally notifiable disease in the United States, the degree of spread has been well documented. Initially, spread was due to importation followed by limited localized transmission. However, most contemporary cases have been acquired through localized transmission, and clinical cases continue to increase 46. Of great concern, the annual percentage increase in clinical cases of C. auris was 44% in 2019 and 95% in 2021 (Ref. 47).
Evolution.
The sudden, simultaneous emergence of C. auris on multiple continents has led to speculation on its origin 13,14,48. The two clades that have not yet spread outwardly, clade II (East Asian) and clade V (Iran), are primarily associated with the skin of the ear canal, a clue to the possible origin of emergence 33,49. It is unlikely that both clades independently found their way to this highly specialized niche following emergence, especially because neither clade seems to be spreading globally; rather, the ear canal may be a natural niche for these clades and could have been the original skin reservoir for all of the C. auris clades before emergence33,35,37. C. auris has been identified in the environment both from coastal wetlands in the Andaman Islands and Colombia, and from apples that had been stored and processed in India 50–52. What we cannot discern from these environmental sources is whether these are the natural habitat of C. auris or a secondary niche following contamination of the environment following colonization of humans. Whether C. auris emerged from the environment or from an obscure human commensal niche, selective pressure from antifungal usage, the disruption of microbiomes, and climate change are all possible drivers of the emergence.
Pathogenicity and clinical manifestations.
As a colonizer of the skin, C. auris does not cause any symptoms to the colonized patient. This lack of symptoms enables C. auris colonization to go undetected and likely plays a substantial role in its ability to spread within healthcare facilities. At this point there is no evidence that Candida auris colonizes individuals without healthcare exposures. Outside of its detection in the ear, C. auris is not yet found in or on medically naïve persons41,42. Because of these factors, infection prevention and control measures [https://www.cdc.gov/fungal/candida-auris/c-auris-infection-control.html] are critical to preventing the spread of C. auris
The clinical manifestations seen with C. auris are similar to those seen with infections caused by other Candida species 53. Candidemia, urinary tract infections, wound infections, otitis and skin abscesses are the most common infection types, but C. auris can also cause disseminated disease 54. C. auris has not been implicated as a frequent pathogen in other more common types of candidiasis (for example, vulvovaginal candidiasis or oral thrush). Risk factors for C. auris infection mirror those for infections by other Candida species including recent surgery, immunosuppression, neutropenia, extended healthcare exposure and use of antibiotics55. One of the primary risk factors for C. auris infection is the presence of indwelling medical devices including ventilator tubes, central venous catheters, feeding tubes and urinary catheters, as they provide a mechanism for C. auris to disseminate from the skin. The ability of C. auris to form biofilms both on these devices as well as robust biofilms on the skin may prolong colonization and increase the risk of a disseminated infection56–58. C. auris infections made a leap forward during the first few years of the COVID-19 pandemic. Increased medical intervention, limited personal protective equipment due to supply chain issues, and increased corticosteroid use for COVID-19 treatment are likely to have contributed to the increase in cases59.
In a mouse model of infection C. auris is less virulent than C. albicans but more virulent than C. glabrata and C. haemulonii60. This decreased overall virulence may be related to the fact that C. auris is only able to form rudimentary hyphae, which is notable because hyphae formation is a major virulence factor of other Candida species61. One of the more intriguing virulence factors of C. auris may be its ability to evade phagocytosis by neutrophils62. In a zebrafish model of disseminated infection, neutrophils were twice as responsive to C. albicans as to C. auris62. Further work in this area has revealed that mannans in the C. auris cell wall may mask critical components recognized by neutrophils such as β-glucan, and chitin63. There is also evidence that C. auris can replicate within and escape from macrophages 64. Under in vitro conditions, C. auris may be able to kill macrophages as well through the mechanism of rapid replication and glucose depletion64.
Resistance mechanisms and treatment options.
Aside from intrinsically resistant species, such as Candida krusei, fluconazole resistance among Candida species is rare. Before the emergence of C. auris, the Candida species with the highest frequency of acquired resistance was C. glabrata, a pathogen whose overall worldwide resistance rate is approximately 10–12% (Ref. 65). This changed with the emergence of C. auris, for which acquired resistance to fluconazole is observed in most clinical isolates 31,66,67. Resistance seems to be clade-specific, with isolates of clades I and III being almost universally resistant to fluconazole, isolates of clade II being almost universally susceptible to fluconazole, and isolates of clade IV showing variable resistance, depending on the circulating clone36.
The most common mechanism of azole resistance in C. auris is a mutation in the drug target lanosterol 14-a-demethylase (ERG11) such as Y132F, K143R and F126L (Figure 2). Increased expression of efflux pumps, including those due to mutations in the gene encoding the transcription factor TAC1b, which cause an increase in the expression of the efflux pump Cdr1p, have also been determined to increase resistance68,69. Resistance to echinocandins is uncommon. However, resistance can develop during echinocandin therapy and is caused by several different mutations at amino acid S639 (S639Y, S639F or S639P) in hotspot1 of the drug target 1,3-β-D-glucan synthase (FKS1)66,70,71. A breakpoint for amphotericin B against C. auris does not exist, but based on pharmacokinetic–pharmacodynamic values in animal models, a tentative breakpoint of 2 μg/ml was set 72. Using this value, resistance to amphotericin B has been reported in about 30% of isolates; however, resistance seems to vary based on the clone or the clade being tested 31,32. Because resistance to amphotericin B is difficult to define, it is unknown whether this represents clinical or in vitro resistance. To date, only a single mechanism, a mutation in the ERG6 gene, has been definitively associated with amphotericin B resistance 73.
Figure 2.
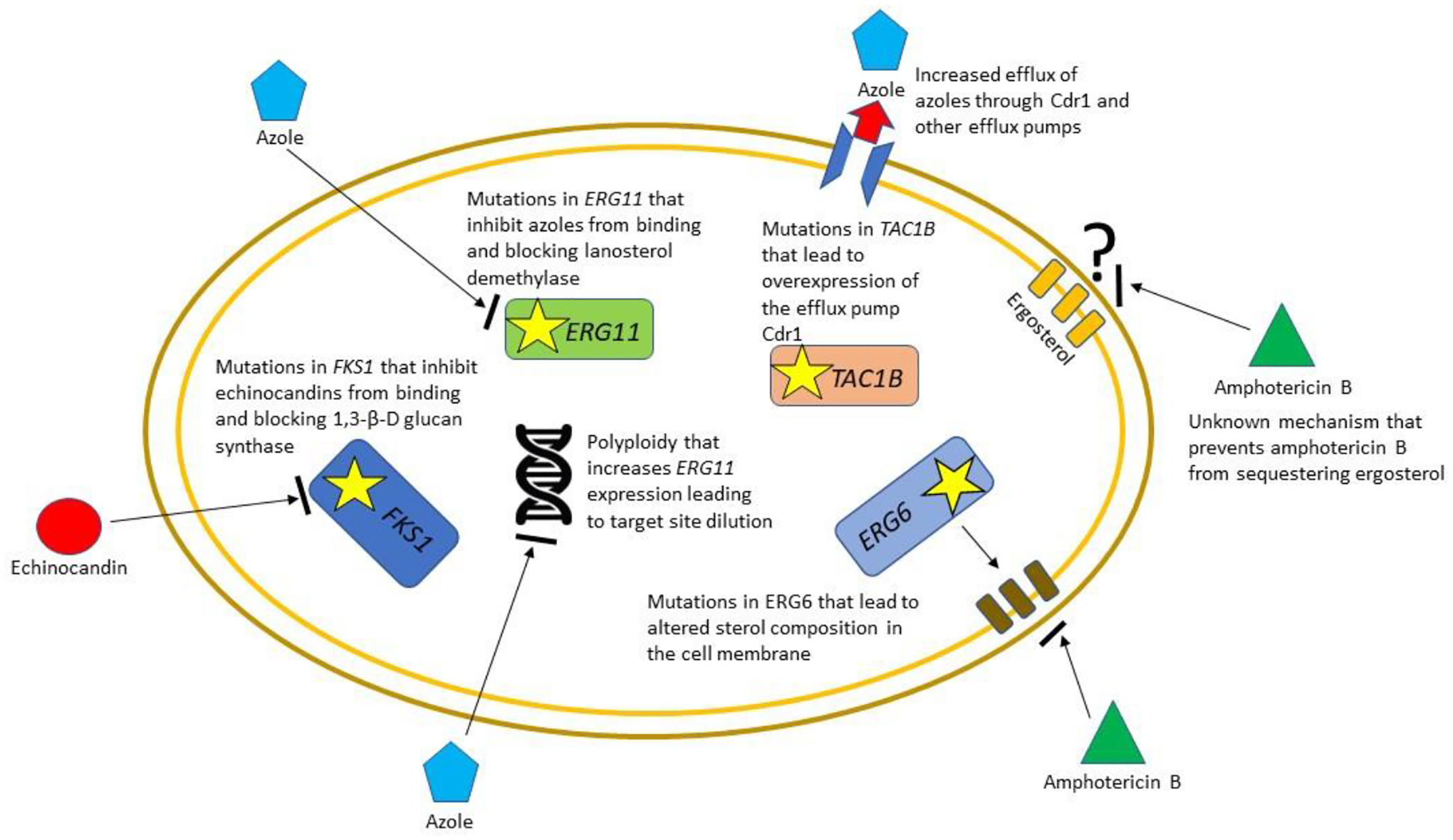
The more common mechanisms of acquired antifungal resistance of Candida auris. One of the most remarkable aspects of C. auris is the number of different mechanisms of acquired antifungal resistance that have been identified. For the azoles, especially fluconazole, these include mutations in the target enzyme lanosterol demethylase (ERG11), mutations in transcription factors such as TAC1B that lead to the over-expression of efflux pumps, and changes in ploidy that lead to overexpression of ERG11 and target site dilution. For the echinocandins, the predominant mechanism of resistance is a change in the amino acid sequence at the target site hotspot in Fks1p the mechanism that is conserved across Candida species. For amphotericin B, a specific amino acid change in Ergp6 that alters membrane sterol composition has been identified, but other, as yet unidentified, mechanism that prevent ergosterol sequestration have been identified phenotypically but not molecularly.
Multidrug resistance in most Candida species is extremely rare and pan-antifungal resistance has rarely been reported in any Candida species74. As C. auris began to emerge, several pan-antifungal resistant isolates were reported. Fortunately, further spread of these isolates did not occur, and it was speculated that pan-resistance in these organisms was associated with a fitness cost75. However, documented transmission of pan-resistant and echinocandin-resistant C. auris isolates was later reported from the United States, which suggests that high transmissibility of C. auris can lead to spread of drug-resistant clones in healthcare settings despite any actual or perceived fitness cost 70.
Several societies, including the Australasian Society for Infectious Disease and the Federation of Infectious Disease Societies of Southern Africa, have released guidelines that include treatment recommendations for C. auris76–78. The first option for treating C. auris infection in adults and children aged >2 months is an echinocandin, which is in accordance with the Infectious Diseases Society of America guidelines for the treatment of candidiasis79. For a central nervous system infection, for infants aged <2 months or when echinocandins are unavailable, amphotericin B is the recommended treatment. Because antifungal resistance is common, susceptibility testing is recommended for all isolates for which treatment is intended. The use of antifungals in patients who are only colonized is not recommended.
Treatment options for pan-antifungal resistant isolates of C. auris may soon become available. Two new antifungals in development, ibrexafungerp and manogepix, have shown good in-vitro activity against multi-drug resistant isolates of C. auris80,81. Ibrexafungerp has an FDA-approved indication for the treatment of vulvovaginal candidiasis, but both ibrexafungerp (ClinicalTrials.gov Identifier: NCT03059992) and manogepix (ClinicalTrials.gov Identifier: NCT05421858) are undergoing clinical trials for the treatment of systemic infections, including the treatment of patients with multi-drug resistant C. auris.
Azole-resistant Aspergillus fumigatus
Emergence and epidemiology.
Aspergillus fumigatus, a globally distributed mold found throughout the environment, is an important opportunistic pathogen in humans. It is the primary cause of invasive aspergillosis, an infection responsible for substantial morbidity (>14,000 annual U.S. hospitalizations) and mortality (>1,200 annual U.S. deaths) 82–84 and requiring treatment with systemic antifungal medications. Globally, there may be as many as 250,000 cases of invasive aspergillosis and 3 million cases of chronic pulmonary aspergillosis each year 85. In addition to invasive disease, A. fumigatus is also responsible for various clinical syndromes, including chronic pulmonary aspergillosis and allergic bronchopulmonary aspergillosis, especially in patients with cystic fibrosis or asthma, that cause considerable morbidity and frequently require treatment with antifungal therapy 86,87.
Isolates of azole-resistant A. fumigatus have been recognized almost since the use of azoles for treatment began a few decades ago, but during the last decade or so resistance has noticeably increased 88–90. Of public health concern, isolates of A. fumigatus are increasingly identified that are resistant to some or all azole antifungals 12,91. Patients acquire azole-resistant A. fumigatus infections through two routes: repeated exposure to azoles for the treatment of chronic Aspergillus infections or direct inhalation of resistant A. fumigatus spores from the environment 92 (Figure 3). Even though active surveillance for drug-resistant molds is absent or minimal in most countries, researchers have identified azole-resistant A. fumigatus across the globe 93,94. As of this report, 20 countries have reported clinical isolates demonstrating azole resistance, and it has been identified in environmental specimens in an even larger number of countries (Figures 1b and 1c). In Dutch studies focused primarily on patients at high risk, >20% of patients with invasive aspergillosis harbored a triazole-resistant strain, with rates varying based on the population studied 95.
Figure 3.
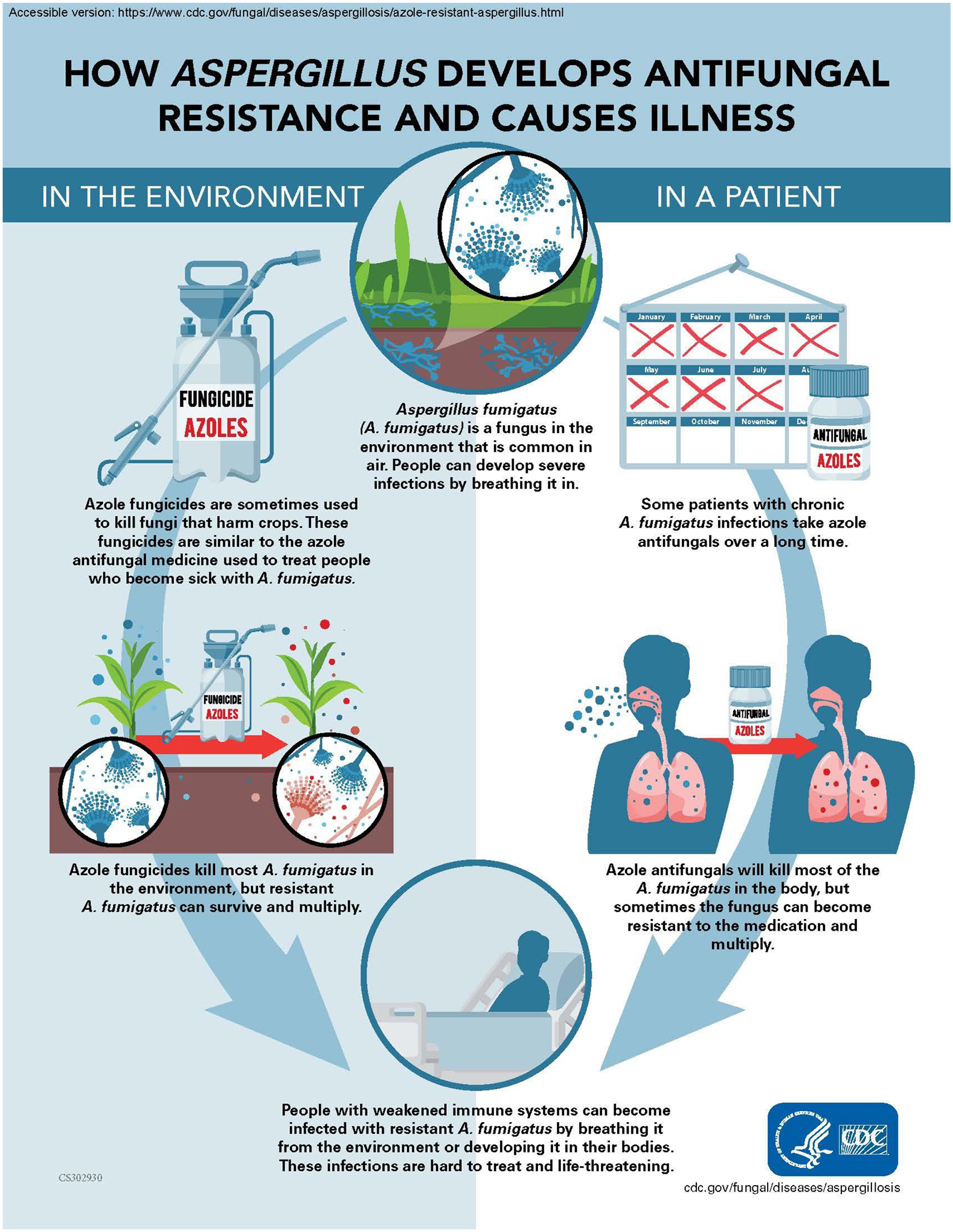
Diagram of possible routes of acquisition of antifungal resistant Aspergillus fumigatus.
Pathogenicity and clinical manifestation.
Aspergillus is a saprophytic fungus whose spores are found throughout the environment. A. fumigatus is the most common Aspergillus species encountered by humans. Humans inhale Aspergillus spores every day, but because the innate immune system can clear these spores, most immunocompetent persons do not develop illness96. However, some immunocompetent hosts, especially those with cavitary lesions of the lung, can develop chronic pulmonary aspergillosis, and other immunocompetent hosts can suffer from allergic bronchopulmonary aspergillosis97,98. In immunocompromised patients, Aspergillus species can colonize the lungs, potentially leading to the development of invasive pulmonary aspergillosis97–99. Invasive aspergillosis is the most common infection among patients who received hematopoietic stem cell transplant and the second most common infection among patients who received solid organ transplant 100–102. Aspergillus species colonization and infection are also associated with other chronic conditions such as cystic fibrosis and chronic granulomatous disease103, and invasive aspergillosis can develop as a secondary complication in patients recovering from tuberculosis, viral influenza and COVID-19 (Refs. 104,105). In severely immunocompromised patients, mortality from A. fumigatus infection can be 40–50% despite the availability of several treatment options97,98.
Data to understand the clinical significance of azole-resistant A. fumigatus infections have generally come from small case series and several larger cohort studies, primarily from Europe 106. In one cohort study, patients with azole-resistant invasive aspergillosis had a 90-day mortality rate approximately 25% higher than those with azole-susceptible infections 107. That study also found a higher 42-day mortality among patients with voriconazole-resistant aspergillosis who were started on inappropriate antifungal therapy (for example, voriconazole monotherapy) versus patients receiving appropriate therapy 107. In another study involving patients with hematologic malignancies, the 6-week mortality was 2.7-times higher among patients with azole resistance-associated mutations in the Cyp51A gene 108. These studies point out that two of the outstanding features of infections due to azole-resistant A. fumigatus are resistant infections in azole-naïve patients and treatment failure. Azole-resistant infections not only occur in patients with invasive aspergillosis, they also occur in patients with chronic pulmonary aspergillosis.
Resistance mechanisms.
Recent research has linked the use of azole fungicides, which are chemically similar to azole antifungal compounds used in medicine, to the development of azole-resistant infections in patients 109–111. Azole fungicides are applied in numerous agricultural and non-agricultural settings around the world. Although A. fumigatus is not a plant pathogen, it is found throughout the environment, especially associated with plant debris. The use of azole fungicides, such as tebuconazole, propiconazole, epoxiconazole and difenoconazole, can select for A. fumigatus strains that are resistant to antifungal drugs used in clinical medicine to treat aspergillosis 92. Aspergillus fumigatus strains that have developed resistance through exposure to azole fungicides in the environment harbor unique mutations in the target gene CYP51A, which encodes lanosterol demethylase, an essential enzyme in the ergosterol pathway (Figure 4). The two most widely dispersed CYP51A mutations are TR34/L98H (a 34 base-pair tandem repeat in the promoter sequence and a leucine to histidine mutation at amino acid 98) and TR46/Y121F/T289A (a 46 base-pair tandem repeat in the promoter sequence and mutations at amino acids 121 and 289) 112,113. These mutations confer resistance to itraconazole and voriconazole, respectively, but isolates carrying these mutations are often, but not always, pan-azole resistant when tested in vitro113–116. The tandem repeats in the promoter region cause an upregulation of gene transcription while the amino acid mutations decrease the efficacy of the antifungals by altering the target binding site 117. The TR34 and TR46 mutations have been shown to be associated with environmental fungicide exposure, with a recent study confirming that A. fumigatus strains that became drug resistant in the environment can cause resistant infections in humans 109. Although these two mutations are predominant, surveillance of A. fumigatus isolates collected during environmental sampling indicates that some of the other CYP51A mutations that were long thought to be exclusively associated with long-term azole use in patients, such as mutations at M220 and G54, may also be associated with environmental fungicide exposure 118–120. In addition, the mutations in the CYP51A promoter are evolving both through sexual recombination and random mutation 121,122. How these newer mutations will affect drug resistance and organism fitness remains to be seen.
Figure 4.
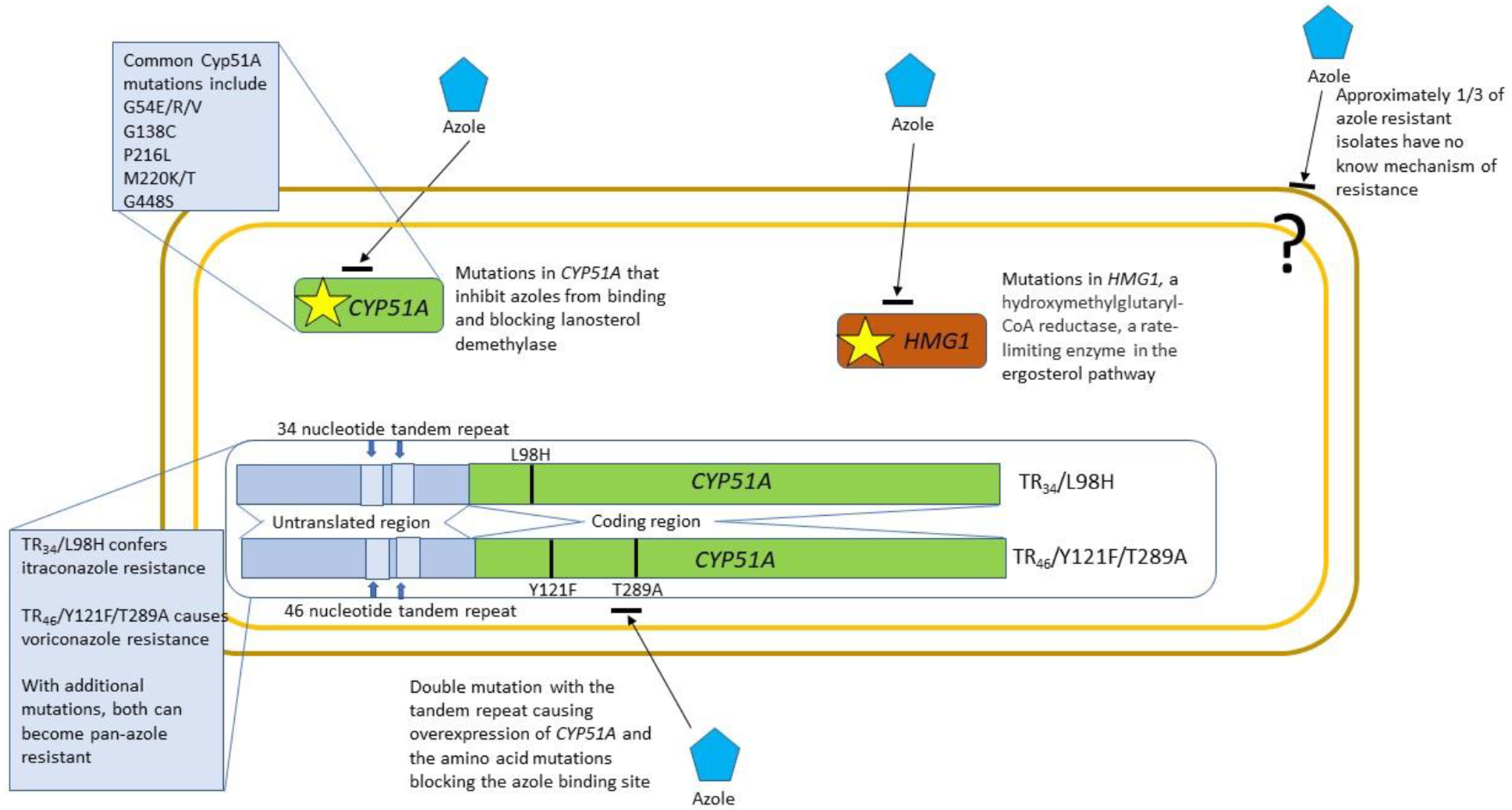
Mechanisms of acquired resistance of azole-resistant Aspergillus fumigatus. Azole resistance in A. fumigatus appeared almost as soon as azoles became available. Approximately 30% of isolates have no identified mechanism of resistance, but the other 70% have changes either in the azole target enzyme lanosterol demethylase (CYP51A), or in HMG1, the gene encoding hydroxymethylglutaryl-CoA reductase, a rate-limiting enzyme in the ergosterol pathway. Some target site mutations, such as those at amino acids G54, G138, P216, M220 and G448, are associated predominantly with acquired resistance following long-term azole treatment. There is a unique set of targets, TR34/L98H (TR34) and TR46/Y121F/T289A (TR46) that are specifically associated with environmental acquisition of resistance due to exposure to agricultural fungicides. These mutations consist of both a duplication in the 5’ untranslated region that causes increased transcription and a single or double amino acid change in the target binding site. The TR34 mutation leads to pan-azole resistance while the TR46 mutation is specific for voriconazole resistance.
Azole resistance acquired during long-term treatment is generally associated with either specific amino-acid substitutions in the CYP51A gene or mutations in HMG1, a hydroxymethylglutaryl-CoA reductase, which is a rate-limiting enzyme in the ergosterol pathway (Figure 4). Approximately 30% of isolates of A. fumigatus that exhibit in vitro resistance to azoles have no defined mechanism of resistance. Whether these isolates also exhibit in vivo resistance in not known. There are no breakpoints for echinocandins and A. fumigatus, but increased minimal effective concentration values are rare123. The European Commission on Antimicrobial Susceptibility Testing (EUCAST) has developed breakpoints for A. fumigatus, and amphotericin B resistance is rare but not unprecedented124. Resistance to multiple classes of medical antifungals in A. fumigatus is exceedingly rare, but resistance to multiple classes of fungicides has recently been reported in environmental A. fumigatus isolates from the United States 111.
Evolution.
Recent use of whole-genome sequencing that compares TR34 isolates collected around the globe to azole susceptible A. fumigatus isolates indicates that the TR34 isolates are tightly clustered in a single clade 109,111,125. These data suggest that the TR34 and TR46 mutations arose only once or, at most, a few times and that the global spread is due to one or a limited number of closely related strains and not the repeated de novo generation of this resistance mechanism 125. Of concern, in the United States, azole fungicide use has drastically increased in recent years (four-fold increase during 2006–2016), and in Europe, azole fungicide use is also increasing 17,126. As fields are sprayed year after year, continued selection for azole-resistant strains is a concern. Although not specifically a plant pathogen, the association with agriculture is likely to have a role in the global dispersal of these resistant strains, as TR34 A. fumigatus has been isolated from flower bulbs and other agricultural products destined for global distribution and are likely to be globally dispersed as aerosols as well 127. The current distribution of these mutations in the environment is not known because most countries, including much of the Americas, Africa and Asia, do not have robust surveillance mechanisms in place and have simply not looked for it.
The CYP51A gene, the target of azole resistance, is highly divergent across individual isolates, with many reported amino acid polymorphisms that have not been linked to azole resistance 88,128. This pleomorphism, in combination with sexual recombination and selective pressure of environmental fungicides may drive the future selection of other resistance mutations 122,129.
Azoles are currently the only class of antifungal used in both human medicine and as agricultural fungicides. However, this may soon be changing with a new class of antifungal, the orotomides, which act by inhibiting the dihydroorotate dehydrogenase (DHODH) enzyme. A new mold-active DHODH inhibitor, olorofim, is active against azole-resistant Aspergillus species and is currently in phase 3 clinical trials. Recently, the fungicide ipflufenoquin, also a DHODH inhibitor, has been approved by the U.S. Environmental Protection Agency (EPA) for agricultural use. A recent study shows that resistance mechanisms between the antifungal and the fungicide are similar and therefore may lead to a similar circumstance of environmentally induced resistance to a medically important antifungal130.
Treatment options.
Azole antifungal drugs (for example, voriconazole, itraconazole, posaconazole and isavuconazole) are the first-line treatment for invasive aspergillosis and the only orally available therapeutic options. The emergence of azole-resistant A. fumigatus could limit the clinical use of these life-saving drugs 87,92. Data to guide the optimal treatment for patients with azole-resistant aspergillosis are limited, and consensus guidelines do not exist 131. However, expert opinions on the management of patients with azole-resistant infections have been published and emphasize the importance of choosing initial antifungal therapy based on the prevalence of environmental azole resistance. For example, in areas where azole resistance related to environmental mechanisms is more than 10%, clinicians might reconsider voriconazole monotherapy for primary treatment of invasive aspergillosis, pending the availability of susceptibility testing results. For patients with aspergillosis confirmed to be resistant to azoles, therapy could be adjusted to include a drug to which the A. fumigatus is susceptible such as amphotericin B 131.
Three new antifungals, ibrexafungerp, fosmanogepix and olorofim, have shown good in vitro activity against azole-resistant isolates132–134. Although these new agents are not yet available, clinical trials are underway. Neither trial specifically includes treatment of resistant isolates, but neither excludes them either. The SCYNERGIA trial is a phase 2 randomized double-blind study of ibrexafungerp in combination with voriconazole for the treatment of invasive pulmonary aspergillosis versus voriconazole alone (ClinicalTrials.gov Identifier: NCT03672292). The OASIS study is a phase 3 randomized blinded study to compare treatment of proven invasive aspergillosis with either olorofim or liposomal amphotericin B (ClinicalTrials.gov Identifier: NCT05101187).
Trichophyton indotineae
Emergence and epidemiology.
Dermatophytosis, also known as ringworm or by the syndromic name tinea, is a contagious superficial infection caused by keratinophilic fungi called dermatophytes 135. The most common causative organisms are Trichophyton spp. A substantial public health burden, dermatophytosis affects an estimated 20–25% of the world’s population 136. Patients may acquire dermatophyte infections from fomites or through direct contact with infected persons or animals. Risk factors include living in humid climates and overcrowded conditions, participating in activities involving skin-to-skin contact (for example, certain sports) and having a weakened immune system 137. Though generally considered a mild, treatable condition, dermatophytosis can cause stigma, debilitating pruritis, immune reactions, and bacterial superinfections, particularly in immunocompromised patients 138.
Historically, clinicians could successfully treat most patients with dermatophytosis using available antifungal drugs, but dermatophyte resistance to conventional antifungal drugs, including terbinafine and itraconazole, is an emerging public health threat 139. During the past decade, researchers from India have noted an alarming increase in the incidence of difficult-to-treat dermatophytosis cases 140. These infections can persist for years, spread easily within households, and cause substantial suffering 139. Researchers identified Trichophyton interdigitale/Trichophyton mentagrophytes ITS (internal transcribed spacer) Type VIII as the etiologic agent behind the epidemic of antifungal resistant tinea in India. In 2020, researchers determined that these highly terbinafine-resistant Trichophyton strains were different enough from the T. mentagrophytes/T. interdigitale complex to be a new species, Trichophyton indotineae 11. Several lines of evidence support this separation into distinct species. Trichophyton indotineae clusters separately from T. mentagrophytes sensu stricto and T. interdigitale at several unlinked DNA loci141. Also, T. mentagrophytes is zoophilic and has a sexual cycle, whereas T. indotineae has evolved to be a predominantly clonal anthropophilic fungus141. And finally, T. indotineae is frequently associated with terbinafine resistance, conferred by mutations in the squalene epoxidase gene 142. A large study of patients from eight geographically dispersed regions in India found that T. indotineae was the dominant causative agent for dermatophytosis in all regions studied, representing an epidemiologic shift in the predominant pathogen causing dermatophytosis from Trichophyton rubrum 139.
Even though T. indotineae as a recognized species is only a few years old, it has already become a major public health concern. Cases of T. indotineae infection have been identified in numerous countries outside of India, including Germany, Belgium, Denmark, France, Greece, Italy, Switzerland, Canada, the United States Australia, United Arab Emirates, Oman, Iran, China and Vietnam 143–157 (Figure 1d). The extent of spread is likely to be underestimated because speciation of dermatophytes and antifungal susceptibility testing are not routinely performed in most countries. Most cases of T. indotineae in Europe and North America have been linked to international travel, but because the pathogen can be acquired from contact with infected individuals or fomites, there is the potential for domestic spread. T. indotineae infections have been identified on dogs in India, which suggests the pathogen could start to spread zoonotically as well158.
The total burden of T. indotineae infections is unknown, as identification of dermatophytes to the species level is not routinely performed. Further, identification of T. indotineae is challenging because unequivocal identification can only be performed using DNA sequencing 141. T. indotineae has become widespread; therefore, it is important to recognize the risk of local transmission of T. indotineae in non-endemic countries that may give rise to a worsening worldwide epidemic scenario of difficult-to-treat infections.
Pathogenicity and clinical manifestations.
As mentioned previously, various underlying conditions (for example, diabetes mellitus, immune‐suppressive conditions, Cushing syndrome, atopic dermatitis and systemic corticosteroid therapy) may increase host susceptibility for dermatophytosis135. Dermatophytes possess virulence factors that facilitate infection in patients, regardless of host immune status, but immunocompromising conditions may lead to more severe infection. For example, the dermatophytic fungi produce several enzymes, including proteases, lipases, elastases, collagenases, phosphatases and esterases, that have been implicated in host invasion and assimilation of nutrition159. The dermatophytic hyphae invades the uppermost, non-living, keratinized layer of the skin, namely the stratum corneum, produce exo-enzyme keratinase, and induce inflammatory reaction at the site of infection. Although studies pertaining to T. indotineae-specific immune responses are lacking, the cell-mediated immune response is responsible for the control of dermatophytosis160.
Typical manifestations of T. indotineae include extensive chronic relapsing infection of the body (tinea corporis) and the groin (tinea cruris). Clinical presentation commonly involves multiple lesions in different anatomical locations exhibiting varying degrees of inflammation with peripherally spreading, flat, whitish or brownish pigmentation. One of the hallmarks of infection by T. indotineae is the severity of infection compared with those caused by the closely related species T. mentagrophytes and T. interdigitale. Many infections with T. indotineae are highly inflammatory, cover much or all the lower body and groin, and can extend to the extremities and head. Because of the inflammatory nature of the lesions, they are often associated with painful burning and itching and can cause substantial morbidity161. It has been speculated that a large proportion of the most severe cases present with steroid-modified dermatophytosis after having used combination creams with steroids161,162.
Inappropriate use of antifungal drugs, particularly the use of over-the-counter antifungal creams containing high potency steroid clobetasol is likely to have been and is likely to continue to be a driver of dermatophyte resistance and selection in India162. These creams are widely available without a prescription in India and are frequently used by patients to self-treat skin conditions162. Creams with antifungal medications are available in other countries to treat both dermatophyte infections as well as mucocutaneous candidiasis. These creams do not contain steroids and there is no indication that these creams have led to an increase in antifungal resistant organisms. Although corticosteroids like clobetasol may temporarily relieve symptoms of dermatophytosis, these medications do not treat the underlying infection and the steroid-associated immunosuppression may ultimately worsen the dermatophytosis163.
Evolution.
Internal transcribed spacer sequencing, multilocus sequence typing and whole-genome sequencing clearly delineate T. indotineae from the T. mentagrophytes/T. interdigitale complex11,141,164. Multilocus sequence data from T. mentagrophytes species complex isolates from humans and animals shows that T. indotineae has evolved to be distinct from T. mentagrophytes, with every genotype unique only to the new species. Although the genomes are clearly different across species, T. indotineae is still so young as a clonal species that the overall structural architecture of its genomes is quite similar to the parental species T. mentagrophytes. A full analysis of the differences between T. indotineae and the T. mentagrophytes/interdigitale complex has not yet been performed. Trichophyton indotineae may still be in the early stages of separation as a species from T. mentagrophytes. As it moves across the world reproducing as an anthropophilic rather than a zoonotic fungus, its genome may undergo more changes that cannot be repaired in a predominantly clonal population, and this may further separate it from T. mentagrophytes141. Although the literature on this new fungus dates from only the past few years, it may have arrived in Europe as early as 2011, and a search of Genbank for sequences corresponding to T. mentagrophytes type VIII (the previous designation) indicates it was in Indian, Oman, Iran and Australia by 2013 and possibly as early as 2004 (Refs. 144,145).
Resistance mechanisms.
Terbinafine is a first-line drug for treatment of dermatophytosis. High terbinafine resistance rates, reaching 80% in at least one study, were one of the first indicators that T. indotineae strains from India were different from the usual T. mentagrophytes/interdigitale complex 165,166. More alarming, T. indotineae isolates resistant to itraconazole have also been identified, including isolates that are resistant to both itraconazole and terbinafine167–169.
The mechanisms of resistance to both terbinafine and itraconazole have been determined. Terbinafine-resistant T. indotineae isolates exhibit single point mutations in the target gene squalene epoxidase (also known as squalene monooxygenase and designated either as SQLE or ERG1; which is a key enzyme in the ergosterol biosynthetic pathway), which lead to single amino-acid substitutions, Leu393Phe, Phe397Leu, or Phe415Val169 (Figure 5). In addition, azole resistance is frequently identified in T. indotineae. With the emphasis on terbinafine resistance, few studies have been performed to determine the mechanism of azole resistance144,167,168. However, one study identified tandem repeats of the lanosterol demethylase gene CYP51B, another study identified specific point mutations associated with resistance, and an early study found a point mutation in SQLE that caused itraconazole resistance while the same isolate was susceptible to terbinafine144,167,168 Interestingly, neither study identified mutations in the primary lanosterol demethylase CYP51A.
Figure 5.
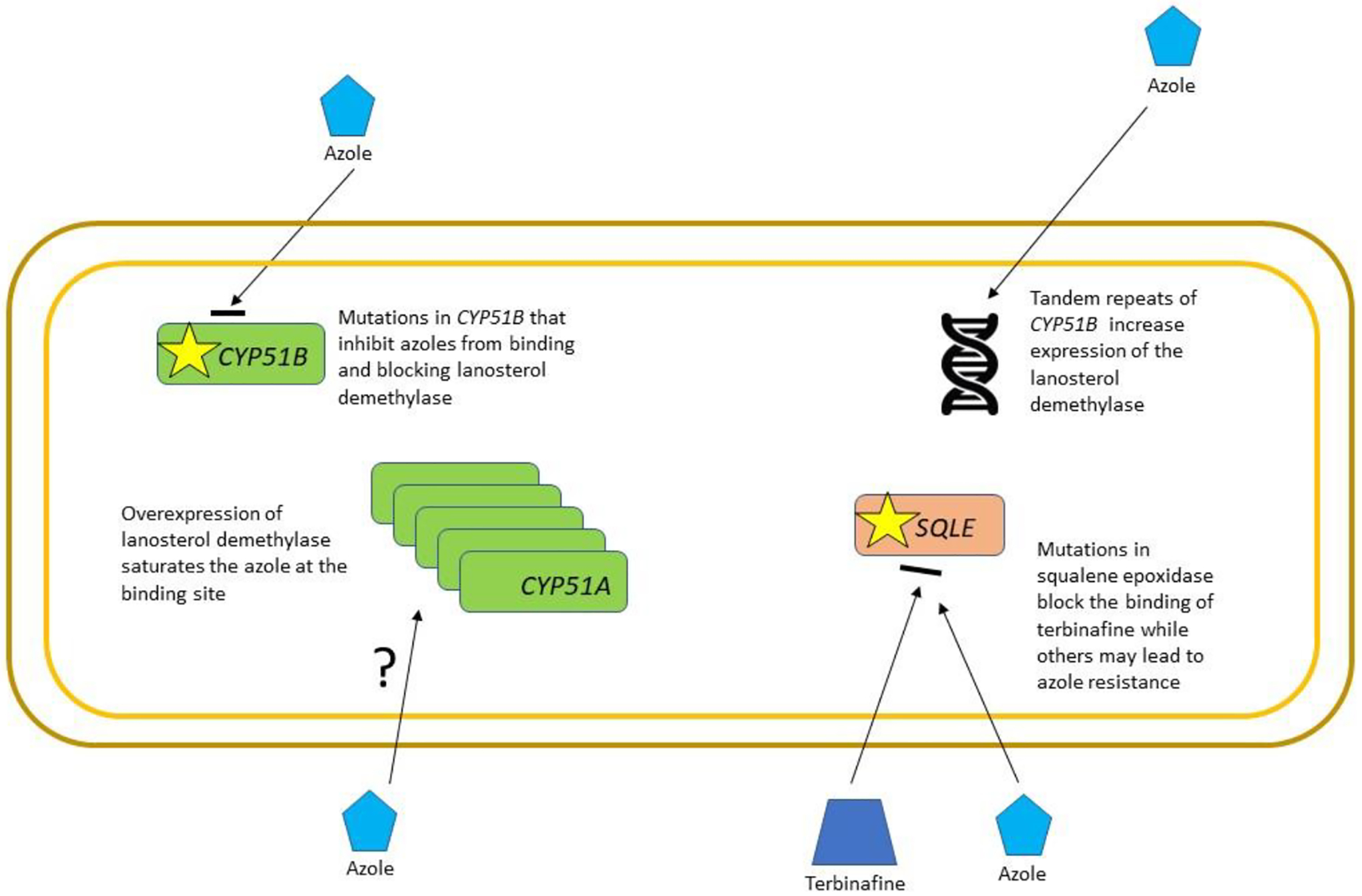
Mechanisms of acquired resistance of Trichophyton indotineae. T. indotineae is a newly emerging species so our current knowledge of the mechanisms of resistance is limited. Resistance to terbinafine is caused by specific mutations in the target enzyme, squalene epoxidase, which may also lead to azole resistance as it is a component of the ergosterol pathway. Azole resistance has been linked to changes in the target enzyme lanosterol demethylase (CYP51B) either through amino acid mutation, changes in ploidy, or mutations in transcription factors that result in overexpression.
Treatment options.
There are no guidelines specifically for the treatment of Trichophyton infections, but a frequent and effective choice is terbinafine170. However, given the proportion of T. indotineae isolates that are resistant to terbinafine and the general lack of availability of susceptibility testing of dermatophytes, other treatment options require consideration. In one randomized control trial of dermatophyte infection, itraconazole had a higher success rate than terbinafine171. There are reports of successful treatment of T. indotineae specifically using other topical antifungals such as ciclopirox olamine, bifonazole and miconazole, but these case reports are anecdotal147. Topical voriconazole has also been used successfully for treatment of terbinafine-resistant T. indotineae, and the newer topical azole antifungal luliconazole exhibits high in vitro activity against T. indotineae and could be considered as a treatment option172,173.
Several public health actions might help prevent further spread of T. indotineae. Such actions include increasing laboratory capacity to detect and monitor the spread of resistance, redoubling antifungal stewardship efforts with an emphasis on appropriate diagnostic testing and proper antifungal utilization, and performing further research to quantify the overall burden of disease and identify potential drivers of infection. In addition, guidelines for therapy, partially based on new animal models for infection are warranted.
Summary and outlook
This Review highlights three antifungal-resistant fungi that have recently emerged and are spreading across the globe, posing an ongoing threat to public health. C. auris is acquired and spread almost exclusively though the healthcare community. Medical tourism, global migration and the international transfer of patients has promoted the spread of C. auris to six continents in a little over a decade. T. indotineae is carried on the skin by otherwise healthy individuals and is spread within families and across communities, a situation that is likely to be exacerbated by the overuse and misuse of over-the-counter medications. The spread of azole-resistant A. fumigatus is driven by the use of fungicides in agricultural and other settings and is likely facilitated by the international trade in agricultural products127.
Although each pathogen emerged differently and affects distinct populations, the three fungi share the common thread of antifungal resistance, the potential to become a worsening public health threat, and the likelihood of spreading further through international travel and trade. However, despite the need, fungal disease surveillance is still rare in most countries, and antifungal susceptibility testing is still the exception rather than the norm in most clinical laboratories across the globe, even in resource-rich countries. Because C. auris is transferred from person to person within healthcare settings, rigorous infection control practices, screening and surveillance are essential for stopping or slowing the spread. Likewise, robust surveillance for azole-resistant A. fumigtus and T. indotineae infections is critical to monitor the spread of these pathogens and inform public health actions and policy.
Although this Review focused on three specific examples of fungi that are both spreading globally and carry antifungal resistance, other resistant fungi are quickly becoming a global concern. C. glabrata has long had the highest proportion of multidrug-resistant isolates, but fortunately, it does not seem to be spreading and outbreaks of multidrug-resistant isolates have been rare174. However, aside from C. auris, we may be seeing the emergence of other antifungal resistant Candida species. Azole-resistant C. parapsilosis has recently caused outbreaks in several different countries including South Africa, Turkey, Mexico, Brazil and Spain, and we are starting to see a similar emergence of azole-resistant C. tropicalis in Asia and South America 175–181. Similar to what has been seen with T. indotineae, reports of terbinafine- and azole-resistant T. rubrum have been increasing 143,182,183. Although species and dissemination may vary, antifungal resistance is a recurring theme. The World Health Organization (WHO) has recently released the WHO fungal priority pathogens list. Two of the fungi in this Review, C. auris and A. fumigatus, are in the critical group primarily because the WHO recognized that resistance poses such a high public health risk184. A cross-sector emphasis on antifungal and fungicide stewardship is urgently needed to protect human health in addition to preventing the emergence of fungicide-resistant plant pathogens185. Multiple barriers exist to antifungal stewardship programs. The first is a paucity of effective fungal expertise and fungal diagnostics. Most fungal infections are treated empirically until diagnosis is confirmed, which can take up to a few weeks, if this occurs at all. The second barrier is the lack of availability on a global scale of rapid susceptibility testing. Susceptibility testing can facilitate step-down of therapy to generic or more easily tolerated antifungals. The third barrier is the that only three antifungal classes are currently approved for the treatment of systemic fungal infections. With the availability of three new classes of antifungals in the foreseeable future, the development of antifungal susceptibility testing capacity and effective stewardship programs are of paramount importance. Preventing the emergence of resistant human-pathogenic fungi hinges on the improvement of clinical and environmental fungal surveillance, increasing clinical capacity for fungal speciation and antifungal susceptibility testing, and encouraging policies that improve antifungal stewardship in both the clinical and agricultural realm.
Acknowledgements:
The authors wish to acknowledge Maryann McCloskey for her assistance with data collection. A.C. is a fellow of the CIFAR program Fungal Kingdom: Threats & Opportunities.
Footnotes
Disclaimer: The findings and conclusions of this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control (CDC).
Competing Interests: The authors declare no competing interests.
Related links:
CDC Infection Prevention and Control for Candida auris: https://www.cdc.gov/fungal/candida-auris/c-auris-infection-control.html
Supplementary information
Supplementary information is available for this paper at https://doi.org/10.1038/s415XX-XXX-XXXX-X
References
- 1.Rigling D & Prospero S Cryphonectria parasitica, the causal agent of chestnut blight: invasion history, population biology and disease control. Mol Plant Pathol 19, 7–20, doi: 10.1111/mpp.12542 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dita M, Barquero M, Heck D, Mizubuti ESG & Staver CP Fusarium Wilt of Banana: Current Knowledge on Epidemiology and Research Needs Toward Sustainable Disease Management. Front Plant Sci 9, 1468, doi: 10.3389/fpls.2018.01468 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fisher MC & Garner TWJ Chytrid fungi and global amphibian declines. Nat Rev Microbiol 18, 332–343, doi: 10.1038/s41579-020-0335-x (2020). [DOI] [PubMed] [Google Scholar]
- 4.Hoyt JR, Kilpatrick AM & Langwig KE Ecology and impacts of white-nose syndrome on bats. Nat Rev Microbiol 19, 196–210, doi: 10.1038/s41579-020-00493-5 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Martin-Urdiroz M, Oses-Ruiz M, Ryder LS & Talbot NJ Investigating the biology of plant infection by the rice blast fungus Magnaporthe oryzae. Fungal Genet Biol 90, 61–68, doi: 10.1016/j.fgb.2015.12.009 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Singh RP et al. Emergence and Spread of New Races of Wheat Stem Rust Fungus: Continued Threat to Food Security and Prospects of Genetic Control. Phytopathology 105, 872–884, doi: 10.1094/phyto-01-15-0030-fi (2015). [DOI] [PubMed] [Google Scholar]
- 7.Schorling SR, Kortinga HC, Froschb M & Mühlschlegel FA The role of Candida dubliniensis in oral candidiasis in human immunodeficiency virus-infected individuals. Crit Rev Microbiol 26, 59–68, doi: 10.1080/10408410091154183 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Maziarz EK & Perfect JR Cryptococcosis. Infect Dis Clin North Am 30, 179–206, doi: 10.1016/j.idc.2015.10.006 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dellière S, Gits-Muselli M, Bretagne S & Alanio A Outbreak-Causing Fungi: Pneumocystis jirovecii. Mycopathologia 185, 783–800, doi: 10.1007/s11046-019-00408-w (2020). [DOI] [PubMed] [Google Scholar]
- 10.Satoh K et al. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiology and immunology 53, 41–44, doi: 10.1111/j.1348-0421.2008.00083.x (2009). [DOI] [PubMed] [Google Scholar]
- 11.Kano R et al. Trichophyton indotineae sp. nov.: A New Highly Terbinafine-Resistant Anthropophilic Dermatophyte Species. Mycopathologia 185, 947–958, doi: 10.1007/s11046-020-00455-8 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Snelders E, Melchers WJ & Verweij PE Azole resistance in Aspergillus fumigatus: a new challenge in the management of invasive aspergillosis? Future Microbiol 6, 335–347, doi: 10.2217/fmb.11.4 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Casadevall A, Kontoyiannis DP & Robert V On the Emergence of Candida auris: Climate Change, Azoles, Swamps, and Birds. mBio 10, doi: 10.1128/mBio.01397-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Casadevall A, Kontoyiannis DP & Robert V Environmental Candida auris and the Global Warming Emergence Hypothesis. mBio 12, doi: 10.1128/mBio.00360-21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nnadi NE & Carter DA Climate change and the emergence of fungal pathogens. PLoS Pathog 17, e1009503, doi: 10.1371/journal.ppat.1009503 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gadre A, Enbiale W, Andersen L & Coate S The effects of climate change on fungal diseases with cutaneous manifestations: a report from the International Society of Dermatology Climate Change Committee. The Journal of Climate Change and Health, doi:j.joclim.2022.100156 (2022). [Google Scholar]
- 17.Fisher MC, Hawkins NJ, Sanglard D & Gurr SJ Worldwide emergence of resistance to antifungal drugs challenges human health and food security. Science 360, 739–742, doi: 10.1126/science.aap7999 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Guo X et al. Climate warming accelerates temporal scaling of grassland soil microbial biodiversity. Nat Ecol Evol 3, 612–619, doi: 10.1038/s41559-019-0848-8 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Meena M et al. Multifarious Responses of Forest Soil Microbial Community Toward Climate Change. Microb Ecol, doi: 10.1007/s00248-022-02051-3 (2022). [DOI] [PubMed] [Google Scholar]
- 20.Gillings MR, Paulsen IT & Tetu SG Ecology and Evolution of the Human Microbiota: Fire, Farming and Antibiotics. Genes (Basel) 6, 841–857, doi: 10.3390/genes6030841 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang X et al. Skin Metagenomic Sequence Analysis of Early Candida auris Outbreaks in U.S. Nursing Homes. mSphere 6, e0028721, doi: 10.1128/mSphere.00287-21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Proctor DM et al. Integrated genomic, epidemiologic investigation of Candida auris skin colonization in a skilled nursing facility. Nat Med 27, 1401–1409, doi: 10.1038/s41591-021-01383-w (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chien AL et al. Association of Systemic Antibiotic Treatment of Acne With Skin Microbiota Characteristics. JAMA Dermatol 155, 425–434, doi: 10.1001/jamadermatol.2018.5221 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xue P, Liu X, Zhao L, Zhang J & He Z Integrating high-throughput sequencing and metabolomics to investigate the stereoselective responses of soil microorganisms to chiral fungicide cis-epoxiconazole. Chemosphere 300, 134198, doi: 10.1016/j.chemosphere.2022.134198 (2022). [DOI] [PubMed] [Google Scholar]
- 25.Han L, Kong X, Xu M & Nie J Repeated exposure to fungicide tebuconazole alters the degradation characteristics, soil microbial community and functional profiles. Environ Pollut 287, 117660, doi: 10.1016/j.envpol.2021.117660 (2021). [DOI] [PubMed] [Google Scholar]
- 26.Zhang H et al. Exposure to fungicide difenoconazole reduces the soil bacterial community diversity and the co-occurrence network complexity. J Hazard Mater 405, 124208, doi: 10.1016/j.jhazmat.2020.124208 (2021). [DOI] [PubMed] [Google Scholar]
- 27.Baćmaga M, Wyszkowska J, Borowik A & Kucharski J Effects of Tebuconazole Application on Soil Microbiota and Enzymes. Molecules 27, doi: 10.3390/molecules27217501 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee WG et al. First three reported cases of nosocomial fungemia caused by Candida auris. Journal of clinical microbiology 49, 3139–3142, doi: 10.1128/jcm.00319-11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chowdhary A et al. New clonal strain of Candida auris, Delhi, India. Emerging infectious diseases 19, 1670–1673, doi: 10.3201/eid1910.130393 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Magobo RE, Corcoran C, Seetharam S & Govender NP Candida auris-associated candidemia, South Africa. Emerging infectious diseases 20, 1250–1251, doi: 10.3201/eid2007.131765 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lockhart SR et al. Simultaneous Emergence of Multidrug-Resistant Candida auris on 3 Continents Confirmed by Whole-Genome Sequencing and Epidemiological Analyses. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America 64, 134–140, doi: 10.1093/cid/ciw691 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rhodes J & Fisher MC Global epidemiology of emerging Candida auris. Curr Opin Microbiol 52, 84–89, doi: 10.1016/j.mib.2019.05.008 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Jung J et al. Candida auris colonization or infection of the ear: A single-center study in South Korea from 2016 to 2018. Med Mycol 58, 124–127, doi: 10.1093/mmy/myz020 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Desnos-Ollivier M, Fekkar A & Bretagne S Earliest case of Candida auris infection imported in 2007 in Europe from India prior to the 2009 description in Japan. J Mycol Med 31, 101139, doi: 10.1016/j.mycmed.2021.101139 (2021). [DOI] [PubMed] [Google Scholar]
- 35.Iguchi S et al. Candida auris: A pathogen difficult to identify, treat, and eradicate and its characteristics in Japanese strains. J Infect Chemother 25, 743–749, doi: 10.1016/j.jiac.2019.05.034 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Chow NA et al. Tracing the Evolutionary History and Global Expansion of Candida auris Using Population Genomic Analyses. mBio 11, doi: 10.1128/mBio.03364-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chow NA et al. Potential Fifth Clade of Candida auris, Iran, 2018. Emerging infectious diseases 25, 1780–1781, doi: 10.3201/eid2509.190686 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chowdhary A, Sharma C & Meis JF Candida auris: A rapidly emerging cause of hospital-acquired multidrug-resistant fungal infections globally. PLoS Pathog 13, e1006290, doi: 10.1371/journal.ppat.1006290 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hallen-Adams HE & Suhr MJ Fungi in the healthy human gastrointestinal tract. Virulence 8, 352–358, doi: 10.1080/21505594.2016.1247140 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kühbacher A, Burger-Kentischer A & Rupp S Interaction of Candida Species with the Skin. Microorganisms 5, doi: 10.3390/microorganisms5020032 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schelenz S et al. First hospital outbreak of the globally emerging Candida auris in a European hospital. Antimicrobial resistance and infection control 5, 35, doi: 10.1186/s13756-016-0132-5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruiz-Gaitan A et al. An outbreak due to Candida auris with prolonged colonisation and candidaemia in a tertiary care European hospital. Mycoses 61, 498–505, doi: 10.1111/myc.12781 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Biswal M et al. Controlling a possible outbreak of Candida auris infection: lessons learnt from multiple interventions. J Hosp Infect 97, 363–370, doi: 10.1016/j.jhin.2017.09.009 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Sexton DJ et al. Positive Correlation Between Candida auris Skin-Colonization Burden and Environmental Contamination at a Ventilator-Capable Skilled Nursing Facility in Chicago. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America 73, 1142–1148, doi: 10.1093/cid/ciab327 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adams E et al. Candida auris in Healthcare Facilities, New York, USA, 2013–2017. Emerging infectious diseases 24, 1816–1824, doi: 10.3201/eid2410.180649 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chow NA et al. Multiple introductions and subsequent transmission of multidrug-resistant Candida auris in the USA: a molecular epidemiological survey. Lancet Infect Dis 18, 1377–1384, doi: 10.1016/s1473-3099(18)30597-8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lyman M et al. Worsening Spread of Candida auris in the United States, 2019 to 2021. Ann Intern Med, doi: 10.7326/m22-3469 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jackson BR et al. On the Origins of a Species: What Might Explain the Rise of Candida auris? Journal of fungi (Basel, Switzerland) 5, doi: 10.3390/jof5030058 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abastabar M et al. Candida auris otomycosis in Iran and review of recent literature. Mycoses 62, 101–105, doi: 10.1111/myc.12886 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Arora P et al. Environmental Isolation of Candida auris from the Coastal Wetlands of Andaman Islands, India. mBio 12, doi: 10.1128/mBio.03181-20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yadav A et al. Candida auris on Apples: Diversity and Clinical Significance. mBio 13, e0051822, doi: 10.1128/mbio.00518-22 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Escandón P Novel Environmental Niches for Candida auris: Isolation from a Coastal Habitat in Colombia. Journal of fungi (Basel, Switzerland) 8, doi: 10.3390/jof8070748 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chakrabarti A & Singh S Multidrug-resistant Candida auris: an epidemiological review. Expert Rev Anti Infect Ther 18, 551–562, doi: 10.1080/14787210.2020.1750368 (2020). [DOI] [PubMed] [Google Scholar]
- 54.Chowdhary A, Voss A & Meis JF Multidrug-resistant Candida auris: ‘new kid on the block’ in hospital-associated infections? J Hosp Infect 94, 209–212, doi: 10.1016/j.jhin.2016.08.004 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Toda M et al. Population-Based Active Surveillance for Culture-Confirmed Candidemia - Four Sites, United States, 2012–2016. MMWR Surveill Summ 68, 1–15, doi: 10.15585/mmwr.ss6808a1 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kean R et al. Transcriptome Assembly and Profiling of Candida auris Reveals Novel Insights into Biofilm-Mediated Resistance. mSphere 3, doi: 10.1128/mSphere.00334-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Singh R, Kaur M, Chakrabarti A, Shankarnarayan SA & Rudramurthy SM Biofilm formation by Candida auris isolated from colonising sites and candidemia cases. Mycoses 62, 706–709, doi: 10.1111/myc.12947 (2019). [DOI] [PubMed] [Google Scholar]
- 58.Horton MV et al. Candida auris Forms High-Burden Biofilms in Skin Niche Conditions and on Porcine Skin. mSphere 5, doi: 10.1128/mSphere.00910-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hoenigl M et al. COVID-19-associated fungal infections. Nat Microbiol 7, 1127–1140, doi: 10.1038/s41564-022-01172-2 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fakhim H et al. Comparative virulence of Candida auris with Candida haemulonii, Candida glabrata and Candida albicans in a murine model. Mycoses 61, 377–382, doi: 10.1111/myc.12754 (2018). [DOI] [PubMed] [Google Scholar]
- 61.Yue H et al. Filamentation in Candida auris, an emerging fungal pathogen of humans: passage through the mammalian body induces a heritable phenotypic switch. Emerg Microbes Infect 7, 188, doi: 10.1038/s41426-018-0187-x (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Johnson CJ, Davis JM, Huttenlocher A, Kernien JF & Nett JE Emerging Fungal Pathogen Candida auris Evades Neutrophil Attack. mBio 9, doi: 10.1128/mBio.01403-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Horton MV et al. Candida auris Cell Wall Mannosylation Contributes to Neutrophil Evasion through Pathways Divergent from Candida albicans and Candida glabrata. mSphere 6, e0040621, doi: 10.1128/mSphere.00406-21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weerasinghe H et al. Candida auris uses metabolic strategies to escape and kill macrophages while avoiding robust activation of the NLRP3 inflammasome response. Cell Rep 42, 112522, doi: 10.1016/j.celrep.2023.112522 (2023). [DOI] [PubMed] [Google Scholar]
- 65.Lamoth F, Lockhart SR, Berkow EL & Calandra T Changes in the epidemiological landscape of invasive candidiasis. The Journal of antimicrobial chemotherapy 73, i4–i13, doi: 10.1093/jac/dkx444 (2018). [DOI] [PubMed] [Google Scholar]
- 66.Chowdhary A et al. A multicentre study of antifungal susceptibility patterns among 350 Candida auris isolates (2009–17) in India: role of the ERG11 and FKS1 genes in azole and echinocandin resistance. The Journal of antimicrobial chemotherapy 73, 891–899, doi: 10.1093/jac/dkx480 (2018). [DOI] [PubMed] [Google Scholar]
- 67.Arendrup MC, Prakash A, Meletiadis J, Sharma C & Chowdhary A Comparison of EUCAST and CLSI Reference Microdilution MICs of Eight Antifungal Compounds for Candida auris and Associated Tentative Epidemiological Cutoff Values. Antimicrobial agents and chemotherapy 61, doi: 10.1128/aac.00485-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rybak JM et al. Mutations in TAC1B: a Novel Genetic Determinant of Clinical Fluconazole Resistance in Candida auris. mBio 11, doi: 10.1128/mBio.00365-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ben-Ami R et al. Multidrug-Resistant Candida haemulonii and C. auris, Tel Aviv, Israel. Emerging infectious diseases 23, 195–203, doi: 10.3201/eid2302.161486 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lyman M et al. Notes from the Field: Transmission of Pan-Resistant and Echinocandin-Resistant Candida auris in Health Care Facilities - Texas and the District of Columbia, January-April 2021. MMWR Morb Mortal Wkly Rep 70, 1022–1023, doi: 10.15585/mmwr.mm7029a2 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Berkow EL & Lockhart SR Activity of CD101, a long-acting echinocandin, against clinical isolates of Candida auris. Diagnostic microbiology and infectious disease 90, 196–197, doi: 10.1016/j.diagmicrobio.2017.10.021 (2018). [DOI] [PubMed] [Google Scholar]
- 72.Lepak AJ, Zhao M, Berkow EL, Lockhart SR & Andes DR Pharmacodynamic Optimization for Treatment of Invasive Candida auris Infection. Antimicrobial agents and chemotherapy 61, doi: 10.1128/aac.00791-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rybak JM et al. In vivo emergence of high-level resistance during treatment reveals the first identified mechanism of amphotericin B resistance in Candida auris. Clin Microbiol Infect 28, 838–843, doi: 10.1016/j.cmi.2021.11.024 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Asner SA, Giulieri S, Diezi M, Marchetti O & Sanglard D Acquired Multidrug Antifungal Resistance in Candida lusitaniae during Therapy. Antimicrobial agents and chemotherapy 59, 7715–7722, doi: 10.1128/aac.02204-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ostrowsky B et al. Candida auris Isolates Resistant to Three Classes of Antifungal Medications - New York, 2019. MMWR Morb Mortal Wkly Rep 69, 6–9, doi: 10.15585/mmwr.mm6901a2 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Alastruey-Izquierdo A et al. [GEMICOMED/GEIRAS-SEIMC recommendations for the management of Candida auris infection and colonization]. Rev Iberoam Micol 36, 109–114, doi: 10.1016/j.riam.2019.06.001 (2019). [DOI] [PubMed] [Google Scholar]
- 77.Govender NP et al. Federation of Infectious Diseases Societies of Southern Africa guideline: Recommendations for the detection, management and prevention of healthcare-associated Candida auris colonisation and disease in South Africa. S Afr J Infect Dis 34, 163, doi: 10.4102/sajid.v34i1.163 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ong CW et al. Diagnosis, management and prevention of Candida auris in hospitals: position statement of the Australasian Society for Infectious Diseases. Intern Med J 49, 1229–1243, doi: 10.1111/imj.14612 (2019). [DOI] [PubMed] [Google Scholar]
- 79.Pappas PG et al. Clinical Practice Guideline for the Management of Candidiasis: 2016 Update by the Infectious Diseases Society of America. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America 62, e1–50, doi: 10.1093/cid/civ933 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Berkow EL & Lockhart SR Activity of novel antifungal compound APX001A against a large collection of Candida auris. The Journal of antimicrobial chemotherapy 73, 3060–3062, doi: 10.1093/jac/dky302 (2018). [DOI] [PubMed] [Google Scholar]
- 81.Berkow EL, Angulo D & Lockhart SR In Vitro Activity of a Novel Glucan Synthase Inhibitor, SCY-078, against Clinical Isolates of Candida auris. Antimicrobial agents and chemotherapy 61, doi: 10.1128/aac.00435-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Benedict K, Jackson BR, Chiller T & Beer KD Estimation of Direct Healthcare Costs of Fungal Diseases in the United States. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America 68, 1791–1797, doi: 10.1093/cid/ciy776 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gold JAW et al. Increased deaths from fungal infections during the COVID-19 pandemic-National Vital Statistics System, United States, January 2020-December 2021. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America, doi: 10.1093/cid/ciac489 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vallabhaneni S, Benedict K, Derado G & Mody RK Trends in Hospitalizations Related to Invasive Aspergillosis and Mucormycosis in the United States, 2000–2013. Open Forum Infect Dis 4, ofw268, doi: 10.1093/ofid/ofw268 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bongomin F, Gago S, Oladele RO & Denning DW Global and Multi-National Prevalence of Fungal Diseases-Estimate Precision. Journal of fungi (Basel, Switzerland) 3, doi: 10.3390/jof3040057 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Denning DW, Pleuvry A & Cole DC Global burden of allergic bronchopulmonary aspergillosis with asthma and its complication chronic pulmonary aspergillosis in adults. Med Mycol 51, 361–370, doi: 10.3109/13693786.2012.738312 (2013). [DOI] [PubMed] [Google Scholar]
- 87.Patterson TF et al. Executive Summary: Practice Guidelines for the Diagnosis and Management of Aspergillosis: 2016 Update by the Infectious Diseases Society of America. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America 63, 433–442, doi: 10.1093/cid/ciw444 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bueid A et al. Azole antifungal resistance in Aspergillus fumigatus: 2008 and 2009. The Journal of antimicrobial chemotherapy 65, 2116–2118, doi: 10.1093/jac/dkq279 (2010). [DOI] [PubMed] [Google Scholar]
- 89.Verweij PE, Snelders E, Kema GH, Mellado E & Melchers WJ Azole resistance in Aspergillus fumigatus: a side-effect of environmental fungicide use? Lancet Infect Dis 9, 789–795, doi: 10.1016/s1473-3099(09)70265-8 (2009). [DOI] [PubMed] [Google Scholar]
- 90.Howard SJ et al. Frequency and evolution of Azole resistance in Aspergillus fumigatus associated with treatment failure. Emerging infectious diseases 15, 1068–1076, doi: 10.3201/eid1507.090043 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bradley K et al. Fatal Fungicide-Associated Triazole-Resistant Aspergillus fumigatus Infection, Pennsylvania, USA. Emerging infectious diseases 28, 1904–1905, doi: 10.3201/eid2809.220517 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Verweij PE, Chowdhary A, Melchers WJ & Meis JF Azole Resistance in Aspergillus fumigatus: Can We Retain the Clinical Use of Mold-Active Antifungal Azoles? Clinical infectious diseases : an official publication of the Infectious Diseases Society of America 62, 362–368, doi: 10.1093/cid/civ885 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Resendiz Sharpe A et al. Triazole resistance surveillance in Aspergillus fumigatus. Med Mycol 56, 83–92, doi: 10.1093/mmy/myx144 (2018). [DOI] [PubMed] [Google Scholar]
- 94.Burks C, Darby A, Gómez Londoño L, Momany M & Brewer MT Azole-resistant Aspergillus fumigatus in the environment: Identifying key reservoirs and hotspots of antifungal resistance. PLoS Pathog 17, e1009711, doi: 10.1371/journal.ppat.1009711 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.van der Linden JW, Arendrup MC, Melchers WJ & Verweij PE Azole Resistance of Aspergillus fumigatus in Immunocompromised Patients with Invasive Aspergillosis. Emerging infectious diseases 22, 158–159, doi: 10.3201/eid2201.151308 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rosowski EE et al. Macrophages inhibit Aspergillus fumigatus germination and neutrophil-mediated fungal killing. PLoS Pathog 14, e1007229, doi: 10.1371/journal.ppat.1007229 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Earle K et al. Pathogenicity and virulence of Aspergillus fumigatus. Virulence 14, 2172264, doi: 10.1080/21505594.2023.2172264 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Latgé JP & Chamilos G Aspergillus fumigatus and Aspergillosis in 2019. Clin Microbiol Rev 33, doi: 10.1128/cmr.00140-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mortensen KL et al. A prospective survey of Aspergillus spp. in respiratory tract samples: prevalence, clinical impact and antifungal susceptibility. Eur J Clin Microbiol Infect Dis 30, 1355–1363, doi: 10.1007/s10096-011-1229-7 (2011). [DOI] [PubMed] [Google Scholar]
- 100.Pappas PG et al. Invasive fungal infections among organ transplant recipients: results of the Transplant-Associated Infection Surveillance Network (TRANSNET). Clinical infectious diseases : an official publication of the Infectious Diseases Society of America 50, 1101–1111, doi: 10.1086/651262 (2010). [DOI] [PubMed] [Google Scholar]
- 101.Kontoyiannis DP et al. Prospective surveillance for invasive fungal infections in hematopoietic stem cell transplant recipients, 2001–2006: overview of the Transplant-Associated Infection Surveillance Network (TRANSNET) Database. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America 50, 1091–1100, doi: 10.1086/651263 (2010). [DOI] [PubMed] [Google Scholar]
- 102.Sehgal IS, Muthu V & Agarwal R Aspergillus infection is an important complication of post-TB bronchiectasis. Int J Tuberc Lung Dis 27, 89a–89, doi: 10.5588/ijtld.22.0526 (2023). [DOI] [PubMed] [Google Scholar]
- 103.Warris A Immunopathology of Aspergillus Infections in Children With Chronic Granulomatous Disease and Cystic Fibrosis. Pediatr Infect Dis J 38, e96–e98, doi: 10.1097/inf.0000000000002265 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Prattes J et al. Risk factors and outcome of pulmonary aspergillosis in critically ill coronavirus disease 2019 patients-a multinational observational study by the European Confederation of Medical Mycology. Clin Microbiol Infect 28, 580–587, doi: 10.1016/j.cmi.2021.08.014 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Schauwvlieghe A et al. Invasive aspergillosis in patients admitted to the intensive care unit with severe influenza: a retrospective cohort study. Lancet Respir Med 6, 782–792, doi: 10.1016/s2213-2600(18)30274-1 (2018). [DOI] [PubMed] [Google Scholar]
- 106.Lestrade PPA, Meis JF, Melchers WJG & Verweij PE Triazole resistance in Aspergillus fumigatus: recent insights and challenges for patient management. Clin Microbiol Infect 25, 799–806, doi: 10.1016/j.cmi.2018.11.027 (2019). [DOI] [PubMed] [Google Scholar]
- 107.Lestrade PP et al. Voriconazole Resistance and Mortality in Invasive Aspergillosis: A Multicenter Retrospective Cohort Study. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America 68, 1463–1471, doi: 10.1093/cid/ciy859 (2019). [DOI] [PubMed] [Google Scholar]
- 108.Chong GM et al. PCR-based detection of Aspergillus fumigatus Cyp51A mutations on bronchoalveolar lavage: a multicentre validation of the AsperGenius assay® in 201 patients with haematological disease suspected for invasive aspergillosis. The Journal of antimicrobial chemotherapy 71, 3528–3535, doi: 10.1093/jac/dkw323 (2016). [DOI] [PubMed] [Google Scholar]
- 109.Rhodes J et al. Population genomics confirms acquisition of drug-resistant Aspergillus fumigatus infection by humans from the environment. Nat Microbiol 7, 663–674, doi: 10.1038/s41564-022-01091-2 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gonzalez-Jimenez I et al. Multiresistance to Nonazole Fungicides in Aspergillus fumigatus TR(34)/L98H Azole-Resistant Isolates. Antimicrobial agents and chemotherapy 65, e0064221, doi: 10.1128/aac.00642-21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kang SE et al. Evidence for the agricultural origin of resistance to multiple antimicrobials in Aspergillus fumigatus, a fungal pathogen of humans. G3 (Bethesda) 12, doi: 10.1093/g3journal/jkab427 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mellado E et al. A new Aspergillus fumigatus resistance mechanism conferring in vitro cross-resistance to azole antifungals involves a combination of cyp51A alterations. Antimicrobial agents and chemotherapy 51, 1897–1904, doi: 10.1128/aac.01092-06 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.van der Linden JW et al. Aspergillosis due to voriconazole highly resistant Aspergillus fumigatus and recovery of genetically related resistant isolates from domiciles. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America 57, 513–520, doi: 10.1093/cid/cit320 (2013). [DOI] [PubMed] [Google Scholar]
- 114.Chowdhary A, Kathuria S, Xu J & Meis JF Emergence of azole-resistant aspergillus fumigatus strains due to agricultural azole use creates an increasing threat to human health. PLoS Pathog 9, e1003633, doi: 10.1371/journal.ppat.1003633 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lestrade PPA et al. Paradoxal Trends in Azole-Resistant Aspergillus fumigatus in a National Multicenter Surveillance Program, the Netherlands, 2013–2018. Emerging infectious diseases 26, 1447–1455, doi: 10.3201/eid2607.200088 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Snelders E et al. Emergence of azole resistance in Aspergillus fumigatus and spread of a single resistance mechanism. PLoS Med 5, e219, doi: 10.1371/journal.pmed.0050219 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Paul S, Verweij PE, Melchers WJG & Moye-Rowley WS Differential Functions of Individual Transcription Factor Binding Sites in the Tandem Repeats Found in Clinically Relevant cyp51A Promoters in Aspergillus fumigatus. mBio 13, e0070222, doi: 10.1128/mbio.00702-22 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Riat A, Plojoux J, Gindro K, Schrenzel J & Sanglard D Azole Resistance of Environmental and Clinical Aspergillus fumigatus Isolates from Switzerland. Antimicrobial agents and chemotherapy 62, doi: 10.1128/aac.02088-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sharma C, Hagen F, Moroti R, Meis JF & Chowdhary A Triazole-resistant Aspergillus fumigatus harbouring G54 mutation: Is it de novo or environmentally acquired? J Glob Antimicrob Resist 3, 69–74, doi: 10.1016/j.jgar.2015.01.005 (2015). [DOI] [PubMed] [Google Scholar]
- 120.Bader O et al. Environmental isolates of azole-resistant Aspergillus fumigatus in Germany. Antimicrobial agents and chemotherapy 59, 4356–4359, doi: 10.1128/aac.00100-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhang J et al. The Medical Triazole Voriconazole Can Select for Tandem Repeat Variations in Azole-Resistant Aspergillus Fumigatus Harboring TR(34)/L98H Via Asexual Reproduction. Journal of fungi (Basel, Switzerland) 6, doi: 10.3390/jof6040277 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhang J et al. A Novel Environmental Azole Resistance Mutation in Aspergillus fumigatus and a Possible Role of Sexual Reproduction in Its Emergence. mBio 8, doi: 10.1128/mBio.00791-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Satish S et al. Stress-Induced Changes in the Lipid Microenvironment of β-(1,3)-d-Glucan Synthase Cause Clinically Important Echinocandin Resistance in Aspergillus fumigatus. mBio 10, doi: 10.1128/mBio.00779-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fakhim H et al. Trends in the Prevalence of Amphotericin B-Resistance (AmBR) among Clinical Isolates of Aspergillus Species. J Mycol Med 32, 101310, doi: 10.1016/j.mycmed.2022.101310 (2022). [DOI] [PubMed] [Google Scholar]
- 125.Etienne KA et al. Genomic Diversity of Azole-Resistant Aspergillus fumigatus in the United States. mBio 12, e0180321, doi: 10.1128/mBio.01803-21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Toda M, Beer KD, Kuivila KM, Chiller TM & Jackson BR Trends in Agricultural Triazole Fungicide Use in the United States, 1992–2016 and Possible Implications for Antifungal-Resistant Fungi in Human Disease. Environ Health Perspect 129, 55001, doi: 10.1289/ehp7484 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Dunne K, Hagen F, Pomeroy N, Meis JF & Rogers TR Intercountry Transfer of Triazole-Resistant Aspergillus fumigatus on Plant Bulbs. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America 65, 147–149, doi: 10.1093/cid/cix257 (2017). [DOI] [PubMed] [Google Scholar]
- 128.Howard SJ & Arendrup MC Acquired antifungal drug resistance in Aspergillus fumigatus: epidemiology and detection. Med Mycol 49 Suppl 1, S90–95, doi: 10.3109/13693786.2010.508469 (2011). [DOI] [PubMed] [Google Scholar]
- 129.Alvarez-Moreno C et al. Azole-resistant Aspergillus fumigatus harboring TR(34)/L98H, TR(46)/Y121F/T289A and TR(53) mutations related to flower fields in Colombia. Sci Rep 7, 45631, doi: 10.1038/srep45631 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Verweij PE et al. Dual use of antifungals in medicine and agriculture: How do we help prevent resistance developing in human pathogens? Drug Resist Updat 65, 100885, doi: 10.1016/j.drup.2022.100885 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Verweij PE et al. International expert opinion on the management of infection caused by azole-resistant Aspergillus fumigatus. Drug Resist Updat 21–22, 30–40, doi: 10.1016/j.drup.2015.08.001 (2015). [DOI] [PubMed] [Google Scholar]
- 132.Shaw KJ & Ibrahim AS Fosmanogepix: A Review of the First-in-Class Broad Spectrum Agent for the Treatment of Invasive Fungal Infections. Journal of fungi (Basel, Switzerland) 6, doi: 10.3390/jof6040239 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Buil JB et al. In vitro activity of the novel antifungal compound F901318 against difficult-to-treat Aspergillus isolates. The Journal of antimicrobial chemotherapy 72, 2548–2552, doi: 10.1093/jac/dkx177 (2017). [DOI] [PubMed] [Google Scholar]
- 134.Pfaller MA et al. In vitro activity of a novel broad-spectrum antifungal, E1210, tested against Aspergillus spp. determined by CLSI and EUCAST broth microdilution methods. Antimicrobial agents and chemotherapy 55, 5155–5158, doi: 10.1128/aac.00570-11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rinaldi MG Dermatophytosis: epidemiological and microbiological update. J Am Acad Dermatol 43, S120–124, doi: 10.1067/mjd.2000.110378 (2000). [DOI] [PubMed] [Google Scholar]
- 136.Havlickova B, Czaika VA & Friedrich M Epidemiological trends in skin mycoses worldwide. Mycoses 51 Suppl 4, 2–15, doi: 10.1111/j.1439-0507.2008.01606.x (2008). [DOI] [PubMed] [Google Scholar]
- 137.Leung AK, Lam JM, Leong KF & Hon KL Tinea corporis: an updated review. Drugs Context 9, doi: 10.7573/dic.2020-5-6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Urban K et al. The global, regional, and national burden of fungal skin diseases in 195 countries and territories: A cross-sectional analysis from the Global Burden of Disease Study 2017. JAAD Int 2, 22–27, doi: 10.1016/j.jdin.2020.10.003 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ebert A et al. Alarming India-wide phenomenon of antifungal resistance in dermatophytes: A multicentre study. Mycoses 63, 717–728, doi: 10.1111/myc.13091 (2020). [DOI] [PubMed] [Google Scholar]
- 140.Dogra S & Uprety S The menace of chronic and recurrent dermatophytosis in India: Is the problem deeper than we perceive? Indian Dermatol Online J 7, 73–76, doi: 10.4103/2229-5178.178100 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Tang C et al. Taxonomy of the Trichophyton mentagrophytes/T. interdigitale Species Complex Harboring the Highly Virulent, Multiresistant Genotype T. indotineae. Mycopathologia 186, 315–326, doi: 10.1007/s11046-021-00544-2 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Khurana A et al. Correlation of In Vitro Susceptibility Based on MICs and Squalene Epoxidase Mutations with Clinical Response to Terbinafine in Patients with Tinea Corporis/Cruris. Antimicrobial agents and chemotherapy 62, doi: 10.1128/aac.01038-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Astvad KMT et al. Increasing Terbinafine Resistance in Danish Trichophyton Isolates 2019–2020. Journal of fungi (Basel, Switzerland) 8, doi: 10.3390/jof8020150 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Brasch J et al. “Indian” strains of Trichophyton mentagrophytes with reduced itraconazole susceptibility in Germany. J Dtsch Dermatol Ges 19, 1723–1727, doi: 10.1111/ddg.14626 (2021). [DOI] [PubMed] [Google Scholar]
- 145.Jabet A et al. Extensive Dermatophytosis Caused by Terbinafine-Resistant Trichophyton indotineae, France. Emerging infectious diseases 28, 229–233, doi: 10.3201/eid2801.210883 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Klinger M, Theiler M & Bosshard PP Epidemiological and clinical aspects of Trichophyton mentagrophytes/Trichophyton interdigitale infections in the Zurich area: a retrospective study using genotyping. J Eur Acad Dermatol Venereol 35, 1017–1025, doi: 10.1111/jdv.17106 (2021). [DOI] [PubMed] [Google Scholar]
- 147.Posso-De Los Rios CJ, Tadros E, Summerbell RC & Scott JA Terbinafine Resistant Trichophyton Indotineae Isolated in Patients with Superficial Dermatophyte Infection in Canadian Patients. J Cutan Med Surg, 12034754221077891, doi: 10.1177/12034754221077891 (2022). [DOI] [PubMed] [Google Scholar]
- 148.Dellière S et al. Emergence of Difficult-to-Treat Tinea Corporis Caused by Trichophyton mentagrophytes Complex Isolates, Paris, France. Emerging infectious diseases 28, 224–228, doi: 10.3201/eid2801.210810 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sacheli R et al. Belgian National Survey on Tinea Capitis: Epidemiological Considerations and Highlight of Terbinafine-Resistant T. mentagrophytes with a Mutation on SQLE Gene. Journal of fungi (Basel, Switzerland) 6, doi: 10.3390/jof6040195 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Saunte DML et al. Emerging antifungal treatment failure of dermatophytosis in Europe: take care or it may become endemic. J Eur Acad Dermatol Venereol 35, 1582–1586, doi: 10.1111/jdv.17241 (2021). [DOI] [PubMed] [Google Scholar]
- 151.Siopi M, Efstathiou I, Theodoropoulos K, Pournaras S & Meletiadis J Molecular Epidemiology and Antifungal Susceptibility of Trichophyton Isolates in Greece: Emergence of Terbinafine-Resistant Trichophytonmentagrophytes Type VIII Locally and Globally. Journal of fungi (Basel, Switzerland) 7, doi: 10.3390/jof7060419 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Fattahi A et al. Multidrug-resistant Trichophyton mentagrophytes genotype VIII in an Iranian family with generalized dermatophytosis: report of four cases and review of literature. Int J Dermatol 60, 686–692, doi: 10.1111/ijd.15226 (2021). [DOI] [PubMed] [Google Scholar]
- 153.Uhrlaß S et al. Trichophyton indotineae-An Emerging Pathogen Causing Recalcitrant Dermatophytoses in India and Worldwide-A Multidimensional Perspective. Journal of fungi (Basel, Switzerland) 8, doi: 10.3390/jof8070757 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ngo TMC et al. First detection of Trichophyton indotineae causing tinea corporis in Central Vietnam. Med Mycol Case Rep 36, 37–41, doi: 10.1016/j.mmcr.2022.05.004 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bortoluzzi P et al. Report of terbinafine resistant Trichophyton spp. in Italy: Clinical presentations, molecular identification, antifungal susceptibility testing and mutations in the squalene epoxidase gene. Mycoses, doi: 10.1111/myc.13597 (2023). [DOI] [PubMed] [Google Scholar]
- 156.Caplan AS et al. Notes from the Field: First Reported U.S. Cases of Tinea Caused by Trichophyton indotineae - New York City, December 2021-March 2023. MMWR Morb Mortal Wkly Rep 72, 536–537, doi: 10.15585/mmwr.mm7219a4 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Jia S et al. The epidemic of the multiresistant dermatophyte Trichophyton indotineae has reached China. Front Immunol 13, 1113065, doi: 10.3389/fimmu.2022.1113065 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kumar M et al. Molecular epidemiology of Trichophyton infections among canines from Northern India. J Mycol Med 33, 101352, doi: 10.1016/j.mycmed.2022.101352 (2022). [DOI] [PubMed] [Google Scholar]
- 159.Achterman RR & White TC Dermatophyte virulence factors: identifying and analyzing genes that may contribute to chronic or acute skin infections. Int J Microbiol 2012, 358305, doi: 10.1155/2012/358305 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Celestrino GA, Verrinder Veasey J, Benard G & Sousa MGT Host immune responses in dermatophytes infection. Mycoses 64, 477–483, doi: 10.1111/myc.13246 (2021). [DOI] [PubMed] [Google Scholar]
- 161.Gupta AK, Venkataraman M, Hall DC, Cooper EA & Summerbell RC The emergence of Trichophyton indotineae: Implications for clinical practice. Int J Dermatol, doi: 10.1111/ijd.16362 (2022). [DOI] [PubMed] [Google Scholar]
- 162.Verma SB & Vasani R Male genital dermatophytosis - clinical features and the effects of the misuse of topical steroids and steroid combinations - an alarming problem in India. Mycoses 59, 606–614, doi: 10.1111/myc.12503 (2016). [DOI] [PubMed] [Google Scholar]
- 163.Sardana K, Gupta A & Mathachan SR Immunopathogenesis of Dermatophytoses and Factors Leading to Recalcitrant Infections. Indian Dermatol Online J 12, 389–399, doi: 10.4103/idoj.IDOJ_503_20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kumar P et al. Whole genome sequences of two Trichophyton indotineae clinical isolates from India emerging as threats during therapeutic treatment of dermatophytosis. 3 Biotech 11, 402, doi: 10.1007/s13205-021-02950-1 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kong X et al. Antifungal Susceptibility and Mutations in the Squalene Epoxidase Gene in Dermatophytes of the Trichophyton mentagrophytes Species Complex. Antimicrobial agents and chemotherapy 65, e0005621, doi: 10.1128/aac.00056-21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Singh A et al. High terbinafine resistance in Trichophyton interdigitale isolates in Delhi, India harbouring mutations in the squalene epoxidase gene. Mycoses 61, 477–484, doi: 10.1111/myc.12772 (2018). [DOI] [PubMed] [Google Scholar]
- 167.Burmester A, Hipler UC, Elsner P & Wiegand C Point mutations in the squalene epoxidase erg1 and sterol 14-α demethylase erg11 gene of T indotineae isolates indicate that the resistant mutant strains evolved independently. Mycoses 65, 97–102, doi: 10.1111/myc.13393 (2022). [DOI] [PubMed] [Google Scholar]
- 168.Yamada T et al. Gene Amplification of CYP51B: a New Mechanism of Resistance to Azole Compounds in Trichophyton indotineae. Antimicrobial agents and chemotherapy 66, e0005922, doi: 10.1128/aac.00059-22 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Singh A et al. A unique multidrug-resistant clonal Trichophyton population distinct from Trichophyton mentagrophytes/Trichophyton interdigitale complex causing an ongoing alarming dermatophytosis outbreak in India: Genomic insights and resistance profile. Fungal Genet Biol 133, 103266, doi: 10.1016/j.fgb.2019.103266 (2019). [DOI] [PubMed] [Google Scholar]
- 170.van Zuuren EJ, Fedorowicz Z & El-Gohary M Evidence-based topical treatments for tinea cruris and tinea corporis: a summary of a Cochrane systematic review. Br J Dermatol 172, 616–641, doi: 10.1111/bjd.13441 (2015). [DOI] [PubMed] [Google Scholar]
- 171.Singh SK, Subba N & Tilak R Efficacy of Terbinafine and Itraconazole in Different Doses and in Combination in the Treatment of Tinea Infection: A Randomized Controlled Parallel Group Open Labeled Trial with Clinico-Mycological Correlation. Indian J Dermatol 65, 284–289, doi: 10.4103/ijd.IJD_548_19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Shaw D et al. MIC and Upper Limit of Wild-Type Distribution for 13 Antifungal Agents against a Trichophyton mentagrophytes-Trichophyton interdigitale Complex of Indian Origin. Antimicrobial agents and chemotherapy 64, doi: 10.1128/aac.01964-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Gueneau R et al. Extensive dermatophytosis caused by terbinafine-resistant Trichophyton indotineae, successfully treated with topical voriconazole. Int J Antimicrob Agents 60, 106677, doi: 10.1016/j.ijantimicag.2022.106677 (2022). [DOI] [PubMed] [Google Scholar]
- 174.Alexander BD et al. Increasing echinocandin resistance in Candida glabrata: clinical failure correlates with presence of FKS mutations and elevated minimum inhibitory concentrations. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America 56, 1724–1732, doi: 10.1093/cid/cit136 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Magobo RE, Lockhart SR & Govender NP Fluconazole-resistant Candida parapsilosis strains with a Y132F substitution in the ERG11 gene causing invasive infections in a neonatal unit, South Africa. Mycoses 63, 471–477, doi: 10.1111/myc.13070 (2020). [DOI] [PubMed] [Google Scholar]
- 176.Corzo-Leon DE, Peacock M, Rodriguez-Zulueta P, Salazar-Tamayo GJ & MacCallum DM General hospital outbreak of invasive candidiasis due to azole-resistant Candida parapsilosis associated with an Erg11 Y132F mutation. Med Mycol 59, 664–671, doi: 10.1093/mmy/myaa098 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Alcoceba E et al. Fluconazole-resistant Candida parapsilosis clonally related genotypes: first report proving the presence of endemic isolates harbouring the Y132F ERG11 gene substitution in Spain. Clin Microbiol Infect, doi: 10.1016/j.cmi.2022.02.025 (2022). [DOI] [PubMed] [Google Scholar]
- 178.Arastehfar A et al. First Report of Candidemia Clonal Outbreak Caused by Emerging Fluconazole-Resistant Candida parapsilosis Isolates Harboring Y132F and/or Y132F+K143R in Turkey. Antimicrobial agents and chemotherapy 64, doi: 10.1128/aac.01001-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Castanheira M, Deshpande LM, Messer SA, Rhomberg PR & Pfaller MA Analysis of global antifungal surveillance results reveals predominance of Erg11 Y132F alteration among azole-resistant Candida parapsilosis and Candida tropicalis and country-specific isolate dissemination. Int J Antimicrob Agents 55, 105799, doi: 10.1016/j.ijantimicag.2019.09.003 (2020). [DOI] [PubMed] [Google Scholar]
- 180.Thomaz DY et al. Environmental Clonal Spread of Azole-Resistant Candida parapsilosis with Erg11-Y132F Mutation Causing a Large Candidemia Outbreak in a Brazilian Cancer Referral Center. Journal of fungi (Basel, Switzerland) 7, doi: 10.3390/jof7040259 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Zhou ZL et al. Genetic relatedness among azole-resistant Candida tropicalis clinical strains in Taiwan from 2014 to 2018. Int J Antimicrob Agents 59, 106592, doi: 10.1016/j.ijantimicag.2022.106592 (2022). [DOI] [PubMed] [Google Scholar]
- 182.Kano R, Kimura U, Noguchi H & Hiruma M Clinical Isolate of a Multi-Antifungal-Resistant Trichophyton rubrum. Antimicrobial agents and chemotherapy 66, e0239321, doi: 10.1128/aac.02393-21 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Gu D, Hatch M, Ghannoum M & Elewski BE Treatment-resistant dermatophytosis: A representative case highlighting an emerging public health threat. JAAD Case Rep 6, 1153–1155, doi: 10.1016/j.jdcr.2020.05.025 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.WHO. WHO Fungal Priority Pathogens List, <https://www.who.int/publications/i/item/9789240060241> (2023).
- 185.Price CL, Parker JE, Warrilow AG, Kelly DE & Kelly SL Azole fungicides - understanding resistance mechanisms in agricultural fungal pathogens. Pest Manag Sci 71, 1054–1058, doi: 10.1002/ps.4029 (2015). [DOI] [PubMed] [Google Scholar]
- 186.Fisher MC et al. Tackling the emerging threat of antifungal resistance to human health. Nat Rev Microbiol 20, 557–571, doi: 10.1038/s41579-022-00720-1 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Pfaller MA, Marco F, Messer SA & Jones RN In vitro activity of two echinocandin derivatives, LY303366 and MK-0991 (L-743,792), against clinical isolates of Aspergillus, Fusarium, Rhizopus, and other filamentous fungi. Diagnostic microbiology and infectious disease 30, 251–255, doi: 10.1016/s0732-8893(97)00246-0 (1998). [DOI] [PubMed] [Google Scholar]
- 188.Fera MT, La Camera E & De Sarro A New triazoles and echinocandins: mode of action, in vitro activity and mechanisms of resistance. Expert Rev Anti Infect Ther 7, 981–998, doi: 10.1586/eri.09.67 (2009). [DOI] [PubMed] [Google Scholar]
- 189.Orozco AS et al. Mechanism of fluconazole resistance in Candida krusei. Antimicrobial agents and chemotherapy 42, 2645–2649, doi: 10.1128/aac.42.10.2645 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Guinea J, Sánchez-Somolinos M, Cuevas O, Peláez T & Bouza E Fluconazole resistance mechanisms in Candida krusei: the contribution of efflux-pumps. Med Mycol 44, 575–578, doi: 10.1080/13693780600561544 (2006). [DOI] [PubMed] [Google Scholar]
- 191.Prevention, C. f. D. C. a. (2019).
- 192.Revie NM, Iyer KR, Robbins N & Cowen LE Antifungal drug resistance: evolution, mechanisms and impact. Curr Opin Microbiol 45, 70–76, doi: 10.1016/j.mib.2018.02.005 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Whaley SG et al. Azole Antifungal Resistance in Candida albicans and Emerging Non-albicans Candida Species. Front Microbiol 7, 2173, doi: 10.3389/fmicb.2016.02173 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Mba IE, Nweze EI, Eze EA & Anyaegbunam ZKG Genome plasticity in Candida albicans: A cutting-edge strategy for evolution, adaptation, and survival. Infect Genet Evol 99, 105256, doi: 10.1016/j.meegid.2022.105256 (2022). [DOI] [PubMed] [Google Scholar]
- 195.Zafar H, Altamirano S, Ballou ER & Nielsen K A titanic drug resistance threat in Cryptococcus neoformans. Curr Opin Microbiol 52, 158–164, doi: 10.1016/j.mib.2019.11.001 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Singh-Babak SD et al. Global analysis of the evolution and mechanism of echinocandin resistance in Candida glabrata. PLoS Pathog 8, e1002718, doi: 10.1371/journal.ppat.1002718 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Duxbury SJN, Bates S, Beardmore RE & Gudelj I Evolution of drug-resistant and virulent small colonies in phenotypically diverse populations of the human fungal pathogen Candida glabrata. Proc Biol Sci 287, 20200761, doi: 10.1098/rspb.2020.0761 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Fan S, Li C, Bing J, Huang G & Du H Discovery of the Diploid Form of the Emerging Fungal Pathogen Candida auris. ACS Infect Dis 6, 2641–2646, doi: 10.1021/acsinfecdis.0c00282 (2020). [DOI] [PubMed] [Google Scholar]
- 199.Weil T et al. Adaptive Mistranslation Accelerates the Evolution of Fluconazole Resistance and Induces Major Genomic and Gene Expression Alterations in Candida albicans. mSphere 2, doi: 10.1128/mSphere.00167-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Zhang H et al. Global Screening of Genomic and Transcriptomic Factors Associated with Phenotype Differences between Multidrug-Resistant and -Susceptible Candida haemulonii Strains. mSystems 4, doi: 10.1128/mSystems.00459-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]


