Mutations in ovarian cancer risk genes are found in women of diverse ancestries, supporting genetic testing for ALL
Abstract
PURPOSE
To evaluate rates of germline pathogenic/likely pathogenic variants (PVs) and genetic counseling by ancestry in patients with epithelial ovarian cancer (EOC).
METHODS
Patients with pathologically confirmed EOC who underwent clinical tumor-normal sequencing from January 1, 2015, to December 31, 2020, inclusive of germline analysis of ≥76 genes were included. Patients with newly identified PVs were referred for Clinical Genetics Service (CGS) counseling. Ancestry groups were defined using self-reported race/ethnicity and Ashkenazi Jewish (AJ) heritage. Genetic ancestry was inferred computationally using validated algorithms. Logistic regression models were built.
RESULTS
Of 1,266 patients, self-reported ancestry (AJ, 17%; Asian, 10%; Black/African American, 5.4%; Hispanic, 6.2%; non-Hispanic White, 57%; other, 0.16%; unknown, 4.0%) correlated with genetic ancestry (AJ ancestry, 18%; admixed, 10%; African, 4%; East Asian [EAS], 6%; European, 56%; Native American, 0.2%; South Asian [SAS], 4%; unknown, 2%). Germline PVs were observed in 313 (25%) patients, including 195 (15%) with PVs in EOC-associated genes. Those with PVs were younger at diagnosis (59 v 62 years; P < .001) and more likely to have high-grade serous ovarian cancer (83% v 72%; P = .009). PV prevalence varied between ancestry groups (P < .001), with highest rates in the AJ (39.9%) and Asian (26.5%) groups and similar rates (>10%) across other ancestry groups. Use of genetic ancestry demonstrated similar findings and further characterized high rates of PV in EAS/SAS groups. Younger age, high-grade serous histology, and self-reported AJ or Asian ancestry were associated with PV in an EOC-associated gene. Rates of CGS counseling for newly identified PVs were high (80%) across ancestry groups.
CONCLUSION
Rates of PV, particularly in EOC-associated genes, were high regardless of ancestry, with similar rates of counseling between groups, emphasizing the importance of universal genetic testing in all patients with EOC.
INTRODUCTION
Ovarian cancer (OC) is the leading cause of gynecologic cancer–related death, and 15%-20% of women with epithelial ovarian cancer (EOC) harbor a germline pathogenic/likely pathogenic variant (PV) in a hereditary cancer predisposition gene.1,2 The majority occur in BRCA1 and BRCA2,1,3 as well as other genes involved in homologous recombination (RAD51C, RAD51D, BRIP1, PALB2, and ATM) or DNA mismatch repair (MLH1, MSH2, MSH6, and PMS2).4,5 Accordingly, universal germline testing for all women with newly diagnosed EOC is recommended3,6-9; however, there are disparities in genetic testing and counseling uptake on the basis of race, ethnicity, and insurance status, with potential implications for treatment, cancer prevention, and at-risk family members.10,11
CONTEXT
Key Objective
Although universal germline testing is recommended in epithelial ovarian cancer (EOC), much of the knowledge about rates of pathogenic/likely pathogenic variants (PV) and genetic counseling are derived from those of European ancestry, and less is known about those of other ancestries. This study evaluated rates of germline PV and subsequent genetic counseling by self-reported ancestry and genetic ancestry in a large and diverse cohort of patients with EOC.
Knowledge Generated
Rates of PV in EOC-associated genes were high across all self-reported and genetic ancestries (10%-24%) and were highest in the Ashkenazi Jewish and Asian cohorts. Integration of germline testing with clinical tumor-normal sequencing resulted in high levels of post-test genetic counseling across all ancestry groups.
Relevance
These findings validate the need for universal genetic testing in all patients with EOC, regardless of background, given the implications on both oncologic treatment and at-risk family members via cascade testing.
Most of the knowledge surrounding inherited predisposition to EOC is derived from non-Hispanic White and Ashkenazi Jewish (AJ) cohorts.12-14 Consequently, less is known about the prevalence of EOC-associated PVs in more diverse cohorts, potentially contributing to disparities in genetic testing and counseling uptake.15 Data from preliminary studies suggest high rates of germline PVs in BRCA1/216,17 and other EOC-associated genes18 in minority groups in the United States and in other non-European (EUR) countries19-21; however, more studies are needed to evaluate rates of PV and subsequent counseling for EOC-associated genes in diverse populations.
We hypothesize that rates of germline PV in EOC-associated genes may vary between ancestry groups and influence subsequent genetic testing, counseling, and care. We sought to evaluate differences in PV rates and subsequent genetic counseling by self-reported and genetic ancestry in a diverse cohort of patients with EOC who underwent germline assessment as part of clinical care.
METHODS
Patient Selection
We included all patients with pathologically confirmed EOC at a single institution between January 1, 2015, and December 31, 2020, who underwent tumor-normal sequencing via a Food and Drug Administration-approved targeted next-generation sequencing (NGS) panel (Memorial Sloan Kettering Cancer Center-Integrated Mutation Profiling of Actionable Cancer Targets [MSK-IMPACT]) inclusive of germline analysis of ≥76 genes. The assay uses DNA from formalin-fixed, paraffin-embedded tumors and patient-matched blood samples to assess for PVs in coding exons.22,23 Testing was ordered by the primary oncologist per standardized workflow (Data Supplement, Fig S1), as previously described.24
Germline Analysis and Protocol for Genetic Counseling
PVs were independently assessed and manually curated using standards for variant classification by the American College of Medical Genetics and Genomics/Association of Molecular Pathology at the time of data collection.25 Variants of uncertain significance (VUS) were not reported in this study, given limited data. PVs were classified as high (relative risk [RR], >4), moderate (RR, 2-4), or low (RR, <2) penetrance, recessive, or of uncertain clinical actionability on the basis of previous modeling.6,26,27 EOC-associated genes included BRCA1, BRCA2, RAD51C, RAD51D, BRIP1, PALB2, ATM, MLH1, MSH2, MSH6, and PMS2.3,28,29
All patients with newly identified PVs, defined as germline findings that were not previously known by the patient or their provider, received expedited referrals to our institutional Clinical Genetics Service (CGS) for additional counseling and identification of at-risk family members. Patients with known PVs who desired additional counseling were also referred to CGS. For patients who declined CGS appointments, failed to return at least three phone calls to schedule appointments, or did not have written documentation of a results discussion with their treating provider in the medical record, letters were sent to disclose the results. Patients who did not speak English received telephone calls via an interpreter or letters translated into their preferred language.24 Nondisclosure rate was calculated as the percentage of patients with newly diagnosed PV who had no documentation of results disclosure. Data regarding CGS follow-up and counseling rates were available from clinical databases up until December 31, 2019; patients enrolled in 2020 were not included in these analyses because of conversion to a telemedicine format during the COVID-19 pandemic.24,30
Data Collection
Self-reported ancestry was defined using patient-reported race/ethnicity and AJ heritage from the electronic medical record, and patients were categorized into mutually exclusive ancestry groups: AJ, Asian, Black/African American (AA), Hispanic, non-Hispanic White, other, or unknown, with patients who self-identified as AJ or Hispanic classified as such, regardless of race. Patients who self-identified as American Indian, Alaskan Native, Native Hawaiian, or Other Pacific Islander were classified as other.24
Genetic ancestry was inferred from MSK-IMPACT as previously described.31 Briefly, we ran ADMIXTURE v1.332 in supervised mode using the 1000 Genomes Project33 cohort as reference to infer ancestral proportions of African (AFR), EUR, East Asian (EAS), Native American (NAM), and South Asian (SAS) populations; AJ genetic ancestry (ASJ) was added recently. Patients who had an ancestral fraction of >0.8 for any single population were assigned that population label, otherwise they were considered admixed (ADM). Patients with no sequencing data available for genetic ancestry were labeled unknown.
Statistical Analysis
Clinical characteristics, including age at diagnosis, stage, BMI, and histologic type (ie, high-grade serous, low-grade serous, endometrioid, clear cell, carcinosarcoma, mucinous, mixed, and poorly differentiated/undifferentiated), were collected. Descriptive statistics were provided for clinical characteristics by PV status. The association between the clinical features and PV status were tested using the Fisher exact test for categorical variables and the Wilcoxon rank sum test for continuous variables. Logistic regression was applied to examine the associations between clinical features and PV status. Variables shown to be statistically significant in univariate analyses were entered into the multivariable models. Two outcomes (PV in any gene and PV in EOC-associated genes) were examined. Analyses were performed using R 4.2.2 (The R Project for Statistical Computing34). All tests are two-sided and P < .05 was considered statistically significant. All patients were consented to MSK-IMPACT per protocol, and this study was approved under MSK institutional review board #12-245 (ClinicalTrials.gov identifier: NCT01775072).
RESULTS
Clinical Characteristics
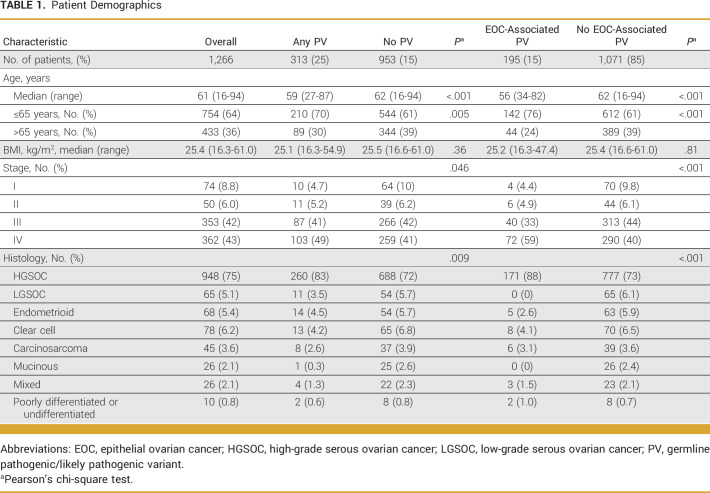
Between January 1, 2015, and December 31, 2020, 1,266 patients with EOC underwent germline assessment and were included in this study (Fig 1). Median age at EOC diagnosis was 61 years (range, 16-94 years; Table 1). Most patients had stage III (42%) or IV (43%) EOC; 75% of tumors were high-grade serous ovarian cancers (HGSOCs), 6.2% were clear cell, 5.4% were endometrioid, and 5.1% were low-grade serous (Table 1).
FIG 1.
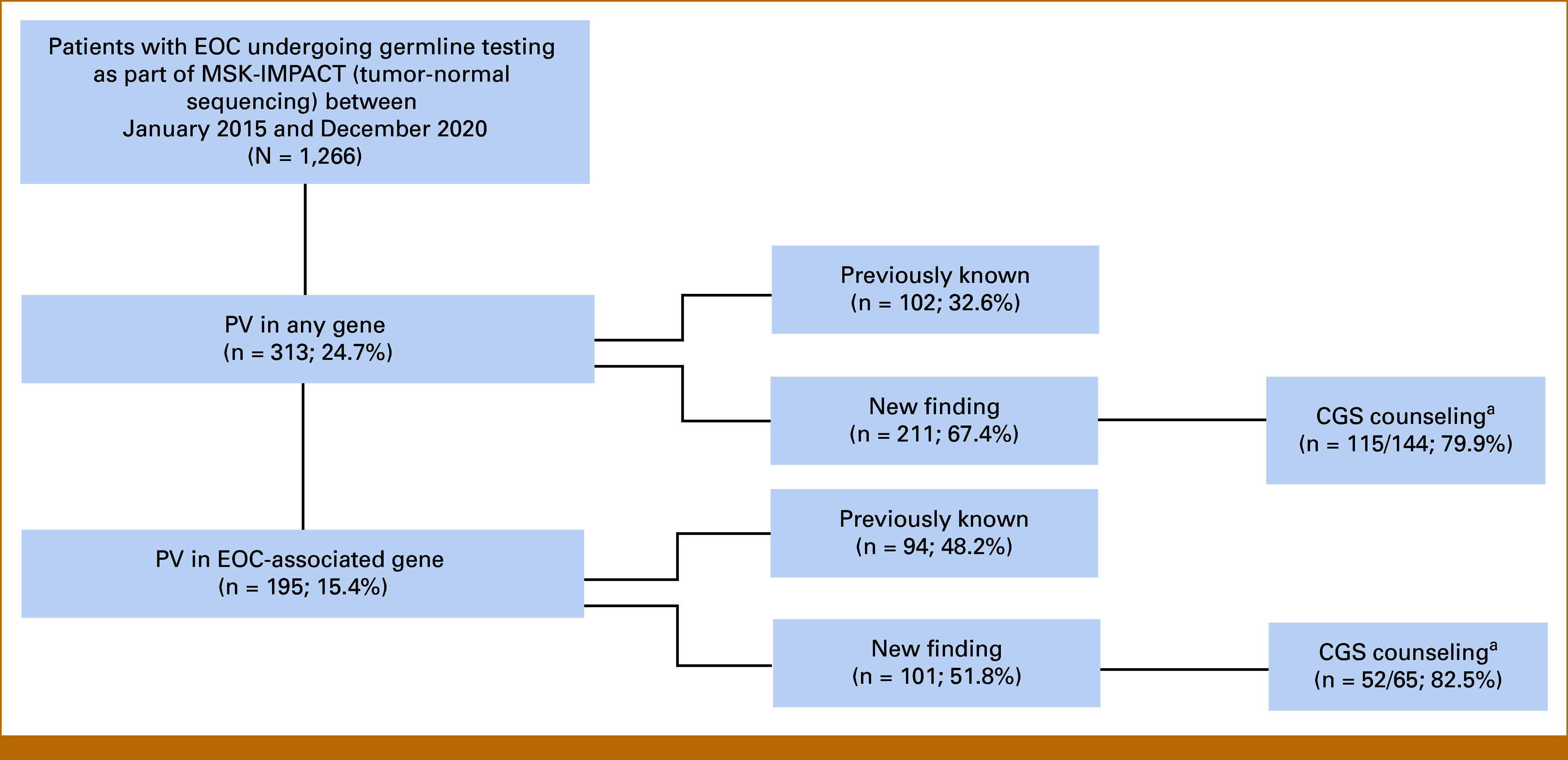
Patient flow diagram depicting patients with epithelial ovarian cancer undergoing MSK-IMPACT and the proportion with PV, new and previously known, as well as those undergoing subsequent genetic counseling. EOC-associated genes: BRCA1, BRCA2, RAD51C, RAD51D, BRIP1, PALB2, ATM, MLH1, MSH2, MSH6, PMS2. aPercentages are based upon 2015-2019 data only as CGS follow-up data are available only for this time period. CGS, Clinical Genetics Service; EOC, epithelial ovarian cancer; MSK-IMPACT, Memorial Sloan Kettering Cancer Center-Integrated Mutation Profiling of Actionable Cancer Targets; PV, germline pathogenic/likely pathogenic variant.
TABLE 1.
Patient Demographics
PVs were identified in 313 (25%) patients, including 195 (15%) with PVs in an EOC-associated gene (Table 1; Data Supplement, Tables S1 and S2). The most common PVs were in BRCA1 (n = 94) and BRCA2 (n = 54), representing 7.4% and 4.3% of the group, respectively (Data Supplement, Tables S1 and S3). The median age at diagnosis was 59 years for patients with PVs compared with 62 years for those with sporadic EOC (P < .001; Table 1). Patients with PVs compared with those without were more likely to have HGSOC (83% v 72%, respectively; P = .009). Compared with those without, patients with a PV in an EOC-associated gene were more likely to have stage III/IV disease at diagnosis (92% v 84%; P < .001).
Self-Reported Ancestry and Genetic Ancestry
Using self-reported ancestry, patients were classified as AJ (17%; n = 218), Asian (10%; n = 132), Black/AA (5%; n = 68), Hispanic (6%; n = 79), non-Hispanic White (57%; n = 716), other (0.2%; n = 2), or unknown (4%; n = 51; Fig 2; Data Supplement, Fig S2A). Using genetic ancestry, patients were classified as ADM (10%; n = 127), AFR (4%; n = 53), ASJ (18%; n = 223), EAS (6%; n = 72), EUR (56%; n = 713), SAS (4%; n = 46), NAM (0.2%; n = 2), or unknown (2%; n = 30; Fig 2; Data Supplement, Fig S2B). In patients self-identifying as unknown (n = 51), genetic ancestry was able to further classify these patients into ADM (25%), ASJ (8%), AFR (8%), EAS (6%), EUR (43%), or SAS (8%; Data Supplement, Table S4). In those of self-reported Asian ancestry (n = 132), genetic ancestry calculations further classified these patients as EAS (52%), SAS (30%), ADM (14%), or unknown (4%). Patients who were classified to have a genetic ancestry of ADM (n = 127), for which no one ancestry group met the prespecified 80% threshold, corresponded to self-reported ancestry groups of AJ (0.8%), Asian (15%), Black/AA (14%), Hispanic (43%), non-Hispanic White (15%), other (2%), or unknown (10%) (Data Supplement, Table S4).
FIG 2.
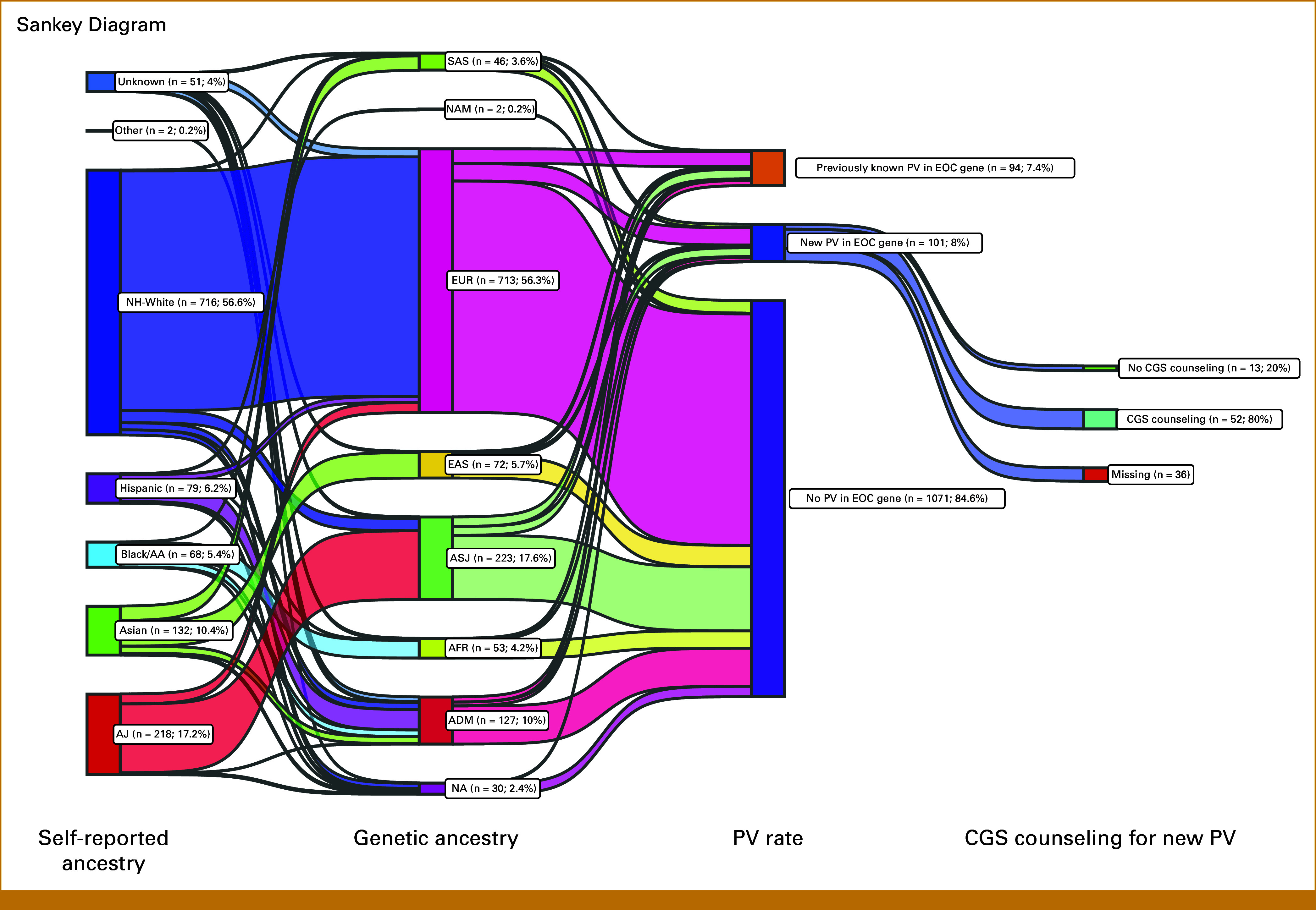
Sankey diagram depicting self-reported ancestry, genetic ancestry, presence of germline pathogenic variants, and clinical genetics counseling. Using self-reported ancestry, patients were classified into AJ, Asian, Black/AA, Hispanic, NH-White, other, and unknown groups. Use of genetic ancestry further classified patients into ADM, AFR, ASJ, EAS, EUR, NAM, and SAS groups. A small subset (2.4%) of patients were unable to be classified into a genetic ancestry category. The rate of newly diagnosed PV in an EOC-related gene was 8%, compared with 7.4% of patients who previously knew about their PV in an EOC-related gene. Of patients with a new PV finding in an EOC-related gene for whom we have CGS data available, 80% underwent CGS counseling. AA, African American; ADM, admixed; AFR, African; AJ, Ashkenazi Jewish; ASJ, AJ genetic ancestry; CGS, Clinical Genetics Service; EAS, East Asian; EOC, epithelial ovarian cancer; EUR, European; NA, not applicable; NAM, Native American; NH, non-Hispanic; PV, germline pathogenic/likely pathogenic variant; SAS, South Asian.
Germline Findings by Ancestry
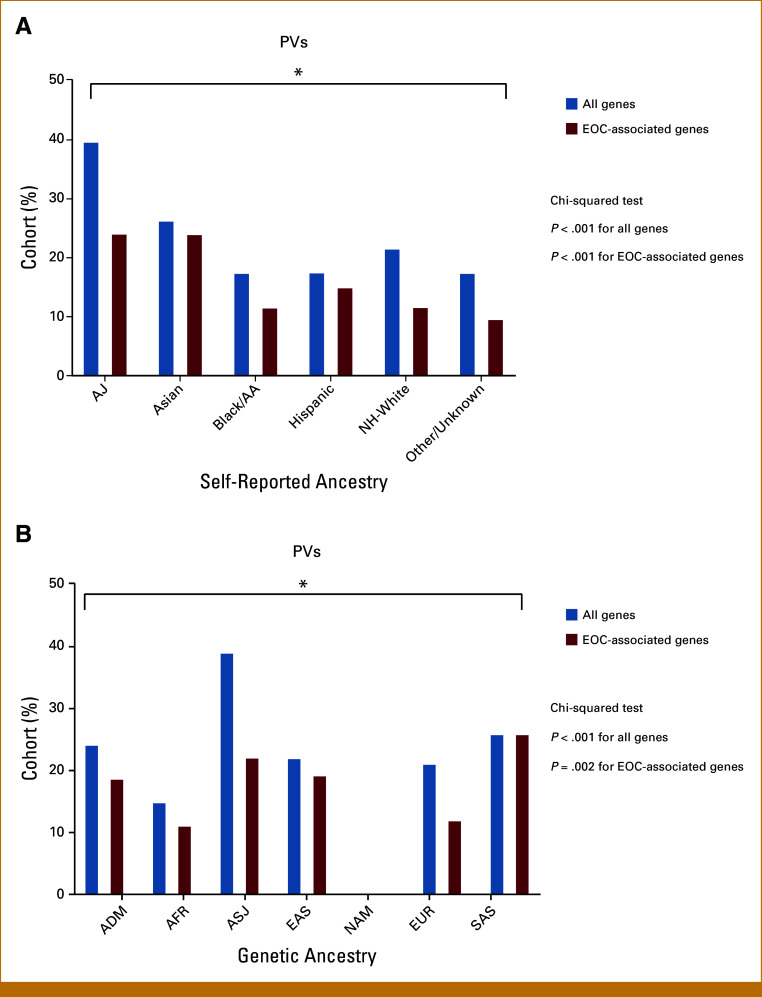
The prevalence of PVs differed by self-reported ancestry (AJ, 39.9%; Asian, 26.5%; Black/AA, 17.7%; Hispanic, 17.7%, non-Hispanic White 21.8%; and other/unknown, 17.7%; P < .001; Fig 3A; Data Supplement, Table S5). Most PVs occurred in either high- or moderate-penetrance genes. However, there was significant variation in rates of uncertain/low/recessive PVs among self-reported ancestry groups (AJ, 34%; Asian, 8.6%; Black/AA, 25%; Hispanic, 14%; non-Hispanic White, 37%; and unknown, 33%; P = .019; Data Supplement, Fig S3A), with the highest rates in the non-Hispanic White and AJ cohorts. Similar patterns were seen when using genetic ancestry, with high rates of PV in all groups, mostly in high-penetrance genes (Data Supplement, Table S6 and Fig S3B). This method also allowed further elucidation of high PV rates in both EAS and SAS groups (Data Supplement, Table S6).
FIG 3.
Germline pathogenic variant by (A) self-reported ancestry and (B) genetic ancestry. (A) Rates of PVs in all assessed genes were high across all self-reported ancestries but were highest in the AJ and Asian populations (blue). When assessing EOC-associated genes only (red), rates of PVs continued to be highest in the AJ and Asian populations (24% each), and lowest in the Black/AA and NH-White groups (11% each). Rates of pathogenic/likely pathogenic variants differed between all groups for all genes and EOC-associated genes (P < .001). Other includes patients who self-identified as American Indian, Alaskan Native, Native Hawaiian, or Other Pacific Islander. (B) When evaluating rates of PVs across genetic ancestry admixture, similar patterns were seen, with high rates of EOC-associated genes in the ASJ, East Asian, and South Asian groups, and lowest in the African and non-AJ EUR groups. Notably, no pathogenic/likely pathogenic variants were identified in the two patients classified as NAM. Rates of pathogenic/likely pathogenic variants differed between all groups for all genes (P < .001) and EOC-associated genes (P = .002). AA, African American; ADM, admixed; AFR, African; AJ, Ashkenazi Jewish; ASJ, AJ genetic ancestry; EAS, East Asian; EOC, epithelial ovarian cancer; EUR, European; NAM, Native American; NH, non-Hispanic; PV, germline pathogenic/likely pathogenic variant; SAS, South Asian.
When assessing specifically for EOC-associated genes, rates of PV were high in all self-reported ancestry groups (AJ, 24.3%; Asian, 24.2%; Hispanic, 15.2%; non-Hispanic White, 11.9%; Black/AA, 11.8%; and other/unknown, 9.8%; P < .001; Fig 3A; Data Supplement, Table S5). When evaluating rates of PV by genetic ancestry, similar patterns were identified (Fig 3B; Data Supplement, Table S6), with high rates of EOC-associated PV in both the SAS and EAS groups.
Within EOC-associated genes, BRCA1 (n = 94; 48%) and BRCA2 (n = 54; 28%) comprised most PVs (Figs 4A and 4B; Data Supplement, Table S3). This was particularly notable in the AJ group, in whom the prevalence of BRCA2 almost equaled BRCA1 because of AJ founder mutations (Data Supplement, Table S1). PVs in other genes related to homologous recombination deficiency (RAD51C, RAD51D, BRIP1, PALB2, and ATM) comprised 18% of all PVs and were primarily found in the non-Hispanic White and Asian groups, although numbers were limited (Fig 4A; Data Supplement, Table S3). When using genetic ancestry, rates of PVs in other homologous recombination deficiency genes were highest in the EUR population (Fig 4B). PVs in Lynch syndrome–associated mismatch repair genes accounted for 6.7% of all PVs and were primarily found within the non-Hispanic White and EUR groups (Figs 4A and 4B).
FIG 4.
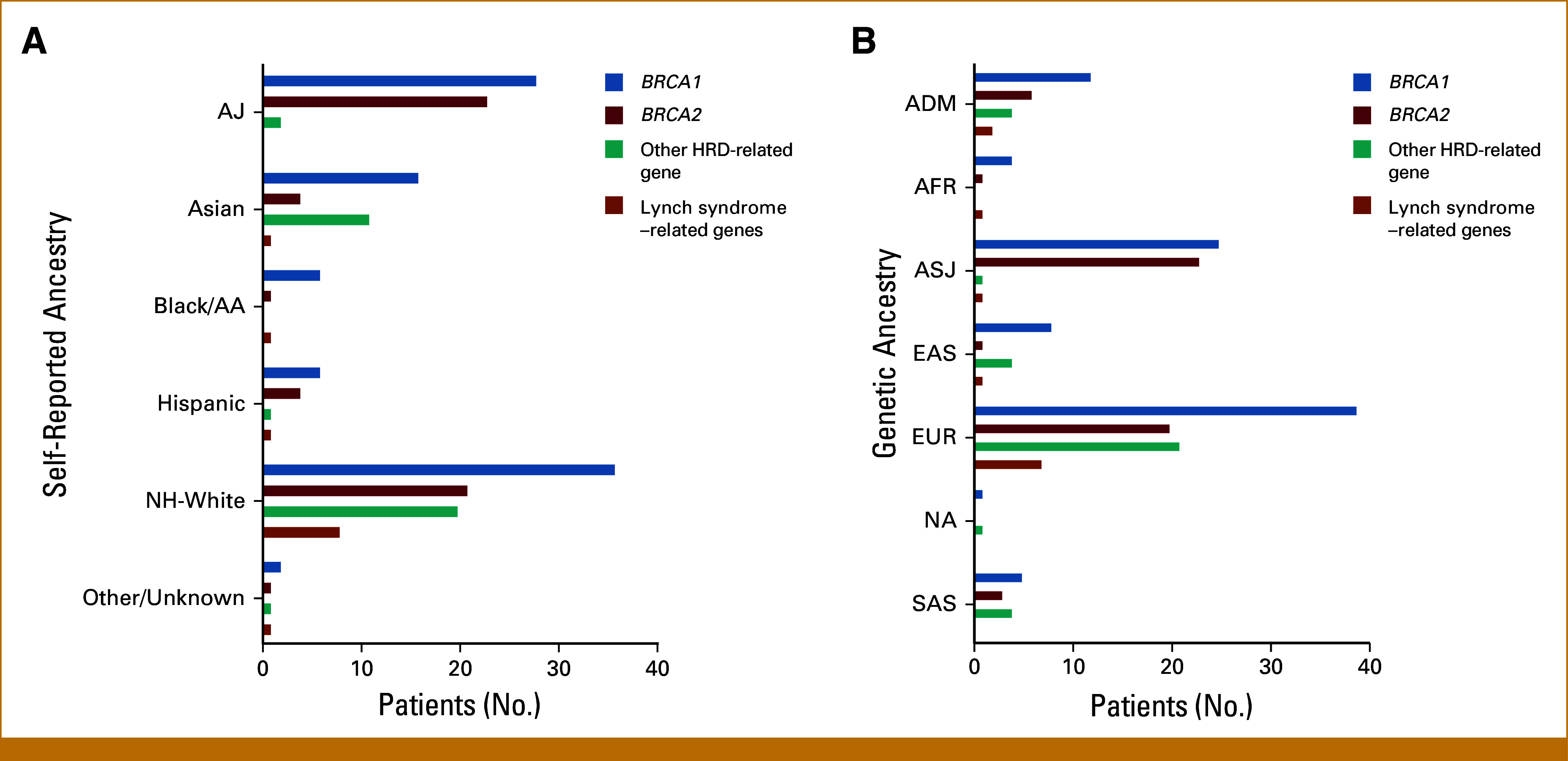
Distribution of the genes with germline pathogenic variants found in patients with epithelial ovarian cancer by (A) self-reported ancestry and (B) genetic ancestry. Other HRD-related gene includes the following genes: ATM, BRIP1, PALB2, RAD51C, and RAD51D. Lynch syndrome–related genes includes the following genes: MLH1, MLH2, MSH6, and PMS2. Other includes patients who self-identified as American Indian, Alaskan Native, Native Hawaiian, or Other Pacific Islander. As the number of patients in the other and unknown groups were low, these groups were combined. AA, African American; ADM, admixed; AFR, African; AJ, Ashkenazi Jewish; ASJ, AJ genetic ancestry; EAS, East Asian; EUR, European; HRD, homologous recombination deficiency; NA, genetic ancestry not available; NH, non-Hispanic; SAS, South Asian.
Predictors of PVs
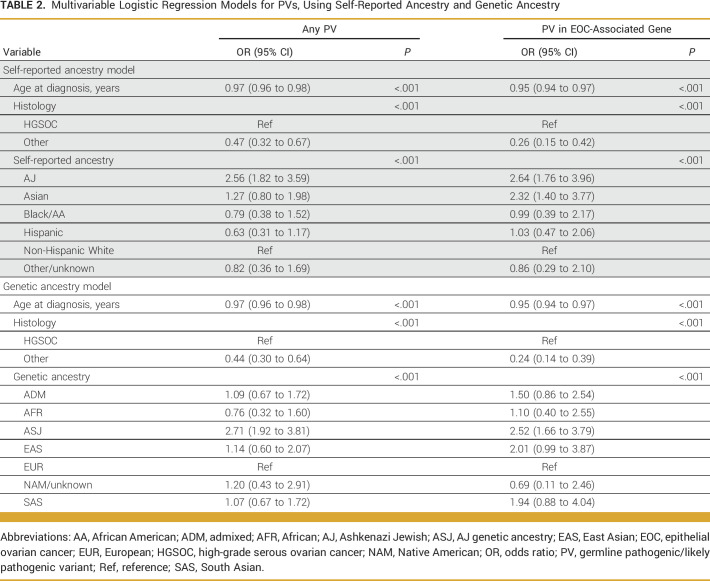
On univariate analyses, age of diagnosis, HGSOC histology, self-reported ancestry, and genetic ancestry were all predictors of presence of PV, overall and for EOC-associated genes. On multivariable models, self-reported ASJ (odds ratio [OR], 2.56; 95% CI, 1.82 to 3.59) was associated with increased odds of PV overall, even after adjustment for age at diagnosis and histology (Table 2). When examining PVs specifically in EOC-associated genes, self-reported ASJ (OR, 2.64; 95% CI, 1.76 to 3.96) and Asian ancestry (OR, 2.32; 95% CI, 1.40 to 3.77) were associated with increased odds of PV, even after adjustment for age at diagnosis and histology (Table 2). Similar findings were noted using genetic ancestry for PVs overall and EOC-associated PVs (Table 2).
TABLE 2.
Multivariable Logistic Regression Models for PVs, Using Self-Reported Ancestry and Genetic Ancestry
Genetic Counseling
Of the 313 patients with PVs, 102 (32.6%) knew about this result before MSK-IMPACT germline assessment (Fig 1). Among the 144 patients with a newly identified PV and available CGS follow-up information, 115 (79.9%) completed CGS counseling, including 69 (47.9%) via in-person appointment and 46 (31.9%) via phone call. Close-out letters were sent to 29 patients (20.1%), and the nondisclosure rate was 5 patients (2.4%). Although limited in numbers, rates of CGS counseling for those with new PV were high across all self-reported ancestry groups (Data Supplement, Table S5).
Of the 195 patients with PVs in an EOC-associated genes, 94 (48.2%) previously knew about the result (Fig 1). Of the 65 patients for whom this was a new finding with available CGS follow-up information, 52 (80%) completed CGS counseling. Rates of CGS follow-up for EOC-associated genes were high across all self-reported ancestry groups (Fig 2; Data Supplement, Table S5).
DISCUSSION
We evaluated rates of germline PV and subsequent genetic counseling in a large cohort of patients with EOC to assess for potential differences across populations. Rates of PV in EOC-associated genes, mostly BRCA1/2, were high across all self-reported ancestries (10%-24%) and were highest in the AJ and Asian cohorts (24%), even in multivariable logistic regression models. Using a novel algorithm to infer genetic ancestry admixture from MSK-IMPACT, we found similar results, further strengthening our findings. For patients with newly identified PVs in EOC-associated genes, rates of follow-up counseling were high across all ancestry groups. This highlights the importance of universal genetic testing in OC, regardless of ancestry, and supports integration of germline assessment into routine oncologic care given implications for treatment, cancer prevention, and at-risk family members.
We found that 25% of patients with EOC within our diverse cohort had a PV and 15% of patients had a PV in an EOC-associated gene, which is consistent with other studies demonstrating rates between 14% and 22%.1,12,35 Although rates of PV varied between ancestry groups, much of the variation was found within the low/uncertain/recessive gene penetrance group, and rates of PV in EOC-associated genes were high across all groups. A study of 6,000 diverse women with OC from California and Georgia found the highest PV rates among Hispanic patients (27.6%) compared with non-Hispanic White (12.3%), Black/AA (13.2%), and Asian (13.3%) patients.17 Somasegar et al35 studied rates of germline PV in EOC-associated genes in 51 self-reported Black/AA patients with OC and reported a PV rate of 25.5%, the majority of which were in BRCA1 (13.7%) and BRCA2 (7.8%). In a study of germline genetic testing results of patients from the Caribbean diagnosed with breast cancer and/or OC, the rate of germline PV was 14.2%.36 In our study, we observed a high PV rate for all patients, particularly Asian patients (24%), whereas the PV rates in our Hispanic and Black/AA cohorts were lower at 18% each. These differences likely reflect ascertainment bias, size of multigene panels, and variations within specific populations and geographical locations; however, overall rates of PV were high across all groups. These findings further highlight the importance of testing in all women with OC and the need for continued studies in large, diverse OC cohorts.
Despite recommendations for universal germline testing in all patients with EOC since 2010, rates of actual testing in practice have remained low (10%-30%).17 Our study found high rates of genetics follow-up across all ancestry groups (64%-100%), which may be influenced by our institutional practice to integrate germline and tumor assessment in oncology clinics, a form of mainstreaming that is becoming more common.10 The testing is offered directly by oncologists to expand access and decrease barriers and is particularly important in EOC, given implications on treatment with PARP inhibitor therapies.37-39 However, rates are not 100%, and reasons may be complex and involve various social determinants of health at multiple levels of care. Studies have observed that disparities in referrals to genetic counseling/testing persist, and vulnerable patients including racial/ethnic minorities, low-income patients, and non–English-speaking patients are at increased risk for not receiving either recommended genetics care17,37,38,40 or guideline-concordant treatment.41 Other reasons include lack of availability of genetic counselors, language and cultural discordance, fear of potential retribution from insurance carriers, and lack of awareness about genetic testing and prevalence of PVs across demographic groups because many studies and educational campaigns focus on non-Hispanic White and AJ communities.11,42,43 Additional work investigating the social determinants of health outside of ancestry that influence tumor-normal genetic testing in EOC are ongoing, and recent work has demonstrated progress with the use of NGS in historically underserved ethnic groups.44
Our study has several strengths. Our population contained a large and diverse cohort of patients with EOC, with >25% of patient self-reporting ancestries that were not non-Hispanic White or AJ, a group we separated out, given high rates of PV, to avoid bias. Additionally, patients underwent germline assessment of ≥76 cancer-associated genes, which encompassed BRCA1/2 and other moderate-penetrance genes. Although the most common PVs across all groups were found in BRCA1/2, we observed that PVs in other homologous recombination deficiency genes (ie, PALB2 and RAD51D) may be more prevalent within Asian compared with other ancestry groups. This is hypothesis-generating and should be further explored. We acknowledge the limitations of race/ethnicity measures, which are social constructs and closely tied to identity.45,46 To address this, we used a second measure of ancestry inferred from sequencing of normal tissue, which correlated well with self-reported measures and added additional insights, particularly in patients of self-reported Asian and unknown ancestry. Importantly, high rates of PV were observed across all groups using both methods, further highlighting the need for universal genetic testing.
A limitation of our study was the lack of VUS data, which may occur more frequently in Black/AA, Asian, and Hispanic patients,25 as genetic counseling and ongoing follow-up for these results are critical to equitable care. Reclassification of PV and VUS findings is ongoing, and our results published here reflect classification designations at the time of data analysis. Additionally, patients undergoing genetic testing through an outside laboratory were not included in our cohort, leading to potential underestimation of PV rates. We also acknowledge the possibility of ascertainment bias, given our tertiary cancer center with a large AJ population. However, our New York City patient cohort is racially/ethnically diverse, with >25% of patients identifying as non-White. Finally, our CGS follow-up data are limited for each ancestry group and should be interpreted with caution. Additional analyses of our telemedicine experience are ongoing as studies have demonstrated disparities in access to and use of telemedicine platforms for health care delivery, with lowest rates of uptake in Hispanic, Asian, and non–English-speaking groups.47-49 Notably, disparities among Black/AA patients found on pan-cancer analysis were not observed in patients with EOC,24 which may reflect smaller sample size or differences by tumor types.
In conclusion, we found that the prevalence of germline PVs in patients of diverse ancestries with EOC was high across all groups, particularly among AJ and Asian patients, using both self-reported and genetic ancestry to define populations. Integration of germline assessment with clinical tumor-normal sequencing resulted in high levels of post-test genetic counseling, with no differences between ancestry groups. These findings highlight the need for universal genetic testing in all patients with EOC, regardless of background. We hope these data increase public awareness and improve health equity in genetic testing and counseling of patients with EOC, particularly given the implications on oncologic treatment and family members via cascade testing.
SUPPORT
Supported by the Robert and Kate Niehaus Center for Inherited Cancer Genomics and the Precision, Interception and Prevention Program at Memorial Sloan Kettering, and in part by a Cancer Center Support Grant by the National Institutes of Health/National Cancer Institute (P30 CA008748). Britta Weigelt, PhD, is funded in part by Cycle for Survival and Breast Cancer Research Foundation grants.
PRIOR PRESENTATION
Presented as a poster at the 2022 Society of Gynecologic Oncology Meeting, Phoenix, AZ, March 19, 2022.
AUTHOR CONTRIBUTIONS
Conception and design: Tiffany Y. Sia, Alexia Iasonos, Carol L. Brown, Kenneth Offit, Jada G. Hamilton, Carol Aghajanian, Britta Weigelt, Zsofia K. Stadler, Ying L. Liu
Financial support: Carol L. Brown, Kenneth Offit, Carol Aghajanian, Zsofia K. Stadler, Ying L. Liu
Administrative support: Ying L. Liu
Provision of study materials or patients: Tiffany Y. Sia, Ryan M. Kahn, Margaret A. Sheehan, Kara Long Roche, Yukio Sonoda, Ginger J. Gardner, Dennis S. Chi, Alicia J. Latham, Kenneth Offit, Ying L. Liu
Collection and assembly of data: Tiffany Y. Sia, Anna Maio, Yelena M. Kemel, Sushmita B. Gordhandas, Ryan M. Kahn, Erin E. Salo-Mullen, Margaret A. Sheehan, Prince Rainier Tejada, Chaitanya Bandlamudi, Rachel N. Grisham, William P. Tew, Kara Long Roche, Oliver Zivanovic, Yukio Sonoda, Ginger J. Gardner, Dennis S. Chi, Alicia J. Latham, Maria I. Carlo, Marie Will, Michael F. Walsh, Kenneth Offit, Ying L. Liu
Data analysis and interpretation: Tiffany Y. Sia, Kanika S. Arora, Sushmita B. Gordhandas, Ryan M. Kahn, Margaret A. Sheehan, Chaitanya Bandlamudi, Qin Zhou, Alexia Iasonos, Rachel N. Grisham, William P. Tew, Yukio Sonoda, Alicia J. Latham, Yonina R. Murciano-Goroff, Michael F. Walsh, Mark E. Robson, Diana L. Mandelker, Michael F. Berger, Nadeem R. Abu-Rustum, Kenneth Offit, Britta Weigelt, Zsofia K. Stadler, Ying L. Liu
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Tiffany Y. Sia
Employment: Memorial Sloan-Kettering Cancer Center
Honoraria: OBG Management
Research Funding: Department of Defense (Inst)
Travel, Accommodations, Expenses: OncLive
Kanika S. Arora
Employment: Ro (I)
Rachel N. Grisham
Consulting or Advisory Role: GlaxoSmithKline, AstraZeneca, Signatera, Corcept Therapeutics, Intellisphere, SpringWorks Therapeutics, Verastem, Myriad Genetics
Research Funding: Context Therapeutics (Inst), Verastem (Inst), SpringWorks Therapeutics (Inst), Bayer (Inst), Novartis (Inst)
Travel, Accommodations, Expenses: EMD Serono
Other Relationship: Prime Oncology, MCM Education, OncLive, Aptitude Health, Cardinal Health
Uncompensated Relationships: Verastem
Roisin E. O'Cearbhaill
Honoraria: GlaxoSmithKline, Curio Science, MJH Life Sciences, MJH/PER
Consulting or Advisory Role: Seagan, Aptitude Health, Fresenius Kabi, GlaxoSmithKline, Bayer, Regeneron, Carina Biotech, Immunogen, R-Pharm, GOG Foundation, Miltenyi Biotec, 2seventy bio
Research Funding: Juno Therapeutics (Inst), Sellas Life Sciences (Inst), Ludwig Institute for Cancer Research (Inst), TapImmune Inc (Inst), TCR2 Therapeutics (Inst), Regeneron (Inst), Genmab (Inst), Atara Biotherapeutics (Inst), GlaxoSmithKline (Inst), AstraZeneca/Merck (Inst), Syndax (Inst), Genentech (Inst), Kite/Gilead (Inst), GOG Foundation (Inst), Merck/Genentech (Inst), Acrivon Therapeutics (Inst), Bristol Myers Squibb (Inst)
Travel, Accommodations, Expenses: Hitech Health, Gathering Around Cancer, Society of Gynecologic Oncology
Other Relationship: JAMA Oncology
Uncompensated Relationships: Children's Medical Research Foundation (CMRF)
Open Payments Link: https://openpaymentsdata.cms.gov/physician/785539
Kara Long Roche
Stock and Other Ownership Interests: Doximity
Yukio Sonoda
Patents, Royalties, Other Intellectual Property: Pending patent for a surgical instrument (uterine manipulator)
Ginger J. Gardner
Consulting or Advisory Role: Eisai
Dennis S. Chi
Employment: Memorial Sloan-Kettering Cancer Center
Stock and Other Ownership Interests: Doximity
Honoraria: Biom'Up, AstraZeneca, UpToDate
Consulting or Advisory Role: Verthermia, Biom'up, Apyx Medical
Travel, Accommodations, Expenses: Biom'Up
Maria I. Carlo
Honoraria: OncLive/MJH Life Sciences
Other Relationship: Prostate Cancer Foundation, Robert Wood Johnson Foundation
Yonina R. Murciano-Goroff
Employment: Memorial Sloan-Kettering Cancer Center
Honoraria: Virology Education, Amgen
Research Funding: Loxo/Lilly (Inst), Elucida Oncology (Inst), Taiho Oncology (Inst), Hengrui Pharmaceutical (Inst), Jiangsu Hengrui Pharmaceuticals (Inst), Luzsana Biotechnology (Inst), Endeavor BioMedicines (Inst), Conquer Cancer, The ASCO Foundation, National Cancer Institute, Druckenmiller Center for Lung Cancer Research, Andrew Sabin Family Foundation, The Society of Memorial Sloan Kettering, Mirati Therapeutics (Inst), Loxo Oncology/Eli Lilly (Inst)
Patents, Royalties, Other Intellectual Property: Wolters Kluwer, Rutgers University Press
Travel, Accommodations, Expenses: AstraZeneca, Loxo Oncology/Eli Lilly
Other Relationship: Endeavor Biomedicines
Mark E. Robson
Consulting or Advisory Role: Change HealthCare
Research Funding: AstraZeneca (Inst), Pfizer (Inst), Merck (Inst), Zenith Pharmaceuticals (Inst)
Travel, Accommodations, Expenses: AstraZeneca
Other Relationship: Research to Practice, Clinical Care Options, Physicans' Education Resource, Pfizer, MyMedEd, Clinical Education Alliance, MJH Healthcare Holdings, LLC, AstraZeneca
Uncompensated Relationships: Merck, Pfizer, Artios, Tempus, Zenith Pharmaceuticals, GlaxoSmithKline
Open Payments Link: https://openpaymentsdata.cms.gov/physician/612669
Michael F. Berger
This author is a member of the JCO Precision Oncology Editorial Board. Journal policy recused the author from having any role in the peer review of this manuscript.
Consulting or Advisory Role: Lilly, PetDx, AstraZeneca
Patents, Royalties, Other Intellectual Property: Provisional patent pending for Systems and Methods for Detecting Cancer via cfDNA Screening
Nadeem R. Abu-Rustum
Honoraria: NCCN
Research Funding: Grail (Inst)
Carol L. Brown
Uncompensated Relationships: American College of Surgeons, President's Cancer Panel
Kenneth Offit
Patents, Royalties, Other Intellectual Property: Patent pending on therapeutic applications of targeting ERCC3 mutations in cancer. Diagnosis & treatment of ercc3-mutant cancer US20210137850A1
Other Relationship: AnaNeo Therapeutics
Carol Aghajanian
Leadership: GOG Foundation
Consulting or Advisory Role: Blueprint Medicines
Research Funding: Genentech/Roche (Inst), AbbVie (Inst), Clovis Oncology (Inst), AstraZeneca (Inst)
Britta Weigelt
Stock and Other Ownership Interests: Repare Therapeutics
Consulting or Advisory Role: Volition RX, Paige, Goldman Sachs, Repare Therapeutics, Bain Capital Life Sciences, SAGA Diagnostics
Research Funding: Repare Therapeutics
Zsofia K. Stadler
This author is an Associate Editor for JCO Precision Oncology. Journal policy recused the author from having any role in the peer review of this manuscript.
Consulting or Advisory Role: Adverum, Neurogene, Genentech/Roche, Regeneron, Outlook Therapeutics, Optos, Novartis
Ying L. Liu
Research Funding: AstraZeneca (Inst), Tesaro/GSK (Inst), Repare Therapeutics (Inst)
Travel, Accommodations, Expenses: AstraZeneca
No other potential conflicts of interest were reported.
REFERENCES
- 1. Norquist BM, Harrell MI, Brady MF, et al. Inherited mutations in women with ovarian carcinoma. JAMA Oncol. 2016;2:482–490. doi: 10.1001/jamaoncol.2015.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 3. Konstantinopoulos PA, Norquist B, Lacchetti C, et al. Germline and somatic tumor testing in epithelial ovarian cancer: ASCO guideline. J Clin Oncol. 2020;38:1222–1245. doi: 10.1200/JCO.19.02960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pennington KP, Swisher EM. Hereditary ovarian cancer: Beyond the usual suspects. Gynecol Oncol. 2012;124:347–353. doi: 10.1016/j.ygyno.2011.12.415. [DOI] [PubMed] [Google Scholar]
- 5. Walsh T, Casadei S, Lee MK, et al. Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc Natl Acad Sci. 2011;108:18032–18037. doi: 10.1073/pnas.1115052108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hampel H, Bennett RL, Buchanan A, et al. A practice guideline from the American College of Medical Genetics and Genomics and the National Society of Genetic Counselors: Referral indications for cancer predisposition assessment. Genet Med. 2015;17:70–87. doi: 10.1038/gim.2014.147. [DOI] [PubMed] [Google Scholar]
- 7. Lancaster JM, Powell CB, Chen L-M, et al. Society of Gynecologic Oncology statement on risk assessment for inherited gynecologic cancer predispositions. Gynecol Oncol. 2015;136:3–7. doi: 10.1016/j.ygyno.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 8. The American College of Obstetricians and Gynecologists ACOG Practice Bulletin No. 103: Hereditary breast and ovarian cancer syndrome. Obstet Gynecol. 2009;113:957–966. doi: 10.1097/AOG.0b013e3181a106d4. [DOI] [PubMed] [Google Scholar]
- 9.National Comprehensive Cancer Network Genetic/familial high-risk assessment: Breast, ovarian, and pancreatic, version 2.2022, 2022.
- 10. Lin J, Sharaf RN, Saganty R, et al. Achieving universal genetic assessment for women with ovarian cancer: Are we there yet? A systematic review and meta-analysis. Gynecol Oncol. 2021;162:506–516. doi: 10.1016/j.ygyno.2021.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chapman-Davis E, Zhou ZN, Fields JC, et al. Racial and ethnic disparities in genetic testing at a hereditary breast and ovarian cancer center. J Gen Intern Med. 2021;36:35–42. doi: 10.1007/s11606-020-06064-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Moslehi R, Chu W, Karlan B, et al. BRCA1 and BRCA2 mutation analysis of 208 Ashkenazi Jewish women with ovarian cancer. Am J Hum Genet. 2000;66:1259–1272. doi: 10.1086/302853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Modan B, Hartge P, Hirsh-Yechezkel G, et al. Parity, oral contraceptives, and the risk of ovarian cancer among carriers and noncarriers of a BRCA1 or BRCA2 mutation. N Engl J Med. 2001;345:235–240. doi: 10.1056/NEJM200107263450401. [DOI] [PubMed] [Google Scholar]
- 14. Kauff ND, Perez-Segura P, Robson ME, et al. Incidence of non-founder BRCA1 and BRCA2 mutations in high risk Ashkenazi breast and ovarian cancer families. J Med Genet. 2002;39:611–614. doi: 10.1136/jmg.39.8.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hinchcliff EM, Bednar EM, Lu KH, et al. Disparities in gynecologic cancer genetics evaluation. Gynecol Oncol. 2019;153:184–191. doi: 10.1016/j.ygyno.2019.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kurian AW. BRCA1 and BRCA2 mutations across race and ethnicity: Distribution and clinical implications. Curr Opin Obstet Gynecol. 2010;22:72–78. doi: 10.1097/GCO.0b013e328332dca3. [DOI] [PubMed] [Google Scholar]
- 17. Kurian AW, Ward KC, Howlader N, et al. Genetic testing and results in a population-based cohort of breast cancer patients and ovarian cancer patients. J Clin Oncol. 2019;37:1305–1315. doi: 10.1200/JCO.18.01854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kurian AW, Ward KC, Abrahamse P, et al. Time trends in receipt of germline genetic testing and results for women diagnosed with breast cancer or ovarian cancer, 2012-2019. J Clin Oncol. 2021;39:1631–1640. doi: 10.1200/JCO.20.02785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Alhuqail A-J, Alzahrani A, Almubarak H, et al. High prevalence of deleterious BRCA1 and BRCA2 germline mutations in arab breast and ovarian cancer patients. Breast Cancer Res Treat. 2018;168:695–702. doi: 10.1007/s10549-017-4635-4. [DOI] [PubMed] [Google Scholar]
- 20. Wu X, Wu L, Kong B, et al. The first nationwide multicenter prevalence study of germline BRCA1 and BRCA2 mutations in Chinese ovarian cancer patients. Int J Gynecol Cancer. 2017;27:1650–1657. doi: 10.1097/IGC.0000000000001065. [DOI] [PubMed] [Google Scholar]
- 21. Kim SI, Lee M, Kim HS, et al. Germline and somatic BRCA1/2 gene mutational status and clinical outcomes in epithelial peritoneal, ovarian, and fallopian tube cancer: Over a decade of experience in a single institution in Korea. Cancer Res Treat. 2020;52:1229–1241. doi: 10.4143/crt.2020.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-integrated mutation profiling of actionable cancer targets (MSK-IMPACT): A hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015;17:251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schrader KA, Cheng DT, Joseph V, et al. Germline variants in targeted tumor sequencing using matched normal DNA. JAMA Oncol. 2016;2:104–111. doi: 10.1001/jamaoncol.2015.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu YL, Maio A, Kemel Y, et al. Disparities in cancer genetics care by race/ethnicity among pan-cancer patients with pathogenic germline variants. Cancer. 2022;128:3870–3879. doi: 10.1002/cncr.34434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Srinivasan P, Bandlamudi C, Jonsson P, et al. The context-specific role of germline pathogenicity in tumorigenesis. Nat Genet. 2021;53:1577–1585. doi: 10.1038/s41588-021-00949-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mandelker D, Zhang L, Kemel Y, et al. Mutation detection in patients with advanced cancer by universal sequencing of cancer-related genes in tumor and normal DNA vs guideline-based germline testing. JAMA. 2017;318:825–835. doi: 10.1001/jama.2017.11137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Domchek SM, Robson ME. Update on genetic testing in gynecologic cancer. J Clin Oncol. 2019;37:2501–2509. doi: 10.1200/JCO.19.00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu YL, Breen K, Catchings A, et al. Risk-reducing bilateral salpingo-oophorectomy for ovarian cancer: A review and clinical guide for hereditary predisposition genes. JCO Oncol Pract. 2022;18:201–209. doi: 10.1200/OP.21.00382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Breen KE, Tuman M, Bertelsen CE, et al. Factors influencing patient preferences for telehealth cancer genetic counseling during the COVID-19 pandemic. JCO Oncol Pract. 2022;18:e462–e471. doi: 10.1200/OP.21.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Arora K, Tran TN, Kemel Y, et al. Genetic ancestry correlates with somatic differences in a real-world clinical cancer sequencing cohort. Cancer Discov. 2022;12:2552–2565. doi: 10.1158/2159-8290.CD-22-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Auton A, Abecasis GR, Altshuler DM, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.R Core Team: R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria, 2021. https://www.R-project.org
- 35. Somasegar S, Weiss AS, Norquist BM, et al. Germline mutations in Black patients with ovarian, fallopian tube and primary peritoneal carcinomas. Gynecol Oncol. 2021;163:130–133. doi: 10.1016/j.ygyno.2021.08.017. [DOI] [PubMed] [Google Scholar]
- 36. George SHL, Donenberg T, Alexis C, et al. Gene sequencing for pathogenic variants among adults with breast and ovarian cancer in the caribbean. JAMA Netw Open. 2021;4:e210307. doi: 10.1001/jamanetworkopen.2021.0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ademuyiwa FO, Salyer P, Tao Y, et al. Genetic counseling and testing in African American patients with breast cancer: A nationwide survey of US breast oncologists. J Clin Oncol. 2021;39:4020–4028. doi: 10.1200/JCO.21.01426. [DOI] [PubMed] [Google Scholar]
- 38. Levy DE, Byfield SD, Comstock CB, et al. Underutilization of BRCA1/2 testing to guide breast cancer treatment: Black and Hispanic women particularly at risk. Genet Med. 2011;13:349–355. doi: 10.1097/GIM.0b013e3182091ba4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Beard C, Monohan K, Cicciarelli L, et al. Mainstream genetic testing for breast cancer patients: Early experiences from the Parkville Familial Cancer Centre. Eur J Hum Genet. 2021;29:872–880. doi: 10.1038/s41431-021-00848-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Muessig KR, Zepp JM, Keast E, et al. Retrospective assessment of barriers and access to genetic services for hereditary cancer syndromes in an integrated health care delivery system. Hered Cancer Clin Pract. 2022;20:7. doi: 10.1186/s13053-022-00213-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Montes de Oca MK, Wilson LE, Previs RA, et al. Healthcare access dimensions and guideline-concordant ovarian cancer treatment: SEER-Medicare analysis of the ORCHiD study. J Natl Compr Cancer Netw. 2022;20:1255–1266.e11. doi: 10.6004/jnccn.2022.7055. [DOI] [PubMed] [Google Scholar]
- 42. Frey MK, Finch A, Kulkarni A, et al. Genetic testing for all: Overcoming disparities in ovarian cancer genetic testing. Am Soc Clin Oncol Educ Book. 2022;42:1–12. doi: 10.1200/EDBK_350292. [DOI] [PubMed] [Google Scholar]
- 43. Hann KEJ, Freeman M, Fraser L, et al. Awareness, knowledge, perceptions, and attitudes towards genetic testing for cancer risk among ethnic minority groups: A systematic review. BMC Public Health. 2017;17:503. doi: 10.1186/s12889-017-4375-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mata DA, Rotenstein LS, Ramos MA, et al. Disparities according to genetic ancestry in the use of precision oncology assays. N Engl J Med. 2023;388:281–283. doi: 10.1056/NEJMc2213457. [DOI] [PubMed] [Google Scholar]
- 45. Deyrup A, Graves JL. Racial biology and medical misconceptions. N Engl J Med. 2022;386:501–503. doi: 10.1056/NEJMp2116224. [DOI] [PubMed] [Google Scholar]
- 46. Kaufman JS, Merckx J, Cooper RS. Use of racial and ethnic categories in medical testing and diagnosis: Primum non nocere. Clin Chem. 2021;67:1456–1465. doi: 10.1093/clinchem/hvab164. [DOI] [PubMed] [Google Scholar]
- 47. Qian AS, Schiaffino MK, Nalawade V, et al. Disparities in telemedicine during COVID-19. Cancer Med. 2022;11:1192–1201. doi: 10.1002/cam4.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Eberly LA, Kallan MJ, Julien HM, et al. Patient characteristics associated with telemedicine access for primary and specialty ambulatory care during the COVID-19 pandemic. JAMA Netw open. 2020;3:e2031640. doi: 10.1001/jamanetworkopen.2020.31640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen EM, Andoh JE, Nwanyanwu K. Socioeconomic and demographic disparities in the use of telemedicine for ophthalmic care during the COVID-19 pandemic. Ophthalmology. 2022;129:15–25. doi: 10.1016/j.ophtha.2021.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]