Abstract
Aging and age‐associated disease are a major medical and societal burden in need of effective treatments. Cellular reprogramming is a biological process capable of modulating cell fate and cellular age. Harnessing the rejuvenating benefits without altering cell identity via partial cellular reprogramming has emerged as a novel translational strategy with therapeutic potential and strong commercial interests. Here, we explore the aging‐related benefits of partial cellular reprogramming while examining limitations and future directions for the field.
Keywords: aging, aging hallmarks, lifespan, partial cellular reprogramming, rejuvenation
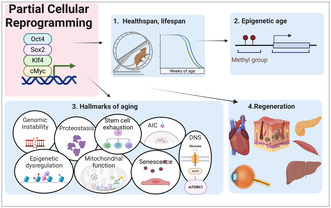
Partial cellular reprogramming is able to improve or restore multiple age‐related phenotypes including lifespan, healthspan, epigenetic age, aging hallmarks, and tissue regeneration.
Abbreviations
- AAD
age‐associated diseases
- HGPS
hutchinson gilford progeria syndrome
- IVPR
in vivo partial reprogramming
- OSKM
Oct4, Sox2, Klf4, cMyc
1. AGE‐RELATED DISEASES
Significant increases in lifespan worldwide have shifted demographics dramatically over the last 100 years to the point; there is now an estimated 125 million people 80 years and older. Elderly individuals over 65 years currently outnumber those under 5 years old, demonstrating a “graying” of the human population (Partridge et al., 2020). The unintended consequence of this increase in lifespan has been the exponential growth of age‐associated disease that accompany aging (Garmany et al., 2021). Although more people can now expect to live longer, their healthspan, also known as healthy life expectancy, shows little or no improvement based on morbidity and disability (Crimmins, 2015). The top four age‐associated diseases (AADs) including cardiovascular diseases, cancer, respiratory diseases, and diabetes now account for more than 2/3 of all mortality annually worldwide with related health care costs in the trillions of dollars (Beard et al., 2016; Nugent, 2019). On top of this, in 2015, the World Health Organization announced the rise of AADs as a global epidemic (Kassebaum et al., 2017). It is now clear that significant improvement to healthspan and treatment of aging and AADs represents an important medical and societal goal but success requires a clear understanding of the aging processes involved.
2. AGING PHENOMENA
Ordinarily, molecular and cellular aging experienced by living organisms is counteracted through evolutionarily derived processes including for example the DNA damage repair response or autophagy (Ferrucci et al., 2010; Tian et al., 2017). The process of aging results in a progressive failure in these critical repair and homeostatic mechanisms subsequently inducing a decline in physiological function, increased susceptibility to disease, and eventual mortality (Rando & Chang, 2012). In order to describe and quantify this process, several categories of aging drivers, the so‐called hallmarks of aging, have been identified including epigenetic alterations, genomic instability, loss of proteostasis, telomere attrition, deregulated nutrient sensing, mitochondrial dysfunction, cellular senescence, stem cell exhaustion, and altered intercellular communication (López‐Otín et al., 2013). Recently, additional hallmarks have been proposed including cell size alterations, stochastic non‐enzymatic modification of long lived molecules, and dysbiosis (Davies et al., 2022; Fedintsev & Moskalev, 2020; López‐Otín et al., 2023). Inclusion as a hallmark of aging requires that the phenotype manifests during aging, and if manipulated can accelerate aging or slow down its progress. Although these age sensitive phenotypes are important readouts to enable identification of novel interventions, the underlying processes, mechanisms, and potential interrelationships to which they contribute are still under investigation (Keshavarz et al., 2022).
August Weismann first noted over a hundred years ago the immortal germline exists in a privileged state, uncoupled from aging, in order to transmit hereditary information across generations while somatic cells lack this selective pressure and are therefore finite (Larocca et al., 2021; West et al., 2019). In this line, the somatic restriction theory of aging proposes that aging in multicellular animals is a result of the confinement of pluripotent traits, like unlimited self‐renewal, to the germ line (West et al., 2019). It is not until these cells exit the pluripotent state and differentiate into somatic cells that aging ensues, resulting in replicative senescence (Hayflick & Moorhead, 1961). In multicellular organisms, the global aging process is currently proposed to be a multi‐causal and interrelated phenomenon9. Indeed, the majority of programmed and damage‐related aging theories are not mutually exclusive and are often proposed to overlap by modern theorists (Bjorksten, 1968; Gladyshev, 2016; Harman, 1992; Jin, 2010; Kane & Sinclair, 2019; Kirkwood, 2005; Szilard, 1959; Weismann, 1893; West et al., 2019; Williams, 2001).
3. CELLULAR REPROGRAMMING
In the mid‐20th century, the epigenetic landscape model was first proposed to describe the unidirectional process of development following oocyte fertilization (Waddington, 1957). John Gurdon overturned this concept when he first demonstrated nuclear reprogramming. Specifically, he showed a cell fate could be reversed into a viable zygote via the transfer of an adult intestinal epithelial cell nucleus into an oocyte using somatic cell nuclear transfer (SCNT; Gurdon, 1962). This process of converting a somatic cell into a dedifferentiated toti‐ or pluripotent cell is called cellular reprogramming (Saitou et al., 2012). During development, cellular reprogramming occurs following fertilization and is driven by epigenetic remodeling, reorganization of the chromatin state, extinguishment of the somatic identity, and finally expression of the pluripotent gene regulatory network (Koche et al., 2011; Morgan et al., 2005; Polo et al., 2012). Importantly, as cell identity is reset, all age accrued defects are restored and a minimal biological age, referred to as ground zero, is reached during early embryogenesis (Kerepesi et al., 2021; Seisenberger et al., 2013). Confirmation of these profound observations are demonstrated in cloning experiments. Specifically, clones produced with adult somatic cells via SCNT are born young, with restored telomere length, live a full lifespan, and can be serially cloned further, suggesting complete rejuvenation has occurred (Lanza et al., 2000; Sinclair et al., 2016; Wakayama et al., 2013; Wilmut et al., 1997). Still, SCNT is technically challenging and reliant on endogenous maternal factors and chromatin modifying enzymes within the egg to replace the somatic proteins and machinery to induce reprogramming (Jullien et al., 2011). Importantly, this process was streamlined in 2006 when Shinya Yamanaka's laboratory screened 24 genes associated with embryonic stem cell identity and discovered four transcription factors; Oct4, Sox2, Klf4, and cMyc (OSKM) sufficient to induce pluripotency in mouse fibroblasts (Takahashi & Yamanaka, 2006). Together, Gurdon and Yamanaka were awarded the 2012 Nobel Prize for the discovery of cellular reprogramming and thus significantly advanced research into the field of regenerative medicine.
Initial investigations into the age‐related effects of cellular reprogramming using pluripotent transcription factors in vitro demonstrated a restoration of telomere size, gene expression profile, oxidative stress, and mitochondrial metabolism in both human centenarian fibroblasts and senescent cells (Lapasset et al., 2011; Lee et al., 2020; Yagi et al., 2012). Still, important questions remained, namely, can the restoration of cellular age be separated from the loss of cell fate. A solution to this problem, first proposed by Singh and Zacouto (2010), presented the concepts of partial reprogramming and partial cloning in which reprogramming would be induced only until the cells had undergone signs of rejuvenation. These ideas were later confirmed by Manukyan and Singh (2014) when they first observed epigenetic rejuvenation in senescent human fibroblasts following 9 days of reprogramming without loss of cell identity. Specifically, 5 factor expression (Oct 4, Sox 2, Klf4, cMyc, and Lin28) via a piggybac transposase vector restored the mobility of heterochromatin protein 1 (HP1) to young levels without progression through a dedifferentiated state (Manukyan & Singh, 2014). Follow‐up work has since demonstrated that the dose of reprogramming is linked to the amount of age rejuvenation. Notably, when aged human cells are reprogrammed for up to 20 days, the epi‐age based on the DNA methylation clock is completely reverted to zero suggesting a dose dependent effect (Olova et al., 2019). By demonstrating that the loss of cell identity due to cellular reprogramming could be separated from the impacts on aging, the next stage was set for translational investigations in vivo.
4. IN VIVO PARTIAL REPROGRAMMING
In vivo cellular reprogramming via OSKM expression provides an important strategy to investigate cellular reprogramming based phenomena, including regeneration and aging, within a biologically relevant multicellular organism. Initial attempts at in vivo reprogramming using a mouse model with a doxycycline inducible OSKM transgene cassette generated pluripotent cells capable of forming the three germ layers and mouse chimeras with germline contribution. At the same time, full pluripotency induction in vivo proved problematic due to dysplastic cell proliferation and teratoma formation across multiple organs (Abad et al., 2013; Ohnishi et al., 2014). To avoid any deleterious oncogenic changes associated with acquisition of pluripotency, a novel treatment strategy was developed using a short term and cyclic induction protocol called in vivo partial reprogramming (IVPR). IVPR in mice successfully prevented loss of cell identity while still extending lifespan and ameliorating multiple hallmarks of aging (Ocampo, Reddy, Martinez‐Redondo, et al., 2016). Since this important proof of principle, multiple laboratories have further demonstrated that IVPR can increase lifespan and restore tissue dysfunction across multiple tissues and organs including the dentate gyrus, optic nerve, liver, skeletal muscle, skin, intervertebral disc, and heart, although with some limitations and hurdles to yet be overcome (Figure 1; Alle et al., 2022; Chen et al., 2021; Cheng et al., 2022; de Lázaro et al., 2019; Hishida et al., 2022; Lu et al., 2020; Rodríguez‐Matellán et al., 2020). In order to validate future clinical development, here we review the key partial reprogramming studies, both in vivo and in vitro, in detail as related to lifespan, epigenetic age, hallmarks of aging, and regenerative effects (Figure 2).
FIGURE 1.
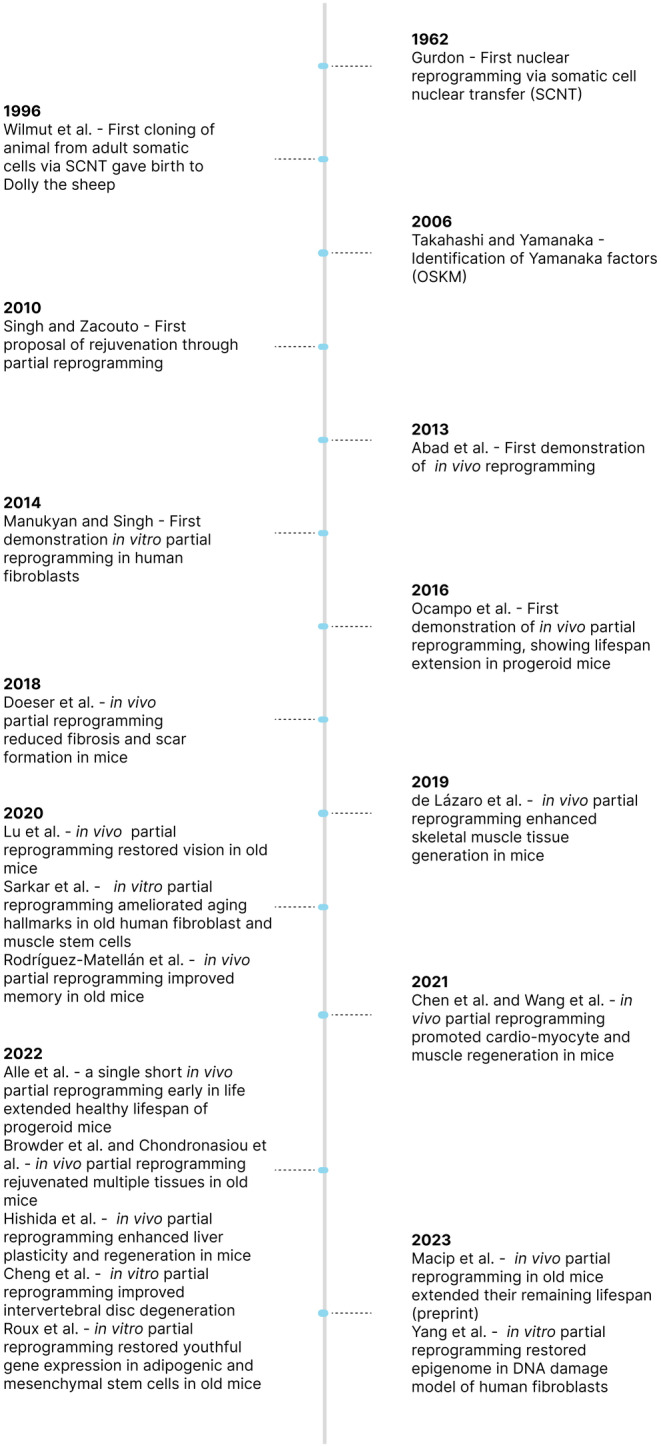
Timeline of key partial reprogramming studies. Key historical milestones in the development of partial reprogramming for rejuvenation beginning with John Gurdon first demonstrating nuclear reprogramming is possible using the somatic cell nuclear transfer technique.
FIGURE 2.
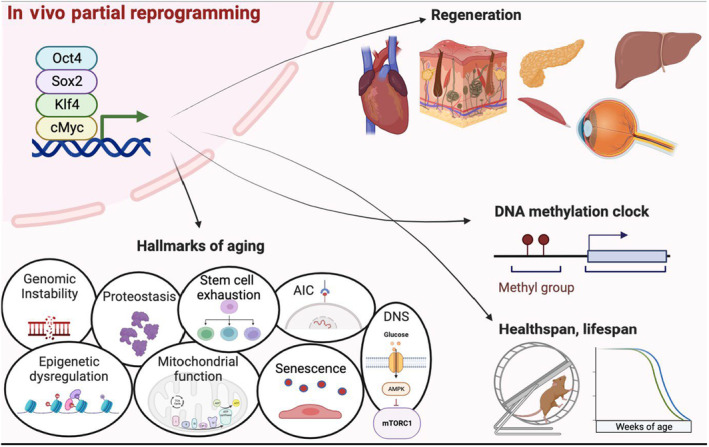
Multiparameter rejuvenation following in vivo partial reprogramming (IVPR). In vivo partial reprogramming drives rejuvenated phenotypes in tissue repair and function, hallmarks of aging, DNAm clock, and extends healthspan and lifespan. (created with biorender.com).
5. LIFESPAN EXTENSION
Lifespan extension is one of the key readouts to demonstrate the effects of anti‐aging interventions based on chronological time. Several groups have tested the benefits of IVPR on lifespan with varying results (Table 1). At this point, two groups have observed an increase in median and maximum lifespan following IVPR in a short‐lived LAKI progeria model of premature aging. The LAKI mouse is an accelerated model of aging with a truncated lamin A protein, called progerin, which is responsible for Hutchinson Gilford progeria syndrome in humans (HGPS; Osorio et al., 2011).
TABLE 1.
Lifespan.
| Model | Partial reprogramming protocol | Lifespan increase | Ref |
|---|---|---|---|
| LAKI (homozygous) 4F mice | 2 days on, 5 days off, starting at 2 months old until death |
33% median 18% maximum |
Ocampo, Reddy, Martinez‐Redondo, et al. (2016) |
| LAKI (heterozygous) 4F mice | 2 days on, 5 days off, starting at 2 months old until death |
26% median 16% maximum |
Alle et al. (2022) |
| LAKI (heterozygous) 4F mice | Single short induction, starting at 2 months old for 2.5 weeks |
No change median 18% maximum |
Alle et al. (2022) |
| Wildtype 4F mice | Single short induction, starting at 2 months old for 2.5 weeks |
No change median No change maximum 13% increase third quartile |
Alle et al. (2022) |
| Wildtype mice | 1 week on, 1 week off, starting at 124 weeks old until death | 109% increase in remaining median lifespan | Macip et al. (2023) |
Abbreviation: 4F, 4 factors.
In 2016, seminal work from Ocampo, Reddy, Martinez‐Redondo, et al. (2016) first demonstrated that IVPR, via a cyclic induction of OSKM (2 days on and 5 days off), could extend median and maximum lifespan in a homozygous LAKI mouse model by 33% and 18%, respectively. Alternatively, the Lemaitre group tested the effects of IVPR in both wildtype mice and heterozygous LAKI mice with a less severe progeroid phenotype as an intermediary model (Alle et al., 2022). First, they explored dosage effects in the LAKI mouse using either a high dose of doxycycline cyclically or a low dose continuously beginning at 2 months of age. Both approaches resulted in a similar increase in median and maximal lifespan. At the same time, they documented that a single short burst of OSKM induction with an intermediate dose of doxycycline in 8‐week‐old LAKI mice significantly increased the third quartile lifespan by almost 10 weeks and maximal lifespan by 11 weeks. Potentially, this single burst of IVPR was effective due to a deeper alteration of the epigenome that can occur after 2.5 weeks of reprogramming (Alle et al., 2022; Gill et al., 2022). In line with this observation, Macip et al. (2023) observed a significant effect of partial reprogramming on late stage median lifespan when applied to aged wild type mice. Using an adeno‐associated virus 9 (AAV9) vector to systemically deliver OSK to 124‐week‐old mice, the group showed that IVPR could extend the remaining median lifespan of treated mice by 109% (Macip et al., 2023).
These improvements to the lifespan of homozygotic and heterozygotic LAKI mice along with wild type mice following IVPR are important demonstrations of reprogramming induced rejuvenation. Notably, the benefits are most robust in the most severe premature aging model perhaps due to a rescue of the complex pathology specific to the homozygous mutant (Ocampo, Reddy, & Belmonte, 2016). A key limitation to the current research using transgenic mouse models is the variability in the expression of the reprogramming factors, and therefore the total amount of reprogramming, among different cells, tissue types, and organs (Alle et al., 2021; Chondronasiou et al., 2022; Parras et al., 2022). In this line, it has been demonstrated that different cell types reprogram at different rates and follow specific reprogramming trajectories (Hussein et al., 2014; Nefzger et al., 2017; Tonge et al., 2014; Zunder et al., 2015). For this reason, whole‐body OSKM induction is confounded by excess reprogramming and loss of function in some cells while under reprogramming without rejuvenation in others. This could impact lifespan studies in several ways, for example, a loss of digestive function and therefore an unintended caloric restriction effect could overstate reprogramming effects. In fact, improved whole‐body reprogramming models are an ongoing pursuit and at the same time have shifted emphasis into tissue‐specific models of reprogramming, targeted reprogramming, or enhanced reprogramming methods (Chen et al., 2021; de Lázaro et al., 2019; Hishida et al., 2022; Lu et al., 2020; Parras et al., 2022; Roux et al., 2022; Wang et al., 2021).
6. EPIGENETIC AGE
The ability to determine epigenetic age based on gain or loss of methylation at specific CpG loci is a recent important discovery for accurately measuring biological age in cells and tissues (Hannum et al., 2013; Horvath, 2013). In vitro experiments demonstrated the epigenetic age of fully reprogrammed aged cells can be completely restored to zero (Horvath, 2013; Olova et al., 2019). In addition, several groups (Table 2) have now shown an improvement to epigenetic age following partial reprogramming in vivo and in vitro using mouse and human cells (Browder et al., 2022; Chondronasiou et al., 2022; Gill et al., 2022; Lu et al., 2020; Sarkar et al., 2020; Yang et al., 2023). Recently, the Belmonte group observed that in 15‐month‐old wildtype mice given 7 months of IVPR, 2 days on and 5 days off, the epigenetic clock was improved in the kidney and skin but not the liver, lung, muscle, or spleen (Browder et al., 2022). Simultaneously, they observed no improvement to the epigenetic clock in 25‐month‐old wildtype mice following IVPR for 1 month within those tissues. In line with this, Chondronasiou et al. (2022) observed rejuvenation of differentially methylated promoters and enhancers in wildtype 55‐week‐old mice pancreas, liver, and the spleen, after only 1 week of IVPR. Lu et al. (2020) performed an intra‐ocular injection with an AAV vector containing 3 factors; Oct 4, Sox 2, and Klf4 (OSK) and observed a decrease in DNA methylation age based on ribosomal DNA in retinal ganglion cells from 12‐month‐old wildtype mice after 4 weeks of continuous reprogramming. Using a unique double strand break accelerated aging mouse model, Yang et al. (2023) also showed this 3 factor AAV vector reversed epigenetic age in fibroblasts up to 57% based on four different epigenetic clocks. The Sebastiano group pioneered a novel IVPR technique using modified RNA (modRNA) of 6 pluripotency factors including Oct 4, Sox 2, cMyc, Klf 4, Lin 28, and Nanog. Transient reprogramming with this 6F modRNA cocktail for 4 days in vitro restored epigenetic and transcriptomic age in old human fibroblasts and endothelial cells in vitro (Sarkar et al., 2020). Gill et al. (2022) observed aged human fibroblasts transduced with a 4F cassette underwent a morphological shift associated with a mesenchymal to epithelial transition and restored epigenetic age during reprogramming. Specifically, they observed 13 days of reprogramming reduced median DNA methylation clock age by approximately 30 years and was superior to 10, 15, or 17 days of reprogramming (Gill et al., 2022).
TABLE 2.
Epigenetic age.
| Model | Partial reprogramming protocol | Clock type | Epigenetic age | Ref |
|---|---|---|---|---|
| Human Fb (60–90 yo) | modRNA delivery of OSKMLN, in vitro for 4 days | Horvath pan‐tissue clock. Skin and blood clock | −1.84 years, −1.07 years | Sarkar et al. (2020) |
| Human EC (50–65 yo) | mRNA delivery of OSKMLN, in vitro for 4 days | Horvath pan‐tissue clock. Skin and blood clock | −4.94 years, −1.62 years | Sarkar et al. (2020) |
| RGC 12‐month‐old WT mice | intra‐orbital injection of AAV OSK, in vivo for 4 weeks | Ribosomal DNA methylation clock | Significant reduction | Lu et al. (2020) |
| MEFs DNA damage model | In vitro AAV OSK, induced for 10 days | 4 clocks: Thompson, Petkovich, Meer, and Stubbs | Up to 57% decrease | Yang et al. (2023) |
| Human Fb (38–53 yo) | lentiviral delivery of OSKM, in vitro for 13 days | Horvath pan‐tissue clock, Skin and blood clock | −30 years | Gill et al. (2022) |
| Mouse WT 4F 12‐month‐old, 15‐month‐old | In vivo OSKM, 2 days on 5 days off, for 10 months | Lifespan Uber Correlation clock | Significant reduction in skin and kidney no changes in liver, lung, muscle, spleen | Browder et al. (2022) |
| Mouse WT 4F 25‐month‐old | In vivo OSKM, 2 days on 5 days off, for 1 months | Lifespan Uber Correlation clock | No change | Browder et al. (2022) |
| Mouse WT 4F 55‐week‐old | In vivo low dose OSKM, 1 week burst | No clock. Methylation profile | Restored in pancreas, liver, and spleen | Chondronasiou et al. (2022) |
Abbreviations: AAV, adeno‐associated virus; C, chondrocyte; EC, endothelial cell; Fb, fibroblast; LV, lentivirus; NPC, nucleus pulposus cells; OSKMLN, Oct4, Sox2, Klf4, cMyc, Lin28, Nanog; RGC, retinal ganglion cell; SC, satellite cells.
In summary, multiple laboratories have shown reprogramming can induce reversal of epigenetic aging in both mice and human cells, in vivo and in vitro, and across multiple cell types although with some caveats. Browder et al. (2022) for example saw improvement in several tissues, but not all, following IVPR in wildtype mice even though expression of the 4Fs was similar. Browder also did not see reduction to the epigenetic clock in aged mice after 1 month of IVPR (Browder et al., 2022). At the same time, in vitro data demonstrated improvements to the epigenetic age with partial reprogramming but individual results varied and in several specific instances no restoration was observed (Sarkar et al., 2020). Possible reasons for these discrepancies could be due to the low efficiency of reprogramming, irreversibly aged phenotypes in specific tissues, variance in rates of epigenetic aging, or inherent differences in reprogramming potential due to cell type of origin and environmental cues (Browder et al., 2022; Gill et al., 2022; Nefzger et al., 2017). Notably, the results produced by an epigenetic clock analysis are strongly dependent on the tissue type and the clock selected, suggesting the alterations to cellular identity during reprogramming could be confounding at times (Liu et al., 2020).
7. AGING HALLMARKS
Currently, partial reprogramming has been demonstrated to effectively ameliorate 8 of 9 hallmarks of aging, although specific results vary depending on the assay, protocol, and model used (Table 3). Interestingly, some hallmarks of aging in different models are improved following a short treatment of 2 or 4 days while others require a more substantial reprogramming duration. As a key driver of aging, it is notable that DNA damage shows improvement following short‐term reprogramming in fibroblasts of wildtype and progeria mouse models along with nucleus pulposus cells from naturally aged mice (Cheng et al., 2022; Chondronasiou et al., 2022; Ocampo, Reddy, Martinez‐Redondo, et al., 2016). Epigenetic dysregulation is a critical driver of aging according to the Information Loss Theory of aging (Kane & Sinclair, 2019). Cellular reprogramming has been shown to improve a variety of epigenetic markers including the DNA methylome, H3K9me3, H4K20me3, H3k36me2, HP1a, and nuclear abnormalities (Browder et al., 2022; Cheng et al., 2022; Chondronasiou et al., 2022; Lu et al., 2020; Manukyan & Singh, 2014; Ocampo, Reddy, Martinez‐Redondo, et al., 2016; Rodríguez‐Matellán et al., 2020; Sarkar et al., 2020; Yang et al., 2023). Improvements to proteostasis after partial reprogramming have been observed based on increased autophagic flux or proteasomal enzyme activity in wildtype mouse and human fibroblasts (Alle et al., 2021; Sarkar et al., 2020). Mitochondrial dysregulation appears restored after partial cellular reprogramming in human and mouse models based on decreased levels of reactive oxygen species, mitochondrial membrane function, and mitochondrial number (Alle et al., 2021; Cheng et al., 2022; Sarkar et al., 2020). Intercellular communication alterations that occur with aging have been ameliorated following reprogramming as demonstrated by decreased inflammaging markers and senescent associated secretory proteins in human chondrocytes and mouse wildtype and progeria models (Alle et al., 2021; Cheng et al., 2022; Chondronasiou et al., 2022; Doeser et al., 2018; Ocampo, Reddy, Martinez‐Redondo, et al., 2016; Sarkar et al., 2020). A decrease in senescence has been demonstrated following short‐term reprogramming based on lower levels of senescence‐associated beta‐Galactosidase (SABG) staining and decreased expression of p16, p21, and p53 in both human and mouse cell types (Alle et al., 2022; Browder et al., 2022; Cheng et al., 2022; Ocampo, Reddy, Martinez‐Redondo, et al., 2016; Sarkar et al., 2020; Wang et al., 2021). Notably, improvement to telomere attrition is a hallmark of aging that remains unimproved with IVPR. This is expected since the increase in telomere length does not appear until late stage reprogramming during conversion to pluripotency (Marión et al., 2017).
TABLE 3.
Effects of partial reprogramming on aging hallmarks.
| Model | Protocol | Cell, tissue | Hallmark of aging ameliorated | Ref | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ref tissue gen | Epi | Prot | Mito | Nutr | Sen | Stem | Inter | ||||
| WT mouse | OSKML, 9 days in vitro | Senescent Fb | Yes | Manukyan and Singh (2014) | |||||||
| 4F LAKI Mouse | In vitro, 2+4 day | Fb | Yes | Yes | – | Yes | – | Yes | – | – | Ocampo, Reddy, Martinez‐Redondo, et al. (2016) |
| 6F A Human | In vitro, 4 days | EC, Fb, Ch, SC | Yes | Yes | Yes | Yes | Yes | yes | Yes | Yes | Sarkar et al. (2020) |
| 4F Mouse | In vitro, 2+4 day | NPC, Intervertebral disc | Yes | Yes | – | Yes | Yes | Yes | Yes | – | Cheng et al. (2022) |
| 4F topical Mouse | In vivo, pre‐, post‐injury | Skin | – | – | – | – | – | No | – | – | Doeser et al. (2018) |
| 4F Mouse | In vivo, 4m | Brain | – | Yes | – | – | – | – | – | – | Rodríguez‐Matellán et al. (2020) |
| 3F AAV Mouse | In vivo | RGC, Optic nerve, | – | Yes | – | – | – | – | – | – | Lu et al. (2020) |
| 4F LV Human | In vitro, 10‐13 day | Fb | – | Yes | – | – | – | – | Yes | Roux et al. (2022) | |
| 4F Mouse | In vivo, cyclic, 2.5 week | Multi‐tissue | – | Yes | – | Yes | Yes | Yes | Yes | Yes | Browder et al. (2022) |
| 4F Mouse | In vivo, 1 week | Multi‐tissue | – | Yes | – | – | Yes | – | – | Yes | Alle et al. (2022) |
| 4F Mouse | In vivo, 1 week | Liver, spleen, pancreas | No | Yes | – | – | – | No | – | – | Chondronasiou et al. (2022) |
| 3F AAV mouse (ICE) | In vivo, in vitro | Fb, liver, kidney | – | Yes | – | – | – | – | – | – | Yang et al. (2023) |
Abbreviations: AAV, adeno‐associated virus; Ch, chondrocyte; EC, endothelial cell; Epi, epigenetic dysregulation; Gen, genomic instability; ICE, inducible changes to the epigenome; Inter, altered intercellular communication. Fb, fibroblast; LV, lentivirus; Mito, mitochondrial dysfunction; NPC, nucleus pulposus cells; Nutri, dysregulated nutrient sensing; Prot, loss of proteostasis; RGC, retinal ganglion cell; SC, satellite cells; Sen, senescence; Stem, stem cell exhaustion.
There are a few exceptions that indicate the amelioration of aging hallmarks via IVPR are context dependent. For example, senescent marker genes were not improved after 1 week of IVPR nor in a skin wound healing model (Chondronasiou et al., 2022; Doeser et al., 2018). In another example, ROS levels were improved in human fibroblasts and chondrocytes but not in endothelial cells (Sarkar et al., 2020). A key technical limitation to some HOA readouts is due to a reprogramming induced decrease in cell and nucleus size (Samavarchi‐Tehrani et al., 2010; van den Hurk et al., 2016). For certain epigenetic changes, this is potentially a confounding factor for observed signal increases based solely on immunostaining and likely requires further validation (Mattout et al., 2011). Generally, these data support a case for IVPR being an effective and reproducible intervention for the majority of key hallmarks of aging with future work needed to address the associated mechanisms that drive the rejuvenated phenotypes.
8. REGENERATION AND FUNCTION
Impaired tissue regeneration and fibrosis are well defined aging characteristics with a large clinical burden (Rando & Jones, 2021). In early mammalian development, scarless wound healing occurs until the late gestational fetus and then is lost (Bullard et al., 2003). Interestingly, in some highly regenerative vertebrates including salamanders and fish, wound repair follows a process of dedifferentiation and proliferation mimicking aspects of cellular reprogramming (Jopling et al., 2010; Wang & Simon, 2016). Currently, improvements to proliferation, regeneration, and/or fibrosis following reprogramming have been observed in multiple organs and associated cell types including the skin, muscle, optic nerve, heart, liver, lung, brain, and pancreas (Table 4; Browder et al., 2022; Chen et al., 2021; de Lázaro et al., 2019; Doeser et al., 2018; Guo et al., 2018; Hishida et al., 2022; Lu et al., 2020; Ocampo, Reddy, Martinez‐Redondo, et al., 2016; Sarkar et al., 2020; Seo et al., 2016; Wang et al., 2021).
TABLE 4.
Effects of partial reprogramming on regeneration.
| Tissue | Regeneration | Phenotypes | Function | Model | Protocol | Ref |
|---|---|---|---|---|---|---|
| Muscle | Yes | Increase Pax7 cells | – | 4F WT Mouse | Intramuscular injection | Ocampo, Reddy, Martinez‐Redondo, et al. (2016) |
| Muscle | Yes | Increase proliferation | Increase tetanic force | 6F mRNA Ms, Hu | Ex vivo SC | Sarkar et al. (2020) |
| Muscle | Yes | Increase proliferation, decrease fibrosis | Non‐significant increase to tetanic force | 4F plasmid Mouse | In situ | de Lázaro et al. (2019) |
| Muscle | Yes | Increase Pax7 cells | Grip strength in 2‐4mo | 4F myofiber specific Mouse | In vivo, pre‐, post‐ injury | Wang et al. (2021) |
| Skin | Yes | Increase proliferation, decrease fibrosis | – | 4F Mouse | Topical | Doeser et al. (2018) |
| Skin | Yes | Decrease fibrosis, Increase epidermal proliferation and thickness | – | 4F Mouse | In vivo | Browder et al. (2022) |
| Liver | Yes | Increased proliferation | Increased survival after lethal dose acetaminophen | 4F liver specific mouse | In vivo | Hishida et al., 2022 |
| Optic nerve | Yes | Enhanced RGC survival | Improved vision in 11mo after 4wk trx | OSK AAV Mouse | In vivo, | Lu et al. (2020) |
| Heart | Yes | Increased proliferation, reduced scar | Improved contractile force pre and during injury | 4F heart specific Mouse | In vivo, 1 week | Chen et al. (2021) |
| Lung | Yes | Reduced fibrosis in bleomycin model of pulmonary fibrosis | Improved mechanical function | 4F alveolar epithelial cells | Ex vivo, 2 week on, 2 week off | Guo et al. (2018) |
| Brain | Yes | Increase proliferation, neovascularization | Restored motor function in cerebral ischemia model | 4F, mouse | In situ induction | Seo et al. (2016) |
| Pancreas | Yes | Increase pancreatic islets | Improved glucose tolerance test | 4F WT Mouse | In vivo | Ocampo, Reddy, and Belmonte (2016), Ocampo, Reddy, Martinez‐Redondo, et al. (2016) |
Abbreviation: RGC, retinal ganglion cell.
As an example, loss of muscle mass, decline in function, and decreased muscle stem cell potency is observed with aging (Blau et al., 2015; Naranjo et al., 2017). Although four groups have demonstrated partial reprogramming can improve muscle regeneration, specific results varied depending on the method of induction and model tested (de Lázaro et al., 2019; Ocampo, Reddy, Martinez‐Redondo, et al., 2016; Sarkar et al., 2020; Wang et al., 2021). For example, Sarkar et al. demonstrated enhanced regeneration and muscle stem cell potency following cardiotoxin injury and transplantation with partially reprogrammed satellite cells from aged humans (60–80 year) or aged mice (20–24 month; Sarkar et al., 2020). At the same time, muscle function based on tetanic force was also restored with this 6 factor modRNA ex vivo method in aged mouse cells (20–24 month; Sarkar et al., 2020). In contrast, muscle regeneration and function was improved in a myofiber specific but not a satellite cell specific transgenic model of 4 factor IVPR in adult wildtype mice (12–15 month; Wang et al., 2021). Previously, it was shown that IVPR with a systemic 4 factor cassette enhanced muscle regeneration and satellite cell proliferation in adult wildtype mice (12 month) after 3 weeks of cyclic induction via intramuscular injection of doxycycline (Ocampo, Reddy, Martinez‐Redondo, et al., 2016). Oppositely, whole‐body cyclic IVPR for 7 months did not significantly improve regeneration in 22 month wild type mice perhaps due to low factor expression (Browder et al., 2022). Interestingly, in situ reprogramming with a 4 factor plasmid vector enhanced regeneration and proliferation with decreased fibrosis following muscle injury to young adult mice (8 week) but cell specific contributions were undetermined (de Lázaro et al., 2019). These results support IVPR as a viable therapeutic route to accelerate muscle repair and function albeit with method and model dependent effects.
Alternatively, skin reprogramming is another example where multiple groups have demonstrated improvements to regeneration and age‐related phenotypes (Alle et al., 2022; Browder et al., 2022; Doeser et al., 2018; Ocampo, Reddy, Martinez‐Redondo, et al., 2016). An Increase in rate of proliferation in skin epidermal cells has been observed following 4F IVPR in both aged wildtype and LAKI mice (Browder et al., 2022; Ocampo, Reddy, Martinez‐Redondo, et al., 2016). Associated increases to epidermal thickness were noted in wild type (female) along with increased dermal thickness in the progeria model (Browder et al., 2022; Ocampo, Reddy, Martinez‐Redondo, et al., 2016). At the same time, improved regeneration capacity by in situ IVPR via local application of doxycycline to an excisional wound model demonstrated decelerated wound closure, reduced fibrosis and scarring, along with a decrease in fibrosis associated TGFb1 expression (Doeser et al., 2018). In wild type aged mice, long‐term IVPR reduced fibrosis and increased epidermal cell proliferation in an excisional wound model, in line with previous results (Browder et al., 2022).
Other individual examples of enhanced regeneration have been noted in several tissues including the liver, optic nerve, heart, and pancreas (Chen et al., 2021; Hishida et al., 2022; Lu et al., 2020; Ocampo, Reddy, Martinez‐Redondo, et al., 2016). In the liver, regenerative capacity and hepatocyte proliferation is enhanced with IVPR following a normally lethal dose of acetaminophen (Hishida et al., 2022). Using an AAV2 vector to deliver OSK directly to retinal ganglion cells (RGCs), Lu et al. (2020) first demonstrated IVPR was able to regenerate the optic nerve and improve RGC survival in a crush injury model. Short‐term cardiomyocyte (CM) specific OSKM expression significantly improved heart regeneration and stimulated CM proliferation following myocardial infarction (Chen et al., 2021). Lastly, in wild‐type aged mice, the size of pancreatic isles and glucose tolerance were increased with IVPR following injury with the beta cell toxin SZT (Ocampo, Reddy, Martinez‐Redondo, et al., 2016). Importantly, improvements to tissue functionality have also been demonstrated in the intervertebral disc, dentate gyrus, kidney, bone, and lung (Alle et al., 2022; Cheng et al., 2022; Rodríguez‐Matellán et al., 2020). In sum, the data show a promising, albeit, context and tissue dependent regenerative effect of partial reprogramming likely confounded at times due to multiple issues including timing, delivery, and dosage along with cell type specific effects of the reprogramming trajectory.
9. COMMERCIAL LANDSCAPE
Novel therapeutics able to successfully treat age‐associated chronic disease while restoring the underlying aged phenotype hold immense value for human society. It is estimated that increasing healthspan in the elderly by just 1 year is valued at over 38 trillion dollars (Scott et al., 2021). In the last few years, numerous companies have been established to explore the use of partial reprogramming for the treatment of AADs (Table 5). Among them, Altos Labs stands out with a $3 billion investment from tech investor Yuri Milner and Amazon founder Jeff Bezos, easily making it the largest biotech startup investment in history (Fleming, 2023). Despite the size of investment and number of companies involved, IVPR is still far from being commercially viable. All of the companies listed in Table 5 are currently in preclinical stage, and only Turn Biotechnologies has announced near‐term clinical trials. As the Yamanaka factors are pro‐oncogenic with an associated risk of tumor formation, their therapeutic applications are currently limited and will require improvements to partial reprogramming methods and safety prior to significant expansion of the commercial sector (Klimczak, 2015).
TABLE 5.
Partial reprogramming in commercial development.
| Company | Founded | Funding |
|---|---|---|
| Calico | 2013 | 3.5B a |
| Shift Bioscience | 2017 | <10M |
| Rejuvenate Bio | 2017 | 17M a |
| Turn Biotechnologies | 2018 | <20M |
| Reverse Bioengineering (AgeX subsidiary) | 2019 | 63M a |
| Iduna Therapeutics (Life Biosciences subsidiary) | 2020 | 206M a |
| YouthBio Therapeutics | 2020 | <10M |
| Retro Biosciences | 2021 | 180M a |
| Altos Labs | 2021 | 3B |
| New Limit | 2021 | 105M |
Subsidiary/working on other programs besides reprogramming.
10. LIMITATIONS AND FUTURE PERSPECTIVES FOR IVPR
Multiple publications on partial reprogramming have confirmed a wide variety of restorative effects on lifespan, epigenetic age, aging hallmarks, and tissue regeneration, yet hurdles to successful clinical translation remain (Goya et al., 2018; Singh & Zhakupova, 2022). In order to specifically address these hurdles, such as teratoma formation or context dependent effects, a better understanding of the mechanisms and control over the reprogramming process combined with technical advances in delivery and targeted expression are required.
Developmental models both highlight and provide some insights to address these limitations. For example, during development, cellular reprogramming in mice is a direct and efficient process with a well‐orchestrated gain of pluripotency after only 3.5 days (Paranjpe & Veenstra, 2015). Similarly, SCNT also follows a relatively direct reprogramming process driven by maternal factors (Han et al., 2015; Jullien et al., 2011). Alternately, current transcription factor based reprogramming rejuvenation methods follows a stochastic process that suffer from the inability to universally reprogram target cells along the same trajectory at the same rate and to the same degree in a timely manner. Further delineation of the barriers specific to subpopulations within a tissue that result in this asynchronous process with altered reprogramming potential are needed. One way to address this problem was demonstrated by the Brunet laboratory in 2019. Using a single‐cell RNA‐seq and multi‐omic profiling approach, they identified key contributors to reprogramming variability in aged fibroblasts such as inflammatory cytokines (Mahmoudi et al., 2019). At the same time, groups have used a bioinformatic approach to mine reprogramming data sets in order to enable identification of shared gene targets and enriched cellular pathways that could be utilized to develop enhanced reprogramming (Kareta, 2016; Knyazer et al., 2021). In this line, Calico recently performed a screening of reprogramming factors to better characterize reprogramming induced rejuvenation while simultaneously identifying a novel multipotent strategy derived from amphibian regeneration and the MSX1 transcription factor (Roux et al., 2022). Performing screenings via RNA interference, CRISPR knockdowns, or reprogramming compound libraries also present a viable way forward to better understand and optimize cellular reprogramming (Lee et al., 2019). Together, these examples demonstrate the innovative routes and improved methods of reprogramming needed in order to allow the development of future clinical translation.
An optimized and synchronous process of cellular reprogramming will only exacerbate the need for improved tunability and delivery to avoid deleterious effects. Consequently, cell‐specific reprogramming via targetable delivery vectors might be the next step for a safe and effective clinical therapy. Currently, several groups have demonstrated translational methods of ectopic factor expression utilizing either naked plasmid DNA in situ, localized AAV injection, or ex vivo modified mRNA transfection to induce rejuvenation (de Lázaro et al., 2019; Lu et al., 2020; Sarkar et al., 2020). Although promising, in certain cases these methods can suffer from poor delivery efficiency, immunogenicity, and poor specificity. Clinical use of gene therapy has surged over the last couple decades100. Interesting improvements in next generation gene insertion strategies and RNA gated payload expression could be potential game changers for the field, although they also require improvements to delivery (Burgess, 2022; Jiang et al., 2022; Srivastava & DeWitt, 2016; Yarnall et al., 2022). In several cases, groups have observed effective delivery using a variety of different nanoparticles for in vivo reprogramming (Ofenbauer & Tursun, 2019; Romanazzo et al., 2020). Preliminary work from Huang et al., Tang et al., and our laboratory (under review) have demonstrated treatment with reprogramming small molecules can enhance regeneration in vivo, induce multiparameter rejuvenation in vitro, and extend lifespan in C. elegans (Huang et al., 2018; Schoenfeldt et al., 2022; Tang & Cheng, 2017). For these reasons, chemical induced partial reprogramming (ciPR) could be a particularly intriguing method of IVPR, especially if combined with prodrug targeting strategies such as antibody directed enzyme prodrug therapy (ADEPT; Sharma & Bagshawe, 2017; Tang & Cheng, 2017).
In summary, the partial reprogramming data reviewed here present a currently limited rejuvenation strategy capable of ameliorating aging phenotypes, increasing healthspan or lifespan, restoring epigenetic age, and improving tissue regeneration and tissue function (Tables 1, 2, 3 and 4). As detailed above, specific cases only show limited or no improvements across similar aging readouts and models, potentially due to the stochastic nature of reprogramming along with cell intrinsic and microenvironment dependent effects. To address these lingering questions, improvements in mechanistic understanding, delivery, safety, tunability, and efficiency will be required. Though the challenges for this endeavor are considerable, collective improvements could pave the way to a future where effective treatment of AADs is attainable and hopefully no longer a serious burden to society.
AUTHOR CONTRIBUTIONS
P.T.P., A.N., and A.O. conceived the idea for the review. P.T.P. and A.N. wrote the manuscript and created the figures and tables with contributions and editing from A.O.
ACKNOWLEDGEMENTS
We would like to acknowledge all members of the Ocampo laboratory for support during writing.
FUNDING INFORMATION
No funding information provided.
CONFLICT OF INTEREST STATEMENT
A.O. is co‐founder and shareholder of EPITERNA SA (non‐financial interests) and co‐founder of Longevity Consultancy Group (non‐financial interests). The rest of the authors declare no competing interests.
Paine, P. T. , Nguyen, A. , & Ocampo, A. (2024). Partial cellular reprogramming: A deep dive into an emerging rejuvenation technology. Aging Cell, 23, e14039. 10.1111/acel.14039
REFERENCES
- Abad, M. , Mosteiro, L. , Pantoja, C. , Cañamero, M. , Rayon, T. , Ors, I. , Graña, O. , Megías, D. , Domínguez, O. , Martínez, D. , Manzanares, M. , Ortega, S. , & Serrano, M. (2013). Reprogramming in vivo produces teratomas and iPS cells with totipotency features. Nature, 502(7471), 340–345. 10.1038/nature12586 [DOI] [PubMed] [Google Scholar]
- Alle, Q. , Le Borgne, E. , Bensadoun, P. , Lemey, C. , Béchir, N. , Gabanou, M. , Estermann, F. , Bertrand‐Gaday, C. , Pessemesse, L. , Toupet, K. , Desprat, R. , Vialaret, J. , Hirtz, C. , Noël, D. , Jorgensen, C. , Casas, F. , Milhavet, O. , & Lemaitre, J.‐M. (2022). A single short reprogramming early in life initiates and propagates an epigenetically related mechanism improving fitness and promoting an increased healthy lifespan. Aging Cell, 21(11), e13714. 10.1111/acel.13714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alle, Q. , Le Borgne, E. , Milhavet, O. , & Lemaitre, J.‐M. (2021). Reprogramming: Emerging strategies to rejuvenate aging cells and tissues. International Journal of Molecular Sciences, 22(8), 3990. 10.3390/ijms22083990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beard, J. R. , Officer, A. , de Carvalho, I. A. , Sadana, R. , Pot, A. M. , Michel, J.‐P. , Lloyd‐Sherlock, P. , Epping‐Jordan, J. E. , Peeters, G. M. E. E. G. , Mahanani, W. R. , Thiyagarajan, J. A. , & Chatterji, S. (2016). The world report on ageing and health: A policy framework for healthy ageing. Lancet, 387(10033), 2145–2154. 10.1016/S0140-6736(15)00516-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorksten, J. (1968). The Crosslinkage theory of aging. Journal of the American Geriatrics Society, 16(4), 408–427. 10.1111/j.1532-5415.1968.tb02821.x [DOI] [PubMed] [Google Scholar]
- Blau, H. M. , Cosgrove, B. D. , & Ho, A. T. V. (2015). The central role of muscle stem cells in regenerative failure with aging. Nature Medicine, 21(8), 854–862. 10.1038/nm.3918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browder, K. C. , Reddy, P. , Yamamoto, M. , Haghani, A. , Guillen, I. G. , Sahu, S. , Wang, C. , Luque, Y. , Prieto, J. , Shi, L. , Shojima, K. , Hishida, T. , Lai, Z. , Li, Q. , Choudhury, F. K. , Wong, W. R. , Liang, Y. , Sangaraju, D. , Sandoval, W. , … Izpisua Belmonte, J. C. (2022). In vivo partial reprogramming alters age‐associated molecular changes during physiological aging in mice. Nature Aging, 2(3), 243–253. 10.1038/s43587-022-00183-2 [DOI] [PubMed] [Google Scholar]
- Bullard, K. M. , Longaker, M. T. , & Lorenz, H. P. (2003). Fetal wound healing: Current biology. World Journal of Surgery, 27(1), 54–61. 10.1007/s00268-002-6737-2 [DOI] [PubMed] [Google Scholar]
- Burgess, D. J. (2022). RADARs and READRs for programmable RNA sensing. Nature Reviews. Genetics, 23(12), 711. 10.1038/s41576-022-00548-8 [DOI] [PubMed] [Google Scholar]
- Chen, Y. , Lüttmann, F. F. , Schoger, E. , Schöler, H. R. , Zelarayán, L. C. , Kim, K.‐P. , Haigh, J. J. , Kim, J. , & Braun, T. (2021). Reversible reprogramming of cardiomyocytes to a fetal state drives heart regeneration in mice. Science, 373(6562), 1537–1540. 10.1126/science.abg5159 [DOI] [PubMed] [Google Scholar]
- Cheng, F. , Wang, C. , Ji, Y. , Yang, B. , Shu, J. , Shi, K. , Wang, L. , Wang, S. , Zhang, Y. , Huang, X. , Zhou, X. , Xia, K. , Liang, C. , Chen, Q. , & Li, F. (2022). Partial reprogramming strategy for intervertebral disc rejuvenation by activating energy switch. Aging Cell, 21(4), e13577. 10.1111/acel.13577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chondronasiou, D. , Gill, D. , Mosteiro, L. , Urdinguio, R. G. , Berenguer‐Llergo, A. , Aguilera, M. , Durand, S. , Aprahamian, F. , Nirmalathasan, N. , Abad, M. , Martin‐Herranz, D. E. , Stephan‐Otto Attolini, C. , Prats, N. , Kroemer, G. , Fraga, M. F. , Reik, W. , & Serrano, M. (2022). Multi‐omic rejuvenation of naturally aged tissues by a single cycle of transient reprogramming. Aging Cell, 21(3), e13578. 10.1111/acel.13578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crimmins, E. M. (2015). Lifespan and Healthspan: Past, present, and promise. The Gerontologist, 55(6), 901–911. 10.1093/geront/gnv130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, D. M. , van den Handel, K. , Bharadwaj, S. , & Lengefeld, J. (2022). Cellular enlargement—A new hallmark of aging? Frontiers in Cell and Developmental Biology, 10, 1036602. 10.3389/fcell.2022.1036602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lázaro, I. , Yilmazer, A. , Nam, Y. , Qubisi, S. , Razak, F. M. A. , Degens, H. , Cossu, G. , & Kostarelos, K. (2019). Non‐viral, tumor‐free induction of transient cell reprogramming in mouse skeletal muscle to enhance tissue regeneration. Molecular Therapy, 27(1), 59–75. 10.1016/j.ymthe.2018.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doeser, M. C. , Schöler, H. R. , & Wu, G. (2018). Reduction of fibrosis and scar formation by partial reprogramming in vivo. Stem Cells, 36(8), 1216–1225. 10.1002/stem.2842 [DOI] [PubMed] [Google Scholar]
- Fedintsev, A. , & Moskalev, A. (2020). Stochastic non‐enzymatic modification of long‐lived macromolecules—A missing hallmark of aging. Ageing Research Reviews, 62, 101097. 10.1016/j.arr.2020.101097 [DOI] [PubMed] [Google Scholar]
- Ferrucci, L. , Hesdorffer, C. , Bandinelli, S. , & Simonsick, E. (2010). Frailty as a nexus between the biology of aging, environmental conditions and clinical geriatrics. Public Health Reviews, 32, 475–488. 10.1007/BF03391612 [DOI] [Google Scholar]
- Fleming, S. (2023). Altos Labs' Mega‐Startup: Wretched Excess Or The Future Of Biotech…Or Both? Retrieved January 29, 2023, from https://www.forbes.com/sites/stanfleming/2022/03/09/alta‐labs‐mega‐startup‐wretched‐excess‐or‐the‐future‐of‐biotechor‐both/
- Garmany, A. , Yamada, S. , & Terzic, A. (2021). Longevity leap: Mind the healthspan gap. NPJ Regenerative Medicine, 6(1), 1–7. 10.1038/s41536-021-00169-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill, D. , Parry, A. , Santos, F. , Okkenhaug, H. , Todd, C. D. , Hernando‐Herraez, I. , Stubbs, T. M. , Milagre, I. , & Reik, W. (2022). Multi‐omic rejuvenation of human cells by maturation phase transient reprogramming. eLife, 11, e71624. 10.7554/eLife.71624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladyshev, V. N. (2016). Aging: Progressive decline in fitness due to the rising deleteriome adjusted by genetic, environmental, and stochastic processes. Aging Cell, 15(4), 594–602. 10.1111/acel.12480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goya, R. G. , Lehmann, M. , Chiavellini, P. , Canatelli‐Mallat, M. , Hereñú, C. B. , & Brown, O. A. (2018). Rejuvenation by cell reprogramming: A new horizon in gerontology. Stem Cell Research & Therapy, 9(1), 349. 10.1186/s13287-018-1075-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, L. , Karoubi, G. , Duchesneau, P. , Aoki, F. G. , Shutova, M. V. , Rogers, I. , Nagy, A. , & Waddell, T. K. (2018). Interrupted reprogramming of alveolar type II cells induces progenitor‐like cells that ameliorate pulmonary fibrosis. NPJ Regenerative Medicine, 3(1), 1–13. 10.1038/s41536-018-0052-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdon, J. B. (1962). Adult frogs derived from the nuclei of single somatic cells. Developmental Biology, 4(2), 256–273. 10.1016/0012-1606(62)90043-X [DOI] [PubMed] [Google Scholar]
- Han, F. , Li, X. , Song, D. , Jiang, S. , Xu, Q. , & Zhang, Y. (2015). SCNT versus iPSCs: Proteins and small molecules in reprogramming. International Journal of Developmental Biology, 59(4‐5‐6), 179–186. 10.1387/ijdb.150042fh [DOI] [PubMed] [Google Scholar]
- Hannum, G. , Guinney, J. , Zhao, L. , Zhang, L. , Hughes, G. , Sadda, S. , Klotzle, B. , Bibikova, M. , Fan, J. B. , Gao, Y. , Deconde, R. , Chen, M. , Rajapakse, I. , Friend, S. , Ideker, T. , & Zhang, K. (2013). Genome‐wide methylation profiles reveal quantitative views of human aging rates. Molecular Cell, 49(2), 359–367. 10.1016/j.molcel.2012.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harman, D. (1992). Free radical theory of aging. Mutation Research/DNAging, 275(3), 257–266. 10.1016/0921-8734(92)90030-S [DOI] [PubMed] [Google Scholar]
- Hayflick, L. , & Moorhead, P. S. (1961). The serial cultivation of human diploid cell strains. Experimental Cell Research, 25(3), 585–621. 10.1016/0014-4827(61)90192-6 [DOI] [PubMed] [Google Scholar]
- Hishida, T. , Yamamoto, M. , Hishida‐Nozaki, Y. , Shao, C. , Huang, L. , Wang, C. , Shojima, K. , Xue, Y. , Hang, Y. , Shokhirev, M. , Memczak, S. , Sahu, S. K. , Hatanaka, F. , Ros, R. R. , Maxwell, M. B. , Chavez, J. , Shao, Y. , Liao, H. K. , Martinez‐Redondo, P. , … Izpisua Belmonte, J. C. (2022). In vivo partial cellular reprogramming enhances liver plasticity and regeneration. Cell Reports, 39(4), 110730. 10.1016/j.celrep.2022.110730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath, S. (2013). DNA methylation age of human tissues and cell types. Genome Biology, 14(10), 3156. 10.1186/gb-2013-14-10-r115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, C. , Tu, W. , Fu, Y. , Wang, J. , & Xie, X. (2018). Chemical‐induced cardiac reprogramming in vivo. Cell Research, 28(6), 686–689. 10.1038/s41422-018-0036-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein, S. M. I. , Puri, M. C. , Tonge, P. D. , Benevento, M. , Corso, A. J. , Clancy, J. L. , Mosbergen, R. , Li, M. , Lee, D. S. , Cloonan, N. , Wood, D. L. , Munoz, J. , Middleton, R. , Korn, O. , Patel, H. R. , White, C. A. , Shin, J. Y. , Gauthier, M. E. , Cao, K. A. , … Nagy, A. (2014). Genome‐wide characterization of the routes to pluripotency. Nature, 516(7530), 198–206. 10.1038/nature14046 [DOI] [PubMed] [Google Scholar]
- Jiang, K. , Koob, J. , Chen, X. D. , Krajeski, R. N. , Zhang, Y. , Volf, V. , Zhou, W. , Sgrizzi, S. R. , Villiger, L. , Gootenberg, J. S. , Chen, F. , & Abudayyeh, O. O. (2022). Programmable eukaryotic protein synthesis with RNA sensors by harnessing ADAR. Nature Biotechnology, 41, 698–707. 10.1038/s41587-022-01534-5 [DOI] [PubMed] [Google Scholar]
- Jin, K. (2010). Modern biological theories of aging. Aging and Disease, 1(2), 72–74. [PMC free article] [PubMed] [Google Scholar]
- Jopling, C. , Sleep, E. , Raya, M. , Martí, M. , Raya, A. , & Belmonte, J. C. I. (2010). Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature, 464(7288), 606–609. 10.1038/nature08899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien, J. , Pasque, V. , Halley‐Stott, R. P. , Miyamoto, K. , & Gurdon, J. B. (2011). Mechanisms of nuclear reprogramming by eggs and oocytes: A deterministic process? Nature Reviews Molecular Cell Biology, 12(7), 453–459. 10.1038/nrm3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane, A. E. , & Sinclair, D. A. (2019). Epigenetic changes during aging and their reprogramming potential. Critical Reviews in Biochemistry and Molecular Biology, 54(1), 61–83. 10.1080/10409238.2019.1570075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kareta, M. S. (2016). Bioinformatic and genomic analyses of cellular reprogramming and direct lineage conversion. Current Pharmacology Reports, 2(3), 103–112. 10.1007/s40495-016-0054-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassebaum, N. J. , Smith, A. G. C. , Bernabé, E. , Fleming, T. D. , Reynolds, A. E. , Vos, T. , Murray, C. J. L. , Marcenes, W. , Yonemoto, N. , & GBD 2015 Oral Health Collaborators . (2017). Global, regional, and National Prevalence, incidence, and disability‐adjusted life years for Oral conditions for 195 countries, 1990–2015: A systematic analysis for the global burden of diseases, injuries, and risk factors. Journal of Dental Research, 96(4), 380–387. 10.1177/0022034517693566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerepesi, C. , Zhang, B. , Lee, S.‐G. , Trapp, A. , & Gladyshev, V. N. (2021). Epigenetic clocks reveal a rejuvenation event during embryogenesis followed by aging. Science Advances, 7(26), eabg6082. 10.1126/sciadv.abg6082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshavarz, M. , Xie, K. , Schaaf, K. , Bano, D. , & Ehninger, D. (2022). Targeting the “hallmarks of aging” to slow aging and treat age‐related disease: Fact or fiction? | molecular psychiatry. Molecular Psychiatry, 28, 242–255. 10.1038/s41380-022-01680-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkwood, T. B. L. (2005). Understanding the odd science of aging. Cell, 120(4), 437–447. 10.1016/j.cell.2005.01.027 [DOI] [PubMed] [Google Scholar]
- Klimczak, M. (2015). Oncogenesis and induced pluripotency – Commonalities of signalling pathways. Contemporary Oncology/Współczesna Onkologia, 2015(1), 16–21. 10.5114/wo.2014.47133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knyazer, A. , Bunu, G. , Toren, D. , Mracica, T. B. , Segev, Y. , Wolfson, M. , Muradian, K. K. , Tacutu, R. , & Fraifeld, V. E. (2021). Small molecules for cell reprogramming: A systems biology analysis. Aging (Albany NY), 13(24), 25739–25762. 10.18632/aging.203791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koche, R. P. , Smith, Z. D. , Adli, M. , Gu, H. , Ku, M. , Gnirke, A. , Bernstein, B. E. , & Meissner, A. (2011). Reprogramming factor expression initiates widespread targeted chromatin remodeling. Cell Stem Cell, 8(1), 96–105. 10.1016/j.stem.2010.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanza, R. P. , Cibelli, J. B. , Blackwell, C. , Cristofalo, V. J. , Francis, M. K. , Baerlocher, G. M. , Mak, J. , Schertzer, M. , Chavez, E. A. , Sawyer, N. , Lansdorp, P. M. , & West, M. D. (2000). Extension of cell life‐span and telomere length in animals cloned from senescent somatic cells. Science, 288(5466), 665–669. 10.1126/science.288.5466.665 [DOI] [PubMed] [Google Scholar]
- Lapasset, L. , Milhavet, O. , Prieur, A. , Besnard, E. , Babled, A. , Aït‐Hamou, N. , Leschik, J. , Pellestor, F. , Ramirez, J. M. , De Vos, J. , Lehmann, S. , & Lemaitre, J.‐M. (2011). Rejuvenating senescent and centenarian human cells by reprogramming through the pluripotent state. Genes & Development, 25(21), 2248–2253. 10.1101/gad.173922.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larocca, D. , Lee, J. , West, M. D. , Labat, I. , & Sternberg, H. (2021). No time to age: Uncoupling aging from chronological time. Genes, 12(5), 611. 10.3390/genes12050611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J. , Bignone, P. A. , Coles, L. S. , Liu, Y. , Snyder, E. , & Larocca, D. (2020). Induced pluripotency and spontaneous reversal of cellular aging in supercentenarian donor cells. Biochemical and Biophysical Research Communications, 525(3), 563–569. 10.1016/j.bbrc.2020.02.092 [DOI] [PubMed] [Google Scholar]
- Lee, J. , Park, Y.‐J. , & Jung, H. (2019). Protein kinases and their inhibitors in pluripotent stem cell fate regulation. Stem Cells International, 2019, e1569740. 10.1155/2019/1569740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z. , Leung, D. , Thrush, K. , Zhao, W. , Ratliff, S. , Tanaka, T. , Schmitz, L. L. , Smith, J. A. , Ferrucci, L. , & Levine, M. E. (2020). Underlying features of epigenetic aging clocks in vivo and in vitro. Aging Cell, 19(10), e13229. 10.1111/acel.13229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López‐Otín, C. , Blasco, M. A. , Partridge, L. , Serrano, M. , & Kroemer, G. (2013). The hallmarks of aging. Cell, 153(6), 1194–1217. 10.1016/j.cell.2013.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López‐Otín, C. , Blasco, M. A. , Partridge, L. , Serrano, M. , & Kroemer, G. (2023). Hallmarks of aging: An expanding universe. Cell, 186, 243–278. 10.1016/j.cell.2022.11.001 [DOI] [PubMed] [Google Scholar]
- Lu, Y. , Brommer, B. , Tian, X. , Krishnan, A. , Meer, M. , Wang, C. , Vera, D. L. , Zeng, Q. , Yu, D. , Bonkowski, M. S. , Yang, J. H. , Zhou, S. , Hoffmann, E. M. , Karg, M. M. , Schultz, M. B. , Kane, A. E. , Davidsohn, N. , Korobkina, E. , Chwalek, K. , … Sinclair, D. A. (2020). Reprogramming to recover youthful epigenetic information and restore vision. Nature, 588(7836), 124–129. 10.1038/s41586-020-2975-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macip, C. C. , Hasan, R. , Hoznek, V. , Kim, J. , Metzger, L. E. , Sethna, S. , & Davidsohn, N. (2023). Gene therapy mediated partial reprogramming extends lifespan and reverses age‐related changes in aged mice. bioRxiv. p. 2023.01.04.522507. 10.1101/2023.01.04.522507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoudi, S. , Mancini, E. , Xu, L. , Moore, A. , Jahanbani, F. , Hebestreit, K. , Srinivasan, R. , Li, X. , Devarajan, K. , Prélot, L. , Ang, C. E. , Shibuya, Y. , Benayoun, B. A. , Chang, A. L. S. , Wernig, M. , Wysocka, J. , Longaker, M. T. , Snyder, M. P. , & Brunet, A. (2019). Heterogeneity in old fibroblasts is linked to variability in reprogramming and wound healing. Nature, 574(7779), 553–558. 10.1038/s41586-019-1658-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manukyan, M. , & Singh, P. B. (2014). Epigenome rejuvenation: HP1β mobility as a measure of pluripotent and senescent chromatin ground states. Scientific Reports, 4(1), 4789. 10.1038/srep04789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marión, R. M. , López de Silanes, I. , Mosteiro, L. , Gamache, B. , Abad, M. , Guerra, C. , Megías, D. , Serrano, M. , & Blasco, M. A. (2017). Common telomere changes during in vivo reprogramming and early stages of tumorigenesis. Stem Cell Reports, 8(2), 460–475. 10.1016/j.stemcr.2017.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattout, A. , Biran, A. , & Meshorer, E. (2011). Global epigenetic changes during somatic cell reprogramming to iPS cells. Journal of Molecular Cell Biology, 3(6), 341–350. 10.1093/jmcb/mjr028 [DOI] [PubMed] [Google Scholar]
- Morgan, H. D. , Santos, F. , Green, K. , Dean, W. , & Reik, W. (2005). Epigenetic reprogramming in mammals. Human Molecular Genetics, 14(suppl_1), R47–R58. 10.1093/hmg/ddi114 [DOI] [PubMed] [Google Scholar]
- Naranjo, J. D. , Dziki, J. L. , & Badylak, S. F. (2017). Regenerative medicine approaches for age‐related muscle loss and sarcopenia: A mini‐review. Gerontology, 63(6), 580–589. 10.1159/000479278 [DOI] [PubMed] [Google Scholar]
- Nefzger, C. M. , Rossello, F. J. , Chen, J. , Liu, X. , Knaupp, A. S. , Firas, J. , Paynter, J. M. , Pflueger, J. , Buckberry, S. , Lim, S. M. , Williams, B. , Alaei, S. , Faye‐Chauhan, K. , Petretto, E. , Nilsson, S. K. , Lister, R. , Ramialison, M. , Powell, D. R. , Rackham, O. J. L. , & Polo, J. M. (2017). Cell type of origin dictates the route to pluripotency. Cell Reports, 21(10), 2649–2660. 10.1016/j.celrep.2017.11.029 [DOI] [PubMed] [Google Scholar]
- Nugent, R. (2019). Preventing and managing chronic diseases. BMJ, 364, l459. 10.1136/bmj.l459 [DOI] [PubMed] [Google Scholar]
- Ocampo, A. , Reddy, P. , & Belmonte, J. C. I. (2016). Anti‐aging strategies based on cellular reprogramming. Trends in Molecular Medicine, 22(8), 725–738. 10.1016/j.molmed.2016.06.005 [DOI] [PubMed] [Google Scholar]
- Ocampo, A. , Reddy, P. , Martinez‐Redondo, P. , Platero‐Luengo, A. , Hatanaka, F. , Hishida, T. , Li, M. , Lam, D. , Kurita, M. , Beyret, E. , Araoka, T. , Vazquez‐Ferrer, E. , Donoso, D. , Roman, J. L. , Xu, J. , Rodriguez Esteban, C. , Nuñez, G. , Nuñez Delicado, E. , Campistol, J. M. , … Izpisua Belmonte, J. C. (2016). In vivo amelioration of age‐associated hallmarks by partial reprogramming. Cell, 167(7), 1719.e12–1733.e12. 10.1016/j.cell.2016.11.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofenbauer, A. , & Tursun, B. (2019). Strategies for in vivo reprogramming. Current Opinion in Cell Biology, 61, 9–15. 10.1016/j.ceb.2019.06.002 [DOI] [PubMed] [Google Scholar]
- Ohnishi, K. , Semi, K. , Yamamoto, T. , Shimizu, M. , Tanaka, A. , Mitsunaga, K. , Okita, K. , Osafune, K. , Arioka, Y. , Maeda, T. , Soejima, H. , Moriwaki, H. , Yamanaka, S. , Woltjen, K. , & Yamada, Y. (2014). Premature termination of reprogramming in vivo leads to cancer development through altered epigenetic regulation. Cell, 156(4), 663–677. 10.1016/j.cell.2014.01.005 [DOI] [PubMed] [Google Scholar]
- Olova, N. , Simpson, D. J. , Marioni, R. E. , & Chandra, T. (2019). Partial reprogramming induces a steady decline in epigenetic age before loss of somatic identity. Aging Cell, 18(1), e12877. 10.1111/acel.12877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio, F. G. , Navarro, C. L. , Cadiñanos, J. , López‐Mejía, I. C. , Quirós, P. M. , Bartoli, C. , Rivera, J. , Tazi, J. , Guzmán, G. , Varela, I. , Depetris, D. , de Carlos, F. , Cobo, J. , Andrés, V. , De Sandre‐Giovannoli, A. , Freije, J. M. , Lévy, N. , & López‐Otín, C. (2011). Splicing‐directed therapy in a new mouse model of human accelerated aging. Science Translational Medicine, 3(106), 106ra107. 10.1126/scitranslmed.3002847 [DOI] [PubMed] [Google Scholar]
- Paranjpe, S. S. , & Veenstra, G. J. C. (2015). Establishing pluripotency in early development. Biochimica et Biophysica Acta, 1849(6), 626–636. 10.1016/j.bbagrm.2015.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parras, A. , Vílchez‐Acosta, A. , Desdín‐Micó, G. , Picó, S. , Mrabti, C. , Montenegro‐Borbolla, E. , Maroun, C. Y. , Haghani, A. , Brooke, R. , Del Carmen Maza, M. , Rechsteiner, C. , Battiston, F. , Branchina, C. , Perez, K. , Horvath, S. , Bertelli, C. , Sempoux, C. , & Ocampo, A. (2023). In vivo reprogramming leads to premature death linked to hepatic and intestinal failure. Nature Aging. 10.1038/s43587-023-00528-5 [DOI] [PubMed] [Google Scholar]
- Partridge, L. , Fuentealba, M. , & Kennedy, B. K. (2020). The quest to slow ageing through drug discovery | nature reviews drug discovery. Nature Reviews Drug Discovery, 19(8), 513–532. 10.1038/s41573-020-0067-7 [DOI] [PubMed] [Google Scholar]
- Polo, J. M. , Anderssen, E. , Walsh, R. M. , Schwarz, B. A. , Nefzger, C. M. , Lim, S. M. , Borkent, M. , Apostolou, E. , Alaei, S. , Cloutier, J. , Bar‐Nur, O. , Cheloufi, S. , Stadtfeld, M. , Figueroa, M. E. , Robinton, D. , Natesan, S. , Melnick, A. , Zhu, J. , Ramaswamy, S. , & Hochedlinger, K. (2012). A molecular roadmap of reprogramming somatic cells into iPS cells. Cell, 151(7), 1617–1632. 10.1016/j.cell.2012.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando, T. A. , & Chang, H. Y. (2012). Aging, rejuvenation, and epigenetic reprogramming: Resetting the aging clock. Cell, 148(1), 46–57. 10.1016/j.cell.2012.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando, T. A. , & Jones, D. L. (2021). Regeneration, rejuvenation, and replacement: Turning Back the clock on tissue aging. Cold Spring Harbor Perspectives in Biology, 13(9), a040907. 10.1101/cshperspect.a040907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez‐Matellán, A. , Alcazar, N. , Hernández, F. , Serrano, M. , & Ávila, J. (2020). In vivo reprogramming ameliorates aging features in dentate gyrus cells and improves memory in mice. Stem Cell Reports, 15(5), 1056–1066. 10.1016/j.stemcr.2020.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanazzo, S. , Lin, K. , Srivastava, P. , & Kilian, K. A. (2020). Targeting cell plasticity for regeneration: From in vitro to in vivo reprogramming. Advanced Drug Delivery Reviews, 161–162, 124–144. 10.1016/j.addr.2020.08.007 [DOI] [PubMed] [Google Scholar]
- Roux, A. E. , Zhang, C. , Paw, J. , Zavala‐Solorio, J. , Malahias, E. , Vijay, T. , Kolumam, G. , Kenyon, C. , & Kimmel, J. C. (2022). Diverse partial reprogramming strategies restore youthful gene expression and transiently suppress cell identity. Cell Systems, 13(7), 574.e11–587.e11. 10.1016/j.cels.2022.05.002 [DOI] [PubMed] [Google Scholar]
- Saitou, M. , Kagiwada, S. , & Kurimoto, K. (2012). Epigenetic reprogramming in mouse pre‐implantation development and primordial germ cells. Development, 139(1), 15–31. 10.1242/dev.050849 [DOI] [PubMed] [Google Scholar]
- Samavarchi‐Tehrani, P. , Golipour, A. , David, L. , Sung, H. , Beyer, T. A. , Datti, A. , Woltjen, K. , Nagy, A. , & Wrana, J. L. (2010). Functional genomics reveals a BMP‐driven mesenchymal‐to‐epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell, 7(1), 64–77. 10.1016/j.stem.2010.04.015 [DOI] [PubMed] [Google Scholar]
- Sarkar, T. J. , Quarta, M. , Mukherjee, S. , Colville, A. , Paine, P. , Doan, L. , Tran, C. M. , Chu, C. R. , Horvath, S. , Qi, L. S. , Bhutani, N. , Rando, T. A. , & Sebastiano, V. (2020). Transient non‐integrative expression of nuclear reprogramming factors promotes multifaceted amelioration of aging in human cells. Nature Communications, 11(1), 1545. 10.1038/s41467-020-15174-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfeldt, L. , Paine, P. T. , Kamaludeen, M. N. H. , Phelps, G. B. , Mrabti, C. , Perez, K. , & Ocampo, A. (2022). Chemical reprogramming ameliorates cellular hallmarks of aging and extends lifespan. bioRxiv. p. 2022.08.29.505222. 10.1101/2022.08.29.505222 [DOI] [Google Scholar]
- Scott, A. J. , Ellison, M. , & Sinclair, D. A. (2021). The economic value of targeting aging. Nature Aging, 1(7), 616–623. 10.1038/s43587-021-00080-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seisenberger, S. , Peat, J. R. , Hore, T. A. , Santos, F. , Dean, W. , & Reik, W. (2013). Reprogramming DNA methylation in the mammalian life cycle: Building and breaking epigenetic barriers. Philosophical Transactions of the Royal Society B: Biological Sciences, 368(1609), 20110330. 10.1098/rstb.2011.0330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, J. H. , Lee, M.‐Y. , Yu, J. H. , Kim, M.‐S. , Song, M. , Seo, C. H. , Kim, H. H. , & Cho, S.‐R. (2016). In situ pluripotency factor expression promotes functional recovery from cerebral ischemia. Molecular Therapy, 24(9), 1538–1549. 10.1038/mt.2016.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, S. K. , & Bagshawe, K. D. (2017). Translating antibody directed enzyme prodrug therapy (ADEPT) and prospects for combination. Expert Opinion on Biological Therapy, 17(1), 1–13. 10.1080/14712598.2017.1247802 [DOI] [PubMed] [Google Scholar]
- Sinclair, K. D. , Corr, S. A. , Gutierrez, C. G. , Fisher, P. A. , Lee, J.‐H. , Rathbone, A. J. , Choi, I. , Campbell, K. H. , & Gardner, D. S. (2016). Healthy ageing of cloned sheep. Nature Communications, 7(1), 12359. 10.1038/ncomms12359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, P. B. , & Zacouto, F. (2010). Nuclear reprogramming and epigenetic rejuvenation. Journal of Biosciences, 35(2), 315–319. 10.1007/s12038-010-0034-2 [DOI] [PubMed] [Google Scholar]
- Singh, P. B. , & Zhakupova, A. (2022). Age reprogramming: Cell rejuvenation by partial reprogramming. Development, 149(22), dev200755. 10.1242/dev.200755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava, D. , & DeWitt, N. (2016). In vivo cellular reprogramming: The next generation. Cell, 166(6), 1386–1396. 10.1016/j.cell.2016.08.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szilard, L. (1959). On the nature of the aging process. Proceedings of the National Academy of Sciences of the United States of America, 45(1), 30–45. 10.1073/pnas.45.1.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, K. , & Yamanaka, S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell, 126(4), 663–676. 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- Tang, Y. , & Cheng, L. (2017). Cocktail of chemical compounds robustly promoting cell reprogramming protects liver against acute injury. Protein & Cell, 8(4), 273–283. 10.1007/s13238-017-0373-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, X. , Seluanov, A. , & Gorbunova, V. (2017). Molecular mechanisms determining lifespan in short‐ and long‐lived species. Trends in Endocrinology & Metabolism, 28(10), 722–734. 10.1016/j.tem.2017.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonge, P. D. , Corso, A. J. , Monetti, C. , Hussein, S. M. I. , Puri, M. C. , Michael, I. P. , Michael, I. P. , Li, M. , Lee, D. S. , Mar, J. C. , Cloonan, N. , Wood, D. L. , Gauthier, M. E. , Korn, O. , Clancy, J. L. , Preiss, T. , Grimmond, S. M. , Shin, J. Y. , Seo, J. S. , … Nagy, A. (2014). Divergent reprogramming routes lead to alternative stem‐cell states. Nature, 516(7530), 192–197. 10.1038/nature14047 [DOI] [PubMed] [Google Scholar]
- van den Hurk, M. , Kenis, G. , Bardy, C. , van den Hove, D. L. , Gage, F. H. , Steinbusch, H. W. , & Rutten, B. P. (2016). Transcriptional and epigenetic mechanisms of cellular reprogramming to induced pluripotency. Epigenomics, 8(8), 1131–1149. 10.2217/epi-2016-0032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington, C. H. (1957). The strategy of the genes. A discussion of some aspects of theoretical biology . With an Appendix by H. Kacser. https://www.cabdirect.org/cabdirect/abstract/19580101706
- Wakayama, S. , Kohda, T. , Obokata, H. , Tokoro, M. , Li, C. , Terashita, Y. , Mizutani, E. , Nguyen, V. T. , Kishigami, S. , Ishino, F. , & Wakayama, T. (2013). Successful serial Recloning in the mouse over multiple generations. Cell Stem Cell, 12(3), 293–297. 10.1016/j.stem.2013.01.005 [DOI] [PubMed] [Google Scholar]
- Wang, C. , Rabadan Ros, R. , Martinez‐Redondo, P. , Ma, Z. , Shi, L. , Xue, Y. , Guillen‐Guillen, I. , Huang, L. , Hishida, T. , Liao, H. K. , Nuñez Delicado, E. , Rodriguez Esteban, C. , Guillen‐Garcia, P. , Reddy, P. , & Izpisua Belmonte, J. C. (2021). In vivo partial reprogramming of myofibers promotes muscle regeneration by remodeling the stem cell niche. Nature Communications, 12(1), 3094. 10.1038/s41467-021-23353-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. , & Simon, A. (2016). Skeletal muscle dedifferentiation during salamander limb regeneration. Current Opinion in Genetics & Development, 40, 108–112. 10.1016/j.gde.2016.06.013 [DOI] [PubMed] [Google Scholar]
- Weismann, A. (1893). The germ‐plasm: A theory of heredity. Scribner's. [Google Scholar]
- West, M. D. , Sternberg, H. , Labat, I. , Janus, J. , Chapman, K. B. , Malik, N. N. , de Grey, A. D. , & Larocca, D. (2019). Toward a unified theory of aging and regeneration. Regenerative Medicine, 14(9), 867–886. 10.2217/rme-2019-0062 [DOI] [PubMed] [Google Scholar]
- Williams, G. C. (2001). Pleiotropy, natural selection, and the evolution of senescence. Science of Aging Knowledge Environment, 2001(1), cp13. 10.1126/sageke.2001.1.cp13 [DOI] [Google Scholar]
- Wilmut, I. , Schnieke, A. E. , McWhir, J. , Kind, A. J. , & Campbell, K. H. S. (1997). Viable offspring derived from fetal and adult mammalian cells. Nature, 385(6619), 810–813. 10.1038/385810a0 [DOI] [PubMed] [Google Scholar]
- Yagi, T. , Kosakai, A. , Ito, D. , Okada, Y. , Akamatsu, W. , Nihei, Y. , Nabetani, A. , Ishikawa, F. , Arai, Y. , Hirose, N. , Okano, H. , & Suzuki, N. (2012). Establishment of induced pluripotent stem cells from centenarians for neurodegenerative disease research. PLoS One, 7(7), e41572. 10.1371/journal.pone.0041572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J.‐H. , Hayano, M. , Griffin, P. T. , Amorim, J. A. , Bonkowski, M. S. , Apostolides, J. K. , Salfati, E. L. , Blanchette, M. , Munding, E. M. , Bhakta, M. , Chew, Y. C. , Guo, W. , Yang, X. , Maybury‐Lewis, S. , Tian, X. , Ross, J. M. , Coppotelli, G. , Meer, M. V. , Rogers‐Hammond, R. , … Sinclair, D. A. (2023). Loss of epigenetic information as a cause of mammalian aging. Cell, 186(2), 305.e27–326.e27. 10.1016/j.cell.2022.12.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarnall, M. T. N. , Ioannidi, E. I. , Schmitt‐Ulms, C. , Krajeski, R. N. , Lim, J. , Villiger, L. , Zhou, W. , Jiang, K. , Garushyants, S. K. , Roberts, N. , Zhang, L. , Vakulskas, C. A. , Walker, J. A., 2nd , Kadina, A. P. , Zepeda, A. E. , Holden, K. , Ma, H. , Xie, J. , Gao, G. , … Gootenberg, J. S. (2022). Drag‐and‐drop genome insertion of large sequences without double‐strand DNA cleavage using CRISPR‐directed integrases. Nature Biotechnology, 41(4), 500–512 10.1038/s41587-022-01527-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zunder, E. R. , Lujan, E. , Goltsev, Y. , Wernig, M. , & Nolan, G. P. (2015). A continuous molecular roadmap to iPSC reprogramming through progression analysis of single‐cell mass cytometry. Cell Stem Cell, 16(3), 323–337. 10.1016/j.stem.2015.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]


