Abstract
The mesophilic purple sulfur phototrophic bacterium Allochromatium (Alc.) vinosum (bacterial family Chromatiaceae) has been a favored model for studies of bacterial photosynthesis and sulfur metabolism, and its core light-harvesting (LH1) complex has been a focus of numerous studies of photosynthetic light reactions. However, despite intense efforts, no high-resolution structure and thorough biochemical analysis of the Alc. vinosum LH1 complex have been reported. Here we present cryo-EM structures of the Alc. vinosum LH1 complex associated with reaction center (RC) at 2.24 Å resolution. The overall structure of the Alc. vinosum LH1 resembles that of its moderately thermophilic relative Alc. tepidum in that it contains multiple pigment-binding α- and β-polypeptides. Unexpectedly, however, six Ca ions were identified in the Alc. vinosum LH1 bound to certain α1/β1- or α1/β3-polypeptides through a different Ca2+-binding motif from that seen in Alc. tepidum and other Chromatiaceae that contain Ca2+-bound LH1 complexes. Two water molecules were identified as additional Ca2+-coordinating ligands. Based on these results, we reexamined biochemical and spectroscopic properties of the Alc. vinosum LH1–RC. While modest but distinct effects of Ca2+ were detected in the absorption spectrum of the Alc. vinosum LH1 complex, a marked decrease in thermostability of its LH1–RC complex was observed upon removal of Ca2+. The presence of Ca2+ in the photocomplex of Alc. vinosum suggests that Ca2+-binding to LH1 complexes may be a common adaptation in species of Chromatiaceae for conferring spectral and thermal flexibility on this key component of their photosynthetic machinery.
Subject terms: Bioenergetics, Cryoelectron microscopy
The cryo-EM structure of the partially Ca2 + -bound light-harvesting 1–reaction center (LH1–RC) complex from Alc. vinosum, the best-studied model purple sulfur bacterium, is presented, characterized and compared to other photosynthetic bacteria.
Introduction
The phototrophic bacterium Allochromatium (Alc.) vinosum is the most-investigated purple sulfur bacterium and has been widely used as a model for studies of anoxygenic photosynthesis and associated sulfur metabolism1,2. Alc. vinosum uses light energy to drive photophosphorylation and extract electrons from reduced sulfur compounds to produce the ATP and NADH needed for autotrophic growth. The photosynthetic apparatus of Alc. vinosum consists of two light-harvesting (LH) complexes, LH1 and LH2, and the reaction center (RC) complex. Both LH1 and LH2 contain multiple α- and β-polypeptides3–6 with variable compositions7,8, making it difficult for definitive structural analysis. A preliminary crystallographic study of the Alc. vinosum LH1–RC core complex was reported at 4.5–5.15 Å resolutions9; however, further efforts could not improve the resolution10.
Alc. vinosum is a mesophilic species of the family Chromatiaceae and is phylogenetically related to the mild thermophile Alc. tepidum11 and the thermophile Thermochromatium (Tch.) tepidum12,13, with average nucleotide identities (ANI) of 90.8% and 85.7%11, respectively. As a result, Alc. vinosum has been used as a reference for comparison of the thermostability of many isolated proteins with their counterparts from Tch. tepidum and Alc. tepidum, including RuBisCO14,15, high-potential iron–sulfur protein (HiPIP)16,17, cytochrome c′ and flavocytochrome c18, and the LH1–RC complex itself19,20. In addition to thermostability, spectroscopic properties of the photocomplexes from Alc. vinosum have been studied21–26, and the organism’s LH1–RC complex has been used as a reference point for the spectral changes observed in the Ca2+-induced, redshifted LH1–RC complexes of other purple sulfur bacteria20,27–30. Surprisingly, however, despite extensive reference to Alc. vinosum photocomplexes in the literature, no high-resolution structures of these complexes have been reported.
Following structural determinations of the LH1–RC complexes from Tch. tepidum31,32 and Alc. tepidum33, here we report a high-resolution cryo-EM structure of the Alc. vinosum LH1–RC complex. A key discovery in our work is that metal ions unequivocally identified as Ca2+ are bound in the LH1 complex. Based on this unexpected result and because the presence of Ca2+ was neither suspected nor revealed in previous studies of the Alc. vinosum LH1–RC complex11,20,33, we have reinvestigated this complex and determined the effects of Ca2+ on its biochemical and spectroscopic properties. Moreover, we also report the effects of variations in illumination on the spectral characteristics and polypeptide composition of the purified Alc. vinosum LH1–RC complexes. After five decades of research on Alc. vinosum, our results provide a fresh new insight into the key photocomplex of this model purple sulfur bacterium and provide the necessary foundation for exploring the mechanisms by which it captures and converts solar energy.
Results
Structural overview of the Alc. vinosum LH1–RC
The Alc. vinosum LH1–RC complexes used for cryo-EM analysis were purified using two different methods: (i) one-step extraction using n-dodecyl β-D-maltopyranoside (DDM) followed by sucrose density gradient centrifugation (hereafter designated as sucrose-density), and (ii) two-step solubilization by lauryldimethylamine N-oxide (LDAO) and n-octyl β-D-glucopyranoside (OG) followed by DEAE chromatography using DDM and CaCl2 (see Methods for details). Both purification methods yielded essentially the same absorption spectra with an LH1-Qy maximum at 889 nm (Supplementary Fig. 1a); this differs from that (884 nm) reported for the Alc. vinosum LH1–RC purified using n-dodecyl phosphocholine (DDPC, an ionic detergent) and NaCl20,28,29, and the reason for these differences in Qy maxima is described in the Discussion.
The cryo-EM structures of the Alc. vinosum LH1–RC complexes were determined at 2.24 Å resolution for both sucrose-density and Ca-DEAE purified samples (Fig. 1, Table 1, Supplementary Figs. 2–5). The two structures are virtually identical in both protein conformation and cofactor arrangement with a root-mean-square deviation (RMSD) of 0.37 Å for the mainchain Cα carbons (Supplementary Fig. 6b) but they differ in the number of divalent cations bound to the RC Cyt-subunit (Supplementary Fig. 7). Hereafter, we describe only the structure of the LH1–RC complex purified by the sucrose-density method because it was purified under physiological conditions that retained all natively bound Ca ions.
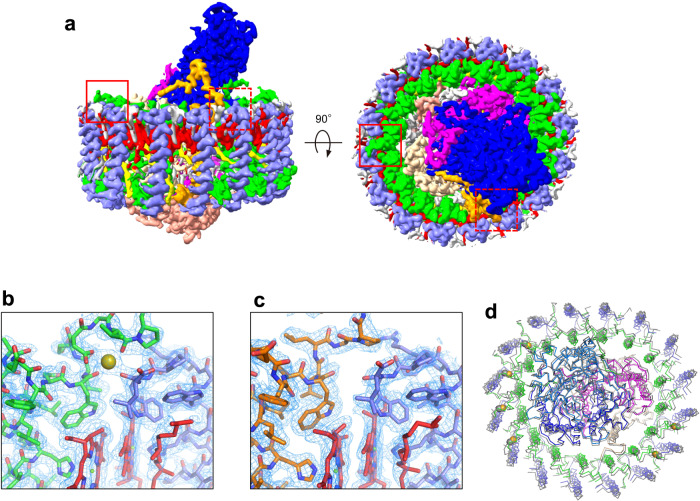
Fig. 1. Overall structure and cofactor arrangement of the Alc. vinosum LH1–RC complex.
a Side and top views of surface representations of the LH1–RC parallel to the membrane and from the periplasmic side of the membrane, respectively. b A typical Ca2+-binding site (marked in a by the red rectangle) with the density map around the C-termini of LH1 α1(green)- and β3(slate blue)-polypeptides. Ca2+ bound to LH1 is shown by an olive sphere. c A typical Ca2+-free site (marked in a by the red dashed rectangle) with the density map around the C-termini of LH1 α3(orange)- and β1(slate blue)-polypeptides. d Overlap view of the Alc. vinosum LH1–RC (colored) and that of Alc. tepidum (gray, PDB: 7VRJ) by superposition of Cα carbons of the RC-M subunits. Color scheme: LH1-α, green; LH1-β, slate blue; RC-L, wheat; RC-M, magenta; RC-H, salmon; RC-C, blue; BChls, red; carotenoids, yellow; lipids, gray.
Table 1.
Cryo-EM data collection, refinement and validation statistics of the Alc. vinosum LH1–RC complexes.
| Sucrose-density purified LH1–RC complex (EMDB-37465, PDB-8WDU) | Ca2+-DEAE purified LH1–RC complex (EMDB-37466, PDB-8WDV) | |
|---|---|---|
| Data collection and processing | ||
| Microscope | JEOL CRYO-ARM300II | TF Titan Krios |
| Camera | K3 | Falcon III |
| Magnification | 80,000 | 96,000 |
| Voltage (kV) | 300 | 300 |
| Electron exposure (e–/Å2) | 50 | 40 |
| Defocus range (μm) | −0.7 to −3.2 | −0.5 to −2.6 |
| Calibrated pixel size (Å) | 0.606 | 0.82 |
| Detector physical pixel size (μm) | 5 | 14 |
| Symmetry imposed | C1 | C1 |
| Initial particle images (no.) | 361,654 | 833,998 |
| Final particle images (no.) | 252,230 | 219,233 |
| Map resolution (Å) | 2.2 | 2.2 |
| FSC threshold | 0.143 | 0.143 |
| Map resolution range (Å) | 3.2–2.2 | 4.3–2.1 |
| Refinement | ||
| Initial model used (PDB code) | 7VRJ | 7VRJ |
| Model resolution (Å) | 2.3 | 2.3 |
| FSC threshold | 0.5 | 0.5 |
| Model resolution range (Å) | 135–2.2 | 135–2.2 |
| Map sharpening B factor (Å2) | –44 | –63 |
| Model composition | ||
| Non-hydrogen atoms | 26,274 | 26,261 |
| Protein residues | 2575 | 2585 |
| Ligands | 113 | 111 |
| Waters | 334 | 354 |
| B factors (Å2) | ||
| Protein | 27.4 | 45.9 |
| Ligand | 30.3 | 51.2 |
| Water | 23.6 | 47.8 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.006 | 0.006 |
| Bond angles (°) | 2.821 | 2.796 |
| Validation | ||
| MolProbity score | 1.77 | 1.61 |
| Clashscore | 12.15 | 12.55 |
| Poor rotamers (%) | 1.63 | 0.98 |
| Ramachandran plot | ||
| Favored (%) | 97.95 | 98.2 |
| Allowed (%) | 2.05 | 1.8 |
| Disallowed (%) | 0 | 0 |
The overall structure of the Alc. vinosum LH1–RC resembles that of Alc. tepidum LH1–RC33 with an RMSD of 1.62 Å for the mainchain Cα carbons (Fig. 1d) and shares some features with the LH1–RCs of Tch. tepidum32 and the spectrally unusual Thiorhodovibrio (Trv.) strain 97034 LH1–RCs. The Alc. vinosum LH1 is a closed ring structure composed of 16 pairs of αβ-polypeptides, 32 bacteriochlorophylls (BChl) a, and 16 all-trans spirilloxanthins that are uniformly distributed around the RC (Fig. 1a, Fig. 2a, Fig. 3a, Supplementary Fig. 6). Multiple forms of LH1 α- and β-polypeptides were confirmed in the high-resolution cryo-EM structure (see next section). The Alc. vinosum RC contains a tetraheme-cytochrome (Cyt) subunit (Fig. 1a) and four BChls a, two bacteriopheophytins a, one 15-cis-spirilloxanthin, a menaquinone (MQ)-8 at the QA site and a ubiquinone (UQ)-8 at the QB site (Fig. 3a). Light-induced P+/P absorption difference spectra showed that the reduced form of the Alc. vinosum RC special pair has an absorption maximum at 885 nm (Supplementary Fig. 8a).
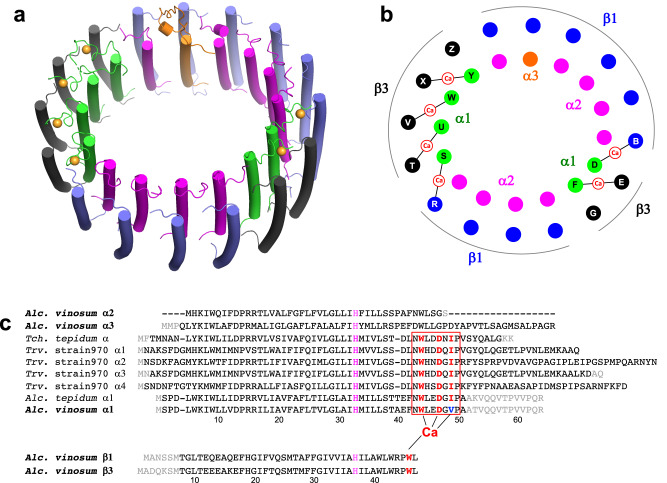
Fig. 2. Arrangement of the Alc. vinosum LH1 multiple polypeptides and Ca2+-binding motifs.
a Tilted view of the LH1–RC from the periplasmic side of the membrane. Color scheme: α1, green; α2, magenta; α3, orange; β1, blue; β3, black; Ca2+, orange ball. b Illustration of the arrangement of the LH1 polypeptides. Letters in the colored circles denote chain IDs. Color scheme as in (a). c Sequence alignment of the Alc. vinosum LH1 polypeptides with those of Tch. tepidum, Trv. strain 970 and Alc. tepidum relative to the BChl a-coordinating histidine residues. Alignment of the α-polypeptides shows that the Ca2+-binding motif WxxDxV is only present in the Alc. vinosum α1-polypeptides but is similar to the WxxDxI motif present in the Ca2+-binding LH1 polypeptides from other purple sulfur bacteria.
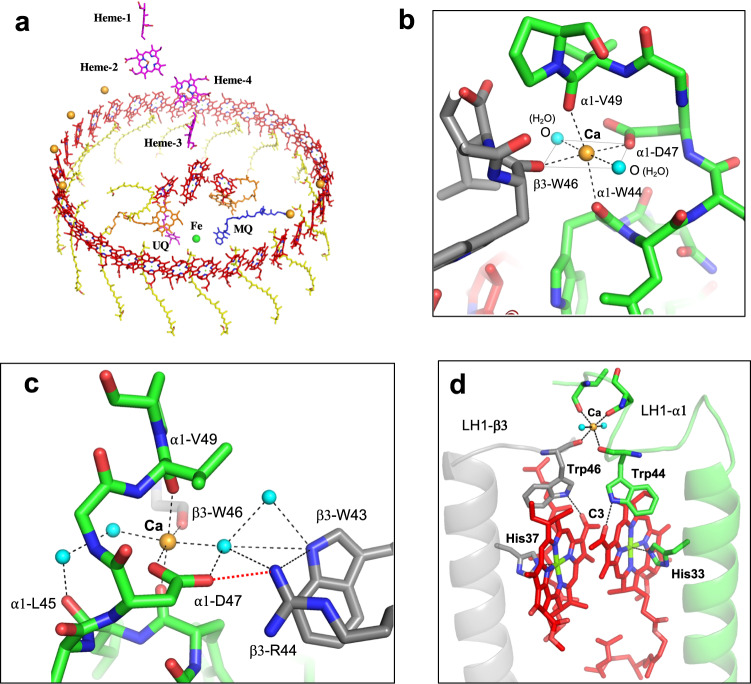
Fig. 3. Ca2+- and BChl a-binding sites in the Alc. vinosum LH1 complex.
a Tilted view of the cofactor arrangement with the periplasm above and the cytoplasm below. b A typical Ca2+-binding site in the pair of α1/β3-polypeptides. c Hydrogen-bonding network (black dashed lines) formed by water molecules (cyan balls) around the Ca2+-binding site. A salt-bridge is formed between α1-Asp47 and β3-Arg44 (red dotted line). d A typical BChl a-binding site in an α1/β3 subunit showing that α1-Trp44 and β3-Trp46 coordinate to Ca2+ with their mainchain oxygens and form hydrogen bonds through their sidechains with the BChl a C3-acetyl groups.
Six metal ions bound to the Alc. vinosum LH1 α- and β-polypeptides were detected in the density map (Fig. 1b, c, Fig. 2, Supplementary Fig. 7a) and identified as Ca2+ by inductively coupled plasma atomic emission spectroscopy (ICP-AES)20 (Supplementary Fig. 9). Further quantitative measurements by ICP-AES placed the Ca2+ stoichiometry at approximately 7 Ca2+ per LH1–RC, which is comparable with the number observed from the cryo-EM density map (Supplementary Fig. 7) and very close to the actual number of 6 (see next two sections). The high-resolution structure of the Alc. vinosum LH1–RC also identified a total of 333 water molecules that are mainly distributed on the transmembrane surfaces and the surface of the membrane-extruded Cyt-subunit (Supplementary Fig. 6c).
Arrangement of the multiple Alc. vinosum LH1 polypeptides
Three pairs of pufBA genes (pufB1A1, pufB2A2, and pufB3A3) encoding LH1 β- and α-polypeptides are present in the Alc. vinosum genome4–6. Previous work detected two forms of α-polypeptides (α1 and α2) and two forms of β-polypeptides (β1 and β3) in the purified LH1 complex7. The cryo-EM structure of the Alc. vinosum LH1–RC complex identified three forms of α-polypeptides (six α1, nine α2, and one α3) and two forms of β-polypeptides (ten β1 and six β3) (Fig. 2a, b). Based on this result, we reexamined the LH1 polypeptide composition using reverse-phase HPLC analysis33,35 and confirmed the presence of the single α3-polypeptide (Supplementary Fig. 10b). The α1-polypeptides of Alc. vinosum form face-to-face dimeric subunits specifically with β3-polypeptides through interactions in their N-terminal domains, whereas α2-polypeptides specifically pair with β1-polypeptides (Fig. 2b), a feature also observed in the Alc. tepidum LH133. The single α3-polypeptide forms a subunit with the β1-polypeptide, likely due to its higher sequence similarity with the α2-polypeptide (Supplementary Fig. 11b). One of the α1-polypeptides and the α3-polypeptide interact extensively with the RC Cyt-subunit through their membrane-extruded C-terminal domains (Supplementary Fig. 6d), strongly stabilizing the entire structure of the LH1–RC complex. Comparisons of the arrangement of Alc. vinosum LH1 αβ-polypeptides with those of Alc. tepidum, Trv. strain 970, and Tch. tepidum are shown in Supplementary Fig. 12.
Among the Alc. vinosum LH1 α-polypeptides, only α1-polypeptides are capable of binding Ca2+ with β1- or β3-polypeptides (Fig. 2a, b), a feature also observed in the Alc. tepidum LH133. Inspection of amino acid sequences revealed that the Alc. vinosum LH1 α1-polypeptide has a region of highly similar sequence to that of the Ca2+-binding motif reported for the α-polypeptides in the Ca2+-bound LH1 complexes (Fig. 2c) from Tch. tepidum32, Trv. strain 97034, and Alc. tepidum33. However, one Ca2+-ligating residue (Ile) in the previously reported motifs is replaced by Val in the Alc. vinosum α1-polypeptide. By contrast, no such region is present in the Alc. vinosum α2- and α3-polypeptides (Fig. 2c). In addition, the shortest α2-polypeptides of Alc. vinosum exhibited a conformation in the C-terminus that would disrupt the Ca2+-binding site and the longest α3-polypeptide adopted a different conformation from that of the α1-polypeptide around the Ca2+-binding site, as is also the case in Alc. tepidum33 (Supplementary Fig. 13). As a result, both α2- and α3-polypeptides of Alc. vinosum LH1 are unable to bind Ca2+.
The Ca2+-binding sites and pigment arrangement
A total of six Ca ions are located on the periplasmic side of the Alc. vinosum LH1 complex and are divided into two groups: four on one side of the LH1 ring and two on the opposite side (Fig. 3a). All Ca2+ are positioned close to the LH1 BChl a molecules, and each Ca2+ is ligated by the mainchain oxygens of Trp44 and Val49, the sidechain of Asp47 in α1-polypeptides, and the mainchain oxygen of β3-Trp46 (or β1-Trp45) (Fig. 3b). Because of the high resolution of our structure, two water molecules were identified as additional Ca2+ ligands (Fig. 3b); this leads to a hexa-coordinated octahedral structure, in which each Ca2+ resides at the center of a plane formed by α1-Asp47, β3-Trp46 (or β1-Trp45) and oxygens of the two waters with the mainchain oxygen atoms of α1-Trp44 and α1-Val49 as axial ligands (Fig. 3b). In addition, several water molecules were identified near the Ca2+-binding sites forming a hydrogen-bonding network with nearby residues and the Ca2+-ligating waters (Fig. 3c) that stabilizes the Ca2+-binding sites. The octahedral Ca2+-binding pattern is similar to that of Tch. tepidum32 but differs from that of Alc. tepidum33. In the latter case, both oxygen atoms in the carboxyl group of the α1-Asp47 are ligands to Ca2+, whereas one of the carboxyl oxygens in the α-Asp (α1-Asp47 for Alc. tepidum and α-Asp49 for Tch. tepidum) forms a salt-bridge with a nearby guanidino group of β-Arg in the subunit (Fig. 3c).
Of the Ca2+-ligating residues, sidechains of the α1-Trp44 and β3-Trp46 (or β1-Trp45) also form hydrogen bonds with the BChl a C3-acetyl group (Fig. 3d), implying that Ca2+-binding may affect hydrogen-bonding for these residues. In fact, torsion angles of the C3-acetyl group tend to have smaller values for the Ca2+-binding residues compared with those that do not bind Ca2+ (Supplementary Fig. 14), despite relatively large deviations. The significance of this is that the torsion angle is thought to correlate with the Qy excitation energy36,37, larger values resulting in more blue-shifted Qy peaks. Overall, the Ca2+-binding effect on LH1 BChl a organization is likely limited because there are no apparent differences in the Mg–Mg distances between BChls a, the length of the His–Mg(BChl a) coordination, and the length of the hydrogen bond for the Trp and BChl a C3-acetyl group between the Ca2+-bound and free LH1-αβ pairs. The latter was supported by the same wavenumber (1637 cm−1) being observed for the C = O stretching mode of the C3-acetyl groups in resonance Raman spectra (Supplementary Fig. 14c) used as a measure for estimating the strength of hydrogen bonding30. There were no apparent differences in average Mg–Mg and His–Mg distances in the Alc. vinosum LH1 complex from those of other purple bacteria (Supplementary Table 1). The long and short Mg–Mg distances and their difference well correlate with the Alc. vinosum LH1-Qy transition (889 nm, Fig. 4a), a correlation reported previously30 showing that Mg–Mg distance differences larger than 0.8 Å tend to yield an LH1-Qy in the 870–890 nm range, whereas differences smaller than 0.8 Å (more homogeneous distances) typically yield an LH1-Qy beyond 900 nm for both BChl a- and BChl b-containing LH1 complexes.
Fig. 4. Absorption spectra and thermostability of the Alc. vinosum LH1–RC complex.
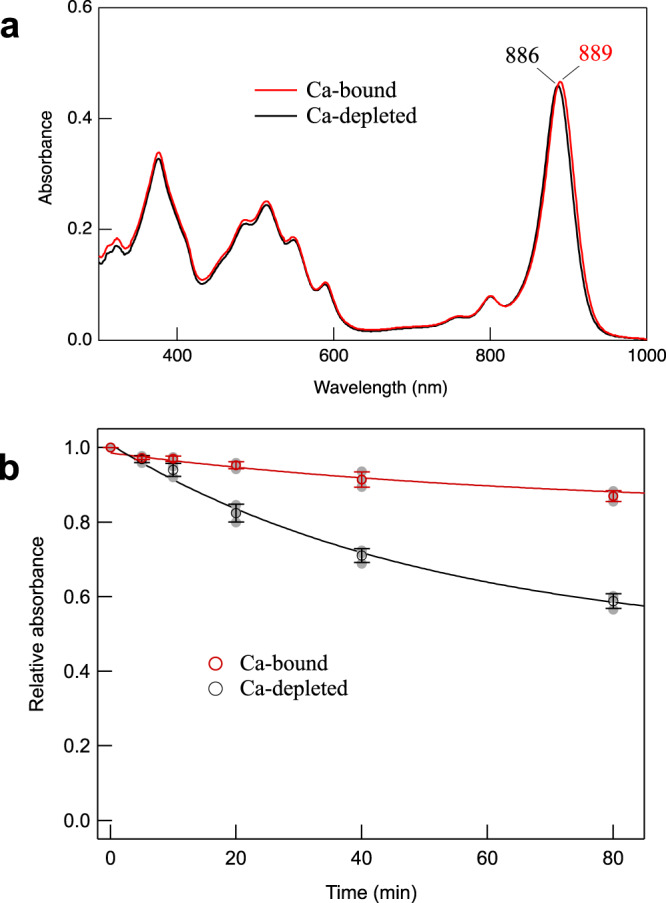
a Absorption spectra of the sucrose-density purified LH1–RC complex treated with 20 mM EDTA in 20 mM Tris-HCl (pH7.5) buffer containing 0.08% w/v DDM followed by removing the EDTA (Ca2+-depleted, black curve) and then adding 20 mM CaCl2 (Ca2+-bound, red curve). b Time course of the relative LH1-Qy intensities upon incubation of the Ca2+-depleted and Ca2+-bound LH1–RC complexes at 60 ˚C. Gray points indicate raw data.
Biochemical and spectroscopic properties of the Alc. vinosum LH1–RC complex
Because Ca2+-binding to select polypeptides in the Alc. vinosum LH1 complex was identified from the cryo-EM structure (Fig. 1, Supplementary Fig. 7), we examined the effects of Ca2+ on biochemical and spectroscopic properties of the complex. Removal of Ca2+ from the sucrose-density purified LH1–RC using EDTA resulted in a complex with an LH1-Qy at 886 nm, and subsequent addition of Ca2+ to the Ca2+-depleted LH1–RC restored its LH1-Qy to the original position (889 nm) at room temperature (Fig. 4a), revealing small but distinct differences in the absorption spectra of Ca2+-containing versus Ca2+-free complexes. Similar spectral differences were observed at 77 K (Supplementary Fig. 1b) and in room temperature spectra of the Alc. vinosum LH1–RC purified using NaCl instead of CaCl2 in the DEAE chromatography step (Supplementary Fig. 1c); the latter indicates that Ca2+ can be replaced either partially or completely by Na+ during Alc. vinosum LH1–RC purification. These Ca2+ effects were more remarkable on thermostability of the Alc. vinosum LH1–RC complex. Incubation of the Alc. vinosum complex in the presence of Ca2+ at 60 ˚C—well above the maximum growth temperature of this species—for 80 min resulted in only a slight reduction of the LH1-Qy intensity, whereas similar treatment for the Ca2+-depleted LH1–RC led to loss of nearly half the absorbance (Fig. 4b), indicating that the Ca2+-bound Alc. vinosum LH1–RC is significantly more heat stable than the Ca2+-depleted complex.
Our previous study demonstrated that cultures of Alc. vinosum cells grew in the absence of Ca ions29, and so here we examined the effects of Ca2+ on the spectroscopy of the isolated LH1–RC. The LH1–RC purified from cells of Alc. vinosum taken from a 7th serial subculture grown without Ca2+ supplementation showed an absorption maximum at 887 nm (Fig. 5a) that did not change upon addition of Ca2+. This indicates that polypeptide conformations around the Ca2+-binding sites in the LH1 complex of Ca2+-starved cells were irreversibly altered during growth, possibly as a mechanism to accommodate metal ions other than Ca2+.
Fig. 5. Effects of growth condition on the Alc. vinosum LH1–RC complex.
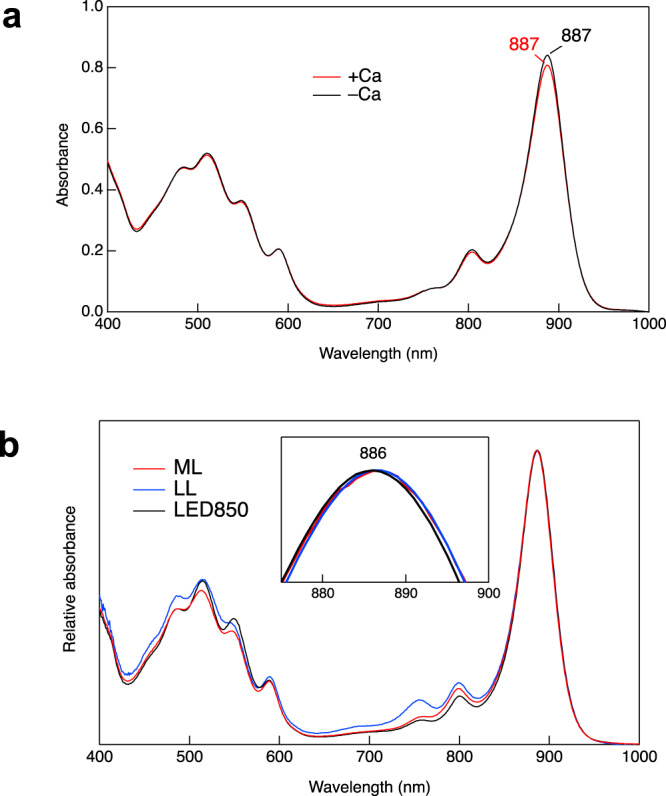
a Absorption spectra of the sucrose-density purified LH1–RC complexes from cells grown in Ca2+-free media (−Ca) followed by addition of 20 mM CaCl2 (+Ca) in 20 mM Tris-HCl (pH7.5) buffer containing 0.08% w/v DDM. b Absorption spectra of the LH1–RC complexes purified using Na-DEAE chromatography from cells grown under incandescent illumination at middle light (ML) and low light (LL) intensities, or under LED illumination (LED850).
We also investigated the effects of light intensity and quality on absorption spectra and polypeptide composition of the Alc. vinosum LH1 complex because different spectral forms and polypeptide compositions of the Alc. vinosum LH2 complex have been reported in cells grown under different illumination regimens8. Alc. vinosum intracytoplasmic membranes (ICM) contained slightly more LH2 than LH1 at low incandescent light intensity (LL, 10 μmol m−2 s−1) compared with that at middle light intensity (ML, 52 μmol m−2 s−1) (Supplementary Fig. 10a), and the two LH2 peaks at 804 nm and 848 nm showed similar intensities. By contrast, when cells were grown using an LED lamp (LED850, peak at 850 nm), although no apparent changes were evident in the proportion of LH1 expression in ICM compared with that at incandescent ML intensity, the two LH2 peaks (804 nm and 852 nm in this case) showed significantly different intensities, with a more intense 852-nm peak than 804-nm peak; this signals a marked change in the polypeptide composition of the Alc. vinosum LH2 between the two illumination regimens. Despite these differences, the purified Alc. vinosum LH1–RC complexes exhibited essentially identical absorption spectra (Fig. 5b) and similar polypeptide compositions within the bounds of experimental errors (Supplementary Fig. 10b). These data suggest that changes induced by variations in light intensity and quality in Alc. vinosum mainly induce changes in the organism’s abundant LH2 complexes rather than LH1–RC complexes.
Discussion
The high-resolution structure of the Alc. vinosum LH1–RC complex revealed many details previously unsuspected, including in particular the presence of bound Ca2+ and several bound water molecules. The overall structural features of the Alc. vinosum LH1–RC complex resemble those of the Alc. tepidum LH1–RC in both composition and arrangement of the multiple LH1 polypeptides (Fig. 1d, Fig. 2), likely due to their genetic similarity11. Nevertheless, Ca ions were unexpected in the Alc. vinosum LH1 complex but indeed were present and in the same number and sites as those for Alc. tepidum LH133. The Ca2+-binding motif in Alc. vinosum LH1 is composed of a region (WxxDxV) present only in the α1-polypeptides and a Trp residue in the C-terminal region of β1- or β3-polypeptides. Moreover, the WxxDxV Ca2+-binding motif in Alc. vinosum differs from the WxxDxI motif in the LH1 α-polypeptides of Alc. tepidum33, Tch. tepidum32 and Trv. strain 97034 (Fig. 2c, Table 2), indicating that a substitution for Ile in the motif is not as critical for Ca2+-binding as was previously thought33. Similar to the Ca2+-ligating pattern in Tch. tepidum LH132, the Ca ions in Alc. vinosum LH1 are hexa-coordinated (including two water ligands) and form an octahedral structure, which differs from the pattern seen in Alc. tepidum33. Thus, the Ca2+-binding motif in the LH1 complexes of purple sulfur bacteria can now be updated to a sequence of “WxxDx(I/V)”.
Table 2.
Characteristics of Ca2+-binding and its effects on the LH1 complexes of different purple sulfur bacteria.
| Number of Ca2+ | Ca2+-binding motif | αβ-polypeptide | LH1-Qy (nm) | Ca2+ effect on LH1-Qy | Ca2+ effect on stability | |
|---|---|---|---|---|---|---|
| Alc. vinosum | 6 (partial) | WxxDxV | Multiple | 889 | 2–3 nm | Moderate |
| Alc. tepidum | 6 (partial) | WxxDxI | Multiple | 890 | 8 nm | Moderate |
| Tch. tepidum | 16 (full) | WxxDxI | Single | 915 | 35 nm | Significant |
| Trv. strain 970 | 16 (full) | WxxDxI | Multiple | 959 | 84 nm | Significant |
Discovery of Ca2+ in the Alc. vinosum LH1 prompted us to investigate the effects of Ca2+ on biochemical and spectroscopic properties of its LH1–RC complex in more detail. Absorption maxima (Qy) of certain purple bacterial LH complexes are sensitive to the detergents used for solubilization and/or treatment of the complexes38–41, and the effects of detergents on Qy can be influenced by the presence of metal ions41. Generally, non-ionic detergents (such as DDM and OG) are milder and less affected by salts and tend to yield LH complexes with an identical or very close Qy to that observed in ICM. By contrast, ionic detergents (such as LDAO, DDPC, and sodium cholate) tend to be influenced by salts, resulting in LH complexes with different spectral forms and/or Qy from those in ICM. Detergent-induced spectral changes in a photocomplex signal an alteration of its tertiary structure that modifies pigment–pigment interactions39. However, the spectral changes caused by ionic detergents are often reversible upon removal or exchange of the detergents. Differing from the LH1 complexes of Alc. tepidum, Tch. tepidum and Trv. strain 970, the Alc. vinosum LH1 exhibited a detergent-sensitive Qy that was further affected by metal ions (Table 2, Supplementary Fig. 15). Because the Ca2+ effect on the Alc. vinosum LH1-Qy is small (2–3 nm) and similar to that ( ~ 3 nm) of the detergent, it had been overlooked in several experiments conducted previously using DDPC as a detergent, including the absorption spectrum, thermostability (Fig. 4) and a metal-exchanging FTIR measurement42. With this in mind, the subtle differences observed in the perfusion-induced FTIR difference spectra for the Alc. vinosum LH1–RC upon Sr2+/Ca2+ exchange42 can now be considered significant changes. To confirm this, we further measured the Sr2+/Ca2+ (or Ca2+/Sr2+) FTIR difference spectra of the Alc. vinosum LH1–RC with increased acquisition numbers to improve the S/N ratio (Supplementary Fig. 8b). The results are compared with those reported for Tch. tepidum where, based on their isotopic shifts, the characteristic FTIR difference bands have been assigned to the vibrational modes for polypeptide backbones, amino acid side chains and BChl a surrounding the Ca ions42,43. Among these, two intensive differential bands at 1015/1007 cm−1 and at 891/883 cm−1 ascribed to the ν C = C and δ NCCm of BChl a, respectively, are useful marker bands for the conformational changes of the LH1 BChl a. In the Alc. vinosum LH1–RC FTIR spectra (Supplementary Fig. 8b), these differential bands were largely reduced in intensity compared with those of Tch. tepidum, indicating that no significant conformational changes are induced in Alc. vinosum LH1 BChl a upon Ca2+-to-Sr2+ substitution. These results strongly support the conclusion that the Alc. vinosum LH1-Qy band should be, as was found to be the case, only slightly blue-shifted upon removal of Ca2+ (Fig. 4a).
Compared with the fully Ca2+-bound LH1 complexes of Tch. tepidum and Trv. strain 970, the partially Ca2+-bound LH1 complexes of Alc. vinosum and Alc. tepidum exhibit only small redshifts, and these may be attributed to the following factors. First, based on the results of resonance Raman spectroscopy, hydrogen-bonding interactions between BChl and LH1 polypeptides have been shown to correlate with the LH1-Qy30, and such interactions in the LH1 of Alc. vinosum and Alc. tepidum are much weaker than those in the Tch. tepidum and Trv. strain 970 LH120,28–30. Second, the partial Ca2+-binding networks formed in the Alc. vinosum and Alc. tepidum LH1 are divided into two parts (Fig. 2b) and are thus much less extensive than those of the fully Ca2+-bound LH1 complexes from Tch. tepidum and Trv. strain 970. Ca ions function to “lock” the LH1 structure, and different network patterns formed in LH1 complexes have been shown to affect their Qy transitions44. Third, the BChl-coordinating C-terminal domains of α-polypeptides are either much shorter or more mobile in Alc. vinosum and Alc. tepidum than in Tch. tepidum and Trv. strain 970. This is supported by the fact that the C-terminal residues in the α1-polypeptides (6 copies) of Alc. vinosum and Alc. tepidum immediately following the Ca2+-binding sites are invisible in the cryo-EM density maps (gray letters in Fig. 2c, Supplementary Fig. 16), and this indicates that large fluctuations and/or disorders are present that could destabilize Ca2+-binding sites. Such structural fluctuations and static disorders influence LH1-Qy and cause an inhomogeneous broadening of the absorption band45–47, a result also observed with the Alc. vinosum LH1 where a large Qy redshift and band narrowing occurred at 77 K (Supplementary Fig. 1b). Fourth, it has been shown that point charge and protein polarity can induce a large Qy redshift of the BChl in a hydrophobic environment30,36,48,49. And finally, the number of Ca ions present in an LH1 and their close proximity to BChl a (Fig. 3d) also contributes to the differences observed in the LH1-Qy of partial and fully Ca2+-bound complexes. It is unlikely that any of the above factors affect LH1 absorbance independently but instead function in combination to confer the spectral changes observed in the Alc. vinosum LH1.
Despite Ca2+-incorporation into LH1 when Ca2+ is available, Alc. vinosum cells are also capable of growing phototrophically in a Ca2+-free medium29 with repeated serial subcultures without a noticeable effect on either growth rate or growth yield. This is in sharp contrast to Tch. tepidum, Trv. strain 970 and Alc. tepidum, all of which exhibited a strong Ca2+-dependence for phototrophic growth20,29. Beside Ca2+, Tch. tepidum cells can grow when Ca2+ is replaced by Sr2+, but no other divalent cations support growth in the absence of Ca2+50. The LH1 of Sr2+-grown Tch. tepidum shows a Qy at 888 nm that shifts to 908 nm (rather than to the 915 nm of wild-type) by addition of Ca2+, implying irreversible changes in the protein conformation around Ca2+-binding pockets during Sr2+-dependent growth50. A subsequent study51 on Sr2+-grown Tch. tepidum revealed preferential truncations of C-terminal residues in the LH1 α-polypeptides that play important roles in stabilizing the LH1 structure and modifying its spectral properties. Such polypeptide modifications may also explain the irreversible feature observed for LH1-Qy from Alc. vinosum cells grown in the absence of Ca2+ (Fig. 5a).
To date, Ca2+ has been identified in the LH1 complexes of all purple sulfur phototrophic bacteria with known LH1 structures. Thus, it is possible that Ca2+-binding to LH1 is a universal feature of Chromatiaceae, the variable Ca2+ amounts and their topology in the complex reflecting a flexible strategy for inhabiting ecological niches that differ in temperature, light availability and quality, or levels of nutritional resources.
Methods
Cultivation of Alc. vinosum cells
Alc. vinosum strain D (DSM 180 T) was used in this study. Unless otherwise stated, cells of Alc. vinosum were cultivated phototrophically (anoxic/light) in a mineral medium11 containing 0.34 mM CaCl2 at room temperature for 7 days under incandescent illumination at middle-light intensity (ML, 52 μmol m−2 s−1). For light experiments, Alc. vinosum cultures were also cultivated under low-light intensity incandescent light (LL, 10 μmol m-2 s-1) or with an LED panel (ISL-150 × 150II85, CCS Inc., Japan) having peak wavelength emission at 850 nm (LED850). To investigate Ca2+ effects, Alc. vinosum cells were cultivated in a Ca2+-free mineral medium. The LED850-illuminated and Ca2+-free cells were harvested after 6 and 7 subcultures, respectively.
Preparation of LH1–RC complexes
The Alc. vinosum LH1–RC complexes for cryo-EM analysis were purified using two different methods. In the first method, LH1–RC complexes were prepared by one-step extraction from ICM using 1.0% w/v DDM, followed by sucrose density gradient centrifugation (five-stepwise concentrations: 10, 17.5, 25, 32.5, and 40% w/v) in 20 mM Tris-HCl (pH 8.0) buffer containing 0.05% w/v DDM. The LH1–RC band was collected and concentrated for cryo-EM measurements. In the second method, ICM were first treated with 0.35% LDAO to remove excess LH2, followed by solubilization using 1.0% w/v OG. Then, crude LH1–RC complexes were loaded onto a DEAE column equilibrated with 20 mM Tris-HCl buffer (pH 7.5) containing 0.1% w/v DDM. The LH1–RC fractions were eluted by a linear gradient of CaCl2 from 0 mM to 100 mM and collected for cryo-EM analysis. Both purification methods yielded essentially the same absorption spectra with the LH1-Qy maxima at 889 nm (Supplementary Fig. 1a). The major difference between the two isolation methods is that no Ca2+ or other salts were used in the sucrose-density purification. LH1–RC complexes from Alc. vinosum cells grown in a Ca2+-free medium were purified by sucrose-density. LH1–RC complexes from cells grown at different light intensities and LED850 were purified by DEAE chromatography with a linear gradient (0 mM to 200 mM) of NaCl.
Cryo-EM data collection
Proteins for cryo-EM were concentrated to 5.2 and 4.0 mg/ml of the sucrose-density and Ca-DEAE purified samples, respectively. Three microliters of the protein solution were applied on glow-discharged holey carbon grids (200 mesh Quantifoil R1.2/1.3 and R2/2 molybdenum) that had been treated in a PIB-20 (Shinku Device) and Solarus-2 (Gatan) for 90 s and 30 s for the sucrose-density and Ca-DEAE purified samples, respectively, and then plunged into liquid ethane at −182 °C using an EM GP2 plunger (Leica, Microsystems, Vienna, Austria). The applied parameters were a blotting time of 6 s and humidity of 90% at 4 °C. Data from the sucrose-density sample were collected on a CRYO-ARM300II (JEOL) electron microscope at 300 kV equipped with a K3 camera (Gatan). An in-column energy filter with a slit width of 20 eV was inserted for acquisition of movie frames. Data from the Ca-DEAE sample were collected on a Titan Krios (Thermo Fisher Scientific) electron microscope at 300 kV equipped with a Falcon-III camera (Thermo Fisher Scientific). Movies were recorded using SerialEM52 and EPU at a nominal magnification of 80 K and 96 K in CDS and counting mode and a pixel size of 0.606 and 0.820 Å at the specimen level for the sucrose-density and Ca-DEAE purified samples, respectively. The dose rate was 25.0 and 1.32 e- per Å2 per second at the specimen level, and the exposure time was 2.0 and 30.15 s, resulting in an accumulated dose of 50.0 and 40.0 e− per Å2 for the sucrose-density and Ca-DEAE purified samples, respectively. Each movie includes 50 and 40 fractioned frames for the sucrose-density and Ca-DEAE purified samples, respectively.
Image processing
All of the stacked frames were subjected to motion correction with MotionCor253, and defocus was estimated using CTFFIND454. A total of 361,654 and 833,998 particles were selected from 7704 and 3114 micrographs for the sucrose-density and Ca-DEAE purified samples, respectively, using crYOLO55 and RELION3.156. All of the picked particles were further analyzed with RELION, and 359,604 and 338,772 particles were selected by 2-D classification and 1st 3D classification for the sucrose-density and Ca-DEAE purified samples, respectively. They were divided into four classes by 3-D classification resulting in only one good class containing 252,230 and 219,233 particles for the sucrose-density and Ca-DEAE purified samples, respectively. The initial 3-D model was generated in RELION. The 3-D auto refinement without any imposed symmetry (C1) produced two maps at 2.24 Å resolution after contrast transfer function refinement, Bayesian polishing, masking, and post-processing. These particle projections were then subjected to subtraction of the detergent micelle density followed by 3-D auto refinement to yield final maps with a resolution of 2.24 Å according to the gold-standard Fourier shell correlation using a criterion of 0.143. The local resolution maps were calculated on RELION.
Model building and refinement of the LH1–RC complex
The atomic model of the Alc. tepidum LH1–RC (PDB code 7VRJ) was fitted to the cryo-EM map obtained for the Alc. vinosum LH1-RC using Chimera57. Amino acid substitutions and real space refinement for the peptides and cofactors were performed using COOT58. The C-terminal regions of the LH1 α3-subunit were modelled ab-initio based on their density. The manually modified model was real-space-refined on PHENIX59, and the COOT/PHENIX refinement was iterated until the refinements converged. Finally, the statistics calculated using MolProbity60 were checked. Figures were drawn with the Pymol Molecular Graphic System (Schrödinger)61, UCSF Chimera57 and Chimera-X62.
Biochemical analyses of the Alc. vinosum LH1–RC complex
Metal ion identification in the Alc. vinosum LH1–RC complex purified by sucrose-density was carried out on an ICP-AES spectrophotometer (ICPS-7510, Shimadzu). Qualitative measurements detected three metal ions (Ca, Mg and Fe) above 0.1 ppm in the LH1–RC at a concentration of A889 = 1.3 cm−1. Further quantitative measurements on the same LH1–RC sample were conducted using Multielement Standard Solution W-II (Wako Pure Chemical Industries, Ltd.) that contains 1000 mg/L for each Ca, Fe, and Mg in 1 M HNO3 solution and was diluted to appropriate concentrations used for calibration. Detection wavelengths were 393.366 nm for Ca, 279.553 nm for Mg and 259.940 nm for Fe. The Alc. vinosum LH1–RC complex purified by sucrose-density was treated by 20 mM EDTA in 20 mM Tris-HCl (pH7.5) buffer containing 0.08% w/v DDM on ice, followed by removing EDTA to yield Ca-depleted LH1–RC (Fig. 4a), and then adding 20 mM CaCl2 to yield Ca-bound LH1–RC (Fig. 4a). Thermal degradations of the Ca-bound and Ca-depleted LH1–RCs were monitored via the LH1-Qy intensity after incubation at 60 °C for 0–80 min (Fig. 4b). The LH1–RC complex purified by sucrose-density from cells grown in a Ca2+-free medium was treated with 20 mM CaCl2 in 20 mM Tris-HCl (pH7.5) buffer containing 0.08% w/v DDM.
Statistics and reproducibility
Thermal stability measurements (Fig. 4b) were performed in triplicate (three individual assays) independently to verify reproducibility. Experimental errors for all absorption spectra were within 1 nm.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
This research was partially supported by Platform Project for Supporting Drug Discovery and Life Science Research (Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS)) from AMED under Grant Numbers JP21am0101118 and JP21am0101116, and JP23ama121004. R.K., E.R.P., M.H., and B.M.H. acknowledge the generous support of the Okinawa Institute of Science and Technology (OIST), Scientific Computing & Data Analysis Section and Scientific Imaging Section at OIST and the Japanese Cabinet Office. R.K. acknowledges the support from Prof. Tsumoru Shintake. L.-J.Y. acknowledges support of the National Key R&D Program of China (Nos. 2021YFA0909600 and 2019YFA0904600). M.T.M. was supported in part by NASA Cooperative Agreement 80NSSC21M0355. This work was supported in part by JSPS KAKENHI Grant Numbers 20H05086, 20H02856, and 22K06111.
Author contributions
Z.-Y.W.-O., Y.Ki., and K.T. designed the work. K.T., R.K., A.H., Y.Ko., A.Min., N.N., X.-C.J., E.R.P. and M.H. performed the experiments, K.T., R.K., S.T., L.-J.Y., M.T.M., A.Miz., K.I., B.M.H., Y.Ki. and Z.-Y.W.-O. analyzed data, Z.-Y.W.-O., K.T., Y.Ki. and M.T.M. wrote the paper.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Isabelle Lucet and Tobias Goris. A peer review file is available.
Data availability
Maps and models have been deposited in the EMDB and PDB with the accession codes: EMD-37465 and PDB-8WDU for the Alc. vinosum LH1–RC purified by sucrose-density; EMD-37466 and PDB-8WDV for the Alc. vinosum LH1–RC purified by Ca-DEAE. The numerical source values underlying Fig. 4 can be found in Supplementary Data 1. All other data are available from the authors upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Kazutoshi Tani, Ryo Kanno, Ayaka Harada.
These authors jointly supervised this work: Kazutoshi Tani, Yukihiro Kimura, Zheng-Yu Wang-Otomo.
Contributor Information
Kazutoshi Tani, Email: ktani@ccs.tsukuba.ac.jp.
Yukihiro Kimura, Email: ykimura@people.kobe-u.ac.jp.
Zheng-Yu Wang-Otomo, Email: wang@ml.ibaraki.ac.jp.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-024-05863-w.
References
- 1.Brune, D. C. Sulfur compounds as photosynthetic electron donors. in Anoxygenic Photosynthetic Bacteria (eds Blankenship, R. E., Madigan, M. T. & Bauer, C. E.) 847–870 (Kluwer Academic Publishers, Dordrecht, The Netherlands, 1995).
- 2.Weissgerber T, et al. Genome-wide transcriptional profiling of the purpler sulfur bacterium Allochromatium vinosum DSM 180T during growth on different reduced sulfur compounds. J. Bacteriol. 2013;195:4231–4245. doi: 10.1128/JB.00154-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bissig, I., Wagner-Huber, R.V., Brunisholz, R. A. & Zuber, H. Multiple antenna complexes in various purple photosynthetic bacteria. in Molecular Biology of Membrane-Bound Complexes in Phototrophic Bacteria (eds Drews, G. & Dawes, E. A.) 199–210 (Plenum Press, New York, 1990).
- 4.Corson GE, et al. Genes encoding light-harvesting and reaction center proteins from Chromatium vinosum. Photosynth. Res. 1999;59:39–52. doi: 10.1023/A:1006182818010. [DOI] [Google Scholar]
- 5.Nagashima S, Shimada K, Matsuura K, Nagashima KVP. Transcription of three sets of genes coding for the core light-harvesting proteins in the purple sulfur bacterium, Allochromatium vinosum. Photosynth. Res. 2002;74:269–280. doi: 10.1023/A:1021280104053. [DOI] [PubMed] [Google Scholar]
- 6.Weissgerber T, et al. Complete genome sequence of Allochromatium vinosum DSM 180T. Stand. Genomic Sci. 2011;5:311–330. doi: 10.4056/sigs.2335270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Z-Y, Shimonaga M, Suzuki H, Kobayashi M, Nozawa T. Purification and characterization of the polypeptides of core light-harvesting complexes from purple sulfur bacteria. Photosynth. Res. 2003;78:133–141. doi: 10.1023/B:PRES.0000004328.11219.79. [DOI] [PubMed] [Google Scholar]
- 8.Carey A-M, et al. Characterisation of the LH2 spectral variants produced by the photosynthetic purple sulphur bacterium Allochromatium vinosum. Biochim. Biophys. Acta. 2014;1837:1849–1860. doi: 10.1016/j.bbabio.2014.07.022. [DOI] [PubMed] [Google Scholar]
- 9.Mulvaney, R. M., Roszak, A. W., Gardiner, A. T. & Cogdell, R. J. Structural analysis of the LH1:RC core complex from Allochromatium vinosum strain D. in The 16th International Congress onPhotosynthesis Research (St. Louis, MO, USA, 2013).
- 10.Nakamura, N. Master Degree Thesis, Ibaraki University (2016).
- 11.Madigan MT, et al. Allochromatium tepidum, sp. nov., a hot spring species of purple sulfur bacteria. Arch. Microbiol. 2022;204:115. doi: 10.1007/s00203-021-02715-7. [DOI] [PubMed] [Google Scholar]
- 12.Madigan MT. A novel photosynthetic bacterium isolated from a Yellowstone hot spring. Science. 1984;225:313–315. doi: 10.1126/science.225.4659.313. [DOI] [PubMed] [Google Scholar]
- 13.Sattley WM, et al. Complete genome of the thermophilic purple sulfur bacterium Thermochromatium tepidum compared to Allochromatium vinosum and other Chromatiaceae. Photosynth. Res. 2022;151:125–142. doi: 10.1007/s11120-021-00870-y. [DOI] [PubMed] [Google Scholar]
- 14.Heda GD, Madigan MT. Thermal properties and oxygenase activity of ribulose-1,5-bisphosphate carboxylase from the thermophilic purple bacterium, Chromatium tepidum. FEMS Microbiol. Lett. 1988;51:45–50. doi: 10.1111/j.1574-6968.1988.tb02966.x. [DOI] [Google Scholar]
- 15.Heda GD, Madigan MT. Purification and characterization of the thermostable ribulose-1,5-bisphosphate carboxylate/oxygenase from the thermophilic purple bacterium Chromatium tepidum. Eur. J. Biochem. 1989;184:313–319. doi: 10.1111/j.1432-1033.1989.tb15021.x. [DOI] [PubMed] [Google Scholar]
- 16.Moulis J-M, et al. Primary structure of Chromatium tepidum high-potential iron-sulfur protein in relation to thermal denaturation. Arch. Biochem. Biophys. 1993;305:186–192. doi: 10.1006/abbi.1993.1409. [DOI] [PubMed] [Google Scholar]
- 17.Kobayashi M, Saito T, Takahashi K, Wang Z-Y, Nozawa T. Electronic properties and thermal stability of soluble redox proteins from a thermophilic purple sulfur photosynthetic bacterium, Thermochromatium tepidum. Bull. Chem. Soc. Jpn. 2005;78:2164–2170. doi: 10.1246/bcsj.78.2164. [DOI] [Google Scholar]
- 18.Hirano Y, Kimura Y, Suzuki H, Miki K, Wang Z-Y. Structure analysis and comparative characterization of the cytochrome c’ and flavocytochrome c from thermophilic purple photosynthetic bacterium Thermochromatium tepidum. Biochemistry. 2012;51:6556–6567. doi: 10.1021/bi3005522. [DOI] [PubMed] [Google Scholar]
- 19.Kimura Y, Yu L-J, Hirano Y, Suzuki H, Wang Z-Y. Calcium ions are required for the enhanced thermal stability of the light-harvesting-reaction center core complex from thermophilic purple sulfur bacterium Thermochromatium tepidum. J. Biol. Chem. 2009;284:93–99. doi: 10.1074/jbc.M806840200. [DOI] [PubMed] [Google Scholar]
- 20.Kimura Y, et al. Effects of calcium ions on the thermostability and spectroscopic properties of the LH1-RC complex from a new thermophilic purple bacterium Allochromatium tepidum. J. Phys. Chem. B. 2017;121:5025–5032. doi: 10.1021/acs.jpcb.7b03341. [DOI] [PubMed] [Google Scholar]
- 21.Thornber JP, Sokoloff MK. Photochemical reactions of purple bacteria as revealed by studies of three different carotenobacteriochlorophyll-protein complexes isolated from Chromatium, strain D. Biochemistry. 1970;9:2688–2698. doi: 10.1021/bi00815a017. [DOI] [PubMed] [Google Scholar]
- 22.Kennel SJ, Kamen MD. Iron-containing proteins in Chromatium. II. Purification and properties of cholate-solubilized cytochrome complex. Biochim. BIophys. Acta. 1971;253:153–166. doi: 10.1016/0005-2728(71)90241-6. [DOI] [PubMed] [Google Scholar]
- 23.Mechler B, Oelze J. Differentiation of the photosynthetic apparatus of Chromatium vinosum, strain D III. Analyses of spectral alterations. Arch. Microbiol. 1978;118:109–114. doi: 10.1007/BF00406082. [DOI] [Google Scholar]
- 24.Hayashi H, Morita S. Near-infrared absorption spectra of light harvesting bacteriochlorophyll protein complexes from Chromatium vinosum. J. Biochem. 1980;88:1251–1258. doi: 10.1093/oxfordjournals.jbchem.a133093. [DOI] [PubMed] [Google Scholar]
- 25.Niedzwiedzki DM, et al. Spectroscopic studies of two spectral variants of light-harvesting complex 2 (LH2) from the photosynthetic purple sulfur bacterium Allochromatium vinosum. Biochim. Biophys. Acta. 2012;1817:1576–1587. doi: 10.1016/j.bbabio.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 26.Kell A, et al. Conformational complexity in the LH2 antenna of the purple sulfur bacterium Allochromatium vinosum revealed by Hole-burning spectroscopy. J. Phys. Chem. A. 2017;121:4435–4446. doi: 10.1021/acs.jpca.7b03188. [DOI] [PubMed] [Google Scholar]
- 27.Kimura Y, et al. Calcium ions are involved in the unusual red shift of the light-harvesting 1 Qy transition of the core complex in thermophilic purple sulfur bacterium Thermochromatium tepidum. J. Biol. Chem. 2008;283:13867–13873. doi: 10.1074/jbc.M800256200. [DOI] [PubMed] [Google Scholar]
- 28.Kimura Y, et al. Metal cations modulate the bacteriochlorophyll-protein interaction in the light-harvesting 1 core complex from Thermochromatium tepidum. Biochim. Biophys. Acta. 2012;1817:1022–1029. doi: 10.1016/j.bbabio.2012.03.016. [DOI] [PubMed] [Google Scholar]
- 29.Imanishi M, et al. A dual role for Ca2+ in expanding the spectral diversity and stability of light-harvesting 1 reaction center photocomplexes of purple phototrophic bacteria. Biochemistry. 2019;58:2844–2852. doi: 10.1021/acs.biochem.9b00351. [DOI] [PubMed] [Google Scholar]
- 30.Kimura Y, Tani K, Madigan MT, Wang-Otomo Z-Y. Advances in the spectroscopic and structural characterization of core light-harvesting complexes from purple phototrophic bacteria. J. Phys. Chem. B. 2023;127:6–17. doi: 10.1021/acs.jpcb.2c06638. [DOI] [PubMed] [Google Scholar]
- 31.Niwa S, et al. Structure of the LH1-RC complex from Thermochromatium tepidum at 3.0 Å. Nature. 2014;508:228–232. doi: 10.1038/nature13197. [DOI] [PubMed] [Google Scholar]
- 32.Yu L-J, Suga M, Wang-Otomo Z-Y, Shen J-R. Structure of photosynthetic LH1-RC supercomplex at 1.9 Å resolution. Nature. 2018;556:209–213. doi: 10.1038/s41586-018-0002-9. [DOI] [PubMed] [Google Scholar]
- 33.Tani K, et al. A Ca2+-binding motif underlies the unusual properties of certain photosynthetic bacterial core light–harvesting complexes. J. Biol. Chem. 2022;298:101967. doi: 10.1016/j.jbc.2022.101967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tani K, et al. Cryo-EM structure of a Ca2+-bound photosynthetic LH1-RC complex containing multiple αβ-polypeptides. Nat. Commun. 2020;11:4955. doi: 10.1038/s41467-020-18748-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Z-Y, Shimonaga M, Muraoka Y, Kobayashi M, Nozawa T. Methionine oxidation and its effect on the stability of reconstituted subunit of light-harvesting complex from Rhodospirillum rubrum. Eur. J. Biochem. 2001;268:3375–3382. doi: 10.1046/j.1432-1327.2001.02234.x. [DOI] [PubMed] [Google Scholar]
- 36.Gudowska-Nowak E, Newton MD, Fajer J. Conformational and environmental effects on bacteriochlorophyll optical spectra: correlations of calculated spectra with structural results. J. Phys. Chem. 1990;94:5795–5801. doi: 10.1021/j100378a036. [DOI] [Google Scholar]
- 37.McLuskey K, Prince SM, Cogdell RJ, Isaac NW. The crystallographic structure of the B800-820 LH3 light-harvesting complex from the purple bacteria Rhodopseudomonas acidophila strain 7050. Biochemistry. 2001;40:8783–8789. doi: 10.1021/bi010309a. [DOI] [PubMed] [Google Scholar]
- 38.Ghosh R, Hauser H, Bachofen R. Reversible dissociation of the B873 light-harvesting complex from Rhodospirillum rubrum G9. Biochemistry. 1988;27:1004–1014. doi: 10.1021/bi00403a025. [DOI] [Google Scholar]
- 39.Doi M, Shioi Y, Grad’on N, Golecki JR, Drews G. Spectroscopic studies of the light-harvesting pigment-protein complex II from dark-aerobic and light-anaerobic grown cells of Rhodobacter sulfidophilus. Biochimi. Biophys. Acta. 1991;1058:235–241. doi: 10.1016/S0005-2728(05)80242-7. [DOI] [Google Scholar]
- 40.de Zarate IO, Picorel R. Spectral changes of the B800-850 antenna complex from Ectothiorhodospira sp. induced by detergent and salt treatment. Photosynth. Res. 1994;41:339–347. doi: 10.1007/BF00019411. [DOI] [PubMed] [Google Scholar]
- 41.Sekine F, et al. Gene sequencing and characterization of the light-harvesting complex 2 from thermophilic purple sulfur bacterium Thermochromatium tepidum. Photosynth. Res. 2012;111:9–18. doi: 10.1007/s11120-011-9658-9. [DOI] [PubMed] [Google Scholar]
- 42.Li Y, Kimura Y, Arikawa T, Wang-Otomo Z-Y, Ohno T. ATR-FTIR detection of metal-sensitive structural changes in the light-harvesting 1 reaction center complex from the thermophilic purple sulfur bacterium Thermochromatium tepidum. Biochemistry. 2013;52:9001–9008. doi: 10.1021/bi401033y. [DOI] [PubMed] [Google Scholar]
- 43.Kimura Y, et al. Identification of metal-sensitive structural changes in the Ca2+-binding photocomplex from Thermochromatium tepidum by isotope-edited vibrational spectroscopy. J. Chem. Phys. 2022;156:105101. doi: 10.1063/5.0075600. [DOI] [PubMed] [Google Scholar]
- 44.Yu L-J, Kawakami T, Kimura Y, Wang-Otomo Z-Y. Structural basis for the unusual Qy red-shift and enhanced thermostability of the LH1 complex from Thermochromatium tepidum. Biochemistry. 2016;55:6495–6504. doi: 10.1021/acs.biochem.6b00742. [DOI] [PubMed] [Google Scholar]
- 45.van Mourik F, Visschers RW, van Grondelle R. Energy transfer and aggregate size effects in the inhomogeneously broadened core light-harvesting complex of Rhodobacter sphaeroides. Chem. Phys. Lett. 1992;193:1–7. doi: 10.1016/0009-2614(92)85674-Y. [DOI] [Google Scholar]
- 46.Monshouwer R, Visscher RW, van Mourik F, Freiberg A, van Grondelle R. Low-temperature absorption and site-selected fluorescence of the light-harvesting antenna of Rhodopseudomonas viridis. Evidence for heterogeneity. Biochim. Biophys. Acta. 1995;1229:373–380. doi: 10.1016/0005-2728(95)00020-J. [DOI] [Google Scholar]
- 47.Robert, B. Spectroscopic properties of antenna complexes from purple bacteria. in The purple phototrophic bacteria (eds Hunter, C. N., Daldal, F. & Beatty, J. T.) 199–212 (Springer, 2009).
- 48.Eccles J, Honig B. Charged amino acids as spectroscopic determinants for chlorophyll in vivo. Proc. Natl. Acad. Sci. USA. 1983;80:4959–4962. doi: 10.1073/pnas.80.16.4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Renger T. Theory of excitation energy transfer: from structure to function. Photosynth. Res. 2009;102:471–485. doi: 10.1007/s11120-009-9472-9. [DOI] [PubMed] [Google Scholar]
- 50.Kimura Y, Inada Y, Yu L-J, Wang Z-Y, Ohno T. A spectroscopic variant of the light-harvesting 1 core complex from the thermophilic purple sulfur bacterium Thermochromatium tepidum. Biochemistry. 2011;50:3638–3648. doi: 10.1021/bi200278u. [DOI] [PubMed] [Google Scholar]
- 51.Kimura Y, et al. C-terminal cleavage of the LH1 α-polypeptide in the Sr2+-cultured Thermochromatium tepidum. Photosynth. Res. 2018;135:23–31. doi: 10.1007/s11120-017-0393-8. [DOI] [PubMed] [Google Scholar]
- 52.Zhang J, et al. JADAS: a customizable automated data acquisition system and its application to ice-embedded single particles. J. Struct. Biol. 2009;165:1–9. doi: 10.1016/j.jsb.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rohou A, Grigorieff N. Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wagner T, Raunser S. The evolution of SPHIRE-crYOLO particle picking and its application in automated cryo-EM processing workflows. Commun. Biol. 2020;3:61. doi: 10.1038/s42003-020-0790-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zivanov J, et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife. 2018;7:e42166. doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 58.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Cryst. 2010;D66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst. 2010;D66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.DeLano, W. L. The PyMOL Molecular Graphics System. (DeLano Scientific, LCC, San Carlos, CA USA, 2004).
- 62.Pettersen EF, et al. UCSF ChimeraX: structure visualization for researchers, editors, and developers. Protein Sci. 2021;30:70–82. doi: 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
Maps and models have been deposited in the EMDB and PDB with the accession codes: EMD-37465 and PDB-8WDU for the Alc. vinosum LH1–RC purified by sucrose-density; EMD-37466 and PDB-8WDV for the Alc. vinosum LH1–RC purified by Ca-DEAE. The numerical source values underlying Fig. 4 can be found in Supplementary Data 1. All other data are available from the authors upon reasonable request.