Abstract
Escherichia coli strains carrying recombinant plasmids encoding either the type 1 fimbria of Salmonella enterica serovar Typhimurium or the G fimbria of E. coli exhibited binding of human 125I-Glu-plasminogen and enhanced the tissue-type plasminogen activator-catalyzed conversion of plasminogen to plasmin. Purified type 1 or G fimbriae similarly bound plasminogen and enhanced its activation. The binding of plasminogen did not involve the characteristic carbohydrate-binding property of the fimbriae but was inhibited at low concentrations by the lysine analog ɛ-aminocaproic acid. Because these fimbrial types bind to laminin of basement membranes (M. Kukkonen et al., Mol. Microbiol. 7:229–237, 1993; S. Saarela et al., Infect. Immun. 64:2857–2860, 1996), the results demonstrate a structural unity in the creation and targeting of bacterium-bound proteolytic plasmin activity to basement membranes.
Plasminogen is the precursor of the human protease plasmin and is abundant in human plasma and extracellular fluids. It is converted to the active, proteolytic form plasmin by eukaryotic activators such as tissue-type plasminogen activator (tPA) and urokinase. tPA is the principal activator in plasma and intercellular fluid, and its action is due to proteolytic cleavage of plasminogen. Plasminogen activation by tPA proceeds poorly in solution but is dramatically enhanced by immobilization of plasminogen on fibrin, eukaryotic tissue surfaces, or certain bacterial cells. Immobilization of plasminogen is a key determinant in the control of plasminogen activation as well as of the formed plasmin activity (reviewed in reference 37). The immobilization is mediated by five kringle domains of plasminogen which bind to lysine-containing domains on the target proteins; immobilization on receptor proteins is associated with dramatic conformational changes in the plasminogen molecule (27). The bound plasminogen is more susceptible to tPA-mediated activation and, on the other hand, is more resistant to the physiological inhibitors of activation and of the formed plasmin activity. Plasmin is a trypsin-like serine protease with a broad substrate specificity and functions in, e.g., the degradation of fibrin (fibrinolysis) and of noncollagenous proteins of the extracellular matrix (ECM), in activation of latent procollagenases, and in penetration of basement membranes (BM) by metastatic cancer cells (for reviews, see references 4, 30 and 37).
Several gram-positive and gram-negative invasive bacterial pathogens have been found to express a plasminogen receptor (PlgR) function (1, 8, 19, 20, 32, 41–43). These bacteria immobilize plasminogen on their cell surfaces and enhance the tPA-catalyzed plasminogen activation. In essence, the bacterial PlgRs function to generate proteolytic activity on the bacterial surface by utilizing a host-derived proteolytic system. To date, only a few bacterial PlgR molecules have been identified. The S fimbriae in Escherichia coli and the curli, or thin aggregative fimbriae, in Salmonella enterica have been identified as PlgRs (32, 39). The M53 protein of group A streptococci and flagellar antigens of E. coli have been found to have plasminogen-binding characteristics (1, 23). An outer cell surface lipoprotein functions as a PlgR in Borrelia burgdorferi (8). In addition, a plasmin receptor of group A streptococci has been identified as a surface-bound glyceraldehyde 3-phosphate dehydrogenase (24, 31).
Bacterial enzymes acting directly on mammalian ECM or activating latent procollagenases exist but are not frequently expressed by invasive, pathogenic species (9, 11). Because plasmin degrades noncollagenous proteins of ECM, such as laminin, and activates latent procollagenases, it has been proposed that one function of bacterial PlgRs is to potentiate bacterial damage to and bacterial spread through tissue barriers, such as BM. This hypothesis (17) was based on findings that adhesiveness to BM (reviewed in reference 45) and expression of the PlgR function (reviewed in references 2 and 25) are shared by a number of invasive bacterial pathogens. Furthermore, it is supported by experimental evidence recently obtained in vitro with S. enterica (21) and with Haemophilus influenzae (44). These two bacteria penetrate in vitro through a reconstituted BM with the help of bacterium-bound plasmin activity. Plasminogen has also been shown to potentiate bacterial transcytosis across an epithelial cell monolayer (47) and, in experimental infections, to enhance spirochetemia by B. burgdorferi in mice (3). S. enterica serovar Typhimurium SH401, used in this study, adheres to the high-mannose chains of laminin as well as to reconstituted BM, and the adherence is mediated by the type 1 fimbriae (18). Typhimurium SH401 also expresses PlgR activity, and it has been demonstrated that plasmin bound on the surfaces of SH401 cells degrades laminin and potentiates bacterial penetration through BM (21). Other studies have described the G (34) or F17 (6) fimbriae on uropathogenic and septicemic enterotoxigenic E. coli that bind to terminal N-acetyl-d-glucosamine residues on glycoproteins, such as laminin of BM (36). We report here that both fimbrial types also exhibit the PlgR function.
MATERIALS AND METHODS
Bacteria.
S. enterica serovar Typhimurium SH401 (18, 21) and the nonfimbriated recipient E. coli strain LE392 (5) were available from previous work. E. coli IHE11088(pRR-5), expressing the G fimbria gene cluster from a uropathogenic E. coli O2 isolate, and IHE11088(pHUB110), expressing a mutated gene cluster, have been described (34, 36). pHUB110 contains a 6-bp deletion within the coding region of the gafD gene, resulting in G-fimbrial filaments lacking the GlcNAc-binding property. The G fimbriae were expressed in the IHE11088 background because in K-12 derivatives the lectin activity of the G fimbria causes autoaggregation of E. coli cells (34). The strains were cultivated in Luria broth or on Luria agar plates, which were supplemented with antibiotics in the case of the recombinant strains.
DNA techniques.
Construction of a cosmid library in plasmid pHC79 from partially Sau3AI-digested Typhimurium genomic DNA, DNA hybridization, subcloning of the fim gene cluster, and DNA manipulations were performed by standard procedures (38).
Plasminogen binding and activation assays.
The binding of plasminogen to purified fimbriae was measured by time-resolved fluorometry (essentially as described in references 19, 21, and 23). The type 1 fimbriae were purified from S. enterica SH401 and the recombinant E. coli and the G fimbriae were purified from E. coli IHE11088(pRR-5) by using deoxycholate and concentrated urea (16). We used the fimbriae at a concentration of 10 μg per ml, human Glu-plasminogen (Biopool, Umeå, Sweden) at 10 or 50 μg/ml, and, in inhibition tests, ɛ-aminocaproic acid (EACA) (Sigma) at 10 mM. To assess the binding of plasminogen to bacterial cells, Glu-plasminogen was labeled with 125I (Amersham, Little Chalfont, Buckinghamshire, United Kingdom) by the Iodogen method (28). The activities obtained were 6 × 105 cpm/μg of plasminogen (the preparation used with the type 1 fimbriae) and 2 × 106 cpm/μg (the preparation used with the G fimbriae). Binding of 125I-plasminogen to bacterial cells was assessed as described previously (19, 23). We used 2 × 109 cells and 0.2 μg of plasminogen in the assay; the inhibition assays were performed with 4 mM EACA.
The ability of the purified fimbriae to enhance plasmin formation by tPA was measured as detailed earlier (23). Purified fimbriae were tested at 10 to 100 μg/ml; bovine serum albumin (BSA), a negative control, was tested at 100 μg/ml; and laminin (Upstate Biotechnology, Lake Placid, N.Y.), a positive control, was tested at 10 μg/ml. Control assays included tests from which plasminogen or tPA was omitted. The ability of bacterial cells to enhance plasmin formation was measured as detailed elsewhere (21, 23, 32). We used a bacterial density of 4 × 108 per ml, plasminogen was tested at 20 μg per ml, tPA was tested at 50 ng per ml, the chromogenic substrate S-2251 was tested at 0.45 mM, and EACA was tested at 0.4 mM, in a test volume of 200 μl. Results from the activation assays are given as data from a representative assay with duplicate independent samples; the range of individual test results was within ±10% of the mean.
Other tests.
Mannose-sensitive yeast cell agglutination (14), agglutination tests with an antiserum raised against purified type 1 fimbriae of S. enterica serovar Typhimurium (15), and electron microscopy of negatively stained recombinant cells (16) were used to assess expression of the type 1 fimbriae of S. enterica SH401. Bacterial agglutination in an antiserum raised against the type 1 fimbriae of E. coli (16) was assessed as a negative control. The expression of the G fimbriae was confirmed by hemagglutination of endo-β-galactosidase-treated human erythrocytes and by bacterial agglutination in an antiserum raised against the purified G fimbriae (34).
RESULTS
Binding of plasminogen to the type 1 fimbriae of S. enterica.
It has been shown that strain SH401 of S. enterica serovar Typhimurium expresses PlgR activity (21). A recombinant strain harboring a cosmid with a 40-kb fragment of SH401 DNA and strongly enhancing tPA-mediated plasminogen activation was isolated and named E. coli LE392(pMK1). The strain agglutinated yeast cells in a mannose-reversible manner, expressed fimbriae as determined by electron microscopy, and was agglutinated in an antiserum raised against the type 1 fimbriae of S. enterica but not in an antiserum against the type 1 fimbriae of E. coli (data not shown), indicating that the recombinant strain LE392(pMK1) expressed the type 1 fimbriae of Typhimurium SH401. Furthermore, the restriction map of pMK1 (data not shown) showed the presence of a 12.5-kb SphI fragment similar to that identified by Purcell et al. (33) and shown to carry the fim cluster of Typhimurium. To test the function of the type 1 fimbria of S. enterica as a PlgR, we subcloned the SphI fragment from pMK1 into plasmid pUC19, obtaining plasmid pMK25, and expressed the SH401 fimbriae for binding and activation experiments in the nonfimbriated strain E. coli LE392.
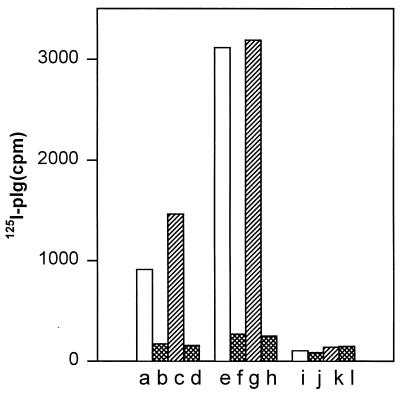
Binding of plasminogen to E. coli LE392(pMK25) is shown in Fig. 1. The strain LE392(pMK25) bound plasminogen three times more efficiently than did E. coli LE392(pUC19) carrying the vector plasmid alone (Fig. 1). It has been shown (23) that the flagella of E. coli LE392 bind plasminogen, and it is likely that the activity seen with the nonfimbriated strain LE392(pUC19) resulted from the binding of plasminogen to the flagella. Binding of plasminogen to both strains was inhibited by 4 mM EACA to the background level seen in tests performed without added bacteria; EACA is a lysine analog and a well-characterized binding inhibitor of the kringle domains of plasminogen (37). Plasmin has a higher affinity than plasminogen for lysine-containing target proteins (19). To exclude the possibility that the observed binding to LE392(pMK25) cells was induced by plasmin possibly present in the plasminogen preparation or formed during the assay, we also performed the binding experiments in the presence of 150 kIU of aprotinin, an inhibitor of plasmin activity (25). The presence of aprotinin did not decrease the binding of plasminogen to LE392(pMK25) or LE392(pUC19) cells; on the contrary, a slight increase was observed (Fig. 1).
FIG. 1.
Binding of 125I-plasminogen (125I-plg) onto recombinant E. coli expressing the type 1 fimbriae from S. enterica. Bars a to d show the nonfimbriated strain E. coli LE392(pUC19); bars e to h show E. coli LE392(pMK25) cells expressing the type 1 fimbriae. The background binding to BSA-coated plastic is shown in bars i to l. The binding in the absence of EACA and aprotinin is shown by bars a, e, and i, and the binding in the presence of 4 mM EACA is shown by bars b, f, and j. Bars c, g, and k show the binding in the presence of 150 kIU of aprotinin, and bars d, h, and l show the binding in the presence of both EACA and aprotinin.
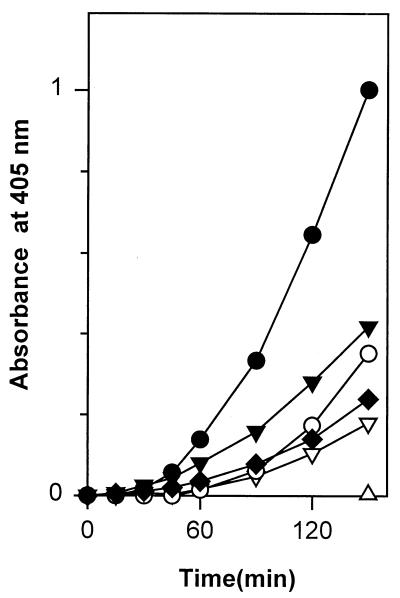
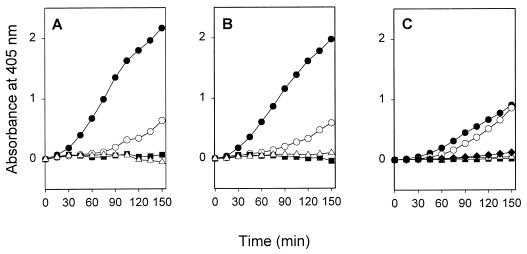
We next assessed whether the fimbriated recombinant E. coli strain enhanced tPA-catalyzed plasminogen activation. In different experiments, strain LE392(pMK25) caused a 5- to 10-fold enhancement in plasmin formation by tPA, compared to the activation seen in buffer alone, whereas the strain LE392(pUC19) caused only a 2-fold increase in the activation (Fig. 2). With both bacterial strains, EACA decreased plasmin formation close to the level seen in buffer without bacteria. Control experiments in the presence of LE392(pMK25) or LE392(pUC19) cells showed that no plasmin was formed if either plasminogen or tPA was omitted from the suspensions (Fig. 2).
FIG. 2.
Enhancement of tPA-catalyzed plasmin formation in the presence of recombinant E. coli expressing the type 1 fimbriae of S. enterica. Kinetic measurements of tPA-catalyzed plasmin formation in the presence of LE392(pUC19) cells (▾), LE392(pUC19) cells and 0.4 mM EACA (▿), LE392(pMK25) cells (•), and LE392(pMK25) cells and 0.4 mM EACA (○) and in the absence of bacterial cells (⧫) are shown. ▵, plasmin formation when tPA or plasminogen was omitted in the tests performed with strain LE392(pMK25).
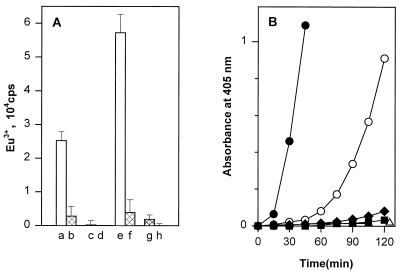
To confirm the function of the type 1 fimbria of S. enterica SH401 as a PlgR, we tested the capacity of purified fimbriae to bind plasminogen. The purified fimbriae efficiently bound plasminogen (Fig. 3). The binding was dependent on the concentration of plasminogen used in the assay and was inhibited by EACA to a level close to the background level seen with immobilized BSA. At 10 μg/ml, the type 1 fimbriae caused a 100-fold enhancement of the tPA-catalyzed activation compared to the activation in phosphate-buffered saline containing only plasminogen and tPA (Fig. 3). The enhancement was dependent on the concentration of the type 1 fimbriae (data not shown). BSA at 100 μg/ml caused only a marginal enhancement of the tPA-catalyzed activation. The formation of plasmin activity in the presence of the fimbriae was greatly decreased when 0.8 mM EACA was added to the protein mixture, again indicating that the immobilization of plasminogen on the purified fimbriae was necessary for the observed enhancement. No difference was observed between the type 1 fimbriae purified from the wild-type strain S. enterica SH401 and those purified from the recombinant E. coli strains (data not shown). The formation of plasmin on the bacterial cells or on the purified fimbriae was not affected by the addition of 2.5 mM α-methyl-d-mannoside into the suspension (data not shown).
FIG. 3.
Binding of plasminogen (A) and enhancement of tPA-catalyzed plasmin formation (B) on purified type 1 fimbriae of S. enterica. (A) Plasminogen was added at two concentrations, 10 μg/ml (bars a to d) and 50 μg/ml (bars e to h), to plastic coated with fimbriae, and the binding was measured by time-resolved fluorometry. Binding was assessed in the absence (bars a, c, e, and g) or presence (bars b, d, f, and h) of 10 mM EACA. Bars a, b, e, and f show the binding to type 1 fimbriae, and bars c, d, g, and h show the binding to plastic coated with BSA. (B) Enhancement of tPA-catalyzed plasmin formation was measured at a fimbria concentration of 10 μg/ml in the absence (•) or presence (○) of 0.8 mM EACA. Levels of plasmin formation in the presence of BSA (100 μg/ml) (⧫) or in buffer alone (■) are also shown. ▵, plasmin activity when tPA or plasminogen was omitted in the tests performed with type 1 fimbriae.
Function of the G fimbria of E. coli as a PlgR.
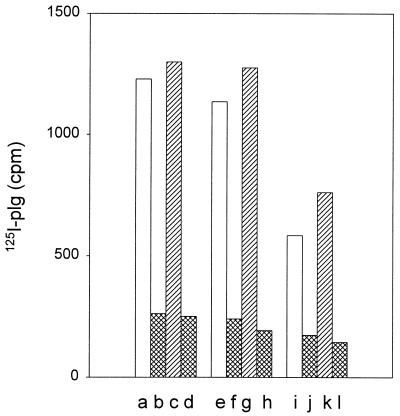
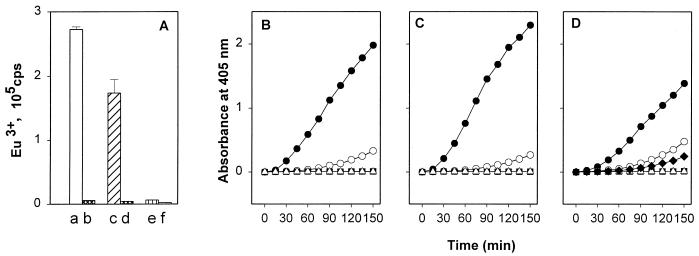
Because the G-fimbriated, uropathogenic E. coli strain IHE11165 expresses PlgR activity (data not shown), we tested the binding of plasminogen to the G-fimbriated recombinant strain E. coli IHE11088(pRR-5). To evaluate the role of the fimbrial lectin activity, the test also included the strain IHE11088(pHUB110), expressing G-fimbrial filaments deleted in frame for the amino acids Gly-94 and Thr-95 of the GafD lectin subunit and lacking the GlcNAc-binding property (34). Both G-fimbriated strains bound plasminogen and the plasminogen-aprotinin complex more efficiently than did the nonfimbriated strains IHE11088 (Fig. 4) and IHE11088(pACYC184) (data not shown). Again, the binding of plasminogen was inhibited by a small amount of EACA.
FIG. 4.
Binding of 125I-plasminogen (125I-plg) onto G-fimbriated bacterial cells. Bars a to d show E. coli IHE11088(pRR-5) with the G fimbria gene cluster, bars e to h show E. coli IHE11088(pHUB110) expressing G fimbriae that lack the GlcNAc-binding property, and bars i to l show the nonfimbriated E. coli strain IHE11088. Binding in the absence of EACA and aprotinin is shown in bars a, e, and i, and binding in the presence of 4 mM EACA is shown in bars b, f, and j. Bars c, g, and k show the binding in the presence of 150 kIU of aprotinin, and bars d, h, and l show the binding in the presence of both EACA and aprotinin.
The E. coli strain IHE11088(pRR-5), expressing G fimbriae, and the strain IHE11088(pHUB110), expressing nonadhesive G fimbriae, enhanced plasmin formation compared to the activation seen in the presence of IHE11088 (Fig. 5) or IHE11088(pACYC184) (data not shown) cells lacking fimbriae. In the presence of EACA, the formation of plasmin was inhibited and no endogenous plasminogen activation was detected. The wild-type G fimbriae from E. coli IHE11088(pRR-5) and the mutated G fimbriae from IHE11088(pHUB110) both bound plasminogen and enhanced the tPA-catalyzed plasmin formation in an EACA-inhibitable manner (Fig. 6). Tested at 10 μg/ml, the ability of the G fimbriae to enhance the tPA-catalyzed plasminogen activation was similar to that shown by laminin, a characterized ECM target for plasminogen binding (Fig. 6).
FIG. 5.
Enhancement of tPA-catalyzed plasmin formation in the presence of E. coli IHE11088(pRR-5) expressing G fimbriae (A), of E. coli IHE11088(pHUB110) expressing G fimbriae devoid of GlcNAc-binding activity (B), and of the nonfimbriated strain IHE11088 (C). Also shown are plasmin formation in the presence of bacteria, plasminogen, and tPA (•); plasmin formation when 0.4 mM EACA was added to the suspension (○); plasmin formation when tPA was omitted (▵); plasmin formation when plasminogen was omitted (■); and tPA-catalyzed plasmin formation without added bacteria (⧫).
FIG. 6.
Binding of plasminogen and enhancement of tPA-catalyzed plasmin formation on purified G fimbriae of E. coli. (A) The binding of plasminogen to purified fimbriae was measured by time-resolved fluorometry. Bars a and b show binding to the wild-type G fimbriae, bars c and d show binding to the mutated G fimbriae, and bars e and f show binding to BSA. Binding in the absence of EACA is shown in bars a, c, and e; binding in the presence of 10 mM EACA is shown in bars b, d, and f. Levels of plasmin formation in the presence of the G fimbriae from E. coli IHE11088(pRR-5) (B) and of the mutated G fimbriae from E. coli IHE11088(pHUB110) (C) are shown. Plasmin formation in the presence of fimbriae (•), in the presence of fimbriae and 0.4 mM EACA (○), without added tPA (▵), and without added plasminogen (■) is shown. (D) For comparison, plasmin formation in the presence of laminin (•), laminin and 0.4 mM EACA (○), and BSA (⧫) is shown.
DISCUSSION
Our results demonstrate that the type 1 fimbriae of S. enterica serovar Typhimurium as well as the G fimbriae of E. coli function as PlgRs. This was inferred from two lines of evidence. First, the purified fimbriae efficiently bound plasminogen and enhanced tPA-mediated activation of plasminogen. Second, expression of both fimbria gene clusters in E. coli resulted in enhanced plasmin formation and plasminogen binding. Our results also give evidence that the carbohydrate-binding activity of the fimbriae is not involved in the interaction with plasminogen.
A similar enhancement of plasminogen binding and of tPA-mediated activation was observed with the complete G fimbriae and with the mutated fimbriae lacking GlcNAc binding. This indicates that the plasminogen binding and acquisition of a correct formation for tPA-mediated activation are not dependent on the lectin activity of the G fimbria. The mutated GafD lectin subunit encoded by pHUB110 is expressed in a few copies in the G-fimbria filament, similarly to the wild-type GafD lectin (35). Furthermore, we found that the receptor analog for the type 1 fimbriae, α-methyl-d-mannoside, had no effect on plasmin formation. In contrast, the binding and activation processes observed with both fimbrial types were efficiently inhibited by a low concentration of the lysine analog EACA. This indicates that the fimbria-plasminogen interactions follow the general mechanism of plasminogen-receptor interactions that involve the lysine-binding kringle domains of plasminogen (27, 37). The results thus indicate that the fimbrial filaments are functioning as targets in their interaction with plasminogen.
The expression of S. enterica fim genes on the plasmid pMK25 as well as of the gaf gene cluster on the plasmids pRR-5 and pHUB110 enhanced binding and activation of plasminogen on recombinant E. coli cells. These results indicate that the fim and the gaf gene clusters are sufficient to confer plasminogen-binding capacity on bacterial cells. The variable and seemingly lower PlgR activity exhibited by the recombinant strains, compared to that shown by the purified fimbriae, most likely resulted from fimbrial phase variation, which rendered the bacterial cell population heterogeneous in regard to fimbriation. The type 1 and the G fimbriae are not the only surface appendages of Salmonella or E. coli that have the capacity to bind plasminogen and enhance its activation by tPA. Curli, or thin aggregative fimbriae, have also been identified as PlgRs (39). We have not detected any expression of curli by Typhimurium strain SH401 under the growth conditions we used. Additionally, other research (23) indicates that the flagella of S. enterica also function as PlgRs. Finally, PlgR activity is associated with the S fimbriae of meningitis-associated E. coli (32). It is notable that many of the identified bacterial PlgRs are filamentous structures, such as fimbriae, flagella, or M proteins (1), which bear morphological similarity to fibrin, a major eukaryotic target for plasminogen binding and plasmin activity (37). On the other hand, it is apparent that not all fimbrial filaments express PlgR activity (e.g., PlgR activity in P-fimbrial variants occurring on human uropathogenic E. coli has not been detected [13]). A detailed comparison of the various enterobacterial fimbrial types in regard to PlgR function, however, has not been performed. Furthermore, our knowledge of the fimbrial subunits and of amino acid sequences recognized by plasminogen is very limited (32). Overall, the binding of plasminogen to lysine and lysine analogs is well characterized (27), but other structural requirements for PlgR activity have remained poorly defined. A plasminogen-binding 13- to 16-mer repeat domain has been identified in the PAM surface protein of group A streptococci (46); this sequence, however, exhibits no significant identity to the sequences of the major and minor fimbrial subunits of this study. Our present results indicate that the lectin activity of fimbriae is not involved in the PlgR function; however, this does not rule out the possibility that certain regions of the FimH or the GafD adhesins are physically involved in plasminogen binding.
It has been shown that PlgRs on the surfaces of Typhimurium SH401 cells function under physiological conditions to create bacterium-bound plasmin activity that can be targeted at laminin and BM (22), which are adhesion targets for the fimbriae used in this study (18, 36). Adhesion to laminin and activation of plasminogen and other latent proteases are important in tumor cell invasion through tissue barriers (4, 30). In vitro evidence has been presented in favor of the hypothesis that these characteristics also contribute to bacterial penetration through BM; this process has been termed bacterial metastasis in an analogy to the behavior of metastatic tumor cells (21, 44). The contribution to virulence of our present findings still needs to be assessed in vivo. To date, the role of plasminogen activation in vivo has been tested in two animal models. The plasminogen activator surface protein was found to enhance the spread of Yersinia pestis into circulation (40), and plasminogen activation was required for dissemination of B. burgdorferi in the tick vector and for enhancement of spirochetemia in mice (3). Interestingly, both of these bacterial species also express ECM- and BM-binding properties (10, 12, 21). In particular, research that associates the expression of the plasminogen activator of Y. pestis with an increased adhesiveness to laminin (22) stresses the common occurrence of laminin adherence and PlgR function in invasive enterobacterial pathogens. BMs are considered to function as reservoirs for plasminogen, metalloproteinases (collagenases), and plasminogen activators (7, 26, 29). Hence, a close association of bacteria with BM in tissues might bring them to a microenvironment where formation and targeting of plasmin activity can effectively promote bacterial metastasis. The major finding of the present study is a physical link between adhesiveness to tissues and immobilization of plasminogen on a surface filament of two invasive bacterial pathogens.
ACKNOWLEDGMENTS
This work has been supported by grants from the University of Helsinki, the Sigrid Jusélius Foundation, and the Academy of Finland (grant numbers 29346, 42103, and 42107).
We thank Raili Lameranta for technical assistance.
REFERENCES
- 1.Berge A, Sjöbring U. PAM, a novel plasminogen-binding protein from Streptococcus pyogenes. J Biol Chem. 1993;268:25417–25424. [PubMed] [Google Scholar]
- 2.Boyle M D P, Lottenberg R. Plasminogen activation by invasive human pathogens. Thromb Haemostasis. 1997;77:1–10. [PubMed] [Google Scholar]
- 3.Coleman J L, Gebbia J A, Piesman J, Degen J L, Buggs T H, Benach J L. Plasminogen is required for efficient dissemination of B. burgdorferi in ticks and for enhancement of spirochetemia in mice. Cell. 1997;89:1111–1119. doi: 10.1016/s0092-8674(00)80298-6. [DOI] [PubMed] [Google Scholar]
- 4.Danø K, Andreasen P A, Grøndahl-Hansen J, Kristensen P, Nielsen L S, Skriver L. Plasminogen activators, tissue degradation and cancer. Adv Cancer Res. 1985;44:139–266. doi: 10.1016/s0065-230x(08)60028-7. [DOI] [PubMed] [Google Scholar]
- 5.Elliot S J, Nandapalan N, Chang B J. Production of type 1 fimbriae by Escherichia coli HB101. Microb Pathog. 1991;10:481–486. doi: 10.1016/0882-4010(91)90114-p. [DOI] [PubMed] [Google Scholar]
- 6.El Mazouari K, Oswald E, Hernalsteens J-P, Lintermans P, De Greve H. F17-like fimbriae from an invasive Escherichia coli strain producing cytotoxic necrotizing factor type 2 toxin. Infect Immun. 1994;62:2633–2638. doi: 10.1128/iai.62.6.2633-2638.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farina A R, Tiberio A, Tacconelli A, Cappabianca L, Guline A, Mackay A R. Identification of plasminogen in Matrigel and its activation by reconstitution of this basement membrane extract. BioTechniques. 1997;21:904–909. doi: 10.2144/96215rr03. [DOI] [PubMed] [Google Scholar]
- 8.Fuchs H, Wallich R, Simon M M, Kramer M D. The outer surface protein A of the spirochete Borrelia burgdorferi is a plasmin(ogen) receptor. Proc Natl Acad Sci USA. 1994;91:12594–12598. doi: 10.1073/pnas.91.26.12594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goguen J D, Hoe N P, Subrahmanyam Y V B K. Proteases and bacterial virulence: a view from the trenches. Infect Agents Dis. 1995;4:47–54. [PubMed] [Google Scholar]
- 10.Guo B P, Norris S J, Rosenberg L C, Höök M. Adherence of Borrelia burgdorferi to the proteoglycan decorin. Infect Immun. 1995;63:3467–3472. doi: 10.1128/iai.63.9.3467-3472.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harrington D J. Bacterial collagenases and collagen-degrading enzymes and their potential role in human disease. Infect Immun. 1996;64:1885–1891. doi: 10.1128/iai.64.6.1885-1891.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kienle Z, Emödy L, Svanborg C, O’Toole P W. Adhesive properties conferred by the plasminogen activator of Yersinia pestis. J Gen Microbiol. 1992;138:1679–1687. doi: 10.1099/00221287-138-8-1679. [DOI] [PubMed] [Google Scholar]
- 13.Korhonen, T. K. Unpublished data.
- 14.Korhonen T K. Yeast cell agglutination by purified enterobacterial pili. FEMS Microbiol Lett. 1979;6:421–425. [Google Scholar]
- 15.Korhonen T K, Lounatmaa K, Ranta H, Kuusi N. Characterization of type 1 pili of Salmonella typhimurium LT2. J Bacteriol. 1980;144:800–805. doi: 10.1128/jb.144.2.800-805.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korhonen T K, Nurmiaho E-L, Ranta H, Svanborg Edén C. New method for isolation of immunologically pure pili from Escherichia coli. Infect Immun. 1980;27:569–575. doi: 10.1128/iai.27.2.569-575.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Korhonen T K, Virkola R, Lähteenmäki K, Björkman Y, Kukkonen M, Raunio T, Tarkkanen A-M, Westerlund B. Penetration of fimbriate enteric bacteria through basement membranes: a hypothesis. FEMS Microbiol Lett. 1992;100:307–312. doi: 10.1111/j.1574-6968.1992.tb14057.x. [DOI] [PubMed] [Google Scholar]
- 18.Kukkonen M, Raunio T, Virkola R, Lähteenmäki K, Mäkelä P H, Klemm P, Clegg S, Korhonen T K. Basement membrane carbohydrate as a target for bacterial adhesion: binding of type 1 fimbriae of Salmonella enterica and Escherichia coli to laminin. Mol Microbiol. 1993;7:229–237. doi: 10.1111/j.1365-2958.1993.tb01114.x. [DOI] [PubMed] [Google Scholar]
- 19.Kuusela P, Saksela O. Binding and activation of plasminogen at the surface of Staphylococcus aureus. Eur J Biochem. 1990;193:759–765. doi: 10.1111/j.1432-1033.1990.tb19397.x. [DOI] [PubMed] [Google Scholar]
- 20.Kuusela P, Ullberg M, Saksela O, Kronvall G. Tissue-type plasminogen activator-mediated activation of plasminogen on the surface of group A, C, and G streptococci. Infect Immun. 1992;60:196–201. doi: 10.1128/iai.60.1.196-201.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lähteenmäki K, Virkola R, Pouttu R, Kuusela P, Kukkonen M, Korhonen T K. Bacterial plasminogen receptors: in vitro evidence for a role in degradation of the mammalian extracellular matrix. Infect Immun. 1995;63:3659–3664. doi: 10.1128/iai.63.9.3659-3664.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lähteenmäki, K., R. Virkola, A. Sarén, L. Emödy, and T. K. Korhonen. Expression of the plasminogen activator Pla of Yersinia pestis enhances bacterial attachment to the mammalian extracellular matrix. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 23.Lähteenmäki K, Westerlund B, Kuusela P, Korhonen T K. Immobilization of plasminogen on Escherichia coli flagella. FEMS Microbiol Lett. 1993;106:309–314. doi: 10.1111/j.1574-6968.1993.tb05981.x. [DOI] [PubMed] [Google Scholar]
- 24.Lottenberg R, Broder C C, Boyle M D P, Kain S J, Schroeder B L, Curtiss R., III Cloning, sequence analysis, and expression in Escherichia coli of a streptococcal plasmin receptor. J Bacteriol. 1992;174:5204–5210. doi: 10.1128/jb.174.16.5204-5210.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lottenberg R, Minning-Wenz D, Boyle M D P. Capturing host plasmin(ogen): a common mechanism for invasive pathogens? Trends Microbiol. 1994;2:20–24. doi: 10.1016/0966-842x(94)90340-9. [DOI] [PubMed] [Google Scholar]
- 26.Mackay A R, Gomez D E, Cottam D W, Rees R C, Nason A M, Thorgeirsson U P. Identification of the 72-kDa (MMP-2) and 92-kDa (MMP-9) gelatinase/type IV collagenase in preparations of laminin and Matrigel. BioTechniques. 1993;15:1048–1051. [PubMed] [Google Scholar]
- 27.Mangel W F, Lin B, Ramakrishnan V. Characterization of an extremely large, ligand-induced conformational change in plasminogen. Science. 1990;248:69–73. doi: 10.1126/science.2108500. [DOI] [PubMed] [Google Scholar]
- 28.Markwell M A K, Fox C F. Surface-specific iodination of membrane proteins of viruses and eukaryotic cells using 1,3,4,6-tetrachloro-3α,6α-diphenylglycouril. Biochemistry. 1978;17:4807–4817. doi: 10.1021/bi00615a031. [DOI] [PubMed] [Google Scholar]
- 29.McGuire A R, Seed N W. The interaction of plasminogen activator with reconstituted basement matrix and extracellular macromolecules produced by cultured epithelial cells. J Biol Chem. 1989;40:215–227. doi: 10.1002/jcb.240400210. [DOI] [PubMed] [Google Scholar]
- 30.Mignatti P, Rifkin D B. Biology and biochemistry of proteinases in tumor invasion. Physiol Rev. 1993;73:161–195. doi: 10.1152/physrev.1993.73.1.161. [DOI] [PubMed] [Google Scholar]
- 31.Pancholi V, Fischetti V A. A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate-dehydrogenase with multiple binding activity. J Exp Med. 1992;176:415–426. doi: 10.1084/jem.176.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parkkinen J, Hacker J, Korhonen T K. Enhancement of tissue plasminogen activator-catalyzed plasminogen activation by Escherichia coli S fimbriae associated with neonatal septicaemia and meningitis. Thromb Haemostasis. 1991;65:483–486. [PubMed] [Google Scholar]
- 33.Purcell B K, Pruckler J, Clegg S. Nucleotide sequences of the genes encoding type 1 fimbrial subunits of Klebsiella pneumoniae and Salmonella typhimurium. J Bacteriol. 1987;169:5831–5834. doi: 10.1128/jb.169.12.5831-5834.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saarela S, Taira S, Nurmiaho-Lassila E-L, Makkonen A, Rhen M. The Escherichia coli G-fimbrial lectin protein participates both in fimbrial biogenesis and in recognition of the receptor N-acetyl-d-glucosamine. J Bacteriol. 1995;177:1477–1484. doi: 10.1128/jb.177.6.1477-1484.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saarela, S., J. Tanskanen, B. Westerlund-Wikström, and T. K. Korhonen. Unpublished data.
- 36.Saarela S, Westerlund-Wikström B, Rhen M, Korhonen T K. The GafD protein of the G (F17) fimbrial complex confers adhesiveness of Escherichia coli to laminin. Infect Immun. 1996;64:2857–2860. doi: 10.1128/iai.64.7.2857-2860.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saksela O, Rifkin D B. Cell-associated plasminogen activation: regulation and physiological functions. Annu Rev Cell Biol. 1988;4:93–126. doi: 10.1146/annurev.cb.04.110188.000521. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 39.Sjöbring U, Pohl G, Olsén A. Plasminogen, absorbed by Escherichia coli expressing curli or by Salmonella enteritidis expressing thin aggregative fimbriae, can be activated by simultaneously captured tissue-type plasminogen activator (t-PA) Mol Microbiol. 1994;14:443–452. doi: 10.1111/j.1365-2958.1994.tb02179.x. [DOI] [PubMed] [Google Scholar]
- 40.Sodeinde O A, Subrahmanyam Y V B K, Stark K, Quan T, Bao Y, Goguen J D. A surface protease and the invasive character of plague. Science. 1992;258:1004–1007. doi: 10.1126/science.1439793. [DOI] [PubMed] [Google Scholar]
- 41.Ullberg M, Kronvall G, Karlsson I, Wiman B. Receptors for human plasminogen on gram-negative bacteria. Infect Immun. 1990;58:21–25. doi: 10.1128/iai.58.1.21-25.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ullberg M, Kronvall G, Wiman B. New receptor for human plasminogen on gram positive cocci. APMIS. 1989;97:996–1002. doi: 10.1111/j.1699-0463.1989.tb00508.x. [DOI] [PubMed] [Google Scholar]
- 43.Ullberg M, Kuusela P, Kristiansen B-E, Kronvall G. Binding of plasminogen to Neisseria meningitidis and Neisseria gonorrhoeae and formation of surface-associated plasmin. J Infect Dis. 1992;166:1329–1334. doi: 10.1093/infdis/166.6.1329. [DOI] [PubMed] [Google Scholar]
- 44.Virkola R, Lähteenmäki K, Eberhard T, Kuusela P, van Alphen L, Ullberg M, Korhonen T K. Interaction of Haemophilus influenzae with the mammalian extracellular matrix. J Infect Dis. 1996;173:1137–1147. doi: 10.1093/infdis/173.5.1137. [DOI] [PubMed] [Google Scholar]
- 45.Westerlund B, Korhonen T K. Bacterial proteins binding to the mammalian extracellular matrix. Mol Microbiol. 1993;9:687–694. doi: 10.1111/j.1365-2958.1993.tb01729.x. [DOI] [PubMed] [Google Scholar]
- 46.Wistedt A C, Ringdahl U, Müller-Esterl W, Sjöbring U. Identification of a plasminogen-binding motif in PAM, a bacterial surface protein. Mol Microbiol. 1995;18:569–578. doi: 10.1111/j.1365-2958.1995.mmi_18030569.x. [DOI] [PubMed] [Google Scholar]
- 47.Zavizion B, White J H, Bramley A J. Staphylococcus aureus stimulates urokinase-type plasminogen activator expression by bovine mammary cells. J Infect Dis. 1997;176:1637–1640. doi: 10.1086/517345. [DOI] [PubMed] [Google Scholar]