Abstract
Despite the importance of methicillin-resistant Staphylococcus aureus (MRSA) as a priority nosocomial pathogen, the genome sequences of Malaysian MRSA isolates are currently limited to a small pool of samples. Here, we present the genome sequence analyses of 88 clinical MRSA isolates obtained from the main tertiary hospital in Terengganu, Malaysia in 2016–2020, to obtain in-depth insights into their characteristics. The EMRSA-15 (ST22-SCCmec IV) clone of the clonal complex 22 (CC22) lineage was predominant with a total of 61 (69.3%) isolates. Earlier reports from other Malaysian hospitals indicated the predominance of the ST239 clone, but only two (2.3%) isolates were identified in this study. Two Indian-origin clones, the Bengal Bay clone ST772-SCCmec V (n = 2) and ST672 (n = 10) were also detected, with most of the ST672 isolates obtained in 2020 (n = 7). Two new STs were found, with one isolate each, and were designated ST7879 and ST7883. From the core genome phylogenetic tree, the HSNZ MRSA isolates could be grouped into seven clades. Antimicrobial phenotype-genotype concordance was high (> 95%), indicating the accuracy of WGS in predicting most resistances. Majority of the MRSA isolates were found to harbor more than 10 virulence genes, demonstrating their pathogenic nature.
Subject terms: Antimicrobial resistance, Bacterial genomics, Pathogens
Introduction
As a major nosocomial pathogen, methicillin-resistant Staphylococcus aureus (MRSA) sparks considerable interest and has been the focus of multiple whole genome sequencing (WGS) studies. Many WGS studies on MRSA entail outbreak investigations1–3, indicating the high transmissibility of this microbe and the consequent danger of outbreak eruption. In addition, the highly virulent and multidrug-resistant nature of MRSA imposes a considerable burden on the medical community, emphasizing its importance as a bacterial pathogen. Infections caused by MRSA strains are usually associated with prolonged hospitalization and high treatment costs. A positive MRSA test can result in an additional 6.6 days of hospitalization and a USD 9,237 post-discharge expense4. A nationwide study in Japan involving data from inpatients at 1,133 acute care hospitals between April 1, 2014 and March 31, 2015, projected the overall incremental burden of MRSA to be around USD 2 billion, with a length of hospitalization of 4.34 million days5. Compared to methicillin-susceptible S. aureus (MSSA), patients with MRSA infections have higher morbidity and mortality rates. For example, a cohort study conducted in India found that MSSA patients fared better in terms of survival than MRSA patients6. Additionally, MRSA patients also have a much greater incidence of sequelae such as sepsis and multiorgan dysfunction 6. In Malaysia, the mortality rate of MRSA infection was reported at 11.76% (6/51) among patients admitted to the general surgical wards of Hospital Tuanku Jaafar (HTJ)7 and 37.3% (25/67) among patients with MRSA bacteremia at University Malaya Medical Centre (UMMC)8.
The National Surveillance of Antimicrobial Resistance (NSAR) program in Malaysia tracks and reports on nine pathogen species every year, including MRSA. The MRSA prevalence rates typically varied between 15 and 20% in years 2011–2020, but in 2021 they only reached 7%9. Although the incidence of MRSA in Malaysian hospitals appears to be on the decline, vigilant surveillance of this pathogen is still required. Even in non-outbreak circumstances, MRSA has a number of potential transmission events involving healthcare personnels, patients, and the environment that could ultimately result in an outbreak10. Thus, regular surveillance of MRSA is required to maintain control over this infection. Whole genome research involving MRSA strains is still limited in Malaysia and typically includes a small number of samples11. In this study, we used WGS to examine 88 clinical MRSA isolates collected over a period of five years (2016 – 2020) from a tertiary hospital in the state of Terengganu, Malaysia, to provide a comprehensive genomic insight of these isolates as well as to determine their population structure and strain relatedness. A further 18 MRSA genomes from Malaysia available in GenBank were also included in the analyses.
Results
Antimicrobial susceptibility of the HSNZ MRSA isolates (2016–2020)
A total of 197 MRSA isolates were collected from the Microbiology Laboratory of Hospital Sultanah Nur Zahirah (HSNZ), the main public tertiary hospital in the state of Terengganu, Malaysia from July 2016 to December 2020 (but with no collection in 2018). The antimicrobial resistance rates along with the prevalence of the macrolide-lincosamide-streptogrammin B (MLSB) resistance phenotypes and multidrug resistance (MDR) among these isolates are indicated in Table 1. The MRSA isolates exhibited high levels of phenotypic resistance to the following antimicrobial classes: beta-lactams (> 90% for cefoxitin, oxacillin, penicillin, and cefoperazone), fluoroquinolones (78.7% for ciprofloxacin, and 74.6% for moxifloxacin), and MLSB (62.9% for erythromycin, and 58.9% for clindamycin). All isolates were susceptible to glycopeptides, linezolid, and ceftaroline, whereas resistance rates for other antibiotics ranged from 1 to 19% (Table 1). Inducible clindamycin resistance (iMLSB or D-test positive) was the most prevalent MLSB phenotype, occurring in 52.3% (103/197) of the isolates, whereas constitutive resistance (cMLSB or resistance to both erythromycin and clindamycin) and the MS phenotype (D-test negative) were observed much less frequently at 6.6% (13/197) and 4.0% (8/197), respectively.
Table 1.
Antimicrobial resistance characteristics of the HSNZ MRSA isolates obtained in 2016–2020.
| Antimicrobial | No. of isolates per year | Total no. of isolates (n = 197) (%) | |||
|---|---|---|---|---|---|
| 2016 (n = 69) (%) | 2017 (n = 69) (%) | 2019 (n = 43) (%) | 2020 (n = 16) (%) | ||
| Beta-lactam | |||||
| Cefoxitin | 69 (100) | 69 (100) | 43 (100) | 16 (100) | 197 (100) |
| Oxacillin | 69 (100) | 65 (94.2) | 38 (88.4) | 14 (87.5) | 186 (94.4) |
| Penicillin | 69 (100) | 68 (98.6) | 43 (100) | 16 (100) | 196 (99.5) |
| Cefoperazone | 65 (94.2) | 64 (92.8) | 36 (83.7) | 14 (87.5) | 179 (90.9) |
| Fluoroquinolone | |||||
| Ciproloxacin | 52 (75.4) | 54 (78.3) | 35 (81.4) | 14 (87.5) | 155 (78.7) |
| Moxifloxacin | 52 (75.4) | 54 (78.3) | 28 (65.1) | 13 (81.3) | 147 (74.6) |
| Macrolide | |||||
| Erythromycin | 48 (69.6) | 51 (73.9) | 17 (39.5) | 8 (50.0) | 124 (62.9) |
| Lincosamide | |||||
| Clindamycin | 46 (66.7) | 46 (66.7) | 17 (39.5) | 7 (43.8) | 116 (58.9) |
| Aminoglycoside | |||||
| Gentamicin | 15 (21.7) | 11 (15.9) | 5 (11.6) | 6 (37.5) | 37 (18.8) |
| Amikacin | 11 (15.9) | 4 (5.8) | 0 | 0 | 15 (7.6) |
| Folate inhibitor | |||||
| Trimethoprim-sulfamethoxazole | 8 (11.6) | 2 (2.9) | 1 (2.3) | 0 | 11 (5.6) |
| Fucidane | |||||
| Fusidic acid | 7 (10.1) | 12 (17.4) | 3 (7.0) | 1 (6.3) | 23 (11.7) |
| Tetracycline | |||||
| Tetracycline | 9 (13.0) | 4 (5.8) | 2 (4.7) | 0 | 15 (7.6) |
| Doxycycline | 6 (8.7) | 1 (1.4) | 1 (2.3) | 0 | 8 (4.1) |
| Minocycline | 3 (4.3) | 0 | 0 | 0 | 3 (1.5) |
| Glycylcycline | |||||
| Tigecycline | 1 (1.4) | 4 (5.8) | 0 | 5 (31.3) | 10 (5.1) |
| Phenicol | |||||
| Chloramphenicol | 1 (1.4) | 6 (8.7) | 1 (2.3) | 1 (6.3) | 9 (4.6) |
| Monoxycarbolic acid | |||||
| Mupirocin | 1 (1.4) | 1 (1.4) | 0 | 0 | 2 (1.0) |
| Ansamycin | |||||
| Rifampin | 2 (2.9) | 0 | 1 (2.3) | 0 | 3 (1.5) |
| Glycopeptide | |||||
| Vancomycin | 0 | 0 | 0 | 0 | 0 |
| Teicoplanin | 0 | 0 | 0 | 0 | 0 |
| Oxazolidinone | |||||
| Linezolid | 0 | 0 | 0 | 0 | 0 |
| Streptogramin | |||||
| Quinupristin-dalfopristin | 0 | 0 | 0 | 0 | 0 |
| Anti-MRSA cephalosporin | |||||
| Ceftaroline | 0 | 0 | 0 | 0 | 0 |
| MLSB resistance phenotype | |||||
| cMLSB | 7 (10.1) | 5 (7.2) | 1 (2.3) | 0 | 13 (6.6) |
| iMLSB | 39 (56.5) | 41 (59.4) | 16 (37.2) | 7 (43.8) | 103 (52.3) |
| MS | 2 (2.9) | 5 (7.2) | 0 | 1 (6.3) | 8 (4.1) |
| Susceptible | 21 (30.4) | 18 (26.1) | 26 (60.5) | 8 (50.0) | 73 (37.1) |
| No. of MDR isolates | 54 (78.3) | 57 (82.6) | 21 (48.8) | 14 (87.5) | 146 (74.1) |
| No. of non-MDR isolates | 15 (21.7) | 12 (17.4) | 22 (51.2) | 2 (12.5) | 51 (25.9) |
| No. of MDR isolates selected for whole genome sequencing in this study | 25 | 29 | 20 | 14 | 88 |
The majority of the MRSA isolates (146/197; 74.1%) were multidrug-resistant (MDR), exhibiting resistance primarily against four antibiotic classes. One MDR isolate, SauR65, had a notable level of resistance with up to 10 antibiotic classes. The rest of the isolates (25.9%, 51/197) showed resistance only to beta-lactams (n = 15) or beta-lactams in combination with another antibiotic class (n = 36) (Table 1). It should be noted that the isolates obtained in 2020 were considerably fewer in quantity (n = 16) due to the Covid-19 pandemic which restricted our access to the hospital laboratory.
Genome assembly quality
Out of the 146 MDR MRSA isolates that were collected, a total of 88 MDR MRSA isolates were randomly chosen to be sequenced on the short-read Illumina or DNBSeq platforms with the mean sequencing coverage of 205 × . The average number of contigs among the sequenced isolates was 53 (Min = 24; Max = 173). The contigs had an average total length of 2,819,047 bp, while the GC content averaged at 32.7%. Additionally, the average N50 value was 189,136, while the average L50 value was six. The details of the assembly qualities and statistics are available in Supplementary Table S2.
Epidemiological characteristics of the sequenced MRSA isolates
Molecular typing using whole genome data identified three main SCCmec types, 11 sequence types (STs) and 19 spa types (Fig. 1). ST22-SCCmec IV was identified as the predominant clone in 61 isolates (69.3%), followed by ST672-SCCmec V in seven isolates (7.9%). The in silico SCCmec typing was unable to type 10 isolates belonging to ST1 (n = 5), ST672 (n = 3), ST9 (n = 1), and ST573 (n = 1). SauR7 and SauR54 isolates showed new allelic profiles, which ST7879 and ST7883 were respectively assigned to them by PubMLST. These STs could be grouped into four different clonal complexes (CCs), with ST22 clustered into CC22 which constituted the largest group (n = 61). ST1, ST9, ST573 and ST772 belonged to CC1, ST1178 belonged to CC5, and ST239 along with the new ST7879 belonged to CC8. ST7879 only differed by a single locus to ST239, i.e., the pta allele. However, ST88, ST672 and the new ST7883 do not belong to any known CCs. Of the 19 identified spa types, t032 was the most common with 38 (43.2%) isolates, all of which were found exclusively in ST22. This was followed by t3841 (n = 10, 11.4%) and t379 (n = 8, 9.1%), which were found in ST672 and ST22 isolates, respectively. Two isolates were spa untypeable, one of which was the novel ST7883 isolate.
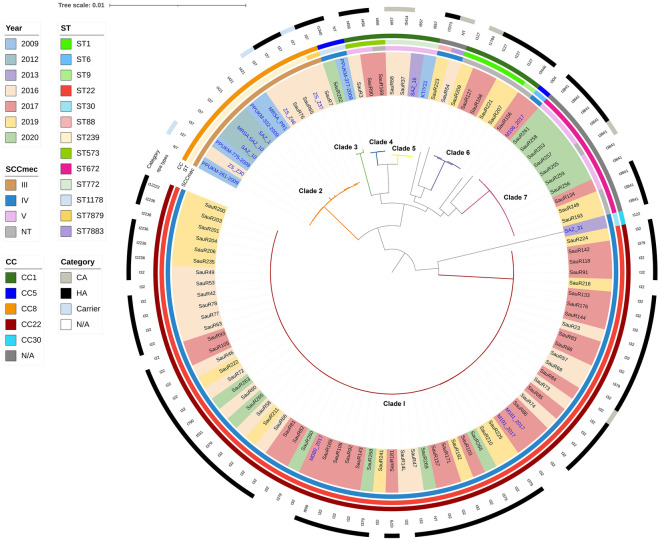
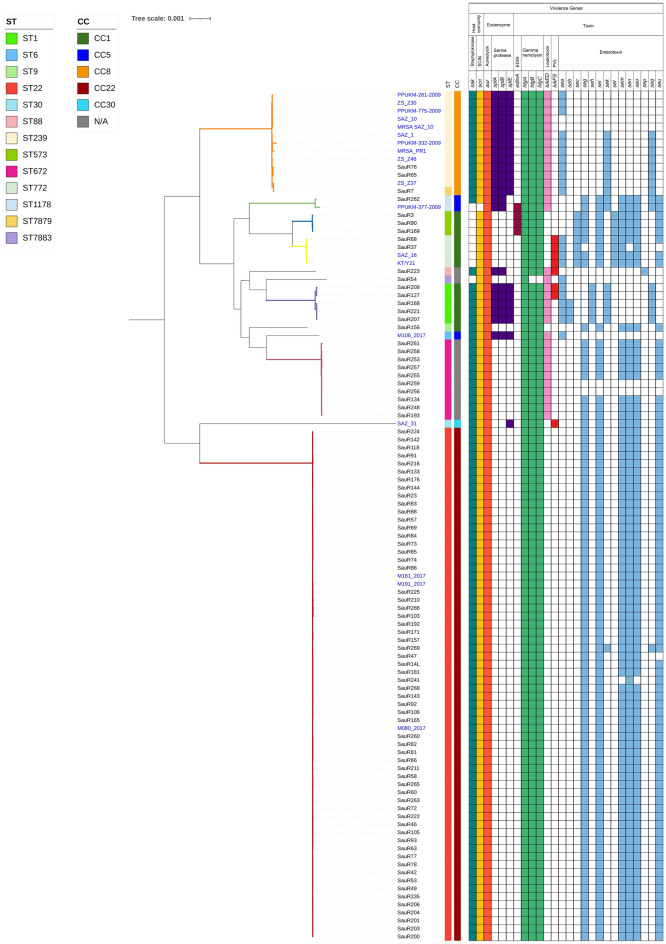
Figure 1.
Core genome maximum-likelihood phylogenetic tree of the 88 MRSA isolates obtained from HSNZ (from 2016 to 2020) along with 18 other Malaysian MRSA isolates that were available in GenBank (labelled in blue fonts; see Supplementary Table S1 for details of these isolates). Seven clades could be identified from the phylogenetic tree and these are represented by the following color-coded branches: Clade I (in red), Clade II (orange), Clade III (green), Clade IV (blue), Clade V (yellow), Clade VI (purple), and Clade VII (magenta). The color blocks with the names of the isolates indicate the year of isolation as in the top left legend. Other information included in the diagram are (from inner to outer circle): SCCmec type, sequence type (ST), clonal complex (CC), spa type, and category (i.e., whether they are hospital-acquired (HA), community-acquired (CA), or carrier). N/A = not available.
Based on the clinical records, the 88 MRSA isolates were classified either as hospital-associated (HA-MRSA) or community-associated MRSA (CA-MRSA) following the criteria defined by the Centers for Disease Control and Prevention (CDC)12. HA-MRSA isolates made up the majority with 70 isolates, while CA-MRSA accounted for only six isolates, whereas 12 isolates could not be categorized (as these isolates originated from samples provided to the HSNZ laboratory from district clinics and hospitals and were thus without detailed clinical data). Two of the six CA-MRSA isolates belonged to ST772, and the remaining four CA-MRSA isolates belonged to ST1, ST22, ST672, and ST7883 each. The isolates harboring SCCmec III and those belonging to ST9, ST88, ST573, and ST1178 were all classified as HA-MRSA.
Pan genome and phylogenetic analysis
The pan genome analysis of the 88 MRSA isolates from HSNZ revealed a total of 4,742 genes, which consisted of 1,973 core genes, 157 soft core genes, 783 shell genes and 1,829 cloud genes. The 88 MRSA isolates from the present study along with the 18 Malaysian database isolates could be grouped into seven clades based on their STs (Fig. 1). Clade I was the largest and was represented by the 61 ST22 isolates from HSNZ, and three other ST22 Malaysian isolates obtained from GenBank. Most of these HSNZ isolates were HA-MRSA strains. In Clade II, the novel ST7879 isolate was clustered with 12 ST239 isolates, demonstrating a close relationship between the two STs, which belonged to CC8. All these CC8 isolates also harbored SCCmec type III. Ten of the remaining 18 Malaysian MRSA database isolates were clustered in Clade II (ST239), one in Clade III (ST1178), two in Clade V (ST772), while two did not cluster in any clade. Clades IV (ST573), VI (ST1), and VII (ST672) were fully represented by our HSNZ isolates. Although ST1, ST9, ST573, and ST772 belonged to CC1, they appeared to be separate lineages and were not closely related. Notably, while the ST239-SCCmec III clone dominated the Malaysian MRSA isolates collected in 2009 and 2012, only two of our isolates belonged to this clone, both of which were obtained in 2016.
Among the isolates collected in 2020, there was a sudden increase in the number of ST672 isolates, with half of them (n = 7/14) belonging to this clone (Fig. 1). In contrast, only one ST672 isolate was found out of the 29 isolates that were sequenced in 2017, and two out of 20 in 2019. Even though only 14 MRSA isolates from 2020 were sequenced due to the small pool of isolates that could be obtained that year, this sudden increase in the proportion of ST672 isolates that were obtained in 2020 is certainly noteworthy. Seven of these ST672 isolates harbored SCCmec type V whereas the remaining three were non-typeable (Fig. 1).
Distribution of antimicrobial resistance genes
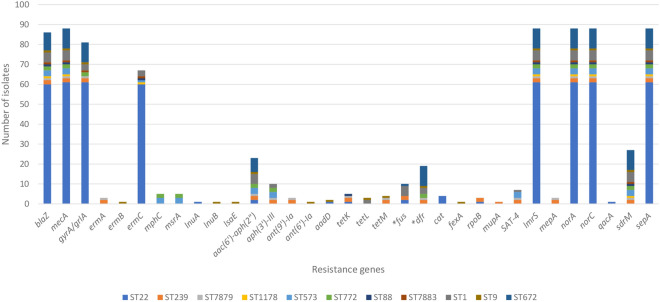
The distribution of antibiotic resistance genes is shown in Fig. 2. A total of 35 genes causing resistance to different antibiotic agents including beta lactams (blaZ and mecA), MLSB antibiotics (ermABC, msrA, mphC, lnuAB, and lsaE), aminoglycosides [aac(6’)-aph(2″), aph(3’)-III, ant(9)-Ia, ant(6)-Ia, and aadD], streptothricin (SAT-4), tetracyclines (tetK, tetL, and tetM), fusidic acid (fusC), trimethoprim (dfrG), chloramphenicol (cat and fexA) and mupirocin (mupA) were identified (Fig. 3). Chromosomal point mutations including gyrA/grlA (fluoroquinolone), fusA (fusidic acid), dfrB (trimethoprim), rpoB (rifampin) and genes encoding multidrug efflux pumps such as lmrS, mepA, norA, norC, qacA, sdrM, and sepA were also identified.
Figure 2.
Distribution of antimicrobial resistance determinants amongst the 88 MRSA isolates from HSNZ according to their STs. Genes marked with asterisks represent genes with variations in the form of mutations that were predicted to cause resistance and are grouped together.
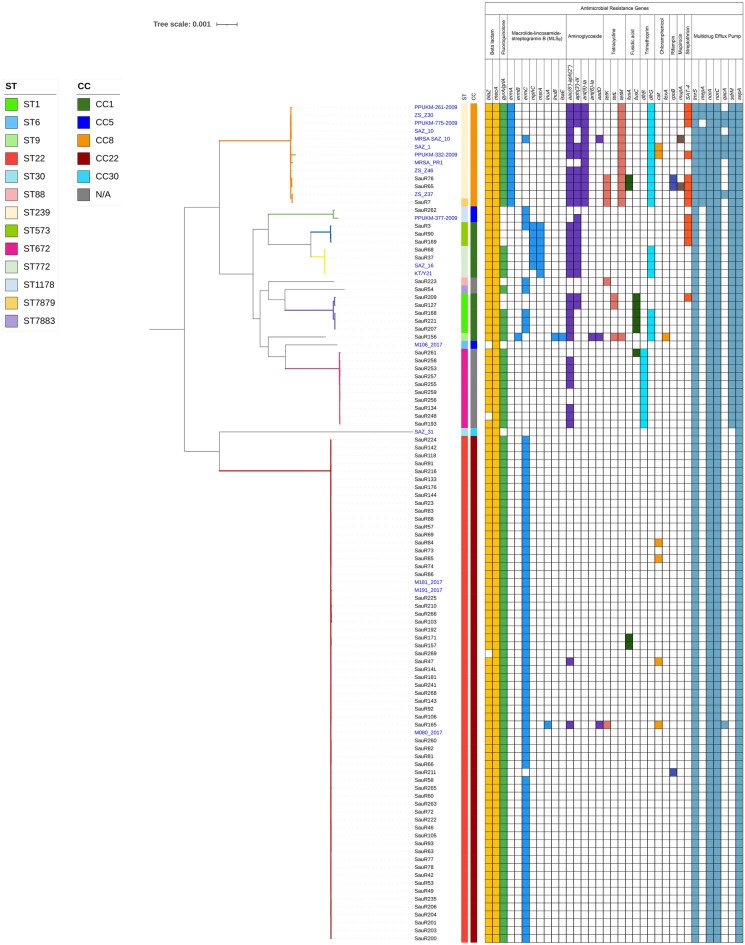
Figure 3.
Antimicrobial resistance determinants amongst the 88 MRSA isolates sequenced in this study along with another 18 Malaysian MRSA isolates obtained from GenBank (labelled in blue fonts). The core genome maximum-likelihood phylogenetic tree of the isolates is shown on the left and the branches are colored according to the seven clades identified in Fig. 1. The presence of a particular resistance determinant is indicated by a colored box. Genes marked with asterisks (*) represent host genes in which mutations were detected that would lead to resistance.
In general, ST22 isolates appeared to be more susceptible. ermC was detected in the ST22 isolates at a high frequency (n = 60). This gene is located on a small RepL-type plasmid (2.4–2.7 kb) in the ST22 isolates13 as well as other isolates. The cat gene that mediates resistance to chloramphenicol was identified only in four isolates of ST22 (Fig. 3). SauR165 (ST22 cat-positive isolate) carried more resistance genes (n = 14) compared to other ST22 isolates (n = 7–10). SauR165 was found to harbor a 28.6 kb RepA_N-type plasmid, pSauR165-1, which encode six antibiotic resistance genes including cat, tetK, ant(4’’)-Ib, aac(6’)-Ie-aph(2’’)-Ia, lnuA and qacA, along with the cadDX cadmium resistance genes13.
ST88, ST1178, and ST7883 isolates had almost similar resistomes as the ST22 isolates but the ST88 isolate, SauR223, had an additional tetK gene. Among our isolates, those belonging to ST239 and ST7879 were predicted to be highly resistant compared to other isolates. They carried ermA, mepA, and unique combinations of aminoglycoside and tetracycline resistance genes that were not found in other STs. One of the ST239 isolates, SauR65, was the only isolate harboring the mupA gene in our collection. The other mupA-positive isolate, MRSA SAZ_10 (accession no. SWED00000000), was also ST239 and from HSNZ in 2012 but was sequenced in an earlier study.
ST573 (n = 3) and ST772 (n = 2) isolates carried more than 10 resistance genes including msrA and mphC macrolide resistance genes and aminoglycoside resistance genes as a combination of aac(6’)-aph(2″) and aph(3’)-III. All ST573 and ST772 isolates harbored the SAT-4 and dfrG resistance genes, respectively. dfrG was also found in two other ST772 isolates that were obtained from HSNZ previously (KT/Y21 in 2009 and SAZ_16 in 2013). We recently reported the complete genome sequence of one of the ST573 isolates, SauR3 (CP098727), by hybrid assembly of Illumina and Oxford Nanopore sequence reads14. SauR3 carried a 42.9 kb plasmid (pSauR3-1) with an approximately 14 kb genomic island containing resistance genes including the bla operon in a full-length Tn552, mphC, msrA, SAT-4, and aadE-aph(3’’)-IIIa. Besides, SauR3 also harbored the small, 2.5 kb RepL-type plasmid encoding ermC (designated pSauR3-3) which was responsible for its inducible iMLSB phenotype. Another interesting feature of the SauR3 genome is the presence of a SCCmec type V (5C2&5) variant which also encodes the aac(6’)-aph(2’’) aminoglycoside-resistance genes flanked by IS43114. However, tracing the presence of such genetic features in the other isolates of ST573 (SauR90 and SauR169) was not possible using short-read sequences alone. Nevertheless, both SauR90 and SauR169 did not harbor the ermC-encoding RepL-type plasmid and displayed the MS phenotype13.
The fusC gene was primarily associated with ST1, with five out of six fusC-positive isolates belonging to this ST. The other isolate harboring fusC, SauR261, was ST672 (Fig. 3). Three of the ST1 isolates (SauR168, SauR207, and SauR221) had a combination of the ermC and dfrG genes. The aac(6’)-aph(2’’) genes were identified in all ST1 isolates and in two of these isolates (SauR127 and SauR209), an additional aph(3’)-III gene in combination with tetL were found. SauR209 also had an additional SAT-4 streptothricin resistance gene which was not found in the other ST1 isolates.
The findings revealed that SauR156 (the only ST9 isolate) carried 18 resistance genes, including some genes that were not found in other STs such as ermB, lnuB, lsaE, and fexA. Furthermore, an 11.3 kb PriCT_1 plasmid (pSauR156-1), which encodes ermB, tetL, and aadD was detected in SauR15613.
Similar to ST22 isolates, the ST672 isolates also appeared to be more susceptible to antimicrobials, with resistance genes detected in the range of nine to ten genes. Nine of the ten ST672 isolates harbored the blaZ gene in the 6,545 bp transposon Tn552 which was encoded on a 20.7 kb plasmid that was highly similar to pMW2, a 20,654 bp Rep_3-type plasmid initially described in MRSA strain MW2 isolated in USA in 199813,15. The sole blaZ-negative ST672 isolate, SauR248, was plasmid-free. The dfrB gene was identified strictly among these isolates, with all 10 of them carrying this gene. Additionally, seven isolates possessed the aac(6’)-aph(2″) aminoglycoside resistance gene. In contrast to the ST22 isolates, the ST672 isolates do not harbor the ermC MLSB resistance gene.
Correspondence of phenotypic and genotypic resistance
Phenotypic resistance of the 88 MRSA isolates from HSNZ was compared with genotype prediction from WGS data for 10 antibiotic classes. Generally, the concordance rate was determined at > 95% for all antibiotics tested in this study (Table 2). In isolates demonstrating phenotypic resistance to trimethoprim-sulfamethoxazole (SXT) that were sequenced (n = 9), WGS analysis using ResFinder and CARD databases identified the dfrG gene as the resistance determinant for trimethoprim but the acquired sulfonamide resistance genes sul1, sul2 and sul3 were absent. To elucidate the resistance mechanism associated with sulfamethoxazole, we conducted BLAST searches and found mutations in the folP gene, which encodes the dihydropteroate synthase (DHPS) enzyme. The resistant ST239 (n = 2) and ST7879 (n = 1) isolates exhibited five mutations (F17L, T28S, T59S, L64M, and KE257_Dup), while the remaining resistant isolates (n = 6) had three mutations (T28S, T59S, and L64M). Previous studies have shown that these mutations confer resistance to sulfamethoxazole, thereby explaining the observed SXT resistance phenotype16,17.
Table 2.
Concordance rate between antimicrobial phenotypes and predicted genotypes.
| Antibiotic class | Antibiotic | Resistant isolates | False positive | False negative | Concordance (%) | Resistance determinants |
|---|---|---|---|---|---|---|
| Beta-lactam | Penicillin | 88 | 0 | 2 | 97.7 | blaZ |
| Cefoxitin | 88 | 0 | 0 | 100 | mecA | |
| MLSB | Erythromycin/ clindamycin/ streptogramin | 76 | 0 | 1 | 98.9 | erm, msr |
| Fluoroquinolone | Ciprofloxacin/ moxifloxacin | 82 | 0 | 0 | 100 | Mutations in gyrA/grlA, norA |
| Aminoglycoside | Amikacin/ gentamicin | 26 | 0 | 2 | 97.7 | aac(6’)-aph(2″), aph(3’)-III, ant(9)-Ia, ant(6)-Ia, aadD |
| Tetracycline | Tetracycline/ doxycycline/ minocycline | 9 | 0 | 1 | 98.9 | tet |
| Folate inhibitor | Trimethoprim-sulfamethoxazole | 8 | 1 | 0 | 98.9 | dfrG |
| Phenicol | Chloramphenicol | 5 | 0 | 0 | 100 | cat |
| Fucidane | Fusidic acid | 13 | 0 | 3 | 96.6 | fus |
| Ansamycin | Rifampicin | 3 | 0 | 0 | 100 | Mutation in rpoB |
| Monoxycarbolic acid | Mupirocin | 1 | 0 | 0 | 100 | mupA |
Distribution of virulence genes
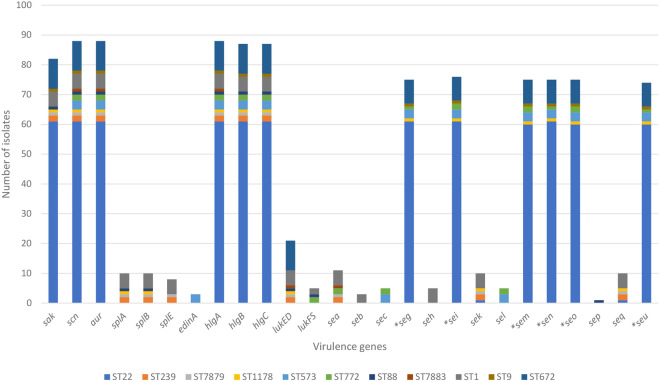
VirulenceFinder identified a total of 26 distinct virulence genes among the 88 MRSA isolates, which consisted of host immunity genes (sak and scn), exoenzyme genes (aur and splABE), and toxin genes such as epidermal cell differentiation (edinA), gamma hemolysins (hlgABC), leukotoxin (lukED), Panton-Valentine leukocidin (lukFS) and enterotoxins (sea—sec, seg—sei, sek—seq, and seu). The majority of the isolates (96.6%, 85/88) carried more than 10 virulence genes in their genomes.
Genes such as sak, scn, aur, hlgABC, and genes of the enterotoxin gene cluster (egc) including seg, sei, sem, sen, seo, and seu, were widely present among the HSNZ as well as the other Malaysian isolates (Figs. 4 and 5, respectively). These genes constituted the virulence profiles of most (n = 58) ST22 isolates and the single ST9 isolate. Of the 61 ST22 isolates, only three differed slightly in their enterotoxin gene profile. One isolate (SauR269) had additional sek and seq genes, one (SauR47) lacked the seu gene, and another (SauR241) lacked the sem and seo genes. The virulence profiles of ST239 (SauR65 and SauR76) and ST7879 (SauR7) isolates were comparable, with the exoenzyme splABE and leukotoxin lukED genes present in all three isolates. These three isolates also contained the combination of sea, sek and seq genes in place of the missing egc genes. In contrast, the sek and seq genes were less common among the Malaysian ST239 isolates in the database, with five of them (i.e., PPUKM-261-2009, PPUKM-775-2009, ZS_Z30, SAZ_10 and MRSA SAZ_10) lacking these genes. Furthermore, the sea gene was also absent in both SAZ_10 and MRSA SAZ_10.
Figure 4.
Distribution of virulence genes amongst the 88 MRSA isolates from HSNZ that were sequenced in this study according to their STs. Genes marked with asterisks (*) represent the genes from the enterotoxin gene cluster (egc).
Figure 5.
Virulence genes among the 88 MRSA isolates from HSNZ (2016–2020) along with another 18 Malaysian MRSA isolates obtained from GenBank (labelled in blue fonts). The core genome maximum-likelihood phylogenetic tree of the isolates is shown on the left with the branches colored according to the seven clades identified in Fig. 1. The presence of a particular virulence gene is marked by a colored box. SCIN = staphylococcal complement inhibitor; EDIN = epidermal cell differentiation inhibitor; PVL = Panton-Valentine leucocidin.
The lukED, splAB, sek, and seq genes were also present in the genome of the ST1178 isolate, SauR262. However, unlike ST239 and ST7879, this isolate contained the egc genes. Notably, SauR262 carried the highest number of virulence genes in this study, with a total of 17 genes identified in its genome. The only other Malaysian ST1178 genome which was obtained from GenBank, PPUKM-377-2009, was different from SauR262 in that it contained the edinA epidermal cell differentiation gene but lacked the sak and scn genes.
Both ST573 and ST772 isolates lacked the sak gene, which was found in most (n = 82) of the other MRSA isolates. In addition to the egc genes, these isolates also contained the sec and sel genes, which were not detected in any of the other isolates. Among the HSNZ isolates, the edinA gene was identified exclusively in ST573, whereas ST772 isolates also had the lukFS Panton-Valentine leukocidin and sea genes. The presence of enterotoxin genes was rare in the HSNZ ST88 isolate, which only possessed the single sep gene discovered in this study. This isolate was also one of the few to contain both lukED and lukFS genes.
Similar to ST88, the ST7883 isolate (SauR54) also lacked enterotoxin genes. SauR54 carried the least number of virulence genes, including scn, aur, hlgA, lukED, and sea. ST1 isolates displayed a diverse virulome, with up to 15 types of virulence genes identified across all five isolates. Their enterotoxin gene profiles were mostly similar to ST239 and ST7879, but with an additional seh gene. The seh gene was a unique feature of ST1 isolates, as it was not identified in other STs.
The splABE and lukED genes were also found in all five ST1 isolates. Two isolates (i.e., SauR127 and SauR209 also contained lukFS, while the remaining three lukFS-negative isolates (i.e., SauR168, SauR207, and SauR221) had the seb gene instead.
The virulome of ST672 isolates was comparable to that of ST22 and ST9 isolates with the addition of lukED. Except SauR256 and SauR259 isolates, which carried seven virulence genes and lacked the egc genes, most (n = 8) ST672 isolates carried 13 virulence genes (Fig. 5).
Discussion
WGS has emerged as a successful approach for identifying the transmission and outbreaks of bacterial pathogens such as MRSA. Due to its multiple resistances and high pathogenicity, MRSA has remained a persistent threat since the 1960s, having significant detrimental effect on public health. Therefore, continuous surveillance of this pathogen is essential to contain its spread. In this study, 88 clinical MDR MRSA isolates were analyzed using WGS to obtain information about the population and clonal composition of circulating MRSA isolates in HSNZ, the main tertiary hospital in Terengganu in 2016–2020.
Molecular epidemiology of the 88 MRSA isolates in this study revealed that the ST22-SCCmec IV, also known as epidemic MRSA-15 (EMRSA-15) was the most prevalent clone, with 61 (69.3%) isolates. EMRSA-15 is a major clone that has exhibited considerable success on a global scale and was previously highly prevalent in the UK and Ireland18. Studies in Singapore showed that in the 1980s and 1990s, the ST239-SCCmec III clone was predominant, but starting from 2003 when the first ST22-SCCmec IV clone was introduced, the ST239-SCCmec III clone began to be slowly displaced by ST22-SCCmec IV which by the mid-2010’s had established itself in most Singapore hospitals19,20. In Malaysia, a similar clonal displacement event had also likely occurred. During the mid-2000’s, the ST239-SCCmec III clone was predominant in Malaysia21,22 but from early 2010’s onwards, there were reports of the growing proportion of ST22-SCCmec IV in Malaysian hospitals23. Interestingly, most of the Malaysian ST239-SCCmec III genomes that were obtained from GenBank (n = 10) originated in 2009 and 2012, perhaps reflecting their predominance during that time. By the mid 2010’s, the ST22-SCCmec IV clone appeared to predominate with a study from the University of Malaya Medical Centre (UMMC) in Kuala Lumpur, Malaysia, indicating that 55.6% of the 99 MRSA isolates collected between 2014–2015 belonging to this particular clone24. The predominance of the ST22-SCCmec IV clone was also clearly demonstrated in the MRSA isolates from HSNZ, Terengganu in 2016–2020. Despite the ST22-SCCmec IV clone being more susceptible to antibiotics compared to ST239-SCCmec III19,23, the success of the ST22 clone could be due to its physiological characteristics. In vitro studies indicate that ST22 is a fit clone relative to ST239, with a higher growth rate and resistance to desiccation25. Besides, the smaller size of SCCmec IV as compared to SCCmec III could enable it to be more efficiently transmitted via horizontal gene transfer26,27.
Among the MRSA isolates in this study, the presence of two Indian-origin clones, ST672-SCCmec V and the Bengal Bay clone ST772-SCCmec V, is noteworthy. Since the majority of the Bengal Bay clone in Norway was isolated from patients with ties to the Indian subcontinent28, it is possible that these two clones made its way to Malaysia through travelers from the same region. Although the ST772 Bengal Bay clone has previously been observed in Malaysian isolates29,30 including from HSNZ31, to the best of our knowledge, the detection of the ST672 clone has not been documented. However, it has been reported in neighboring Indonesia32. Interestingly, the ST672 clone was identified in just one isolate in 2017, two isolates in 2019, and seven in 2020, with majority of these isolates (7/10) harboring SCCmec V. This increasing prevalence of the ST672 clone suggests a potential upward trend. Continued surveillance is needed to confirm this and assess if another clonal displacement, like the ST239-SCCmec IV in the early 2010s, is occurring. The possible reason for this apparent prevalence of ST672 in HSNZ is currently unknown.
The detection of resistance determinants uncovered a total of 35 resistance genes associated with resistances to antiseptics and 12 antibiotics classes. Methicillin resistance among the MRSA isolates in this study is mediated by mecA, while the mecA homologue, mecC, was not detected. The mecC gene has so far been found primarily in European isolates of varying clonal lineages such as CC130, CC49, CC599, and CC194333. To date, mecC has not been detected among human MRSA isolates in Malaysia. However, one recent study reported the detection of 15 mecC-positive livestock-associated MRSA (LA-MRSA) isolates in milk and nasal swab samples from dairy cattle farms34. Despite being a successful pandemic clone, ST22-SCCmec IV isolates in this study were among the most susceptible, exhibiting resistance to only beta lactams, fluoroquinolones, and MLSB antibiotics, which is consistent with previous studies19,23,35. Conversely, ST239-SCCmec III and ST7879-SCCmec III isolates, which shared similar resistomes, were predicted that they would be the most resistant (resistance against aminoglycosides, tetracyclines, and nucleoside antibiotics). The Vienna/Hungarian/Brazilian clone ST239-SCCmec III, which has been around since the 1970s, is the oldest pandemic strain and is known to be multidrug-resistant36,37. The larger size of SCCmec III in comparison to SCCmec IV, enables it to carry a plethora of resistance genes38, thus explaining the observed multidrug resistance.
The solitary ST9 isolate in this study, SauR156, is particularly intriguing as ST9 strains are commonly LA-MRSA. Our finding that the ST9 isolate carried a high number of resistance genes corroborated previous reports that strains of this ST are exceptionally resistant39,40.
We found 95% concordance between phenotypic resistance and genotypic prediction, indicating the high accuracy of WGS in detecting antimicrobial resistance, as reported by other studies41,42. In addition to resistance genes, the MRSA isolates were also found to harbor numerous virulence genes. Across the 88 MRSA genomes, as many as 26 distinct genes were discovered, with sak, scn, aur, egc, and hlg genes being among the most frequently encountered. The sak gene encodes staphylokinase, a plasminogen activator with fibrinolytic function43, while the scn gene encodes staphylococcal complement inhibitor (SCIN), an immune modulating protein implicated in the prevention of phagocytosis44. Both sak and scn genes are located on the ϕSa3 prophage, and are components of the immune evasion cluster (IEC), which has been demonstrated to play an essential role in the colonization of human hosts40,45. The IEC is usually absent in animal isolates; hence the presence of these genes typically indicates human adaptation. Notably, the absence of the scn gene is considered a hallmark for LA-MRSA strains46. However, we found that this gene was present in our ST9 isolate, which is commonly associated with LA-MRSA. This indicates that the isolate has successfully adapted to and colonize the human host, eventually leading to clinical infection47. The high prevalence of egc genes observed in this study is consistent with findings from earlier studies48,49. Similarly, the widespread nature of gamma hemolysin across our isolates is also explained by the fact that it is a core genome-encoded toxin found in virtually all strains of S. aureus50. On the contrary, the occurrence of the leukotoxin genes, lukED encoding leukocidin ED and lukFS encoding Panton-Valentine leukocidin (PVL), was observed to be relatively rare. A previous review has indicated that the lukED genes are widely prevalent among S. aureus, particularly in epidemic strains51. However, in the present study, only a total of 21 isolates (23.9%) harbored the lukED genes, which were distributed among various sequence types (ST1: n = 5, ST88: n = 1, ST239: n = 2, ST672: n = 10, ST1178: n = 1, ST7879: n = 1, ST7883: n = 1). In addition, the lukED genes were not found in the ST22 isolates, which was consistent with a previous study52. Despite recent findings which described the emergence of hypervirulent ST22 strains carrying both PVL and tst genes53,54, the HSNZ ST22 isolates notably lacked these virulence factors. Interestingly, PVL- and TSST-1-positive ST22 strains were associated with SCCmec IVa53,54, whereas the HSNZ isolates belonged to the SCCmec IVh subtype, suggesting a genetic makeup distinctive from the hypervirulent strains despite their shared ST22 lineage. The PVL-encoded gene lukFS were identified only in the genomes of ST88 (SauR233), two ST772 (SauR37 and SauR68), and two ST1 (SauR127 and SauR209) isolates. Three Malaysian isolates from earlier studies were found to be PVL-positive, and these were SAZ_31 belonged to ST30, and SAZ_16 and KT/Y21 belonged to ST772. Earlier research had documented the carriage of PVL in ST88 strains55, while the ST772 Bengal Bay clone is known to be a PVL-positive clone28. Because PVL has been linked with severe disease56,57, carriage of this gene might indicate the high pathogenicity of ST88 and ST772 isolates.
Conclusions
WGS of 88 MDR MRSA isolates obtained from 2016 to 2020 from HSNZ in Terengganu has given us important insights into their genome characteristics, carriage of antibiotic resistance and virulence genes, and their clonal lineages. The ST22-SCCmec IV (EMRSA-15) clone was predominant in HSNZ (n = 61/88; 69.3%), reflecting other studies done in Malaysia and Singapore in the past ten years11,19,20,23,24. Despite their relative susceptibility to antibiotics, the success of the ST22-SCCmec IV clone in displacing the more resistant ST239-SCCmec III clone warrants vigilance, particularly with the recent rise of hypervirulent ST22-SCCmec IV subclones that harbor the pvl and tst toxin genes in China and Japan53,54. The emergence of the Indian-origin clones ST772, and particularly ST672 in 2020 highlights their potential to spread and affirms the need for continuous monitoring by WGS.
Methods
Collection and selection of isolates
From July 2016 until December 2020, a total of 197 MRSA isolates were collected from various clinical specimens from the Microbiology Laboratory of Hospital Sultanah Nur Zahirah (HSNZ), which is the main referral and tertiary hospital of Terengganu, a state located in the eastern coast of Peninsular Malaysia. The collection of bacterial isolates was carried out with ethical approval from the Medical Research and Ethics Committee (MREC) of the Ministry of Health Malaysia (National Medical Research Registry nos. NMRR-15-2369-28,130 [IIR] and NMRR-19-3702-52104[IIR]). All methods were performed in accordance with the relevant guidelines and regulations.
All MRSA isolates were validated by the presence of the thermonuclease (nuc) gene58 and methicillin resistance (mecA) gene59 through PCR-amplification. Phenotypic resistance profiles against 24 antibiotics from 16 antibiotic classes using disk diffusion method and E-test strips, as well as double disk diffusion test (D-test) for macrolide-lincosamide-streptogramin B (MLSB) resistance phenotypes, i.e., inducible (iMLSB), constitutive (cMLSB), and macrolide-streptogramin (MS), were characterized as previously described60. From this collection, 88 MDR isolates, which were categorized as resistant to at least one agent in three or more antibiotic categories61 were selected for whole genome sequencing.
DNA extraction and whole genome sequencing
Genomic DNA was extracted using Presto™ Mini gDNA Bacteria Kit (Geneaid Biotech Ltd., New Taipei City, Taiwan) according to the manufacturer’s instruction with lysostaphin (300 µg/mL) as an additional cell lysis reagent13. Construction of DNA libraries and whole genome sequencing using short-read, paired-end 150 bp (PE150) sequencing strategy were performed by commercial WGS service providers. Thirty-one isolates were sequenced on the Illumina platform using Illumina HiSeq 2500 (average sequencing coverage 250 ×) (Novogene, Singapore), and the remaining 49 isolates were sequenced on the DNBSEQ platform with BGISEQ-500 (average sequencing coverage 200 ×) (BGI, Shenzhen, China). An additional eight samples were sequenced using the Illumina MiSeq (average sequencing coverage 64 ×) under a collaborative program with the Kyushu Institute of Technology, Japan. Sequences were subjected to de novo assembly using Unicycler v0.4.8 (Wick et al., 2017). The quality of the assembled genomes was evaluated using QUAST v5.0.262, where assemblies with > 200 contigs and N50 < 40,000 bp were excluded63. The assembled genomes were annotated using Prokka v1.13.364.
Identification of epidemiological markers, antimicrobial resistome and virulome
Molecular characterization of epidemiological marker elements such as staphylococcal cassette chromosome mec (SCCmec) types, multilocus sequence typing (MLST) sequence types and staphylococcal protein A (spa) types were determined from the assembled genomes using web-based databases, i.e., SCCmecFinder 1.265, MLST 2.066 and spaTyper 1.067, respectively, which are available at the Center for Genomic Epidemiology (CGE) (http://www.genomicepidemiology.org/services/). Presence of acquired antimicrobial resistance genes and chromosomal mutations was identified using Resistance Gene Identifier (RGI) at the Comprehensive Antibiotic Resistance Database (CARD) (Alcock et al., 2020)] and ResFinder 4.168,69, while VirulenceFinder 2.070 was used to identify virulence genes.
Determination of concordance rate between antimicrobial phenotypes and genotypes
The phenotypic resistance of the MRSA isolates was compared with the genomic prediction of antibiotic resistance determined from their respective assembled genomes. The concordance rate was calculated as the percentage of concordant isolates from the total number of isolates, with concordant isolates defined as those having phenotypic resistance and the corresponding resistance determinants in their genomes41.
Pan genome and phylogenetic tree analysis
The annotated general feature format (GFF) files derived from Prokka were used as input for the pan genome pipeline using Roary71 with parameters set at 95% amino acid sequence identity and presence in 99% isolates to define a core gene. A maximum likelihood phylogenetic tree was constructed from the derived core genome alignment using FastTree 2.1.1072, with 100 bootstraps under the generalized time reversible (GTR) model. The phylogenetic tree also included the genome sequences of 18 Malaysian MRSA isolates available in the NCBI GenBank database. Among these isolates, 15 were clinical MRSA isolates obtained from various specimens, while three isolates were obtained from the nasal swabs of healthy student volunteers. The collection of these isolates spanned from 2009 to 2017. Further information about these isolates is included in Supplementary Table S1. The resulting phylogenetic tree was visualized and edited using iTOL v6 (https://itol.embl.de/)73. The accession numbers for all 88 MRSA genomes that were sequenced in this study are available in GenBank under BioProject accession number PRJNA722830.
Supplementary Information
Acknowledgements
This research was funded by Fundamental Research Grant Scheme FRGS/1/2019/SKK11/UNISZA/02/1 from the Malaysian Ministry of Higher Education. Our thanks to Dr Fatimah Haslina Abdullah, Dr Norlela Othman and medical laboratory technologists of the Microbiology Laboratory of Hospital Sultanah Nur Zahirah for their help in the collection of MRSA isolates and the corresponding clinical data used in this study. Our gratitude also goes to Dr Pakorn Aiewsakun and Bharkbhoom Jaemsai of CENMIG, Mahidol University, for their valuable insights and suggestions in the construction of the phylogenetic trees.
Author contributions
C.C.Y. and C.H.C.: conceptualization; A.M.C.H. and E.I.A.-T.: investigation and data curation; A.M.C.H., C.H.C., P.P., and C.C.Y.: analysis and interpretation; A.M.C.H., C.H.C. and C.C.Y.: preparation of original draft; C.H.C., C.C.Y., S.M.P., K.H.C., N.I.A.R., S.I., T.M., and P.P.: review and editing; C.H.C., C.C.Y., S.M.P., K.H.C., N.I.A.R., S.I.: funding acquisition. All authors have read, reviewed, and approved the manuscript.
Data availability
The genomic data of this study were submitted and deposited in NCBI GenBank database under BioProject No. PRJNA722830. Details regarding the 18 Malaysian database isolates are provided in Supplementary Table S1 while details regarding assembly qualities and statistics are provided in Supplementary Table S2.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ching Hoong Chew, Email: chewch@unisza.edu.my.
Chew Chieng Yeo, Email: chewchieng@gmail.com.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-54182-x.
References
- 1.Madigan T, et al. Whole-genome sequencing for methicillin-resistant Staphylococcus aureus (MRSA) outbreak investigation in a neonatal intensive care unit. Infect. Control Hosp. Epidemiol. 2018;39:1412–1418. doi: 10.1017/ice.2018.239. [DOI] [PubMed] [Google Scholar]
- 2.Durand G, et al. Routine whole-genome sequencing for outbreak investigations of Staphylococcus aureus in a national reference center. Front. Microbiol. 2018;9:511. doi: 10.3389/fmicb.2018.00511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kong Z, et al. Whole-genome sequencing for the investigation of a hospital outbreak of MRSA in China. PLoS One. 2016;11:e0149844. doi: 10.1371/journal.pone.0149844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nelson RE, et al. The impact of healthcare-associated methicillin-resistant Staphylococcus aureus infections on postdischarge health care costs and utilization across multiple health care systems. Health Serv. Res. 2018;53:5419–5437. doi: 10.1111/1475-6773.13063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uematsu H, Yamashita K, Kunisawa S, Fushimi K, Imanaka Y. Estimating the disease burden of methicillin-resistant Staphylococcus aureus in Japan: Retrospective database study of Japanese hospitals. PLoS One. 2017;12:e0179767. doi: 10.1371/journal.pone.0179767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chatterjee A, Rai S, Guddattu V, Mukhopadhyay C, Saravu K. Is methicillin-resistant Staphylococcus aureus infection associated with higher mortality and morbidity in hospitalized patients? A cohort study of 551 patients from south western India. Risk Manag. Healthc. Policy. 2018;11:243–250. doi: 10.2147/RMHP.S176517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zainol Abidin N, et al. MRSA infection in general surgical wards in a Malaysian tertiary hospital: A retrospective study. Ann. Clin. Surg. 2020;1:1008. [Google Scholar]
- 8.Sit PS, Teh CSJ, Idris N, Ponnampalavanar S. Methicillin-resistant Staphylococcus aureus (MRSA) bacteremia: Correlations between clinical, phenotypic, genotypic characteristics and mortality in a tertiary teaching hospital in Malaysia. Infect. Genet. Evol. 2018;59:132–141. doi: 10.1016/j.meegid.2018.01.031. [DOI] [PubMed] [Google Scholar]
- 9.Ministry of Health Malaysia. National Surveillance of Antibiotic Resistance 2021. https://www.imr.gov.my/images/uploads/NSAR/2021/NSAR-2021_to-be-uploaded.pdf (2022).
- 10.Kinnevey PM, et al. Meticillin-resistant Staphylococcus aureus transmission among healthcare workers, patients and the environment in a large acute hospital under non-outbreak conditions investigated using whole-genome sequencing. J. Hosp. Infect. 2021;118:99–107. doi: 10.1016/j.jhin.2021.08.020. [DOI] [PubMed] [Google Scholar]
- 11.Che Hamzah AM, Yeo CC, Puah SM, Chua KH, Chew CH. Staphylococcus aureus infections in Malaysia: A review of antimicrobial resistance and characteristics of the clinical isolates, 1990–2017. Antibiotics. 2019 doi: 10.3390/antibiotics8030128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.CDC. Active Bacterial Core Surveillance Report, Emerging Infections Program Network, Methicillin-Resistant Staphylococcus aureus. https://www.cdc.gov/abcs/reports-findings/survreports/mrsa14.html (2014).
- 13.Al-Trad EI, et al. The plasmidomic landscape of clinical methicillin-resistant Staphylococcus aureus isolates from Malaysia. Antibiotics. 2023 doi: 10.3390/antibiotics12040733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Al-Trad EI, et al. Complete genome sequence and analysis of a ST573 multidrug-resistant methicillin-resistant Staphylococcus aureus SauR3 clinical isolate from Terengganu, Malaysia. Pathogens. 2023 doi: 10.3390/pathogens12030502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baba T, et al. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet. 2002;359:1819–1827. doi: 10.1016/S0140-6736(02)08713-5. [DOI] [PubMed] [Google Scholar]
- 16.Nurjadi D, Zizmann E, Chanthalangsy Q, Heeg K, Boutin S. Integrative analysis of whole genome sequencing and phenotypic resistance toward prediction of trimethoprim-sulfamethoxazole resistance in Staphylococcus aureus. Front. Microbiol. 2021 doi: 10.3389/fmicb.2020.607842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reeve SM, et al. MRSA isolates from United States hospitals carry dfrG and dfrK resistance genes and succumb to propargyl-linked antifolates. Cell Chem. Biol. 2016;23:1458–1467. doi: 10.1016/j.chembiol.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grundmann H, et al. Geographic distribution of Staphylococcus aureus causing invasive infections in Europe: A molecular-epidemiological analysis. PLoS Med. 2010;7:e1000215. doi: 10.1371/journal.pmed.1000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu L-Y, et al. Evolutionary dynamics of methicillin-resistant Staphylococcus aureus within a healthcare system. Genome Biol. 2015;16:81. doi: 10.1186/s13059-015-0643-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chow A, et al. MRSA transmission dynamics among interconnected acute, intermediate-term, and long-term healthcare facilities in Singapore. Clin. Infect. Dis. 2017;64:S76–S81. doi: 10.1093/cid/cix072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neela V, et al. First report on methicillin-resistant Staphylococcus aureus of Spa type T037, Sequence type 239, SCCmec type III/IIIA in Malaysia. Eur. J. Clin. Microbiol. Infect. Dis. 2010;29:115–117. doi: 10.1007/s10096-009-0813-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ehsanollah G-R, et al. Predominance and emergence of clones of hospital-acquired methicillin-resistant Staphylococcus aureus in Malaysia. J. Clin. Microbiol. 2010;48:867–872. doi: 10.1128/JCM.01112-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim, K. T., Hanifah, Y. A., Mohd Yusof, M. Y., Ito, T. & Thong, K. L. Comparison of methicillin-resistant Staphylococcus aureus strains isolated in 2003 and 2008 with an emergence of multidrug resistant ST22: SCCmec IV clone in a tertiary hospital, Malaysia. J. Microbiol. Immunol. Infect.46, 224–233 (2013). [DOI] [PubMed]
- 24.Niek WK, Teh CSJ, Idris N, Thong KL, Ponnampalavanar S. Predominance of ST22-MRSA-IV clone and emergence of clones for methicillin-resistant Staphylococcus aureus clinical isolates collected from a tertiary teaching hospital over a two-year period. Jpn. J. Infect. Dis. 2019;72:228–236. doi: 10.7883/yoken.JJID.2018.289. [DOI] [PubMed] [Google Scholar]
- 25.Knight GM, et al. Shift in dominant hospital-associated methicillin-resistant Staphylococcus aureus (HA-MRSA) clones over time. J. Antimicrob. Chemother. 2012;67:2514–2522. doi: 10.1093/jac/dks245. [DOI] [PubMed] [Google Scholar]
- 26.Sit PS, et al. Prevalence of methicillin-resistant Staphylococcus aureus (MRSA) infection and the molecular characteristics of MRSA bacteraemia over a two-year period in a tertiary teaching hospital in Malaysia. BMC Infect. Dis. 2017;17:274. doi: 10.1186/s12879-017-2384-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hiramatsu K, et al. Genomic basis for methicillin resistance in Staphylococcus aureus. Infect. Chemother. 2013;45:117–136. doi: 10.3947/ic.2013.45.2.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blomfeldt A, et al. Emerging multidrug-resistant Bengal Bay clone ST772-MRSA-V in Norway: Molecular epidemiology 2004–2014. Eur. J. Clin. Microbiol. Infect. Dis. 2017;36:1911–1921. doi: 10.1007/s10096-017-3014-8. [DOI] [PubMed] [Google Scholar]
- 29.Neela VK, Ehsanollah GR, Zamberi S, Van Belkum A, Mariana NS. Prevalence of Panton-Valentine leukocidin genes among carriage and invasive Staphylococcus aureus isolates in Malaysia. Int. J. Infect. Dis. 2009;13:e131–e132. doi: 10.1016/j.ijid.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 30.Ghasemzadeh-Moghaddam H, Van Belkum A, Hamat RA, Van Wamel W, Neela V. Methicillin-susceptible and -resistant Staphylococcus aureus with high-level antiseptic and low-level mupirocin resistance in Malaysia. Microb. Drug Resist. 2014;20:472–477. doi: 10.1089/mdr.2013.0222. [DOI] [PubMed] [Google Scholar]
- 31.Suhaili, Z. et al. Draft genome sequence of methicillin-resistant Staphylococcus aureus KT/Y21, a sequence type 772 (ST772) strain isolated from a pediatric blood sample in Terengganu, Malaysia. Genome Announc.2, (2014). [DOI] [PMC free article] [PubMed]
- 32.Santosaningsih D, et al. Characterisation of clinical Staphylococcus aureus isolates harbouring mecA or Panton-Valentine leukocidin genes from four tertiary care hospitals in Indonesia. Trop. Med. Int. Heal. 2016;21:610–618. doi: 10.1111/tmi.12692. [DOI] [PubMed] [Google Scholar]
- 33.Lozano C, et al. Human mecC-carrying MRSA: Clinical implications and risk factors. Microorganisms. 2020 doi: 10.3390/microorganisms8101615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aklilu E, Chia HY. First mecC and mecA positive livestock-associated methicillin resistant Staphylococcus aureus (mecC MRSA/LA-MRSA) from dairy cattle in Malaysia. Microorganisms. 2020;8:147. doi: 10.3390/microorganisms8020147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silva V, et al. High frequency of the EMRSA-15 clone (ST22-MRSA-IV) in hospital wastewater. Microorganisms. 2022;10:147. doi: 10.3390/microorganisms10010147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jain S, Chowdhury R, Datta M, Chowdhury G, Mukhopadhyay AK. Characterization of the clonal profile of methicillin resistant Staphylococcus aureus isolated from patients with early post-operative orthopedic implant based infections. Ann. Clin. Microbiol. Antimicrob. 2019;18:8. doi: 10.1186/s12941-019-0307-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ben Said, M. et al. Widespread of the Vienna/Hungarian/Brazilian CC8-ST239-SCCmec III MRSA clone in patients hospitalized in the Tunisian Burn and Traumatology Center. Lett. Appl. Microbiol.76, ovac001 (2023). [DOI] [PubMed]
- 38.Shokravi Z, Haseli M, Mehrad L, Ramazani A. Distribution of Staphylococcal cassette chromosome mecA (SCCmec) types among coagulase-negative Staphylococci isolates from healthcare workers in the North-West of Iran. Iran. J. Basic Med. Sci. 2020;23:1489–1493. doi: 10.22038/ijbms.2020.48481.11127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu S, et al. Prevalence and characterization of food-related methicillin-resistant Staphylococcus aureus (MRSA) in China. Front. Microbiol. 2019;10:304. doi: 10.3389/fmicb.2019.00304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu F, et al. Molecular evolution and adaptation of livestock-associated methicillin-resistant Staphylococcus aureus (LA-MRSA) sequence type 9. mSystems. 2021;6:e00492–21. doi: 10.1128/mSystems.00492-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Masim ML, et al. Genomic surveillance of methicillin-resistant Staphylococcus aureus in the Philippines, 2013–2014. West. Pacific Surveill. Response J. 2021;12:6–16. doi: 10.5365/wpsar.2020.11.1.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Babiker A, et al. Use of online tools for antimicrobial resistance prediction by whole-genome sequencing in methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant enterococci (VRE) J. Glob. Antimicrob. Resist. 2019;19:136–143. doi: 10.1016/j.jgar.2019.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bokarewa MI, Jin T, Tarkowski A. Staphylococcus aureus: Staphylokinase. Int. J. Biochem. Cell Biol. 2006;38:504–509. doi: 10.1016/j.biocel.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 44.Rooijakkers SHM, et al. Early expression of SCIN and CHIPS drives instant immune evasion by Staphylococcus aureus. Cell. Microbiol. 2006;8:1282–1293. doi: 10.1111/j.1462-5822.2006.00709.x. [DOI] [PubMed] [Google Scholar]
- 45.Ahmadrajabi R, Khavidaki SL, Kalantar-Neyestanaki D, Fasihi Y. Molecular analysis of immune evasion cluster (IEC) genes and intercellular adhesion gene cluster (ICA) among methicillin-resistant and methicillin-sensitive isolates of Staphylococcus aureus. J. Prev. Med. Hyg. 2017;58:E308–E314. doi: 10.15167/2421-4248/jpmh2017.58.4.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wardyn SE, et al. Swine farming is a risk factor for infection with and high prevalence of carriage of multidrug-resistant Staphylococcus aureus. Clin. Infect. Dis. 2015;61:59–66. doi: 10.1093/cid/civ234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boswihi SS, et al. Livestock-associated methicillin-resistant Staphylococcus aureus in patients admitted to Kuwait hospitals in 2016–2017. Front. Microbiol. 2019 doi: 10.3389/fmicb.2019.02912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fischer AJ, et al. High prevalence of Staphylococcus aureus enterotoxin gene cluster superantigens in cystic fibrosis clinical isolates. Genes. 2019 doi: 10.3390/genes10121036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kwapisz E, et al. Presence of egc-positive major clones ST 45, 30 and 22 among methicillin-resistant and methicillin-susceptible oral Staphylococcus aureus strains. Sci. Rep. 2020;10:18889. doi: 10.1038/s41598-020-76009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alonzo F, Torres VJ. The bicomponent pore-forming leucocidins of Staphylococcus aureus. Microbiol. Mol. Biol. Rev. 2014;78:199–230. doi: 10.1128/MMBR.00055-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seilie ES, Bubeck Wardenburg J. Staphylococcus aureus pore-forming toxins: The interface of pathogen and host complexity. Semin. Cell Dev. Biol. 2017;72:101–116. doi: 10.1016/j.semcdb.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ullah N, et al. Comparative genomic analysis of a Panton-Valentine leukocidin-positive ST22 community-acquired methicillin-resistant Staphylococcus aureus from Pakistan. Antibiotics. 2022 doi: 10.3390/antibiotics11040496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huilin Z, et al. Phenotypic and genomic analysis of the hypervirulent ST22 methicillin-resistant Staphylococcus aureus in China. mSystems. 2023;8:e01242–22. doi: 10.1128/msystems.01242-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kaneko H, et al. The emerging threat of methicillin-resistant Staphylococcus aureus (MRSA) clone ST22-PT, carrying both Panton-Valentine leucocidin and toxic shock syndrome toxin 1 genes. J. Antimicrob. Chemother. 2023;78:1023–1027. doi: 10.1093/jac/dkad039. [DOI] [PubMed] [Google Scholar]
- 55.Hu Q, et al. Panton-Valentine leukocidin (PVL)-positive health care-associated methicillin-resistant Staphylococcus aureus isolates are associated with skin and soft tissue infections and colonized mainly by infective PVL-encoding bacteriophages. J. Clin. Microbiol. 2015;53:67–72. doi: 10.1128/JCM.01722-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haider, S. & Wright, D. Panton-Valentine leukocidin Staphylococcus causing fatal necrotising pneumonia in a young boy. BMJ Case Rep.2013, bcr2012007655 (2013). [DOI] [PMC free article] [PubMed]
- 57.Hoppe, P. A. et al. Severe infections of Panton-Valentine leukocidin positive Staphylococcus aureus in children. Medicine (Baltimore).98, e17185 (2019). [DOI] [PMC free article] [PubMed]
- 58.Brakstad OG, Aasbakk K, Maeland JA. Detection of Staphylococcus aureus by polymerase chain reaction amplification of the nuc gene. J. Clin. Microbiol. 1992;30:1654–1660. doi: 10.1128/jcm.30.7.1654-1660.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang K, et al. New quadriplex PCR assay for detection of methicillin and mupirocin resistance and simultaneous discrimination of Staphylococcus aureus from coagulase-negative staphylococci. J. Clin. Microbiol. 2004;42:4947–4955. doi: 10.1128/JCM.42.11.4947-4955.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Che Hamzah, A. M. et al. Tigecycline and inducible clindamycin resistance in clinical isolates of methicillin-resistant Staphylococcus aureus from Terengganu, Malaysia. J. Med. Microbiol.68, 1299–1305 (2019). [DOI] [PubMed]
- 61.Magiorakos AP, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 62.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bruce SA, et al. Shared antibiotic resistance and virulence genes in Staphylococcus aureus from diverse animal hosts. Sci. Rep. 2022;12:4413. doi: 10.1038/s41598-022-08230-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 65.Kaya H, et al. SCC mec Finder, a web-based tool for typing of staphylococcal cassette chromosome mec in Staphylococcus aureus using whole-genome sequence data. mSphere. 2018;3:e00612–17. doi: 10.1128/mSphere.00612-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Larsen MV, et al. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 2012;50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bartels MD, et al. Comparing whole-genome sequencing with Sanger sequencing for spa typing of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 2014;52:4305–4308. doi: 10.1128/JCM.01979-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bortolaia V, et al. ResFinder 40 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020;75:3491–3500. doi: 10.1093/jac/dkaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zankari E, et al. PointFinder: A novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J. Antimicrob. Chemother. 2017;72:2764–2768. doi: 10.1093/jac/dkx217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Joensen KG, et al. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 2014;52:1501–1510. doi: 10.1128/JCM.03617-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Page AJ, et al. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Price MN, Dehal PS, Arkin AP. FastTree 2 - Approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Letunic I, Bork P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genomic data of this study were submitted and deposited in NCBI GenBank database under BioProject No. PRJNA722830. Details regarding the 18 Malaysian database isolates are provided in Supplementary Table S1 while details regarding assembly qualities and statistics are provided in Supplementary Table S2.