Significance
Bacterial large and highly structured ncRNA classes contribute to fundamental processes such as protein biosynthesis, protein localization, RNA processing, and gene regulation. Intriguingly, additional structured ncRNA classes exist in bacteria, but the functions of several of the most widespread classes have yet to be established. Here, we describe initial studies on raiA motif RNAs, which represent a remarkably well-conserved ncRNA class found in many Gram-positive species. Bioinformatic, biochemical, and genetic analyses reveal that the Clostridium acetobutylicum raiA motif RNA is important for cellular differentiation to form spores and aggregates, implicating this ncRNA as an important contributor to major physiological changes.
Keywords: cell differentiation, Clostridium acetobutylicum, ComFC, RNA World, ribozyme
Abstract
Several structured noncoding RNAs in bacteria are essential contributors to fundamental cellular processes. Thus, discoveries of additional ncRNA classes provide opportunities to uncover and explore biochemical mechanisms relevant to other major and potentially ancient processes. A candidate structured ncRNA named the “raiA motif” has been found via bioinformatic analyses in over 2,500 bacterial species. The gene coding for the RNA typically resides between the raiA and comFC genes of many species of Bacillota and Actinomycetota. Structural probing of the raiA motif RNA from the Gram-positive anaerobe Clostridium acetobutylicum confirms key features of its sophisticated secondary structure model. Expression analysis of raiA motif RNA reveals that the RNA is constitutively produced but reaches peak abundance during the transition from exponential growth to stationary phase. The raiA motif RNA becomes the fourth most abundant RNA in C. acetobutylicum, excluding ribosomal RNAs and transfer RNAs. Genetic disruption of the raiA motif RNA causes cells to exhibit substantially decreased spore formation and diminished ability to aggregate. Restoration of normal cellular function in this knock-out strain is achieved by expression of a raiA motif gene from a plasmid. These results demonstrate that raiA motif RNAs normally participate in major cell differentiation processes by operating as a trans-acting factor.
Structured noncoding RNAs (ncRNAs) are known to perform various roles in fundamental biological processes in bacteria. Some ncRNA classes, such as RNA-processing ribozymes (1–4) and riboswitches (5, 6), are likely representative of functions that predate the emergence of proteins. Others, such as ribosomal RNAs (rRNAs), transfer RNAs (tRNAs), signal recognition particle (SRP) RNAs, and transfer-messenger RNAs (tmRNAs) participate in the production and manipulation of proteins (7–9), whose evolutionary emergence presumably triggered the eventual loss of RNA-based organisms from the RNA World (10, 11). Although the RNAs noted above are ubiquitous, or at least very common, additional large, structured ncRNA classes have been reported to be well conserved and widespread in certain bacterial lineages (12–14). These RNAs provide opportunities to uncover additional functions for structured ncRNAs whose origins might date back to the RNA World.
Recently, we described the biochemical function of a ~1,000-nucleotide ncRNA class called CP group II ribozymes, which catalyze successive self-splicing reactions to make small RNA circles (15). This finding continues a trend that most large ncRNA classes in bacteria whose functions have been experimentally established operate as catalytic RNAs. We think this trend is justified by the fact that RNAs performing sophisticated chemical reactions must use complex and highly conserved structures to form effective active sites. Thus, we speculate that other large and well-conserved bacterial ncRNAs (14) might also function as ribozymes.
Two other large, structured ncRNAs in bacteria that are relatively common but whose biochemical and biological functions remain to be fully defined are ornate, large, extremophilic (OLE) RNAs (16, 17) and raiA motif RNAs (13). OLE RNA participates as a component of a large ribonucleoprotein (RNP) complex that localizes to the membrane of Gram-positive organisms where it is proposed to serve as a multifunctional stress-responsive particle (17). If true, then OLE RNAs fit the pattern that large ncRNAs in bacteria often perform complex and fundamental biochemical roles.
The raiA motif class of bacterial ncRNAs also exhibits sequence and structural features that suggest it performs one or more biochemical functions that are relevant to an important and perhaps ancient cellular process. Each ~200-nucleotide raiA RNA forms a complex-folded structure interspersed with numerous highly conserved nucleotides (13), which is indicative that it forms a sophisticated three-dimensional (3D) architecture essential for its biochemical function. Its presence in more than 2,500 bacterial species (predominantly in Bacillota and Actinomycetota) whose genomes have been sequenced suggests that raiA motif RNAs perform a broadly important function.
To investigate the possible biological and biochemical functions of the raiA motif RNA, we conducted a series of bioinformatic, biochemical, and genetic analyses to seek clues regarding its roles. Gene associations often provide clues regarding the functions of ncRNAs. The gene for the raiA motif RNA is often found adjacent to the raiA open reading frame, which codes for a protein that acts to place ribosomes in “hibernation” as cells enter stationary phase (18, 19). In addition, the raiA motif frequently resides adjacent to the comFC gene, which codes for a protein involved in competence—a process wherein cells can take up exogenous DNA for genomic integration (20). Furthermore, we demonstrate that the RNA adopts a strikingly well-folded structure and that it is expressed as an independent transcript in abundance when cells approach stationary phase. A Clostridium acetobutylicum strain wherein the raiA motif gene is deleted exhibits strong defects in aggregation and sporulation functions, indicating that this highly structured ncRNA plays an essential role in these fundamental physiological processes.
Results and Discussion
Consensus Sequence and Structural Model for raiA Motif RNAs Reflect an Intricate Structure.
Since the initial report of raiA motif RNAs (13), bacterial genomic sequence databases have substantially expanded. Therefore, we conducted computational searches based on comparative sequence analysis (13, 21–23) to identify additional members of this ncRNA class. Using the original consensus model (13), we queried all representative genomes from the Genome Taxonomy Database (GTDB) R07-RS207 (24). This search strategy yielded 2,377 distinct-sequence raiA motif examples distributed across 2,720 representative species (SI Appendix, raiA Sequence Alignments), which is a greater than fivefold increase of the previously reported collection (13).
Based on the expanded collection of raiA motif RNAs, a revised consensus sequence and structural model was prepared (Fig. 1A). Key features include six major base-paired regions (called P1 through P6) strongly supported by nucleotide sequence covariation that retains Watson–Crick base pairing, two pseudoknots that are similarly supported by covariation, and 51 nucleotide positions whose base identities are highly conserved. Additionally, the P6 region has two major variations, including a small hairpin loop (32% of the representatives) or a larger multi-stem domain (68%) that often carries a highly conserved E-loop structure (25, 26). These characteristics suggest that the raiA motif uses a sophisticated secondary and tertiary architecture to perform its biochemical function(s) and that formation of this structure demands the presence of a large number of exceedingly well-conserved nucleotides. In rare instances (~20 representatives), the RNA motif includes only P1, P3, and P4 but still carries most of the highly conserved nucleotide positions (SI Appendix, raiA Sequence Alignments).
Fig. 1.
Consensus model and phylogenetic distribution of raiA motif RNAs. (A) Updated consensus sequence and structural model for raiA motif RNAs. Predicted base-paired regions are labeled P1 through P8 or Pk1 and Pk2 for pseudoknots. Percentages indicate the abundances of variant substructures (boxed) residing at the P2 or P6 regions. (B) Phylogenetic species tree (27) of Bacillota and Actinomycetota derived from the GTDB R07-RS207 (24) with organisms containing the raiA motif gene marked in green. Scale bar length represents an average phylogenetic distance of one amino acid substitution per site.
The various raiA motif hits were mapped on a phylogenetic species tree (Fig. 1B), indicating that this ncRNA class is well represented in species of the phyla Bacillota and Actinomycetota. Most organisms with raiA motifs are found in a few well-defined clades among the Clostridia, Actinomycetia, Coriobacteriia, Negativicutes, and Acidimicrobiia taxonomic classes. These observations indicate that the raiA motif is a genetically stable feature of these bacteria. However, several major classes such as Bacilli and Anaerolineae have few, if any, organisms with a raiA motif gene. This distribution pattern appears consistent with the hypothesis that raiA motif RNAs have an ancient origin and that some bacterial lineages have since adapted to the absence of this otherwise well-conserved ncRNA. Alternative explanations for the distribution of raiA motif RNAs seem far less likely. For example, multiple evolutionary inventions of the raiA motif seem unlikely due to the long length, high percentage of conserved nucleotides, and complex structure of this ncRNA class. Also, horizontal gene transfer of a late-emerging raiA motif RNA is unlikely because this would have led to the more scattered distribution of species that carry the ncRNA, rather than its clustering among species of certain lineages.
Location of the raiA Motif Gene Implies Links to Diverse Cellular Processes.
Bacterial genome organization often exploits the colocalization of genes whose protein products are related to a specific biochemical pathway or biological process (28). This organizational strategy is also used for ncRNAs, such as RNAs involved in the translation apparatus (29) and OLE RNA (16, 17). Exploiting information regarding gene associations has been particularly useful for establishing candidate ligands for newly found riboswitch candidates. Specifically, the function of the protein whose gene is located immediately downstream and in the same orientation as a riboswitch candidate provides strong clues regarding the ligand that regulates riboswitch function (6, 30). Therefore, we sought clues regarding the biological and biochemical functions of raiA motif RNAs by conducting a thorough analysis of genes located near to the raiA motif gene.
To achieve this objective, a search of 887 annotated genomes carrying a raiA motif gene was performed. We evaluated the annotated functions of gene products ten positions upstream and ten positions downstream of the raiA motif gene (Dataset S1). Several observations from this analysis are notable. Importantly, the raiA motif does not exhibit genomic location characteristics indicative of riboswitch function. The motif is commonly separated in the genome from the downstream ORF by hundreds of nucleotides, and it is often encoded in the opposite orientation relative to the same gene in different species. These distance and orientation characteristics are highly atypical for riboswitches, indicating that a cis-regulatory role for raiA RNAs is unlikely.
As the distance from the raiA motif gene increased, the gene annotations became more numerous and less consistent at each location. Therefore, the distribution of genes located only up to four positions before and after the gene for the raiA motif RNA were graphed (Fig. 2). Despite this noted variability in location, orientation, and gene association, the motif commonly resides downstream of comFC genes (74%) and upstream of raiA genes (66%). This latter gene association was the motivation for choosing the raiA motif name (13). As noted above, the comFC gene codes for a protein that participates in DNA uptake (20) and therefore is relevant to bacterial competence (31, 32), or the ability of cells to take up foreign DNA and integrate these molecules into their genome.
Fig. 2.
Genes commonly residing in proximity to the gene for raiA motif RNA. Plot of the most common genes located at gene positions −4 to −1 (upstream) and +1 to +4 (downstream) of the gene for raiA motif RNAs (position 0). The genes that are most frequently located at each position are represented by the colored portions of the bars, where the color identifies categories of biological functions. Genes present with an abundance of less than 10% were grouped with other rare gene associations and depicted as a white bar. RR indicates a gene for a Response Regulator portion of a two-component signaling complex (33). ABC indicates a gene for an adenosine triphosphate (ATP)-binding cassette transporter (34).
As noted earlier, the raiA gene codes for a ribosome hibernation factor, which stabilizes the 70S ribosome in an inactive state when cells are experiencing low nutrient conditions (18, 19, 35). In addition, the prfB gene resides near the raiA motif gene. The prfB gene codes for Release Factor 2 (36), which is involved in disengaging the ribosome from the mRNA and the completed protein product once the ribosome reaches UAA or UGA stop codons (37). Although this gene association is less common than the association with the raiA gene, it further strengthens the possible connection between the raiA motif RNA and translation control.
Other common gene associations imply that raiA motif RNAs might also be relevant to processes involving signaling (mtrA, mtrB, lpqB, and RR), and membrane transporter functions (ABC transporters), protein secretion (secA), and membrane composition (degV). These gene associations are rather disparate, and there appears to be no unifying theme regarding the functions of their protein products. Thus, we cannot precisely define a strong hypothesis for the function of raiA motif RNAs based only on gene associations. Furthermore, we do not detect a strong candidate for a protein component of a possible RNP complex that might be formed by the raiA motif RNA. Such partnerships between ncRNAs and proteins are evident in some cases, such as for group II ribozymes (3) and OLE RNAs (16, 38).
The raiA Motif RNA from C. acetobutylicum Conforms to the Predicted Secondary Structure Model.
To conduct biochemical and genetic analyses, we chose to investigate the raiA motif representative from C. acetobutylicum ATCC 824 (SI Appendix, Table S1). This Gram-positive species is an obligate anaerobe from the Bacillota phyla and is known to conduct Acetone-Butanol-Ethanol fermentation (39). This metabolism feature is exploited for its basic science value and for its industrial utility regarding the large-scale production of certain biofuel and bulk chemicals (40). Thus, previous work has provided a range of genetic tools (41) that are useful for the investigation of raiA motif RNAs.
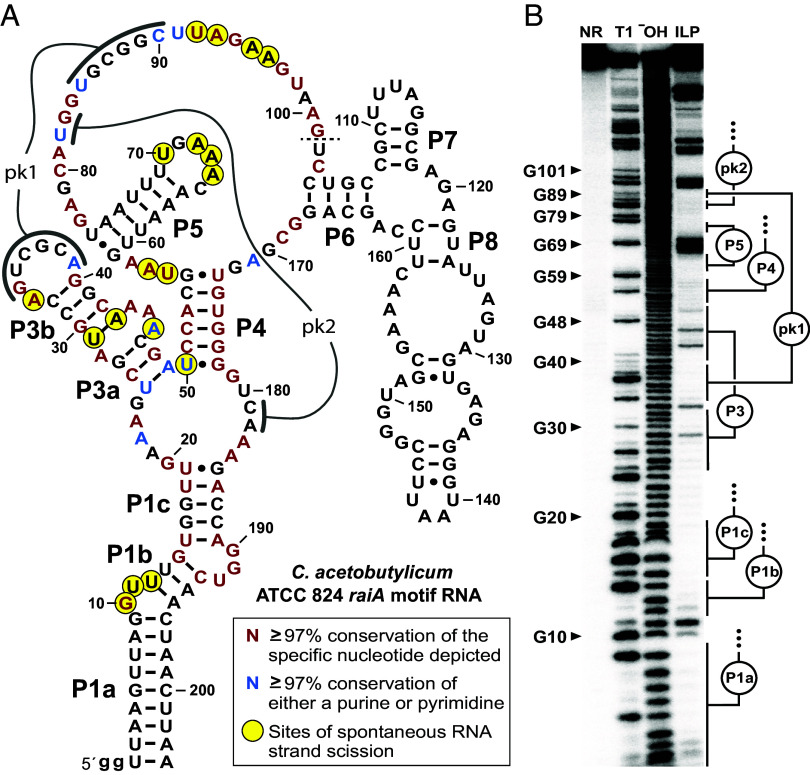
The raiA motif RNA from C. acetobutylicum (Fig. 3A) adopts a folded form in vitro that is entirely consistent with the consensus structure model predicted previously (13) and of that derived from our updated comparative sequence analysis (Fig. 1A). Nucleotides predicted to form base-paired regions P1 through P7 indeed exhibit expected base-pairing potential for this representative. However, the initial stem area of P8 appears to lack the four base pairs typical of this region. Although the structure of the P8 stem appears to be weakened in the C. acetobutylicum representative (Fig. 3A), this region is the least-conserved portion of the raiA motif as observed by comparative sequence analysis.
Fig. 3.
Evidence that raiA motif RNAs form complex-folded structures. (A) Sequence and secondary structure model for the raiA motif representative from C. acetobutylicum. Predicted base-paired stems are labeled P1 through P8. Nucleotides involved in predicted pseudoknots (pk1 and pk2) are also identified. Colored nucleotides match highly conserved nucleotides of the consensus model as indicated. Sites of spontaneous RNA strand scission were derived from in-line probing assay data depicted in (B). The dashed line at position 101 indicates that the in-line probing data was not mapped beyond this point. Lowercase “g” letters indicate nucleotides added to the construct to facilitate preparation by in vitro transcription. (B) Autoradiogram of a denaturing polyacrylamide gel electrophoresis (PAGE) separation of the products of an in-line probing reaction using trace amounts of 5′ 32P-labeled C. acetobutylicum raiA motif RNA. RNAs were not reacted (NR), subjected to partial digestion with RNase T1 (T1) or alkaline conditions (−OH), or were incubated under in-line probing conditions at room temperature (~20 °C) for 72 h. Selected bands corresponding to products resulting from RNase T1 digestion (cleavage after G nucleotides) are labeled accordingly.
The proposed structure is also strongly supported by structural probing data. Specifically, we used in-line probing (42, 43) to map the locations of robust spontaneous RNA strand scission. Rapid strand scission typically occurs at nucleotides either undergoing substantial structural motion or at sites wherein the internucleotide linkage is held in an in-line geometry that facilitates RNA chain cleavage by internal phosphoester transfer (42). As expected for a well-structured RNA, in-line probing results reveal that most internucleotide linkages of raiA motif RNA are remarkably stable (Fig. 3B). This indicates that these linkages join nucleotides that are participating in stable structures that do not permit the nucleophilic attack of a ribose 2′-oxyanion on the adjacent phosphorus center (42).
In contrast, some linkages do undergo relatively robust scission (Fig. 3B). These labile sites often map to loops, bulges, or other unpaired regions that are likely to be relatively unstructured. Intriguingly, there are numerous nucleotides residing in regions outside of the predicted base-paired regions that also resist spontaneous cleavage. These findings strongly indicate that raiA motif RNAs form complex tertiary interactions that provide global stability of the RNA even when the nucleotides are not in conventional base-paired stems. Many of these stable regions encompassing bulges or unpaired linkers carry highly conserved nucleotides, which are likely necessary for forming a sophisticated 3D RNA structure.
The raiA Motif RNA Is One of the Most Abundant RNAs in C. acetobutylicum.
To initiate the analysis of raiA motif RNAs in cells, we established or exploited methods for the manipulation and detection of the RNA in C. acetobutylicum. To conduct genetic alterations, we used a CRISPR/Cas9-based editing system that enables the scarless insertion or removal of target genes (44). First, we introduced a cas9 gene under a xylose-inducible promoter at a genomic location where insertions are known not to perturb normal cell function (44). The resulting strain carrying the cas9 gene was named CAS9. Beginning with the CAS9 strain, a raiA motif RNA knockout (KO) variant strain was prepared by deletion of the raiA motif RNA gene to create a strain called “ΔraiA motif”. Successful removal of the natural raiA motif locus was confirmed by PCR analysis (SI Appendix, Fig. S1).
Furthermore, the ΔraiA motif strain was evaluated by qRT-PCR assays to confirm that the mRNA levels of the flanking genes were not substantially altered. The gene located immediately upstream of the raiA motif gene in C. acetobutylicum is comFC, which is typical of most species (Fig. 2 and SI Appendix, Fig. S2). However, the gene located immediately downstream of the raiA motif gene is annotated as gntR, which encodes a member of a large family of transcription factors (45). Regardless of the functions of the protein products of these genes, the levels of their mRNAs are largely unchanged when the raiA motif gene is deleted (SI Appendix, Fig. S2).
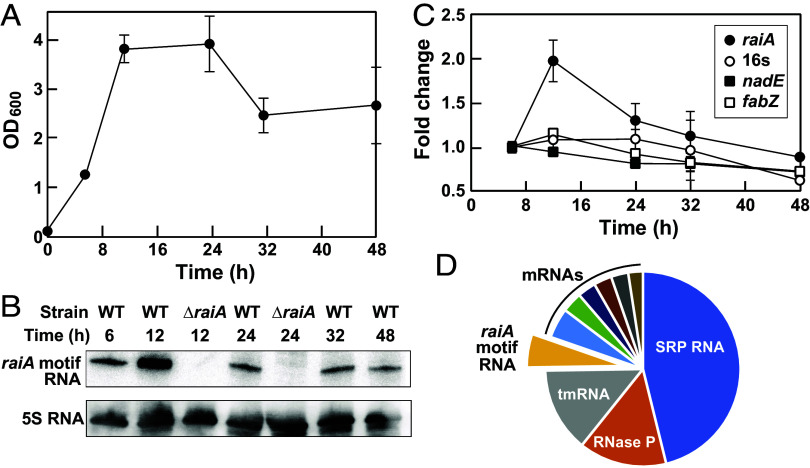
Detection of raiA motif RNA transcripts was achieved initially by northern blot analysis. A single raiA motif RNA transcript band was identified using a complementary oligonucleotide probe in wild-type (WT) C. acetobutylicum cells (Fig. 4A), whereas two transcripts differing in their 3′ termini have been observed for the homologous representative from the bacterium Clostridioides difficile (46, 47). No transcript is observed with the ΔraiA motif strain, which further demonstrates that the gene for the raiA motif RNA has been deleted in these cells. This analysis also reveals that the raiA motif RNA peaks in abundance at the 12-h time point, which suggests that the RNA is in greatest demand as cells transition from exponential growth phase to stationary phase. These results were confirmed by qRT-PCR assays (Fig. 4B).
Fig. 4.
Detection and quantitation of the raiA motif RNA transcript. (A) Growth curve for C. acetobutylicum under anaerobic conditions in 2× YTG medium (48). (B) Northern blot analysis of raiA motif RNAs from WT and ΔraiA motif strains of C. acetobutylicum. 5S rRNA is used as a loading control. (C) Plot of the quantitation of raiA motif RNA levels from WT C. acetobutylicum at various time points using qPCR. Controls include 16S RNA, nadE mRNA, and fabZ mRNA. Fold change values were established relative to the ct numbers obtained at 6 h, which were set to 1. (D) Pie chart of the relative abundances of the top 10 RNA transcripts in WT C. acetobutylicum cells upon exclusion of tRNAs and rRNAs, as determined by analysis of whole-transcriptome sequencing (RNAseq) data published previously (49).
To establish the abundance of raiA motif RNAs relative to other transcripts, we analyzed previously published C. acetobutylicum whole-transcriptome RNAseq data (49). Transcriptomics data were derived from cells collected 75 min past an OD600 measurement of 1 and therefore represent cells at or near stationary phase. On exclusion of tRNAs and rRNAs, raiA motif RNAs represent the fourth most abundant transcript in cells (Fig. 4C and Dataset S2). Thus, all RNAs that are more abundant than raiA motif RNA are exclusively (tRNAs, rRNAs, SRP RNA, RNase P) or substantively (tmRNA) ncRNAs. The broad phylogenetic distribution of raiA motif RNA, its extraordinary sequence and structure conservation, and its abundance are all consistent with a biochemical function as a structured ncRNA. This conclusion is also consistent with the lack of a protein-coding region among the various other conserved features of the molecule.
The C. acetobutylicum ΔraiA Motif Strain Is Defective for Sporulation and Aggregation.
Although the raiA motif RNA is present throughout the growth phases of C. acetobutylicum, its abundance peaks when cells transition to the stationary phase. This transitional period requires that cells undergo multiple physiological changes depending on environmental conditions. Cells need to cease growth, initiate spore formation, and/or produce large-scale macromolecular aggregate structures on which cells can maintain a vegetative stage, including biofilm formation (50).
The persistent abundance of raiA motif RNA (Fig. 4) suggests that it might have functional roles throughout the life cycle of the cell. Because this ncRNA experiences its highest concentration at the transitional phase between exponential growth and stationary phase, it must serve one or more important roles during this period. Therefore, we searched for effects on major physiological processes relevant to the transitional period when the raiA motif RNA is deleted.
First, we assessed the abilities of both the CAS9 and the ΔraiA motif cells to undergo sporulation. The ΔraiA motif strain yields ~1/10th the number of viable spores compared to the CAS9 strain based on bacterial colony abundances on solid media (Fig. 5A). Poorer spore formation by ΔraiA motif cells was confirmed by determining sporulation efficiency, which accounts for the number of vegetative cells in each culture (Fig. 5B). This deficiency in spore formation can be restored when a gene for the raiA motif RNA is reintroduced under its natural promoter via a plasmid (Fig. 5B, “Rescue”; SI Appendix, Table S2).
Fig. 5.
C. acetobutylicum ΔraiA motif cells exhibit a sporulation deficiency phenotype. (A) Agar growth assays of untreated and heat-treated (80 °C, 10 min) cultures of CAS9, ΔraiA motif, and raiA motif rescue strains of C. acetobutylicum. (B) Plot of sporulation efficiency as determined by comparing CFU values of untreated cell cultures to heat-treated cultures [(CFUheat treated/CFUuntreated) × 100]. The asterisk indicates the significance between CAS9 and ΔraiA motif values (t test): P < 0.05.
Compared to the C. acetobutylicum CAS9 strain, the ΔraiA motif strain also exhibits reduced levels of aggregation when cultured in Clostridium growth medium (CGM) for 48 h. This phenotype is observed by visual inspection of culture tubes (Fig. 6A), by measuring OD600 before and after mixing liquid cultures (Fig. 6B) and by calculating aggregation index values (Fig. 6C). Again, a rescue strain carrying the raiA motif gene on a plasmid reverses this aggregation deficiency phenotype. The ΔraiA motif strain also manifests reduced overall growth (Fig. 6B), which might be due to adverse effects of poor biofilm production (51). Together, the sporulation and aggregation deficiency phenotypes reveal that the raiA motif plays important roles in cell differentiation in C. acetobutylicum.
Fig. 6.
C. acetobutylicum ΔraiA motif cells exhibit an aggregation deficiency phenotype. (A) Photographs of microfuge tubes containing cultures of CAS9, ΔraiA motif, and raiA motif rescue cells in liquid CGM. Unmixed: 48 h cultures without shaking. Mixed: the resulting unmixed cultures were mixed by vortex for 5 min. (B) Plot of cell densities for replicate cultures as depicted in (A). (C) Plot of the aggregation index as computed using measured OD600 values for unmixed and mixed cultures from B based on the following equation: [{(mixed-unmixed)/mixed} × 100]. The asterisk indicates the significance between CAS9 and ΔraiA motif values (t test): P < 0.0005.
Concluding Remarks.
The large size and highly conserved sequence and structural features of raiA motif RNAs strongly indicate that members of this unusual ncRNA class perform one or more biochemical functions that require the formation of a complex 3D architecture. Some features of raiA motif RNAs are like those of riboswitches that form selective binding pockets for their target ligands. However, other attributes differ markedly from those of riboswitches. While riboswitch structures often include gene control substructures called expression platforms to modulate gene expression (5, 6), such as intrinsic terminator stems (52, 53) and Shine-Dalgarno occlusion structures (54), these elements are typically not associated with raiA motif RNAs. Furthermore, despite appearing in various gene contexts, the raiA motif RNA gene exists as a single copy in bacterial chromosomes. These and other characteristics are inconsistent with a cis-regulatory role for the raiA ncRNA.
Because most large ncRNAs with conserved structures in bacteria are ribozymes (12, 14), we speculate that raiA motif RNAs most likely catalyze a chemical transformation. Again, based on its established characteristics, the range of possible ribozyme functions can be somewhat bounded. For example, unlike group I and group II self-splicing introns that commonly participate as components of mobile genetic elements (2, 3), and therefore are often present in multiple copies in bacterial chromosomes, raiA motif RNAs are encoded at single genetic loci. Moreover, raiA motif RNA sequences have never been observed to interrupt ORFs, nor have they been observed to undergo self-processing reactions in vitro. It therefore seems certain that raiA motif RNAs do not function as self-splicing RNAs, or as ribozyme components of transposable elements. Similar reasoning can be used to exclude the possibility that raiA motif RNAs function as self-cleaving ribozymes (4, 55).
Although the raiA motif ncRNA class is more narrowly distributed in bacteria compared to the RNase P ribozyme class (1), both these RNAs are encoded by single-copy genes in bacteria. Therefore, we speculate that raiA motif RNAs might also catalyze an important chemical transformation in a manner that, like RNase P, does not require multiple variants in the cell to achieve its biological objective. Notably, members of these two RNA classes differ substantially in the proportion of nucleotides that are highly conserved. Over 50 positions throughout the approximately 200 nt (25%) are 97% conserved in raiA motif RNAs (Fig. 1A). In contrast, only 24 of ~400 nt (6%) are conserved at a similarly high level in RNase P RNA (56). Bacterial RNase P, an RNP consisting of one protein and one catalytic RNA subunit, is an endonuclease that cleaves a wide array of RNA substrates, including tRNA precursors and other RNAs (1). The relatively low percentage of conserved sequences in RNase P RNA can probably be attributed in part to its vastly broader phylogenetic distribution and the diversity of its RNA substrates. The far greater extent of sequence conservation displayed by the raiA motif RNA might indicate that it recognizes a substrate molecule that is invariant in structure.
The observation that genes for raiA motif RNAs often reside near genes involved in competence, translation regulation, sporulation, cyclic dinucleotide signaling, and motility (Fig. 2 and SI Appendix, Fig. S2) is consistent with our observations that the representative from C. acetobutylicum participates in major lifestyle transitions and developmental programs relating to these genes. This juxtaposition of genes involved in natural competence and translation arrest is broadly conserved, even among distantly related bacteria that lack the raiA motif RNA, and is consistent with competence as a developmental program that is often concomitant with growth arrest or a decrease in rRNA production (57, 58). Given the importance of cyclic dinucleotide signaling to bacterial physiological changes (59–61), and given that these signaling molecules likely emerged during the RNA World (62, 63), an intriguing possibility is that raiA motif RNAs serve as receptors or as ribozymes involved in cyclic dinucleotide signaling processes.
Materials and Methods
Bioinformatics Analyses.
Genomic assemblies for representative GTDB species (64), version r207, were retrieved from the NCBI Genome Assembly FTP (65). A covariance model for raiA motif RNAs was built from the originally published alignment (13) and queried against the GTDB bacterial representative genome assemblies using Infernal (66). The secondary structure was analyzed in RALEE (67) and the consensus diagram was generated with R2R (68), after removing hits with three or more ambiguous nucleotides.
Genes in the vicinity of the raiA motif were identified using a custom Python script implementing the Biopython package (69). The script uses NCBI RefSeq (70) chromosome accessions and coordinates from the raiA motif STOCKHOLM file to access NCBI GenBank records using Entrez queries and identify genes upstream and downstream of each motif. The gene product name of the coding sequence at each position upstream and downstream of each raiA motif as specified in the GenBank record was taken as each gene name.
A custom R script utilizing the ape (27) and TreeTools (71) packages was used to prune a bacterial tree of life from GTDB R07-RS207 (24) to generate the smallest tree containing all genome assemblies containing the raiA motif. The resulting phylogenetic tree was visualized and annotated using iTOL v6 (72).
Bacterial Strains and Media.
C. acetobutylicum ATCC 824 cells for all experiments were cultured in an anaerobic chamber (Bactron 300) containing an atmosphere of 90% nitrogen, 5% hydrogen, and 5% carbon dioxide. Growth media used for each experiment are noted and were composed as follows: 2× YTG medium (48) includes 16 g L−1 tryptone, 10 g L−1 yeast extract, 5 g L−1 NaCl, and 10 g L−1 glucose. The pH was adjusted to 5.2 for liquid media and 5.8 for solid media. All solid media were prepared with 15 g L−1 agar. Clostridial Growth Medium (CGM) (73) includes 30 g L−1 glucose, 6.25 g L−1 yeast extract, 2.5 g L−1 ammonium sulfate, 1.25 g L−1 NaCl, 2.5 g L−1 asparagine, 0.95 g L−1 monobasic potassium phosphate, 0.95 g L−1 dibasic potassium phosphate, 0.5 g L−1 magnesium sulfate heptahydrate, 13 mg L−1 manganese sulfate heptahydrate, and 13 mg L−1 iron sulfate heptahydrate. The pH was adjusted to 6.4. Clostridial basal medium (CBM) (74) includes 10 g L−1 glucose, 0.5 g L−1 monobasic potassium phosphate, 0.5 g L−1 dibasic potassium phosphate, 4 g L−1 tryptone, 0.2 g L−1 magnesium sulfate heptahydrate, 10 mg L−1 manganese sulfate heptahydrate, 10 mg L−1 ferrous sulfate heptahydrate, 1 mg L−1 para-aminobenzoic acid, 1 mg L−1 thiamin hydrochloride, and 2 μg L−1 biotin. The pH was adjusted to 6.9. Escherichia coli TOP10 cells (Thermo Fischer Scientific) were grown in Luria-Bertani medium at 37 °C. For the appropriate Clostridium strains, culture media was supplemented with thiamphenicol (Th, 15 μg mL−1). For the appropriate E. coli strains, kanamycin (Kan, 60 μg mL−1) or chloramphenicol (Cm, 25 μg mL−1) were added. C. acetobutylicum ATCC 824 and E. coli strains were maintained as 20% v/v glycerol stocks stored at −80 °C. Spore suspension stocks of C. acetobutylicum ATCC 824 used in this work were maintained at −20 °C as previously described (75).
Chemicals and Oligonucleotides.
All synthetic oligonucleotides (SI Appendix, Table S3) were supplied by Integrated DNA Technologies. [γ-32P] ATP (specific activity: 6,000 Ci mmol−1) was purchased from PerkinElmer.
RNA Oligonucleotide Preparation.
In vitro transcription from synthetic DNA templates was used to prepare raiA motif RNAs for analysis following a protocol described previously (76). The resulting RNAs were purified by denaturing 10% polyacrylamide gel electrophoresis (PAGE). The bands corresponding to the desired products were excised, the RNAs were eluted using crush-soak solution (200 mM NH4Cl, 10 mM Tris-HCl [pH 7.5 at ∼20 °C], 1 mM ethylenediaminetetraacetic acid [EDTA]), and the RNAs were recovered by precipitation with ethanol. Then, 60 pmoles of the RNA transcripts were dephosphorylated using rAPid alkaline phosphatase (Roche Life Sciences) following the manufacturer’s protocol. Five pmoles of dephosphorylated RNAs were 5′ 32P-radiolabeled using T4 polynucleotide kinase (New England Biolabs). Radiolabeled RNAs were purified using denaturing 10% PAGE, recovered as described above, and resuspended in dH2O for storage at −20 °C until use.
In-Line Probing Assays.
Following the protocol described previously (42, 43), 5′ 32P-labeled C. acetobutylicum raiA motif RNA was subjected to ILP (in-line probing) analysis in a 10-µL reaction mixture containing 100 mM Tris-HCl (pH 8.3 at 20 °C), 100 mM KCl, and 20 mM MgCl2. The mixture was incubated at room temperature for ∼40 h. Marker lane preparation included NR (no reaction): RNA was kept at −20 °C; T1 (RNase T1 treatment): partial digestion with RNase T1; −OH (partial alkaline degradation): incubation with sodium carbonate (pH 9). RNA products were separated using denaturing 10% PAGE and visualized using a Typhoon FLA 9500.
Northern Blot Analyses.
Northern blots were prepared by using a 32P-labeled DNA probe (SI Appendix, Table S3) according to a previously published protocol (77). Hybridization was carried out in GE Rapid-Hyb buffer at 42 °C according to the manufacturer’s protocol. Signals were normalized to 16S rRNA.
Cellular RNA Isolation and cDNA Production.
Total cellular RNA samples were prepared from 5 to 10 mL of cell culture by pelleting cells by centrifugation at 3,500 × g for 15 min at 4 °C, washing pellets once with RNAprotect (Qiagen), and storing the resulting cells at −80 °C until use. For RNA isolation, cell pellets were processed with the Quick-RNA Fungal/Bacterial Miniprep kit (Zymo Research). Isolated nucleic acid material was subjected to DNase treatment using a TURBO™ DNase kit (ThermoFisher). The resulting RNA was subjected to cDNA synthesis using an iScript cDNA Synthesis Kit (Bio-Rad Laboratories). All kits were used following the manufacturers’ directions.
qRT-PCR.
One μL of each cDNA synthesis preparation (see above) was combined with appropriate target-specific primers (SI Appendix, Table S3), SYBR Green (Applied Biosystems), and dH2O to generate 10 µL qRT-PCR mixtures. Samples were subjected to amplification and analysis on an iCycler iQ Real-Time PCR Detection System (Bio-Rad Laboratories) following the manufacturer’s directions. To reduce data variability, each assay was run in triplicate on the same 96-well plate along with three negative control wells per target (containing all qRT-PCR components with 1 µL dH2O substituted for the cDNA product) to verify that the signatures are not due to misamplification. For each target, a PCR standard curve was generated to calculate the PCR efficiency and to verify that all fluorescence curves were linear. Analyses of control targets (16S rRNA, nadE mRNA, and fabZ mRNA) were conducted on each plate and used as a baseline to establish fold change values.
Transcriptomics Data Analysis.
Total transcriptomics sequence data analyzed were published previously (49) and were obtained in the Sequence Read Archive under the tag SRP052867. Raw FASTQ files were processed for trimming and ribosomal RNA removal following a processing pipeline provided by the authors (https://github.com/MatthewRalston/NGS_scripts). The resulting file was subjected to final alignment to the genome and expression analysis using Rockhopper (78).
Plasmid Construction.
Plasmids used in this study (SI Appendix, Table S2) were generated using the following methods. Isolation of genomic DNA from C. acetobutylicum ATCC 824 was achieved by growing cells overnight to stationary phase in 5 mL of CGM and employing the Quick-DNA Fungal/Bacterial Kit (Zymo Research) according to the manufacturer’s instructions. Plasmid pINTcas9 (44) was reconstructed via synthesis (GenScript). Similarly, the plasmid pgRNAraiAKO was constructed based on the plasmid pgRNA (44) and included the appropriate guide RNA sequence targeting the raiA motif RNA gene sequence plus appropriate sequences that are homologous to the C. acetobutylicum genome flanking the raiA motif gene.
The praiARescue plasmid was prepared by using the pgRNAraiAKO plasmid as a PCR template to create a linear construct containing the C. acetobutylicum and E. coli origin of replication, and the chloramphenicol resistance gene (catP). DreamTaq green PCR master mix (ThermoFisher) was used for all PCR reactions relevant to plasmid manipulation. DpnI restriction enzyme was added to the resulting PCR mixture to fragment the naturally methylated pgRNAraiAKO plasmid DNA and the desired linear PCR product was purified using a QIAquick PCR Purification Kit (Qiagen) according to the manufacturer’s instructions. The PCR product was then used for Gibson assembly together with a 500 base-pair PCR product carrying the raiA motif gene, its promoter, and its terminator. This PCR product was generated by using the gDNA of C. acetobutylicum as a template.
The sequences described above were incubated together with Gibson Assembly® Master Mix (New England Biolabs) at a 3:1 molar ratio of insert to vector, following the manufacturer’s instructions. The resulting circularized DNA product was transformed into E. coli TOP10 cells. The desired transformed clones were identified by PCR. Nanopore sequencing was used to confirm the sequence of the final praiARescue plasmid. Plasmid pN3 used for methylation of pINTcas9, pgRNAraiAKO, and praiARescue, necessary for C. acetobutylicum transformation, was generously provided by the Papoutsakis laboratory at the University of Delaware.
Electro-Transformation of C. acetobutylicum.
All plasmids transformed into C. acetobutylicum were subjected to methylation by co-transformation of plasmid pN3 into E. coli TOP10 cells facilitated by heat shock. This yielded plasmids that are protected against the C. acetobutylicum native restriction-modification system (79). Purification of co-transformed plasmids was achieved by using QIAprep Spin Miniprep Kit (Qiagen), and the resulting plasmid mixture was used for electroporation of C. acetobutylicum as previously reported (79) with minor modifications as detailed below.
Spore suspensions (see above) were heated at 80 °C for 10 min and inoculated in 10 mL 2× YT. Germination and growth were allowed to proceed overnight at 37 °C. Aliquots of these cultures (OD600 of 1.0 to 2.0) were used to initiate fresh 80 mL 2× YT cultures at an OD600 of 0.15. Once an OD600 of 0.6 to 0.9 was reached, indicating mid-log phase, tubes were briefly placed on ice and then centrifuged at 3,500 × g for 15 min at 4 °C. Following removal of the supernatant, the cell pellet was resuspended in 30 mL ice-cold electroporation buffer (EPB: 270 mM sucrose, 5 mM NaH2PO4, pH 7.4). The resuspended cells were centrifuged at 3,500 × g for 15 min at 4 °C, the supernatant was discarded, and the cell pellet was resuspended in 1.1 mL of ice-cold EPB, resulting in electrocompetent cells. Each electrotransformation was performed by mixing 550 μL competent cells and 50 μL of plasmid solution (~2 μg total). The mixture was prepared in a 0.4 cm electroporation cuvette and electrotransformation was conducted with the following parameters: 2,000 V, 25 μF, and infinite Ω. Immediately following electroporation, samples were resuspended in 1 mL 2× YT and transferred to a 15-mL culture tube containing 9 mL 2× YT. Cultures were then allowed to recover at 37 °C for 4 h. The recovery cultures were centrifuged at 3,500 × g for 15 min at room temperature, and the supernatant was discarded. The cell pellet was resuspended in 50 μL 2× YT and plated onto solid 2× YT plates supplemented with the appropriate antibiotic. Plates were incubated at 37 °C for 2 to 3 d. Transformant colonies were subjected to PCR verification.
Construction of CAS9, ΔraiA Motif, and raiA Motif Rescue Strains.
Genetic manipulation of C. acetobutylicum was done as previously described (44). For the construction of the CAS9 strain, the pINTcas9 plasmid was introduced by the method described above, and transformants were inoculated into CGM containing 5% xylose for 2 d to allow cas9 gene integration into the C. acetobutylicum chromosome. Colonies were subject to PCR verification and whole-genome sequencing to confirm precise insertion. Multiple passages using solid agar without antibiotic were performed for plasmid curing. The resulting CAS9 strain was then used as a control instead of WT for subsequent experiments. For the construction of the ΔraiA motif strain, CAS9 was subjected to electroporation of the plasmid pgRNAraiAKO and followed all steps as described above. Lastly, production of the raiA motif rescue strain was achieved by electroporation of the ΔraiA strain with the praiARescue plasmid, which was retained throughout all experiments through continuous passage in thiamphenicol-containing media.
Phenotype Assays.
Sporulation assays were performed as previously described (80) with minor modifications. Spore suspensions were heated at 80 °C for 10 min and inoculated in 10 mL CGM, where they were allowed to germinate overnight at 37 °C. Overnight cultures were plated on solid CBM, and thiamphenicol (15 μg mL−1) was included for plasmid maintenance when necessary. Sampling of plated cultures was conducted after 2, 3, 4, and 6 d by resuspending 3 individual colonies in 80 μL liquid CGM. Then, 40 μL of the resuspended colony mixture was heated at 80 °C for 10 min. Serial dilutions (10−1 to 10−6) were prepared from both the heated and unheated samples. From each strain, 10 μL of the dilution series were spotted onto 2× YT agar and incubated for 30 h. Sporulation efficiency percentages were determined by counting the colony-forming units per ml (CFU mL−1) and dividing the CFU values of total vegetative cells by the heat-resistant spores × 100.
Aggregation assays were performed by heating triplicate spore suspensions at 80 °C for 10 min and inoculating 5 mL 2× YT, where they were allowed to germinate overnight at 37 °C. Overnight culture samples were used to inoculate 10 mL CGM at a final OD600 of 0.1, and these cultures were incubated at 37 °C for 48 h. Thiamphenicol was included for plasmid maintenance when necessary. Cells were removed from the anaerobic chamber and their OD600 values were measured either before mixing (unmixed) or after subjecting them to strong mixing for 5 min (maximum speed, Vortex-Genie 2). The aggregation index was then calculated according to the following equation: aggregation index = [(OD600mixed – OD600unmixed)/OD600mixed] × 100. Statistical analysis was performed on GraphPad Prism v. 10.0.0 using a nonparametric t test at P < 0.05.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (TXT)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Acknowledgments
We thank members of the Breaker laboratory for helpful discussions regarding the project and comments on the manuscript. We also thank the Papoutsakis laboratory at the University of Delaware for supplying plasmid pN3. Graduate studies by L.W.S. were funded by the CAPES Foundation of the Ministry of Education of Brazil. R.R.B. is an Investigator with the Howard Hughes Medical Institute, which also funded this research.
Author contributions
L.W.S., A.R., and R.R.B. designed research; L.W.S., C.G.K., and C.M.F. performed research; L.W.S. contributed new reagents/analytic tools; L.W.S., C.G.K., C.M.F., A.R., and R.R.B. analyzed data; R.R.B. obtained funding; and L.W.S. and R.R.B. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
Reviewers: N.G.W., University of Michigan; and J.D.W., University of Wisconsin-Madison.
Data, Materials, and Software Availability
All experimental data are included in the main text and supporting information.
Supporting Information
References
- 1.Phan H. D., Lai L. B., Zahurancik W. J., Gopalan V., The many faces of RNA-based RNase P, an RNA-world relic. Trends Biochem. Sci. 46, 976–991 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hausner G., Hafez M., Edgell D. R., Bacterial group I introns: Mobile RNA catalysts. Mobile DNA 5, 8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lambowitz A. M., Zimmerly S., Group II introns: Mobile ribozymes that invade DNA. Cold Spring Harb. Perspect. Biol. 3, a003616 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jimenez R. M., Polanco J. A., Lupták A., Chemistry and biology of self-cleaving ribozymes. Trends Biochem. Sci. 40, 648–661 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCown P. J., Corbino K. A., Stav S., Sherlock M. E., Breaker R. R., Riboswitch diversity and distribution. RNA 23, 995–1011 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kavita K., Breaker R. R., Discovering riboswitches: The past and the future. Trends Biochem. Sci. 48, 119–141 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steitz T. A., Moore P. B., RNA, the first macromolecular catalyst: The ribosome is a ribozyme. Trends Biochem. Sci. 28, 411–418 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Akopian D., Shen K., Zhang X., Shan S. O., Signal recognition particle: An essential protein-targeting machine. Annu. Rev. Biochem. 82, 693–721 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keiler K. C., Biology of trans-translation. Annu. Rev. Microbiol. 62, 133–151 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Benner S. A., Ellington A. D., Tauer A., Modern metabolism as a palimpsest of the RNA world. Proc. Natl. Acad. Sci. U.S.A. 86, 7054–7058 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen X., Li N., Ellington A. D., Ribozyme catalysis of metabolism in the RNA world. Chem. Biodivers. 4, 633–655 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Weinberg Z., Perreault J., Meyer M. M., Breaker R. R., Exceptional structured noncoding RNAs revealed by bacterial metagenome analysis. Nature 462, 656–659 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weinberg Z., et al. , Detection of 224 candidate structured RNAs by comparative analysis of specific subsets of intergenic regions. Nucleic Acids Res. 45, 10811–10823 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harris K. A., Breaker R. R., Large noncoding RNAs in bacteria. Microbiol. Spectr. 6, 10.1128/microbiolspec.RWR-0005-2017 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roth A., Weinberg Z., Vanderschuren K., Murdock M. H., Breaker R. R., Natural circularly permuted group II introns in bacteria produce RNA circles. iScience 24, 22 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Puerta-Fernandez E., Barrick J. E., Roth A., Breaker R. R., Identification of a large noncoding RNA in extremophilic eubacteria. Proc. Natl. Acad. Sci. U.S.A. 103, 19490–19495 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Breaker R. R., Harris K. A., Lyon S. E., Wencker F. D. R., Fernando C. M., Evidence that OLE RNA is a component of a major stress-responsive ribonucleoprotein particle in extremophilic bacteria. Mol. Microbiol. 120, 324–340 (2023). [DOI] [PubMed] [Google Scholar]
- 18.Agafonov D. E., Kolb V. A., Spirin A. S., Ribosome-associated protein that inhibits translation at the aminoacyl-tRNA binding stage. EMBO Rep. 2, 399–402 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lang M., et al. , Sleeping ribosomes: Bacterial signaling triggers RaiA mediated persistence to aminoglycosides. iScience 24, 103128 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Damke P. P., et al. , ComFC mediates transport and handling of single-stranded DNA during natural transformation. Nat. Commun. 13, 1961 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michel F., Westhof E., Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J. Mol. Biol. 216, 585–610 (1990). [DOI] [PubMed] [Google Scholar]
- 22.Pace N. R., et al. , “Probing RNA structure, function, and history by comparative analysis” in The RNA World (Cold Spring Harbor Laboratory Press, NY, ed. 2, 1999), pp. 113–141. [Google Scholar]
- 23.Eddy S. R., Durbin R., RNA sequence analysis using covariance models. Nucleic Acids Res. 22, 2079–2088 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parks D. H., et al. , A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 36, 996–1004 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Leontis N. B., Westhof E., A common motif organizes the structure of multi-helix loops in 16 S and 23 S ribosomal RNAs. J. Mol. Biol. 283, 571–583 (1998). [DOI] [PubMed] [Google Scholar]
- 26.Leontis N. B., Westhof E., The 5S rRNA loop E: Chemical probing and phylogenetic data versus crystal structure. RNA 4, 1134–1153 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paradis E., Schliep K., ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinf. 35, 526–528 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Rocha E. P. C., The organization of the bacterial genome. Annu. Rev. Genet. 42, 211–233 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Espejo R. T., Plaza N., Multiple ribosomal RNA operons in bacteria; their concerted evolution and potential concequences on the rate of evolution of their 16S rRNA. Front. Microbiol. 9, 1232 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sherlock M. E., Breaker R. R., Former orphan riboswitches reveal unexplored areas of bacterial metabolism, signaling, and gene control processes. RNA 26, 675–693 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mell J. C., Redfield R. J., Natural competence and the evolution of DNA uptake specificity. J. Bacteriol. 196, 1471–1483 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Connell L. M., et al. , Natural transformation in Gram-positive bacteria and its biotechnological relevance to lactic acid bacteria. Annu. Rev. Food Sci. Technol. 13, 409–431 (2022). [DOI] [PubMed] [Google Scholar]
- 33.Galperin M. Y., Nikolskaya A. N., Identification of sensory and signal-transducing domains in two-component signaling systems. Methods Enzymol. 422, 47–74 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davidson A. L., Dassa E., Orelle C., Chen J., Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol. Mol. Biol. Rev. 72, 317–364 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agafonov D. E., Kolb V. A., Nazimov I. V., Spirin A. S., A protein residing at the subunit interface of the bacterial ribosome. Proc. Natl. Acad. Sci. U.S.A. 96, 12345–12349 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petry S., et al. , Crystal structures of the ribosome in complex with release factors RF1 and RF2 bound to a cognate stop codon. Cell 123, 1255–1266 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Youngman E. M., McDonald M. E., Green R., Peptide release on the ribosome: Mechanism and implications for translational control. Annu. Rev. Microbiol. 62, 353–373 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Block K. F., Puerta-Fernandez E., Wallace J. G., Breaker R. R., Association of OLE RNA with bacterial membranes via an RNA-protein interaction. Mol. Microbiol. 79, 21–34 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones D. T., Woods D. R., Acetone-butanol fermentation revisited. Microbiol. Rev. 50, 484–524 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moon H. G., et al. , One hundred years of clostridial butanol fermentation. FEMS Microbiol. Lett. 363, fnw001 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Joseph R. C., Kim N. M., Sandoval N. R., Recent developments of the synthetic biology toolkit for Clostridium. Front. Microbiol. 9, 154 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soukup G. A., Breaker R. R., Relationship between internucleotide linkage geometry and the stability of RNA. RNA 5, 1308–1325 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Regulski E. E., Breaker R. R., “In-line probing analysis of riboswitches” in Post-Transcriptional Gene Regulation, Wilusz J., Eds. (Humana Press, Totowa, NJ, 2008), pp. 53–67. [DOI] [PubMed] [Google Scholar]
- 44.Wilding-Steele T., Ramette Q., Jacottin P., Soucaille P., Improved CRISPR/Cas9 tools for the rapid metabolic engineering of Clostridium acetobutylicum. Int. J. Mol. Sci. 22, 3704 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suvorova I. A., Korostelev Y. D., Gelfand M. S., GntR family of bacterial transcription factors and their DNA binding motifs: Structure, positioning and co-evolution. PLoS One 10, e0132618 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lamm-Schmidt V., et al. , Grad-seq identifies KhpB as a global RNA-binding protein in Clostridiodies difficile that regulates toxin production. Microlife 2, uqab004 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fuchs M., et al. , An RNA-centric global view of Clostridioides difficile reveals broad activity of Hfq in a clinically important gram-positive bacterium. Proc. Natl. Acad. Sci. USA 118, e2103579118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oultram J. D., et al. , Introduction of plasmids into whole cells of Clostridium acetobutylicum by electroporation. FEMS Microbiol. Lett. 56, 83–88 (1988). [Google Scholar]
- 49.Ralston M. T., Papoutsakis E. T., RNAseq-based transcriptome assembly of Clostridium acetobutylicum for functional genome annotation and discovery. AlChE J. 64, 4271–4280 (2018). [Google Scholar]
- 50.Zhang H. F., et al. , Clostridium acetobutylicum biofilm: Advances in understanding the basis. Front. Bioeng. Biotechnol. 9, 658568 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu D., et al. , Enhanced butanol production by modulation of electron flow in Clostridium acetobutylicum B3 immobilized by surface adsorption. Biores. Technol. 129, 321–328 (2013). [DOI] [PubMed] [Google Scholar]
- 52.Wilson K. S., von Hippel P. H., Transcription termination at intrinsic terminators: The role of the RNA hairpin. Proc. Natl. Acad. Sci. U.S.A. 92, 8793–8797 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yarnell W. S., Roberts J. W., Mechanism of intrinsic transcription termination and antitermination. Science 284, 611–615 (1999). [DOI] [PubMed] [Google Scholar]
- 54.Breaker R. R., Riboswitches and translation control. Cold Spring Harb. Perspect. Biol. 10, a032797 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.A. R. Ferré-D’Amaré, W. G. Scott, Small self-cleaving ribozymes. Cold Spring Harb. Perspect. Biol. 2, a003574 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kazantsev A. V., Pace N. R., Bacterial RNase P: A new view of an ancient enzyme. Nat. Rev. Microbiol. 4, 729–740 (2006). [DOI] [PubMed] [Google Scholar]
- 57.Johnston C., Martin B., Fichant G., Polard P., Claverys J. P., Bacterial transformation: Distribution, shared mechanisms and divergent control. Nat. Rev. Microbiol. 12, 181–196 (2014). [DOI] [PubMed] [Google Scholar]
- 58.Zarrella T. M., Yang J., Metzger D. W., Bai G., Bacterial second messenger cyclic di-AMP modulates the competence state in Streptococcus pneumoniae. J. Bacteriol. 202, e00691-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stülke J., Krüger L., Cyclic di-AMP signaling in bacteria. Annu. Rev. Microbiol. 74, 159–179 (2020). [DOI] [PubMed] [Google Scholar]
- 60.da Purificação A. D., de Azevedo N. M., de Araujo G. G., de Souza R. F., Guzzo C. R., The world of cyclic dinucleotides in bacterial behavior. Molecules 25, 2462 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Römling U., Cyclic di-GMP signaling—Where did you come from and where will you go? Mol. Microbiol. 120, 564–574 (2023). [DOI] [PubMed] [Google Scholar]
- 62.Nelson J. W., Breaker R. R., The lost language of the RNA World. Sci. Signal. 10, eaam8812 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hernández-Morales R., Becerra A., Lazcano A., Alarmones as vestiges of a bygone RNA World. J. Mol. Evol. 87, 37–51 (2019). [DOI] [PubMed] [Google Scholar]
- 64.Parks D. H., et al. , A complete domain-to-species taxonomy for Bacteria and Archaea. Nat. Biotechnol. 38, 1079–1086 (2020). [DOI] [PubMed] [Google Scholar]
- 65.Kitts P. A., et al. , Assembly: A resource for assembled genomes at NCBI. Nucleic Acids Res. 44, D73–D80 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nawrocki E. P., Eddy S. R., Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29, 2933–2935 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Griffiths-Jones S., RALEE—RNA ALignment Editor in Emacs. Bioinformatics 21, 257–259 (2005). [DOI] [PubMed] [Google Scholar]
- 68.Weinberg Z., Breaker R. R., R2R—Software to speed the depiction of aesthetic consensus RNA secondary structures. BMC Bioinf. 12, 3 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cock P. J. A., et al. , Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 25, 1422–1423 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.O’Leary N. A., et al. , Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Smith M. R., TreeTools: Create, Modify and Analyse Phylogenetic Trees (Comprehensive R Archive Network, 2019). GitHub. https://ms609.github.io/TreeTools/. Accessed 19 August 2019. [Google Scholar]
- 72.Letunic I., Bork P., Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dusseaux S., Croux C., Soucaille P., Meynial-Salles I., Metabolic engineering of Clostridium acetobutylicum ATCC 824 for the high-yield production of a biofuel composed of an isopropanol/butanol/ethanol mixture. Metab. Eng. 18, 1–8 (2013). [DOI] [PubMed] [Google Scholar]
- 74.Baer S. H., Blaschek H. P., Smith T. L., Effect of butanol challenge and temperature on lipid composition and membrane fluidity of butanol-tolerant Clostridium acetobutylicum. Appl. Environ. Microbiol. 53, 2854–2861 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Roos J. W., McLaughlin J. K., Papoutsakis E. T., The effect of pH on nitrogen supply, cell lysis, and solvent production in fermentations of Clostridium acetobutylicum. Biotechnol. Bioeng. 27, 681–694 (1985). [DOI] [PubMed] [Google Scholar]
- 76.Sherlock M. E., Sadeeshkumar H., Breaker R. R., Variant bacterial riboswitches associated with nucleotide hydrolase genes sense nucleoside diphosphates. Biochemistry 58, 401–410 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brown T., Mackey K., Du T., Analysis of RNA by northern and slot blot hybridization. Curr. Protoc. Mol. Biol. Ch. 4, Unit 4.9 (2004). [DOI] [PubMed] [Google Scholar]
- 78.McClure R., et al. , Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res. 41, e140 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mermelstein L. D., Papoutsakis E. T., In vivo methylation in Escherichia coli by the Bacillus subtilis phage phi 3T I methyltransferase to protect plasmids from restriction upon transformation of Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 59, 1077–1081 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ehsaan M., et al. , Mutant generation by allelic exchange and genome resequencing of the biobutanol organism Clostridium acetobutylicum ATCC 824. Biotechnol. Biofuels 9, 4 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (TXT)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Data Availability Statement
All experimental data are included in the main text and supporting information.