Significance
In this study, we have identified a functional role of the m6A demethylase, ALKBH5, in regulating trigeminal neuropathic pain. Our findings demonstrate that nerve injury–induced downregulation of HDAC11 facilitates the binding of the FOXD3 transcription factor to the Alkbh5 gene promoter, leading to the increased protein expression of ALKBH5. This upregulation of ALKBH5 promotes neuropathic pain through erasing m6A in Htr3a mRNA and stabilizing the increased Htr3a mRNA. This mechanistic understanding of nociceptive sensory neurons may offer insights into potential targets for the treatment of neuropathic pain.
Keywords: neuropathic pain, ALKBH5, histone acetylation, N6-methyladenosine, trigeminal ganglion neurons
Abstract
The N6-methyladenosine (m6A) modification of RNA is an emerging epigenetic regulatory mechanism that has been shown to participate in various pathophysiological processes. However, its involvement in modulating neuropathic pain is still poorly understood. In this study, we elucidate a functional role of the m6A demethylase alkylation repair homolog 5 (ALKBH5) in modulating trigeminal-mediated neuropathic pain. Peripheral nerve injury selectively upregulated the expression level of ALKBH5 in the injured trigeminal ganglion (TG) of rats. Blocking this upregulation in injured TGs alleviated trigeminal neuropathic pain, while mimicking the upregulation of ALKBH5 in intact TG neurons sufficiently induced pain-related behaviors. Mechanistically, histone deacetylase 11 downregulation induced by nerve injury increases histone H3 lysine 27 acetylation (H3K27ac), facilitating the binding of the transcription factor forkhead box protein D3 (FOXD3) to the Alkbh5 promoter and promoting Alkbh5 transcription. The increased ALKBH5 erases m6A sites in Htr3a messenger RNA (mRNA), resulting in an inability of YT521-B homology domain 2 (YTHDF2) to bind to Htr3a mRNA, thus causing an increase in 5-HT3A protein expression and 5-HT3 channel currents. Conversely, blocking the increased expression of ALKBH5 in the injured TG destabilizes nerve injury–induced 5-HT3A upregulation and reverses mechanical allodynia, and the effect can be blocked by 5-HT3A knockdown. Together, FOXD3-mediated transactivation of ALKBH5 promotes neuropathic pain through m6A-dependent stabilization of Htr3a mRNA in TG neurons. This mechanistic understanding may advance the discovery of new therapeutic targets for neuropathic pain management.
Neuropathic pain caused by somatosensory nerve damage or disease is a common chronic pain that has a major impact on quality of life (1). Current analgesic medications, including opioids and nonsteroidal anti-inflammatory drugs (NSAIDs), are ineffective and/or have severe side effects in most patients with neuropathic pain, making therapeutic management for this disorder difficult (2). Therefore, understanding the mechanisms of chronic neuropathic pain at both the molecular and cellular levels is crucial for the creation of cutting-edge therapies for this condition. Accumulating studies in first-order sensory neurons such as trigeminal ganglion (TG) neurons have shown that peripheral nerve injury can cause alterations in the expression of pain-associated genes at both the transcriptional and translational levels (3, 4). These changes may possess some pivotal genomic signatures/regulation and result in the initiation and maintenance of neuropathic pain (3–5). Understanding the molecular mechanisms of how these changes occur in the TGs following peripheral nerve injury may enable us to develop an avenue for trigeminal-mediated neuropathic pain management.
Emerging as the most prevalent type of mRNA methylation in eukaryotes, N6-methyladenosine (m6A) modification has attracted increasing attention (6). m6A, enriched particularly in the 3′-untranslated region (3′-UTR) and near stop codons, is installed reversibly by the multisubunit methyltransferase complex (writers), removed by demethylases (erasers), and recognized by m6A-binding proteins (readers) (7). Among the demethylases, ALKBH5 is identified as a key catalytic component of the m6A “eraser” complex, and evidence has identified ALKBH5-induced regulation of m6A RNA modification as resulting in the control of gene expression (8) and modulation of several physiological processes, such as γδ T cell development (9), and pathological processes, such as cardiac differentiation and cancer (10, 11). Limited evidence has also demonstrated a potential regulatory function of ALKBH5 in nervous system disorders (12). For instance, ALKBH5 plays a crucial role in cerebral ischemia–reperfusion injury by demethylating Bcl2 transcripts (13). Moreover, ALKBH5 was shown to be involved in neural tube development and regulation by decreasing the protein expression of cyclin D1 (14). Nonetheless, to date, the role of ALKBH5 in pain regulation remains unknown.
In this study, we elucidated a critical role of ALKBH5 in contributing to the maintenance and development of trigeminal-mediated neuropathic pain by targeting 5-HT3A. Downregulation of histone deacetylase 11 (HDAC11) induced by nerve injury increased histone H3 lysine 27 acetylation (H3K27ac), facilitating the binding of the transcription factor FOXD3 to the promoter of the Alkbh5 gene and increasing the protein expression of ALKBH5 in injured TGs. The increased ALKBH5 contributed to the onset and maintenance of nerve injury–induced neuropathic pain by erasing m6A in Htr3a mRNA and stabilizing the increased Htr3a mRNA. This mechanistic understanding may offer important insights into the discovery of therapeutic targets for neuropathic pain management.
Results
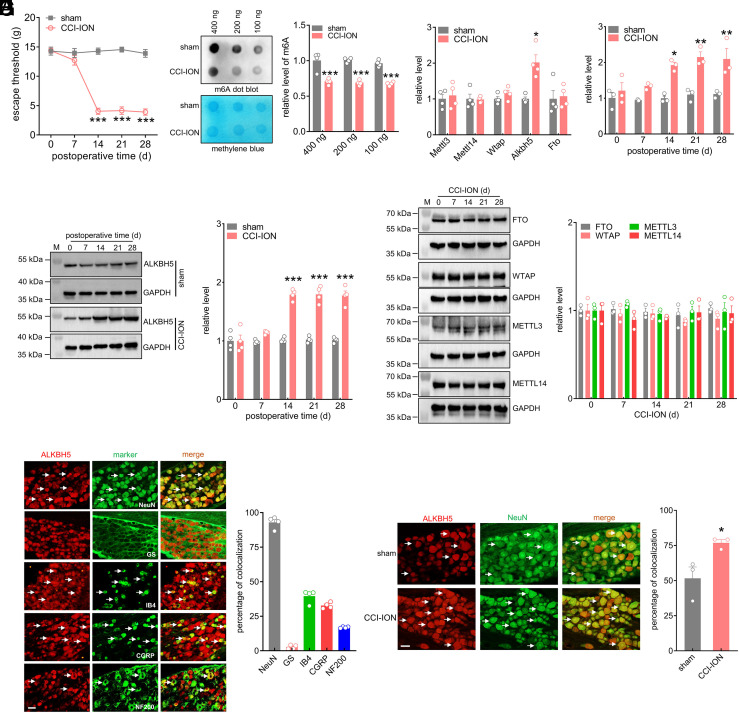
ALKBH5 Is Upregulated in Ipsilateral TG Neurons after Peripheral Nerve Injury.
An experimental model of trigeminal neuropathic pain was established using unilateral chronic constriction injury to the infraorbital nerve (CCI-ION). Rats exhibited a dramatic reduction in escape threshold to mechanical stimuli on Day 14 following CCI-ION compared with that following sham-operated groups, and this reduction was maintained on Day 28 (Fig. 1A). Dot blot assays suggested that the global level of m6A was markedly decreased in the ipsilateral TG post-CCI-ION (Fig. 1B and SI Appendix, Fig. S1). To identify the key molecules that are critical for the changes in m6A levels, we examined the expression of methyltransferases and demethylases in the injured TG after CCI-ION operation. Unilateral CCI-ION increased the level of Alkbh5 mRNA in the ipsilateral TG, whereas Fto, Wtap, Mettl3, and Mettl14 remained unchanged (Fig. 1C). We subsequently investigated the time course of changes in Alkbh5 expression in the ipsilateral TG. The Alkbh5 mRNA level in CCI-ION rats significantly increased on Day 14 and was maintained on Day 28, while there were no significant differences in the levels of Alkbh5 mRNA in rats following sham surgery (Fig. 1D). Consistently, ALKBH5 protein expression was markedly increased in CCI-ION-operated rats in a time-dependent manner (Fig. 1E and SI Appendix, Fig. S2), while the protein abundance of FTO, WTAP, METTL3, and METTL14 did not show significant changes over the testing period (Fig. 1F and SI Appendix, Fig. S3). As expected, following sham surgery, basal ALKBH5 expression in the ipsilateral TG remained unchanged (Fig. 1E and SI Appendix, Fig. S2). Additionally, the protein expression of ALKBH5 did not display any change in the contralateral TGs following CCI-ION operation (SI Appendix, Fig. S4). Next, we investigated the cellular localization of ALKBH5. Double-staining assays revealed that ALKBH5 primarily coexisted with the neuronal marker NeuN, while exhibiting rarely colocalization with the satellite glial cell-specific marker glutamine synthetase (GS), indicating that ALKBH5 was predominantly expressed in TG neurons (Fig. 1G). Statistical analysis revealed that ~93.3% of the ALKBH5+ TG neurons were labeled by NeuN, and ~2.7% were labeled by GS (Fig. 1G). Further determination of the expression pattern of ALKBH5 in peripheral nociceptors of intact rats demonstrated that ALKBH5-positive TG neurons strongly colocalized with calcitonin gene-related peptide (CGRP) and isolectin B4 (IB4) but showed comparatively lower coexistence with neurofilament 200 (NF200) (Fig. 1G). Approximately 32.7% of the ALKBH5-positive neurons were labeled by CGRP, ~39.5% by IB4, and 16.9% by NF200 (Fig. 1G). Moreover, consistent with the qPCR results, the number of ALKBH5-positive neurons in the ipsilateral TG on Day 14 after CCI-ION increased notably compared with that in the corresponding sham group (increased by 49.2% ± 3.8%; Fig. 1H).
Fig. 1.
ALKBH5 is upregulated in the ipsilateral TG following nerve injury. (A) Escape threshold in rats following CCI-ION operation or sham surgery. ***P < 0.001 vs. sham (two-way ANOVA). n = 7 rats/group. (B) Representative dot blots and summary data of m6A levels in the ipsilateral TG at 14 d following CCI-ION operation or sham surgery. Representative blots from four independent experiments are shown. ***P < 0.001 vs. sham (unpaired t test). n = 8 rats/ group. (C) qPCR analysis of Alkbh5, Fto, Mettl3, Mettl14 and Wtap. *P < 0.05 vs. sham (unpaired t test). n = 8 rats/group. (D) Time course of changes in Alkbh5 mRNA expression. *P < 0.05 and **P < 0.01 vs. the corresponding Day 0 (two-way ANOVA). n = 6 rats/time point/group. (E) Immunoblot analysis of ALKBH protein expression. Representative blots from four independent experiments are shown. ***P < 0.001 vs. the corresponding Day 0 (two-way ANOVA). n = 8 rats/time point/group. (F) Protein abundance of ALKBH5, FTO, METTL3, METTL14, and WTAP in the ipsilateral TG after CCI-ION. Representative blots from three independent experiments are shown. n = 6 rats/time point/group. (G) Co-staining of ALKBH5 (red) with NeuN, GS, IB4, CGRP, or NF200 (green) in the intact TG. The bar graph shows the percentage of double-stained TG neurons among all ALKBH5+-labeled neurons. Data from four independent experiments. (Scale bar, 50 μm.) White arrows indicate colocalization. (H) Number of neurons labeled with ALKBH5 (red) and NeuN (green). *P < 0.05 vs. sham (unpaired t test). Arrows show colocalization. Data from three independent experiments. (Scale bar, 50 μm.)
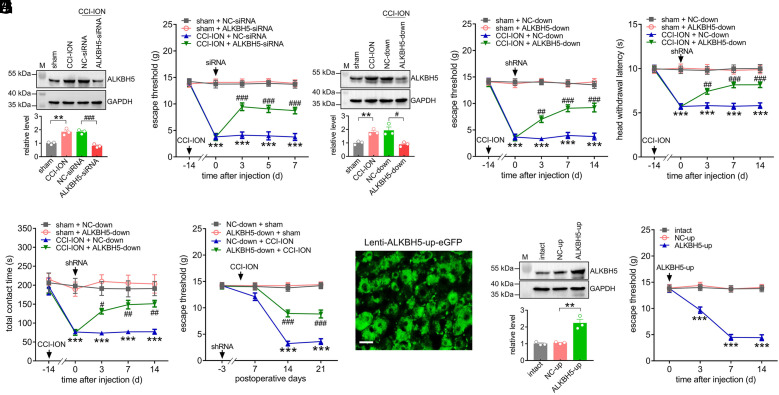
ALKBH5 Regulates Trigeminal Neuropathic Pain Behaviors.
To examine whether ALKBH5 contributes to chronic neuropathic pain behaviors, we unilaterally intra-TG injected chemically modified ALKBH5-siRNA (or the corresponding negative control NC-siRNA) 14 d after CCI-ION. Our results revealed that administration of ALKBH5-siRNA, but not NC-siRNA, dramatically suppressed the increased protein abundance of ALKBH5 in the injured TG (Fig. 2A and SI Appendix, Fig. S5) and markedly alleviated CCI-ION-induced mechanical allodynia (Fig. 2B). Subsequently, we employed lentivirus-mediated short hairpin RNAs (shRNAs) to investigate the effect of ALKBH5 knockdown (ALKBH5-down) on neuropathic pain behaviors. Injection of ALKBH5-down at 14 d post-CCI-ION suppressed the protein abundance of ALKBH5 in the injured TG (Fig. 2C and SI Appendix, Fig. S6) and significantly attenuated mechanical allodynia (Fig. 2D) and heat hyperalgesia (Fig. 2E) from Day 3 through Day 14 following ALKBH5-down administration. As a complimentary verification of our hypothesis, further orofacial operant behavioral assessment revealed that administration of ALKBH5-down reversed the CCI-ION-induced reduction in the total contact time, while the NC-down-treatment elicited no such effects (Fig. 2F). Furthermore, following the injection of ALKBH5-down prior to CCI-ION operation, we assessed the participation of ALKBH5 in the development of neuropathic pain. In comparison with the NC-down-treated groups, CCI-ION rats pretreated with ALKBH5-down exhibited a significant reduction in mechanical allodynia on Days 14 to 21 postsurgery (Fig. 2G). Next, we investigated whether mimicking the increased ALKBH5 following nerve injury in intact TGs alters nociceptive thresholds. We induced ALKBH5 expression specifically in TG neurons utilizing the eGFP-labeled lentiviral vector lenti-hSyn-ALKBH5-up (ALKBH5-up) for neuron-specific gene delivery. From Day 3 after intra-TG injection, eGFP expression in intact TG neurons was observed following direct injection of ALKBH5-up (Fig. 2H) and was maintained for up to 21 d (SI Appendix, Fig. S7). Similarly, the protein expression of ALKBH5 was dramatically upregulated on Day 3 postinjection for ALKBH5-up (Fig. 2I and SI Appendix, Fig. S8). In intact rats, unilateral intra-TG administration of ALKBH5-up markedly induced mechanical hypersensitivity on Day 3, which lasted for at least 14 d after injection, while the NC-up-treatment elicited no such effects (Fig. 2J). Together, these behavioral results demonstrated that ALKBH5 played pivotal roles in the pathogenesis of trigeminal-mediated neuropathic pain.
Fig. 2.
ALKBH5 regulates nociceptive behaviors. (A) Intra-TG administration of ALKBH5-siRNA reduced the nerve injury–induced increase in the protein abundance of ALKBH5. **P < 0.01 vs. sham, ###P < 0.001 vs. CCI-ION + NC-siRNA (one-way ANOVA). Representative blots from three independent experiments are shown. n = 6 rats/group. (B) Injection of ALKBH5-siRNA attenuated nerve injury–induced mechanical allodynia. ***P < 0.001 vs. sham + NC-siRNA, ###P < 0.001 vs. CCI-ION + NC-siRNA (two-way ANOVA), n = 7 rats/group. (C) Injection of lentiviral-mediated shRNA against ALKBH5 (ALKBH5-down) reduced the CCI-ION-induced increase in the protein abundance of ALKBH5. **P < 0.01 vs. sham, #P < 0.05 vs. CCI-ION + NC-shRNA (one-way ANOVA). Representative blots from three independent experiments are shown. n = 6 rats/group. (D and E) Intra-TG administration of ALKBH5-down alleviated CCI-ION-induced mechanical allodynia (D) and heat hyperalgesia (E). ***P < 0.001 vs. sham + NC-down, ##P < 0.01 and ###P < 0.001 vs. CCI-ION + NC-down (two-way ANOVA), n = 7 rats/group. (F) Injection of ALKBH5-down reversed the CCI-ION-induced decrease in total contact time assessed by the orofacial operant test. ***P < 0.001 vs. sham + NC-down, #P < 0.05 and ##P < 0.01 vs. CCI-ION + NC-down (two-way ANOVA), n = 8 rats/group. (G) Preliminary administration of ALKBH5-down reversed the nerve injury–induced reduction in the escape threshold. ***P < 0.001 vs. NC-down + sham, ###P < 0.001 vs. NC-down + CCI-ION (two-way ANOVA), n = 7 to 8 rats/group. (H) Representative images of GFP-expressing TG neurons on Day 3 after intra-TG administration of lenti-hSyn-ALKBH5-up (ALKBH5-up). (Scale bar, 25 μm.) (I) Protein abundance of ALKBH5 in the TGs of the ALKBH5-up-treated groups. **P < 0.01 vs. NC-up (one-way ANOVA). Representative blots from three independent experiments are shown. n = 6 rats/group. (J) Injection of ALKBH5-up in intact rats resulted in mechanical hypersensitivity. ***P < 0.001 vs. NC-up (two-way ANOVA). n = 8 rats/group.
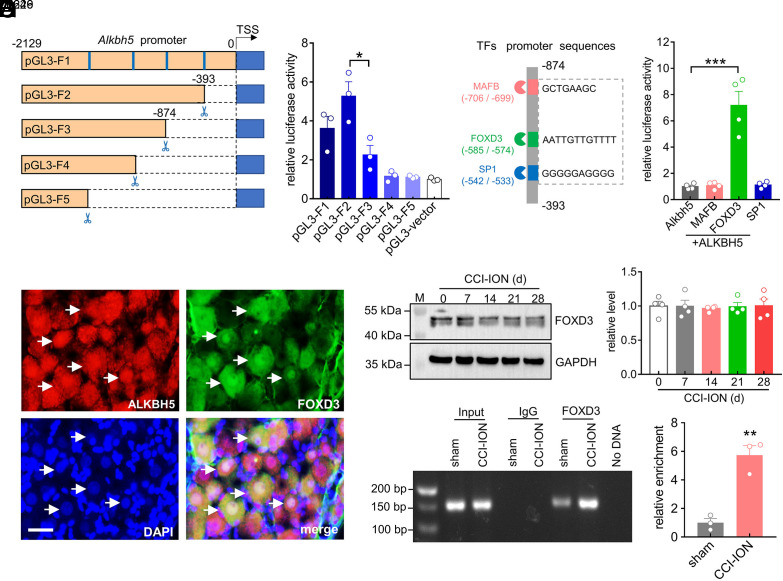
FOXD3 Promotes TG Alkbh5 Gene Transcriptional Activity after CCI-ION.
Gene transcription is controlled by sequence-specific transcription factors that bind to promoters (15). To elucidate the transcription factors that potentially regulate ALKBH5 expression, we carried out deletion analysis of the approximately 2,000 bp (−2,129 bp to 0 bp) 5′ upstream region of the Alkbh5 transcription start site. We constructed a series of pGL3 luciferase reporter plasmids that incorporate distinct fragments from the promoter region of Alkbh5 (pGL3-F1 to pGL3-F5) (Fig. 3A). Dual-luciferase reporter assays revealed that only the luciferase activity of the pGL3-F3 plasmid (−2,129 bp to −874 bp) was significantly decreased when compared with that of pGL3-F2 (Fig. 3B), which suggests that the promoter region from −874 bp to −393 bp (ΔF) contains cis-regulatory elements (CREs) that are critical for Alkbh5 gene expression. Thereafter, the JASPAR database (https://jaspar.genereg.net) was utilized to identify transcription factor binding sites within the ΔF region of the Alkbh5 gene promoter. A total of three transcription factors were identified: MAF bZIP transcription factor B (MAFB), Forkhead Box D3 (FOXD3) and specificity protein 1 (SP1) (Fig. 3C). To determine the interaction between the transcription factors and the ΔF region of the Alkbh5 promoter, a luciferase reporter plasmid carrying the ΔF region was generated and transfected alone or with MAFB, FOXD3, or SP1 plasmids (Fig. 3D). Among them, only cotransfection of the ΔF region with FOXD3, but not with MAFB or SP1, dramatically enhanced the luciferase activity (Fig. 3D). Thus, we further examined whether the FOXD3 levels were correlated with increased ALKBH5 expression. Immunostaining analysis revealed that FOXD3 was strongly coexpressed with ALKBH5 in TG neurons of intact rats (Fig. 3E). Surprisingly, FOXD3 expression was not markedly affected in the injured TG from Day 0 to Day 28 after nerve injury (Fig. 3F and SI Appendix, Fig. S9). Similar results were also observed in the sham groups (SI Appendix, Fig. S10). Given the above, we hypothesized that the nerve injury–driven changes in ALKBH5 might result from the change of FOXD3 binding to the promoter region of the Alkbh5 gene. Indeed, Chromatin Immunoprecipitation (ChIP) analysis revealed a significantly higher enrichment of the Alkbh5 promoter from anti-FOXD3 complexes at 14 d following CCI-ION compared to sham operation (Fig. 3G and SI Appendix, Fig. S11).
Fig. 3.
FOXD3 promotes TG Alkbh5 gene transcriptional activity after CCI-ION. (A) Schematic representation of pGL3 luciferase reporter constructs (pGL3-F1 to pGL3-F5). (B) Dual luciferase assays showing the transcriptional activity indicated in panel (A). Data are expressed as the mean ± SEM of three independent experiments. *P < 0.05 vs. pGL3-F2 (unpaired t test). (C) Schematic diagram showing the firefly luciferase reporter constructs containing MAFB-, FOXD3-, and SP1-binding sites in the ΔF region (−874 bp to −393 bp) of the Alkbh5 gene promoter. (D) Luciferase activities of ΔF of the Alkbh5 gene promoter with/without overexpression of MAFB, FOXD3, or SP1. Data are expressed as the mean ± SEM of four independent experiments. ***P < 0.001 vs. Alkbh5 (one-way ANOVA). (E) Colocalization of ALKBH5 (red) with FOXD3 (green) and DAPI (blue) in intact TGs. Arrows show colocalization. (Scale bar, 25 μm.) (F) Time course of FOXD3 protein expression in the ipsilateral TG following CCI-ION. The blots shown are representative of four independent experiments. n = 8 rats/group. (G) ChIP–qPCR analysis demonstrating the FOXD3 binding to the Alkbh5 promoter. Data were normalized to input and are expressed as the mean ± SEM of three independent experiments. **P < 0.01 vs. sham (unpaired t test). n = 9 rats/group.
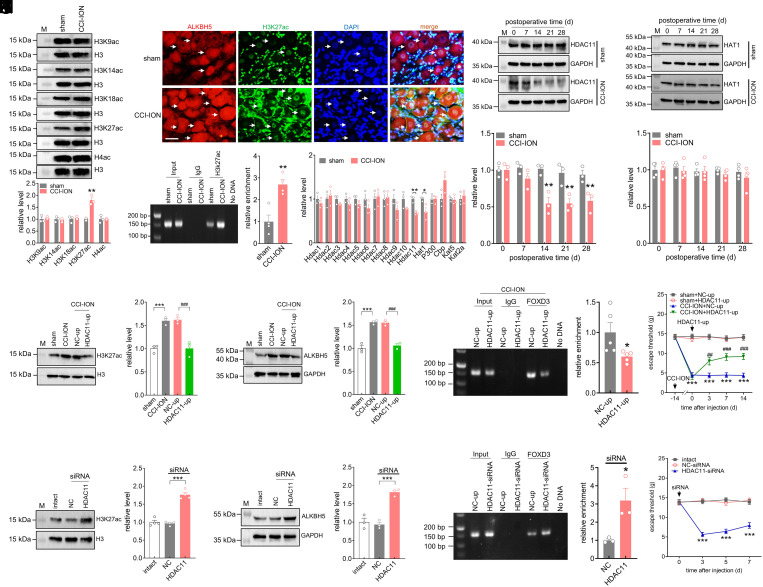
HDAC11 Downregulation Enhances the Binding of FOXD3 to ALKBH5.
Histone acetylation is an important epigenetic mechanism that controls gene expression (16). Notably, acetylation of histone H3 lysine, including H3K27ac, H3K18ac, H3K14ac, and H3K9ac, as well as acetylation of histone H4 (H4ac), are correlated with transcriptional activation (17). Therefore, we detected the protein abundance of these acetylated histone isoforms in TG tissues. Compared with sham surgery, only the H3K27ac protein levels were significantly upregulated 14 d after CCI-ION operation, while other acetylated histones H3 and H4 exhibited no significant changes (Fig. 4A and SI Appendix, Fig. S12). Subsequently, immunostaining analysis revealed that H3K27ac was expressed exclusively in the nucleus of TG cells and colocalized with ALKBH5 in TG neurons of either CCI-ION or sham surgery (Fig. 4B). In particular, most TG neurons with high expression levels of H3K27ac in CCI-ION rats also showed strong ALKBH5 staining (Fig. 4B). Furthermore, ChIP–PCR analysis revealed the amplification of an Alkbh5 promoter fragment from anti-H3K27ac complexes (Fig. 4C and SI Appendix, Fig. S13). Compared with sham surgery, CCI-ION markedly enhanced the H3K27ac binding to the Alkbh5 gene promoter (~2.7-fold) 14 d following CCI-ION (Fig. 4C). Moreover, compared with the CCI-ION group 7 d after surgery, the binding activity of H3K27ac to the Alkbh5 gene promoter was significantly increased in the ipsilateral TG 14 d following CCI-ION (SI Appendix, Fig. S14). In contrast, the enrichment of H3K27ac occupancy in the Alkbh5 gene promoter remained unchanged in injured TGs 7 d post-CCI-ION compared with the corresponding sham-operated groups (SI Appendix, Fig. S15). Histone acetylases, including CBP, EP300, HAT1, KAT2A, and KAT5, are known to acetylate H3K27, while histone deacetylases, including HDAC1 to HDAC11, are known to deacetylate H3K27 (18). qPCR analysis indicated that both Hat1 and Hdac11 mRNA expression were significantly decreased at 14 d post-CCI-ION operation (Fig. 4D), while other deacetylases or acetylases did not exhibit any significant changes (Fig. 4D). Notably, at the protein level, only the abundance of HDAC11 was consistently downregulated in a time-dependent manner following CCI-ION (Fig. 4E and SI Appendix, Fig. S16), while the abundance of the HAT1 protein did not exhibit any significant change (Fig. 4F and SI Appendix, Fig. S17). The protein abundance of both HDAC11 (Fig. 4E and SI Appendix, Fig. S16) and HAT1 (Fig. 4F and SI Appendix, Fig. S17) remained unchanged in sham surgery over the testing period. These results suggested that the increased H3K27ac may be attributed to the downregulation of HDAC11 in the injured TG. Moreover, immunostaining of TG sections revealed that HDAC11 colocalized with ALKBH5 in both CCI-ION and sham-operated rats (SI Appendix, Fig. S18). In both the CCI-ION and sham-operated groups, HDAC11 was primarily found in the cell nuclei (labeled by DAPI), and the majority of the neurons with high levels of ALKBH5 displayed a relatively low level of HDAC11 in the injured TG (SI Appendix, Fig. S18). We further examined whether HDAC11 functionally participated in CCI-ION-mediated mechanical allodynia. Intra-TG injection of lenti-hSyn-HDAC11-up (HDAC11-up) prevented the increased protein abundance of either H3K27ac (Fig. 4G and SI Appendix, Fig. S19) or ALKBH5 (Fig. 4H and SI Appendix, Fig. S20) induced by CCI-ION. Intra-TG administration of HDAC11-up in the injured TG significantly reduced the binding of FOXD3 to the Alkbh5 promoter (Fig. 4I and SI Appendix, Fig. S21) and markedly alleviated mechanical allodynia induced by CCI-ION (Fig. 4J). Furthermore, we explored whether mimicking the nerve injury–induced decrease in HDAC11 in intact TGs would alter nociceptive behaviors. Administration of HDAC11-siRNA in intact rats significantly increased the protein abundance of both H3K27ac (Fig. 4K and SI Appendix, Fig. S22) and ALKBH5 (Fig. 4L and SI Appendix, Fig. S23) and enhanced the binding of FOXD3 to the Alkbh5 promoter (Fig. 4M and SI Appendix, Fig. S24). Moreover, intact rats receiving unilateral intra-TG injections of HDAC11-siRNA developed marked mechanical hypersensitivity from day 3 to day 7 following injection (Fig. 4N).
Fig. 4.
HDAC11 downregulation enhances the binding of FOXD3 to ALKBH5. (A) Immunoblot analysis of H4ac, H3K27ac, H3K18ac, H3K14ac, and H3K9ac protein expression in the ipsilateral TG 14 d following CCI-ION operation or sham surgery. Representative blots from three independent experiments are shown. Histone modifications were normalized to total histone H3 protein expression (H3), and then compared to the corresponding sham groups. **P < 0.01 vs. sham (unpaired t test). n = 6 rats/group. (B) Colocalization of ALKBH5 (red) with H3K27ac (green) and DAPI (blue). (Scale bar, 25 μm.) Arrows indicate colocalization. (C) ChIP–qPCR analysis showing that the binding activity of H3K27ac to the Alkbh5 promoter was increased in the ipsilateral TG 14 d following CCI-ION. Data were normalized to input and are expressed as the mean ± SEM of four independent experiments. **P < 0.01 vs. sham (unpaired t test). n = 12 rats/group. (D) qPCR analysis of Hdac1 to Hdac11, Hat1, P300, Cbp, Kat5 and Kat2a. n = 3 rats/group. *P < 0.05 and **P < 0.01 vs. sham (unpaired t test). (E and F) Protein abundance of HDAC11 (E, n = 6 rats/time point/group) or HAT1 (F, n = 8 rats/time point/group). Representative blots from three independent experiments are shown. **P < 0.01 vs. the corresponding Day 0 (two-way ANOVA). (G and H) Intra-TG injection of lenti-hSyn-HDAC11-up (HDAC11-up) attenuated the increased protein abundance of H3K27ac (G) or ALKBH5 (H) in CCI-ION rats. Representative blots from three independent experiments are shown. ***P < 0.001 vs. sham, ###P < 0.001 vs. NC-up + CCI-ION (one-way ANOVA). n = 6 rats/group. (I) ChIP–PCR analysis showing that administration of HDAC11-up suppressed the binding of FOXD3 to the Alkbh5 promoter in the ipsilateral TG 14 d following CCI-ION. Data were normalized to input and are expressed as the mean ± SEM of five independent experiments. *P < 0.05 vs. NC-up (unpaired t test). n = 15 rats/group. (J) Intra-TG administration of HDAC11-up attenuated mechanical allodynia in CCI-ION-operated groups. ***P < 0.001 vs. sham + NC-up, ###P < 0.001 vs. CCI-ION + NC-up (two-way ANOVA). n = 7 rats/group. (K–M) Administration of HDAC11-siRNA increased the protein expression of H3K27ac (K, n = 8 rats/group) and ALKBH5 (L, n = 6 rats/group) and enhanced the FOXD3 binding to the Alkbh5 promoter (M, n = 9 rats/group) in intact TGs. Representative blots from three or four independent experiments are shown. *P < 0.05 and ***P < 0.001 vs. NC-siRNA (one-way ANOVA). (N) Unilateral intra-TG administration of HDAC11-siRNA increased mechanical sensitivity in intact rats. ***P < 0.001 vs. NC-siRNA (two-way ANOVA). n = 8 rats/group.
ALKBH5-Mediated m6A Modification Enhances the Stability of Htr3a mRNA.
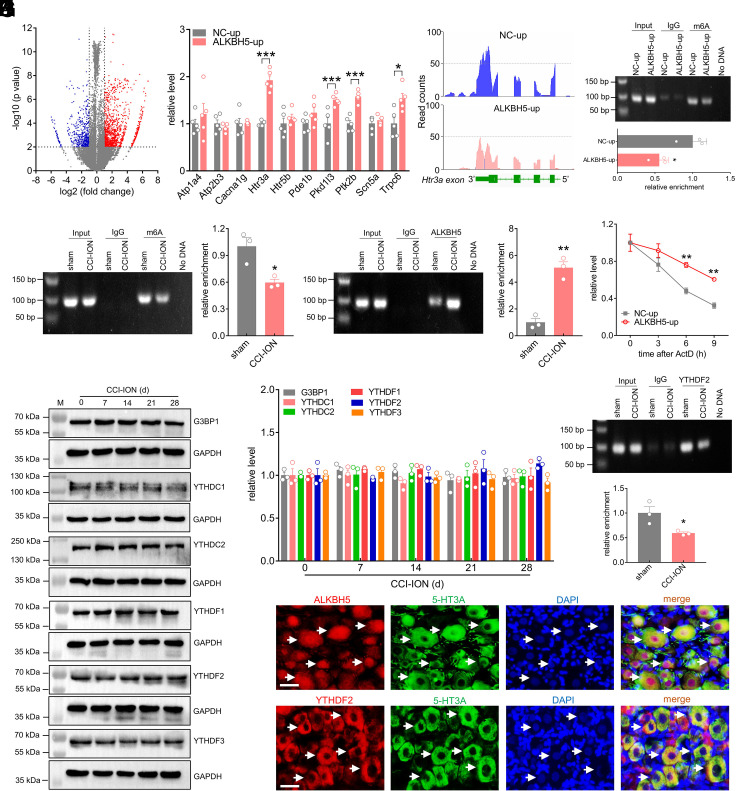
We next explored the underlying molecular mechanisms of ALKBH5 in regulating neuropathic pain. Given that ALKBH5 is a key demethylase that removes m6A on RNAs, we performed m6A methylated RNA immunoprecipitation sequencing (MeRIP-seq) to profile the genome-wide m6A methylation distribution in NC-up- and ALKBH5-up-treated TG cells. In line with previous analyses (19), the consensus motif RRACH (R = A or G; H = A, U or C) was enriched in the rat TGs, and m6A peaks were particularly abundant in the vicinity of 3′-UTRs near stop codons (SI Appendix, Fig. S25). A comparison of the abundance of m6A peaks between the NC-up-treated and ALKBH5-up-treated groups revealed a total of 1,345 m6A-up peaks and 546 m6A-down peaks (Fig. 5A). Gene Ontology (GO) analysis indicated that the transcripts with decreased m6A sites were enriched in ion transporter activity and the cation channel complex (SI Appendix, Fig. S25), and KEGG analysis indicated that they were highly correlated with calcium signaling (SI Appendix, Fig. S25), which plays pivotal roles in pain regulation (20). Intersecting the above GO and KEGG analyses resulted in 10 candidates (Fig. 5B). Among the 10 transcripts, four transcripts—Htr3a, Pkd1l3, Ptk2b, and Trpc6—showed considerably higher expression levels in the ALKBH5-up-treated TGs, with Htr3a being the most robustly upregulated (~191.6%, Fig. 5B). Indeed, as shown in the MeRIP-seq analysis, Htr3a mRNA exhibited a larger loss of m6A sites in the 3′-UTR from ALKBH5-up-treated TGs compared with the NC-up group (Fig. 5C). These results were further verified by the MeRIP-qPCR assay, which showed that administration of ALKBH5-up markedly reduced the m6A level of Htr3a mRNA in intact TGs (Fig. 5D and SI Appendix, Fig. S26). In addition, the immunoprecipitation analysis of TGs in sham surgery revealed m6A enrichment in the Htr3a mRNA fragment (Fig. 5E and SI Appendix, Fig. S27), and the immunoprecipitative activity was markedly decreased in the injured TG 14 d after CCI-ION, demonstrating a loss of m6A sites induced by nerve injury (Fig. 5E). Furthermore, we investigated whether this loss was due to the activity of ALKBH5. Our RNA Immunoprecipitation (RIP) analysis revealed that a fragment of Htr3a mRNA could be amplified with an antibody specific for ALKBH5, and this binding activity was significantly elevated 14 d following CCI-ION (Fig. 5F and SI Appendix, Fig. S28). As overexpression of ALKBH5 might increase the Htr3a mRNA level, we investigated whether m6A modification affects the stability of Htr3a mRNA. Indeed, in the presence of actinomycin D, an inhibitor of transcription, ALKBH5 overexpression induced a lengthened mRNA half-life of Htr3a mRNA, indicating that m6A modification mediated ALKBH5-enhanced Htr3a mRNA stability (Fig. 5G). As “readers” were crucially responsible for the direct effects on m6A-modified transcripts, we further determined potential effectors participating in the process illustrated above. Studies have demonstrated that the identified m6A readers, including the YI521-B homology domain family (YTH) proteins YTHDF1/2/3 and YTHDC1/2 and G3BP1, participate in promoting the instability of their targets (21). Therefore, we examined whether nerve injury would affect the expression of these readers in the injured TG. Surprisingly, the protein abundance of all of these readers showed no discernible change from Day 7 to Day 28 following CCI-ION (Fig. 5 H and I and SI Appendix, Fig. S29). Interestingly, further detection of the binding of Htr3a mRNA to the m6A reader proteins by the RIP assay showed that a fragment of Htr3a mRNA could be amplified with an antibody specific for YTHDF2 (Fig. 5J and SI Appendix, Fig. S30), and 14 d after CCI-ION, this binding was significantly decreased (Fig. 5J). In contrast, other readers, including G3BP1, YTHDC1, YTHDC2, YTHDF1, and YTHDF3, to Htr3a mRNA did not exhibit any binding of Htr3a mRNA in the ipsilateral TG at 14 d following CCI-ION or sham surgery (SI Appendix, Fig. S31). Collectively, ALKBH5-mediated m6A modification repressed Htr3a expression through YTHDF2-dependent mRNA degradation. As complimentary support for this hypothesis, double immunofluorescence analysis of TG sections revealed that either ALKBH5 (Fig. 5K) or YTHDF2 (Fig. 5L) was strongly colocalized with 5-HT3A in rat TG neurons.
Fig. 5.
ALKBH5-mediated m6A modification enhances the stability of Htr3a mRNA. (A) Volcanic plot indicating the increased (red) and decreased (blue) m6A peaks of ALKBH5-up-treated TGs compared with NC-up-treated groups. (B) qPCR analysis demonstrating the mRNA expression levels of Atp1a4, Atp2b3, Cacan1g, Htr3a, Htr5b, Pde1b, Pkd1l3, Ptk2b, Scn5a, and Trpc6 in the ipsilateral TG treated with ALKBH5-up or NC-up. Data are expressed as the mean ± SEM of three independent experiments. *P < 0.05, ***P < 0.001 vs. NC-up (unpaired t test). n = 5 rats/group. (C) Integrative Genomics Viewer tracks MeRIP-seq data displaying the m6A abundance on Htr3a mRNA transcripts in the TGs of ALKBH5-up- or NC-up-treated groups. (D) m6A level of Htr3a mRNA in the ipsilateral TG of intact rats treated with ALKBH5-up or NC-up. Data are expressed as the mean ± SEM of three independent experiments. *P < 0.05 vs. NC-up (unpaired t test). n = 9 rats/group. (E and F) Enrichment level of the Htr3a mRNA fragment immunoprecipitated by anti-m6A antibody (E) or anti-ALKBH5 antibody (F). Data are expressed as the mean ± SEM of three independent experiments. *P < 0.05 and **P < 0.01 vs. sham (unpaired t test). n = 9 rats/group. (G) Degradation rate of Htr3a mRNA in PC12 cells transduced with ALKBH5-up or NC-up. Data are expressed as the mean ± SEM of three independent experiments. **P < 0.01 vs. NC-up (two-way ANOVA). (H and I) Protein abundance of G3BP1, YTHDC1, YTHDC2, YTHDF1, YTHDF2, and YTHDF3 in the ipsilateral TG 14 d following CCI-ION. Representative blots from three independent experiments are shown. n = 6 rats/group. (J) The interaction between YTHDF2 and Htr3a mRNA was examined by RIP analysis in the ipsilateral TG following CCI-ION operation or sham surgery. Representative blots from three independent experiments are shown. *P < 0.05 vs. sham (unpaired t test). n = 9 rats/group. (K and L) Colocalization of ALKBH5 (K) or YTHDF2 (L) (red) with 5-HT3A (green) and DAPI (blue) in intact TGs. Arrows show colocalization. (Scale bar, 25 μm.)
5-HT3A Is Upregulated in the Injured TG after CCI-ION.
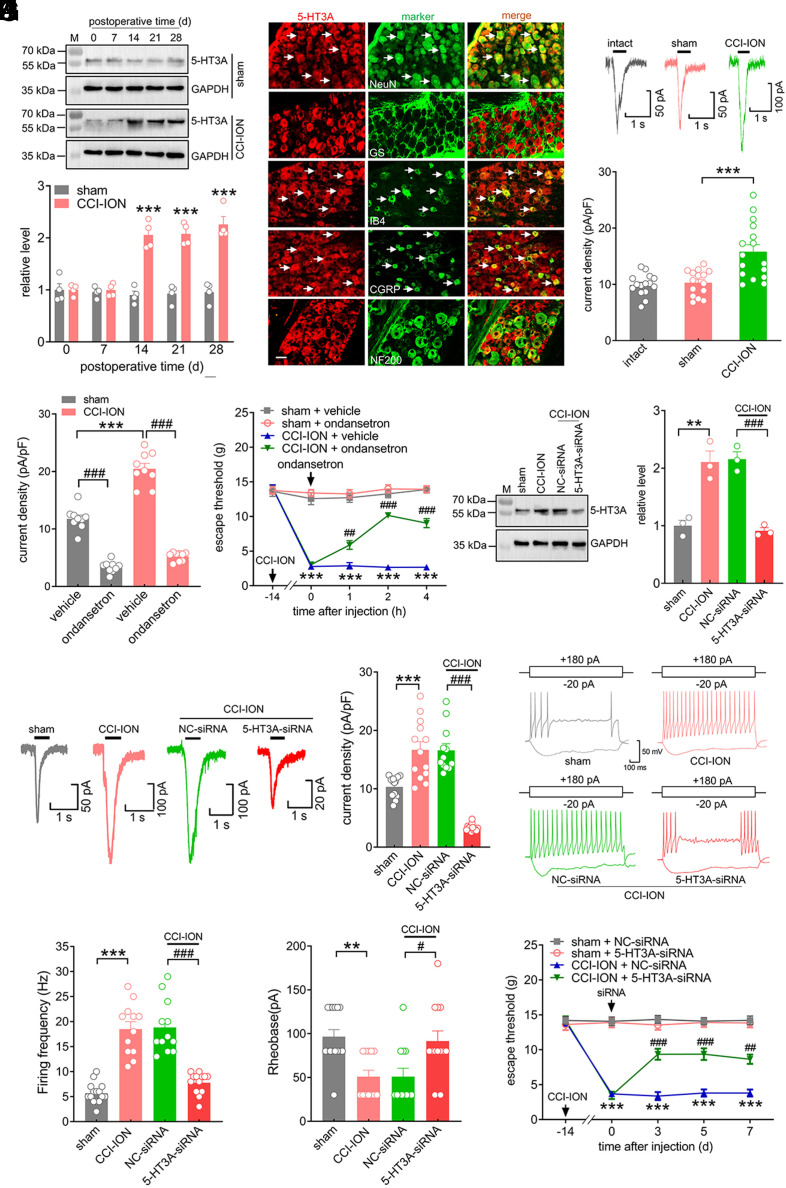
Next, we assessed the potential role of TG 5-HT3A in regulating trigeminal-mediated neuropathic pain behaviors. In comparison with the sham-operated groups, the protein expression level of 5-HT3A was significantly increased in the ipsilateral TG at 14, 21, and 28 d following CCI-ION operation (Fig. 6A and SI Appendix, Fig. S32), while the protein abundance of 5-HT3B showed no change (SI Appendix, Fig. S32). As expected, sham-operated groups also did not exhibit any discernible change in the expression of either the 5-HT3A or 5-HT3B protein (Fig. 6A and SI Appendix, Fig. S33). Next, we examined the cellular localization of 5-HT3A. Double labeling analyses of TG sections of intact rats showed that 5-HT3A primarily coexisted with NeuN but rarely colocalized with GS, suggesting that 5-HT3A is expressed predominantly in TG neurons (Fig. 6B). Furthermore, 5-HT3A showed strong colocalization with CGRP and IB4 but exhibited comparatively little expression in NF200-positive neurons (Fig. 6B). We next determined whether the increased protein abundance of 5-HT3A induced by nerve injury changed 5-HT3 currents in retrograde-labeled small TG neurons (Fig. 6C). Our results revealed that CCI-ION markedly enhanced 5-HT3 currents. The peak current density was increased from 10.8 ± 0.6 pA/pF in the sham group to 15.8 ± 1.2 pA/pF in the CCI-ION group (Fig. 6C). The functional significance of the increased expression of 5-HT3A was further tested at the whole-animal level. Ondansetron (1 μM), a specific 5-HT3 blocker, robustly decreased 5-HT3 currents in both the CCI-ION and sham-operated groups (Fig. 6D). Intra-TG administration of ondansetron (1 nmol) attenuated mechanical allodynia in CCI-ION rats (Fig. 6E). Intra-TG injection of neither ondansetron nor vehicle altered the basal threshold in sham surgery (Fig. 6E). To further determine whether the increased 5-HT3 currents induced by CCI-ION were specifically due to the 5-HT3A isoform, we knocked down 5-HT3A in the injured TG via a chemically modified siRNA. Intra-TG injection of 5-HT3A-siRNA notably decreased the CCI-ION-mediated increase in 5-HT3A protein abundance (Fig. 6F and SI Appendix, Fig. S34) and in 5-HT3 currents (Fig. 6G). Further current-clamp recordings revealed that DiI-labeled small TG neurons from CCI-ION rats exhibited a higher firing frequency of action potentials (Fig. 6 H and I) and a lower rheobase (Fig. 6J) than those from sham groups. Intra-TG injection of chemically modified 5-HT3A-siRNA, but not NC-siRNA, abolished CCI-ION-induced neuronal hyperexcitability (Fig. 6 H–J). Compared to the sham groups, treating rats with 5-HT3A-siRNA attenuated CCI-ION-induced mechanical allodynia during Days 3 to 7 following injection (Fig. 6K).
Fig. 6.
5-HT3A is upregulated in the TGs after CCI-ION. (A) Immunoblot for 5-HT3A in the ipsilateral TG following CCI-ION operation or sham surgery. Blots shown are representative of four independent experiments. ***P < 0.01 vs. the corresponding Day 0 (two-way ANOVA). n = 8 rats/time point/group. (B) Co-staining of 5-HT3A (red) with NeuN, GS, IB4, CGRP, or NF200 (green) in the intact TG. (Scale bar, 25 μm.) Arrows indicate colocalization. (C) Representative traces and summary data showing 5-HT3 currents recorded from retrograde-labeled small-sized TG neurons. n = 15 to 16 neurons from five rats. ***P < 0.001 vs. sham (one-way ANOVA). (D) Bar graph indicating that application of 1 μM ondansetron dramatically decreased 5-HT3 currents in TG neurons at 14 d after CCI-ION or sham operation. ***P < 0.001 vs. sham + vehicle; ###P < 0.001 vs. the corresponding vehicle (unpaired t test). n = 9 neurons from three rats. (E) Intra-TG administration of ondansetron (1 nmol) attenuated mechanical allodynia induced by the CCI-ION operation. ***P < 0.001 vs. sham + vehicle, ###P < 0.001 vs. CCI-ION + vehicle (two-way ANOVA), n = 8 to 9 rats/group. (F) Injection of 5-HT3A-siRNA inhibited the increased protein abundance of 5-HT3A in the ipsilateral TG 14 d following CCI-ION. Representative blots from three independent experiments are shown. **P < 0.01 vs. sham, ###P < 0.05 vs. CCI-ION + NC-siRNA (one-way ANOVA). n = 6 rats/group. (G) Representative traces and summary data indicating that administration of 5-HT3A-siRNA prevented the nerve injury–induced increase in 5-HT3 currents in retrograde-labeled small TG neurons. ***P < 0.001 vs. sham, ###P < 0.05 vs. CCI-ION + NC-siRNA (one-way ANOVA). n = 12 to 13 neurons from four rats. (H and I) Exemplary traces (H) and summary of results (I) indicating that intra-TG administration of 5-HT3A-siRNA prevented the nerve injury–induced increase in the action potential firing frequency. ***P < 0.001 vs. sham, ###P < 0.001 vs. CCI-ION + NC-siRNA (one-way ANOVA). n = 12 to 15 neurons from five rats. (J) The rheobase of TG neurons. **P < 0.01 vs. sham, #P < 0.05 vs. CCI-ION + NC-siRNA (one-way ANOVA). n = 12 to 15 neurons from five rats. (K) Administration of 5-HT3A-siRNA attenuated CCI-ION-induced mechanical allodynia. ***P < 0.001 vs. sham + NC-siRNA, ##P < 0.01 and ###P < 0.001 vs. CCI-ION + NC-siRNA (two-way ANOVA), n = 7 rats/group.
5-HT3A Is Responsible for ALKBH5-Mediated Nociceptive Behaviors.
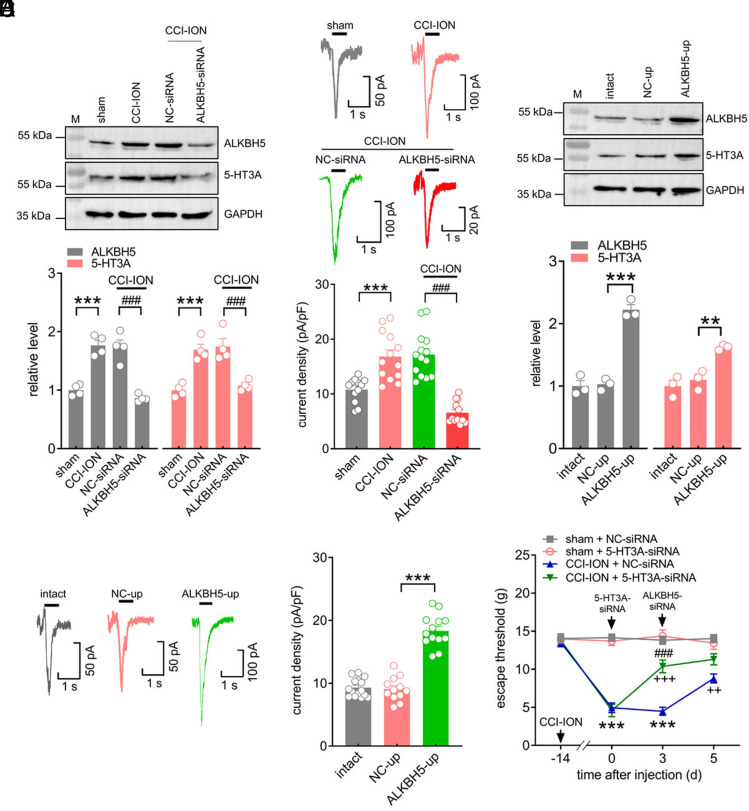
We further investigated whether modulation of ALKBH5 would alter the protein expression and functions of TG 5-HT3A after CCI-ION. Administration of ALKBH5-siRNA significantly suppressed the increased 5-HT3A protein abundance in the injured TG following CCI-ION (Fig. 7A and SI Appendix, Fig. S35). The application of ALKBH5-siRNA also prevented the CCI-ION-induced increase in 5-HT3 currents in retrograde-labeled small TG neurons (Fig. 7B). In addition, unilateral intra-TG injection of ALKBH5-up was performed to explore whether mimicking the CCI-ION-induced increase in ALKBH5 in intact TG neurons would alter the expression of 5-HT3A. Immunoblotting analysis demonstrated that the protein abundance of 5-HT3A was dramatically increased in the ALKBH5-up-treated TGs (Fig. 7C and SI Appendix, Fig. S36). Further recording of small TG neurons showed that ALKBH5 overexpression robustly enhanced 5-HT3 currents (Fig. 7D). To validate whether 5-HT3A actually mediates ALKBH5-induced pain behaviors, we intra-TG injected ALKBH5-siRNA simultaneously with 5-HT3A-siRNA in TG neurons. 5-HT3A-siRNA was administered on Day 0, and ALKBH5-siRNA was then applied on Day 3. In comparison with the NC-siRNA groups, injection of 5-HT3A-siRNA markedly attenuated CCI-ION-induced mechanical allodynia (Fig. 7E). The stable escape threshold from Day 3 to Day 5 following intra-TG administration of ALKBH5-siRNA demonstrated that further injection of ALKBH5-siRNA (Day 3) did not produce any additive effects on the escape threshold in 5-HT3A-siRNA-pretreated CCI-ION rats (Fig. 7E). By contrast, intra-TG injection of ALKBH5-siRNA significantly increased the escape threshold to mechanical stimuli in NC-siRNA-treated CCI-ION rats (Fig. 7E). Thus, 5-HT3A is responsible for ALKBH5-mediated nociceptive behaviors.
Fig. 7.
5-HT3A is responsible for ALKBH5-mediated nociceptive behaviors. (A) Intra-TG injection of ALKBH5-siRNA reduced the increased protein abundance of ALKBH5 or 5-HT3A in the ipsilateral TG 14 d following CCI-ION. The blots shown are representative of four independent experiments. ***P < 0.001 vs. sham, ###P < 0.001 vs. NC-siRNA + CCI-ION (one-way ANOVA). n = 8 rats/group. (B) Representative traces and summary data of 5-HT3 currents recorded from ALKBH5-siRNA-treated TG neurons on Day 14 following CCI-ION operation. ***P < 0.001 vs. sham, ###P < 0.001 vs. CCI-ION + NC-siRNA (one-way ANOVA). n = 12 to 14 neurons from four rats. (C) Injection of ALKBH5-up increased the protein abundance of ALKBH5 and 5-HT3A in the TG of intact rats. Representative blots from three independent experiments are shown. ***P < 0.001 vs. NC-up (one-way ANOVA). n = 6 rats/group. (D) Exemplary traces and summary data showing 5-HT3 currents in ALKBH5-up-treated TG neurons. n = 12 to 15 neurons from three rats. ***P < 0.001 vs. NC-up (one-way ANOVA). (E) Effects of 5-HT3A-siRNA vs. NC-siRNA (Day 0) on ALKBH5-siRNA (Day 3)-mediated relief of mechanical allodynia in CCI-ION-operated rats. ***P < 0.001 vs. sham + NC-siRNA, ###P < 0.001 vs. CCI-ION + 5-HT3A-siRNA (Day 0), ++P < 0.01 and +++P < 0.001 vs. CCI-ION + NC-siRNA (Day 3) (two-way ANOVA). n = 8 to 10 rats/group.
Discussion
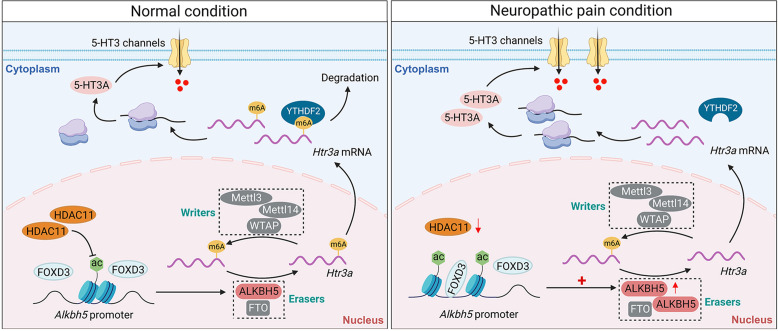
In the current study, we identified ALKBH5 as a pivotal functional m6A modification involved in neuropathic pain regulation. ALKBH5 upregulation appears to causally contribute to trigeminal-mediated neuropathic pain, and inhibition of ALKBH5, either pharmacologically or genetically, effectively alleviated nociceptive behaviors. Mechanistically, HDAC11 downregulation induced by nerve injury led to the accumulation of H3K27ac, which facilitated the binding of FOXD3 to the Alkbh5 promoter and increased ALKBH5 expression in the injured TG. Furthermore, the increased ALKBH5 erased m6A sites in Htr3a mRNA, resulting in an inability of YTHDF2 to bind Htr3a mRNA, in turn maintaining mRNA stability and upregulating the expression of 5-HT3A to promote neuropathic pain behaviors (Fig. 8). Manipulation of ALKBH5 and its relevant epigenetic regulatory factors would provide therapeutic targets for the treatment of neuropathic pain.
Fig. 8.
Schematic diagrams summarizing the molecular mechanisms by which ALKBH5 regulates neuropathic pain. In TG neurons of intact rats, HDAC11 maintains a relatively low expression level of ALKBH5 by decreasing the enrichment of H3K27ac in the Alkbh5 promoter. Both m6A writers and erasers maintain the level of m6A in Htr3a mRNA. The m6A reader YTHDF2 degrades Htr3a mRNA, resulting in normal expression of 5-HT3A. In contrast, downregulation of HDAC11 induced by nerve injury leads to the increased enrichment of H3K27ac, enhancing FOXD3 binding to the Alkbh5 promoter and upregulating the expression level of ALKBH5 in the injured TG. Increased ALKBH5 lowers the level of m6A in Htr3a mRNA, loses its binding to YTHDF2 in the injured TG, and stabilizes the upregulated Htr3a mRNA, thereby increasing TG 5-HT3A protein expression and promoting neuropathic pain behaviors.
Although nociceptors in the trigeminal ganglia (TG) and dorsal root ganglion (DRG) demonstrate a high degree of functional similarity, accumulating studies in both tissues’ first-order sensory neurons have shown that peripheral nerve injury can cause distinct alterations in the expression of pain-associated genes (3, 4). Indeed, we identified in this study that ALKBH5 was selectively increased in the injured TG, while in injured DRGs of the spinal nerve ligation (SNL) model, only another m6A modulator, FTO, was changed (22). However, contradictory results in another study revealed a significant upregulation in ALKBH5, but not FTO and METTL3, in the DRG after SNL (23). Interestingly, no changes in spinal WTAP, FTO, and ALKBH5 were found except for downregulated METTL3 in inflammatory pain (24), although in the same animal model, upregulated expression of METTL3 was demonstrated (25). Thus, these changes may possess some unique genomic signatures/regulation in a tissue-specific and/or pain model-dependent manner and result in the initiation and maintenance of neuropathic pain (3–5). ALKBH5 has been shown to be abundantly expressed in the mammalian peripheral nervous system, but its cellular location remains to be elucidated (4, 22). As it mainly colocalized with NeuN, but not with GS, in the TG, our current study revealed that ALKBH5 was enriched in adult rat TG neurons. In support of this, ALKBH5 was shown to be predominantly expressed in neurons but not glial cells in the adult mouse brain, including the hippocampus and cerebral cortex (26). Interestingly, the potential expression of ALKBH5 in retinal microglia has been demonstrated (27). It was also reported in astrocytes that overexpressed circSTAG1 can capture ALKBH5 and decrease the translocation of ALKBH5 into the nucleus (28). Supposing the tissue-specific expression profiles of ALKBH5 in mammals, morphological observation of ALKBH5-labeled TG neurons in the present study does not support glial expression of ALKBH5 in TGs; rather, our findings further demonstrated that ALKBH5 is coexpressed with IB4, CGRP, and 5-HT3A, all of which are exclusively expressed in neurons. We further revealed that ALKBH5 was localized in both the cytoplasm and the nucleus of TG neurons. Consistent with this, studies have demonstrated nuclear and cytoplasmic localization of ALKBH5 in mammalian central nervous systems, such as the cerebellum, cerebral cortex, and hippocampus (26). However, a recent study reported that ALKBH5 was located in the nucleus of neutrophils (29), while in SH-SY5Y cell lines (26), ALKBH5 protein was detected in both cytosolic and nuclear fractions. Although the discrepancies need further investigation, different tissues/cell types expressing distinct nuclear localization signals (NLSs) may be considered. Indeed, ALKBH5 has been shown to have a putative NLS but no nuclear export signal, which may indicate that ALKBH5 is expressed in the cytoplasm but transported to the cell nucleus by the NLS to play an important role in m6A (26), although ALKBH5 has also been shown to be transported from the nucleus to cytoplasmic synaptic sites during early plasticity (30).
Histone acetylation is an important posttranslational epi-regulatory mechanism that has profound effects on controlling gene expression in a manner dependent on deacetylase and acetyltransferase activities (17, 31). Although the roles of histone deacetylase HDAC11, the most recently identified member of the HDAC family, in neuropathic pain regulation remain unknown, different HDACs may exert diverse or even opposite regulatory effects in different cell types/species or pain models (32). For instance, conditional knockout of HDAC2 in DRG neurons, but not HDAC3, induced long-lasting mechanical pain hypersensitivity (33); meanwhile, in another study, suppression of HDAC2 expression, but not the HDAC1 isoform, alleviated CCI-induced mechanical and thermal hypersensitivity in rats (34). In contrast, inhibition of HDAC1 ameliorates neuropathic pain in a mouse model of trauma-induced peripheral mononeuropathy provoked by spared nerve injury (35). Moreover, neuropathic pain can also modulate mammalian HDAC activities through different mechanisms. Indeed, downregulation of Sp1 decreases the expression of HDAC1/SOX10 within the mouse spinal cord to attenuate neuropathic pain behaviors after SNL (36). Interestingly, in the same animal pain model, phosphorylation of spinal JNK-1 was observed post nerve injury along with a JNK-dependent upregulation in HDAC1 protein levels (37). In addition, cytoplasmic HDAC4 accumulation and SGK1-dependent HDAC4 phosphorylation in dorsal horn neurons play pivotal roles in neuropathic pain maintenance (38). Although the mechanisms underlying the modulation of HDAC11 activity in trigeminal-mediated neuropathic pain need further elucidation, herein, in the injured TG, we revealed that nerve injury upregulated the H3K27ac levels in the promoter region of the Alkbh5 gene by downregulating HDAC11. H3K27ac, but neither H4ac nor other histone H3 acetylation sites, including H3K18ac, H3K14ac, and H3K9ac, was responsible for promoting trigeminal-mediated neuropathic pain. In support of this, it was shown that the number of H3K27ac-positive neurons within the trigeminal root entry zone increased notably in a rat model of trigeminal neuralgia (39); meanwhile, lower H3K9ac expression and little change in H3K27ac expression were reported in TGs using the same animal model (40). In the DRG after sciatic nerve injury, the expression of both H3K9ac and H3K27ac proteins was significantly increased and reported to participate in axonal regeneration and pain modulation (41); however, in a mouse model of chronic postsurgical pain, only the H3K27ac expression level in spinal cord neurons was significantly increased (42). Interestingly, the activity of H3K27ac was shown to be significantly decreased by the brain-derived neutrophilic factor enriched in colorectal carcinoma cells and reported to regulate tumor-induced pain (43). Although further investigation is necessary, the discrepancies may result from histone modifications likely differing in various cell types/tissues that express different histone deacetylases or acetylases (44) and distinct pain models possibly exacerbating this variability. In addition, the equilibrium of methylation and acetylation of H3K27 has been shown to play critical roles in regulating gene transcription (45). While lysine acetylation enhances transcriptional activation by relaxing chromatin, lysine methylation at different positions can have distinct effects (46). For instance, dimethylation and trimethylation of H3K27 (H3K27me2/3) have also been shown to repress gene expression in sensory neurons (17). Although it is beyond the research scope of our present study, the possibility of simultaneous alterations in histone modification (i.e., increased occupancy of H3K27ac and/or decreased H3K27me2/3) contributing to gene transcription cannot be excluded.
Although ALKBH5 may have several molecular targets, our current study mainly focused on Htr3a because 5-HT3A plays pivotal roles in the pathogenesis of pain syndromes (47). As the MeRIP-seq analysis exhibited a large loss of m6A in Htr3a mRNA, given that the RNA m6A reader YTHDF2 facilitates its binding-RNA degradation (22, 48), we proposed that the increased ALKBH5 erased m6A sites in Htr3a mRNA, leading to the loss of Htr3a mRNA binding to YTHDF2 in the injured TG. Therefore, mechanical allodynia induced by the TG ALKBH5 increase in CCI-ION rats resulted from the upregulated expression of 5-HT3A. In line with this, 5-HT3A knockout mice differed from wild-type mice by a dramatic decrease in formalin-induced nociceptive responses (47). 5-HT3 channels in sensory neurons enhance the release of glutamate and substance P, explaining the observed behavioral response of increased 5-HT3-mediated nociception (49). Indeed intraplantar administration of 5-HT3 antagonists produced analgesia against CFA-induced chronic inflammatory pain (50). Interestingly, although inhibiting spinal 5-HT3 by intrathecal injection of ondansetron completely suppressed mechanical allodynia in SNL rats (51), a lack of analgesic efficacy of spinal ondansetron on mechanical hypersensitivity was also reported using the same animal model (52). In contrast, intrathecal application of 5-HT3 receptor agonists induced anti-hypersensitivity in neuropathic pain (53). Although these discrepancies have yet to be explained, the central but not peripheral effects of 5-HT3 may be responsible for the analgesic effect of 5-HT3 agonists, as spinal GABAergic interneurons could mediate the central effects (53). Importantly, the application of antagonists in this study through intra-TG administration is extremely selective and local; thus, the observed behavioral responses are less likely to be the result of confounding influences from other central nuclei. Moreover, different pain models used in the various studies may also be a factor in this variability. Indeed, the regulation of ion channels, receptors, and neurotransmitters at the transcriptional and/or translational levels may vary in distinct pain models, although chronic pain, such as neuropathic pain or inflammation, shares some common signaling messengers (54). Nevertheless, as our MeRIP-seq assay revealed that, in addition to the Htr3a transcript, some other transcripts in the injured TG showed loss and/or gain of m6A sites following CCI-ION as well, an additional putative ALKBH5-mediated mechanism contributing to neuropathic pain cannot be excluded, which still needs further investigation.
Taken together, our present study offers insights into the epigenetic regulatory mechanisms of TG ALKBH5 in neuropathic pain. We demonstrate that downregulation of HDAC11 induced by nerve injury results in H3K27ac enrichment that facilitates the binding of FOXD3 to the Alkbh5 promoter, which in turn upregulates the ALKBH5 expression. This increased ALKBH5 erases m6A sites in Htr3a mRNA, resulting in an inability of YTHDF2 to bind Htr3a mRNA, which maintains mRNA stability and increases the expression of 5-HT3A to promote neuropathic pain. The identification of epigenetic factors such as ALKBH5 in regulating 5-HT3A may provide potential therapeutic targets for neuropathic pain management.
Materials and Methods
Detailed materials and methods are described in the SI Appendix.
Animal Model and Behavioral Tests.
Protocols for animal experiments were approved by the Animal Care and Use Committee of Soochow University strictly according to the NIH guidelines for animal research and the International Association for the Study of Pain. Adult Sprague–Dawley rats (8 to 10 wk, male) were used for experiments. An intraoral approach was used to create the chronic constriction injury to the infraorbital nerve (CCI-ION) model of trigeminal neuropathic pain (55). Mechanical and thermal behavioral analyses were conducted as previously described (55–57). The orofacial stimulation test was assessed by an Orofacial Stimulation Test system (57, 58).
Intra-TG Drug Application.
Intra-TG injection was performed as described previously (55, 56). 2’-O-methyl- and 5’-cholesteryl-modified siRNAs were intra-TG administrated daily for 2 consecutive days. The siRNA sequences are summarized in SI Appendix, Table S1.
Immunoblot Analysis and Immunofluorescence Staining.
Immunoblot analysis and immunofluorescence staining were conducted as previously described (55, 56).
m6A Dot Blot Assay.
The dot blot assay was applied to evaluate the total mRNA m6A levels. Methylene blue staining was applied to verify that equal amounts of RNA samples were spotted on the membrane.
Real-Time Quantitative PCR (qRT-PCR).
Purified RNAs were reverse-transcribed by the PrimeScript RT Reagent Kit (Takara). qPCR was performed using the ROCHE LightCycler® 96 System with SYBR Green qPCR Master Mix. The primers used in the present study are summarized in SI Appendix, Table S2.
RIP-PCR.
The RIP assay was conducted using a Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore) according to the manufacturer’s instructions.
Dual-Luciferase Reporter Assay.
The fragments (pGL3-F1 to pGL3-F5) were amplified from genomic DNA with the primers (SI Appendix, Table S2) to construct the reporter plasmids. Dual-luciferase assays were conducted following the protocol described in the Dual-Luciferase Reporter Assay System (Promega).
ChIP-qPCR.
ChIP assays were conducted with the SimpleChIP® Plus Enzymatic Chromatin IP Kit as described previously (55). The ChIP-specific primers used are shown in SI Appendix, Table S2.
MeRIP and MeRIP Sequencing.
The MeRIP assay was performed using the Magna MeRIP™ m6A Kit (Millipore) according to the manufacturer’s guidelines. Modification of m6A toward particular genes was determined by MeRIP-qPCR analysis with specific primers (SI Appendix, Table S2). MeRIP sequencing and the following data analyses were mainly supported by Shanghai Cloud-Seq Biotech.
RNA Stability Assay.
PC12 cells were transfected with NC-up or ALKBH5-up and then treated with 5 μg/mL actinomycin D. Total RNA was extracted at each time point after treatment for qPCR analysis.
Dissociation of TG Neurons and Electrophysiology.
TG neurons were enzymatically dissociated from adult rats as described previously (55). Whole-cell patch clamp recordings were performed 3 to 7 h after plating in a gap-free pattern.
Data Analysis and Statistics.
All data are expressed as the mean ± SEM. Data acquisition and statistical analysis were performed using Microsoft Excel, ClampFit 10.2, and Prism 8.0.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This work was supported by research grants from the National Natural Science Foundation of China (No. 82071236, No. 82271245, and No. 82371218), Jiangsu Key Laboratory of Neuropsychiatric Diseases (No. BM2013003), Natural Science Foundation of Jiangsu Province (No. BK20211073), Clinical Research Center of Neurological Disease (No. ND2022B03), Science and Technology Bureau of Suzhou (No. SYS2020129), and Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPDJHEI).
Author contributions
Z.H., S.W., R.Q., Y.T., X.J., and J.T. designed research; Z.H., Y.Z., S.W., R.Q., Y.T., and X.J. performed research; Y.S., D.J., and J.T. contributed new reagents/analytic tools; Z.H., Y.Z., S.W., R.Q., and J.T. analyzed data; and Z.H., Y.S., D.J., and J.T. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
The raw m6A RNA sequencing data have been deposited in the NCBI Gene Expression Omnibus (GEO) with accession number GSE226989 (59). All other data are included in the manuscript and/or SI Appendix.
Supporting Information
References
- 1.Finnerup N. B., Kuner R., Jensen T. S., Neuropathic pain: From mechanisms to treatment. Physiol. Rev. 101, 259–301 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Ghazisaeidi S., Muley M. M., Salter M. W., Neuropathic pain: Mechanisms, sex differences, and potential therapies for a global problem. Annu. Rev. Pharmacol. Toxicol. 63, 565–583 (2023). [DOI] [PubMed] [Google Scholar]
- 3.Megat S., et al. , Differences between dorsal root and trigeminal ganglion nociceptors in mice revealed by translational profiling. J. Neurosci. 39, 6829–6847 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kogelman L. J. A., et al. , Whole transcriptome expression of trigeminal ganglia compared to dorsal root ganglia in Rattus Norvegicus. Neuroscience 350, 169–179 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Korczeniewska O. A., et al. , Differential gene expression changes in the dorsal root versus trigeminal ganglia following peripheral nerve injury in rats. Eur. J. Pain 24, 967–982 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Zhao B. S., Roundtree I. A., He C., Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 18, 31–42 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boulias K., Greer E. L., Biological roles of adenine methylation in RNA. Nat. Rev. Genet. 24, 143–160 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meyer K. D., Jaffrey S. R., The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 15, 313–326 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding C., et al. , RNA m(6)A demethylase ALKBH5 regulates the development of gammadelta T cells. Proc. Natl. Acad. Sci. U.S.A. 119, e2203318119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang X., et al. , The role of m6A modification in the biological functions and diseases. Signal Transduct Target Ther. 6, 74 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han Z., et al. , ALKBH5-mediated m(6)A mRNA methylation governs human embryonic stem cell cardiac commitment. Mol. Ther. Nucleic Acids 26, 22–33 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang N., Ding C., Zuo Y., Peng Y., Zuo L., N6-methyladenosine and neurological diseases. Mol. Neurobiol. 59, 1925–1937 (2022). [DOI] [PubMed] [Google Scholar]
- 13.Xu K., et al. , N(6)-methyladenosine demethylases Alkbh5/Fto regulate cerebral ischemia-reperfusion injury. Ther. Adv. Chronic Dis. 11, 2040622320916024 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang L., et al. , Ethionine-mediated reduction of S-adenosylmethionine is responsible for the neural tube defects in the developing mouse embryo-mediated m6A modification and is involved in neural tube defects via modulating Wnt/beta-catenin signaling pathway. Epigenet. Chromatin 14, 52 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boija A., et al. , Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 175, 1842–1855.e1816 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bose D. A., et al. , RNA binding to CBP stimulates histone acetylation and transcription. Cell 168, 135–149.e122 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berger S. L., The complex language of chromatin regulation during transcription. Nature 447, 407–412 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Shahbazian M. D., Grunstein M., Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 76, 75–100 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Garcia-Campos M. A., et al. , Deciphering the “m(6)A Code” via antibody-independent quantitative profiling. Cell 178, 731–747.e716 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Gover T. D., Moreira T. H., Weinreich D., Role of calcium in regulating primary sensory neuronal excitability. Handb. Exp. Pharmacol. 194, 563–587 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Wang X., et al. , N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y., et al. , N(6)-methyladenosine demethylase FTO contributes to neuropathic pain by stabilizing G9a expression in primary sensory neurons. Adv. Sci. (Weinh) 7, 1902402 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu Z., et al. , Positive interaction between GPER and beta-alanine in the dorsal root ganglion uncovers potential mechanisms: Mediating continuous neuronal sensitization and neuroinflammation responses in neuropathic pain. J. Neuroinflamm. 19, 164 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pan Z., et al. , Methyltransferase-like 3 contributes to inflammatory pain by targeting TET1 in YTHDF2-dependent manner. Pain 162, 1960–1976 (2021). [DOI] [PubMed] [Google Scholar]
- 25.Zhang C., Wang Y., Peng Y., Xu H., Zhou X., METTL3 regulates inflammatory pain by modulating m(6)A-dependent pri-miR-365-3p processing. FASEB J. 34, 122–132 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Du T., Li G., Yang J., Ma K., RNA demethylase Alkbh5 is widely expressed in neurons and decreased during brain development. Brain Res. Bull 163, 150–159 (2020). [DOI] [PubMed] [Google Scholar]
- 27.Chen T., et al. , ALKBH5-mediated m(6)A modification of A20 regulates microglia polarization in diabetic retinopathy. Front. Immunol. 13, 813979 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang R., et al. , N(6)-methyladenosine modification of fatty acid amide hydrolase messenger RNA in circular RNA STAG1-regulated astrocyte dysfunction and depressive-like behaviors. Biol. Psychiatry 88, 392–404 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Liu Y., et al. , m(6)A demethylase ALKBH5 is required for antibacterial innate defense by intrinsic motivation of neutrophil migration. Signal Transduct Target Ther. 7, 194 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinez De La Cruz B., et al. , Modifying the m(6)A brain methylome by ALKBH5-mediated demethylation: A new contender for synaptic tagging. Mol. Psychiatry 26, 7141–7153 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shvedunova M., Akhtar A., Modulation of cellular processes by histone and non-histone protein acetylation. Nat. Rev. Mol. Cell Biol. 23, 329–349 (2022). [DOI] [PubMed] [Google Scholar]
- 32.Khangura R. K., Bali A., Jaggi A. S., Singh N., Histone acetylation and histone deacetylation in neuropathic pain: An unresolved puzzle? Eur. J. Pharmacol. 795, 36–42 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Zhang J., et al. , HDAC2 in primary sensory neurons constitutively restrains chronic pain by repressing alpha2delta-1 expression and associated NMDA receptor activity. J. Neurosci. 42, 8918–8935 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Z., et al. , HDAC2, but not HDAC1, regulates Kv1.2 expression to mediate neuropathic pain in CCI rats. Neuroscience 408, 339–348 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Sanna M. D., Guandalini L., Romanelli M. N., Galeotti N., The new HDAC1 inhibitor LG325 ameliorates neuropathic pain in a mouse model. Pharmacol. Biochem. Behav. 160, 70–75 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Xie Y., et al. , Downregulation of Sp1 inhibits the expression of HDAC1/SOX10 to alleviate neuropathic pain-like behaviors after spinal nerve ligation in mice. ACS Chem. Neurosci. 13, 1446–1455 (2022). [DOI] [PubMed] [Google Scholar]
- 37.Sanna M. D., Galeotti N., The HDAC1/c-JUN complex is essential in the promotion of nerve injury-induced neuropathic pain through JNK signaling. Eur. J. Pharmacol. 825, 99–106 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Lin T. B., et al. , Modulation of nerve injury-induced HDAC4 cytoplasmic retention contributes to neuropathic pain in rats. Anesthesiology 123, 199–212 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Lin R., et al. , Immunohistochemical analysis of histone H3 acetylation in the trigeminal root entry zone in an animal model of trigeminal neuralgia. J. Neurosurgery 131, 828–838 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Wei W., et al. , Characterization of acetylation of histone H3 at lysine 9 in the trigeminal ganglion of a rat trigeminal neuralgia model. Oxid. Med. Cell Longev. 2022, 1300387 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palmisano I., et al. , Epigenomic signatures underpin the axonal regenerative ability of dorsal root ganglia sensory neurons. Nat. Neurosci. 22, 1913–1924 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Katsuda Y., et al. , Histone modification of pain-related gene expression in spinal cord neurons under a persistent postsurgical pain-like state by electrocautery. Mol. Brain 14, 146 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cao D. Y., Bai G., Ji Y., Karpowicz J. M., Traub R. J., EXPRESS: Histone hyperacetylation modulates spinal type II metabotropic glutamate receptor alleviating stress-induced visceral hypersensitivity in female rats. Mol. Pain 12, 1744806916660722 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kazantsev A. G., Thompson L. M., Therapeutic application of histone deacetylase inhibitors for central nervous system disorders. Nat. Rev. Drug Discov. 7, 854–868 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Sankar A., et al. , Histone editing elucidates the functional roles of H3K27 methylation and acetylation in mammals. Nat. Genet. 54, 754–760 (2022). [DOI] [PubMed] [Google Scholar]
- 46.Jambhekar A., Dhall A., Shi Y., Roles and regulation of histone methylation in animal development. Nat. Rev. Mol. Cell Biol. 20, 625–641 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kayser V., et al. , Mechanical, thermal and formalin-induced nociception is differentially altered in 5-HT1A−/−, 5-HT1B−/−, 5-HT2A−/−, 5-HT3A−/− and 5-HTT−/− knock-out male mice. Pain 130, 235–248 (2007). [DOI] [PubMed] [Google Scholar]
- 48.Murakami S., Jaffrey S. R., Hidden codes in mRNA: Control of gene expression by m(6)A. Mol. Cell 82, 2236–2251 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Millan M. J., Descending control of pain. Prog. Neurobiol. 66, 355–474 (2002). [DOI] [PubMed] [Google Scholar]
- 50.Giordano J., Rogers L. V., Peripherally administered serotonin 5-HT3 receptor antagonists reduce inflammatory pain in rats. Eur. J. Pharmacol. 170, 83–86 (1989). [DOI] [PubMed] [Google Scholar]
- 51.Suzuki R., Rahman W., Hunt S. P., Dickenson A. H., Descending facilitatory control of mechanically evoked responses is enhanced in deep dorsal horn neurones following peripheral nerve injury. Brain Res. 1019, 68–76 (2004). [DOI] [PubMed] [Google Scholar]
- 52.Peters C. M., et al. , Lack of analgesic efficacy of spinal ondansetron on thermal and mechanical hypersensitivity following spinal nerve ligation in the rat. Brain Res. 1352, 83–93 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hayashida K., et al. , Ondansetron reverses antihypersensitivity from clonidine in rats after peripheral nerve injury: Role of gamma-aminobutyric acid in alpha2-adrenoceptor and 5-HT3 serotonin receptor analgesia. Anesthesiology 117, 389–398 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ji R. R., Strichartz G., Cell signaling and the genesis of neuropathic pain. Sci STKE 2004, reE14 (2004). [DOI] [PubMed] [Google Scholar]
- 55.Qi R., et al. , Histone methylation-mediated microRNA-32-5p down-regulation in sensory neurons regulates pain behaviors via targeting Cav3.2 channels. Proc. Natl. Acad. Sci. U.S.A. 119, e2117209119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang H., et al. , Brain-derived neurotrophic factor stimulation of T-type Ca(2+) channels in sensory neurons contributes to increased peripheral pain sensitivity. Sci. Signal 12, eaaw2300 (2019). [DOI] [PubMed] [Google Scholar]
- 57.Zhang Y., et al. , Neuromedin B receptor stimulation of Cav3.2 T-type Ca(2+) channels in primary sensory neurons mediates peripheral pain hypersensitivity. Theranostics 11, 9342–9357 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Prochazkova M., et al. , Activation of cyclin-dependent kinase 5 mediates orofacial mechanical hyperalgesia. Mol. Pain 9, 66 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang Z., et al. , ALKBH5-mediated m6A modification of 5-HT3A mRNA in primary sensory neurons promotes neuropathic pain behaviors. National Center for Biotechnology Information Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE226989. Deposited 9 March 2023.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
The raw m6A RNA sequencing data have been deposited in the NCBI Gene Expression Omnibus (GEO) with accession number GSE226989 (59). All other data are included in the manuscript and/or SI Appendix.