Significance
The substitution of methionine for isoleucine at codon 148 of PNPLA3 is the most impactful genetic risk factor for fatty liver disease. The mutant isoform accumulates to high levels on lipid droplets, thereby interfering with hepatic triglyceride mobilization. Here, we performed a genetic screen to identify the E3 ubiquitin ligase that targets PNPLA3 for degradation. Inactivation of bifunctional regulator of apoptosis (BFAR) in cultured hepatocytes increased PNPLA3 levels; this observation was replicated in the livers of Bfar−/− mice. The identification of BFAR provides a therapeutic target to enhance PNPLA3 turnover and prevent FLD.
Keywords: ubiquitin proteasome system, steatosis, lipid droplets, mutation, hepatocyte
Abstract
A missense variant in patatin-like phospholipase domain-containing protein 3 [PNPLA3(I148M)] is the most impactful genetic risk factor for fatty liver disease (FLD). We previously showed that PNPLA3 is ubiquitylated and subsequently degraded by proteasomes and autophagosomes and that the PNPLA3(148M) variant interferes with this process. To define the machinery responsible for PNPLA3 turnover, we used small interfering (si)RNAs to inactivate components of the ubiquitin proteasome system. Inactivation of bifunctional apoptosis regulator (BFAR), a membrane-bound E3 ubiquitin ligase, reproducibly increased PNPLA3 levels in two lines of cultured hepatocytes. Conversely, overexpression of BFAR decreased levels of endogenous PNPLA3 in HuH7 cells. BFAR and PNPLA3 co-immunoprecipitated when co-expressed in cells. BFAR promoted ubiquitylation of PNPLA3 in vitro in a reconstitution assay using purified, epitope-tagged recombinant proteins. To confirm that BFAR targets PNPLA3, we inactivated Bfar in mice. Levels of PNPLA3 protein were increased twofold in hepatic lipid droplets of Bfar−/− mice with no associated increase in PNPLA3 mRNA levels. Taken together these data are consistent with a model in which BFAR plays a role in the post-translational degradation of PNPLA3. The identification of BFAR provides a potential target to enhance PNPLA3 turnover and prevent FLD.
Fatty liver disease (FLD) encompasses a spectrum of related disorders that arise from accumulation of triglycerides (TG) in the liver (1). The major causes of excess hepatic TG are obesity/insulin resistance and excessive alcohol consumption, but the relative impact of these factors on hepatic TG content is strongly influenced by genetic factors (2–5). In 2008, we identified a missense variant (I148M) in patatin-like phospholipase domain-containing protein 3 (PNPLA3) that is associated with increased hepatic TG content and elevated levels of serum alanine transaminase (6, 7). This variant, which has a population frequency ranging from 14% in Blacks to 49% in Hispanics, is also strongly associated with the full spectrum of FLD: hepatic steatosis, steatohepatitis, cirrhosis, and hepatocellular carcinoma (6, 8–13). Additional sequence variants have been identified in other genes that are associated with FLD, but PNPLA3(148M) remains the most impactful genetic risk factor for this disorder both in terms of its frequency and effect size (14–16). Despite the primacy of this variant in disease susceptibility, the pathogenic mechanistic basis underlying the PNPLA3(148M)-associated increase in hepatic fat content has not been fully defined.
PNPLA3 most closely resembles in sequence PNPLA2, which is also known as adipose tissue TG lipase (ATGL), the major TG hydrolase in the liver (17). Both proteins localize to lipid droplets (LDs) in hepatocytes (17, 18). Inactivation of ATGL, or of its activator, α/β hydrolase domain containing 5 (ABHD5) causes FLD (19, 20), whereas inactivation of PNPLA3 in mice does not result in hepatic steatosis (21, 22). Either hepatic overexpression of PNPLA3(148M) (23) or substitution of methionine for isoleucine at residue 148 of the mouse gene (Pnpla3148M/M) (24) replicates the FLD phenotype that is observed in humans. Levels of PNPLA3 on hepatic LDs from Pnpla3148M/M mice fed a high sucrose diet (HSD) are ~40-fold higher than those of WT littermates, despite comparable levels of PNPLA3 mRNA in the two groups (24). The amount of ABHD5, but not ATGL, is also increased on hepatic LDs of Pnpla3148M/Mmice (24).
The increased level of PNPLA3(148M) without a concomitant increase in hepatic PNPLA3 mRNA suggests that the impairment in Pnpla3148M/M mice is in PNPLA3 degradation (24). Supporting this notion, treatment of mice with a proteasome inhibitor, bortezomib, causes increased levels of PNPLA3(WT), but not of PNPLA3(148M) on hepatic LD (25). Moreover, the fraction of ubiquitylated PNPLA3 is greater in WT mice than in Pnpla3148M/M mice. Based on these findings, we proposed that PNPLA3(148M) evades ubiquitylation and accumulates on LDs, where it sequesters ABHD5 and prevents activation of ATGL (26, 27).
To elucidate the mechanism by which PNPLA3 is targeted for degradation, we screened an arrayed library of siRNAs targeting transcripts involved in protein ubiquitylation to identify the E3 ubiquitin ligase that promotes ubiquitylation of PNPLA3 in cultured hepatocytes. Here, we show that Bifunctional Apoptosis Regulator (BFAR), an ER-associated E3 ubiquitin ligase (28), contributes to both the ubiquitylation and turnover of PNPLA3.
Results
Proteasome-Mediated Degradation of PNPLA3 in Cultured Hepatocytes.
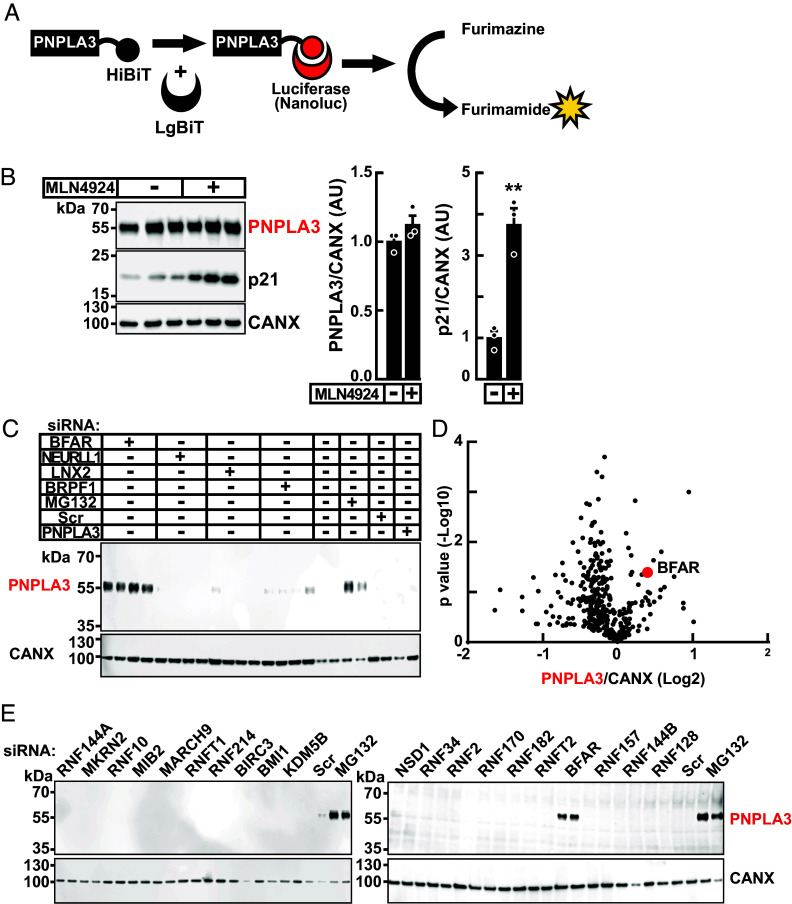
To identify proteins required for proteasome-mediated degradation of PNPLA3, we developed a luciferase reconstitution assay using Nanoluc (SI Appendix, Methods) (29), a small, potent, recombinant luciferase from the deep-sea shrimp Oplophorus gracilirostris (Fig. 1A). Assays were performed in HuH7 cells that stably express human PNPLA3 fused at the C terminus to HiBiT, an 11 residue peptide (PNPLA3-HiBiT). PNPLA3-HiBiT was detected using LgBiT and the NanoBiT blotting assay (SI Appendix). In this manuscript, LgBiT is denoted with red text when used to detect PNPLA3-HiBiT. As reported previously (26, 30), addition of the proteasome inhibitor MG132 and LD-inducer oleate (400 µM) increased levels of PNPLA3 without a concomitant increase in PNPLA3 mRNA levels (SI Appendix, Fig. S1). These experiments confirmed that addition of the small C-terminal HiBiT tag did not interfere with the PNPLA3 stabilization that occurs with either proteasome inhibition or LD formation (26, 30). These results are consistent with our prior observation that increased levels of PNPLA3 protein associated with either MG132 treatment or LD formation are due to a post-translational process (30).
Fig. 1.
Identification and confirmation of BFAR as an E3 ubiquitin ligase for PNPLA3. (A) Schematic of NanoBiT reconstitution assay. HuH7 cells stably expressing recombinant PNPLA3-HiBiT (PNPLA3-HiBiT cells) were lysed and treated with LgBiT to reconstitute luciferase activity. Luciferase cleaves furimazine to generate a luminescent signal (furimamide) that is quantitated using a LI-COR ODYSSEY analyzer. (B) Proteasomal degradation of PNPLA3 is not mediated by a Cullin RING E3 Ligase. HuH7 cells stably expressing PNPLA3 were treated with MLN4924 (1 µM), an inhibitor of Neddylation Activating Enzyme (NAE). Immunoblotting was performed as described in Materials and Methods. Treatment with MLN4924 increased protein levels of p21, a known substrate for Cullin-Ring ligase 4, by fourfold, but had no effect on levels of PNPLA3. (C). A representative blot showing accumulation of PNPLA3 with siRNA knockdown of individual E3 ubiquitin ligase (quadruplicate), compared to treatment with MG-132. Knockdown of BFAR caused a 2.3-fold increase in PNPLA3-HiBiT levels (indicated in red) in comparison to a scrambled non-targeting siRNA (Scr). (D) Volcano plot of relative abundance of PNPLA3-HiBiT after siRNA knockdown of candidate E3 ligases (n = 375). Signals corresponding to PNPLA3 were quantified by LI-COR, normalized to calnexin (CANX) levels, and expressed as arbitrary units (AUs). The x-axis denotes the fold change in normalized PNPLA3 levels (log2), and the y-axis denotes the P value (−log10) of the change in the level of PNPLA3 with siRNA treatment. A list of the 20 siRNAs that resulted in the greatest increase in PNPLA3 levels is provided in SI Appendix, Table S2. (E) Results from NanoBiT immunoblot assay to validate top 20 siRNA pools (duplicate) that caused the greatest increase in PNPLA3 levels in the initial siRNA screen. Values represent means ± SEM. Levels were compared using a Student’s t test. **P < 0.01. The experiments were repeated twice and the results were similar.
Proteasomal Degradation of PNPLA3 Was Not Mediated by Cullin-RING (Really Interesting New Gene) E3 Ligases (CRLs).
The CRLs constitute the largest family of E3 ubiquitin ligases (n > 200) (31, 32). The ubiquitin ligase activity of CRL is activated by the covalent addition of NEDD8 by NEDD8-activating enzyme (NAE) (33, 34), which can be inactivated by a small-molecule, MLN4924 (35). To determine whether the E3 ligase involved in ubiquitylation and degradation of PNPLA3 is a member of the Cullin-RING E3 ligase family, we treated PNPLA3-HiBiT expressing cells with MLN4924 (1 µM). No significant increase in PNPLA3-HiBiT levels was seen in cells treated with the NAE inhibitor (Fig. 1B). In contrast, levels of p21, a target of Cullin4B (36), increased fourfold in the cells, confirming successful NAE inhibition. We concluded from this experiment that the E3 ligase(s) that recognize PNPLA3 is not a CRL.
Identification of a Candidate E3 Ubiquitin Ligase for PNPLA3 Using an siRNA Screen.
If degradation of PNPLA3 is dependent on ubiquitylation by a single E3 ubiquitin ligase, then inactivation of that E3 ligase should increase PNPLA3 levels to the same extent as those observed with MG132 treatment. To identify such an E3 ligase, HuH7 cells stably expressing PNPLA3-HiBiT were treated with pools of four siRNAs targeting each of the known (or putative) E3 ligases. Silencing RNAs were selected from a commercial, arrayed siRNA library (Human ON-TARGETplus) and reverse transfected into PNPLA3-HiBiT cells as described in the SI Appendix. After 48 h, cells were lysed, and PNPLA3 levels were assessed using the NanoBiT blotting assay.
We performed the screen using siRNAs to knockdown expression of two E1 ubiquitin-activating enzymes, 38 E2 conjugating proteins, and 378 E3 ubiquitin ligases, including 350 RING E3 ligases and 28 HECT (Homologous to E6AP carboxy terminus) E3 ligases. A representative blot is shown in Fig. 1C. In this experiment, siRNAs targeting bifunctional apoptosis regulator (BFAR) (37) increased PNPLA3 to levels seen in cells treated with MG132. The results from the entire screen are displayed in a volcano plot in Fig. 1D. The change in the PNPLA3 level relative to baseline [after correction for gel loading using calnexin (CANX) levels] is plotted (x-axis) against the P-value of the change in the PNPLA3 level relative to cells treated with siRNAs encoding a scrambled sequence (scr) (y-axis). The top 20 hits are listed in SI Appendix, Table S2. The screening assay was repeated for these hits and only one siRNA pool, the one targeting BFAR, consistently increased levels of PNPLA3 (Fig. 1E).
Depletion of BFAR Increases PNPLA3 Levels in Cultured Hepatocytes.
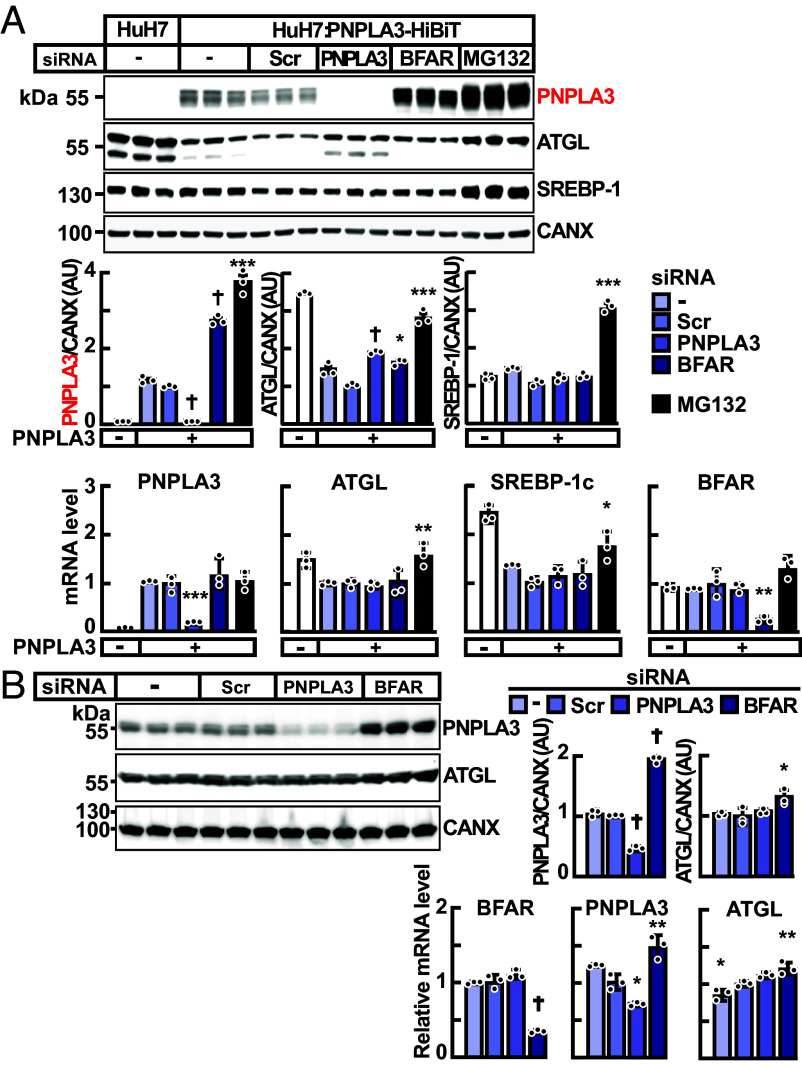
If BFAR is an E3-ligase for PNPLA3, then inactivation of BFAR should increase levels of PNPLA3. We used siRNA to knockdown BFAR in HuH7 cells stably expressing PNPLA3-HiBiT, repeating the conditions used in the siRNA screen. Cells treated with siRNAs targeting BFAR had 2.8-fold higher levels of PNPLA3 protein without any change in PNPLA3 mRNA levels when compared to cells treated with an siRNA containing a scrambled sequence (Scr) (Fig. 2A). Knockdown of BFAR was associated with a modest but significant increase in the levels of ATGL protein but without an increase in ATGL mRNA. The protein and mRNA levels of sterol regulatory element binding protein-1c (SREBP-1c), the major transcriptional activator of PNPLA3 (30), were unchanged (Fig. 2A), consistent with the notion that the increase in the PNPLA3 protein level was due to protein stabilization rather than increased production. The increase in PNPLA3 protein levels was ~30% higher in cells treated MG132 than in cells treated with siRNAs targeting BFAR (Fig. 2A), so BFAR may not be the only E3 ligase that targets PNPLA3. Alternatively, the higher levels of PNPLA3 in MG132-treated cells may be caused by other factors (e.g., incomplete inactivation of BFAR with siRNA treatment, broad effects of MG132 inhibition, etc.).
Fig. 2.
Inactivation of BFAR using siRNAs in HuH7 cells. (A) HuH7 cells stably expressing PNPLA3-HiBiT were treated with pools of four siRNAs (60 pmol) each directed against PNPLA3, BFAR or with scrambled sequences (Scr), or with MG132 (10 µM). Immunoblot analysis was performed using cell lysate, as described in Materials and Methods. Signals were quantified using a LI-COR imaging system, normalized to levels of CANX, and expressed as arbitrary units (AUs). The mRNA levels of indicated genes were measured and normalized as described in Materials and Methods. All mRNA levels were compared to levels seen in cells treated with Scr siRNA using the Student’s t test. (B) Inactivation of endogenous BFAR in HuH7 cells, which express PNPLA3(148M). HuH7 cells were treated with buffer alone, or with Scr-, PNPLA3-, or BFAR-siRNAs (60 pmol). Immunoblot analysis of cell lysates and signal quantification were performed as described in panel (A). Values represent means ± SEM. Means were compared to cells treated with Scr siRNA using the Student’s t test. *P < 0.05, **P < 0.01, ***P < 0.001, †P < 0.0001. The experiments were repeated three times, and the results were similar.
Previously we reported that PNPLA3(148M) was poorly ubiquitylated and accumulated on LDs in vivo in livers of mice (26). To test whether inactivation of BFAR causes changes in PNPLA3(148M) levels, we examined the effect of siRNA-mediated BFAR knockdown on levels of endogenous PNPLA3 in HuH7 cells. HuH7 cells have been shown previously to express only PNPLA3(148M) (38). Treatment with siRNA targeting BFAR increased levels of PNPLA3(148M) protein twofold with an associated 50% increase in levels of PNPLA3 mRNA (Fig. 2B). Similar to what was seen in Fig. 2A, inactivation of BFAR was also associated with a modest but significant increase in levels of both ATGL protein and mRNA. From this experiment, we concluded that BFAR targets PNPLA3(148M) as well as PNPLA3(WT) in HuH7 cells. BFAR may also target ATGL, though the effect appears to be more modest than that seen with PNPLA3.
BFAR Is a Polytopic E3 Ubiquitin Ligase.
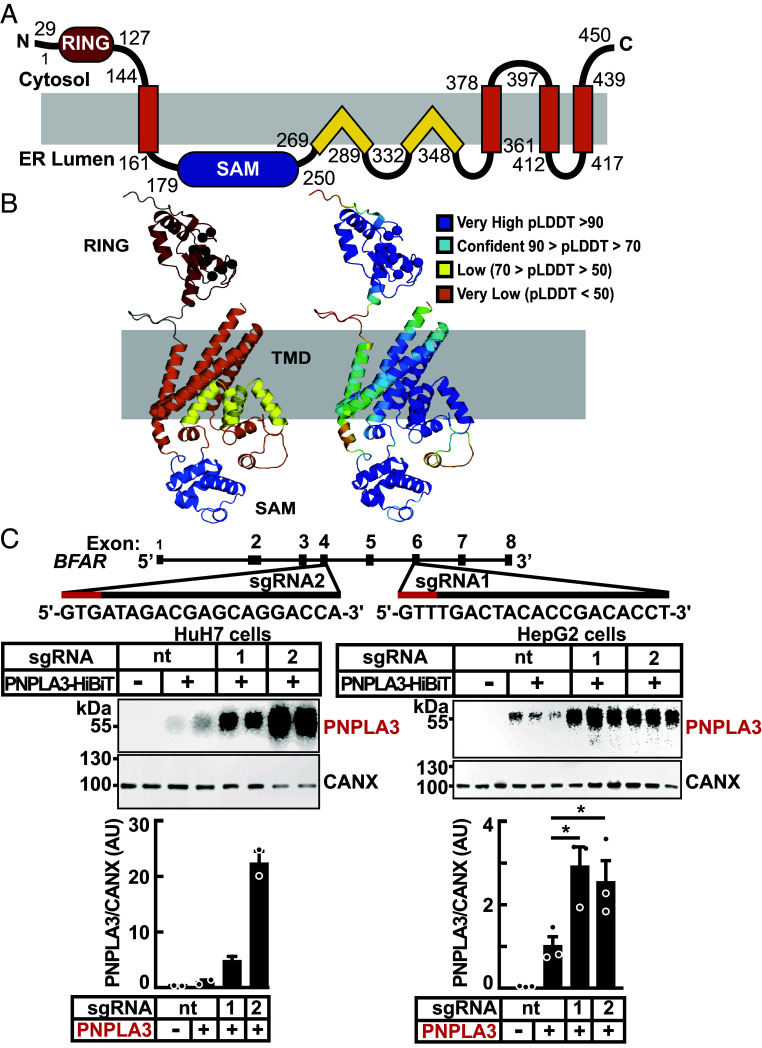
BFAR is an ER-associated protein that has shown in cultured 293 T and Hela cells to promote the degradation of the ER protein Bax inhibitor 1 (BI-1) (28). A linear schematic of the proposed BFAR domain structure is depicted in SI Appendix, Fig. S2A (SI Appendix). The protein has an N-terminal RING domain, which transfers ubiquitin from an E2 ligase to a target substrate, followed by a Sterile Alpha Motif (SAM) (39), a domain that mediates dimerization in other proteins (40). Following the SAM domain, there is a region that is reported to share some features of Death Effector Domains (DED) (28, 37).
To glean further insights into the structure of BFAR, we predicted the positions of transmembrane helices (TMHs) using a suite of programs available on Consensus Constrained TOPology (CCTOP) web server (41) (SI Appendix, Fig. S2B) (37). All nine algorithms predicted two TMHs: TMH1 and TMH2. We then inspected the structure model from AlphaFold (SI Appendix, Fig. S2C) (42–46). Here, the predicted TMHs include TMH1 and TMH2, as well as the split consensus TMHd/e, which forms TMH3 and TMH4 in the structural model.
A schematic of the AlphaFold model is shown in Fig. 3 A and B. TMH1 topology places the N-terminal RING domain on the opposite side of the membrane from the SAM domain. In our modeling, the tertiary structure of the C terminus does not adopt a death domain fold; thus, we do not include a DED in our model. Instead, the sequences in this region form two pairs of re-entrant helices that enter and exit the membrane from the same side. These re-entrant helices interact with each other and with residues from TMH2 with very high confidence according to Local Distance Difference Test (LDDT) estimates from the AlphaFold model (42). The re-entrant helices also interact with residues from TMH3 and TMH4. This model was used to design recombinant forms of BFAR for use in subsequent experiments.
Fig. 3.
Domain organization, structural model, and inactivation of BFAR in cultured hepatocytes. (A) Schematic of the human BFAR domain structure model is colored: RING (red), sterile alpha motif (SAM) (blue), and transmembrane domains with TMH (orange) and re-entrant TM helices (yellow). (B) AlphaFold structure model of BFAR colored and labeled by domains as in (A), with zinc coordinating residues from the ring domain indicated (black sphere for the C-alpha position) and according to pLDDT confidence (Right, scale in legend) colored from very high (blue) to very low (orange). (C) Inactivation of BFAR using CRISPR/Cas9 in cultured hepatocytes results in increased levels of PNPLA3. Schematic of single guide (sg)RNAs used to inactivate BFAR is shown (Top). Two sgRNAs targeting BFAR exons 4 (sgRNA2) and 6 (sgRNA1) were used to introduce frameshift mutations that prematurely terminate the protein at residues 209 and 361, respectively. Red lines denote the Protospacer Adjacent Motif (PAM) sequence. HuH7 cells were transduced with LentiCRISPRv2 containing sgRNA1, sgRNA2, or with a sgRNA with no cognate sequence in the human genome (nontargeting, nt) and cell lines were established from single clones (Materials and Methods). The BFAR−/− HuH7 and HepG2 cells were transfected with PNPLA3-HiBiT (2.5 ng/well) and after 48 h, PNPLA3 levels were measured in cell extracts using the NanoBiT blotting assay. Luminescent signals were quantified using LI-COR, normalized to CANX levels and expressed as arbitrary units (AUs). Values represent means ± SEM. Levels were compared using a Student’s t test. *P < 0.05. The experiments were repeated twice and the results were similar.
Genetic Inactivation of BFAR in Cultured Hepatocytes Increases PNPLA3 Levels.
To generate lines of cells in which expression of BFAR is completely and stably inactivated, we used CRISPR-Cas9 technology. Single guide (sg) RNAs complementary to sequences in exons 4 and 6 of BFAR (Fig. 3 B, Top) were introduced into HuH7 and HepG2 cells. As a control for the experiment, we treated cells with a sgRNA that does not target any human or mouse gene [nontargeting (nt) sgRNA]; clones were propagated, and the inactivating mutations were confirmed by Sanger sequencing. PNPLA3-HiBiT was expressed in the cells. In the cells in which BFAR had been inactivated, the levels of PNPLA3-HiBiT were increased fivefold to 20-fold when compared to cells treated with the nt sgRNA (Fig. 3C).
Overexpression of BFAR Decreases PNPLA3 Levels in Cultured Hepatocytes.
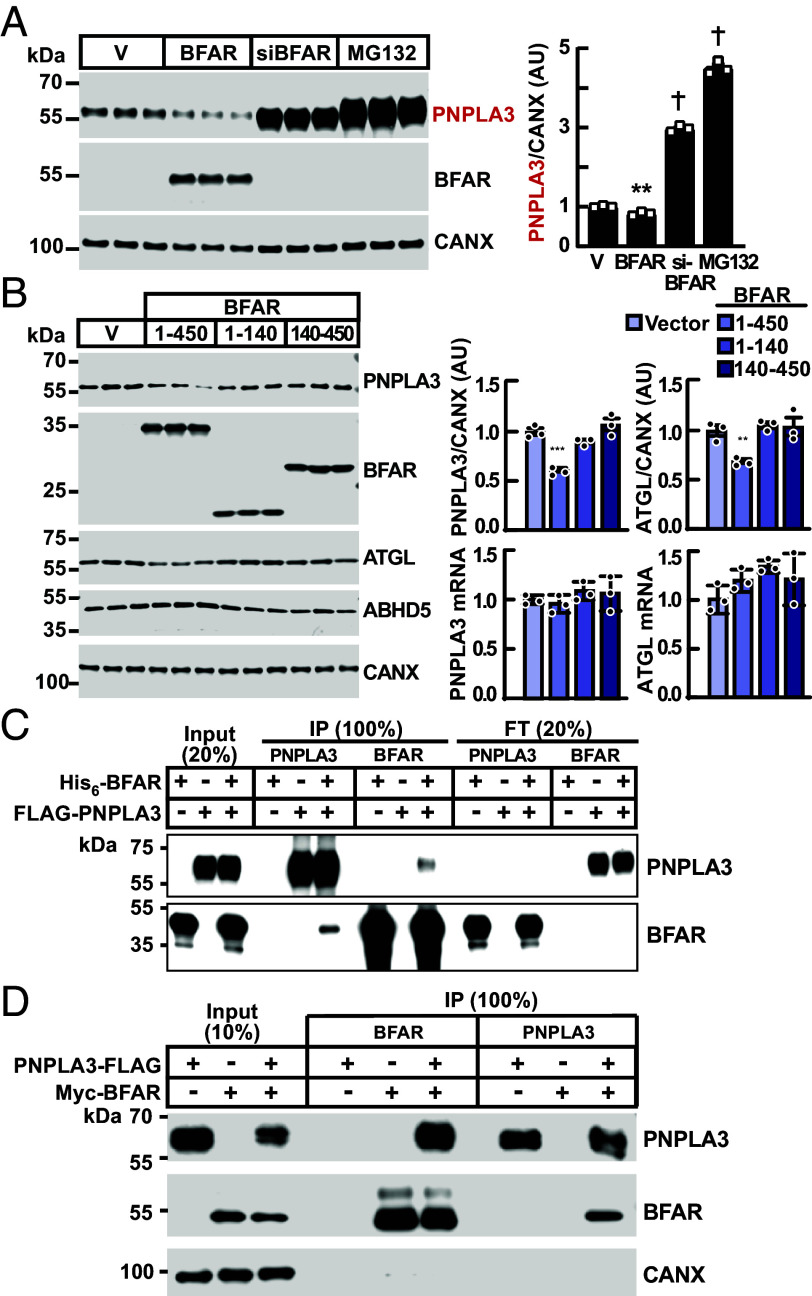
If BFAR directly promotes proteasome-mediated degradation of PNPLA3, it would be expected that overexpression of BFAR would reduce PNPLA3 levels in cells. Expression of epitope-tagged BFAR in cells stably expressing PNPLA3-HiBiT resulted in a 20% reduction in PNPLA3 protein levels when compared to cells expressing vector alone (Fig. 4A). In the same experiment, siRNA-mediated inactivation of BFAR, or treatment of cells with MG132, both increased PNPLA3 levels (Fig. 4A), as seen in previous experiments (Fig. 2). We were unsuccessful in obtaining stable expression of BFAR that contained an inactive RING domain.
Fig. 4.
BFAR expression reduces PNPLA3 protein levels in cultured hepatocytes and interacts with PNPLA3 in vitro and in cells. (A) BFAR overexpression reduces levels of PNPLA3 in HuH7 cells. HuH7 cells stably expressing PNPLA3-HiBiT were transfected with empty vector or a plasmid expressing BFAR (with an N-terminal Myc tag) or with an anti-BFAR siRNA for 48 h. Cells were also treated with MG132 (10 µM) for 16 h. Immunoblot analysis was performed as described in Materials and Methods. (B) HuH7 cells were transfected with pCMV3 plasmids expressing empty vector (V), full-length BFAR (1 to 450), N-terminal BFAR (1 to 140), and C-terminal BFAR (140 to 450), all containing an N-terminal Myc tag. After 48 h, immunoblot analysis was performed, and the signals were quantified using LI-COR and normalized to levels of CANX. The mRNA levels of indicated genes were measured and normalized as described in Materials and Methods. Values represent means ± SEM and all comparisons were with cells treated with empty vector using the Student’s t test. (C) Recombinant His6-BFAR (0.14 nmol) and FLAG-PNPLA3 (0.10 nmol) were incubated in TBST buffer (500 µL) at room temperature for 1 h. The mixture was immunoprecipitated using beads linked with anti-FLAG antibody (M2) or nickel (NTA). Input, flowthrough (both 20% of total), and immunoprecipitates (100%) were subjected to immunoblotting. A small fraction (3%) of Flag-PNPLA3 co-immunoprecipitated with His6-BFAR (and vice versa). (D) PNPLA3-3XFLAG and Myc-BFAR were co-transfected into HuH7 cells in 100 mm dishes. After 48 h, cells were harvested and lysates were immunoprecipitated using FLAG or Myc magnetic beads (50 µL), respectively. **P < 0.01, ***P < 0.001, †P < 0.0001. The experiments were repeated twice, and the results were similar.
Next, we repeated the experiment to examine the effect of BFAR expression on levels of endogenous PNPLA3. In this experiment, we also expressed peptide fragments of BFAR: an N-terminal peptide corresponding to residues 1 to 140 of human BFAR, which includes the RING domain and terminates prior to the TMH1 (Fig. 3A) and a C-terminal peptide (residues 140 to 450), which excludes the RING domain. Expression of full-length BFAR (1 to 450) was associated with a 36% reduction in endogenous immunodetectable PNPLA3, whereas expression of either the N-terminal or C-terminal peptides was not associated with changes in PNPLA3 protein or mRNA levels (Fig. 4B). We observed no change in ABHD5 and a modest reduction in ATGL protein levels with expression of full-length BFAR; mRNA levels of both ABHD5 and ATGL were not changed by BFAR expression (SI Appendix, Fig. S3). Taken together, these data provide evidence that BFAR may promote ATGL degradation, though to a lesser degree than in PNPLA3.
Physical Association of BFAR and PNPLA3 In Vitro and in Cultured Cells.
Next, we tested for physical interaction between the two proteins in vitro. Full-length His6-BFAR was poorly expressed in Escherichia coli; therefore, we generated a series of constructs with C-terminal truncations. Of these, only the peptide fragment corresponding to BFAR residues 1 to 331, which terminates just prior to the second re-entrant helix (Fig. 3A) yielded large quantities of soluble, purified protein. The truncated BFAR(1-331) and full-length PNPLA3 (0.14 and 0.10 nmol, respectively) were co-incubated overnight and then subjected to immunoprecipitation. Only a small proportion of both proteins (2.9% of PNPLA3 and 3.1% of BFAR) co-immunoprecipitated (Fig. 4C). Thus, PNPLA3 and BFAR can physically interact, albeit modestly under these conditions. The interaction may require additional components that were not present in our purified system. It is also possible that the conditions of the experiment failed to simulate adequately those present in cells.
To further test for interaction between the two proteins in a more physiologically relevant setting, we performed co-immunoprecipitation experiments in HuH7 cells transiently transfected with epitope-tagged BFAR (Myc-BFAR) and PNPLA3 (PNPLA3-Flag). After 48 h, we used epitope-specific antibodies to immunoprecipitate the targeted proteins from cell lysates. The level of PNPLA3-Flag was reduced in cells expressing BFAR when compared to cells expressing an empty vector. This result is consistent with our observation that overexpression of BFAR reduces levels of endogenous PNPLA3 (Fig. 4B). Immunoprecipitation of BFAR resulted in robust co-immunoprecipitation of PNPLA3. Essentially, all the PNPLA3 in the lysate co-immunoprecipitated with BFAR when compared to the ratio of PNPLA3/BFAR in input samples. In the converse experiment, an estimated 27% of the BFAR co-immunoprecipitated with the PNPLA3.
In Vitro Ubiquitylation of PNPLA3 by BFAR.
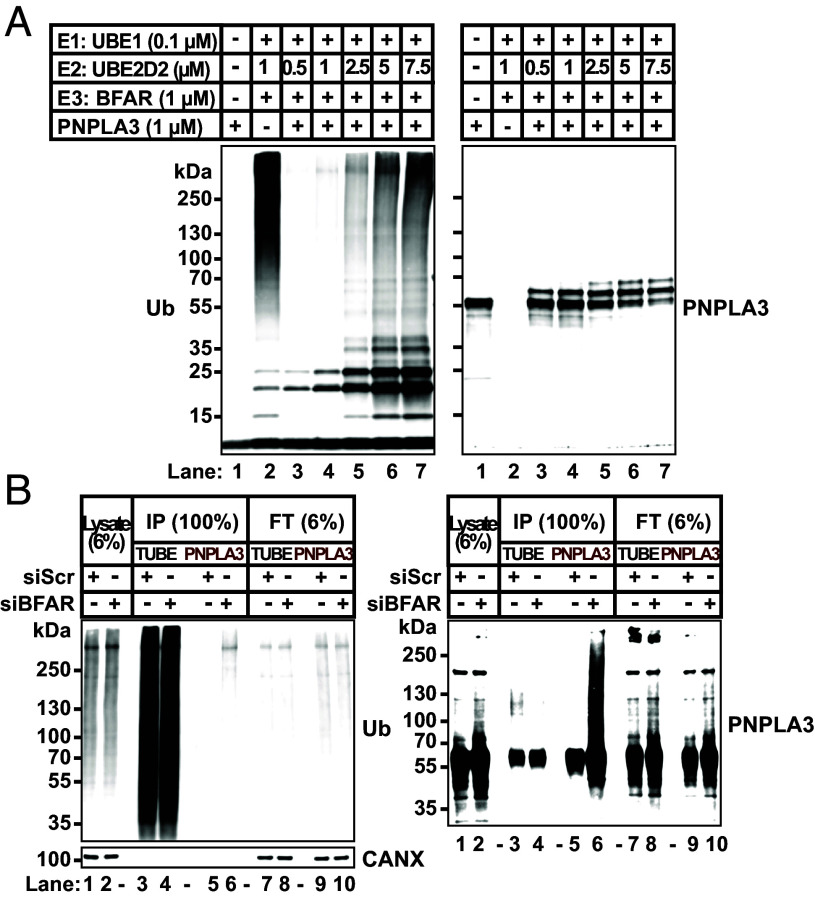
To test whether BFAR can ubiquitylate PNPLA3 directly in vitro, we reconstituted a minimal ubiquitylation reaction using purified recombinant proteins, including ubiquitin, an E1 ubiquitin-activating enzyme (E1), E2 ubiquitin conjugating enzyme (E2), and BFAR (1 to 331 aa). We used UBE2D2 in the assay since it was previously reported that UBE2D2 is the preferred E2 ubiquitin-conjugating enzyme for BFAR (28). When all components except PNPLA3 were incubated together, a dark smear of immunoreactive material migrating with a MW >55 kDa was seen after immunoblotting with an anti-ubiquitin (Ub) antibody (Fig. 5 A, lane 2, Left). The predicted MW of BFAR(1 to 331) is 36 kDa, and we presume that these higher MW forms are ubiquitylated forms of BFAR, a protein previously shown to undergo autoubiquitylation (28). Interestingly, the auto-ubiquitylation of BFAR was dramatically reduced with the addition of PNPLA3 (Fig. 5A, lanes 3); and a band of the expected size of E2 (~15 kDa) was seen. Progressively increasing the amount of E2 to the reaction resulted in a progressive increase in the intensity of higher molecular bands in the blot (lanes 4 and 5). These bands may represent mono-ubiquitylated (~20 kDa) and di-ubiquitylated (~25 kDa) forms of the E2 ligase. When we added even more E2, a smear of higher MW proteins was seen, possibly representing further polyubiquitylated forms of E2 or BFAR (Fig. 5A, lanes 5 to 7).
Fig. 5.
BFAR monoubiquitylates PNPLA3 in vitro. (A) In vitro reconstitution of BFAR-mediated ubiquitylation of PNPLA3. Proteins were incubated in E3 ligase buffer in 50 µL to reconstitute the ubiquitin conjugation reaction as described in Materials and Methods. Immunoblotting was performed on 15% of the volume of each reaction using mAbs to ubiquitin (Left panel) or PNPLA3 (11C5) (Right panel). (B) Inactivation of BFAR in HuH7 cells stably expressing PNPLA3-HiBiT. Cells were treated with a siRNA targeting BFAR or a scrambled sequence (Scr) as described in Materials and Methods. After 32 h, cells were treated with MG132 (10 µM) for 16 h. A total of 1.5 mg of cell lysate was used to immunoprecipitate the ubiquitylated proteins using TUBE beads or to immunoprecipitate PNPLA3 using HiBiT affinity beads. Immunoblotting analysis was performed on 6% of the cell lysates and flow through (22.5 µg), and 100% of the immunoprecipitate (375 µg/lane) using anti-Ub mAb (Left) and anti-PNPLA3 mAb (11C5) (Right). The experiments were repeated three times, and the results were similar.
Next, we examined whether BFAR was associated with a change in the size of PNPLA3. Purified recombinant PNPLA3 migrated at 55 kDa (Fig. 5 A, lane 1, Right). Upon addition of E1, E2, and BFAR to PNPLA3, we saw an additional band that was 8 kDa larger than BFAR: the size of one ubiquitin molecule. When we increased the concentration of E2 (from 0.5 µM to 7.5 µM) in the reaction, the migration of PNPLA3 changed: New bands appeared, corresponding to the expected sizes of mono- (+8 kDa) and di-ubiquitylated PNPLA3 (+16 kDa) (Fig. 5 A, Right). The appearance of these higher MW bands was associated with a reduction in unmodified PNPLA3 (55 kDa). We cannot exclude the possibility that the larger bands correspond to two distinct mono-ubiquitylation events on PNPLA3. We observed migration changes of PNPLA3 only after all components of the ubiquitylation pathway were present: E1, E2, and BFAR (SI Appendix, Fig. S4A, Lane 5 vs. Lanes 1 to 4). We tested the effect of titrating BFAR and E2 on PNPLA3 ubiquitylation (SI Appendix, Fig. S4B). Increasing concentration of BFAR (0.4 to 2.5 µM) did not increase ubiquitination of PNPLA3 (Lanes 4 to 6). However, increasing concentration of E2 (1 to 5 µM) was associated with an increase in ubiquitination of PNPLA3 (Lanes 7 and 8 vs. Lanes 4 and 5), suggesting that E2 activity, not BFAR–PNPLA3 interactions, is the limiting factor in PNPLA3 ubiquitylation.
To examine the type of ubiquitin chain added to PNPLA3, we included ubiquitin that was either lysine-free (K0) or had only a single lysine (either K11, K48, or K63) (SI Appendix, Fig. S4C). Surprisingly, we found no differences in migration of PNPLA3 among the reactions. These data are consistent with the notion that the PNPLA3 bands of higher molecular weight are products of multiple mono-ubiquitination reactions (SI Appendix, Fig. S4C).
Taken together, these in vitro assays confirmed that BFAR can serve as an E3 ubiquitin ligase for mono-ubiquitylation of PNPLA3. We found no evidence for further elongation of the ubiquitin chains on PNPLA3. The smear in Ub-immunoblotting was likely due to polyubiquitylation of BFAR and the E2 protein (SI Appendix, Fig. S4 C, Lower). It is possible that the truncated form of BFAR used in the experiment has reduced E3 ligase activity or that the purification of the protein was associated with a loss of activity. It is also possible that UBE2D2 is a priming E2 that transfers only a single Ub to its substrate, or other E3s are required for elongation of ubiquitin chain on PNPLA3.
Ubiquitylation of PNPLA3 by BFAR in Cultured Cells.
Next, we tested if BFAR promotes ubiquitylation of PNPLA3 in cultured cells. For these experiments, we used Tandem Ubiquitin Binding Entities (TUBEs), which bind to polyubiquitin chains on proteins (47). HuH7 cells stably expressing PNPLA3-HiBiT were treated with either a scrambled siRNA (siScr) or a siRNA targeting BFAR. The cells were treated with MG132 (10 μM) for 16 h to inhibit proteasome-mediated degradation of the ubiquitylated proteins. Proteins were immunoprecipitated using TUBEs (Fig. 5B, lanes 3 and 4) or HiBiT-affinity beads (Fig. 5B, lanes 5 and 6) and subjected to immunoblotting with antibodies recognizing Ub or to PNPLA3, respectively. When blotted with an anti-Ub antibody, a smear of proteins was present in the lysates (lanes 1 and 2) and TUBE immunoprecipitates (lanes 3 and 4) in the absence or presence of BFAR (Fig. 5 B, Left). Therefore, knockdown of BFAR did not cause a general reduction in protein ubiquitylation.
When the TUBE immunoprecipitates were immunoblotted for PNPLA3, a major band of the expected size of PNPLA3, and a faint smear of bands with an apparent size of ~125 kDa was seen in cells that received the siScr but not in cells receiving siBFAR (Fig. 5 B, Right, lane 3 vs. lane 4). This smear may correspond to polyubiquitylated PNPLA3. When PNPLA3 was immunoprecipitated with anti-HiBiT antibody and then immunoblotted with an anti-PNPLA3 antibody, a large smear was seen in cells receiving the BFAR-targeting siRNA (Fig. 5 B, Right, lane 6), presumably due to the 2.5-fold increase in PNPLA3 in these cells. Alternatively, the smear of high molecular weight material detected by anti-PNPLA3 antibodies may be an artifact.
Inactivation of Bfar in Mice Is Associated with Increased Hepatic PNPLA3.
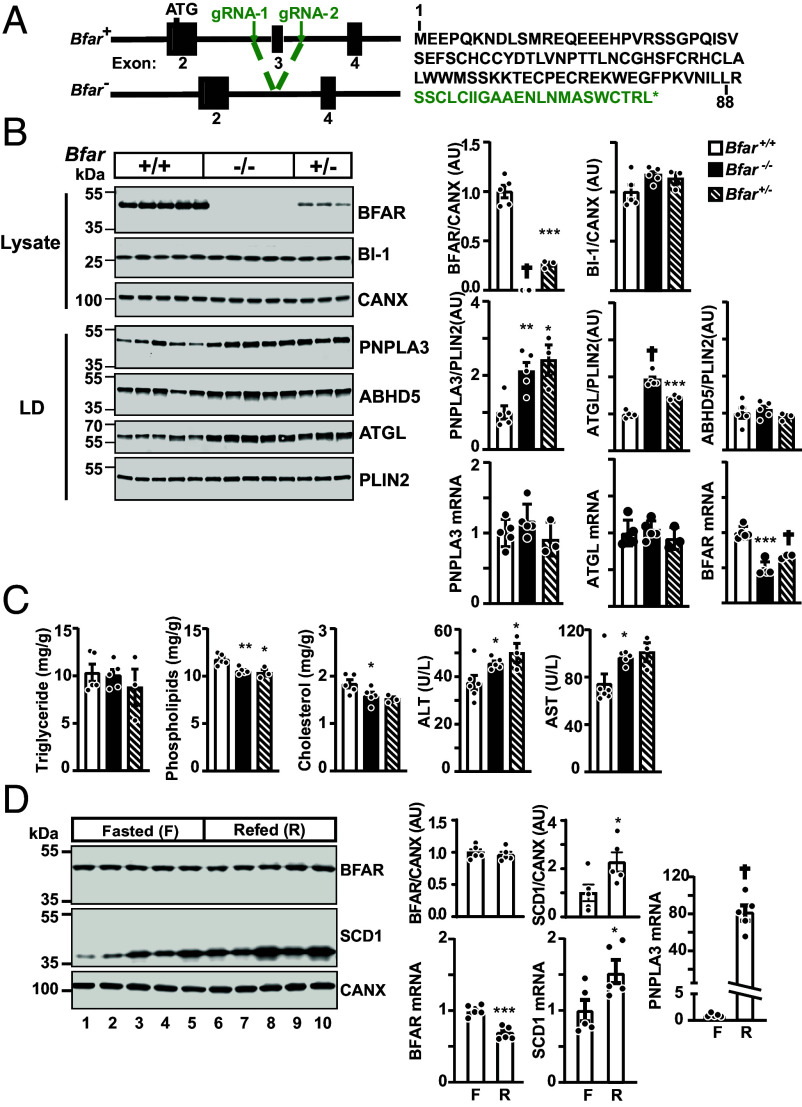
To determine the in vivo relevance of BFAR in PNPLA3 turnover, we used CRISPR/Cas9 to inactivate Bfar in mice (SI Appendix, Methods). Two guide RNAs were designed to delete exon 3 of Bfar (Fig. 6A and SI Appendix, Fig. S5A). The deletion was confirmed by sequencing the genomic DNA of the mice. Exon 2 was spliced to exon 4 of BFAR, resulting in a frameshift at amino acid 88 in the cytoplasmic RING domain of the protein. An additional 22 residues is predicted to be added prior to a premature stop codon (Fig. 6 A, Right). Two independent lines of Bfar knockout mice (with deletions of 794 bp and 773 bp, respectively) were used for further studies, and both produced identical results. F0 founders were crossed with C57BL/6 J mice to obtain Bfar+/− mice. For all animal experiments, Bfar+/+ and Bfar−/− mice produced from intercrosses of Bfar+/− mice were used. The Bfar−/− mice were healthy and no differences between Bfar+/+ and Bfar−/− offspring were apparent. The mice were born in the expected Mendelian ratios (SI Appendix, Table S4). Routine histology of the liver, kidney, and brain in Bfar−/− mice and Bfar+/+ mice (n = 5/group, 8- to 10-wk males) revealed no differences. In chow-fed mice, no differences in body- or liver weights of WT and knockout mice were apparent (SI Appendix, Fig. S5B).
Fig. 6.
PNPLA3 levels are increased in livers of Bfar−/− mice. (A) Generation of Bfar−/− by CRISPR-Cas9 technology. Two guide RNAs (gRNA-1 and gRNA-2) flanking exon 3 (205 nt) were used to delete a ~800 bp region that included exon 3. Sequencing data confirmed that exon 2 is spliced to exon 4, causing a frameshift and a premature stop codon. The predicted sequence of the truncated BFAR contains the first 88 amino acids plus 22 unrelated residues (green). (B) PNPLA3 protein levels are increased in hepatic LDs of Bfar−/− mice. WT male mice, Bfar+−, and Bfar−/− mice (8 wk old) were fed a HSD for 3 wk. After three cycles of fasting and refeeding, mice were killed 6 h after the last refeeding and immunoblotting was performed on liver lysates (20 µg) and LDs (1 µg). BFAR was immunoblotted using a mouse mAb (40H7) against a recombinant fragment of BFAR (1 to 140 aa) that was developed in-house (Materials and Methods). Signals were quantified using LI-COR and normalized to levels of PLIN2 or CANX, respectively. (C) Inactivation of BFAR was not associated with changes in hepatic TG level. Lipids were extracted from livers (∼100 mg) and measured using enzymatic assays (Wako, Richmond, VA). ALT, alanine aminotransferase; AST, aspartate aminotransferase. (D) No change in BFAR protein levels in livers of fasted and refed mice. Livers were collected from chow-fed C57BL/6 J mice (10-wk-old males, five/group) that were fasted 18 h (Fasted) and then refed 6 h (Refed). The results of immunoblot analysis are shown. RT-PCR was used to quantify relative amounts of mRNA with the level in wild-type mice set to 1. Values represent means ± SEM. Levels were compared using a Student’s t test. *P < 0.05, **P < 0.01, ***P < 0.001, †P < 0.0001. The experiments were repeated twice, and the results were similar.
To facilitate detection of endogenous BFAR, we developed monoclonal anti-mouse BFAR antibodies using a recombinant peptide from BFAR (1-140) as the immunogen (SI Appendix, Materials). Immunoblot analysis of liver lysates from offspring of Bfar+/− mice is shown in Fig. 6B. A band of the expected molecular weight (48 kDa) was seen in WT animals that was absent in the Bfar−/− mice. The levels of immunodetectable BFAR were reduced by 74% in Bfar+/− mice. It is unclear why the Bfar+/− mice have a greater than 50% reduction in BFAR, but this finding was consistent.
As mentioned previously, BFAR has been implicated in the regulation of ER stress by promoting degradation of BI-1, a protein that interacts with IRE-1 (Inositol-Requiring Enzyme type 1) (28). All experiments supporting this activity were performed by expressing BFAR in either 293 T cells or HeLa cells (28). We failed to see any changes in BI-1 level in the livers of Bfar−/− mice. This discrepancy may be due to cell type- or species-specific differences in the levels of a protein (or other factors) required for BFAR-mediated degradation of BI-1.
Immunoblot analysis of LDs isolated from livers of mice revealed twofold higher levels of PNPLA3 protein in the Bfar−/− mice than in WT animals (Fig. 6B) with no associated changes in PNPLA3 mRNA levels. Similar increases in PNPLA3 protein levels without changes in its PNPLA3 mRNA levels were seen in the Bfar+/− mice. No significant differences in levels of ABHD5 protein (Fig. 6 B, Right) or mRNA (SI Appendix, Fig. S6A) were found in the Bfar−/− mice. Similar to the changes seen previously in ATGL protein and mRNA levels in our cell culture studies, we found that the Bfar−/− mice had increased levels of hepatic ATGL protein with no change in the corresponding mRNA levels.
Previously, we showed that the high levels of PNPLA3 protein associated with PNPLA3(148M) were related to the accumulation of hepatic TG (26). Therefore, we examined the effect of the increase in the PNPLA3 level on the hepatic TG level. We failed to see any increase in hepatic TG in the Bfar−/−mice (Fig. 6C). A modest reduction in hepatic cholesterol and phospholipids was seen without any changes in plasma lipid levels (SI Appendix, Fig. S6B). The liver enzymes were significantly increased in the knockout mice (Fig. 6C) although we did not appreciate any changes in routine liver histology. We are currently crossing mice to establish Bfar−/−mice that express PNPLA3(148M) to determine whether inactivation of the protein exaggerates the phenotype of these mice.
Hepatic BFAR Levels Do Not Change with Food Intake in Mice.
In WT mice, levels of PNPLA3 mRNA and protein are increased by sucrose feeding (30). To determine whether BFAR expression is also regulated by feeding, we measured levels of BFAR protein and mRNA in livers of fasted (18 h) and refed (6 h) mice (Fig. 6D). No differences in BFAR protein levels were seen in liver lysates from the fasted and fed mice, though BFAR mRNA did decrease slightly in the refed state. In this experiment, stearoyl CoA desaturase (SCD) served as a positive control.
Discussion
The major finding of this study is that genetic manipulation of BFAR, an E3 ubiquitin ligase, leads to changes in levels of PNPLA3 protein in cultured cells and in vivo in the livers of mice. Reducing BFAR expression, either by siRNA or CRISPR/Cas9, increased PNPLA3 levels in two different human hepatocytes cell lines (Figs. 1E, 2 A and B, and 3 B and C) and in the livers of Bfar−/− mice (Fig. 6B). Conversely, over-expression of BFAR was associated with a reduction in PNPLA3 protein, but not mRNA, in cultured hepatocytes (Fig. 4 A and B). These observations are consistent with the PNPLA3-modulating effect of BFAR not being a species-specific phenomenon, nor is it an artifact of cell culture or protein overexpression. Pulldown assays using purified, epitope-tagged recombinant isoforms of PNPLA3 and BFAR (1 to 331) showed a weak, but detectable, interaction between the two proteins (Fig. 4C). A much more robust co-immunoprecipitation of the two proteins was observed in cells expressing epitope-tagged constructs of both proteins (Fig. 4D). These data are consistent with a model in which BFAR ubiquitylates PNPLA3 and targets it for proteasome-mediated degradation.
Ubiquitylation assays using purified constituents showed that BFAR can promote monoubiquitylation, but we found no evidence for polyubiquitylation of PNPLA3 by BFAR in these experiments (Fig. 5B). We do not know whether our failure to demonstrate BFAR-mediated polyubiquitylation of PNPLA3 reflects the biological role of BFAR in vivo, or a limitation of our purified system. Recent studies have shown that some E3 ligases act primarily to transfer the initial ubiquitin onto the target substrate, while others act to extend the chain with additional ubiquitin moieties (48). The consistent increase in PNPLA3 levels in livers of Bfar−/− mice indicates that BFAR is essential for normal turnover and maintenance of hepatic PNPLA3 levels.
BFAR was the only E3 ubiquitin ligase from our screen that was consistently associated with increased PNPLA3 levels. In these experiments, MG132 was used as a positive control for inhibition of proteasome-mediated degradation of PNPLA3. PNPLA3 levels in cells treated with siRNAs directed against BFAR were ~30% lower than cells treated with MG132 (Fig. 2A). We inferred from these results that BFAR is the predominant E3 ligase for PNPLA3 in HuH7 cells and that the collective contributions of other, as yet unidentified E3 ligases account for ~30% of proteasome-mediated PNPLA3 degradation in these cells.
We cannot exclude the possibility that other E3 ligases target PNPLA3 for degradation in other cell types, or under different experimental conditions. The human genome encodes ~600 E3 ubiquitin ligases that mediate the proteasomal degradation of 80% of the ~20,000 proteins expressed in human cells (49). Fewer than 5% of these ligases have been characterized in detail, but if the 16,000 potential substrates are distributed equally among the E3 ligases, then each ligase would be responsible for degradation of ~25 proteins. Accordingly, it seems likely that most proteins are ubiquitylated by multiple E3 ligases, as has been observed for selected other proteins involved in lipid metabolism (50–53). For example, HMG-CoA reductase, the flux-determining enzyme in cholesterol biosynthesis, is targeted for degradation by at least three E3 ligases (GP78, HRD1, and RNF145) (50, 54, 55). Other enzymes in the cholesterol biosynthetic pathway, including SC4MOL (sterol-C4-methyloxidase-like), NSDHL [NAD(P)-dependent steroid dehydrogenase-like], and HSD17B7 (hydroxysteroid 17-beta dehydrogenase 7), are targeted for degradation by MARCHF6, a RING-CH-type finger protein (51–53).
The mechanism(s) by which E3 ligases select proteins to be targeted for degradation is not fully understood (56–58). We speculated that BFAR and PNPLA3 must interact, either directly, or indirectly through accessory proteins. In affinity capture assays performed in vitro, only a small fraction (<3%) of the two proteins co-immunoprecipitated, which is consistent with there being limited, transient direct binding of BFAR and PNPLA3. We cannot exclude the possibility that the bound state may be destabilized by the removal of residues (332 to 450) in the endogenous BFAR protein that were not present in the recombinant BFAR proteins used in this study.
It is also possible that interaction between BFAR and PNPLA3 requires additional factors that were not present in our purified system. A role for accessory proteins has been shown for a number of RING domain ligases (59, 60). For example, Deltex protein DTX3L is a RING-type E3 ligase that requires its binding partner PARP9 [Poly(ADP-) Polymerase Family Member 9] to fulfill its ubiquitylation activity in prostate cancer cells (61). For some E3 ligases, metabolites are required to coordinate E3-substrate recognition, such as in the interaction of Insig-1 and its ER-resident RING E3 ligase, GP78, which is regulated by cholesterol (62).
Co-immunoprecipitation of BFAR and PNPLA3 from cell lysates was much more effective than that obtained using purified proteins. This result is consistent with the notion that BFAR and PNPLA3 interact with high affinity in vivo and that our in vitro studies failed to capture physiologic conditions required for a stable interaction between the two proteins (Fig. 4D). Further studies will be required to identify the factors that promote the formation of a BFAR/PNPLA3 complex (Fig. 4D).
Previously, we found that PNPLA3 on LDs in hepatocytes is polyubiquitylated (25). When we attempted to reconstitute the E3 ubiquitin ligase activity of BFAR on PNPLA3 in vitro, we observed the addition of one or two ubiquitin molecules, not polyubiquitin chains (Fig. 5A). There are several possible explanations for this finding. BFAR may be restricted to ligating short ubiquitin chains to PNPLA3. Alternatively, there may be other components of the reaction that are not present in our reconstitution assay using purified proteins. Some E3 ligases require an additional elongation factor, E4, for chain elongation (63). E4s have been reported to bind to oligo-ubiquitylated substrates and catalyze multiubiquitin-chain assembly in collaboration with E1, E2, and E3 (63–65); if this is the case, BFAR would still act as the rate-limiting factor in ubiquitin-mediated degradation of PNPLA3.
Another possibility is that we have not used the preferred E2(s) in our in vitro reconstitution. Some RING-type E3s catalyze ubiquitination in collaboration with specific priming and elongation E2 enzymes. A well-studied example is Doa10 (E3) in yeast, which employs Ubc6 (priming E2) and Ubc7 (elongation E2) to add the initial ubiquitin and the subsequent ubiquitin chain, respectively (48). Identification of physiological E2(s) for BFAR will require additional studies.
PNPLA3 expression is exquisitely regulated in response to food intake via a feed-forward loop that coordinates transcriptional and post-translational regulation of the protein (30). The ratio of PNPLA3 protein to mRNA levels varies widely during the hours following refeeding, indicating that levels of PNPLA3 protein are strongly influenced by factors beyond mRNA levels (25). To determine whether changes in BFAR levels contributed to the post-translational regulation of PNPLA3, we examined the effect of feeding on BFAR levels. We found no evidence to support the notion that the post-translational increase in PNPLA3 levels seen in the fed state is due to a reduction in BFAR levels. Our results are more consistent with a model in which BFAR is constitutively active, maintaining a high rate of ubiquitylation and degradation of PNPLA3, thus promoting rapid changes in the level of the protein in response to nutritional changes.
A central tenet of our model of PNPLA3 pathogenicity is that the 148M isoform, which evades proteasomal degradation and accumulates on LDs where it interferes with TG hydrolysis, promoting fatty liver (26, 27). Consistent with this scenario is our finding that only a small fraction of PNPLA3(148M) localized on LDs is ubiquitylated (25, 26), possibly because the I148M substitution disrupts the PNPLA3 motif that is recognized by BFAR. To date, we have only tested the activity of BFAR against PNPLA3(148M) in cultured cells and could not distinguish any differences between its effect on the WT and 148M isoforms. When we inactivated BFAR in HuH7 cells, which express only PNPLA3(148M), the resulting increase in PNPLA3 levels was similar to that observed with PNPLA3(WT) (Fig. 2 A and B). It is possible that the factors required to distinguish the two isoforms of PNPLA3 are not maintained in cultured cells. Alternatively, PNPLA3(148M) may associate with LDs in the liver in a manner that differs from what is seen in cultured cells. We are currently crossing the Bfar−/− mice with PNPLA3(148M) knockin mice to determine the effect of inactivation of BFAR on PNPLA3(148M) levels. Studies are underway to determine whether we can reverse the steatosis in livers of Pnpla3148M/M mice by manipulating BFAR expression.
Materials and Methods
Reagents, cell lines and knockouts, plasmids, RNAi screening, lentiviral-mediated transduction of PNPLA3-mCherry and PNPLA3-HiBiT cDNA, NanoBiT Reconstitution Assay, in vitro Ubiquitin Conjugation Reaction, immunoprecipitation, immunoblot analysis, and reproducibility are described in detail in SI Appendix.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Drs. Deepak Nijhawan, Russell Debose Boyd, and George DeMartino for their input and suggestions; Hannah Hudson, Christina Zhao, Tommy Hyatt, and Lisa Beatty for their experimental support; J.C.C. and H.H.H. were supported by grants from the NIH (RO1-DK 090066 and 5PO1 HL160487).
Author contributions
H.C., L.N.K., H.H.H., and J.C.C. designed research; A.D., H.C., Y.W., L.N.K, and S.H. performed research; H.C., L.N.K., G.L., and S.H. contributed new reagents/analytic tools; A.D., H.C., Y.W., L.N.K., G.L., S.H., H.H.H., and J.C.C. analyzed data; and A.D., Y.W., L.N.K., H.H.H., and J.C.C. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
Reviewers: A.J.B., University of New South Wales Sydney; and P.E., Johns Hopkins University School of Medicine.
Contributor Information
Helen H. Hobbs, Email: helen.hobbs@utsouthwestern.edu.
Jonathan C. Cohen, Email: jonathan.cohen@utsouthwestern.edu.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Ludwig J., Viggiano T. R., McGill D. B., Oh B. J., Nonalcoholic steatohepatitis: Mayo Clinic experiences with a hitherto unnamed disease. Mayo Clin. Proc. 55, 434–438 (1980). [PubMed] [Google Scholar]
- 2.Cohen J. C., Horton J. D., Hobbs H. H., Human fatty liver disease: Old questions and new insights. Science 332, 1519–1523 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loomba R., et al. , Heritability of hepatic fibrosis and steatosis based on a prospective twin study. Gastroenterology 149, 1784–1793 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwimmer J. B., et al. , Heritability of nonalcoholic fatty liver disease. Gastroenterology 136, 1585–1592 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tarnoki A. D., et al. , Heritability of non-alcoholic fatty liver disease and association with abnormal vascular parameters: A twin study. Liver Int. 32, 1287–1293 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Romeo S., et al. , Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 40, 1461–1465 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuan X., et al. , Population-based genome-wide association studies reveal six loci influencing plasma levels of liver enzymes. Am. J. Hum. Genet. 83, 520–528 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sookoian S., et al. , A nonsynonymous gene variant in the adiponutrin gene is associated with nonalcoholic fatty liver disease severity. J. Lipid Res. 50, 2111–2116 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Speliotes E. K., Butler J. L., Palmer C. D., Voight B. F., Hirschhorn J. N., PNPLA3 variants specifically confer increased risk for histologic nonalcoholic fatty liver disease but not metabolic disease. Hepatology 52, 904–912 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valenti L., et al. , Homozygosity for the patatin-like phospholipase-3/adiponutrin I148M polymorphism influences liver fibrosis in patients with nonalcoholic fatty liver disease. Hepatology 51, 1209–1217 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Valenti L., Dongiovanni P., Ginanni Corradini S., Burza M. A., Romeo S., PNPLA3 I148M variant and hepatocellular carcinoma: A common genetic variant for a rare disease. Dig. Liver Dis.: Offi. J Italian Soc. Gastroenterol. Italian Assoc. Study of the Liver 45, 619–624 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Trepo E., et al. , Common polymorphism in the PNPLA3/adiponutrin gene confers higher risk of cirrhosis and liver damage in alcoholic liver disease. J. Hepatol. 55, 906–912 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Tian C., Stokowski R. P., Kershenobich D., Ballinger D. G., Hinds D. A., Variant in PNPLA3 is associated with alcoholic liver disease. Nat. Genet 42, 21–23 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Speliotes E. K., et al. , Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 7, e1001324 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kozlitina J., et al. , Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 46, 352–356 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buch S., et al. , A genome-wide association study confirms PNPLA3 and identifies TM6SF2 and MBOAT7 as risk loci for alcohol-related cirrhosis. Nat. Genet. 47, 1443–1448 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Zimmerman R., et al. , Fat mobolization in adipose tissue is promoted by adipose triglyceride lipase. Science 306, 1383–1386 (2004). [DOI] [PubMed] [Google Scholar]
- 18.He S., et al. , A sequence variation (I148M) in PNPLA3 associated with nonalcoholic fatty liver disease disrupts triglyceride hydrolysis. J. Biol. Chem. 285, 6706–6715 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown J. M., et al. , CGI-58 facilitates the mobilization of cytoplasmic triglyceride for lipoprotein secretion in hepatoma cells. J. Lipid Res. 48, 2295–2305 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Fischer J., et al. , The gene encoding adipose triglyceride lipase (PNPLA2) is mutated in neutral lipid storage disease with myopathy. Nat. Genet. 39, 28–30 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Basantani M. K., et al. , Pnpla3/Adiponutrin deficiency in mice does not contribute to fatty liver disease or metabolic syndrome. J. Lipid Res. 52, 318–329 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen W., Chang B., Li L., Chan L., Patatin-like phospholipase domain-containing 3/adiponutrin deficiency in mice is not associated with fatty liver disease. Hepatology 52, 1134–1142 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y., Xing C., Cohen J. C., Hobbs H. H., Genetic variant in PNPLA3 is associated with nonalcoholic fatty liver disease in China. Hepatology 55, 327–328 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smagris E., et al. , Pnpla3I148M knockin mice accumulate PNPLA3 on lipid droplets and develop hepatic steatosis. Hepatology (Baltimore, Md.) 61, 108–118 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.BasuRay S., Smagris E., Cohen J. C., Hobbs H. H., The PNPLA3 variant associated with fatty liver disease (I148M) accumulates on lipid droplets by evading ubiquitylation. Hepatology 66, 1111–1124 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.BasuRay S., Wang Y., Smagris E., Cohen J. C., Hobbs H. H., Accumulation of PNPLA3 on lipid droplets is the basis of associated hepatic steatosis. Proc. Natl. Acad. Sci. U.S.A. 116, 9521–9526 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y., Kory N., Cohen J. C., Hobbs H. H., PNPLA3, CGI-58, and inhibition of hepatic triglyceride hydrolysis in mice. Hepatology 69, 2427–2441 (2019), 10.1002/hep.30583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rong J., et al. , Bifunctional apoptosis regulator (BAR), an endoplasmic reticulum (ER)-associated E3 ubiquitin ligase, modulates BI-1 protein stability and function in ER Stress. J. Biol. Chem. 286, 1453–1463 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hall M. P., et al. , Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem. Biol. 7, 1848–1857 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang Y., et al. , A feed-forward loop amplifies nutritional regulation of PNPLA3. Proc. Natl. Acad. Sci. U.S.A. 107, 7892–7897 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petroski M. D., Deshaies R. J., Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin-RING ubiquitin-ligase complex SCF-Cdc34. Cell 123, 1107–1120 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Petroski M. D., Deshaies R. J., Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6, 9–20 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Osaka F., et al. , A new NEDD8-ligating system for cullin-4A. Genes Dev. 12, 2263–2268 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deshaies R. J., Emberley E. D., Saha A., Control of cullin-ring ubiquitin ligase activity by nedd8. Subcell Biochem. 54, 41–56 (2010). [DOI] [PubMed] [Google Scholar]
- 35.Soucy T. A., et al. , An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458, 732–736 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Abbas T., et al. , PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev. 22, 2496–2506 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang H., et al. , BAR: An apoptosis regulator at the intersection of caspases and Bcl-2 family proteins. Proc. Natl. Acad. Sci. U.S.A. 97, 2597–2602 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ampuero J., et al. , PNPLA3 rs738409 causes steatosis according to viral & IL28B genotypes in hepatitis C. Ann. Hepatol. 13, 356–363 (2014). [PubMed] [Google Scholar]
- 39.Ponting C. P., SAM: A novel motif in yeast sterile and Drosophila polyhomeotic proteins. Protein Sci. 4, 1928–1930 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Denay G., Vachon G., Dumas R., Zubieta C., Parcy F., Plant SAM-domain proteins start to reveal their roles. Trends Plant Sci. 22, 718–725 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Dobson L., Remenyi I., Tusnady G. E., CCTOP: A consensus constrained TOPology prediction web server. Nucleic Acids Res. 43, W408–412 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jumper J., et al. , Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kinch L. N., Schaeffer R. D., Kryshtafovych A., Grishin N. V., Target classification in the 14th round of the critical assessment of protein structure prediction (CASP14). Proteins 89, 1618–1632 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kinch L. N., Pei J., Kryshtafovych A., Schaeffer R. D., Grishin N. V., Topology evaluation of models for difficult targets in the 14th round of the critical assessment of protein structure prediction (CASP14). Proteins 89, 1673–1686 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schaeffer R. D., Kinch L., Kryshtafovych A., Grishin N. V., Assessment of domain interactions in the fourteenth round of the Critical Assessment of Structure Prediction (CASP14). Proteins 89, 1700–1710 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alto N. M., Orth K., Subversion of cell signaling by pathogens. Cold Spring Harb. Perspect. Biol. 4, a006114 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hjerpe R., et al. , Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Rep. 10, 1250–1258 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lips C., et al. , Who with whom: Functional coordination of E2 enzymes by RING E3 ligases during poly-ubiquitylation. Embo J. 39, e104863 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Collins G. A., Goldberg A. L., The logic of the 26S proteasome. Cell 169, 792–806 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jiang L. Y., et al. , Ring finger protein 145 (RNF145) is a ubiquitin ligase for sterol-induced degradation of HMG-CoA reductase. J. Biol. Chem. 293, 4047–4055 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scott N. A., et al. , The cholesterol synthesis enzyme lanosterol 14alpha-demethylase is post-translationally regulated by the E3 ubiquitin ligase MARCH6. Biochem. J. 477, 541–555 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prabhu A. V., Luu W., Sharpe L. J., Brown A. J., Cholesterol-mediated degradation of 7-dehydrocholesterol reductase switches the balance from cholesterol to vitamin D synthesis. J. Biol. Chem. 291, 8363–8373 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Capell-Hattam I. M., et al. , Twin enzymes, divergent control: The cholesterogenic enzymes DHCR14 and LBR are differentially regulated transcriptionally and post-translationally. J. Biol. Chem. 295, 2850–2865 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Song B. L., Javitt N. B., DeBose-Boyd R. A., Insig-mediated degradation of HMG CoA reductase stimulated by lanosterol, an intermediate in the synthesis of cholesterol. Cell Metab. 1, 179–189 (2005). [DOI] [PubMed] [Google Scholar]
- 55.Nguyen A. D., Lee S. H., DeBose-Boyd R. A., Insig-mediated, sterol-accelerated degradation of the membrane domain of hamster 3-hydroxy-3-methylglutaryl-coenzyme A reductase in insect cells. J. Biol. Chem. 284, 26778–26788 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Timms R. T., et al. , Defining E3 ligase-substrate relationships through multiplex CRISPR screening. Nat. Cell Biol. 25, 1535–1545 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cowan A. D., Ciulli A., Driving E3 ligase substrate specificity for targeted protein degradation: Lessons from nature and the laboratory. Annu. Rev. Biochem. 91, 295–319 (2022). [DOI] [PubMed] [Google Scholar]
- 58.Pinch B. J., et al. , A strategy to assess the cellular activity of E3 ligase components against neo-substrates using electrophilic probes. Cell. Ch em. Biol. 29, 57–66.e56 (2022). [DOI] [PubMed] [Google Scholar]
- 59.Karni-Schmidt O., Lokshin M., Prives C., The roles of MDM2 and MDMX in cancer. Annu. Rev. Pathol. 11, 617–644 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wauer T., Simicek M., Schubert A., Komander D., Mechanism of phospho-ubiquitin-induced PARKIN activation. Nature 524, 370–374 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vela-Rodriguez C., Lehtio L., Activities and binding partners of E3 ubiquitin ligase DTX3L and its roles in cancer. Biochem. Soc. Trans. 50, 1683–1692 (2022). [DOI] [PubMed] [Google Scholar]
- 62.Lee J. N., Song B., DeBose-Boyd R. A., Ye J., Sterol-regulated degradation of Insig-1 mediated by the membrane-bound ubiquitin ligase gp78. J. Biol. Chem. 281, 39308–39315 (2006). [DOI] [PubMed] [Google Scholar]
- 63.Koegl M., et al. , A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 96, 635–644 (1999). [DOI] [PubMed] [Google Scholar]
- 64.Hoppe T., Multiubiquitylation by E4 enzymes: “One size” doesn’t fit all. Trends Biochem. Sci. 30, 183–187 (2005). [DOI] [PubMed] [Google Scholar]
- 65.Franz A., Ackermann L., Hoppe T., Ring of change: CDC48/p97 drives protein dynamics at chromatin. Front. Genet. 7, 73 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.