Abstract
Actinobacillus actinomycetemcomitans has been shown to produce a soluble cytotoxic factor(s) distinct from leukotoxin. We have identified in A. actinomycetemcomitans Y4 a cluster of genes encoding a cytolethal distending toxin (CDT). This new member of the CDT family is similar to the CDT produced by Haemophilus ducreyi. The CDT from A. actinomycetemcomitans was produced in Escherichia coli and was able to induce cell distension, growth arrest in G2/M phase, nucleus swelling, and chromatin fragmentation in HeLa cells. The three proteins, CDTA, -B and -C, encoded by the cdt locus were all required for toxin activity. Antiserum raised against recombinant CDTC completely inhibited the cytotoxic activity of culture supernatant and cell homogenate fractions of A. actinomycetemcomitans Y4. These results strongly suggest that the CDT is responsible for the cytotoxic activity present in the culture supernatant and cell homogenate fractions of A. actinomycetemcomitans Y4. This CDT is a new putative virulence factor of A. actinomycetemcomitans and may play a role in the pathogenesis of periodontal diseases.
Actinobacillus actinomycetemcomitans is a gram-negative bacterium belonging to the family Pasteurellaceae. It has been implicated in the pathogenesis of juvenile and adult periodontitis (44, 45, 55). A. actinomycetemcomitans cells adhere to human cells, and some of them eventually invade the attached cells in vitro (8, 25). They produce a variety of virulence factors, including cytotoxic factors (1, 9, 10, 15–17, 24, 41–43, 46, 50, 52), chemotactic inhibitor (49), collagenases (36), and lipopolysaccharide (19, 37), but little is known about the roles of such virulence factors in the pathogenesis of periodontal diseases at the molecular level. Among cytotoxic factors, leukotoxin has been most extensively studied (20–23, 26, 55), but there are several reports concerning the existence of other cytotoxic factors produced by A. actinomycetemcomitans (10, 15, 41–43). Although the possible involvement of these factors in the pathogenesis of periodontitis has been suggested, their exact role has yet to be elucidated.
We have now identified a cluster of genes in A. actinomycetemcomitans that encode proteins belonging to the family of the cytolethal distending toxin (CDT), which is produced by certain pathogenic Escherichia coli strains (29, 31, 40), Campylobacter species (32), Shigella species (28), and Haemophilus ducreyi (7). CDT has recently been shown to block the cell cycle of HeLa cells (29). In this report, we demonstrate that A. actinomycetemcomitans produces a new member of the CDT family which also induced growth arrest in G2/M phase of cultured HeLa cells.
MATERIALS AND METHODS
Materials and chemicals.
All restriction enzymes, T4 DNA ligase, and Klenow fragment of DNA polymerase I were from Boehringer Mannheim, Tokyo, Japan, or New England BioLabs, Inc., Beverly, Mass. Other materials and chemicals used were from commercial sources.
Bacterial strains and culture conditions.
A. actinomycetemcomitans Y4 (serotype b, ATCC 43718) was cultured in Trypticase soy broth (Becton Dickinson Microbiology Systems, Cockeysville, Md.) supplemented with 1% (wt/vol) yeast extract in a 5% CO2 atmosphere. E. coli strains and plasmids used in this study are listed in Table 1. E. coli strains were grown aerobically in Luria-Bertani (LB) medium or on LB agar plates. Ampicillin (50 μg/ml) or kanamycin (50 μg/ml) was used when appropriate. Manipulation of DNA in E. coli was carried out with pUC19 (54), pGEM-T Easy (Promega, Madison, Wis.), or pET-28a(+) (Novagen, Madison, Wis.).
TABLE 1.
Strains and plasmids used
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| E. coli strains | ||
| XL1 Blue | supE44 hsdR17 recA1 endA1 gyrA46 thi relA1 Lac−(F′ [proAB+ lacIqlacZ ΔM15 Tn10 {Tet}]) | 5 |
| HMS174 | recA1 hsdR Rifr | 47 |
| Plasmids | ||
| pUC19 | Cloning vector, Ampr | 54 |
| pGEM-T Easy | Cloning vector, Ampr | Promega |
| pET-28a(+) | Cloning vector, Kanr | Novagen |
| pTK3003 | 5.6-kb EcoRI fragment in pUC19 | This study |
| pTK3005 | Reverse insertion of 5.6-kb EcoRI fragment in pUC19 | This study |
| pTK3022 | 3.4-kb SmaI-EcoRI fragment in pUC19 | This study |
| pTK3034 | 3.7-kb 3′ deletion fragment of pTK3003 EcoRI insert | This study |
| pTK3035 | 3.65-kb 3′ deletion fragment of pTK3003 EcoRI insert | This study |
| pTK3037 | 2.9-kb 3′ deletion fragment of pTK3003 EcoRI insert | This study |
| pTK3043 | 1.9-kb 3′ deletion fragment of pTK3003 EcoRI insert | This study |
| pTK3047 | 1-kb 3′ deletion fragment of pTK3003 EcoRI insert | This study |
| pTK3046 | 5.1-kb 3′ deletion fragment of pTK3005 EcoRI insert | This study |
| pTK3045 | 4.2-kb 3′ deletion fragment of pTK3005 EcoRI insert | This study |
| pTK3251 | cdtA (1,093 bases) PCR fragment in pGEM-T Easy | This study |
| pTK3252 | cdtB (1,211 bases) PCR fragment in pGEM-T Easy | This study |
| pTK3253 | cdtC (969 bases) PCR fragment in pGEM-T Easy | This study |
| pTK3286 | cdtA (636 bases) PCR fragment in pET-28a(+) | This study |
| pTK3287 | cdtB (797 bases) PCR fragment in pET-28a(+) | This study |
| pTK3288 | cdtC (561 bases) PCR fragment in pET-28a(+) | This study |
Cells and culture conditions.
HeLa cells (ATCC CCL2) were grown in Eagle’s minimal essential medium (Nissui) supplemented with 10% fetal bovine serum at 37°C and in a 5% CO2–95% air atmosphere.
Detection of cytodistending activity in bacterial sterile lysates.
Bacterial cells were recovered from cultures of A. actinomycetemcomitans strains or E. coli recombinant strains by centrifugation (10,000 × g, 20 min), and the pellets were resuspended in phosphate-buffered saline (PBS) to an optical density at 660 nm of 0.1. A cell suspension of 1 ml was lysed by periodic sonication for 30 s six times in ice (Ultradysruptor; TOMY SEIKO, Tokyo, Japan). After clarification by centrifugation (10,000 × g, 20 min), lysates or culture supernatants were filtered (0.2-μm-pore-size filter) and placed on HeLa cell monolayers in a 48-well plate (Falcon; Becton Dickinson) (1.6 × 103 cells per well). The occurrence of cytotoxic effects was monitored up to day 5. Cytodistending activity was titrated by using as the end point the highest twofold dilution of toxic material giving 50% transformed cells after 72 h of incubation (50% cytotoxic dose [CD50]).
DNA manipulations.
Routine DNA manipulations, DNA digestion with restriction enzymes, DNA ligations, gel electrophoresis, Southern blotting of DNA and hybridization, and DNA sequencing were performed essentially as described previously (38). Purification of chromosomal DNA from A. actinomycetemcomitans was performed as described previously (38). Restricted genomic DNA was separated by electrophoresis overnight in a 1% agarose gel. DNA fragments of 5 to 10 kb were recovered by electroelution and ligated to EcoRI-digested and alkaline phosphatase-treated pUC19. The ligated DNA was transformed into E. coli XL-1 Blue, and the transformants were selected on LB agar that contained ampicillin (50 μg/ml). Hybridization was performed by means of an enhanced chemiluminescense procedure (ECL direct labelling kit or 3′-oligolabelling kit; Amersham Life Science, Buckinghamshire, United Kingdom). DNA sequences of both strands were determined by the dideoxy-chain termination method (39) with an Auto-Read sequencing kit (Pharmacia Biotech, Tokyo, Japan). A nested set of deletions for sequencing was constructed by using exonuclease III and mung bean nuclease (Kilosequence deletion kit; Takara Biomedicals, Tokyo, Japan) according to the method of Henikoff (11). Extraction of large plasmids from lysed bacteria was carried out by the method of Kado and Liu (14).
PCR.
PCR reagents were from Perkin-Elmer (Norwalk, Conn.), and PCR was performed with the GeneAmp PCR System 2400 (Perkin-Elmer). Primers were supplied by Greiner Japan Co. (Tokyo, Japan). The primer sets used are listed in Table 2.
TABLE 2.
Primers used in this study
| Primer | Sequencea | Position |
|---|---|---|
| MIX5′ | 5′-GAAARYAAATGGARYRYWMRTGTMMG-3′ | |
| MIX3′ | 5′-AAATCWCCWRSAATCATCCAGTTA-3′ | |
| AASPC5′ | 5′-AAAGTAAATGGAATATTAATGTGCG-3′ | 1475–1499 |
| AASPC3′ | 5′-AAATCACCAACAACCATCCAGCTA-3′ | 1926–1949 |
| U257 | 5′-TGCTGAACAAGTATACATTCGTAAAGAA-3′ | 257–284 |
| L1323 | 5′-ATAAACTCCTTAGCTTAATTAACCGCTG-3′ | 1323–1350 |
| U1001 | 5′-ATTCGTAATTGGAAGATAGAACCTGG-3′ | 1001–1026 |
| L2188 | 5′-AGTATTCTCCTTAGCGATCATGAAC-3′ | 2188–2212 |
| U1851 | 5′-TGATGCGGTAAGTTTAATTCGTAATA-3′ | 1851–1875 |
| L2801 | 5′-CCCTCGCCCCACTAAGCATCTTGATT-3′ | 2801–2826 |
| U714 | 5′-GTTCGTCAAATCAACGAATGAGTGACT-3′ | 714–740 |
| U1415 | 5′-CTAACTTGAGTGATTTCAAAGTAGCA-3′ | 1415–1440 |
| U2265 | 5′-GTCATGCAGAATCAAATCCTGATCCG-3′ | 2265–2290 |
R is A or G, Y is C or T, M is A or C, W is A or T, and S is G or C.
Purification of recombinant His6-tagged protein.
E. coli HMS174 (47) carrying a plasmid was grown at 37°C with vigorous shaking until an optical density of 0.5 was reached, and then the expression of His6 protein was induced by addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). After 5 h of incubation, bacteria were precipitated by centrifugation, resuspended in 20 mM Tris-HCl containing 0.5 M NaCl and 50 mM imidazole (pH 7.9) (buffer 1), and disrupted with an Ultrasonic disruptor (TOMY SEIKO). After centrifugation at 9,000 × g for 30 min, the pellet was treated with buffer 1 containing 8 M urea. The recombinant His6-tagged protein was purified by Ni-chelated affinity chromatography. A TSKgel AF-chelate 5PW column (7.5 by 75 mm) was pretreated with 6 column volumes of 50 mM NiSO4 and equilibrated with buffer 1 containing 8 M urea. After removal of the debris from the sonicated sample by centrifugation at 37,500 × g for 30 min, the supernatant was applied to the column until most of the unbound proteins passed through. Bound proteins were eluted with a linear gradient from buffer 1 to 20 mM Tris-HCl buffer containing 0.5 M NaCl and 1 M imidazole (pH 7.9) (buffer 2) at a flow rate of 1 ml/min in 30 min. The protein samples obtained were subjected to disc preparative electrophoresis (NA-1800; Nihon Eido, Tokyo, Japan), and fractions showing a single protein band by analytical sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) were used for immunization with rabbits.
Antiserum.
The purified recombinant His6-tagged proteins were emulsified with either Freund complete or incomplete adjuvant (Difco Laboratories, Detroit, Mich.) (100 μg of protein per ml). For each sample, 2-kg rabbits were immunized at day 1 with sample emulsified with Freund complete adjuvant and at 2 weeks with sample emulsified with Freund incomplete adjuvant. At 4 weeks, the rabbits were injected intravenously with 50 μg of protein from the sample. Antisera were obtained 5 weeks after first injection. Preimmune serum was used as control serum.
In vitro transcription and translation.
In vitro synthesis of proteins from plasmid DNA templates was performed by using the E. coli S30 Extract system (Promega) with EXPRE35S35S protein labelling mix (1,000 Ci/mmol; DuPont-NEN Research Products, Boston, Mass.) according to the manufacturer’s protocol.
Demonstration of HeLa cell morphological and nuclear changes.
At intervals, HeLa cells were monitored by phase-contrast microscopy with photographs taken to document observations. Also, morphological and nuclear changes were demonstrated by means of staining with Giemsa stain or propidium iodide (PI) and Hoechst 33342 as described previously (29).
Flow cytometry analysis.
After trypsinization and washing in PBS (pH 7.2), cells were fixed in 1% formaldehyde in PBS for 15 min on ice. Then, after three washes in PBS, cells were suspended in 70% ice-cold ethanol and immediately transferred to −20°C until ready for use. After fixation, the cells were rehydrated in PBS at room temperature, permeabilized with Triton X-100, and then incubated in the dark at 4°C for 30 min in 1 ml of a PBS solution containing RNase (1 mg/ml) and PI (10 μg/ml). Flow cytometric analysis of the DNA content was performed on a FACScalibur flow cytometer (Becton Dickinson). The data from 2 × 104 cells were collected and analyzed with CellQuest software. In this experiment, flow cytometer parameters were adjusted to obtain G1 (2n DNA content) and G2 (4n DNA content) cell cycle peaks of control cells, centered at 200 and 400 DNA units (DU), respectively. Flow cytometric analysis of the DNA contents from different samples was repeated three times in independent tests.
Other procedures.
SDS-PAGE and Western blotting (immunoblotting) were carried out as described previously (48). Protein was immunodetected by using Renaissance 4CN plus (Dupont-NEN). Protein concentrations were determined with the bicinchoninic acid protein assay reagent (Pierce, Rockford, Ill.), with bovine serum albumin as the standard.
Nucleotide sequence accession number.
The nucleotide sequence data presented in this report will appear in the DDBJ, EMBL, and GenBank nucleotide sequence databases under accession no. AB011405.
RESULTS
A. actinomycetemcomitans Y4 induces distension and cell cycle blockage in G2/M phase in HeLa cells.
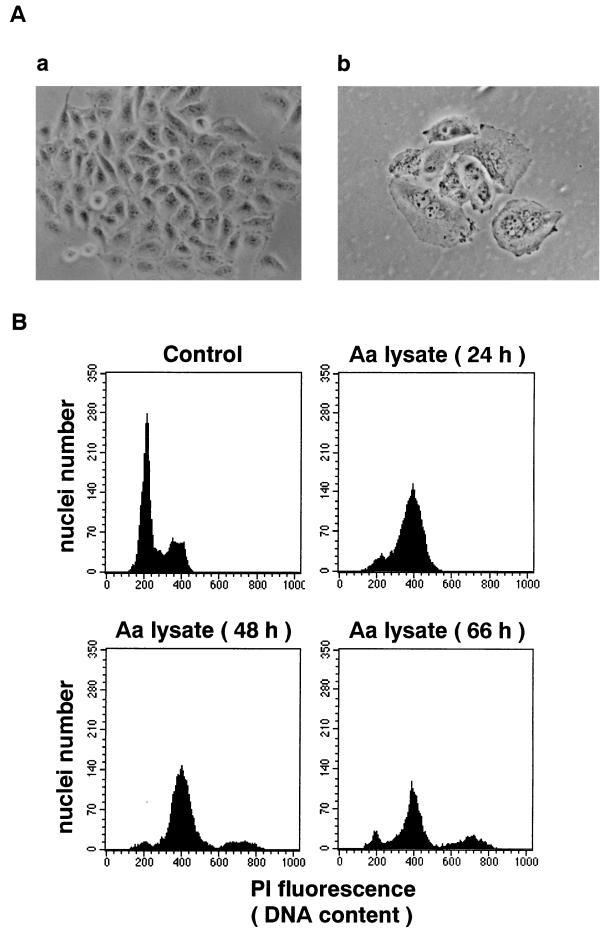
HeLa cells were treated with a sterile sonic lysate of A. actinomycetemcomitans Y4 as described in Materials and Methods. After 3 days of incubation, a cytopathic effect with distension in cell size was observed in treated cells (Fig. 1A, panel b). The mean size of distended cells was four- to fivefold that of control cells. The size of cell nuclei started to increase after 1 day of incubation. The mean diameter of nuclei was 1.4-fold that of control cells. When the incubation time was extended, cells started to detach from the culture dish. Flow cytometry analysis of the DNA content of HeLa cells revealed that the number of cells in G1 decreased while the number of cells in G2/M increased (Fig. 1B). These results suggested that the cells treated with the lysate of A. actinomycetemcomitans Y4 were blocked in the G2/M phase of the cell cycle. After 3 days of incubation, some populations of cells revealed fragmented nuclei with condensed masses of chromatin (Fig. 1A, panel b), which was often observed with apoptotic cells. Similar results were obtained with culture supernatant of A. actinomycetemcomitans Y4 (not shown). The cytodistension titer of the culture supernatant was similar to that of the sonic lysate (18 versus 32 CD50 per mg of protein).
FIG. 1.
Effect of A. actinomycetemcomitans Y4 sonic lysate on cultured HeLa cells. (A) HeLa cells 3 days after incubation with (b) or without (a) 4 CD50 of sterile sonic lysate of A. actinomycetemcomitans Y4. Magnification, ×150. (B) Cell cycle pattern of HeLa cells incubated with 4 CD50 of sterile sonic lysate of A. actinomycetemcomitans Y4 (Aa lysate) for the indicated times. Control, cells without treatment.
Cloning and genetic analysis of the A. actinomycetemcomitans Y4 cdt genes.
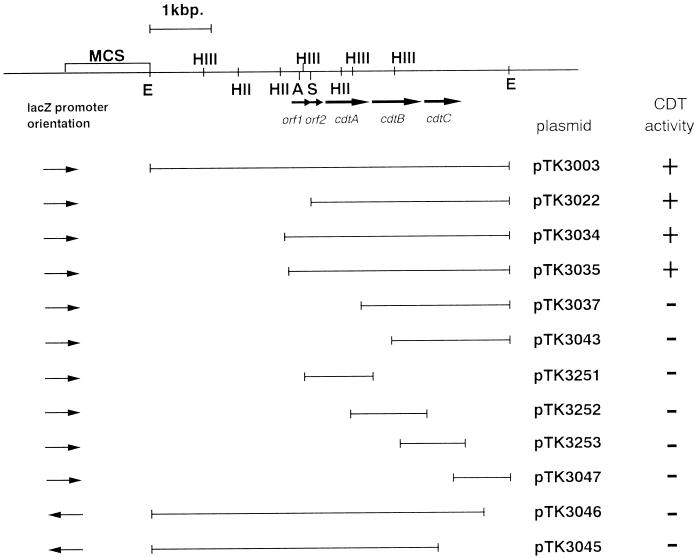
From these observations, we hypothesized that A. actinomycetemcomitans Y4 produces a CDT. Degenerative oligonucleotide primers, MIX5′ and MIX3′, were designed according to the homology observed between the different cdtB genes cloned so far. These primers were then tested in PCR experiments with A. actinomycetemcomitans Y4 template DNA. A single product of approximately 475 bp was amplified. Since this was close to the expected size of 460 bp, the product was cloned into pGEM-T Easy and sequenced. The nucleotide sequence obtained indicated that the relevant portion of the cdtB gene had been amplified. Specific primers, AASPC5′ and AASPC3′, then were generated, and the resulting PCR product was used as a DNA probe to identify one clone containing cdt genes in our A. actinomycetemcomitans Y4 DNA library. This clone carried a recombinant plasmid with a 5.6-kb EcoRI DNA fragment in pUC19. This plasmid was called pTK3003, and a lysate of the pTK3003 clone was found to produce CDT. A restriction map of the 5.6-kb EcoRI fragment was established (Fig. 2). To determine the minimum amount of DNA required for the CDT activity, a deletion series of the EcoRI fragment was constructed. An assay of deletion clones for cytodistending activity revealed that bacteria carrying pTK3022, pTK3034, and pTK3035 expressed cytodistending activity. The other clones listed in Fig. 2 did not show any cytodistending activity. Fragments expressing cytodistending activity were sequenced by using either the universal or the reverse sequencing primer. The locations of the open reading frames (ORFs) are shown in Fig. 2, and the nucleotide sequence for 3,560 bp of DNA, including the entire ORFs, together with the deduced amino acid sequence is shown in Fig. 3.
FIG. 2.
Restriction map of subclones from A. actinomycetemcomitans DNA that contains cdtABC. Arrows represent ORFs and directions of transcription. CDT activity was measured by incubating sterile sonic lysates of recombinant strains with HeLa cell cultures for 3 days. Clones expressing CDT activity are indicated at the right. The restriction sites of relevant endonucleases are indicated: HIII, HindIII; HII, HincII; E, EcoRI; A, AccI; S, SmaI. MCS, multicloning site for cloning vector.
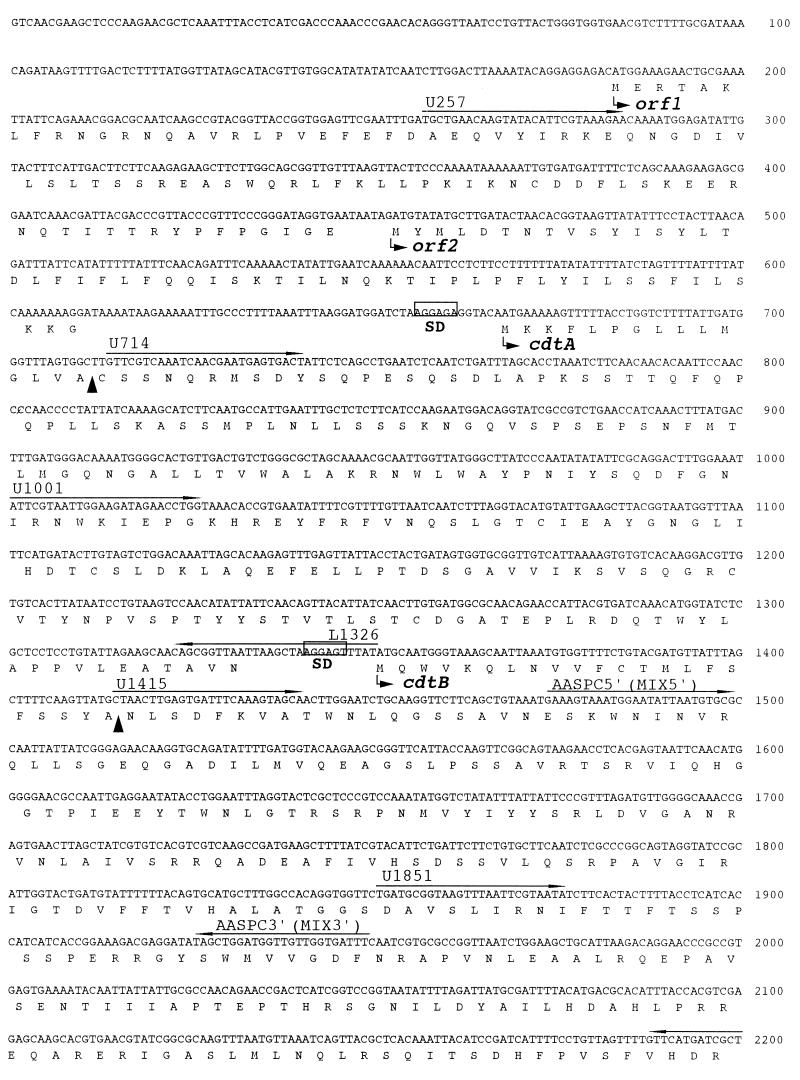
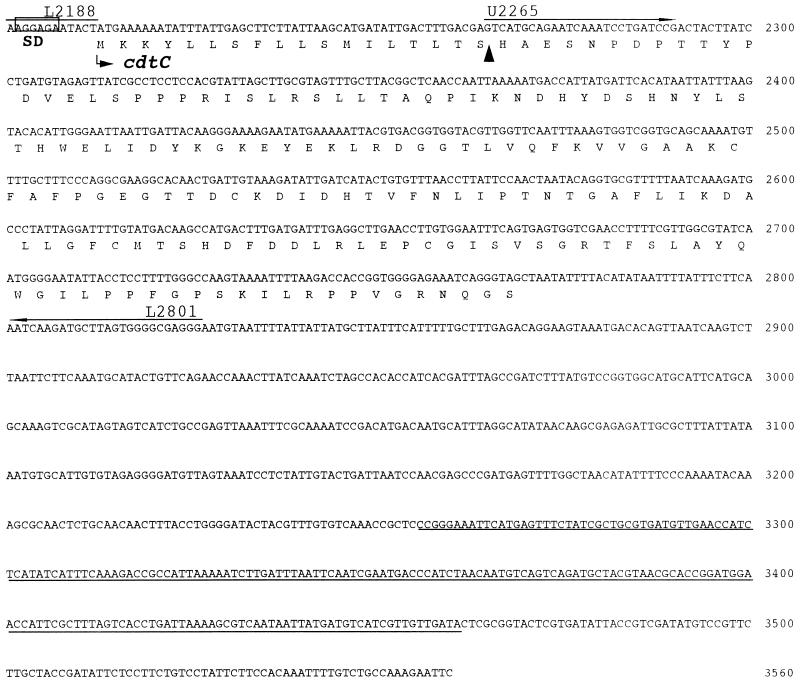
FIG. 3.
DNA sequence analysis in the region of the HincII-EcoRI site of the cloned DNA fragment. ORF1, ORF2, and three complete ORFs defining cdtA, -B, and -C were found. Putative ribosome binding (Shine-Dalgarno [SD]) sites for cdt are boxed. Putative signal peptide cleavage sites are indicated (▴). Oligonucleotide primers used are labelled and are indicated by arrows above the relevant sequences. A DNA sequence similar to that of the integrating plasmid of H. influenzae is underlined.
The cdt genes are arranged in a manner similar to those of other species (7, 28, 29, 31, 32, 40). There are three adjacent ORFs which encode putative CDTA, CDTB, and CDTC proteins. The cdtA and cdtB genes were separated by 17 nucleotides, and the cdtB and cdtC genes were separated by 13 nucleotides. The percent G+C contents of the A. actinomycetemcomitans cdtA, cdtB, and cdtC genes were 38.4, 41.3, and 37.8% respectively, which is similar to the G+C content determined for A. actinomycetemcomitans chromosomal DNA (42.7%) (30). Three putative Shine-Dalgarno sequences were located upstream of the coding sequences.
The deduced molecular masses of the proteins encoded by these three ORFs are 24,512, 31,492 and 20,706 Da, respectively. A possible signal peptide was present in each protein, and each of these had a hydrophobic region of 10 to 12 amino acids at the N terminus that was followed by 1 or 2 basic amino acids. The possible signal sequence for the first ORF revealed the presence of a consensus sequence, LVAC, for prolipoprotein modification and processing by signal peptidase II (4, 53). On the other hand, those for the second and third ORFs were terminated by a sequence resembling that for processing by signal peptidase I. Comparison of the predicted amino acid sequences with known sequences in the DDBJ database revealed that these ORFs code for a new member of the CDT family and are very similar to those of H. ducreyi CDTs (7) (Table 3).
TABLE 3.
Comparison of the predicted amino acid sequences of the three genes of A. actinomycetemcomitans CDT and those of CDT loci previously cloned and sequenced
| A. actinomyce- temcomitans gene product | % Identity witha:
|
||||
|---|---|---|---|---|---|
| CDT-I | CDT-II | CDT-III | CDT-c | CDT-h | |
| CDTA | 29 | 22 | 22 | 27 | 91 |
| CDTB | 48 | 45 | 46 | 48 | 97 |
| CDTC | 19 | 23 | 19 | 20 | 94 |
CDT-I, locus cloned by Scott and Kaper (40) from E. coli E6468/62 (GenBank accession no. U03293); CDT-II, locus cloned by Pickett et al. (31) from E. coli 9142-88 (GenBank accession no. U04208); CDT-III, locus cloned by Pérès et al. (29) from E. coli 1404 (GenBank accession no. U89305); CDT-c, locus cloned by Pickett et al. (32) from C. jejuni 81-176 (GenBank accession no. U51121); CDT-h, locus cloned by Cope et al. (7) from H. ducreyi 35000 (GenBank accession no. U53215).
Besides the cdt locus, we found two putative ORFs, ORF1 and ORF2, upstream of cdtA. Interestingly, ORF1 shows significant homology at the amino acid level with the hypothetical protein HI0321 of Haemophilus influenzae (GenBank accession no. U32717) and with the VagC, VapB, and STBORF1 proteins, which are involved, respectively, in the stability of virulence plasmids in Dichelobacter nodosus (2), Salmonella dublin (33), and Shigella flexneri (35). In addition, we also found downstream of the cdt locus a small DNA stretch homologous to an integrating plasmid of H. influenzae (GenBank accession no. U68467). Taken together, these results raised the possibility that the cdt genes of A. actinomycetemcomitans were located on a plasmid, like the cdtIII genes carried by the Vir plasmid of E. coli (29). Plasmid DNA was therefore extracted from lysed A. actinomycetemcomitans Y4 by the method described by Kado and Liu (14). However, there was no visible band observed in agarose gel electrophoresis. We further tested whether the cdt genes are encoded on a plasmid by Southern hybridization. The PCR product obtained with specific primers, AASPC5′ and AASPC3′, was used as a probe. Southern hybridization of the extract prepared by the Kado-Liu method (14) failed to show any signal, suggesting that the cdt genes were not located on a plasmid (not shown).
Expression and toxic properties of A. actinomycetemcomitans Y4 cdt gene products.
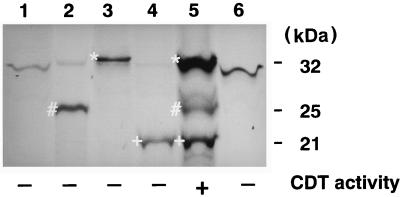
A clone containing all three cdt genes was constructed by inserting a 3.4-kb SmaI-EcoRI fragment into pUC19 (54). The resulting clone, pTK3022, contained intact cdtA, cdtB, and cdtC and produced CDT activity in E. coli XL-1 Blue (5) (Fig. 2). To confirm the presence of the three ORFs and identify protein products for each, we performed in vitro transcription-translation studies with DNA fragments cloned into a plasmid. Each of the cdt genes, containing approximately 250 bp of 5′ flanking DNA from the start codon, was amplified by PCR with primer sets U257 and L1326, U1001 and L2188, and U1851 and L2801, respectively (Table 2), and cloned into pGEM-T Easy. Proteins with apparent molecular masses of 25, 32, and 21 kDa were expressed by cdtA, cdtB, and cdtC, respectively (Fig. 4). These three protein products were present in pTK3022, which includes all three cdt genes, and the estimated molecular masses of the proteins were in good agreement with predicted values. When culture supernatants from these recombinant E. coli strains were tested for CDT activity, only that from the strain carrying pTK3022 had CDT activity against HeLa cells (Fig. 2).
FIG. 4.
Autoradiograph of an SDS–12% polyacrylamide gel containing [35S]methionine-labelled products of in vitro transcription-translation. Lanes: 1, pGEM-T Easy; 2, pTK3251, containing the A. actinomycetemcomitans cdtA gene; 3, pTK3252, containing the A. actinomycetemcomitans cdtB gene; 4, pTK3253, containing the A. actinomycetemcomitans cdtC gene; 5, pTK3022, containing the entire cdtABC gene cluster; 6, pUC19. The CDT activities of sterile sonic lysates from the recombinant strains are indicated at the bottom. Radiolabelled bands marked by #, ∗, and + are putative gene products of cdtA, -B, and -C, respectively.
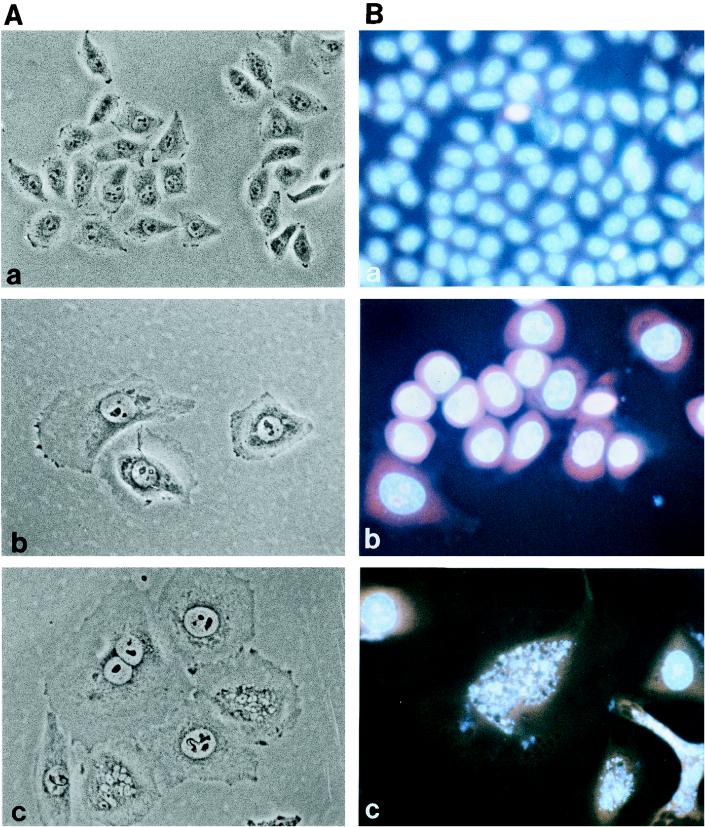
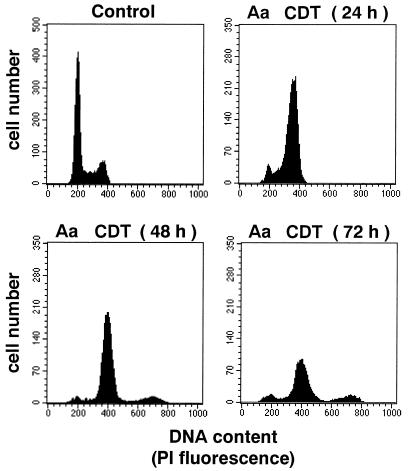
The effect of A. actinomycetemcomitans CDT on cell morphology and on the DNA content of HeLa cells was studied with a sterile sonic lysate of the pTK3022 clone. The HeLa cells were exposed to a dose equivalent to 20 CD50. After 3 days of incubation, treated cells had a distended morphology (Fig. 5A, panel b). The mean size of distended cells was 10- to 18-fold that of control cells. Most of the cells were mononucleated, but a few were binucleated. When the incubation time was extended, most cells started to detach from the culture dish. The DNA content of the HeLa cells treated with A. actinomycetemcomitans CDT was analyzed after staining of nuclei with PI and Hoechst 33342, by fluorescence microscopy, and with PI by flow cytometry (Fig. 5B and 6). PI and Hoechst 33342 staining revealed that the nuclei of A. actinomycetemcomitans CDT-treated cells increased in size after 3 days of incubation (Fig. 5B, panel b). After 4 days, cells with fragmented nuclei were increased in number (Fig. 5B, panel c). Flow cytometry demonstrated that A. actinomycetemcomitans CDT was able to block the cell cycle (Fig. 6). After 24 h, the number of cells in G2 increased, whereas that of cells in G1 decreased. At 48 h, the G2/M peak slightly decreased but was still dominant. A third DNA peak, centered at 700 DU, appeared at 48 h and slightly increased after 72 h. This third peak theoretically suggests the presence of cells with multiple nuclei (nuclei in G1 and/or in G2). Since PI staining suggested the presence of binucleated cells, the third peak in A. actinomycetemcomitans-treated cells is composed mainly of binucleated cells in G2/M and, to a lesser extent, those with four nuclei in G1. Thus, cells exposed to A. actinomycetemcomitans CDT were blocked at the G2/M stage.
FIG. 5.
Effect of A. actinomycetemcomitans Y4 CDT on morphology of cultured HeLa cells. Phase-contrast microscopy (A) and PI and Hoechst 33342 staining (B) of HeLa cells incubated with 20 CD50 of a sterile sonic lysate of E. coli(pTK3022) for 3 days (b) or 4 days (c) are shown. (a) Control HeLa cells treated with a lysate of E. coli XL-1 Blue (5) for 4 days. Magnification, ×200.
FIG. 6.
Effect of A. actinomycetemcomitans Y4 (Aa) CDT on cell cycle pattern of HeLa cells. HeLa cells were incubated with 20 CD50 of a sterile sonic lysate of E. coli(pTK3022) for the indicated times.
Anti-HisCDTC serum inhibits induction of cell distension by A. actinomycetemcomitans Y4 cell lysate.
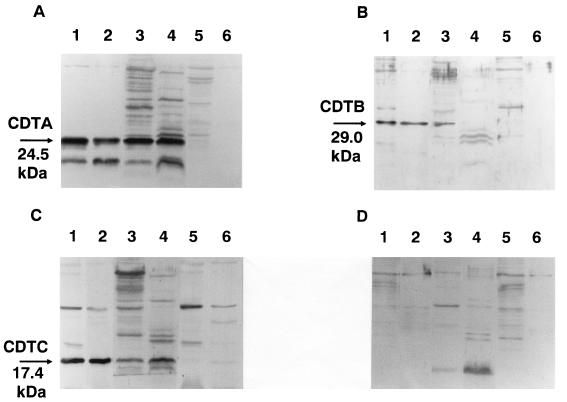
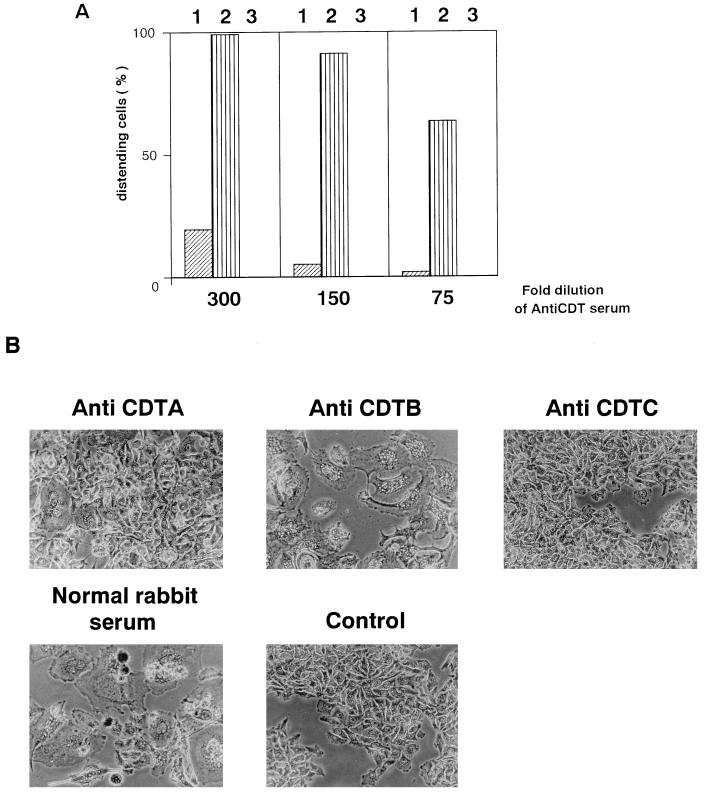
We constructed DNA fragments corresponding to the processed forms of CDTA, CDTB, and CDTC by PCR with primer sets U714 and L1326, U1415 and L2188, and U2265 and L2801, respectively (Table 2). Each of them was placed in frame downstream of the His6 tag sequence of the pET28a vector. After induction with IPTG, each construct yielded a considerable amount of fusion protein in recombinant E. coli HMS174 (47). These fusion proteins, designated HisCDTA, HisCDTB, and HisCDTC, respectively, were purified to homogeneity with an Ni-chelated TSKgel AF-chelate column and subsequent preparative SDS-PAGE (not shown). The purified samples were used for immunization of rabbits. The antisera thus obtained were used for detection of CDT proteins in E. coli XL-1 Blue(pTK3022) and A. actinomycetemcomitans Y4 cell lysates and in culture supernatants by Western blotting. As shown in Fig. 7, anti-HisCDTA, -HisCDTB, and -HisCDTC sera reacted with 24.5-, 29.0-, and 17.4-kDa proteins in the culture supernatant and lysate of E. coli(pTK3022). Similarly, anti-HisCDTA and anti-HisCDTC sera reacted with 24.5- and 17.4-kDa proteins in A. actinomycetemcomitans Y4 cell lysate and culture supernatant. On the other hand, anti-CDTB reacted with a 29.0-kDa protein in sonic lysate but not in culture supernatant of A. actinomycetemcomitans Y4. As shown in Fig. 7A, anti-HisCDTA reacted with additional bands of lower molecular masses in culture supernatant and cell lysate of E. coli(pTK3022) and A. actinomycetemcomitans Y4. These additional bands might represent proteolytic digests of CDTA, but the identities of those cross-reactive bands remain unknown. Anti-CDT sera were then assayed for neutralizing activity against CDT from A. actinomycetemcomitans lysate or culture supernatant. As shown in Fig. 8, a 300-fold dilution of anti-CDTC completely blocked CDT activity of cell lysate from A. actinomycetemcomitans Y4 against HeLa cells. Anti-HisCDTA showed a weaker but clear neutralizing effect. In contrast, anti-HisCDTB failed to show any neutralizing effect at 300-, 150-, and 75-fold dilutions. Similar neutralizing effects were observed with these antisera against culture supernatant of A. actinomycetemcomitans Y4 (not shown). These antisera at effective doses were able to block not only cytodistension but subsequent cell cytotoxicity and morphological changes in the nucleus (not shown).
FIG. 7.
Immunological detection of CDT in culture supernatant or sonic lysate of A. actinomycetemcomitans or recombinant E. coli carrying pTK3022. Western blotting was performed as described previously (49). Immunodetection was performed with antiserum against His-tagged CDTA (A), His-tagged CDTB (B), or His-tagged CDTC (C) or with control nonimmune serum (D). Lanes: 1, sonic lysate of E. coli XL-1 Blue(pTK3022); 2, culture supernatant of E. coli XL-1 Blue(pTK3022); 3, sonic lysate of A. actinomycetemcomitans Y4; 4, culture supernatant of A. actinomycetemcomitans Y4; 5, sonic lysate of E. coli XL-1 Blue(pUC19); 6, culture supernatant of E. coli XL-1 Blue(pUC19).
FIG. 8.
Effect of anti-CDT serum on cytodistending activity of sterile sonic lysate of A. actinomycetemcomitans Y4. (A) HeLa cells were incubated with a sterile sonic lysate of A. actinomycetemcomitans Y4 (17 CD50) in the presence of the indicated dilutions of anti-CDTA (bars 1), anti-CDTB (bars 2), and anti-CDTC (bars 3) for 4 days. The percentage of distended cells was calculated by counting the number of distended cells per 200 cells. (B) Phase-contrast microscopy of HeLa cells incubated with a sterile sonic lysate of A. actinomycetemcomitans Y4 (17 CD50) in the presence of a 300-fold dilution of indicated antiserum for 4 days. Control, cells without treatment. Magnification, ×150.
DISCUSSION
The production of a cytotoxic factor distinct from leukotoxin has been reported by several groups working on A. actinomycetemcomitans. This heat-labile factor, present in the culture supernatant, was shown to inhibit growth of human and murine fibroblasts (41) and human keratinocytes (15) and to have an immunosuppressive activity towards human T and B cells (43). A similar toxic factor has been reported to inhibit human gingival fibroblasts in the G2 phase of the cell cycle (10). In addition, A. actinomycetemcomitans also possesses a cytotoxic or immunosuppressive factor(s) associated with its cell surface (16, 24, 50). In the present study, we have established that A. actinomycetemcomitans produces a new member of the CDT family, which is a previously unrecognized virulence factor of A. actinomycetemcomitans. Amino acid sequences deduced from cloned genes indicate that A. actinomycetemcomitans CDT is very similar to H. ducreyi CDT (Table 3). It was recently shown that the different E. coli CDTs (29) and the CDT of Campylobacter jejuni (51) block the cell cycle in G2 phase. We confirmed unambiguously that A. actinomycetemcomitans CDT also induced cell cycle arrest in G2/M phase. Taken together, our results strongly suggest that CDT is the cytotoxic factor present in the culture supernatant and cell homogenate of A. actinomycetemcomitans Y4.
The CDTs are produced by a variety of bacterial genera and form a heterogeneous family of toxins with similar biological activities (7, 28, 29, 31, 32, 40). The term CDT was coined for activity that induces progressive cell distention and eventual cytotoxicity on cultured cells (12, 13). The CDT activity was first described for cell extracts of Campylobacter spp. and E. coli clinical isolates (12, 13). Now six CDTs have been identified and their genes have been cloned, in addition to A. actinomycetemcomitans CDT: three from E. coli (29, 31, 40), one from C. jejuni (32), one from Shigella dysenteriae (28), and one from H. ducreyi (7). CDTs are encoded by a cluster of three genes, which are separated by a few nucleotides or slightly overlap. The A. actinomycetemcomitans cdt locus also encoded three proteins, CDTA, -B, and -C, which are all required for toxicity to the cells (Fig. 2). CDTA possesses a possible cleavage site for signal peptidase II, a lipoprotein-specific signal peptidase (4, 53). However, the significance of this amino acid sequence motif remains to be determined.
The origin of the cdt genes in A. actinomycetemcomitans remains unknown. The similarity of the amino acid sequences to those of the H. ducreyi CDTs together with the fact that Actinobacillus and Haemophilus are closely related in evolutionary origin suggest that the Actinobacillus and Haemophilus cdt genes originated from the same ancestral genes. Sequence analysis suggested that H. ducreyi cdt was acquired as part of a transposon (7). For E. coli, insertion sequences and phagic elements were found upstream of the cdtIII locus (29). The presence, close to A. actinomycetemcomitans cdt, of both an ORF with a predicted product homologous to VagC/VapB and a DNA sequence homologous to an integrating plasmid of H. influenzae suggests that the cdt locus of A. actinomycetemcomitans is or was on a plasmid. Southern hybridization failed to show any signal suggesting that the cdt genes were still located on a plasmid. Nonetheless, our results further support the notion that the genetic determinants of CDTs have been transferred horizontally among bacterial species (29).
An assay for neutralizing activity against CDT of A. actinomycetemcomitans Y4 lysate showed that anti-CDTC serum had the strongest neutralizing effect (Fig. 8). This was in good agreement with previous observations that a neutralizing monoclonal antibody against H. ducreyi cytotoxin recognizes CDTC (7, 34). By contrast, anti-CDTA showed a weaker neutralizing effect, and anti-CDTB showed only slight inhibition of the CDT activity. It should be noted that Western analysis demonstrated that only CDTA and CDTC were present in the culture supernatant of A. actinomycetemcomitans Y4, while all CDT subunits were present in the sonic lysate of the bacteria (Fig. 8). It is possible that CDTB is a cell-bound protein and is involved in release of other CDTs from A. actinomycetemcomitans Y4, but further study is necessary to understand the secretion mechanism and function of cdt gene products.
The eukaryotic target of A. actinomycetemcomitans CDT is not known at present. Like the CDTs of E. coli and C. jejuni, CDT of A. actinomycetemcomitans induced in HeLa cells growth arrest at the G2/M phase of the cell cycle. Recently, Comayras et al. (6) reported that CDT treatment causes HeLa cells to accumulate the inactive, tyrosine-phosphorylated form of CDC2. This result, recently confirmed by Whitehouse et al. with the CDT produced by C. jejuni (51), indicated that CDT treatment results in a failure to activate CDC2, which leads to cell cycle arrest in G2. Both research groups suggest that the CDTs trigger a mechanism of cell cycle arrest by way of a DNA damage checkpoint system.
The role of A. actinomycetemcomitans CDT in the pathogenesis of periodontal diseases remains to be determined. E. coli and S. dysenteriae CDTs are implicated in inflammatory responses in bacterial infection in the gut (3, 12, 13, 27). As suggested by previous reports of cytotoxic factors other than leukotoxin, cell death or growth arrest of fibroblasts may lead to a decrease in collagen synthesis, which will be manifested as a loss of collagen in periodontal disease. In this regard, elaboration of CDT by A. actinomycetemcomitans may be very relevant. Preliminary observations suggest that most clinical strains investigated possess the cdt gene cluster and produce CDTs extracellularly (unpublished results). Furthermore, CDT may function in concert with other virulence factors, such as leukotoxin and lipopolysaccharide, to induce tissue damage in the periodontal milieu. Recently, Kato et al. (18) reported that A. actinomycetemcomitans infection induced apoptosis in a murine macrophage cell line in vitro. Another possible role of A. actinomycetemcomitans CDT could be suppression of immune cells targeting periodontal lesions. The antiproliferative activity of CDT could account for the pathogenesis of A. actinomycetemcomitans strains by blocking in vivo the expansion of lymphocytes, thus inhibiting the local immune response. Further study is clearly necessary to understand the true importance of this toxin in the pathogenesis of periodontal diseases.
ACKNOWLEDGMENTS
We thank the Research Facility, Hiroshima University School of Dentistry, for the use of their facilities.
This work was supported in part by a grant-in-aid for scientific research (09670282) from the Ministry of Education, Science, Sports and Culture of Japan. S.Y.P. is a recipient of a scholarship from the Ministere de l’Enseignement Superieur et de la Recherche (France).
REFERENCES
- 1.Baehni P, Tsai C-C, McArthur W P, Hammond B F, Taichman N S. Interaction of inflammatory cells and oral microorganisms. VIII. Detection of leukotoxic activity of a plaque-derived gram-negative microorganism. Infect Immun. 1979;24:233–243. doi: 10.1128/iai.24.1.233-243.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Billington S J, Sinistraj M, Cheetham M, Ayres B F, Moses E K, Katz M E, Rood J I. Identification of a native Dichelobacter nodosus plasmid and implications for the evolution of the vap regions. Gene. 1996;172:111–116. doi: 10.1016/0378-1119(96)00032-7. [DOI] [PubMed] [Google Scholar]
- 3.Bouzari S, Vatsala B R, Varghese A. In vitro adherence property of cytolethal distending toxin (CLDT) producing EPEC strains and effect of the toxin on rabbit intestine. Microb Pathog. 1992;12:153–157. doi: 10.1016/0882-4010(92)90118-8. [DOI] [PubMed] [Google Scholar]
- 4.Braun V, Wu H C. Lipoproteins, structure, function, biosynthesis and model for protein export. New Compr Biochem. 1994;27:319–341. [Google Scholar]
- 5.Bullock W O, Fernandez J M, Short J M. XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with beta-galactosidase selection. BioTechniques. 1987;5:376–379. [Google Scholar]
- 6.Comayras C, Tasca C, Pérès S Y, Ducommun B, Oswald E, De Rycke J. Escherichia coli cytolethal distending toxin blocks the HeLa cell cycle at the G2/M transition by preventing Cdc2 protein kinase dephosphorylation and activation. Infect Immun. 1997;65:5088–5095. doi: 10.1128/iai.65.12.5088-5095.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cope L, Lumbley S, Latimer J L, Klesney-Tait J, Stevens M K, Johnson L S, Purven M, Munson J R S, Lagergard T, Radolf J D, Hansen E J. A diffusible cytotoxin of Haemophilus ducreyi. Proc Natl Acad Sci USA. 1997;94:4056–4061. doi: 10.1073/pnas.94.8.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fives-Taylor P, Meyer D, Minz K. Characteristics of Actinobacillus actinomycetemcomitans invasion of and adhesion to cultured epithelial cells. Adv Dent Res. 1995;9:55–62. doi: 10.1177/08959374950090011001. [DOI] [PubMed] [Google Scholar]
- 9.Fives-Taylor, P., D. Meyer, and K. Minz. 1996. Virulence factors of the periodontopathogen Actinobacillus actinomycetemcomitans. J. Periodontol. 67(Suppl.):291–297. [DOI] [PubMed]
- 10.Helgeland K, Nordby Ø. Cell cycle-specific growth inhibitory effect on human gingival fibroblasts of a toxin isolated from the culture medium of Actinobacillus actinomycetemcomitans. J Periodont Res. 1993;28:161–165. doi: 10.1111/j.1600-0765.1993.tb01064.x. [DOI] [PubMed] [Google Scholar]
- 11.Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984;28:351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- 12.Johnson W M, Lior H. A new heat-labile cytolethal distending toxin (CLDT) produced by Escherichia coli isolates from clinical material. Microb Pathog. 1988;4:103–113. doi: 10.1016/0882-4010(88)90052-6. [DOI] [PubMed] [Google Scholar]
- 13.Johnson W M, Lior H. Response of Chinese ovary hamster cells to a cytolethal distending toxin (CDT) of Escherichia coli and possible misinterpretation as heat labile (LT) enterotoxin. FEMS Microbiol Lett. 1987;43:19–23. [Google Scholar]
- 14.Kado C I, Liu S T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamen P R. Inhibition of keratinocyte proliferation by extracts of Actinobacillus actinomycetemcomitans. Infect Immun. 1983;42:1191–1194. doi: 10.1128/iai.42.3.1191-1194.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamin S, Harvey W, Wilson M, Scutt A. Inhibition of fibroblast proliferation and collagen synthesis by capsular material from Actinobacillus actinomycetemcomitans. J Med Microbiol. 1986;22:245–249. doi: 10.1099/00222615-22-3-245. [DOI] [PubMed] [Google Scholar]
- 17.Kataoka M, Kawamura K, Kondoh T, Wakano Y, Ishida H. Purification of a fibroblast-inhibitory factor from Actinobacillus actinomycetemcomitans Y4. FEMS Microbiol Lett. 1993;107:111–114. doi: 10.1111/j.1574-6968.1993.tb06013.x. [DOI] [PubMed] [Google Scholar]
- 18.Kato S, Muro M, Akifusa S, Hanada N, Semba I, Fujii T, Kowashi Y, Nishihara T. Evidence for apoptosis of murine macrophages by Actinobacillus actinomycetemcomitans infection. Infect Immun. 1995;63:3914–3919. doi: 10.1128/iai.63.10.3914-3919.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiley P, Holt S C. Characterization of the lipopolysaccharide from Actinobacillus actinomycetemcomitans Y4 and N27. Infect Immun. 1980;30:862–873. doi: 10.1128/iai.30.3.862-873.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolodrubetz D, Dailey T, Ebersole J, Kraig E. Molecular genetics and the analysis of leukotoxin in Actinobacillus actinomycetemcomitans. J Periodontol. 1996;67:309–316. doi: 10.1902/jop.1996.67.3s.309. [DOI] [PubMed] [Google Scholar]
- 21.Lally E T, Kieba I R. Molecular biology of Actinobacillus actinomycetemcomitans leukotoxin. In: Genco R, Hamada S, Lehner T, McGhee J, Mergenhagen S, editors. Molecular pathogenesis of periodontal disease. Washington, D.C: ASM Press; 1994. pp. 69–82. [Google Scholar]
- 22.Lally E T, Kieba I R, Demuth D R, Rosenbloom J, Golub E E, Taichman N S, Gibson C W. Identification and expression of the Actinobacillus actinomycetemcomitans leucotoxin gene. Biochem Biophys Res Commun. 1989;159:256–262. doi: 10.1016/0006-291x(89)92431-5. [DOI] [PubMed] [Google Scholar]
- 23.Lally E T, Kieba I R, Golub E E, Lear J D, Tanaka J C. Structure/function aspects of Actinobacillus actinomycetemcomitans leukotoxin. J Periodontol. 1996;67:298–308. doi: 10.1902/jop.1996.67.3s.298. [DOI] [PubMed] [Google Scholar]
- 24.Meghji S, Wilson M, Henderson B, Kinane D. Anti-proliferative and cytotoxic activity of surface associated material from periodontopathogenic bacteria. Arch Oral Biol. 1992;37:637–644. doi: 10.1016/0003-9969(92)90126-s. [DOI] [PubMed] [Google Scholar]
- 25.Meyer D H, Sreenivasan P K, Fives-Taylor P M. Evidence for invasion of a human oral cell clone by Actinobacillus actinomycetemcomitans. Infect Immun. 1991;59:2719–2726. doi: 10.1128/iai.59.8.2719-2726.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohta H, Kato K. Leukotoxic activity of Actinobacillus actinomycetemcomitans. In: Hamada S, Holt S C, McGhee J R, editors. Periodontal disease: pathogens & host immune responses. Tokyo, Japan: Quintesence Publishing; 1991. pp. 143–154. [Google Scholar]
- 27.Okuda J, Fukumoto M, Takeda Y, Nishibuchi M. Examination of diarrheagenicity of cytolethal distending toxin: suckling mouse response to the products of the cdtABC genes of Shigella dysenteriae. Infect Immun. 1997;65:428–433. doi: 10.1128/iai.65.2.428-433.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okuda J, Kurazono H, Takeda Y. Distribution of the cytolethal distending toxin A gene (cdtA) among species of Shigella and Vibrio, and cloning and sequencing of the cdt gene from Shigella dysenteriae. Microb Pathog. 1995;18:167–172. doi: 10.1016/s0882-4010(95)90022-5. [DOI] [PubMed] [Google Scholar]
- 29.Pérès S Y, Marchès O, Daigle F, Nougayrède J-P, Hèrault F, Tasca C, De Rycke J, Oswald E. A new cytolethal distending toxin (CDT) from Escherichia coli producing CNF2 blocks HeLa cell division in G2/M phase. Mol Microbiol. 1997;24:1095–1107. doi: 10.1046/j.1365-2958.1997.4181785.x. [DOI] [PubMed] [Google Scholar]
- 30.Phillips J E. Genus III Actinobacillus. In: Krieg N R, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 1. Baltimore: Williams & Wilkins; 1984. pp. 570–575. [Google Scholar]
- 31.Pickett C L, Cottle D L, Pesci E C, Bikah G. Cloning, sequencing, and expression of the Escherichia coli cytolethal distending toxin genes. Infect Immun. 1994;62:1046–1051. doi: 10.1128/iai.62.3.1046-1051.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pickett C L, Pesci E C, Cottle D L, Russell G, Erdem A N, Zeytin H. Prevalence of cytolethal distending toxin production in Campylobacter jejuni and relatedness of Campylobacter sp. cdtB genes. Infect Immun. 1996;64:2070–2078. doi: 10.1128/iai.64.6.2070-2078.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pullinger G D, Lax A J. A Salmonella dublin virulence plasmid locus that affects bacterial growth under nutrient-limited conditions. Mol Microbiol. 1992;6:1631–1643. doi: 10.1111/j.1365-2958.1992.tb00888.x. [DOI] [PubMed] [Google Scholar]
- 34.Purvén M, Frisk A, Lönnroth I, Lagergård T. Purification and identification of Haemophilus ducreyi cytotoxin by use of a neutralizing monoclonal antibody. Infect Immun. 1997;65:3496–3499. doi: 10.1128/iai.65.8.3496-3499.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Radnedge L, Davis M A, Youngren B, Austin S J. Plasmid maintenance functions of the large virulence plasmid of Shigella flexneri. J Bacteriol. 1997;179:3670–3675. doi: 10.1128/jb.179.11.3670-3675.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robertson P M, Lantz M, Marucha P T, Kornman K S, Trummel C L, Holt S C. Collagenolytic activity associated with Bacteroides spp. and Actinobacillus actinomycetemcomitans. J Periodont Res. 1982;17:275–283. doi: 10.1111/j.1600-0765.1982.tb01154.x. [DOI] [PubMed] [Google Scholar]
- 37.Saglie F R, Simon K, Merrill J, Koeffler H P. Lipopolysaccharide from Actinobacillus actinomycetemcomitans stimulates macrophages to produce interleukin-1 and tumor necrosis factor mRNA and protein. Oral Microbiol Immunol. 1990;5:256–262. doi: 10.1111/j.1399-302x.1990.tb00422.x. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 39.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scott D A, Kaper J B. Cloning and sequencing of the genes encoding Escherichia coli cytolethal distending toxin. Infect Immun. 1994;62:244–251. doi: 10.1128/iai.62.1.244-251.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shenker B, Kushner M E, Tsai C-C. Inhibition of fibroblast proliferation by Actinobacillus actinomycetemcomitans. Infect Immun. 1982;38:986–992. doi: 10.1128/iai.38.3.986-992.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shenker B J, McArthur W P, Tsai C C. Immune suppression induced by Actinobacillus actinomycetemcomitans: effects on human peripheral blood lymphocyte responses to mitogens and antigens. J Immunol. 1982;128:148–154. [PubMed] [Google Scholar]
- 43.Shenker B J, Vitale L A, Welham D A. Immune suppression induced by Actinobacillus actinomycetemcomitans: effects on immunoglobulin production by human B cells. Infect Immun. 1990;58:3856–3862. doi: 10.1128/iai.58.12.3856-3862.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Slots J, Reynolds H S, Genco R J. Actinobacillus actinomycetemcomitans in human periodontal disease: a cross-sectional microbiological investigation. Infect Immun. 1980;29:1013–1020. doi: 10.1128/iai.29.3.1013-1020.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Slots J, Schonfeld S E. Actinobacillus actinomycetemcomitans in localized juvenile periodontitis. In: Hamada S, Holt S C, McGhee J R, editors. Periodontal disease: pathogens & host immune responses. Tokyo, Japan: Quintesence Publishing; 1991. [Google Scholar]
- 46.Stevens R H, Gatewood C, Hammond B F. Cytotoxicity of the bacterium Actinobacillus actinomycetemcomitans extracts in human gingival fibroblasts. Arch Oral Biol. 1983;28:981–987. doi: 10.1016/0003-9969(83)90051-1. [DOI] [PubMed] [Google Scholar]
- 47.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 48.Sugai M, Komatsuzawa H, Akiyama T, Hong Y-M, Oshida T, Miyake Y, Yamaguchi T, Suginaka H. Identification of endo-β-N-acetylglucosaminidase and N-acetylmuramyl-l-alanine amidase as cluster-dispersing enzymes in Staphylococcus aureus. J Bacteriol. 1995;177:1491–1496. doi: 10.1128/jb.177.6.1491-1496.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Dyke T E, Bartholomew E, Genco R J, Slots J, Levine M J. Inhibition of neutrophil chemotaxis by soluble bacterial products. J Periodontol. 1982;53:502–508. doi: 10.1902/jop.1982.53.8.502. [DOI] [PubMed] [Google Scholar]
- 50.White P A, Wilson M, Nair S P, Kirby A C, Reddi K, Henderson B. Characterization of an antiproliferative surface-associated protein from Actinobacillus actinomycetemcomitans which can be neutralized by sera from a proportion of patients with localized juvenile periodontitis. Infect Immun. 1995;63:2612–2618. doi: 10.1128/iai.63.7.2612-2618.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Whitehouse C A, Balbo P B, Pesci E C, Cottle D L, Mirabito P M, Pickett C L. Campylobacter jejuni cytolethal distending toxin causes a G2-phase cell cycle block. Infect Immun. 1998;66:1934–1940. doi: 10.1128/iai.66.5.1934-1940.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson M, Henderson B. Virulence factors of Actinobacillus actinomycetemcomitans relevant to the pathogenesis of inflammatory periodontal disease. FEMS Microbiol Rev. 1995;17:365–379. doi: 10.1111/j.1574-6976.1995.tb00220.x. [DOI] [PubMed] [Google Scholar]
- 53.Wu H C. Biosynthesis of lipoproteins. In: Neidhart F F, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1005–1013. [Google Scholar]
- 54.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vector and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 55.Zambon J J, Haraszthy V I, Hariharan G, Lally E T, Demuth D R. The microbiology of early-onset periodontitis: association of highly toxic Actinobacillus actinomycetemcomitans strains with localized juvenile periodontitis. J Periodontol. 1996;67:282–290. doi: 10.1902/jop.1996.67.3s.282. [DOI] [PubMed] [Google Scholar]