Abstract
Nuclear magnetic resonance (NMR) titration and isothermal titration calorimetry can be combined to provide an assessment of how multivalent intrinsically disordered protein (IDP) interactions can involve enthalpy–entropy balance. Here, we describe the underlying technical details and additional methods, such as dynamic light scattering analysis, needed to assess these reactions. We apply this to a central interaction involving the disordered regions of phe–gly nucleoporins (FG-Nups) that contain multiple phenylalanine–glycine repeats which are of particular interest, as their interactions with nuclear transport factors (NTRs) underlie the paradoxically rapid yet also highly selective transport of macromolecules mediated by the nuclear pore complex (NPC). These analyses revealed that a combination of low per-FG motif affinity and the enthalpy–entropy balance prevents high-avidity interaction between FG-Nups and NTRs while the large number of FG motifs promotes frequent FG–NTR contacts, resulting in enhanced selectivity.
Keywords: NMR, IDPs, Isothermal titration calorimetry, Nucleoporins, Nuclear transport factors
1. Introduction
Many cellular processes involve complex, multivalent interactions rather than apparently single “lock-and-key” recognition events [1-9]. It is widely recognized that multivalency (i.e., the presence of multiple interaction motifs on a ligand, possibly complemented by multiple interaction sites on a target) can lead to substantial increases in affinity via “avidity” in antibody recognition [10], signal transduction, and formation of biomolecular condensates [11-12]. Many such interactions form stable, long-lived complexes. In contrast, some intrinsically disordered systems possess multivalent interactions yet maintain rapid exchange [13]. At one extreme of the spectrum of fuzzy, dynamic interactions are those of the lining of the inner portion of the nuclear pore complex (NPC) where varieties of phenylalanine–glycine-rich nucleoporins (FG-Nups) are immobilized at one terminus to the NPC’s rigid structure [14] and provide a barrier to cytoplasmic/nucleoplasmic exchange of materials other than that facilitated by specific nuclear transport receptors (NTRs). The FG-Nups have regular repeat sequence motifs [15] directly implying multivalent interactions, so an obvious question is how avidity is avoided in order to provide fast transit times in the few milliseconds for large cargoes and their NTRs [16]. A general answer involving the modest affinity between FG-Nups and NTRs balanced by the entropic cost of restriction on the intrinsic disorder of the chains has been suggested [17]. This involves a modest affinity between FG-Nups and NTRs, which is offset by the entropic cost of restriction on the intrinsic disorder of the chains, such that optimally the energies of binding and barrier are balanced and a macromolecule neither accumulates at nor is excluded from the NPC but passes rapidly and specifically. Confirmation requires assessment of the thermodynamic properties of FG-Nup/NTR complexes. In this chapter, we describe the technical details of how the balance of multivalent affinities of interactions from a repeated motif and the entropic disorder of connecting linkers can be done, following a shorter description of results, previously published [18].
To address this, several complex issues needed to be resolved.
The most direct assessment of thermodynamic properties of an interaction is the measurement of reaction enthalpy by isothermal titration calorimetry (ITC). However, care needs to be taken as the relatively weak (~millimolar) affinities of individual sites are recognized as being at the limit of ITC sensitivity [19], compromising the precision of measurement. Thus, studying low-affinity interactions requires substantial amounts of material (see Note 1). Other, less direct, methods may be more precise but lack accuracy (see Note 2).
The stoichiometry of the reactions of intrinsically disordered proteins (IDPs) with targets is potentially complicated by the large number of microstates associated with the intrinsic disorder of the linkers between the motifs (FGs for FG nucleoporins), and the possible range of timescales associated with interconversions among states, interacting and noninteracting with the NTR. Can a simple association/dissociation be ascribed to the interaction? Some independent validation of molecular stoichiometry is of significant benefit (see Note 3).
The effect of multivalency needs to be probed by varying the number and position of the interaction motifs with comparable controls.
In the current protocol, we use FG-repeat proteins from yeast NSP1 based on similar sequence lengths and negative controls by substitution of the sequence “FSFG” with “SSSG” (Fig. 1). The direct effect of multiple motifs is then addressed by variation of the number of FSFG motifs with standard adjacent linker separations (upper left, Fig. 1). The role of linker length is probed by increased separation of the active FSFG motifs (upper right, Fig. 1). These variants are then titrated with an NTR, the protein NFT2, which is the principal carrier for Ran GDP required for NPC transport function. ITC provides a direct measure of enthalpy () related directly to the equilibrium constant. Using nuclear magnetic resonance (NMR) spectroscopy, we obtain complementary information with greater precision related to the equilibrium constant. This is complemented by investigation of the stoichiometry of the complexes by dynamic light scattering (DLS).
Fig. 1.
Design of FSFG constructs with varying degrees of valency (left, upper) and with varying distance between two FSFG motifs (right, upper). Full sequence of FSFG6 construct (bottom)
2. Materials
All solutions are prepared with deionized water using Millipore Milli-Q typically with >17 MΩ.cm from the device. Reagents are stored at room temperature.
2.1. Buffers
Buffer A: 20 mM HEPES-KOH, pH 6.8, 150 mM KCl, 2 mM MgCl2.
2.2. Proteins
NMR samples: 7.5% D2O was added to NMR samples described in Methods in order to provide lock signal. There was no additional shift reference material needed because the co-dialysis of components prior to mixing provides a constant environment. As a check, in this system, a significant fraction of peaks is not perturbed in the titration.
2.3. Equipment
ITC: MicroCal Auto-iTC200.
NMR: All NMR experiments were conducted on Bruker spectrometers at 800 MHz.
DLS: DynaPro with plate reader.
2.4. Software
NMRPipe, CCPNMR, GraphPad, Origin, DynaPro DYNAMICS.
3. Methods
All procedures are carried out at 25 °C when temperature is selectable, otherwise at ambient temperature.
3.1. Protein Production
The various sequences of Fig. 1 discussed in 1 were constructed as follows. FSFG6 (identified in Fig. 1) and SSSG6 [20] DNA constructs codon-optimized for bacterial expression were synthesized. Gene fragments from the synthesized plasmids were ligated into either pET21b or pET24a vectors. The FSFG12 plasmid was constructed by inserting a restriction-digested FSFG6 plasmid fragment into a SpeI-digested FSFG6 plasmid. All the other FSFG variants were created by site-directed mutagenesis from those two parent constructs for FSFG6 and SSSG6. For NTF2, the genomic sequence from S. cerevisiae was used. All the proteins in this study are tagged with hexa-histidine on the C-termini, and their primary sequences are listed in the supplemental material p.S-7 of [18]. Samples are prepared and purified by standard methods [21-22], including stable isotope labeling. For FG constructs, [U-15N] labeling was used. For NTF2, [U-2H,15N] was used in order to obtain sufficiently slow transverse relaxation for NMR studies. Proteins were dissolved, dialyzed, and concentrated in Buffer A to typical stock concentration ranges of 0.2–2 mM (ligand FG constructs) and 6 mM for NTF2 for ITC NMR and DLS. Prior to use, diluted or undiluted solutions were centrifuged (~ 20,000 × g, 10 min) and ultra-filtered. The protein concentrations of the filtered samples are measured by bicinchoninic acid assay (BCA) [23] and diluted accordingly to the desired concentration with filtered and degassed dialysate.
3.2. NMR Titration for Affinity Measurement
NMR chemical shift data is analyzed using NMRPipe [24] and CCPNMR Analysis [25]. Titration experiments were performed in Buffer A using a fixed concentration of FG construct 15N-labeled (range 20–120 μM) sample and by preparation of separate samples for each titration point, typically ten in total (see Note 4). These conditions do not uniformly result in saturation but are appropriate for this system (see Note 3). An example of the spectrum is shown in Fig. 2.
- Chemical shifts for both the FSFG residues and all the assigned NTF2 residues were extracted from each titration point, and the chemical shift perturbations (CSP) were calculated as CSP, , where and are 15N and 1H chemical shift changes with respect to the free state. The CSPs were treated as a function of titrant concentration and fit to a standard equation [26].
where is the change in chemical shift at saturation, is the fixed protein concentration, and is the titrant concentration. Global fitting was performed with the above equation to derive . An example of the chemical shift changes of residues Fsfg, fSfg, fsFg, fsfG and their corresponding fits is shown in Fig. 2b.
Fig. 2.
NMR titrations. (a) HSQC spectrum of FSFG6 titrated with NTF2. (b) CSPs for residues with overlapping shifts associated with the six occurrences of FSFG are fitted to determine a global and fitted curves corresponding to the derived value normalized by the expected maximum CSP for each position. In this case, was determined to be 560 ± 10 μM
3.3. ITC Titration for Affinity measument
For each ITC experiment, separate stock solutions of FSFGx (identified in Fig. 1 upper) and NTF2 under go two rounds of dialysis together against the same Buffer A and are then concentrated by centrifugal concentrators. Samples were filtered and the final concnetrations for titration redetermined by BCA [23].
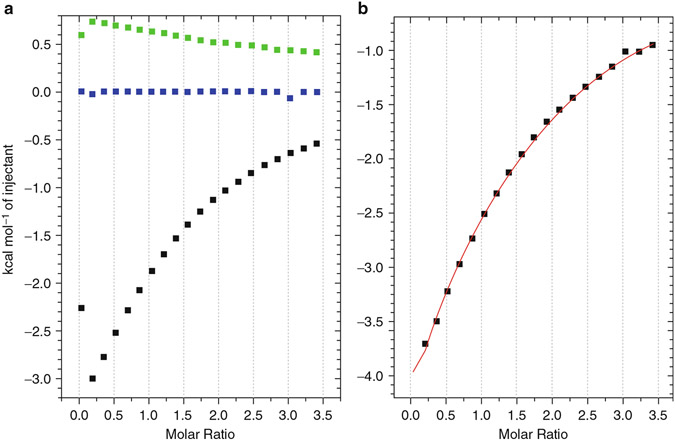
All the ITC experiments may be conducted on a MicroCal Auto-iTC200 (see Notes 1 and 5). FSFG constructs are placed in the cell, and NTF2 is titrated in from the syringe in all the experiments. Each experiment consists of three runs: FSFGx against NTF2, Buffer A against NTF2, and FSFGx against Buffer A (Fig. 3a). To obtain the normalized heat for FSFGx-NTF2 interaction, data from “Buffer against NTF2” and “FSFGx against Buffer” runs are subtracted from the “FSFGx against NTF2” run (Fig. 3b). The titration protocol involves either 19 (19 × 2 μL), 16 (10 × 2 μL followed by 6 × 3 μL), or 15 (15 × 2.5 μL) titration points following a 0.4 μL first injection, which is removed from the analysis. The choice of injection schemes does not affect the gross result, though injections with larger volumes generally improve signal-to-noise ratio toward the end of the titration [19]. Typical cell concentration of FSFGx are in the range 0.3–2.11 mM, while the syringe titrant NTF2 is 3.8–6.2 mM. These values are adjustable to obtain a favorable match of total heat released and of saturation.
The raw heat evolution data are integrated and analyzed by the ITC module within the Origin program, using the Wiseman isotherm, for the heat per moles of ligand added per step where for the step concentration of titrand .
The value is rounded to integer one (FSFG1-FSFG6) or two for (FSGS12) (see Subheading 3.3). For the binding reactions with very low affinity and low enthalpy (i.e., FSFG1–FSFG3, kcal/M), and could not be determined independently by ITC, though their product () was observable as previously reported for other low-affinity systems [19, 27]. could be reliably extracted from those reactions as evidenced by their agreement with NMR measurements, confirming the accuracy of the ITC experiment and its fitting procedure. However, since we cannot reliably determine and independently for FSFG1-FSFG3, only and (total enthalpy of the entire molecule) are reported for those constructs (Table S1 in ref. [18]), and values are derived separately using DLS for others. Of course, in the case of more complex reactions than , there will be complexity of interpretation common to such issues [28].
Enthalpy–entropy compensation curve are constructed by plotting against for constructs with increasing valency.
Fig. 3.
(a) NDH (normalized heat per injection) curves for each of the heat evolutions required for correct ITC measurement (black, lowest, FSFG6-NTF2; green, upper, buffer–NTF2; blue, center, FSFG6-buffer). Note (i) the discrepant first points at left out, arising from the initial step variation of the motor-driven syringe of the titrant which is left out during fitting, and (ii) the significant heat of dilution associated with the NTF2 dilution (green). (b) Reference-adjusted NDH curve for FSFG6 titrated with NTF2 (the green and blue NDH curves, left, were subtracted from the black resulting in the points and the fitted curve (red))
3.4. Determination of Stoichiometry
The formed complexes are characterized by dynamic light scattering (DLS). Experiments can ve run in a 384-well plate format with independent samples in triplicate. For each sample, ten acquisitions of 5 s are acquired on triplicate samples (see Note 6).
The protein complexes formed for ITC or NMR titrations are directly used.
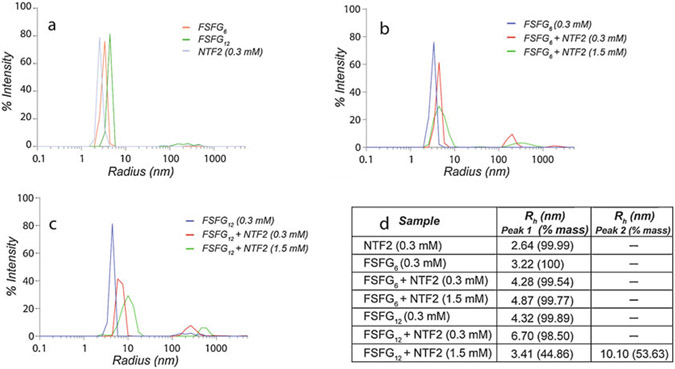
The resulting intensity-weighted regularized autocorrelation data, a decay curve resulting from Brownian motion of the complexes, is averaged over the triplicate data set, and the average radius of hydration calculated (Fig. 4).
Apparent radii of hydration are interpreted as consistent with 1:1 complexes for all ~12 kDa FG mimics and 1:2 for FSFG12/NTF2; that is, two NTF2 dimers interact with one FSFG12.
Fig. 4.
Determination of molecular stoichiometry by dynamic light scattering (DLS). (a) Intensity-weighted DLS of the free forms of FSFG6, FSFG12, and NTF2. (b) Intensity-weighted DLS comparing the free FSFG6 sample to those in the presence of increasing NTF2 concentrations. For the sample at 1:1 molar ratio, the intensity plot is derived from a mixture of free and 1:1 FSFG6:NTF2 molecular stoichiometry. No further increase in the peak position is observed at higher NTF2 concentrations. (c) Intensity-weighted DLS comparing the free FSFG12 sample to those in the presence of increasing NTF2 concentrations. The positions of the peaks in the presence of NTF2 are consistent with a shift from ~1:1 to ~ 2:1 NTF2:FSFG12 molecular stoichiometry at higher NTF2 concentrations. (d) Table reporting the radius of hydration, , and the percentage by mass of the peak. In the case of FSFG12, two separate peaks could be resolved at 1:5 molar ratio with NTF2, where the smaller of the two likely represents the free protein component. Each measurement represents data averaged from independent samples in triplicate
3.5. Estimation of Local Concentration Effect
The local concentration effect can be measured by observation of the change in with variation of the linker length between motifs. In the FG-Nup application, this was done by changing the distance between two FSFG motifs in the natural sequence with SSSG substitutions (Fig. 1, upper right). This results in additional separation equivalent to the insertion of 1, 2, 3, and 4 linked segments separating FSFG motifs.
- To estimate and to illustrate the change in local motif concentration as a function of FSFG motif–motif distance, we calculate [FSFG]local for our designed FSFG2 constructs (Fig. 1, upper right) based on polymer theory. Here, we define the [FSFG]local as the concentration of FSFG motifs within a probing volume defined by the spacer length, . We assumed that the spacer between the two FSFG motifs behaves as a random coil polymer consistent with experimental [21] and simulation [29] results. The calculated local concentration is based on a three-dimensional random flight model which is equivalent to an ideal chain undergoing random walks (rather than self-avoiding random walks) [30].
where is Avogadro’s number and the square distance (dm2) between the ends of the spacer given by where is the intersegment distance in Å and the number of segments. Using the distance distribution between adjacent FSFG motifs obtained from our previous simulation [29] as for adjacent FSFG repeats (), we calculate to be 8.5 Å (see Note 7). For each construct in the bivalent, differentially spaced, FSFG2 series, we calculate by multiplying as above and calculating the range of [FSFG]local concentrations as above. The [FSFG]local calculated above is simply the concentration of FSFG motifs that are within the explorable space of the target protein. The local concentration effect influences the overall binding reaction by either promoting formation of divalently interacting species (two interaction patches on NTF2 bound by two FSFG motifs on a molecule of dual-site NTR) or inducing a rebinding effect [31]. Simulation of this process [29] suggests the latter option.
Based on the calculations for the variant with dual motifs above, we can calculate [FSFG]local for the FSFG3–FSFG12 variants following the model proposed by Gargano et al. [32]. For the case here, we are dealing with a linear polymer and increased local concentration predominantly from vicinal motifs. As intuitively expected, the local concentration effect then increases modestly as the number of motifs increases. Other cases may differ with changes in interaction energy, and . However, the plateauing observed as a function of motif number (Fig. 5) is contrary to a simple local concentration prediction suggesting that a more detailed theory including at least the additions of chain self-avoidance and of excluded volume effect [30] is needed to account quantitatively for experiment.
Fig. 5.
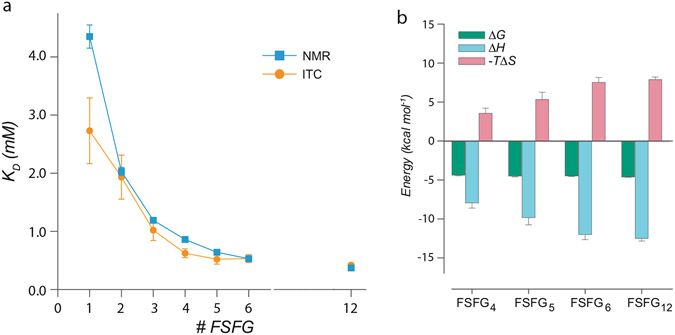
(a) values for each of the FSFG1-FSFG12 constructs (Fig. 1, left) by NMR and ITC. Standard errors of the curve fitting and standard errors of the mean are plotted for NMR and ITC, respectively. (b) Changes in Gibbs free energy (), enthalpy (), and entropy () for the interactions between FSFG4-FSFG12 constructs and NTF2 measured by ITC
3.6. Combining Results from the Different Analyses
As shown in Fig. 5a, there is excellent agreement between dissociation constants measured by the two different methods, strongly supporting the hypothesis that the time-averaged microstates for this interaction, central to the nuclear pore complex’s function, are represented by simple chemical equilibria. The combination of methods has two significant features: (1) the agreement provides a clear underpinning for thermodynamic analysis from ITC to the NMR data that are generally more precise and (2) the ITC provides a direct analysis of enthalpy and entropy as in Fig. 5b not readily available by other methods.
4. Notes
ITC instruments may have cells of different sizes. Larger cells will provide larger heat changes for more precise measurements, but at the cost of significant amounts of material required. In any broad survey study, the number of constructs to test may be large: For example, ref. 18 involved 14 different FG constructs, with each titration requiring two controls as well as repetitions (). Thus, the balance between production scale of materials and precision of measurement will require careful examination. The system used a MicroCal Auto-iTC200 with 200 μL of titrand in the cell. Note also that automated systems operating with sample trays will use significantly more material (~400 μL in our case) because of plumbing dead volumes.
When using ITC, the resulting heat is directly measuring the heat of reaction so that the thermodynamic quantity associated with the equilibrium is obtained without any other consideration. For other methods, there is typically an assumption that the measurable (e.g., chemical shift perturbation, or relaxation property for NMR, intensities of absorption or fluorescence for optical methods) is a direct measure of concentrations of reagents and products. This may be a limited approximation for several reasons. (1) The theoretical underpinning of the measurable/concentration dependence may be incorrect, for example, misanalysis of relaxation properties by NMR [33]. (2) The equilibrium may contain microstates, and the measurable detects a subset, not reflective of the overall reaction. (3) The use of derivative tags (e.g., for fluorescence) may directly affect the equilibrium by providing additional interactions not present in the native case (e.g. [34]).
The conventional analysis of equilibria would rely on observation of saturation at the stoichiometric equivalence point. However, millimolar range affinities frequently preclude direct observation of saturation because of limitations of solubility. Contemporary statistical fitting methods [35] will provide reasonable estimates of stoichiometry (and its significance level) from curvature of titrations without observation of saturation. In the case of FG-Nup/NTRs interaction and similarly non-standard cases, the possibility of separate microstates [36] leads to the need for an independent estimate of stoichiometry from DLS (Subheading 3.3).
In general, a significant quantity of surveying conditions may be needed to establish a range of conditions for observing NMR spectral changes during titration (see Note 2). Considerations will include sufficiently acidic pH such that solvent water exchange with amide 1H’s is minimal (typically, pH ≤ 6.8), and selection of a temperature to permit readily interpretable changes in chemical shifts, or relaxation properties [33], typically .
It is good practice to use the manufacturer’s calibration procedure before extensive work to ensure that the operator can obtain reproducible results. Of many issues, variations of room temperature, dust contamination of solutions, and gas bubbles in the titrant syringe are common. The titration protocol for these low value systems is not as critical as those for high-affinity systems. In brief, the value is the ratio of the titrand (receptor) concentration to the dissociation constant. Turnbull et al. [19] set out the conditions for using low values of as “(1) a sufficient portion of the binding isotherm is used for analysis, (2) the binding stoichiometry is known, (3) the concentrations of both ligand and receptor” conditions met in the study here.
To minimize contaminants, samples for DLS should be centrifuged immediately before pipetting into the sample tray. It is recommended that the sample tray be centrifuged briefly prior to the run to remove interference from air bubbles.
We chose to use a constant value of for simplicity, although the value for FSFG-(linker)-FSFG is obviously slightly shorter than the equivalent one-half of FSFG-(linker)-SSSG-(linker)-FSFG.
References
- 1.Stevers LM, de Vink PJ, Ottmann C et al. (2018) A thermodynamic model for multivalency in 14-3-3 protein-protein interactions. J Am Chem Soc 140(43):14498–14510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harmon TS, Holehouse AS, Rosen MK et al. (2017) Intrinsically disordered linkers determine the interplay between phase separation and gelation in multivalent proteins. elife 6. 10.7554/eLife.30294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gueroussov S, Weatheritt RJ, O’Hanlon D et al. (2017) Regulatory expansion in mammals of multivalent hnrnp assemblies that globally control alternative splicing. Cell 170 (2):324–339 e23 [DOI] [PubMed] [Google Scholar]
- 4.Vonnemann J, Liese S, Kuehne C et al. (2015) Size dependence of steric shielding and multivalency effects for globular binding inhibitors. J Am Chem Soc 137(7):2572–2579 [DOI] [PubMed] [Google Scholar]
- 5.Dubacheva GV, Curk T, Auzely-Velty R et al. (2015) Designing multivalent probes for tunable superselective targeting. Proc Natl Acad Sci U S A 112(18):5579–5584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark SA, Jespersen N, Woodward C et al. (2015) Multivalent IDP assemblies: unique properties of LC8-associated, IDP duplex scaffolds. FEBS Lett 589(19 Pt A):2543–2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li P, Banjade S, Cheng HC et al. (2012) Phase transitions in the assembly of multivalent signalling proteins. Nature 483(7389):336–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fasting C, Schalley CA, Weber M et al. (2012) Multivalency as a chemical organization and action principle. Angew Chem Int Ed Engl 51 (42):10472–10498 [DOI] [PubMed] [Google Scholar]
- 9.Cloninger MJ, Bilgiçer B, Li L et al. (2012) Multivalency. In: Supramolecular chemistry. Wiley, Hoboken, NJ [Google Scholar]
- 10.Koenderman L (2019) Inside-out control of fc-receptors. Front Immunol 10:544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Banani SF, Lee HO, Hyman AA et al. (2017) Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol 18 (5):285–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cable J, Brangwynne C, Seydoux G, et al. (2019) Phase separation in biology and disease—a symposium report. Ann N Y Acad Sci 1452(1):3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu H, Fuxreiter M (2016) The structure and Dynamics of higher-order assemblies: amyloids, signalosomes, and granules. Cell 165 (5):1055–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim SJ, Fernandez-Martinez J, Nudelman I et al. (2018) Integrative structure and functional anatomy of a nuclear pore complex. Nature 555(7697):475–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamada J, Phillips JL, Patel S et al. (2010) A bimodal distribution of two distinct categories of intrinsically disordered structures with separate functions in FG nucleoporins. Mol Cell Proteomics 9(10):2205–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grunwald D, Singer RH (2010) In vivo imaging of labelled endogenous beta-actin mRNA during nucleocytoplasmic transport. Nature 467(7315):604–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rout MP, Aitchison JD, Magnasco MO et al. (2003) Virtual gating and nuclear transport: the hole picture. Trends Cell Biol 13 (12):622–628 [DOI] [PubMed] [Google Scholar]
- 18.Hayama R, Sparks S, Hecht LM et al. (2018) Thermodynamic characterization of the multivalent interactions underlying rapid and selective translocation through the nuclear pore complex. J Biol Chem 293(12):4555–4563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turnbull WB, Daranas AH (2003) On the value of c: can low affinity systems be studied by isothermal titration calorimetry? J Am Chem Soc 125(48):14859–14866 [DOI] [PubMed] [Google Scholar]
- 20.Frey S, Richter RP, Gorlich D (2006) FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science 314(5800):815–817 [DOI] [PubMed] [Google Scholar]
- 21.Hough LE, Dutta K, Sparks S et al. (2015) The molecular mechanism of nuclear transport revealed by atomic-scale measurements. elife 4. 10.7554/eLife.10027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shekhtman A, Ghose R, Goger M et al. (2002) NMR structure determination and investigation using a reduced proton (REDPRO) labeling strategy for proteins. FEBS Lett 524 (1–3):177–182 [DOI] [PubMed] [Google Scholar]
- 23.Pe Smith, Krohn RI, Hermanson G et al. (1985) Measurement of protein using bicinchoninic acid. Analytical biochemistry 150(1):76–85 [DOI] [PubMed] [Google Scholar]
- 24.Delaglio F, Grzesiek S, Vuister GW et al. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6(3):277–293 [DOI] [PubMed] [Google Scholar]
- 25.Vranken WF, Boucher W, Stevens TJ et al. (2005) The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59(4):687–696 [DOI] [PubMed] [Google Scholar]
- 26.Williamson MP (2013) Using chemical shift perturbation to characterise ligand binding. Prog Nucl Magn Reson Spectrosc 73:1–16 [DOI] [PubMed] [Google Scholar]
- 27.Tellinghuisen J (2008) Isothermal titration calorimetry at very low c. Anal Biochem 373 (2):395–397 [DOI] [PubMed] [Google Scholar]
- 28.Weiss JN (1997) The Hill equation revisited: uses and misuses. FASEB J 11(11):835–841 [PubMed] [Google Scholar]
- 29.Raveh B, Karp JM, Sparks S et al. (2016) Slide- and-exchange mechanism for rapid and selective transport through the nuclear pore complex. Proc Natl Acad Sci U S A 113(18): E2489–E2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krishnamurthy VM, Semetey V, Bracher PJ et al. (2007) Dependence of effective molarity on linker length for an intramolecular protein-ligand system. J Am Chem Soc 129 (5):1312–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weber M, Bujotzek A, Haag R (2012) Quantifying the rebinding effect in multivalent chemical ligand-receptor systems. J Chem Phys 137 (5):054111. [DOI] [PubMed] [Google Scholar]
- 32.Gargano JM, Ngo T, Kim JY et al. (2001) Multivalent inhibition of AB(5) toxins. J Am Chem Soc 123(51):12909–12910 [DOI] [PubMed] [Google Scholar]
- 33.Waudby CA, Ramos A, Cabrita LD et al. (2016) Two-dimensional NMR Lineshape analysis. Sci Rep 6:24826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng H, Zhou BR, Bai Y (2018) Binding affinity and function of the extremely disordered protein complex containing human linker histone H1.0 and Its chaperone ProTalpha. Biochemistry 57(48):6645–6648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Motulsky HJ, Ransnas LA (1987) Fitting curves to data using nonlinear regression: a practical and nonmathematical review. FASEB J 1(5):365–374 [PubMed] [Google Scholar]
- 36.Dill K, Bromberg S (2012) Molecular driving forces: statistical thermodynamics in biology, chemistry, physics, and nanoscience. Garland Sci [Google Scholar]