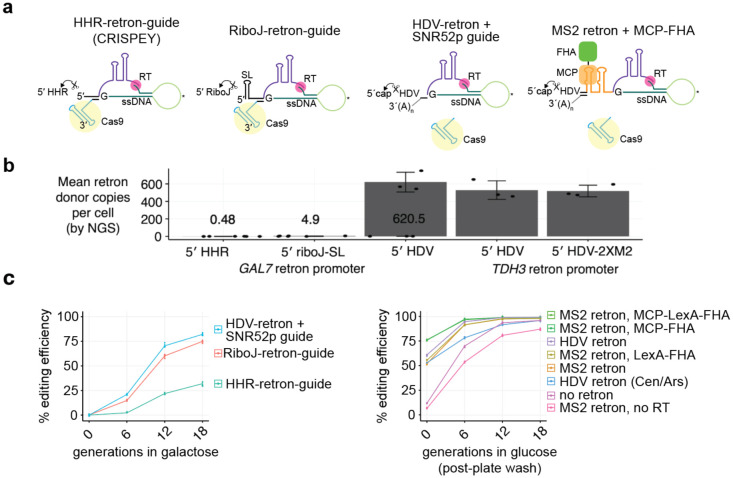
Figure 2.
Recruitment of retron donor DNA using the MS2 system and an MCP-FHA fusion protein.
a, Different arrangement of retrons tested shown from left to right in order of enhanced retron output and editing efficiency.
b, Retron donor cDNA output from each system as measured in Supp Fig 5. The levels for HHR and riboJ retrons are shown above each bar.
C, Editing efficiency for each retron arrangement shown in panel a as a function of generations of Cas9 and retron induction in galactose (left panel) or generations of liquid growth after colony formation on agar plates (right panel). The donor and guide are the same characterized in Sharon et al.4 for targeting the yeast ADE2 gene, where the guide was engineered to have only 18 bp of matching sequence. To give greater sensitivity towards measuring differences in template HDR rates and to simulate the weaker guides which would be observed in a genome-wide library, we artificially weakened this guide with an additional mismatch at position 17. The resulting guide is 5′-cacTTAACGAAATTGCCCCA-3′, where lowercase letters denote mismatches to the target site. All guide-donor plasmids used in the constitutive glucose system are 2μ (high-copy) plasmids, with the exception of the HDV retron (Cen/Ars) shown blue. All systems in the right panel have the guide RNA expressed under the SNR52 promoter, and all have a TDH3 promoter-driven retron donor except for the “no retron” system, which consists of a donor without any promoter or flanking retron elements. All constitutive systems express Cas9 from the TEF1 promoter and the RT from the ADH1 promoter (except for the no RT control), as well as the FHA fusion protein where indicated. Note that the donor transcribed under the retron promoter but without any retron (no RT control) showed reduced editing, suggesting that transcription through the donor is detrimental for plasmid-based template repair.