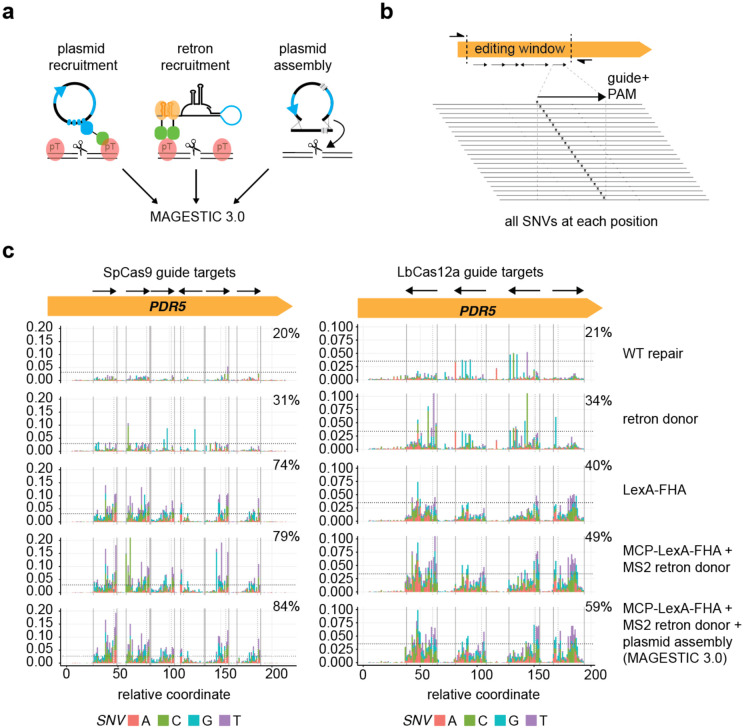
Figure 3.
Saturation genome editing with MAGESTIC 3.0.
a, MAGESTIC 3.0 utilizes three major HDR-enhancing technologies, dsDNA plasmid donor recruitment via LexA sites on the plasmid and the LexA-FHA fusion protein, ssDNA retron donor recruitment via the MS2 system, and plasmid assembly. The use of an MCP-LexA-FHA protein enables simultaneous recruitment of plasmids and retron donor to edit sites.
b, Editing windows with a panel of non-overlapping guides were selected and all possible SNVs across the guide target region and PAM were engineered into the donor DNAs.
c, On-target editing rates were quantified by high-throughput sequencing of target region amplicons. The fraction of reads containing SNVs at each coordinate on the x-axis is plotted on the y-axis with stacked bars with the colors representing the SNV introduced. The arrows in the editing window signify the guide and its orientation relative to the target site, with Cas9 guides containing the PAM 3′ of the arrow head (3′ end of the guide), and LbCas12a guides containing the PAM 5′ of the arrow tail (5′ end of the guide).