Abstract
Tissue development, function, and disease are largely driven by the spatial organization of individual cells and their cell-cell interactions. Precision engineered tissues with single-cell spatial resolution, therefore, have tremendous potential for next generation disease models, drug discovery, and regenerative therapeutics. Despite significant advancements in biofabrication approaches to improve feature resolution, strategies to fabricate tissues with the exact same organization of individual cells in their native cellular microenvironment have remained virtually non-existent to date. Here we report a method to spatially pattern single cells with up to eight cell phenotypes and subcellular spatial precision. As proof-of-concept we first demonstrate the ability to systematically assess the influence of cellular microenvironments on cell behavior by controllably altering the spatial arrangement of cell types in bioprinted precision cell-cell interaction arrays. We then demonstrate, for the first time, the ability to produce high-fidelity replicas of a patient’s annotated cancer biopsy with subcellular resolution. The ability to replicate native cellular microenvironments marks a significant advancement for precision biofabricated in-vitro models, where heterogenous tissues can be engineered with single-cell spatial precision to advance our understanding of complex biological systems in a controlled and systematic manner.
Keywords: Single-Cell, Bioprinting, Spatial Biology, Cell-Cell Interaction, Microfluidics, Tissue Engineering, Tumor Microenvironment
1. Introduction
Recent advances in spatial OMIC technologies have increasingly demonstrated the importance of single-cell spatial organization for the fate and function of a complex biological system.[1–3] The foremost characteristic of heterogenous native tissues and organs is the complexity at which many cell types interact through space and time to enable their biological processes such as morphogenesis and gene expression.[4–10] For example, the tumor microenvironment has been extensively studied for its high cellular heterogeneity and spatial complexity.[11–14] These studies have repeatedly demonstrated that the different cell phenotypes present, their abundance, and spatial organization are key determinants of clinical outcomes.[15–19] Therefore, given the growing knowledge surrounding the significance of cellular organization, precision engineered tissues with single-cell spatial control have extensive potential for next generation disease models, pharmaceutical discovery, and regenerative engineering.
Biofabrication approaches have progressed considerably in the last decade, with the primary goal of recapitulating the key structure and performance of native human tissues.[20] However, to date it has remained virtually impossible to fabricate tissue constructs that display the exact same organization of individual cells in their native cellular microenvironments. Spheroids/organoids rely largely on the stochastic self-assembly of aggregates of cells.[21] Extrusion and light-based 3D bioprinting methods have evolved to enable a remarkable level of material architectural complexity. However, with the exception of a few examples,[22–23] the positioning of cells within these biomaterials have remained limited to an average precision that is typically not higher than 100 μm or more.[24] Also, these methods offer limited ability to dispense the number of cell types required to generate heterogeneity at the levels found in native tissues. Laser and inkjet-based bioprinters have been able to dispense cells with much greater spatial accuracy.[25–30] Yet, their spatial patterning and multiplex dispensing capabilities have remained limited with regards to their ability to controllably recapitulate the exact precision and heterogeneity of single cells in native tissues.[24] Therefore, strategies to truly mimic the complexity of tissues in-vitro, with single-cell spatial resolution and adequate phenotypic heterogeneity, have remained virtually non-existent to date.
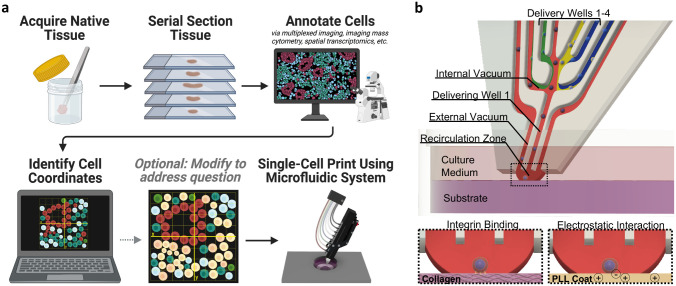
Here we report a method to spatially pattern single cells with an unprecedented level of spatial precision (1.3 μm between cells) and multitypic cell heterogeneity (up to 8 phenotypes in a single print). We demonstrate, for the first time, the feasibility of characterizing a region of interest in a complex patient-derived cell microenvironment, determine the coordinates of each cell from as many as five distinct phenotypes, and create high-fidelity copies of these tissue structures with below single-cell spatial accuracy and biological viability (Figure 1a). We argue that these developments open a new chapter in the field of biofabrication, where heterogenous tissues can be engineered with single-cell spatial precision to replicate native tissue structure.
Figure 1.
Single-cell bioprinting approach to engineer heterogenous tissues replicating native cellular microenvironments. a) Schematic illustration of the printing process from biopsy to print. A sample of native tissue is acquired, sectioned, and annotated to identify the cells present. The annotated image is opened in a custom software to identify the X and Y coordinates of each cell to generate the digital print map. If desired, the print map can be manipulated to investigate a particular question. Using a microfluidic dispenser, single cells are then deposited to match the digital print map. Created using Biorender.com. Annotated breast cancer biopsy adapted with permission.[31] 2020, Springer Nature. b) Microfluidic printhead illustrating the internal fluidic paths and interfaces, demonstrating the delivery of a cell. Cells traveling through the recirculation zone contact the underlying substrate where they immediately bind either through integrin binding to an extracellular matrix protein (i.e., collagen, Matrigel, etc.), or through electrostatic interaction of the negatively charged cell membrane to a positively charged surface (i.e., poly-L-lysine coated tissue culture plastic). If the cell does not immediately bind it is taken to a waste collection well via the external vacuum line.
2. Results and Discussion
Our single-cell bioprinting method builds on a microfluidic dispensing system that deposits cells, one at a time, suspended in their complete cell culture medium (Figure 1b, Figure S1, Supporting Information). The microfluidic device has eight independent wells, four for single-cell suspensions and four for waste collection (Figure S1b, Supporting Information). To enable high precision single-cell patterning, the cell must immediately bind to the substrate after being deposited. This is made possible by submerging the printhead in cell culture medium and positioning it <10 μm from the print surface so that as a cell exits the printhead it comes in contact with the underlying substrate. Upon contact, the cell can immediately bind through one of two mechanisms: integrin binding to an extracellular matrix protein or electrostatic interaction with a positively charged surface (Figure 1b). If the cell does not immediately bind, it is taken up by external vacuums that flank either side of the delivery channel, along with the cell culture medium that the cell was transported in.
Successful cell patterning is contingent upon the integrity of the cell membrane, substrate, printhead distance from the substrate, angle of the printhead, and fluid flow parameters. Dispensing cells with below single-cell spatial resolution and multiple phenotypes requires an additional unique set of optimization parameters. These include optimized microfluidic flow regimes, matrix and substrate conditions, cell-matrix communication parameters, and a complex matrix of parameters, which we report as key determinants to successfully bioprint tissue constructs with subcellular spatial resolution.
The flow of the microfluidic device is controlled by four independent parameters, the non-delivery pressure (0–50 mbar), delivery pressure (0–400 mbar), internal vacuum (0–300 -mbar), and external vacuum (0–300 -mbar), accounting for over 2 billion possible combinations. The non-delivery pressure maintains the flow of cells within all delivery wells where they converge in the crossover zone before interfacing with two internal vacuum lines (Figure S1c, Supporting Information). To dispense cells from a desired well, the delivery pressure is applied only to that well to create a no-slip boundary with the other lanes (Figure S1d, Supporting Information). The cell-laden medium that passes the internal vacuum is then dispensed out of the printhead where it interacts with the external vacuums. The relationship between the delivery pressure and the external vacuum pressures creates a confined recirculation zone of one miscible liquid inside another (Figure S1e, Supporting Information).
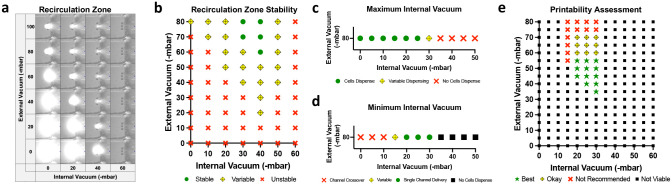
In order to controllably pattern single cells, we first needed to identify the precise balance of flow parameters that would support cell transport and deposition. We began by identifying the conditions that would produce a stable recirculation zone which is required for ideal cell binding during dispensing. We hypothesized that lower delivery pressures would exert less stress on the cells and ultimately lead to greater control over cell placement due to the slower flow speeds. Additionally, we hypothesized the non-delivery pressure would not be necessary and would likely lead to higher incidence of cell crossover (depositing cells from the wrong well). Therefore, we chose to investigate the stability of the recirculation zone at 60, 80, and 100 mbar delivery pressure, with 0 mbar non-delivery pressure, while incrementally varying the internal and external vacuum settings (Figure 2, Figures S2–S5, Supporting Information).
Figure 2.
Fluidics optimization for single-cell delivery at 80 mbar pressure. a) Visualization of recirculation zones and respective no-slip boundary condition by flowing fluorescein through transparent medium. b) Recirculation zone stability across different print heads (n = 3). c) Identification of the maximum internal vacuum which allows for cells to dispense (n = 9). d) Identification of the minimum internal vacuum that prevents unwanted dispensing from other wells (n = 9). e) Single-cell printability success assessment (n = 9).
The recirculation zone was visualized by continuously dispensing fluorescein into an optically clear medium (Figure 2a, Video 1, Supporting Information). Recirculation zones that did not expand beyond a defined boundary were deemed stable (Figure 2b). We found that pressure ranges that were able to consistently produce a stable recirculation zone across printheads varied depending on the combinations of delivery pressure, internal and external vacuums. For instance, 80 mbar of delivery pressure formed a stable recirculation zone for an external vacuum of −80 mbar when the internal vacuum ranged from −30 to −40 mbar. When the external vacuum was adjusted to −60 mbar, a higher internal vacuum was needed to offset the reduced vacuum; only −40 mbar internal vacuum was able to form stable recirculation at −60 mbar external vacuum.
We next sought to determine the pressure conditions that would allow cells to be dispensed from the printhead consistently as single cells, as opposed to doublets, triplets, or clusters. Human umbilical vein endothelial cells (HUVECs) were loaded into the printhead and dispensed, while the internal vacuum setting was varied at 5 mbar increments from 0 to −50 mbar, to identify the maximum viable internal vacuum setting (Figure 2c, Figures S6a and S7a, Supporting Information). As predicted, the threshold to enable cell dispensing was less than the threshold to enable fluid dispensing; at 80 mbar delivery pressure and −80 mbar external vacuum, fluid is consistently dispensed up to −40 mbar internal vacuum, but cells are only able to be consistently dispensed up to −25 mbar internal vacuum.
We then characterized the conditions that would enable controlled dispensing of a single cell type when all the dispensing wells of the device were loaded with different cell populations. GFP+ HUVECs were loaded into the desired dispensing well and GFP+ HUVECs that were co-stained with Hoechst were loaded into the remaining wells. If a co-expressing (GFP + Hoechst) HUVEC was deposited, unwanted crossover from another delivery well occurred and we concluded the internal vacuum setting was too low (Figure 2d, Figures S6b–c and S7b, Supporting Information). We also found that low internal vacuum pressures reduced the overall print precision by allowing more doublets or residual cell debris from culture and harvest to be deposited (Figure S6d, Supporting Information).
The remaining pressure combinations that maintained a stable recirculation zone, enabled cells to be deposited, and prevented unwanted channel crossover were investigated for their ability to controllably pattern single-cells. HUVECs were dispensed onto a collagen substrate and the printability was assessed based on the binding success, external vacuum interaction with underlying substrate, ability to deposit a single cell vs doublet(s), and printing speed (Figure 2e). We found HUVECs were able to immediately bind to the underlying collagen upon contact regardless of pressure combination. However, the lowest viable internal vacuum pressure deposited the most doublets and thus did not result in desirable single-cell printing results. Conversely, the highest viable internal vacuum pressures had the most variability across printheads (Figure S8, Supporting Information), requiring the most tuning per experiment. High internal vacuum settings also had overall slower print speeds as a greater number of cells are intercepted by the internal vacuum causing less cell to be deposited per second. We also found external vacuum pressures >70 -mbar would aspirate the collagen substrate, blocking the external vacuum and destabilizing the recirculation zone. Other substrates, such as rigid tissue culture plastic with a single layer protein coating, would likely support single-cell prints at higher external vacuum pressures.
We next asked whether these pressure conditions would be compatible with other substrates and cell phenotypes. 80 mbar delivery pressure, −25 mbar internal vacuum, and −40 mbar external vacuum were used to controllably deposit single GFP+ HUVECs onto collagen, gelatin, Matrigel, tissue culture plastic, and poly-L-lysine (PLL) coated tissue culture plastic with 50 μm spacing between cells (Figure 3a). Within 1 hour the cells began to migrate, spread, and divide. The prints were imaged again at 24 hours to demonstrate the printing did not grossly affect cell health, with Matrigel supporting a population doubling. We then printed eight of the most common cell phenotypes found in the body (immune, stem, nerve, bone, muscle, fibroblast, epithelial, and endothelial), fluorescently tagged prior to printing to facilitate phenotype identification, onto a collagen substrate using two printheads (Figure 3b). We were surprised to find these pressure conditions were compatible with all cell types tested despite the variability of physical characteristics, such as cell size or tendency for cells to aggregate which would add variability to the fluid regime. Together these results demonstrate our single-cell printing method is compatible with a variety of substrates and agnostic to cell and tissue type or disease state, providing a platform technology for a range of applications such as mechanistic investigation.
Figure 3.
Substrate and cellular compatibility with optimized flow parameters. a) GFP+ human umbilical vein endothelial cells (HUVEC) printed onto collagen, gelatin, Matrigel, tissue culture plastic (TCP), and poly-L-lysine (PLL) coated tissue culture plastic. Cells bind to collagen, gelatin, and Matrigel via integrin binding. Cells bind to TCP and PLL through electrostatic interaction. Scale bar = 100 μm. b) Eight of the most common cell types found in the body patterned in one print. Cells fluorescently tagged prior to printing to facilitate cell phenotype identification. Image taken immediately after printing before cells have spread. Scale bar = 100 μm.
Our driving motivation for developing the single-cell bioprinting method was to be able to create heterogeneous tissues that precisely replicate native cellular microenvironments. We approached this by first identifying and replicating key structural features of a tissue to incrementally build toward tissue level complexity. First, we needed to be able to controllably pattern single cells. We used prostate cancer cells (PC3) to pattern “OHSU” to illustrate controlled, single-cell deposition with complex curvatures (Figure 4a). Next is the variation in cell-cell spacing; most often cells are densely packed in a tissue with physical cell-cell junctions, but others are surrounded by extracellular matrix. To demonstrate our ability to controllably space cells, the Fibonacci sequence (also known as the golden ratio, where each number is the sum of the last two numbers) was printed (Figure 4b). At the smallest point the cells were spaced 1.3 μm apart, which we found to be an 84% improvement from existing inkjet and laser-assisted bioprinting approaches (~8 μm) published in the literature.[25] Being able to spatially pattern cells at this resolution is especially powerful, since the cells rapidly form cell-cell junctions with neighboring cells to create dense, tissue-like structures.
Figure 4.
Single-cell bioprinted constructs using fluorescently tagged cells. a) “OHSU” written using prostate cancer cells. Scale bar = 100 μm. b) Fibonacci sequence illustrating high precision spacing. Scale bar = 200 μm. c) Heterotypic print using four cell populations. Scale bar = 100 μm. d) 3D bioprint via collagen layering. Scale bar = 50 μm. e) Sequence demonstrating controlled single-cell manipulation. Using the controlled recirculation zone, cell detachment solution (TrypLE) was administered to a single cell until it detached. A new MCF10A with a different color was deposited in its place. Scale bar = 100 μm.
Cellular heterogeneity is another prominent feature of most tissues, many containing five cell types (endothelial, epithelial, fibroblast, immune, and specialized functional cell) or more. Unlike other high precision cell patterning strategies which can only support one material/cell type at a time, the most advanced version of this method can simultaneous print up to 8 different cell phenotypes without having to swap the loaded material. To demonstrate our multiplex abilities for heterotypic tissue generation on demand, HUVEC, MSC, red PC3, and yellow PC3 were loaded into the printhead and printed into a small array of cells (Figure 4c). This also serves as confirmation that the fluidics optimization was robust, as the internal vacuum was properly tuned to prevent cellular crossover. We then demonstrate that prints can also be extended into three dimensions (3D) by dispensing a thin hydrogel layer on top of the previously printed layer of cells. Here we printed MCF10A onto a collagen substrate, covered it with more collagen, and printed a subsequent layer of MDA-MB-231 (Figure 4d). 3D cultures hold many advantages over 2D cultures,[32] however, fabricating 3D structures via this approach is limited by different technical challenges relative to cell imaging and position during printing, as well as difficulties due to hydrogel dispensing fluidically or manually.
Although not a key structural feature of tissues, an advantageous fabrication strategy that we also optimized is the controlled manipulation of single cells on site. Using the confined recirculation zone, a dispensable liquid can be selectively delivered to one cell without reaching the neighbor cells.[33] We demonstrated the ability to manipulate a single cell by flowing a cell detachment agent (TrypLE) over one cell until it detached (Figure 4e). The cell was aspirated by the external vacuum along with the flowing TrypLE and brought to the waste collection well. A new cell (note different color) was then deposited into its place and this process was repeated with the remaining cells. Potential applications of this method include manipulation of cell-cell interactions after initial morphogenesis, targeted growth factor/drug delivery to single cells for mechanistic studies, activating a genetic mutation of a single cell within a printed environment after it has stabilized, and more.
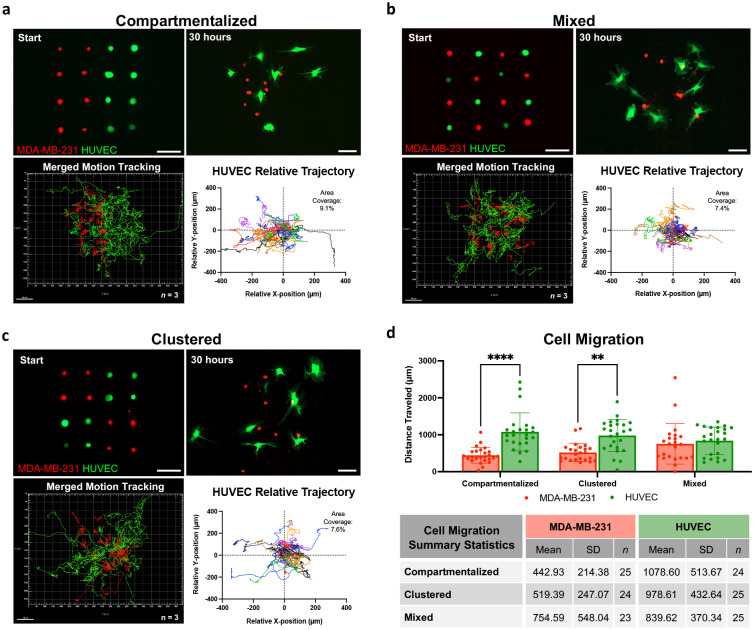
As proof of concept for how the single-cell printing method could be used to systematically assess the influence of cellular microenvironment on cell behavior, we printed cell-cell interaction arrays with varying spatial organization patterns. Recent work analyzing the influence of tumor microenvironment organization on cancer aggressiveness found that patients with compartmentalized cancer cells had longer progression-free survival compared with patients where cancer cells were sparsely distributed throughout the tumor.[17] Inspired by these spatially-determined outcomes, we printed MDA-MB-231 (triple negative breast cancer) and HUVECs into organized cell arrays with either compartmentalized or mixed spatial arrangements (Figure 5a–b). Each array had equal number of cells with 100 μm spacing between each cell. Live cell imaging over 30 hours revealed clear differences in migration pattern relative to starting spatial arrangement (Video 2–3, Supporting Information). Irrespective of patterning, however, HUVECs had a clear tendency to guide MDA-MB-231 cells together. HUVECs traveled less distance in the mixed arrangement compared to compartmentalized (839.6 ± 370.3 μm vs 1,078.6 ± 513.7 μm), which we suspect is because they did not have to travel as far to interact with a cancer cell. This was supported by relative trajectory analysis which revealed HUVECs in the compartmentalized arrangement tended to travel to the left, toward the cancer compartment, and their migration covered a greater area than the mixed arrangement (9.1% vs 7.4% area coverage). MDA-MB-231 migration also aligns with the observed HUVEC herding; MDA-MB-231 traveled the furthest distance in the mixed prints as the HUVECs tried to cluster them together (754.6 ± 548.0 μm vs 442.9 ± 214.4 μm).
Figure 5.
Cell-cell interaction arrays using human umbilical vein endothelial cells (HUVEC) and triple negative breast cancer cells (MDA-MB-231) fluorescently tagged prior to printing. Cells were printed in three spatial arrangements a) compartmentalized, b) mixed, and c) clustered and live cell imaged for 30 hours to track cell migration. Each line of the motion tracking indicates the path a single cell traveled. Replicates merged, n = 3 biological replicates. HUVEC relative trajectory with calculated area coverage. a–c) Scale bar = 100 μm. d) Distance each cell traveled with summary statistics displayed.
We next asked if these trends would be preserved in an arrangement that was less polarized than the compartmentalized vs mixed patterns (Figure 5c). When we clustered like cell types in the four corners, we found HUVEC and MDA-MB-231 migration fell within the range of the other patterns (978.6 ± 432.6 μm and 519.4 ± 247.07 μm respectively) (Figure 5d, Video 4, Supporting Information). Additionally, the HUVEC relative trajectory map of the clustered arrangement showed tight clustering like the mixed arrangement, but also had a few cells that traveled far out from the origin like the compartmentalized print (7.6% area coverage), suggesting a hybrid behavior of the two polarized spatial patterns. Together, these results demonstrate cell-cell interaction arrays can aid our understanding of cell response while accounting for the spatial distribution of cells within the local microenvironment in a highly systematic manner that hasn’t been possible to date. Unlike current approaches to study cell-cell interactions,[34] which rely on bulk populations of cells with little to no cellular spatial control, this approach allows for the direct and quantitative determination of the contributions of individual cells to the overall cellular, molecular, and genetic evolution of a cellular microenvironment.
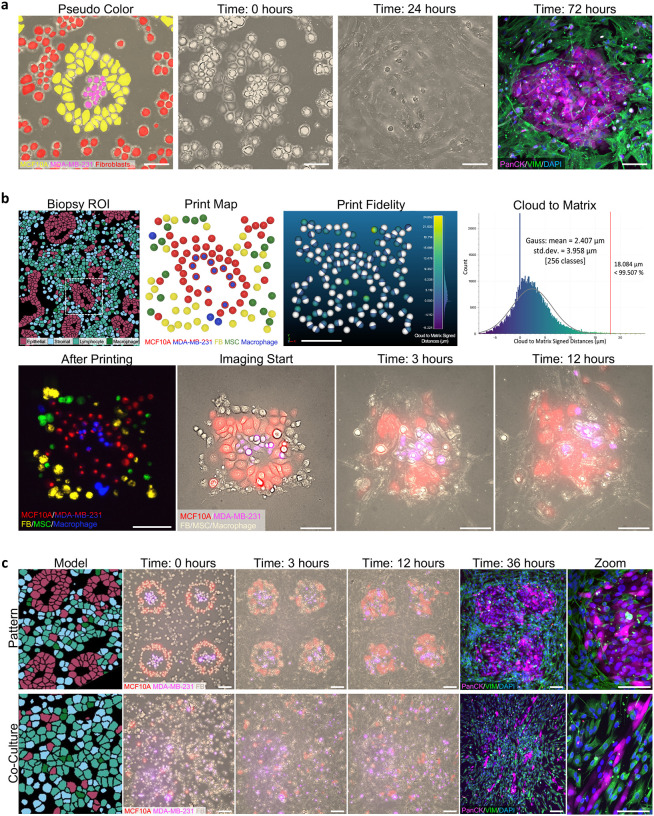
Once we were able to replicate the key structural features of a tissue with preserved biological viability, we then wanted to fabricate a densely populated, heterogenous tissue construct. Following the typical structure of a pre-invasive breast cancer model, we printed an idealized epithelial ring (MCF10A) around triple negative breast cancer cells (MDA-MB-231) and surrounded it with mammary fibroblasts (Figure 6a, Figure S9, Supporting Information). Live cell imaging revealed MCF10A spread to form junctions with their neighbors within 45 minutes and appeared to be actively trying to hold the cancer cells within their confines, which is a reported feature of these tumors.[35] Fibroblasts also quickly spread to fill in the empty spaces of the stromal compartment resulting in a dense, tissue-like morphology. Even after creating a confluent sheet, fibroblasts continued moving in a circulation pattern around the epithelial cells (Video 5, Supporting Information), potentially illustrating patterns of cell migration in the tissue that are lost in conventional static histological analyses. Immunostaining after three days of culture revealed the persistence of epithelial nests with fibroblasts wrapping around the edges. These results suggest heterogenous tissue constructs can be engineered for real-time monitoring of microenvironment evolution following a perturbation (i.e., drug treatment), for up to 72 hours.
Figure 6.
Native cellular microenvironment reconstruction with subcellular resolution. a) Engineered breast tumor microenvironment model pseudo colored to identify cell phenotypes: mammary epithelial (MCF10A), triple negative breast cancer (MDA-MB-231), and mammary fibroblasts. Live cell images at 0 and 24 hours, followed by immunofluorescence staining at 72 hours for pan cytokeratin (PanCK), vimentin (VIM), and nucleus (DAPI). Scale bar = 100 μm. b) High-fidelity replica of a native, patient tumor microenvironment. A region of interest (ROI) was identified from an annotated breast cancer biopsy adapted with permission.[31] 2020, Springer Nature. Digital print map and print result using five cell types: MCF10A, MDA-MB-231, mammary fibroblasts, mesenchymal stem/stromal cells (MSC), and macrophages. White spheres in print fidelity indicate print map and colored spheres are print result. Histogram of cloud to matrix signed distances (μm) with Gaussian distribution. Print result with cells stained prior to printing for phenotype identification with cell tracker. Live cell imaging of high-fidelity replica at imaging time 0, 3, and 12 hours. Scale bar = 100 μm. c) Patterned vs random (“co-culture”) prints containing MCF10A, MDA-MB-231, and mammary fibroblasts. Live cell images at 0, 3, and 12 hours. Immunofluorescence staining for pan cytokeratin (PanCK), vimentin (VIM), and nucleus (DAPI). Model images adapted with permission.[31] 2020, Springer Nature. Scale bar = 100 μm.
We then sought to replicate an exact native cellular microenvironment derived from a patient biopsy. This is a key component to enable the development of personalized cancer avatars,[36] where the precise position of each cell in a patient tumor microenvironment can be replicated to enable drug testing or disease studies without the spatial variability that is inherent to conventional co-culture models. To demonstrate this capability, a region of interest from a previously obtained and annotated breast cancer biopsy,[31] containing cancerous mammary epithelial cells with surrounding stromal compartment, was selected (Figure 6b). The X and Y coordinates of each cell were identified to generate a high-fidelity print map. Any cell that had a diameter less than the size of the cells we would be depositing was digitally removed (Figure S10a, Supporting Information). Most of these small spots represented edges of cells that were predominately in a different Z plane. Regions where multiple cells had been removed (i.e. wall of the duct) were filled in to reconstruct the native cellular environment as closely as possible. The final print map included five distinct cell phenotypes: MCF10A, MDA-MB-231, mammary fibroblasts, mesenchymal stem/stromal cells, and macrophages. We found the average distance between the digitally mapped targets and the resulting position of our tumor replicas was 1.603 ± 0.552 μm (n = 4, biological replicates) with 99.6% of points within one cell distance or less (18 μm), demonstrating for the first time the ability to recreate a native cellular microenvironment containing multiple cell phenotypes with subcellular resolution (Figure 6b, Figure S10b, Supporting Information). Live cell imaging of the tumor replica validated preserved cell viability, as cells were seen quickly taking on their expected morphology, were migratory, and proliferative (Video 6, Supporting Information). MCF10A cells had especially characteristic morphology, closely mirroring the native biopsy, as they quickly formed their junctions with neighboring epithelial cells. Overtime, however, the cells began migrating outward; without physical constraints, such as a wall or surrounding cells, cells will spread, migrate, and continue proliferating until the area is confluent.
To further illustrate the effect of microenvironment spatial organization on cell behavior, we fabricated a larger pre-invasive breast cancer tumor microenvironment, inspired by the cellular arrangement of the original patient biopsy, and compared to randomly deposited cells in a standard co-culture model (Figure 6c). There were clear differences in cellular morphology, organization, and density throughout the 36 hours of culture (Video 7–8, Supporting Information). Most predominate was the fibroblast elongation which guided MCF10A migration and subsequent elongation of these usually cobblestone-like cells. The expected patterns, such as the presence of a circumferential epithelial barrier confining the cancer cells within the core, or the surrounding stroma providing the boundary to this epithelial layer, were both completely absent in the conventional co-culture of plated cells, despite the fact that cells were seeded with the same general densities and on the exact same substrate. Together our results demonstrate the spatial patterning of cells to recreate native tissue structures has profound impacts on cell communication, behavior, and response, and can have major implications for the field of high-precision biofabrication.
3. Conclusion
In summary, here we demonstrate the development and optimization of a novel strategy to multiplex bioprint single cells with subcellular spatial resolution and high heterogeneity. These results demonstrate, for the first time, biofabrication of tissue constructs that match the exact position of cells in a native microenvironment with single cell spatial accuracy. We demonstrate the versatility of the method by printing 10 different cell phenotypes onto a variety of materials, making it a promising platform for a wide range of applications, two of which we illustrated in detail. First, cell-cell interaction arrays were fabricated to study cell behavior relative to their local microenvironment, opening new avenues for understanding and manipulating cellular responses in a systematically controllable spatial context. Secondly, using the breast tumor microenvironment as a heterogenous model tissue, we then demonstrate the ability to replicate native cellular structures with five distinct cell phenotypes, subcellular spatial resolution, and biological viability. These results represent significant advancement in biofabrication in three major directions - resolution, heterogeneity, and recapitulation of native cellular communication. The potential implications for advancing our understanding of complex biological systems and improving therapeutic interventions are vast, thus pointing towards new possibilities in the field of biofabrication.
4. Experimental Section
Human Cell Culture:
Primary green fluorescent protein-expressing human umbilical vein endothelial cells (GFP-HUVEC) were purchased from Angio-Proteomie (Catalog # cAP-0001GFP, Boston, MA) and cultured in vascular endothelial growth factor endothelial growth medium (VascuLife VEGF, Lifeline Cell Technology, Frederick, MD). Primary bone marrow-derived mesenchymal stem/stromal cells (MSC) were purchased from RoosterBio (Catalog # MSC-001, Frederick, MD) and cultured in alphaMEM supplemented with 10% MSC-qualified fetal bovine serum (FBS, Gibco) and 1% penicillin-streptomycin (Gibco). Primary mammary fibroblasts were purchased from ScienCell (Catalog # 7630, Carlsbad, CA) and cultured in DMEM supplemented with 10% FBS and 1% penicillin-streptomycin. Osteoblasts were purchased from ATCC (Catalog # CRL-3602, Manassas, VA) and cultured in osteoblast growth medium (Cell Applications, San Diego, CA). SH-SY5Y were purchased from ATCC (Catalog # CRL-2266, Manassas, VA) and cultured in DMEM supplemented with 10% FBS and 1% penicillin-streptomycin. MCF10A were purchased from Horizon Discovery (Catalog # HD 101–005, Boyertown, PA) and cultured in DMEM/F12 supplemented with 5% horse serum, 20 ng/mL epidermal growth factor (PeproTech, Catalog # AF-100–15, Cranbury, NJ), 0.5 mg/mL hydrocortisone (Sigma, Catalog # H0888), 100 ng/mL cholera toxin (Sigma, Catalog # C8052), 10 μg/mL insulin (Sigma, Catalog # I1882), and 1% penicillin-streptomycin. PC3 were kindly provided by Dr. Ryan Gordon and THP-1 were kindly provided by Dr. Jens Kreth. PC3 and THP-1 were cultured in RPMI 1640 supplemented with 10% FBS and 1% penicillin-streptomycin. Macrophages were derived from THP-1 following a previously established protocol.[37] Briefly, THP-1 were seeded into a 6 well plate at 2 × 105 cells/well and treated with 20 ng/mL phorbol-12-myristate 13-acetate (PMA, Sigma) in RPMI 1640 supplemented with 10% FBS. The media containing PMA was removed after 24 hours and cells were allowed to rest in fresh RPMI without PMA for 24 hours prior to use. Cardiomyocytes were kindly provided by Dr. Christopher Chen, derived from human induced pluripotent stem cells (hiPSCs) via small molecule manipulation of the Wnt signaling pathway.[38–39] Cardiomyocytes were cultured on Matrigel coated plates in RPMI supplemented with 1:50 B-27 (Fisher) and 5% FBS. The culture was maintained until the cardiomyocytes resumed beating before harvesting for printing. All cells were cultured at standard 37 °C and 5% CO2 and primary cells were used between passage 3 and 7. Unless otherwise stated, media was changed every second day and cells were passaged at 70% confluency using TrypLE Express (Gibco).
Cell Preparation for Single-Cell Printing:
Cells were detached using TrypLE for the minimum possible time and pelleted at 300 g for 3–5 minutes. While printing can be done label free, cells were either transduced to include a fluorescent reporter or fluorescently tagged using Hoechst (NucBlue Live ReadyProbes, Invitrogen) or general membrane cell tracker (Cellvue Claret Far Red, PKH26 Red Fluorescent Cell Linker, or PKH67 Green Fluorescent Cell Linker, Sigma) following the manufacture’s protocol to facilitate downstream analysis. Cells were then resuspended at 1 × 106 cells/mL in a 1:1 mixture of complete cell culture medium and polyethylene glycol (PEG, 35,000 m.w., 30 mg/mL, Sigma).
General Single-Cell Printing Setup:
The microfluidic dispenser (Biopixlar, Fluicell, Mölndal, Sweden) configuration was adjusted so that the printhead was angled at 70° and retrofitted with a heating unit to maintain the chamber at 37 °C and custom print map software.[40–41] Next, a collagen I (1.5 mg/mL, rat tail, Corning) receiving substrate was prepared by adding 200 μL of collagen to the inner ring of a 35 mm, low walled petri dish (μ-Dish 35 mm low, Ibidi, Fitchburg, Wisconsin) and placed in the incubator for 30 minutes to crosslink. A 30 μm channeled PDMS printhead (FBPX-30, Fluicell) was primed by adding 25 μL of ultra-pure sterile water (Invitrogen) to each well of the printhead and inserted into the priming pump (Fluicell) at 180 mbar for 3 minutes. Immediately prior to printing, the water was removed from all waste collection wells and any wells which cells will be added to. 25 μL of each cell suspension (1 × 106 cells/mL in a 1:1 mixture of complete cell culture medium and PEG) was added to the desired delivery wells. The printhead was loaded into the Biopixlar and prime 1 pre-set was executed (180 mbar delivery pressure to wells 1–4 simultaneously). 1.5 mL complete medium was added to the 35 mm dish containing the crosslinked collagen substrate and loaded into the printer. Following the completion of prime 1, the tip was gently wiped with the provided lint-free cloth (Fluicell) before lowering the printhead into the dish. Prime 2 pre-set was then executed to remove any bubbles in the fluidics (−220 mbar external and internal vacuums). The print surface was identified by dispensing a few cells onto the surface, bringing them into focus with the onboard microscope (LS620, Etaluma), and lowering the printhead next to the attached cells until a slight deflection was observed. If cells weren’t binding, the printhead was lowered at 5 μm increments. The attached cells were removed to avoid any unwanted print contamination prior to beginning the print by aligning the dispensing channel with the cells and increasing the internal vacuum.
Recirculation Zone Stability Assessment:
To assess the stability of the recirculation zone, fluorescein (200 μM in 1x PBS) was added into well 1 of the printhead and 1x PBS was added to wells 2–4. The printhead was lowered into a petri dish containing 1x PBS and a collagen substrate. Delivery pressure was set to either 60, 80, or 100 mbar, with non-delivery set to 0 mbar, and the internal and external vacuum settings were varied at 10 mbar increments from 0 to −100 mbar. The delivery pressure was applied for 10 seconds to observe the stability of the recirculation zone. If the area of the recirculation zone didn’t expand over the 10 second period, the recirculation was considered stable. A snapshot was taken at the 10 second mark before stopping the delivery pressure. This process was repeated across three different printheads (FBPX-30, Fluicell) to assess printhead to printhead variability.
Maximum Internal Vacuum Assessment:
GFP-HUVECs were added to well 1 and 1x PBS was added to wells 2–4. The delivery pressure was set to either 60, 80, or 100 mbar with equal external vacuum and 0 mbar non-delivery pressure. The internal vacuum was varied at 10 mbar increments from 0 to −50 mbar to determine the range of internal vacuum settings that support cell dispensing. The delivery pressure of the cell containing well was applied for 10 seconds and cell dispensing (yes/no) was recorded. This process was repeated across three different printheads (FBPX-30, Fluicell), with n = 3 per printhead, to assess printhead to printhead variability.
Minimum Internal Vacuum Assessment:
GFP-HUVECs were added to well 1 and GFP-HUVECs co-tagged with Hoechst (NucBlue Live ReadyProbes, Invitrogen) were added to wells 2–4. Delivery pressure was set to either 60, 80, or 100 mbar, with equal external vacuum and 0 mbar non-delivery pressure, and the internal vacuum was varied at 10 mbar increments from 0 to −40 mbar. GFP-HUVECs in well 1 were dispensed in a straight line for 10 seconds. If a Hoechst + GFP-HUVEC was dispensed the internal vacuum setting was determined to be too low. This process was repeated across three different printheads (FBPX-30, Fluicell), with n = 3 per printhead, to assess printhead to printhead variability.
Alternative Receiving Substrates:
35 mm, low walled petri dishes (μ-Dish 35 mm low Grid-500, Ibidi) were prepared by coating the surface with one of the following materials: 0.1% w/v gelatin (from porcine skin, gel strength 300 Type A, Sigma) in sterile water and incubated at room temperature for 5 minutes, Matrigel (Corning) diluted 1:80 in DMEM/F12 and incubated at 37 C for 1 hour, or 2 μg/mL Poly-L-Lysine (PLL, ScienCell) in sterile water and incubated at 37 C for 1 hour. Excess solution was aspirated and replaced with 1.5 mL of the cells’ complete medium minus FBS. PLL coated dishes requires two washes with sterile water before the addition of cell culture medium. To create three-dimensional stacks of collagen, the complete medium was aspirated from the dish following the completion of printing. An additional 25 uL of collagen I was gently pipetted onto the surface and incubated at 37 C and 5% CO2 for 15 minutes. Fresh medium was added to the dish and the printing resumed on the new layer of collagen.
Optimized Single-Cell Printing Settings:
Regardless of cell type, 1 × 106 cells/mL in a 1:1 mixture of complete cell culture medium and polyethylene glycol (PEG, 35,000 m.w., 30 mg/mL) was found to be suitable for single-cell deposition and binding onto a 1.5 mg/mL collagen I substrate. Optimized flow settings were 80 mbar delivery, 0 mbar non-delivery, −25 mbar internal vacuum, and −40 mbar external vacuum. For prints with cells positioned less than 25 μm away from one another, the external vacuum was dropped as low as −25 mbar to avoid disrupting the freshly deposited cells.
Imaging:
Prints were visualized in real-time using the onboard microscope (LS620, Etaluma). Fluorescent images of live cells, pre-labeled with a fluorescent tag prior to printing, were acquired using either EVOS Fl Auto Imaging System (Life Technologies) or spinning disk confocal (Yokogawa CSU-X1 on Zeiss Axio Observer). Live cell imaging of cell-cell interaction arrays was conducted on the Yokogawa spinning disk confocal with onboard environment chamber to maintain 37 °C and 5% CO2. Live cell imaging of tumor microenvironment prints was conducted on a widefield microscope (Nikon Ti2 automated microscope) with Orca Fusion BT camera and Oko-Lab environmental chamber maintained at 37 °C and 5% CO2.
Cell-Cell Interaction Arrays:
GFP+ HUVEC and RFP+ MDA-MB-231 were printed onto a collagen type 1 substrate and in cultured in a 1:1 ratio of HUVEC media (VascuLife VEGF, Lifeline Cell Technology) and MDA-MB-231 media (DMEM + 10% FBS + 1% penicillin-streptomycin). Each print was a 4 × 4 array, with 100 μm between each cell, and contained 8 HUVEC and 8 MDA-MB-231. The spatial arrangement of the cells throughout the array was varied: mixed (alternating cell types), clustered (like cell types in 2×2 clusters in the corners), and compartmentalized (like cell types grouped, half and half). The HUVECs were transduced to be GFP+ and the MDA-MB-231 were stained with red live cell tracker prior to printing (PKH26 Red Fluorescent Cell Linker, Sigma-Aldrich). Immediately following printing, the sample was brought to the spinning disk confocal (Yokogawa CSU-X1 on Zeiss Axio Observer) with environment chamber for live cell imaging at 37 C and 5% CO2. The prints were imaged every 10 minutes for 30 hours. Motion analysis was completed manually using Imaris (Oxford Instruments, Version 9.8). n = 3 biological replicates.
Breast Cancer Biopsy Recreation:
To replicate a tumor biopsy region of interest, a breast cancer biopsy previously annotated using imaging mass cytometry was selected. A region containing a breast duct with surrounding stroma was selected and imported into ImageJ (Fiji, NIH). The image was converted to an 8-bit image and threshold to segment each cell. Cells that were incorrectly fused were manually separated by drawing an eraser line. The particles were then analyzed and any cell with an area less than 254.47 μm2 (18 μm diameter, ~the average size of our detached cells) was removed. The resulting masked image was manually re-colored using Procreate (Savage Interactive) to return the annotations. A 3D print map model was then created using Fusion 360 (Autodesk CAD) by placing 18 μm diameter spheres in the center of each cell of the mask. In regions where cells had been removed due to size limitation, spheres were evenly distributed to fill in the void. The resulting 3D model was exported as the print map with the X,Y location of each cell. Using our custom overlay software,[40] the print map was match using MCF10A, MDA-MB-231, mammary fibroblasts, mesenchymal stem/stromal cells, and THP-1 derived macrophages. The print was cultured in a 1:6 mixture of MCF10A media and DMEM supplemented with 10% FBS and 1% penicillin-streptomycin. Live cell imaging was conducted using a widefield microscope (Nikon Ti2), acquiring an image every 10 minutes over 24 hours.
Biopsy Replica Print Fidelity Analysis:
The fluorescence image of the tumor biopsy replicas was imported into Fusion360 (Autodesk CAD). 18 μm diameter spheres were again placed in the center of each cell of the image. The resulting 3D model of the print and the original print map 3D model were exported as .OBJ files and imported into CloudCompare (v2.13.alpha, 2023). A cloud to mesh (C2M) comparison was computed using the default settings (octree level: auto, signed distances, multi-thread, maximum thread count: 8/8). The points were visualized with a histogram and a Gaussian distribution was fitted to determine print fidelity. n = 4, biological replicates.
Breast Tumor Microenvironment Model:
A pre-invasive breast cancer model was fabricated, modeled after a patient’s breast cancer biopsy, by first printing 200 μm diameter rings of MCF10A onto a collagen substrate. The prints were returned to the incubator while the remaining cell types were harvested. Within 45 minutes of the initial print, MDA-MB-231 were deposited within the MCF10A ring and primary mammary fibroblasts were patterned around the MCF10A rings and throughout the stromal compartment. A thick fibroblast box “wall” was printed around the entire print to serve as physical boundaries to facilitate cells staying within the region instead of migrating to fill in the vast open spaces of the 35 mm petri dish. To simulate standard co-culture approaches, MCF10A, MDA-MB-231, and mammary fibroblasts were mixed at a 2:1:5 ratio respectively and loaded into a single well of the printhead. Cells were deposited in a random distribution and no box perimeter was printed. Immediately following printing, live cell imaging began on a widefield microscope (Nikon Ti2) with environment maintained at 37 °C and 5% CO2. Images were acquired every 10 minutes for 24 hours.
Immunostaining:
Prints were washed three times with 1x PBS and fixed using 10% neutral buffered formalin (Thermo Fisher) for 30 minutes at room temperature. Cells were permeabilized using 0.1% Triton X-100 (Sigma) solution in PBS for 15 minutes at room temperature. The samples were then blocked using 1.5% bovine serum albumin (BSA, Sigma) in PBS for 1 hour at room temperature. Primary antibodies pan cytokeratin (PanCK, 1:100, OriGene Catalog # CF190032, Lot # F003) and vimentin (VIM, 1:200, Novus Biologicals Catalog # NBP1–31327, Lot # 44286) were added in 0.15% BSA and incubated at 4 °C overnight. Primary antibodies were removed, and the samples were washed three times with PBS before adding secondary antibodies (1:250, Invitrogen Alexa Fluor 555 goat anti-mouse, Catalog # A21422, Lot # 2139320, and Invitrogen Alexa Fluor 647 goat anti-rabbit, Invitrogen Catalog # A21244, Lot #2179230). Secondary antibodies were incubated at room temperature for 2 hours, removed, and samples washed twice with 0.1% Tween 20 (Fisher Biotech). Nuclei were then labeled with DAPI (NucBlue Fixed Cell ReadyProbes, Invitrogen) in PBS for 30 minutes at room temperature. Samples were imaged using a spinning disk confocal (Yokogawa CSU-X1 on Zeiss Axio Observer).
Statistical Analysis:
Two-way analysis of variance (ANOVA) with Tukey correction was performed using GraphPad Prism 10. Data are presented as mean ± standard deviation (SD), n = 3 unless otherwise stated. P value less than 0.05 is considered statistically significant. 0.1234 (ns), 0.0332 (*), 0.0021 (**), 0.0002 (***), <0.0001 (****).
Supplementary Material
Acknowledgements
The authors thank the Advanced Light Microscopy Core at Oregon Health and Science University, Michael McLellan of Christopher Chen’s lab for generating the hiPSC derived cardiomyocytes, Ryan Gordon for sharing the prostate cancer cell line PC3, Jens Kreth for sharing the monocyte cell line THP-1, Avathamsa Athirasala for generating the transduced MCF10A cell line, Alexander Davies and Rawan Makkawi for generating the lentiviral reporter used to transduce the MDA-MB-231 cell line, and Fluicell for early access to materials and technical support. This work was supported by funding from the Cancer Early Detection Advanced Research Center (CEDAR) (Project ID# Full 2023-1745) at Oregon Health and Science University, Knight Cancer Institute awarded to H.R.H. and L.E.B, and the OHSU Silver Family Innovation Award, the National Institute of Dental and Craniofacial Research (R01DEO29553), and the National Cancer Institute (R21CA263860) awarded to L.E.B. This work was also supported by fellowships to H.R.H. through the International Alliance for Cancer Early Detection (ACED), the National Institute of Dental and Craniofacial Research T90DE030859, and the Achievement Rewards for College Scientists (ARCS) Foundation Oregon. The International Alliance for Cancer Early Detection is an alliance between Cancer Research UK, Canary Center at Stanford University, the University of Cambridge, Oregon Health and Science University Knight Cancer Institute, University College London, and the University of Manchester. The authors also thank the Cell, Developmental and Cancer Biology Summer Research Internship program at the Knight Cancer Institute, Oregon Health and Science University for supporting K.A.O.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Conflicts of Interest
The authors declare no conflict of interest.
Contributor Information
Haylie R. Helms, Knight Cancer Precision Biofabrication Hub, Knight Cancer Institute, Oregon Health and Science University, Portland, OR 97201, USA Department of Biomedical Engineering, School of Medicine, Oregon Health and Science University, Portland, OR 97201, USA; Cancer Early Detection Advanced Research Center (CEDAR), Knight Cancer Institute, Oregon Health and Science University, Portland, OR 97201, USA.
Kody A. Oyama, Knight Cancer Precision Biofabrication Hub, Knight Cancer Institute, Oregon Health and Science University, Portland, OR 97201, USA
Jason P. Ware, Department of Biomedical Engineering, School of Medicine, Oregon Health and Science University, Portland, OR 97201, USA Cancer Early Detection Advanced Research Center (CEDAR), Knight Cancer Institute, Oregon Health and Science University, Portland, OR 97201, USA.
Stuart D. Ibsen, Department of Biomedical Engineering, School of Medicine, Oregon Health and Science University, Portland, OR 97201, USA Cancer Early Detection Advanced Research Center (CEDAR), Knight Cancer Institute, Oregon Health and Science University, Portland, OR 97201, USA.
Luiz E. Bertassoni, Knight Cancer Precision Biofabrication Hub, Knight Cancer Institute, Oregon Health and Science University, Portland, OR 97201, USA Department of Biomedical Engineering, School of Medicine, Oregon Health and Science University, Portland, OR 97201, USA; Cancer Early Detection Advanced Research Center (CEDAR), Knight Cancer Institute, Oregon Health and Science University, Portland, OR 97201, USA; Division of Oncological Sciences, Knight Cancer Institute, Oregon Health and Science University, Portland, OR 97201, USA; Division of Biomaterials and Biomechanics, Department of Oral Rehabilitation and Biosciences, School of Dentistry, Oregon Health and Science University, Portland, OR 97201, USA; Center for Regenerative Medicine, School of Medicine, Oregon Health and Science University, Portland, OR 97201, USA.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. Printer modifications have been made available at: https://zenodo.org/records/10501249 and https://zenodo.org/records/10511395.
References
- [1].Bressan D., Battistoni G., Hannon G. J., Science 2023, 381, eabq4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Rood J. E., Maartens A., Hupalowska A., Teichmann S. A., Regev A., Nat Med 2022, 28, 2486. [DOI] [PubMed] [Google Scholar]
- [3].Armingol E., Baghdassarian H. M., Lewis N. E., Nat Rev Genet 2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Goh I., Botting R. A., Rose A., Webb S., Engelbert J., Gitton Y., Stephenson E., Quiroga Londono M., Mather M., Mende N., Imaz-Rosshandler I., Yang L., Horsfall D., Basurto-Lozada D., Chipampe N. J., Rook V., Lee J. T. H., Ton M. L., Keitley D., Mazin P., Vijayabaskar M. S., Hannah R., Gambardella L., Green K., Ballereau S., Inoue M., Tuck E., Lorenzi V., Kwakwa K., Alsinet C., Olabi B., Miah M., Admane C., Popescu D. M., Acres M., Dixon D., Ness T., Coulthard R., Lisgo S., Henderson D. J., Dann E., Suo C., Kinston S. J., Park J. E., Polanski K., Marioni J., van Dongen S., Meyer K. B., de Bruijn M., Palis J., Behjati S., Laurenti E., Wilson N. K., Vento-Tormo R., Chedotal A., Bayraktar O., Roberts I., Jardine L., Gottgens B., Teichmann S. A., Haniffa M., Science 2023, 381, eadd7564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zhang B., He P., Lawrence J. E. G., Wang S., Tuck E., Williams B. A., Roberts K., Kleshchevnikov V., Mamanova L., Bolt L., Polanski K., Li T., Elmentaite R., Fasouli E. S., Prete M., He X., Yayon N., Fu Y., Yang H., Liang C., Zhang H., Blain R., Chedotal A., FitzPatrick D. R., Firth H., Dean A., Bayraktar O. A., Marioni J. C., Barker R. A., Storer M. A., Wold B. J., Zhang H., Teichmann S. A., Nature 2023. [Google Scholar]
- [6].Fawkner-Corbett D., Antanaviciute A., Parikh K., Jagielowicz M., Geros A. S., Gupta T., Ashley N., Khamis D., Fowler D., Morrissey E., Cunningham C., Johnson P. R. V., Koohy H., Simmons A., Cell 2021, 184, 810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Sampath Kumar A., Tian L., Bolondi A., Hernandez A. A., Stickels R., Kretzmer H., Murray E., Wittler L., Walther M., Barakat G., Haut L., Elkabetz Y., Macosko E. Z., Guignard L., Chen F., Meissner A., Nat Genet 2023, 55, 1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Sikkema L., Ramirez-Suastegui C., Strobl D. C., Gillett T. E., Zappia L., Madissoon E., Markov N. S., Zaragosi L. E., Ji Y., Ansari M., Arguel M. J., Apperloo L., Banchero M., Becavin C., Berg M., Chichelnitskiy E., Chung M. I., Collin A., Gay A. C. A., Gote-Schniering J., Hooshiar Kashani B., Inecik K., Jain M., Kapellos T. S., Kole T. M., Leroy S., Mayr C. H., Oliver A. J., von Papen M., Peter L., Taylor C. J., Walzthoeni T., Xu C., Bui L. T., De Donno C., Dony L., Faiz A., Guo M., Gutierrez A. J., Heumos L., Huang N., Ibarra I. L., Jackson N. D., Kadur Lakshminarasimha Murthy P., Lotfollahi M., Tabib T., Talavera-Lopez C., Travaglini K. J., Wilbrey-Clark A., Worlock K. B., Yoshida M., Lung Biological Network C., van den Berge M., Bosse Y., Desai T. J., Eickelberg O., Kaminski N., Krasnow M. A., Lafyatis R., Nikolic M. Z., Powell J. E., Rajagopal J., Rojas M., Rozenblatt-Rosen O., Seibold M. A., Sheppard D., Shepherd D. P., Sin D. D., Timens W., Tsankov A. M., Whitsett J., Xu Y., Banovich N. E., Barbry P., Duong T. E., Falk C. S., Meyer K. B., Kropski J. A., Pe’er D., Schiller H. B., Tata P. R., Schultze J. L., Teichmann S. A., Misharin A. V., Nawijn M. C., Luecken M. D., Theis F. J., Nat Med 2023, 29, 1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kumar T., Nee K., Wei R., He S., Nguyen Q. H., Bai S., Blake K., Pein M., Gong Y., Sei E., Hu M., Casasent A. K., Thennavan A., Li J., Tran T., Chen K., Nilges B., Kashikar N., Braubach O., Ben Cheikh B., Nikulina N., Chen H., Teshome M., Menegaz B., Javaid H., Nagi C., Montalvan J., Lev T., Mallya S., Tifrea D. F., Edwards R., Lin E., Parajuli R., Hanson S., Winocour S., Thompson A., Lim B., Lawson D. A., Kessenbrock K., Navin N., Nature 2023, 620, 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zhu K., Bendl J., Rahman S., Vicari J. M., Coleman C., Clarence T., Latouche O., Tsankova N. M., Li A., Brennand K. J., Lee D., Yuan G. C., Fullard J. F., Roussos P., Sci Adv 2023, 9, eadg3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Rozenblatt-Rosen O., Regev A., Oberdoerffer P., Nawy T., Hupalowska A., Rood J. E., Ashenberg O., Cerami E., Coffey R. J., Demir E., Ding L., Esplin E. D., Ford J. M., Goecks J., Ghosh S., Gray J. W., Guinney J., Hanlon S. E., Hughes S. K., Hwang E. S., Iacobuzio-Donahue C. A., Jane-Valbuena J., Johnson B. E., Lau K. S., Lively T., Mazzilli S. A., Pe’er D., Santagata S., Shalek A. K., Schapiro D., Snyder M. P., Sorger P. K., Spira A. E., Srivastava S., Tan K., West R. B., Williams E. H., Human Tumor Atlas N., Cell 2020, 181, 236.32302568 [Google Scholar]
- [12].Lewis S. M., Asselin-Labat M. L., Nguyen Q., Berthelet J., Tan X., Wimmer V. C., Merino D., Rogers K. L., Naik S. H., Nat Methods 2021, 18, 997. [DOI] [PubMed] [Google Scholar]
- [13].Wu Y., Cheng Y., Wang X., Fan J., Gao Q., Clin Transl Med 2022, 12, e696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Johnson B. E., Creason A. L., Stommel J. M., Keck J. M., Parmar S., Betts C. B., Blucher A., Boniface C., Bucher E., Burlingame E., Camp T., Chin K., Eng J., Estabrook J., Feiler H. S., Heskett M. B., Hu Z., Kolodzie A., Kong B. L., Labrie M., Lee J., Leyshock P., Mitri S., Patterson J., Riesterer J. L., Sivagnanam S., Somers J., Sudar D., Thibault G., Weeder B. R., Zheng C., Nan X., Thompson R. F., Heiser L. M., Spellman P. T., Thomas G., Demir E., Chang Y. H., Coussens L. M., Guimaraes A. R., Corless C., Goecks J., Bergan R., Mitri Z., Mills G. B., Gray J. W., Cell Rep Med 2022, 3, 100525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Galon J., Costes A., Sanchez-Cabo F., Kirilovsky A., Mlecnik B., Lagorce-Pages C., Tosolini M., Camus M., Berger A., Wind P., Zinzindohoue F., Bruneval P., Cugnenc P. H., Trajanoski Z., Fridman W. H., Pages F., Science 2006, 313, 1960. [DOI] [PubMed] [Google Scholar]
- [16].Danenberg E., Bardwell H., Zanotelli V. R. T., Provenzano E., Chin S. F., Rueda O. M., Green A., Rakha E., Aparicio S., Ellis I. O., Bodenmiller B., Caldas C., Ali H. R., Nat Genet 2022, 54, 660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Blise K. E., Sivagnanam S., Banik G. L., Coussens L. M., Goecks J., NPJ Precis Oncol 2022, 6, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Risom T., Glass D. R., Averbukh I., Liu C. C., Baranski A., Kagel A., McCaffrey E. F., Greenwald N. F., Rivero-Gutierrez B., Strand S. H., Varma S., Kong A., Keren L., Srivastava S., Zhu C., Khair Z., Veis D. J., Deschryver K., Vennam S., Maley C., Hwang E. S., Marks J. R., Bendall S. C., Colditz G. A., West R. B., Angelo M., Cell 2022, 185, 299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wang X. Q., Danenberg E., Huang C. S., Egle D., Callari M., Bermejo B., Dugo M., Zamagni C., Thill M., Anton A., Zambelli S., Russo S., Ciruelos E. M., Greil R., Gyorffy B., Semiglazov V., Colleoni M., Kelly C. M., Mariani G., Del Mastro L., Biasi O., Seitz R. S., Valagussa P., Viale G., Gianni L., Bianchini G., Ali H. R., Nature 2023, 621, 868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Xie R., Pal V., Yu Y., Lu X., Gao M., Liang S., Huang M., Peng W., Ozbolat I. T., Biomaterials 2024, 304, 122408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zhao Z., Chen X., Dowbaj A. M., Sljukic A., Bratlie K., Lin L., Fong E. L. S., Balachander G. M., Chen Z., Soragni A., Huch M., Zeng Y. A., Wang Q., Yu H., Nat Rev Methods Primers 2022, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].You S., Xiang Y., Hwang H. H., Berry D. B., Kiratitanaporn W., Guan J., Yao E., Tang M., Zhong Z., Ma X., Wangpraseurt D., Sun Y., Lu T. Y., Chen S., Sci Adv 2023, 9, eade7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lim K. S., Levato R., Costa P. F., Castilho M. D., Alcala-Orozco C. R., van Dorenmalen K. M. A., Melchels F. P. W., Gawlitta D., Hooper G. J., Malda J., Woodfield T. B. F., Biofabrication 2018, 10, 034101. [DOI] [PubMed] [Google Scholar]
- [24].Bertassoni L. E., Adv Mater 2022, 34, e2101321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zhang P., Liu C., Modavi C., Abate A., Chen H., Trends Biotechnol 2023. [DOI] [PubMed] [Google Scholar]
- [26].Zhang P., Abate A. R., Adv Mater 2020, 32, e2005346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Park J. A., Yoon S., Kwon J., Now H., Kim Y. K., Kim W. J., Yoo J. Y., Jung S., Sci Rep 2017, 7, 14610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Keriquel V., Oliveira H., Remy M., Ziane S., Delmond S., Rousseau B., Rey S., Catros S., Amedee J., Guillemot F., Fricain J. C., Sci Rep 2017, 7, 1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kang D., Park J. A., Kim W., Kim S., Lee H. R., Kim W. J., Yoo J. Y., Jung S., Adv Sci (Weinh) 2021, 8, 2004990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Jeffries G. D. M., Xu S., Lobovkina T., Kirejev V., Tusseau F., Gyllensten C., Singh A. K., Karila P., Moll L., Orwar O., Sci Rep 2020, 10, 19529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ali H. R., Jackson H. W., Zanotelli V. R. T., Danenberg E., Fischer J. R., Bardwell H., Provenzano E., Team C. I. G. C., Rueda O. M., Chin S. F., Aparicio S., Caldas C., Bodenmiller B., Nat Cancer 2020, 1, 163. [DOI] [PubMed] [Google Scholar]
- [32].Duval K., Grover H., Han L. H., Mou Y., Pegoraro A. F., Fredberg J., Chen Z., Physiology (Bethesda) 2017, 32, 266. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- [33].Ainla A., Jeffries G. D., Brune R., Orwar O., Jesorka A., Lab Chip 2012, 12, 1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Yang B. A., Westerhof T. M., Sabin K., Merajver S. D., Aguilar C. A., Adv Sci (Weinh) 2021, 8, 2002825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Gibson S. V., Roozitalab R. M., Allen M. D., Jones J. L., Carter E. P., Grose R. P., Trends Cancer 2023, 9, 326. [DOI] [PubMed] [Google Scholar]
- [36].Bose S., Barroso M., Chheda M. G., Clevers H., Elez E., Kaochar S., Kopetz S. E., Li X. N., Meric-Bernstam F., Meyer C. A., Mou H., Naegle K. M., Pera M. F., Perova Z., Politi K. A., Raphael B. J., Robson P., Sears R. C., Tabernero J., Tuveson D. A., Welm A. L., Welm B. E., Willey C. D., Salnikow K., Chuang J. H., Shen X., Cancer Cell 2022, 40, 1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Baxter E. W., Graham A. E., Re N. A., Carr I. M., Robinson J. I., Mackie S. L., Morgan A. W., J Immunol Methods 2020, 478, 112721. [DOI] [PubMed] [Google Scholar]
- [38].Javor J., Ewoldt J. K., Cloonan P. E., Chopra A., Luu R. J., Freychet G., Zhernenkov M., Ludwig K., Seidman J. G., Seidman C. E., Chen C. S., Bishop D. J., Microsyst Nanoeng 2021, 7, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Lian X., Zhang J., Azarin S. M., Zhu K., Hazeltine L. B., Bao X., Hsiao C., Kamp T. J., Palecek S. P., Nat Protoc 2013, 8, 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Ware J. P., Zenodo, 2024, 10.5281/zenodo.10511395. [DOI] [Google Scholar]
- [41].Ware J. P., Zenodo, 2024, 10.5281/zenodo.10501249. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. Printer modifications have been made available at: https://zenodo.org/records/10501249 and https://zenodo.org/records/10511395.