Abstract
Together with the molecular knowledge of genes and proteins, biological images promise to significantly enhance the scientific understanding of complex cellular systems and to advance predictive and personalized therapeutic products for human health. For this potential to be realized, quality-assured image data must be shared among labs at a global scale to be compared, pooled, and reanalyzed, thus unleashing untold potential beyond the original purpose for which the data was generated. There are two broad sets of requirements to enable image data sharing in the life sciences. One set of requirements is articulated in the companion White Paper entitled “Enabling Global Image Data Sharing in the Life Sciences,” which is published in parallel and addresses the need to build the cyberinfrastructure for sharing the digital array data (arXiv:2401.13023 [q-bio.OT], https://doi.org/10.48550/arXiv.2401.13023). In this White Paper, we detail a broad set of requirements, which involves collecting, managing, presenting, and propagating contextual information essential to assess the quality, understand the content, interpret the scientific implications, and reuse image data in the context of the experimental details. We start by providing an overview of the main lessons learned to date through international community activities, which have recently made considerable progress toward generating community standard practices for imaging Quality Control (QC) and metadata. We then provide a clear set of recommendations for amplifying this work. The driving goal is to address remaining challenges, and democratize access to common practices and tools for a spectrum of biomedical researchers, regardless of their expertise, access to resources, and geographical location.
Background and Motivation
Biological image data promises to significantly enhance our understanding of complex biological systems, and this promise requires that image data be compared, reanalyzed and shared among labs at a global scale. Over the past decades, advances in sharing of genomics data and protein structure data have revolutionized biology (i.e., the Human Genome Project, https://www.genome.gov/human-genome-project; and the Protein Data Bank - PDB, https://www.rcsb.org) and have been transformative for society. Biological image data sharing promises to have great impact as well because of the unique temporal and spatial data that biological imaging can provide. Together with the molecular knowledge of genes and proteins, biological images are essential for scientific understanding of complex cellular systems. This will make it possible to achieve predictive and personalized therapeutic products for human health and will benefit all sectors of the bioeconomy. Cellular, tissue and medical imaging provides vast amounts of data from the organisms that together hold the answers to disease management (i.e., surveillance, prevention, diagnosis, and treatment), new manufactured products, environmental resilience, and other global issues. The complexity of biological systems and the multidisciplinary nature of the research requirements mean that achieving this vision will require the interoperability, integration, and sharing of image data across laboratories and research studies. Much of this data, while often made publicly accessible in some form through publications, remain largely unexplored, uncurated, siloed and inaccessible, reducing the benefit and value of the tens of billions of dollars that are invested annually in scientific research around the globe.
Data sharing is globally recognized as highly desirable (UNESCO 2022; UNESCO and Canadian Commission for UNESCO 2022). The Open Science Movement (Ramachandran et al. 2021) is motivated by the idea that the production of FAIR (Findable, Accessible, Interoperable and Reusable, or more recently Findable and Artificial Intelligence Ready) (Wilkinson et al. 2016) data unleashes the untold potential for data beyond the original purpose for which it was generated. Many research funding applications now include requirements for Data Management and Sharing plans (NOT-OD-21–013: Final NIH Policy for Data Management and Sharing, Preparing your data management plan; European Research Council - Scientific Council; California Digital Library 2022). Although the intention to reuse image data is widely held, many technical challenges are preventing this vision from being realized. There are two broad requirements for image data sharing. One requirement is the cyberinfrastructure (Andreev et al. 2021) for sharing the digital array data, as articulated in the White Paper on Enabling Global Image Data Sharing in the Life Sciences (Bajcsy et al. 2024) and in other reports (NIH Strategic Plan for Data Science; Nagaraj et al. 2020), which highlight the critical need for infrastructure supporting the collection, analysis, and dissemination of image data in a coordinated, federated manner.
In this companion report, we detail a second class of requirements for image data sharing, which involves collecting, managing, presenting, and propagating contextual information about the data generation process. This information is essential to assess the quality, to understand, interpret, and reuse the image data in the context of the experimental details. These essential provenance and quality control (QC) metadata describe the pre-publication steps in an imaging study, and include details about experimental protocols and reagents, instruments, and image data processing and analysis. These needs are analogous to activities undertaken by users of genomic data: as the collection of genetic and genomic data became more prevalent, the comparison and sharing of data required establishing metrics and protocols for evaluating and assuring the provenance and quality of the data. Two such efforts are the Minimum Information About a Microarray Experiment (MIAME) (Rustici et al. 2008) and Minimum Information about a high-throughput SEQuencing Experiment (MINSEQE) (Brazma et al. 2012) specifications, which include reporting the description of the system, samples and experimental variables, the experimental protocols, the quality scores for sequence data, and the data processing protocols. Image data are more complicated than genomic data because of the variety of experiment types, imaging modalities, and instrumentation that can be used, and the sensitivity of living systems to handling and reagents. Regardless of how convincing published imaging data looks, it often does not convey sufficient information about the conditions in which it was acquired, processed, and analyzed, making its scientific interpretation often difficult, if not impossible (Linkert et al. 2010; Eriksson and Pukonen 2018; Nature Editorial Staff 2018; Sheen et al. 2019; Botvinik-Nezer et al. 2020; Marqués et al. 2020; Pines 2020; Chen et al. 2023; Viana et al. 2023). As is true for genomic data, reporting sufficient information about image data and its appropriate management across the experimental lifecycle enables independent evaluation of the results, engenders confidence in reproducibility, and provides means to assess the suitability (or futility) of data reuse.
A number of international community activities are engaged in addressing the challenges associated with the implementation of FAIR principles during image data generation and post-acquisition processing. The Open Microscopy Environment (OME) (Swedlow et al. 2003, 2006; Goldberg et al. 2005; Linkert et al. 2010; Allan et al. 2012; Moore et al. 2021, 2023) has been active for many years in encouraging metadata collection and in standardizing image data file formats. The global bioimaging community, in particular, the African BioImaging Consortium (ABIC) (African BioImaging Consortium (ABIC) 2020), Association of Biomolecular Resource Facilities (ABRF) (Abrams et al. 2020), BioImaging North America (BINA) (Strambio-De-Castillia et al. 2019), Euro-BioImaging (Kemmer et al. 2023), the European BioInformatics Institute (EMBL-EBI) (Ellenberg et al. 2018), Global BioImaging (Global BioImaging 2015; Eriksson and Pukonen 2018; Swedlow et al. 2021), Quality Assessment and Reproducibility for Instruments & Images in Light Microscopy (QUAREP-LiMi) (Boehm et al. 2021; Nelson et al. 2021), the RTmfm (RTmfm 2022), and the NIH-funded 4D Nucleome (4DN) Project (Dekker et al. 2017, 2023), have recently coalesced around improving community standard practices for instrument QC and metadata (Microscopy Australia 2016; Hammer et al. 2021; Huisman et al. 2021; Montero Llopis et al. 2021; Rigano et al. 2021; Sarkans et al. 2021; Faklaris et al. 2022). Through these community efforts (Supplemental Table 1), individual imaging laboratories and Shared Research Resources (SRR; i.e., commonly known and hereafter referred to as core facilities) are coming together with instrument manufacturers to define shared metadata frameworks and execute inter-laboratory studies to refine and deploy standard methods for QC (Faklaris et al. 2022; Gaudreault et al. 2022; Nelson 2022; Abrams et al. 2023). These groups have demonstrated interest and a willingness to voluntarily commit precious resources, and the funding for these efforts has been sufficient to allow limited but significant headway within small pockets of the broader imaging community. Despite several remaining challenges, this progress is the beginning of a path forward for biomedical researchers to generate and manage reliable and well-documented microscopy data that can be trusted and reused. By satisfying FAIR principles this will then help unlock the vast potential of quantitative image-based research.
In this white paper, we provide an overview of the main lessons learned to date through this community work and provide a clear set of recommendations moving forward on how this work can be amplified. The driving goal is to address remaining challenges and democratize access to common practices and tools so as to involve a wide spectrum of biomedical researchers regardless of their expertise, access to resources, and geographical location.
Data generation challenges lie at the very heart of the image data lifecycle, involve important considerations that are made at the planning phase of the research project, and are relevant often even before the sample hits the image acquisition platform.
We start by describing the challenges connected with sample preparation and image acquisition. We will then move on to describe issues related to reproducible and reliable post-acquisition processing of image data to extract quantitative measurements. Last but not least, we will describe the importance of data stewardship (Steeleworthy 2014; Boeckhout et al. 2018; Demchenko and Stoy 2021), also known as Research Data Management (RDM). Best practices and tools for data stewardship would consist of a broad set of processes that are undertaken to produce organized, well-documented, securely stored, accessible, and reusable high-quality research data both during the course of a research project and in preparation for data sharing. As such, data stewardship is essential to maintain a persistent link between image data and metadata across the entire experimental lifecycle. Ensuring that the origin and lineage (i.e., provenance) of data can be tracked and its quality assessed is an essential prerequisite for guaranteeing FAIR characteristics for the microscopy data. The scientific and sharing value derived from these metadata is the extent to which the data serve its intended scientific purpose and can be shared with other scientists to extract further insights.
While these challenges involve all areas of biomedical imaging, in this white paper, we concentrate specifically on optical microscopy, mass spectrometry imaging and electron microscopy modalities. These modalities are used to perform quantitative biomedical imaging by automated quantification of the spatial distribution of molecules and supramolecular structures at the sub-cellular, cellular, tissue and whole organism level. This choice was made because image data and metadata standards have been established and are routinely used to ensure instrument QC and for the interchange of medical imaging data in clinical settings (Bidgood et al. 1997; Mustra et al. 2008). Even though we are not going to discuss Medical Imaging explicitly, all considerations discussed here broadly apply to several different biomedical imaging techniques.
Data Generation
Good practices in data generation and management that ensure that data are “FAIR from the start” are essential for rigorous and reproducible quantitative cell image-based research and for producing image data that can be interpreted, trusted, and reused through model-based and data-driven mining, aggregation, reanalysis, and integrative modeling.
Although the definition of image quality remains vague and varies significantly depending on the context, what is crucial is that third-party data users have ready access to all data-related information (i.e., metadata) that enables them to evaluate the suitability of given datasets for answering specific scientific questions before accessing or downloading them. In the case of biological imaging, such information is collectively called image metadata and includes details about experimental conditions, sample description and preparation, image acquisition (i.e., hardware description and image acquisition settings) and image processing, visualization, and analysis (i.e., data provenance metadata), as well as system performance recorded through standardized QC protocols and metrics (i.e., QC metadata).
Experimental Conditions and Sample Preparation
Description and interpretation of the results of any microscopy research project requires an extended knowledge of the fundamental experimental factors and the sample itself. Such information should be captured not only in the Methods section of journal articles (Marqués et al. 2020; Montero Llopis et al. 2021; Larsen et al. 2023) but also as machine-readable structured metadata to be associated with any dataset made available for sharing and reuse. These metadata fields should include the following categories: 1) the source or provenance of the biological sample and how it was obtained, processed, cultured, and prepared, 2) the protocols and reagents (e.g., labeling procedures) used to visualize the structure of interest in the sample, 3) the mounting technique and media used to preserve the integrity of the specimen during imaging, and 4) the sample receptacle (e.g., slide and cover slip) used to hold the sample during image acquisition.
To facilitate the appropriate metadata annotation of image datasets to be shared and reused, communities are starting to converge towards shared minimal metadata guidelines such as those provided by the Recommended Metadata for Biological Images (REMBI) framework (Sarkans et al. 2021), which was developed by a 2019 community gathering to address the data stewardship and sharing needs of the light, electron and X-ray microscopy fields (Sarkans et al. 2021). REMBI provides a high-level map of the different metadata topics that have to be covered to ensure interpretability and trust and can serve as a convergence point for other communities working on each individual aspect. Along these lines, important efforts are represented by the multiplexed Tissue Imaging (MITI) minimum information guidelines for highly multiplexed tissue images (Schapiro et al. 2022) and Minimum Information about Cell Migration Experiments (Cell Migration Standardisation Organisation 2021), which provide guidance at multiple levels of the experimental procedure and sample preparation documentation.
Also key to compliance and implementation of effective metadata usage is the utilization of consistent ontologies for knowledge representation (Jupp; Ong et al. 2017; Hotchkiss et al. 2019; Sickle Cell Disease Ontology Working Group 2019), and where possible, the automated capture and annotation of metadata.
Microscope Hardware Specifications, Image Acquisition Settings and QC
Quality assessment, reproducibility, interpretation and reuse of image data are critically dependent on the availability of sufficient information about the hardware specifications, image acquisition settings and performance of the instrument used at the time of the data acquisition (Hammer et al. 2021; Huisman et al. 2021). A full technical description of the configuration of the imaging system can be used to calculate key information about spatiotemporal resolution, the noise associated with the system, as well as the physical and temporal dimensions of the image pixel data it generates. An instrument performance assessment plan, including tracking standardized QC metrics at regular intervals, can be used to quantitatively measure variability, changes in performance over time, and decline in the consistency of measurements (Faklaris et al. 2022; Gaudreault et al. 2022; Nelson 2022; Abrams et al. 2023). These metrics in turn allow us to quantify disparities between expected (theoretical) and observed (empirical) values and compare the results with the values measured during the system installation (t=0 for the microscope lifetime). The metrics can also help to characterize and calibrate derived quantities that can be extracted by image analysis (e.g., coregistration measurements). Ultimately, capturing the overall state (Rigano et al. 2021) and performance of the microscope at the time of data acquisition, and linking this information to the acquired image data in the form of metadata, is essential to identify potential batch effects in large datasets (Viana et al. 2023) Batch effects has been shown to significantly impact the performance of Artificial Intelligence / Machine Learning (AI/ML) algorithms (Arevalo et al. 2023; Cimini et al. 2023; Tromans-Coia et al. 2023), so capturing instrument state and performance is critical for the interpretation of results (Chen et al. 2023; Viana et al. 2023).
Documentation of and Integration with image data processing visualization and analysis
For the assessment of the analysis quality, reproducibility and proper results interpretation, all details pertaining to the image analysis workflow should be provided as described in recently developed community guidelines (Aaron and Chew 2021; Miura and Nørrelykke 2021; Schmied et al. 2023). Additionally, the data size and computing hardware and networking requirements should be provided as part of the metadata.
However, this is not a trivial request, as providing primary software versions does not ensure reproducibility. This is due to potential variabilities introduced by the chain of software packages upon which the primary software depends. Deep dependency graphs are not uncommon in scientific analysis programs written in the Python and R programming languages. As an illustration of this complexity Supplemental Figure 1 displays the dependency graph for the napari image analysis program version 0.4.18 (Ahlers et al. 2023). Thus, if the software tool used to perform a given step of the image processing/analysis pipeline does not explicitly constrain the version of each component of the chain of dependency, installing the original version of the primary software may not ensure the same versions of the components it depends upon were installed. Accurately reporting all dependencies beyond the primary software in a manual fashion is unfeasible and is best performed using automated package managers (e.g., the pip freeze command).
Since image processing pipelines may rely on several tools, one must also address the need to ensure that the intermediate and final results of processing and analysis pipelines and associated metadata are stored in an harmonized and comparable manner across different software tools (Könnecke et al. 2015). Developers have made important strides towards the use of containers (González and Evans 2019; Bajcsy and Hotaling 2020; Mitra-Behura et al. 2021; Schapiro et al. 2021), workflow tools (Wollmann et al. 2017, 2023; Stirling et al. 2021; Berthold 2023; Di Tommaso and Floden 2023; KNIME Community and bioml-konstanz 2023), cloud-ready data exchange formats (Moore et al. 2021, 2023; Swedlow et al. 2021), metadata frameworks (Moore 2022a) and standardized Application Programming Interfaces (APIs) that allow integration of images and results. However, much work is still left to do to make these solutions robust and universally adopted.
If one could recreate the analysis environment in terms of hardware, operating system, image analysis software and all parameter settings, one should obtain the same results. In practice, if the image analysis software utilizes randomness as part of its computations, obtaining exactly the same results is unlikely, and obtaining results within error margins that are considered acceptable in a given experimental context (i.e., similar results) is generally considered sufficient (Registration overview — SimpleITK documentation; PyTorch Consortium 2023; TensorFlow Development Team 2023). Finally, algorithms and implementations utilizing randomness require special care. This entails the fixing and sharing software parameters which are often not fixed, random seed values, or sharing of additional information. For example, when using deep learning, replicating results obtained by a retrained or new model, requires access to the code and model weights which should be shared using an interoperable file format across deep learning frameworks (e.g., the Open Neural Network Exchange, ONNX format, https://onnx.ai/).
Stewardship of image data during the duration of the research project
Data stewardship is an intrinsic and essential aspect of the generation of high-quality image data that is “FAIR from the start.” For this to happen, data stewardship has to involve the entire lifecycle of the data, starting with the planning phase and continuing during experimental design and execution, sample preparation, data acquisition, post-processing, visualization and analysis. In addition, data stewardship has to continue after the conclusion of a research project to ensure that the published data is made available for further re-use as detailed in the companion White Paper on Enabling Global Image Data Sharing in the Life Sciences (Bajcsy et al. 2024). Specifically, correct data stewardship ensures that the conditions used to generate, process, analyze and validate (i.e., assess the appropriateness of all aspects of the imaging pipeline and of the resulting data for a given purpose) data to be shared are transparently documented and propagated alongside the data in both human readable forms (i.e., scientific publications) (Marqués et al. 2020; Heddleston et al. 2021; Montero Llopis et al. 2021; Larsen et al. 2023) and machine readable structured FAIR frameworks (Hammer et al. 2021; Moore et al. 2021, 2023; Rigano et al. 2021; Sarkans et al. 2021; Moore 2022b; Schapiro et al. 2022), and the provenance of the data is automatically reported to downstream users. This, in turn, ensures that data can be trusted, correctly interpreted, reproduced and reused through data aggregation, mining, integrative modeling and further analysis (including AI/ML). In addition, proper data stewardship is crucial to organize data, thus avoiding the waste of time and resources needed to have to re-generate data that has been lost or cannot be interpreted and, as a result, promote efficiency and sustainability (economic, environmental and societal) (Meyn et al. 2022; Budtz Pedersen and Hvidtfeldt 2023).
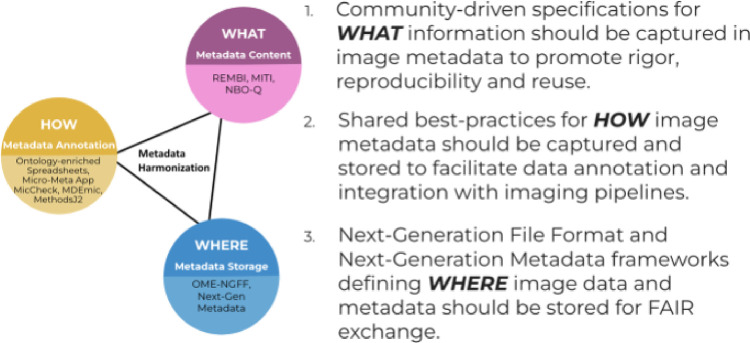
As such, effective data stewardship requires well maintained, enterprise grade to assure scalability, open-source and commonly available cyberinfrastructure (Andreev et al. 2021) leveraging Persistent Identifiers (PIDs) for research resources, individuals, publications and data (Cousijn et al. 2021; Brown et al. 2022a, 2022b; McCafferty et al. 2023), shared file formats such as OME-NGFF and associated APIs (Moore et al. 2021, 2023; Marconato et al. 2023), community-defined ontology-based harmonization (Côté et al. 2010; Lomax 2019)(Ciavotta et al. 2022; Khurana et al. 2023)), Image Metadata specifications (Hammer et al. 2021; Sarkans et al. 2021; Schapiro et al. 2022) and Next Generation Metadata frameworks (Moore 2022b). This cyberinfrastructure should interface with Electronic Lab Notebooks (ELN) and Laboratory Information Management Systems (LIMS) and, whenever possible, automatically capturing and propagating output metadata from all relevant instrumentation (including but not limited to robotic apparatuses, microfluidics and image acquisition hardware) (Marx 2022a, 2022b). In summary, this cyberinfrastructure should cover the following three interconnected aspects (Figure 1):
Figure 1.
The generation and stewardship of FAIR image data requires the harmonization of different aspects of image metadata
WHAT information should be captured in Image Metadata (i.e., develop community-specifications for Experiment description, Sample preparation, Image acquisition, Image Processing, Visualization, and Analysis metadata; in particular, Image acquisition metadata should include hardware specifications, image acquisition settings, and QC protocols and metrics) (Hammer et al. 2021; Huisman et al. 2021; Sarkans et al. 2021; Schapiro et al. 2022).
WHERE Image Metadata should be stored (i.e. OME-NGFF and Next Generation Metadata with shared APIs) (Moore 2022b)(Moore et al. 2021, 2023)
HOW Image Metadata should be captured to facilitate metadata annotation, data curation and seamless integration of all aspects of the imaging pipeline (i.e., integration with LIMS, ELNs and hardware instrumentation; leverage community-specifications and Next Generation Metadata frameworks, ontology enriched, REMBI-based, modular template spreadsheets; incorporate QC protocols and output metrics as image metadata) (Hammer et al. December, 9–12 2019; Kobayashi et al. Dec 10–11 2019; Sansone et al. 2012; Wolstencroft et al. 2012; Bukhari et al. 2018; Kunis et al. 2021; Rigano et al. 2021; Ryan et al. 2021; NFDI4Plants Consortium 2022).
Specific Modalities
Multiplexed RNA, Multiplexed DNA FISH, Protein Optical, and Mass Imaging
Within the last 20 years, the basic principle of imaging—visualizing cells or nucleic acid probes in situ—has expanded to include dozens of methods empowering the granular study of tissues at single and spatial resolution. These techniques include spatial RNA profiling methods capable of resolving hundreds of probes at subcellular resolution using light microscopy, e.g., multiplexed error-robust fluorescence in situ hybridization (MERFISH) (Chen et al. 2015; Moffitt et al. 2018), in situ sequencing (ISS) (Ke et al. 2013), and many others. In addition to imaging-based methods that now allow 100–1000+ RNA probes to be examined in a single tissue section, spatial barcoded techniques empower analysis of the whole transcriptome via capture arrays. Given their considerable promise, these technologies have been the subject of several recent reviews and we refer the reader to these resources (Moffitt et al. 2022; Baysoy et al. 2023; Vandereyken et al. 2023). Multiplexed imaging-based spatial transcriptomic experiments targeting RNAs and their multiplexed ‘spatial genomics’ counterparts targeting DNA (Wang et al. 2016; Takei et al. 2021) rely on sets of bioinformatically designed, oligonucleotide (oligo)-based probes.
Such primary oligo probes can be designed de novo (Rouillard et al. 2003); (Beliveau et al. 2018); (Hu et al. 2020); (Zhang et al. 2021), or by querying genome-scale databases of pre-discovered probes (Gelali et al. 2019); (Hershberg et al. 2021). In either case, care must be taken to ensure that primary probes remain hybridized through many iterative rounds of staining and imaging, have minimal secondary structure, and have sufficient specificity to minimize unwanted background signal. Furthermore, “secondary” or “readout” probes with sequences that are orthogonal to the genome being imaged should be used to facilitate multiplexed imaging. Finally, signal amplification methods (Choi et al. 2014); (Kishi et al. 2019) may be necessary to make FISH signals bright enough to detect, especially in tissue samples.
Several highly multiplexed antibody-based imaging techniques have recently been developed and commercialized (Hickey et al. 2022; Kinkhabwala et al. 2022; Rivest et al. 2023). Methods employing fluorophore-labeled antibodies are the most numerous and include cyclic methods relying on elimination of the fluorescent signal (Gerdes et al. 2013; Lin et al. 2018; Radtke et al. 2020); (Porciani et al. 1992; Kinkhabwala et al. 2022) or antibody removal (Gut et al. 2018; Rivest et al. 2023) to achieve high parameter imaging. Non-cyclic methods such as spectral (Gerner et al. 2012; Lin et al. 2023) and vibrational imaging (Wei et al. 2017) utilize advanced imaging systems with mutliplexed fluorophores to evaluate more than ten markers at once. Methods employing oligo-conjugated antibodies apply a single mixture of antibodies to the tissue and serially reveal markers through fluorescently conjugated oligo-reporters specific to each tagged antibody (Wang et al. 2017; Goltsev et al. 2018; Saka et al. 2019).
An alternative to fluorescence-based approaches is mass spectrometry (MS)-based methods where antibodies are labeled with mass tags instead of fluorophores or chromogens. Whereas antibodies tagged with the latter are quantified using light, MS-based methods create images by mapping the spatial distribution of the mass reporters attached to each antibody. The two most common technologies are multiplexed ion beam imaging (MIBI) (Angelo et al. 2014) and imaging mass cytometry (IMC) (Giesen et al. 2014), which utilize elemental mass tags and differ by the use of an ion beam or laser, respectively, for tag ionization and detection. MIBI and IMC are used routinely to quantify 40 or more biomarkers in a tissue section using a single master mix of metal-conjugated primary antibodies. More recently, methods have emerged that enable highly multiplexed IHC based on matrix-assisted laser desorption/ionization mass spectrometry imaging (MALDI-IHC) (Yagnik et al. 2021). Here antibodies are conjugated with photocleavable synthetic peptides that are released and ionized during laser irradiation. Although spatial resolution is currently limited to ~10 μm pixel sizes, the advantage of this approach is the ability to integrate untargeted metabolomic and lipidomic MALDI imaging mass spectrometry (IMS) collected from the same tissue sections. The major benefit of mass spectrometry-based techniques is the ability to detect and resolve dozens of metal/molecule-labeled antibodies simultaneously. Mass barcodes possess low background signal by circumventing autofluorescence and incorporating high instrumental mass-resolving power (Tideman et al. 2021; Mund et al. 2022).
A majority of these multiplexed imaging techniques have been developed within the past decade. The recent introduction of these methods, combined with their complexity and diversity, result in a variety of workflows related to sample processing, reagent QC, image acquisition, image processing, and image analysis without agreed upon community standards. The establishment and wide adoption of such standards are essential for cross-lab and cross-platform data interoperability and analysis, which is even more critical to the community since these data are expensive to acquire (Hickey et al. 2022; Quardokus et al. 2023; Vandereyken et al. 2023) and are associated with a number of consortia efforts (HuBMAP Consortium 2019; Rozenblatt-Rosen et al. 2020; Jain et al. 2023). For these reasons, critical details related to sample preparation, reagent validation, and platform-specific imaging parameters must be recorded and shared using community-defined metadata. Here we highlight specific challenges around the harmonization of multiplexed image processing and reporting that limit current ability towards FAIR data (Wilkinson et al. 2016).
Harmonization of Sample Preparation Procedures
Despite the diversity of imaging-based spatial omics methods described above, there are several parallels and shared challenges related to imaging tissues. First and foremost, tissues must be optimally prepared to allow downstream profiling of RNA or proteins. This pre-image data generation stage consists of several steps not limited to 1) careful processing of samples into snap frozen, fixed frozen, or formalin-fixed paraffin embedded (FFPE) specimens, 2) optimal placement and orientation into tissue molds, 3) sectioning of thin tissue sections onto slides, imaging chambers, or bar-coded arrays, and 4) antigen retrieval to expose epitopes altered during FFPE preparation (optional). For FISH-based assays, permeabilization with detergents to allow for probe penetration, heat and acid treatment to denature protein structure and promote target accessibility, and denaturation with heat and chemical agents such as formamide to remove secondary structure from the target nucleic acids are critical pre-treatment steps.
Harmonization of Reagent Preparation & Reporting
For multiplexed antibody-based imaging techniques, antibody validation and panel design are resource and time-consuming processes, estimated to take 6–8 months and tens of thousands of dollars to build (Hickey et al. 2022; Quardokus et al. 2023). Recent efforts within the Human BioMolecular Atlas Program Affinity Reagents and Imaging Validation Working Group provided guidelines (HuBMAP Consortium 2019; Jain et al. 2023) and a potential solution (Quardokus et al. 2023) for the tedious process of antibody validation and panel design. Organ Mapping Antibody Panels (OMAPs), accompanying Antibody Validation Reports (AVRs), and resulting datasets are designed to reduce the time, cost, and expertise required for imaging proteins in tissues using antibodies (see Tables in references links; (HuBMAP Consortium 2023a, 2023b). Antibody performance is well known to be context specific. Therefore, it’s important to share the metadata uniquely identifying an antibody, research resource identifier (RRID) (Bandrowski et al. 2016, 2023; Bandrowski 2022), and the experimental details contributing to the success or failure of a particular reagent, e.g., tissue, fixation method, antigen retrieval conditions, imaging method, etc. For these reasons, the sharing of negative data is invaluable as this practice prevents others from wasting time and money on reagents that are known to fail for their particular application. To this end, a community initiative has emerged to support imaging scientists using a variety of methods employing fluorophore-labeled antibodies (Yaniv et al. 2023).
Harmonization of Image Acquisition and Processing
As with traditional microscopy, multiplexed RNA and protein imaging methods report metadata associated with imaging parameters to ensure cross-comparisons and appropriate interpretation of the resulting data. However, there are substantially greater amounts of image processing that go on within multiplexed imaging datasets. This is because tissues need to be imaged iteratively and/or over long periods of time, requiring additional algorithms for tissue registration, stitching, shading correction, deconvolution, and background subtraction. Each of these image processing steps has many different choices of algorithms, each with advantages and disadvantages that continue to grow in number each year. Documenting which algorithm is used at each step of the pipeline is also important for cross-comparison. Moreover, detailing the key parameter choices used when employing image analysis algorithms (e.g., threshold, sigma range, max iterations for Gaussian fitting), recording key output data (e.g., centroid position, precision for Gaussian fitting), and providing example inputs and outputs is essential for reproducibility. Furthermore, it is important to consider how the image analysis workflow is documented and maintained as described above. This focus on reproducibility is a particularly acute need for the multiplexed imaging community because of the size of raw multiplexed imaging datasets that can reach terabytes for even one whole slide image, and processing of datasets can reduce the data to GB sizes. Consequently, what is usually shared with the community are processed images.
Another recent change within the multiplexed imaging community is the number of commercial vendors that now sell multiplexed imaging solutions. Many of these companies provide on-computer image processing. However, with commercial interests, this often precludes sharing of processing steps and also limits users’ ability to interact with raw data. Establishing a framework for standardized reporting of image formats and accompanying metadata will empower cross-comparative studies acquired using either open-source or commercial platforms (Moore et al. 2021, 2023; Swedlow et al. 2021).
Harmonization of Multiplexed Image Analysis
The image analysis pipeline typically involves domain experts initially annotating the image data, bioimage analysts processing the image data, and biologists extracting data-derived insights. Automated image analysis pipelines have been preferred in case of highly multiplexed whole slide imaging because of the size and scale of the images and associated analysis data (Axelrod et al. 2021; Schapiro et al. 2021; Wang et al. 2021; Windhager et al. 2021; Eng et al. 2022). With such large data structures, it is often tempting to assess analysis quality only from final measured metrics and/or proposed regions, but these outputs may often not fully elucidate possible issues with sample preparation and/or the imaging itself (Vierdag and Saka 2024). Thus, there is a need to make QC an integral part of those parts of analysis workflows that deal with the multiplexed imaging data.
Some workflows incorporating QC already exist and are being applied on multiplexed imaging data. Perhaps more so than with other types of image data, QC of any multiplexed image dataset requires a sampling strategy for performing QC as QC of the complete data is extremely time-consuming. The extent of required sampling depends on the research question and the heterogeneity of the data. Additionally, the sampling could be guided by prior information such as annotated regions of interest.
As any sampling strategy still involves visualizing more markers than can be fit on a screen, a multi-canvas view of the data is required in which multiple groups of markers can be visualized at a particular location simultaneously without spectral overlap. While some analysis software such as QuPath (Bankhead et al. 2017) provide such functionality, many tools do not easily permit doing this for dozens of markers (Vierdag and Saka 2024).
However, setting up this kind of view repeatedly is too time-consuming and not easily reproducible. Standardized systems for creating view configurations from metadata could encompass both how images should be rendered as well as the layout for a particular viewer. These could be stored at either the file level and/or within the particular visualization tool, but while OME-Next Generation File Format (NGFF) does specify how images should be rendered it does not allow for storing viewer specific information (Moore et al. 2021, 2023; Swedlow et al. 2021). Additionally, it currently does not provide a specification for storing multiple view configurations.
Given that the multiplexed spatial omics technologies are relatively nascent, and the image analysis workflows share common building blocks, now is the ideal time for the community to agree on data structure and organization, as well as standardized reporting of methods. Such an effort will help prevent multiple methods and formats becoming embedded in different locations or fields.
Cryogenic transmission electron microscopy
Cryogenic transmission electron microscopy (cryo-EM) provides images of biological matter in a frozen-hydrated, near-native state. During the last decade, cryo-EM has grown into the primary technique for high-resolution structure determination with an exponential use trajectory. Advances in the field were recognized with the Nobel Prize in Chemistry in 2017. Cryo-EM in conjunction with Single Particle Analysis (SPA) can provide atomic-resolution volume reconstructions of purified macromolecular assemblies (Nakane et al. 2020; Yip et al. 2020). Furthermore, cryogenic electron tomography (cryo-ET) allows imaging of pleomorphic samples such as cells or tissue samples at the nanometer scale and in three dimensions, in principle enabling the extraction of near-atomic resolution information of in-situ macromolecular assemblies via sub-tomogram averaging (Tegunov et al. 2021; Xue et al. 2022).
The cryo-EM field is recognized as a good example of how to effectively accomplish metadata annotation and stewardship of raw and derived data (Sarkans et al. 2021). To this aim, the field was able to leverage the existing infrastructure of the Protein Data Bank (PDB) (RCSB Consortium 2019) which deals with atomic-level descriptions of protein and other biological structures (e.g, DNA and RNA). The cryo-EM community realized early on that the atomic coordinates must be accompanied by the corresponding volume data used to derive the model to allow appropriate evaluation of the model quality. This was initially driven by the observation that the resolution of cryo-EM volumes generally was well below 1 nm and the assignment of atomic coordinates tended to be quite ambiguous (Volkmann 2014). While since the “resolution revolution” (Kühlbrandt 2014) near-atomic resolution reconstructions for well-behaved samples can be routinely achieved by SPA, the value of density information alongside atomic coordinates is still strongly recognized. Consequently, most journals demand the deposition of atomic coordinates as well as density information to support publication. The cryo-EM community also widely agrees that detailed metadata must be made publicly available alongside the data and that metadata standards need to be reviewed regularly to ensure fitness and relevance to the evolving community needs (Chiu et al. 2021; Sarkans et al. 2021).
Data sharing of cryo-EM derived density maps and associated metadata is implemented in the Electron Microscopy Data Bank (EMDB) (EMBL-EBI; Lawson et al. 2011), which is well established, has consistent data formats and metadata schemas and allows easy examination of the densities in conjunction with the corresponding atomic models via close coupling to the PDB. In turn, the sharing of raw cryo-EM data is implemented in the Electron Microscopy Public Image Archive (EMPIAR) (EMBL-EBI; Ludin et al. 2016) which is linked to the EMDB to allow the association of raw data with the corresponding derived data. As also mentioned in the companion White Paper on Enabling Global Image Data Sharing in the Life Sciences (Bajcsy et al. 2024), much work remains to be done to incorporate in EMPIAR more information, primarily based on feedback from depositors and workshops with community experts (Sarkans et al. 2021). This is particularly important for cellular tomography data, where the nanoscale structural data often needs to be linked to data from light microscopy to gain the full picture in correlative light and electron microscopy (CLEM) experiments. It would also be highly beneficial to enable crosstalk with other spatial information sources such as spatial proteomics or volume EM. However, the main challenge for the cryo-EM field is its growth trajectory and the need for scalability, especially for raw data sharing.
As of November 2023, about 800 high-end cryogenic electron transmission microscopes are in use worldwide (Various Authors 2023). Each of these instruments can produce between 1 to 5 terabytes a day for a total worldwide of 0.7 and 3.5 petabytes of raw data per day. While not all this data will contribute to publications or be otherwise worth preserving, the trajectory of data depositions in the EMDB is exponential with a doubling rate of about two years. By mid-2022, about 5000 cryo-EM structures were deposited in the EMDB (Halfon et al. 2022). Assuming the current growth trend holds, the amount of raw data associated with new EMDB depositions will be between 20 and 100 petabytes by mid-2025, surpassing the exabyte mark as early as 2032. Considering that this estimate constitutes a lower bound it is clear that for the cryo-EM field the main challenge in sharing data lies in the sheer scale and expected exponential growth of the data likely to be produced.
Toward Global Image Data Generation and Stewardship
All stakeholders can play a role In all aspects described in the White Paper. For example, manufacturers can and should actively contribute by making community-defined full technical descriptions of instruments and QC easier and more automated (Marx 2022a, 2022b). In addition, funders can help by providing funds to support bioimaging communities, which are conducting largely unfunded essential tasks, and core facilities personnel and to promote the development of tools, protocols, and metadata standards. As an example, journals can help by requiring all aspects of microscopy metadata (i.e., hardware specifications, image acquisition settings and QC metrics) to be a part of the data package just like control experiments are required for experimental procedures. Finally, if these practices are to be universally adopted, the development of resources in multiple languages need to be encouraged and supported.
In this section we provide detailed steps that could be taken to promote the production and stewardship of image data that is “FAIR from the start” and ready to be shared and reused. These are summarized in a to-do list for various stakeholders presented in a text box at the end.
Data Generation
Overcoming challenges related to the generation of image data that is “FAIR from the start” requires specific solutions that should be planned for and carried out by all interested stakeholders. To guide the development of these solutions, we provide the following specific recommendations:
Promote the widespread adoption of persistent identifiers for institutions (e.g., Research Organization Registry- ROR) (Gould 2023), core facilities (e.g., ROR and Research Resource Identifier - RRID) (Bandrowski 2022), personnel (e.g., Open Researcher and Contributor ID - ORCID) (Haak et al. 2012; Shillum et al. 2021), reagents (e.g., RRID) (Bandrowski 2022), microscope instruments (e.g., PIDINST) (Stocker et al. 2020; Krahl et al. 2021; McCafferty et al. 2023) and datasets (e.g., Digital Object Identifiers - DOI) as a means to enable the FAIR description of all entities used in science, to facilitate reporting and reproducibility and to ensure the recognition of the essential role carried out by research and imaging scientists and core facilities in the research enterprise (Cousijn et al. 2021; Brown et al. 2022a, 2022b; McCafferty et al. 2023). This recommendation is supported by recent cost-benefit analyses (Brown et al. 2022a, 2022b), which demonstrated staggering financial benefits associated with staff salaries, time spent in tedious data entry and potential technological innovation.
Promote the use of existing guidelines such as Recommended Metadata for Biological Images (REMBI) (Sarkans et al. 2021) as a blueprint to guide biomedical scientists on all required components of image metadata. REMBI consists of eight modules, each providing minimal information requirements for each of the phases of a typical imaging experiment. Each module can be extended through consensus-building decision-making processes organized by bioimaging community efforts and involving research scientists, imaging scientists, microscope custodians and manufacturers and image analysts. As an example, pro-bono work being conducted by QUAREP-LiMi (Boehm et al. 2021; Nelson et al. 2021) is expanding the Image “Image acquisition” module of REMBI to include the NBO-Q Microscopy Metadata specifications in close collaboration with instrument manufacturers (Marx 2022a, 2022b). Additionally, workflows that are being developed (https://doi.org/10.1038/s41592-023-01846-7) for reporting the validation and application of complex antibody panels for imaging human tissues could be expanded to other sample types by integrating them in the “Specimen” module of REMBI. Along the same lines, the MITI guidelines (Schapiro et al. 2022) could be used to extend multiple REMBI modules. One solution is for the community to share modular metadata annotation templates based on the REMBI (Sarkans et al. 2021) guidelines to describe different imaging experiments where metadata annotation is semantically enriched (Ciavotta et al. 2022; Khurana et al. 2023) using tools such as ISAtools, RightField, Swate or Cedar (Sansone et al. 2012; Wolstencroft et al. 2012; Bukhari et al. 2018; NFDI4Plants Consortium 2022),
Promote the collection of full technical descriptions of microscope hardware specifications, image acquisition settings and QC protocols and metrics (aka Microscopy Metadata) in compliance with community-defined microscopy metadata 4DN-BINA-OME (NBO-Q) specifications (Hammer et al. 2021; Huisman et al. 2021) that are being developed by consensus by imaging scientists and instrument manufacturers (Marx 2022a, 2022b). These technical descriptions captured in microscopy metadata must become obligatory aspects of the production of any image data because in their absence image data cannot reliably be quantified, reproduced and reused and ultimately loses scientific value even when it is shared. As such Microscopy Metadata to be made transparently available to microscope users, automatically collected using community tools (Kunis et al. 2021; Rigano et al. 2021; Ryan et al. 2021; Kunis and Dohle 2022), and encoded using shared metadata frameworks (Moore et al. 2021, 2023).
Ensure that instrument maintenance and quality assessment are adequately supported to ensure that they become a common practice at all core facilities and individual laboratories utilizing microscopes regardless of local resource availability. Specific funding mechanisms should be considered to provide the necessary instrumentation, training and personnel or traveling metrology services for under-resourced areas. This will allow for the performance of instruments to be evaluated at regular intervals using community-defined metrology standards and QC procedures that are appropriate for each experimental question (Gaudreault et al. 2022; Nelson 2022; Abrams et al. 2023). In addition, specific additional metrics might need to be collected for types of experimental approaches and desired outcomes.
Emphasize large infrastructure investments in core facilities and regional infrastructural hubs (Budtz Pedersen and Hvidtfeldt 2023) employing trained personnel including imaging scientists, data stewards, image analysts and research software engineers. Such shared infrastructure would increase efficiency and reduce costs by maintaining and assessing the performance of instruments, promote the dissemination of technological advances (hardware and software), facilitate user training, provide guidance for experimental procedures, data stewardship and image analysis, and provide image analysis and RDM services to facilitate the deposition of FAIR data packages containing the appropriate image metadata to specialized bioimage repositories. This should include legal support to review data, identify appropriate Creative Commons (CC) and Open-Source Software (OSS) licensing and/or carry out Personal Information (PI) redaction (human subject data).
Invest in the development of open-hardware devices (i.e., robotic devices, fluidics systems, environmental control devices, microscopes, etc.) to carry out all aspects of data generation as the most appropriate way to ensure democratization of access to advanced technology and the efficient use of resources.
Data Processing and Analysis
After acquisition, images often have to undergo complex processing, visualization, and analysis steps to extract quantitative information about the intensity of the signal associated with a given label, as well as the location, morphological characteristics, association and movement of biological entities. To ensure that the results of image processing and analysis pipelines are reproducible and ready for FAIR sharing, community-defined guidelines should be adopted (Aaron and Chew 2021; Miura and Nørrelykke 2021; Schmied et al. 2023). In particular, the following steps should be adopted:
Image processing and analysis workflows should ideally be shared in binary containers such as Docker, Singularity, or Podman that include the complete software environment. This ensures that all aspects of the pipeline remain identical for all users.
The processing and analysis workflow should specify what characteristics of the image data can affect its execution, and should, therefore, be encoded in metadata (i.e., magnification, resolution, signal intensity, QC metrics).
All aspects of the processing and analysis pipeline, including but not limited to data structure and size, rendering and processing steps, algorithm version and input parameters, as well as computing and networking requirements, must be documented in both human and machine-readable manners to ensure interpretability, reproducibility, and downstream reuse. Such metadata should be captured either in the image data file (e.g., information about image rendering and processing) or as part of the documentation of the workflow (e.g., algorithm version and input parameters).
As such, it is imperative that analysis metadata be captured using community-defined metadata specifications and storage frameworks to ensure maximal efficiency with which this information can be extracted and tracked across all steps of the pipeline without the need for time-consuming repeated interactions with the image data itself.
Data Stewardship
The everyday stewardship of data and associated metadata throughout the entire lifecycle of quantitative imaging experiments is essential to ensure rigor, reproducibility and the production of high-quality image data that can be interpreted and is ready to be reused according to FAIR principles.
The generation and stewardship of FAIR image data requires full transparency, management and reporting of all information related to the conditions used for data generation (i.e., experimental conditions, sample preparation, and image acquisition) as well as processing and analysis (i.e., image analysis and visualization).
RDM cyberinfrastructure supporting the generation and pre-publication stewardship of high-quality FAIR image data should be made available to all biomedical researchers using microscopes as an essential prerequisite for image data sharing and reuse.
Such imaging RDM cyberinfrastructure needs to include advanced computing and data repositories that provide integrated data stewardship, metadata annotation, visualization environments, processing pipelines and analysis routines (including AI/ML). Different components have to be connected via high-speed networks to expedite upload and download as needed.
RDM cyberinfrastructure is best supported by easy-to-use, enterprise grade, robust, continually maintained and supported open-source software to carry out all steps of the imaging pipeline from experimental procedures (i.e., Laboratory Information Management Systems - LIMS, Electronic Lab Notebooks - ELNs) to image acquisition (i.e., Micro-Manager, Pycro-Manager, Python Microscope, etc.), to image processing, visualization and analysis and processing pipeline (i.e., CellProfiler, Fiji, napari, CellPose, etc.).
When proprietary software and instrumentation have to be used, community-defined standards must be used to ensure transparency regarding all relevant algorithms and input parameters as well as instruments’ configuration and performance.
- Collection and reporting of metadata has to be based on community-defined standards and it has to occur at two highly interconnected levels:
- Human readable, which is primarily related to Materials and Methods (Marqués et al. 2020; Heddleston et al. 2021; Montero Llopis et al. 2021; Larsen et al. 2023), might represent a subset of the information captured in Image Metadata and has to take into account the capacity of users to understand and describe the imaging experiment.
- Machine-readable, which is captured in Image Metadata (i.e., all information needed to understand the lineage - aka provenance - and quality of image data) and represents the complete technical description to ensure full quality, reproducibility, and reusability (Moore et al. 2021, 2023; Moore 2022b) .
- To ensure Machine Readability:
- Metadata should be encoded in community-specified frameworks to be associated with standardized image data file formats (Moore et al. 2021, 2023; Moore 2022b) or with workflow documentation, and equipped with easily available software API to facilitate transferring the information across the different steps of the imaging pipeline. For example, it should be possible to automatically be put in a SQL database. A potential framework which could fulfill these requirements is LinkML (Solbrig et al. 2023). It allows for easy authoring of metadata schemas in the YAML format, which can be exported into other formats. It is also able to create classes in various programming languages that can serve to validate metadata.
- It is imperative to use specific annotation tools and automation at all aspects of the imaging pipeline to ensure that image processing, visualization and analysis pipelines can leverage metadata. In particular, emphasis should be given to 1) automated processes for microscope systems and peripheral components, including community-defined QC procedures to ensure optimal instrument performance (Hammer et al. 2021; Schapiro et al. 2022). 2) automated metadata annotation at all phases of the image-data life cycle (Kunis et al. 2021; Rigano et al. 2021; Ryan et al. 2021). 3) Integrated image processing, visualization, and analysis pipelines.
- Metadata annotations should be backed by ontologies and knowledge graphs.Ontologies provide descriptions of the hierarchical relationship between concepts and can be used to make metadata machine-readable. Their role in the harmonization of knowledge and data in biomedicine is increasingly recognized especially in the context of AI, where they can provide valuable constraints making ML more efficient (Lomax 2019). As such, tools exist to aid this process such as the Ontology Look Up Service (OLS) (Côté et al. 2010) that can be used to locate appropriate ontological terms although further development is needed to help sort through duplicates and identify which should be used in different contexts.
- Quality, transparency, reproducibility, and reusability require standards defined by the community of all imaging stakeholders.
- Recent advancements have made it clear that the community, when organized in bioimaging organizations, networks, and initiatives, is willing and ready to take on this challenge.
- However, there is a need for deliberate, directed, targeted funding to ensure that ongoing standardization efforts can be expanded to cover all essential aspects of the imaging pipeline.
- In addition to being essential for the generation of standards, the added advantage of community organizations is that they are the ideal forum for guaranteeing the broad community adoption of standards through education, training, and outreach.
- Community organizations are typically run on a volunteer basis
- However, this is clearly not sustainable, as such an essential endeavor for the advancement of quantitative imaging and science as a whole cannot be done in people’s free time
Cyberinfrastructure for RDM is anticipated to have impacts beyond the bioscience research enterprise. Tested and trusted microscope QC protocols and commonly accepted and unambiguous reporting criteria will be beneficial to the many applications of quantitative imaging for cellular analysis, including pathology, cell therapies and regenerative medicine.
Supplementary Material
Text Box - A to-do list for various stakeholders.
Towards Global Image Data Generation and Stewardship
A to-do list for various stakeholders
- Ensure the long term sustainability of national and international bioimaging communities (e.g., ABIC, ABRF, BINA, CBI, GBI, LABI and QUAREP-LiMi; see also Table 1 in supplemental materials) thus enabling recurring gatherings to coordinate (i.e., discuss, recommend, update) the development of:
- Consensus guidelines for quality control procedures and standards to encourage the implementation and reporting of QC protocols and performance benchmarks, including for imaging instrumentation.
- Shared metadata specifications, exchange frameworks, and tools to minimize barriers to metadata guideline adoption by academic, government and industry stakeholders. Specifically, this will empower the annotation of all phases of the image-data lifecycle, including details about samples, reagents and experimental protocols, instrument hardware specifications and image acquisition settings, QC protocols and metrics, image data processing and analysis workflows, and persistent association of metadata and image data.
- Shared computational cyberinfrastructure for image data generation and pre-publication stewardship needs. This consists of well documented and maintained, enterprise-grade, and high-speed software tools, frameworks, computing and storage equipment, and networks to carry out all steps of the imaging pipeline from data annotation to image acquisition, and analysis. To this aim community-defined standards must be used to ensure transparency regarding all relevant instruments and algorithms.
- Invest in core facilities (aka Shared Research Resources] and their Personnel from all backgrounds and regions to:
- Provide expertise on sample preparation, validation of staining protocols, image acquisition, and image analysis.
- Democratize access through shared resources and the promotion of collaborations to facilitate access to advanced technology.
- Serve as pivotal hubs for the dissemination of expertise and user training on all topics essential for the preparation of FAIR image data that is ready to be shared and to engender maximum reuse value, across both resourced and under-resourced regions and communities.
- Develop strong connections with software development centers to ensure the usability, customization, and democratization of cyberinfrastructure for imaging pipeline automation.
Support the career and recognition of imaging scientists specializing in the generation and stewardship of FAIR image data. These include core facility personnel, image data stewards and curators, image analysis experts, and research software engineers.
In collaborations with vendors, develop and deploy automated methods to capture harmonized and consistent metadata documenting all steps of the imaging pipeline from reagents used to generate image data, to microscopy instruments and peripherals.
Promote the use of Persistent Identifier (PID) tor the FAIR description of research resources (i.e., reagents, instruments, core facilities) and outputs (i.e., datasets), to facilitate, reproducibility, and reuse, to democratize access to advanced technologies, improve efficiency, and to ensure that the personnel involved in the research enterprise are appropriately acknowledged.
Develop metrics that describe the qualities of resultant image data
Acknowledgements
Disclaimer: Commercial products are identified in this document in order to specify the experimental procedure adequately. Such identification is not intended to imply recommendation or endorsement by the National Institute of Standards and Technology, nor is it intended to imply that the products identified are necessarily the best available for the purpose.
List of Contributors
| Name | ORCID | Affiliation | Funding Statement | Contribution* | |
|---|---|---|---|---|---|
| Nikki Bialy | nbialy@morgridge.org | https://orcid.org/0000-0001-9681-9632 | BioImaging North America (BINA RRID: SCR_024409), Morgridge Institute for Research, 330 North Orchard Street, Madison, WI 53715, USA. | NB is supported through a grant from the Chan Zuckerberg Initiative DAF, an advised fund of Silicon Valley Community Foundation, to the Morgridge Institute for Research for BioImaging North America (BINA). | Resources, Writing - Original Draft, Writing - Review & Editing, Endorsement, Supervision, Project Administration |
| Frank Alber | falber@g.ucla.edu | https://orcid.org/0000-0003-1981-8390 | Department of Microbiology, Immunology & Molecular Genetics, University of California Los Angeles, USA | Endorsement | |
| Brenda Andrews | brenda.andrews@utoronto.ca | httDs://orcid.org/0000-0001-6427-6493 | The Donnelly Centre, University of Toronto, Toronto Canada M5S 3E1 | Work in the Andrews lab is supported by the NIH (R01HG005853) and the Canadian Institutes of Health Research (CIHR). BA holds at Tier 1 Canada Research Chair in Systems Genetics and Cell Bioloay | Writing - Original Draft, Endorsement |
| Michael Angelo | mangelo0@stanford.edu | Stanford University School of Medicine, Palo Alto, CA, USA | Writing - Original Draft | ||
| Brian Beliveau | beliveau@uw.edu | https://orcid.org/0000-0003-1314-3118 | University of Washington | BJB acknowledaes support from the National Institutes of Health under arant 1R35GM137916 (NIGMS). | Writing - Original Draft |
| Lacramioara (Lacra) Bintu | lbintu@stanford.edu | https://orcid.org/0000-0001-5443-6633 | Stanford University, Stanford, CA, USA | Endorsement | |
| Alistair Boettiger | aboettig@stanford.edu | https://orcid.org/0000-0002-3554-5196 | Stanford University, Stanford, CA, USA | Writing - Review & Editing, Endorsement | |
| Ulrike Boehm | ulrike.boehm@gmail.com | https://orcid.org/0000-0001-7471-2244 | Carl Zeiss AG, Oberkochen, Germany | Writing - Review & Editing, Endorsement | |
| Claire M. Brown | claire.brown@mcgill.ca | https://orcid.org/0000-0003-1622-663X | Advanced Bioimaging Facility (ABIF), McGill University, Montreal, Quebec, H3G 0B1, Canada | This project has been made possible in part by arant number 2020–225398 from the Chan Zuckerbera Initiative DAF, an advised fund of Silicon Valley Community Foundation. | Endorsement |
| Mahmoud Bukar Maina | M.Bukar-Maina@sussex.ac.uk | https://orcid.org/0000-0002-7421-3813 | University of Sussex, Sussex, UK & Biomedical Science Research and Training Centre, Yobe State University, Nigeria | Writing - Review & Editing, Endorsement | |
| James J. Chambers | ichambe@umass.edu | httDs://orcid.org/0000-0003-3883-8215 | Institute for Applied Life Sciences, University of Massachusetts, Amherst, MA 01003, USA | Endorsement | |
| Beth A. Cimini | bcimini@broadinstitute.org | https://orcid.org/0000-0001-9640-9318 | Broad Institute of MIT and Harvard, Imaging Platform, Cambridge, MA, USA | This publication has been made possible in part by CZI grant 2020–225720 (DOI:10.37921/977328 pjvbca) from the Chan Zuckerberg Initiative DAF, an advised fund of Silicon Valley Community Foundation (funder DOI 10.13039/100014989. This work was also supported by the Center for Open Bioimage Analysis (COBA) funded by the National Institute of General Medical Sciences P41 GM135019 | Writing - Review & Editing, Endorsement |
| Kevin Eliceiri | eliceiri@wisc.edu | httDs://orcid.org/0000-0001-8678-670X | Morgridge Institute for Research and the University of Wisconsin-Madis on | This work was supported by the Center for Open Bioimage Analysis (COBA) funded by the National Institute of General Medical Sciences P41 GM135019 (Cimini and Eliceiri) | Writing - Review & Editing, Endorsement |
| Rachel Errington | ErringtonRJ@cardiff.ac.uk | School of Medicine, Cardiff University, Cardiff, UK | Writing - Original Draft, Writing - Review & Editing | ||
| Orestis Faklaris | orestis.faklaris@mri.cnrs.fr | https://orcid.org/0000-0001-5965-5405 | BCM, Univ. Montpellier, CNRS, INSERM, Montpellier 34293, France | OF is supported by the French National Research Agency (ANR-10-INBS-04). | Writing - Review & Editing, Endorsement |
| Nathalie Gaudreault | nathalieg@alleninstitute.org | https://orcid.org/0000-0002-9220-5366 | Allen Institute for Cell Science, Seattle, WA, USA | We wish to thank the Allen Institute for Cell Science Founder, Paul G. Allen, for his vision, encouragement, and support. | Writing - Review & Editing, Endorsement |
| Ronald N. Germain | rgermain@niaid.nih.gov | httDs://orcid.org/0000-0003-1495-9143 | Laboratory of Immune System Biology, NIAID, NIH | Intramural Program of NIAID, NIH. | Writing - Review & Editing, Endorsement |
| Wojtek Goscinski | w.goscinski@anif.org.au | httDs://orcid.org/0000-0001-6587-1016 | National Imaging Facility, Brisbane, Australia | The authors acknowledge the facilities and scientific and technical assistance of the National Imaging Facility, a National Collaborative Research Infrastructure Strategy (NCRIS) capability | Endorsement |
| David Grunwald | David.Grunwald@umassmed.edu | httDs://orcid.org/0000-0001-9067-804X | RNA Therapeutics Institute, UMass Chan Medical School, Worcester MA 01605, USA | Writing - Review & Editing, Endorsement | |
| Michael Halter | michael.halter@nist.gov | httDs://orcid.org/0000-0002-1628-324X | National Institute of Standards and Technology, Gaithersburg, MD, USA | Writing - Review & Editing, Endorsement | |
| Dorit Hanein | dorit@ucsb.edu | https://orcid.org/0000-0002-6072-4946 | Departments of Biochemistry and Chemistry, of Biological Engineering, University of California, Santa Barbara, CA, USA | DH research is in part sponsored by the U.S. Army Research Office and accomplished under contract W911NF-19-D-0001 for the Institute for Collaborative Biotechnologies. | Writing - Original Draft, Writing - Review & Editing, Endorsement |
| John W. Hickey | john.hickey@duke.edu | https://orcid.org/0000-0001-9961-7673 | Department of Biomedical Engineering, Duke University Durham, North Carolina, USA | Writing - Original Draft, Endorsement | |
| Judith Lacoste | jlacoste@miacellavie.com | https://orcid.org/0000-0002-8783-8599 | MIA Cellavie Inc., Montreal, Quebec, H1K 4G6, Canada | Endorsement | |
| Alex Laude | alex.laude@newcastle.ac.uk | https://orcid.org/0000-0002-3853-1187 | BioImaging Unit, Newcastle University, UK | Newcastle University | Endorsement |
| Emma Lundberg | emmalu@stanford.edu | Stanford University, California, USA and SciLifeLab, KTH Royal Institute of Technology, Stockholm, Sweden | Endorsement | ||
| Jian Ma | jianma@cs.cmu.edu | https://orcid.org/0000-0002-4202-5834 | Carnegie Mellon University, Pittsburgh, PA, USA | Endorsement | |
| Leonel Malacrida | lmalacrida@pasteur.edu.uy | https://orcid.org/0000-0001-6253-9229 | Institut Pasteur de Montevideo, & Universidad de la República, Montevideo, Uruguay | Endorsement | |
| Josh Moore | josh@openmicroscopy.org | https://orcid.org/0000-0003-4028-811X | German BioImaging-Ges ellschaft für Mikroskopie und Bildanalyse e.V., Constance, Germany | JM is supported by the Deutsche Forschungsgemeinsch aft (DFG, German Research Foundation) - 501864659 as part of NFDI4BIOIMAGE. | Writing - Review & Editing, Endorsement |
| Glyn Nelson | glyn.nelson@ncl.ac.uk | http://orcid.org/0000-0002-1895-4772 | Bioimaging Unit, Newcastle University, Newcastle upon Tyne, NE2 4HH, UK | Newcastle University, UK | Writing - Review & Editing, Endorsement |
| Elizabeth Kathleen Neumann | ekneumann@ucdavis.edu | https://orcid.org/0000-0002-6078-3321 | Department of Chemistry, University of California, Davis, Davis, California, USA | University of California, Davis | Endorsement |
| Roland Nitschke | Roland.Nitschke@biologie.uni-freiburg.de | https://orcid.org/0000-0002-9397-8475 | Life Imaging Center, Signalling Research Centres CIBSS and BIOSS, University of Freiburg, 79104 Freiburg, Germany | R.N. was supported by grant NI 451/10–1 from the German Research Foundation and grant 03TN0047B ‘FluMiKal’ from the German Federal Ministry for Economic Affairs and Climate Action | Writing - Review & Editing, Endorsement |
| Shuichi Onami | sonami@riken.jp | https://orcid.org/0000-0002-8255-1724 | RIKEN Center for Biosystems Dynamics Research, Kobe, Japan | Endorsement | |
| Jaime A. Pimentel | arturo.pimentel@ibt.unam.mx | https://orcid.org/0000-0001-8569-0466 | Laboratorio Nacional de Microscopía Avanzada, Instituto de Biotecnología, Universidad Nacional Autónoma de México, Cuernavaca, Morelos, 62210, México | Writing - Review & Editing, Endorsement | |
| Anne L. Plant | anne.plant@nist.gov | https://orcid.org/0000-0002-8538-401X | National Institute of Standards and Technology, Gaithersburg, MD, USA | Writing - Review & Editing, Endorsement | |
| Andrea J. Radtke | andrea.radtke@nih.gov | https://orcid.org/0000-0003-4379-8967 | Laboratory of Immune System Biology, Lymphocyte Biology Section and Center for Advanced Tissue Imaging, NIAID, NIH, Bethesda, MD, USA | AJR is supported by the Intramural Research Program of the NIH, National Institute of Allergy and Infectious Diseases (NIAID) and National Cancer Institute (NCI) | Writing - Original Draft, Writing - Review & Editing, Endorsement |
| Bikash Sabata | bsabata@altoslabs.com | NA | Altos Labs, Redwood City, CA, USA | NA | Writing - Review & Editing, Endorsement |
| Denis Schapiro | Denis.Schapiro@uni-heidelberg.de | https://orcid.org/0000-0002-9391-5722 | Institute for Computational Biomedicine, Heidelberg University Hospital, Heidelberg, Germany Institute of Pathology, Heidelberg University Hospital, Heidelberg, Germany | DS is supported by the German Federal Ministry of Education and Research (BMBF 01ZZ2004); the Ministry for Science, Research and Science Baden-Württemberg „AI Health Innovation Cluster“ and "MULTI-SPACE"; and University Hospital, Heidelberg, Germany | Writing - Original Draft, Writing - Review & Editing, Endorsement |
| Johannes Schöneberg | jschoeneberg@health.ucsd.edu | https://orcid.org/0000-0001-7083-1828 | University of California, San Diego, CA, USA | JS is supported by the Hartwell Foundation, the W.M.Keck Foundation, the Brain Research Foundation, The Chan Zuckerberg Foundation, and the NIH (1DP2GM150022–01, 1R01GM148765–01). | Writing - Review & Editing, Endorsement |
| Jeffrey M. Spraggins | jeff.spraggins@Vanderbilt.Edu | https://orcid.org/0000-0001-9198-5498 | Department of Cell & Developmental Biology, Vanderbilt University School of Medicine, Nashville, TN, USA | J.M.S. is supported by the National Institutes of Health U01DK133766 (NIDDK), U54DK134302 (NIDDK), U54EY032442 (NEI), R01AG078803 (NIA), R01AI145992 (NIAID), and R01AI138581 (NIAID). | Writing - Review & Editing |
| Damir Sudar | dsudar@qitissue.com | https://orcid.org/0000-0002-2510-7272 | Quantitative Imaging Systems LLC, Portland, OR, USA | DS is supported by the National Institutes of Health U2CCA23380 (NCI) and 1 R44 CA250861 (NCI). | Endorsement |
| Wouter-Michie l Adrien Maria Vierdag | michiel.vierdag@embl.de | https://orcid.org/0000-0003-1666-5421 | Genome Biology Unit, European Molecular Biology Laboratorium, Heidelberg, Baden-Wurttemb erg, Germany | WMV receives research funding from Cellzome, a GSK company. | Writing - Original Draft, Writing - Review & Editing |
| Niels Volkmann | nvo@ucsb.edu | https://orcid.org/0000-0003-1328-6426 | Departments of Bioengineering, of Biological Engineering, Electrical and Computer Engineering, and Biomolecular Science and Engineering Program, University of California, Santa Barbara, CA, USA | Writing - Original Draft, Writing - Review & Editing, Endorsement | |
| Carolina Wahlby | carolina.wahlby@it.uu.se | https://orcid.org/0000-0002-4139-7003 | Dept. Information Technology and Science for LifeLaboratory, Uppsala University, Uppsala, Sweden | Writing - Review & Editing, Endorsement | |
| Siyuan (Steven) Wang | siyuan.wang@yale.edu | https://orcid.org/0000-0001-6550-4064 | Yale University | Endorsement | |
| Ziv Yaniv | zivyaniv@nih.gov | https://orcid.org/0000-0003-0315-7727 | Bioinformatics and Computational Bioscience Branch, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD, USA | Z.Y. is supported by the Bioinformatics and Computational Biosciences Branch (BCBB) Support Services Contract HHSN316201300006 W/75N93022F00001 to Guidehouse Inc. | Writing - Original Draft, Writing - Review & Editing, Endorsement |
| Caterina Strambio-De- Castillia | caterina.strambio@umass med.edu | https://orcid.org/0000-0002-1069-1816 | Program in Molecular Medicine, UMass Chan Medical School, Worcester MA 01605, USA | Conceptualization, Methodology Resources, Writing - Original Draft, Writing - Review & Editing, Endorsement, Supervision, Funding acquisition |
Author contributions categories comply with the CRediT initiative (Allen et al. 2019)
Footnotes
Conflict of Interest Statements
| Name | Statement |
| Ulrike Boehm | UB’s contribution to this manuscript is a result of her voluntary involvement with QUAREP-LiMi and BINA, and does not reflect the position of Carl Zeiss AG on this matter. |
| Josh Moore | holds equity in Glencoe Software. |
| Denis Schapiro | DS reports funding from GSK and received honorariums from Immunai, Alpenglow and Lunaphore. |
| Damir Sudar | DSu is employed by Quantitative Imaging Systems, a commercial entity developing imaging software. |
| Siyuan (Steven) Wang | Founder, shareholder, consultant of Translura, Inc |
References
- Aaron J, Chew TL. A guide to accurate reporting in digital image processing - can anyone reproduce your quantitative analysis? J Cell Sci [Internet]. 2021. Mar 30;134(6). Available from: 10.1242/jcs.254151 [DOI] [PubMed] [Google Scholar]
- Abrams B, Brown C, Callahan L, Cole R, Pengo T, Wee TL. ABRF light microscopy research group study 3: The development and implementation of new tools for microscope quality assurance testing. J Biomol Tech [Internet]. 2020. Aug;31(Suppl):S33. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7646864/ [Google Scholar]
- Abrams B, Pengo T, Wee TL, Deagle RC, Vuillemin N, Callahan LM, et al. Tissue-like 3D standard and protocols for microscope quality management. Microsc Microanal [Internet]. 2023. Mar 4 [cited 2023 Aug 17];29(2):616–34. Available from: https://academic.oup.com/mam/article-abstract/29/2/616/7069290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- African BioImaging Consortium (ABIC). African BioImaging Consortium (ABIC) [Internet]. African BioImaging Consortium (ABIC). 2020. [cited 2022 Sep 4]. Available from: https://www.africanbioimaging.org [Google Scholar]
- Ahlers J, Althviz Moré D, Amsalem O, Anderson A, Bokota G, Boone P, et al. napari: a multi-dimensional image viewer for Python [Internet]. 2023. Available from: https://zenodo.org/record/8115575 [Google Scholar]
- Allan C, Burel JM, Moore J, Blackburn C, Linkert M, Loynton S, et al. OMERO: flexible, model-driven data management for experimental biology. Nat Methods [Internet]. 2012. Feb 28;9(3):245–53. Available from: 10.1038/nmeth.1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen L, O’Connell A, Kiermer V. How can we ensure visibility and diversity in research contributions? How the Contributor Role Taxonomy (CRediT) is helping the shift from authorship to contributorship. Learn Publ [Internet]. 2019. Jan;32(1):71–4. Available from: 10.1002/leap.1210 [DOI] [Google Scholar]
- Andreev A, Morrell T, Briney K, Gesing S, Manor U. Biologists need modern data infrastructure on campus [Internet]. arXiv [q-bio.OT]. 2021. Available from: http://arxiv.org/abs/2108.07631 [Google Scholar]
- Angelo M, Bendall SC, Finck R, Hale MB, Hitzman C, Borowsky AD, et al. Multiplexed ion beam imaging of human breast tumors. Nat Med [Internet]. 2014. Apr;20(4):436–42. Available from: 10.1038/nm.3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arevalo J, van Dijk R, Carpenter AE, Singh S. Evaluating batch correction methods for image-based cell profiling. bioRxiv [Internet]. 2023. Sep 17; Available from: 10.1101/2023.09.15.558001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelrod S, Cai M, Carr A, Freeman J, Ganguli D, Kiggins J, et al. starfish: scalable pipelines for image-based transcriptomics. J Open Source Softw [Internet]. 2021. May 4;6(61):2440. Available from: 10.21105/joss.02440 [DOI] [Google Scholar]
- Bajcsy P, Hotaling N. Interoperability of Web Computational Plugins for Large Microscopy Image Analyses. NISTIR 8297 [Internet]. 2020. Mar; Available from: 10.6028/NIST.IR.8297 [DOI] [Google Scholar]
- Bajcsy P, Bhattiprolu S, Borner K, Cimini B, Collinson L, Ellenberg J, et al. Enabling Global Image Data Sharing in the Life Sciences. arXiv [q-bioQM]. 2024. Available from: 10.48550/arXiv.2401.13023. [DOI] [Google Scholar]
- Bandrowski A. A decade of GigaScience: What can be learned from half a million RRIDs in the scientific literature? Gigascience [Internet]. 2022. Jun 14;11. Available from: 10.1093/gigascience/giac058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandrowski A, Brush M, Grethe JS, Haendel MA, Kennedy DN, Hill S, et al. The Resource Identification Initiative: A Cultural Shift in Publishing. Neuroinformatics [Internet]. 2016. Apr;14(2):169–82. Available from: 10.1007/s12021-015-9284-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandrowski A, Pairish M, Eckmann P, Grethe J, Martone ME. The Antibody Registry: ten years of registering antibodies. Nucleic Acids Res [Internet]. 2023. Jan 6;51(D1):D358–67. Available from: 10.1093/nar/gkac927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankhead P, Loughrey MB, Fernández JA, Dombrowski Y, McArt DG, Dunne PD, et al. QuPath: Open source software for digital pathology image analysis. Sci Rep [Internet]. 2017. Dec 4;7(1):16878. Available from: 10.1038/s41598-017-17204-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baysoy A, Bai Z, Satija R, Fan R. The technological landscape and applications of single-cell multi-omics. Nat Rev Mol Cell Biol [Internet]. 2023. Oct;24(10):695–713. Available from: 10.1038/s41580-023-00615-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beliveau BJ, Kishi JY, Nir G, Sasaki HM, Saka SK, Nguyen SC, et al. OligoMiner provides a rapid, flexible environment for the design of genome-scale oligonucleotide in situ hybridization probes. Proc Natl Acad Sci U S A [Internet]. 2018. Mar 6;115(10):E2183–92. Available from: 10.1073/pnas.1714530115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthold MR. KNIME: open for innovation [Internet]. Konstanz, Germany; 2023. Available from:http://knime.org/ [Google Scholar]
- Bidgood WD Jr, Horii SC, Prior FW, Van Syckle DE. Understanding and using DICOM, the data interchange standard for biomedical imaging. J Am Med Inform Assoc [Internet]. 1997. May-Jun;4(3):199–212. Available from: 10.1136/jamia.1997.0040199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckhout M, Zielhuis GA, Bredenoord AL. The FAIR guiding principles for data stewardship: fair enough? Eur J Hum Genet [Internet]. 2018. Jul;26(7):931–6. Available from: 10.1038/s41431-018-0160-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm U, Nelson G, Brown CM, Bagley S, Bajcsy P, Bischof J, et al. QUAREP-LiMi: a community endeavor to advance quality assessment and reproducibility in light microscopy. Nature Methods ( 10.1038/s41592-021-01162-y) [Internet]. 2021. May 21;18:1423–6. Available from: 10.1038/s41592-021-01162-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botvinik-Nezer R, Holzmeister F, Camerer CF, Dreber A, Huber J, Johannesson M, et al. Variability in the analysis of a single neuroimaging dataset by many teams. Nature [Internet]. 2020. Jun;582(7810):84–8. Available from: 10.1038/s41586-020-2314-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazma A, Ball C, Bumgarner R, Furlanello C, Miller M. MINSEQE: Minimum Information about a high-throughput Nucleotide SeQuencing Experiment-a proposal for standards in functional genomic data …. Zenodo https://doiorg. 2012; [Google Scholar]
- Brown J, Jones P, Meadows A, Murphy F. Incentives to invest in identifiers: A cost-benefit analysis of persistent identifiers in Australian research systems [Internet]. Zenodo. 2022a. Available from: https://zenodo.org/record/7100578 [Google Scholar]
- Brown J, Jones P, Meadows A, Murphy F. Revised cost-benefit analysis for the UK PID Support Network [Internet]. Zenodo. Zenodo; 2022b. Available from: https://zenodo.org/records/7356219 [Google Scholar]
- Budtz Pedersen D, Hvidtfeldt R. The missing links of research impact. Res Eval [Internet]. 2023. May 12 [cited 2023 Oct 10];rvad011. Available from: 10.1093/reseval/rvad011/7161084 [DOI] [Google Scholar]
- Bukhari SAC, Martínez-Romero M, O’ Connor MJ, Egyedi AL, Willrett D, Graybeal J, et al. CEDAR OnDemand: a browser extension to generate ontology-based scientific metadata. BMC Bioinformatics [Internet]. 2018. Jul 16;19(1):268. Available from: 10.1186/s12859-018-2247-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- California Digital Library. DMPTool [Internet]. The Regents of the University of California; 2022. [cited 2023 Oct 9]. Available from: https://dmptool.org/ [Google Scholar]
- Cell Migration Standardisation Organisation. Minimum Information About Cell Migration Experiments (MIACME 0.2) [Internet]. Cell Migration Standardisation Organisation; 2021. Oct. Available from: https://cmso.science/MIACME/v0.2/ [Google Scholar]
- Chen J, Viana MP, Rafelski SM. When seeing is not believing: application-appropriate validation matters for quantitative bioimage analysis. Nat Methods [Internet]. 2023. Jul;20(7):968–70. Available from: 10.1038/s41592-023-01881-4 [DOI] [PubMed] [Google Scholar]
- Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science [Internet]. 2015. Apr 24;348(6233):aaa6090. Available from: 10.1126/science.aaa6090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu W, Schmid MF, Pintilie GD, Lawson CL. Evolution of standardization and dissemination of cryo-EM structures and data jointly by the community, PDB, and EMDB. J Biol Chem [Internet]. 2021. Mar 18;296:100560. Available from: 10.1016/j.jbc.2021.100560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HMT, Beck VA, Pierce NA. Next-generation in situ hybridization chain reaction: Higher gain, lower cost, greater durability. ACS Nano [Internet]. 2014;8(5):4284–94. Available from: 10.1021/nn405717p [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciavotta M, Cutrona V, De Paoli F, Nikolov N, Palmonari M, Roman D. Supporting Semantic Data Enrichment at Scale. In: Curry E, Auer S, Berre AJ, Metzger A, Perez MS, Zillner S, editors. Technologies and Applications for Big Data Value [Internet]. Cham: Springer International Publishing; 2022. p. 19–39. Available from: 10.1007/978-3-030-78307-5_2 [DOI] [Google Scholar]
- Cimini BA, Chandrasekaran SN, Kost-Alimova M, Miller L, Goodale A, Fritchman B, et al. Optimizing the Cell Painting assay for image-based profiling. Nat Protoc [Internet]. 2023. Jul;18(7):1981–2013. Available from: 10.1038/s41596-023-00840-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côté R, Reisinger F, Martens L, Barsnes H, Vizcaino JA, Hermjakob H. The Ontology Lookup Service: bigger and better. Nucleic Acids Res [Internet]. 2010. Jul;38(Web Server issue):W155–60. Available from: 10.1093/nar/gkq331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousijn H, Braukmann R, Fenner M, Ferguson C, van Horik R, Lammey R, et al. Connected Research: The Potential of the PID Graph. Patterns (N Y) [Internet]. 2021. Jan 8;2(1):100180. Available from: 10.1016/j.patter.2020.100180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Alber F, Aufmkolk S, Beliveau BJ, Bruneau BG, Belmont AS, et al. Spatial and temporal organization of the genome: Current state and future aims of the 4D nucleome project. Mol Cell [Internet]. 2023. Jun 29; Available from: 10.1016/j.molcel.2023.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Belmont AS, Guttman M, Leshyk VO, Lis JT, Lomvardas S, et al. The 4D nucleome project. Nature [Internet]. 2017. Sep 13;549(7671):219–26. Available from: 10.1038/nature23884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demchenko Y, Stoy L. Research data management and data stewardship competences in university curriculum. In: 2021 IEEE Global Engineering Education Conference (EDUCON) [Internet]. IEEE; 2021. Available from: https://ieeexplore.ieee.org/document/9453956/ [Google Scholar]
- Di Tommaso P, Floden E. A DSL for parallel and scalable computational pipelines [Internet]. 2023. [cited 2023 Oct 10]. Available from: https://www.nextflow.io/index.html [Google Scholar]
- Ellenberg J, Swedlow JR, Barlow M, Cook CE, Sarkans U, Patwardhan A, et al. A call for public archives for biological image data. Nat Methods [Internet]. 2018. Nov;15(11):849–54. Available from: 10.1038/s41592-018-0195-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EMBL-EBI. Electron Microscopy Data Bank (EMDB) [Internet]. EMBL European Bioinformatics Institute (EMBL-EBI) - Electron Microscopy Data Bank. [cited 2023a Oct 30]. Available from: https://www.ebi.ac.uk/emdb/ [Google Scholar]
- EMBL-EBI. EMPIAR - Electron Microscopy Public Image Archive [Internet]. EMBL European Bioinformatics Institute (EMBL-EBI) - Electron Microscopy Public Image Archive. [cited 2023b Oct 30]. Available from: https://www.ebi.ac.uk/empiar/ [Google Scholar]
- Eng J, Bucher E, Hu Z, Zheng T, Gibbs SL, Chin K, et al. A framework for multiplex imaging optimization and reproducible analysis. Commun Biol [Internet]. 2022. May 11;5(1):438. Available from: 10.1038/s42003-022-03368-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson J, Pukonen I. D2.3 Common international recommendation for quality assurance and management in open access imaging infrastructures [Internet]. Global BioImaging Project; 2018. Nov. Available from: https://www.globalbioimaging.org/user/pages/05.documents/D2.3_Publication%20of%20common%20recommendation_quality%20assurance%20and%20management%20in%20open%20access%20imaging%20infrastructures.pdf [Google Scholar]
- European Research Council - Scientific Council. Open Research Data and Data Management Plans. In European Commission; [Google Scholar]
- Faklaris O, Bancel-Vallée L, Dauphin A, Monterroso B, Frère P, Geny D, et al. Quality assessment in light microscopy for routine use through simple tools and robust metrics. J Cell Biol [Internet]. 2022. Nov7;221(11). Available from: 10.1083/jcb.202107093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudreault N, Nelson G, Alexopoulos I, Azevedo M, Barachati F, Belyaev Y, et al. Illumination power, stability, and linearity measurements for confocal and Widefield microscopes v2 [Internet]. Protocols.io; 2022. Apr.Available from: https://www.protocols.io/view/illumination-power-stability-and-linearity-measure-b68prhvn [Google Scholar]
- Gelali E, Girelli G, Matsumoto M, Wernersson E, Custodio J, Mota A, et al. iFISH is a publically available resource enabling versatile DNA FISH to study genome architecture. Nat Commun [Internet]. 2019. Apr 9;10(1):1636. Available from: 10.1038/s41467-019-09616-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes MJ, Sevinsky CJ, Sood A, Adak S, Bello MO, Bordwell A, et al. Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. Proc Natl Acad Sci U S A [Internet]. 2013. Jul16;110(29):11982–7. Available from: 10.1073/pnas.1300136110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerner MY, Kastenmuller W, Ifrim I, Kabat J, Germain RN. Histo-cytometry: a method for highly multiplex quantitative tissue imaging analysis applied to dendritic cell subset microanatomy in lymph nodes. Immunity [Internet]. 2012. Aug 24;37(2):364–76. Available from: 10.1016/j.immuni.2012.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giesen C, Wang HAO, Schapiro D, Zivanovic N, Jacobs A, Hattendorf B, et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods [Internet]. 2014. Apr;11(4):417–22. Available from: 10.1038/nmeth.2869 [DOI] [PubMed] [Google Scholar]
- Global BioImaging. Global BioImaging [Internet]. Global BioImaging. 2015. [cited 2022 Sep 4]. Available from: https://globalbioimaging.org/ [Google Scholar]
- Goldberg IG, Allan C, Burel JM, Creager D, Falconi A, Hochheiser H, et al. The Open Microscopy Environment (OME) Data Model and XML file: open tools for informatics and quantitative analysis in biological imaging. Genome Biol [Internet]. 2005. Jan 1;6(5):R47. Available from: http://genomebiology.com/2005/6/5/R47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goltsev Y, Samusik N, Kennedy-Darling J, Bhate S, Hale M, Vazquez G, et al. Deep Profiling of Mouse Splenic Architecture with CODEX Multiplexed Imaging. Cell [Internet]. 2018. Aug 9;174(4):968–81.e15. Available from: 10.1016/j.cell.2018.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González G, Evans CL. Biomedical Image Processing with Containers and Deep Learning: An Automated Analysis Pipeline: Data architecture, artificial intelligence, automated processing, containerization, and clusters orchestration ease the transition from data acquisition to insights in medium-to-large datasets. Bioessays [Internet]. 2019. Jun;41(6):e1900004. Available from: 10.1002/bies.201900004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould M. ROR and organizational identifier interoperability in publishing systems. In: ROR and Organizational Identifier Interoperability in Publishing Systems [Internet]. ScienceOpen; 2023. Available from: 10.14293/S2199-SSP-AM23-01030 [DOI] [Google Scholar]
- Gut G, Herrmann MD, Pelkmans L. Multiplexed protein maps link subcellular organization to cellular states. Science [Internet]. 2018. Aug 3;361(6401). Available from: 10.1126/science.aar7042 [DOI] [PubMed] [Google Scholar]
- Haak LL, Fenner M, Paglione L, Pentz E, Ratner H. ORCID: a system to uniquely identify researchers. Learn Publ [Internet]. 2012. Oct 1;25(4):259–64. Available from: 10.1087/20120404 [DOI] [Google Scholar]
- Halfon Y, Aspinall L, White J, Jackson Hirst I, Wang Y, Darrow MC, et al. Maintaining the momentum in cryoEM for biological discovery. Faraday Discuss [Internet]. 2022. Nov 8;240(0):18–32. Available from: 10.1039/d2fd00129b [DOI] [PubMed] [Google Scholar]
- Hammer M, Huisman M, Rigano A, Boehm U, Chambers JJ, Gaudreault N, et al. Towards community-driven metadata standards for light microscopy: tiered specifications extending the OME model. Nature Methods ( 10.1038/s41592-021-01327-9) [Internet]. 2021;18:1427–40. Available from: 10.1038/s41592-021-01327-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer M, Rigano A, Farzam F, Huisman M, Grünwald D, Strambio-De-Castillia C. The 4DN-OME ontology: an OME-OWL extension with emphasis on usability, minimum information guidelines, and quality control for super-resolution fluorescence microscopy - Poster. In: 12th Annual 2019 SWAT4(HC)LS (Semantic Web Applications and Tools for Healthcare and Life Sciences) Conference [Internet]. figshare.org;December, 9–12 2019. Available from: 10.6084/m9.figshare.12758957.v1 [DOI] [Google Scholar]
- Heddleston JM, Aaron JS, Khuon S, Chew TL. A guide to accurate reporting in digital image acquisition - can anyone replicate your microscopy data? J Cell Sci [Internet]. 2021. Mar 30;134(6). Available from: 10.1242/jcs.254144 [DOI] [PubMed] [Google Scholar]
- Hershberg EA, Camplisson CK, Close JL, Attar S, Chern R, Liu Y, et al. PaintSHOP enables the interactive design of transcriptome- and genome-scale oligonucleotide FISH experiments. Nat Methods [Internet].2021. Aug;18(8):937–44. Available from: 10.1038/s41592-021-01187-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey JW, Neumann EK, Radtke AJ, Camarillo JM, Beuschel RT, Albanese A, et al. Spatial mapping of protein composition and tissue organization: a primer for multiplexed antibody-based imaging. Nat Methods [Internet]. 2022. Mar;19(3):284–95. Available from: 10.1038/s41592-021-01316-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotchkiss J, Manyisa N, Adadey SM, Oluwole OG, Wonkam E, Mnika K, et al. The Hearing Impairment Ontology: A Tool for Unifying Hearing Impairment Knowledge to Enhance Collaborative Research. Genes [Internet]. 2019. Nov 21;10(12). Available from: 10.3390/genes10120960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- HuBMAP Consortium. The human body at cellular resolution: the NIH Human Biomolecular Atlas Program. Nature [Internet]. 2019. Oct;574(7777):187–92. Available from: 10.1038/s41586-019-1629-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- HuBMAP Consortium. HuBMAP Antibody Validation Reports [Internet]. HuBMAP AVR Registry. 2023a. [cited2023 Oct 7]. Available from: https://avr.hubmapconsortium.org/ [Google Scholar]
- HuBMAP Consortium. Human Reference Atlas Portal - Organ Mapping Antibody Panels (OMAPs) 5th HRA Release (v1.4) [Internet]. Human Atlas Portal. 2023b. [cited 2023 Oct 7]. Available from: https://humanatlas.io/omap [Google Scholar]
- Huisman M, Hammer M, Rigano A, Boehm U, Chambers JJ, Gaudreault N, et al. A perspective on Microscopy Metadata: data provenance and quality control. arXiv [q-bioQM]: https://arxiv.org/abs/191011370 [Internet]. 2021. Apr 28; Available from: https://arxiv.org/abs/1910.11370 [Google Scholar]
- Hu M, Yang B, Cheng Y, Radda JSD, Chen Y, Liu M, et al. ProbeDealer is a convenient tool for designing probes for highly multiplexed fluorescence in situ hybridization. Sci Rep [Internet]. 2020. Dec16;10(1):22031. Available from: 10.1038/s41598-020-76439-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Pei L, Spraggins JM, Angelo M, Carson JP, Gehlenborg N, et al. Advances and prospects for the Human BioMolecular Atlas Program (HuBMAP). Nat Cell Biol [Internet]. 2023. Aug;25(8):1089–100. Available from: 10.1038/s41556-023-01194-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jupp S. A new Ontology Lookup Service at EMBL-EBI. In: Malone J, editor. Proceedings of SWAT4LS International Conference 2015. [Google Scholar]
- Kemmer I, Keppler A, Serrano-Solano B, Rybina A, Özdemir B, Bischof J, et al. Building a FAIR image data ecosystem for microscopy communities. Histochem Cell Biol [Internet]. 2023. Sep;160(3):199–209. Available from: 10.1007/s00418-023-02203-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke R, Mignardi M, Pacureanu A, Svedlund J, Botling J, Wählby C, et al. In situ sequencing for RNA analysis in preserved tissue and cells. Nat Methods [Internet]. 2013. Sep;10(9):857–60. Available from: 10.1038/nmeth.2563 [DOI] [PubMed] [Google Scholar]
- Khurana U, Srinivas K, Galhotra S, Samulowitz H. A Vision for Semantically Enriched Data Science [Internet]. arXiv [cs.AI]. 2023. Available from: http://arxiv.org/abs/2303.01378 [Google Scholar]
- Kinkhabwala A, Herbel C, Pankratz J, Yushchenko DA, Rüberg S, Praveen P, et al. MACSima imaging cyclic staining (MICS) technology reveals combinatorial target pairs for CAR T cell treatment of solid tumors. Sci Rep [Internet]. 2022. Feb 3;12(1):1911. Available from: 10.1038/s41598-022-05841-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi JY, Lapan SW, Beliveau BJ, West ER, Zhu A, Sasaki HM, et al. SABER amplifies FISH: enhanced multiplexed imaging of RNA and DNA in cells and tissues. Nat Methods [Internet]. 2019. May 20; Available from: 10.1038/s41592-019-0404-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- KNIME Community, bioml-konstanz. KNIME image processing [Internet]. 2023. [cited 2023 Oct 10]. Available from: https://hub.knime.com/bioml-konstanz/extensions/org.knime.knip.feature/latest [Google Scholar]
- Kobayashi N, Moore J, Onami S, Swedlow JR. OME Core Ontology: An OWL-based Life Science Imaging Data Model. In: Cornet R, Waagmeester A, editors. Proceedings of the 12th SWAT4(HC)LS (Semantic Web Applications and Tools for Healthcare and Life Sciences) Conference [Internet]. http://ceur-ws.org/Vol-2849/paper-25.pdf; Dec 10–11 2019. p. 149–50. Available from: http://ceur-ws.org/Vol-2849/paper-25.pdf [Google Scholar]
- Könnecke M, Akeroyd FA, Bernstein HJ, Brewster AS, Campbell SI, Clausen B, et al. The NeXus data format. J Appl Crystallogr [Internet]. 2015. Feb 1;48(Pt 1):301–5. Available from: 10.1107/S1600576714027575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahl R, Darroch L, Huber R, Devaraju A, Klump J, Habermann T, et al. Metadata schema for the persistent identification of instruments [Internet]. Research Data Alliance; 2021. Available from: https://zenodo.org/record/6396467#.YkQxzRBBwlw [Google Scholar]
- Kühlbrandt W.. Biochemistry. The resolution revolution. Science [Internet]. 2014. Mar 28;343(6178):1443–4.Available from: 10.1126/science.1251652 [DOI] [PubMed] [Google Scholar]
- Kunis S, Dohle J. Structuring of Data and Metadata in Bioimaging: Concepts and technical Solutions in the Context of Linked Data [Internet]. 2022. Available from: https://zenodo.org/record/7018750 [Google Scholar]
- Kunis S, Hänsch S, Schmidt C, Wong F, Strambio-De-Castillia C, Weidtkamp-Peters S. MDEmic: a metadata annotation tool to facilitate FAIR image data management in the bioimaging community. Nature Methods [Internet]. 2021. Oct 11;18:1415–6. Available from: 10.1038/s41592-021-01288-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen DD, Gaudreault N, Gibbs HC. Reporting reproducible imaging protocols. STAR Protoc [Internet]. 2023. Mar 1;4(1):102040. Available from: 10.1016/j.xpro.2022.102040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson CL, Baker ML, Best C, Bi C, Dougherty M, Feng P, et al. EMDataBank.org: unified data resource for CryoEM. Nucleic Acids Res [Internet]. 2011. Jan;39(Database issue):D456–64. Available from: 10.1093/nar/gkq880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JR, Chen YA, Campton D, Cooper J, Coy S, Yapp C, et al. High-plex immunofluorescence imaging and traditional histology of the same tissue section for discovering image-based biomarkers. Nat Cancer [Internet]. 2023. Jul;4(7):1036–52. Available from: 10.1038/s43018-023-00576-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JR, Izar B, Wang S, Yapp C, Mei S, Shah PM, et al. Highly multiplexed immunofluorescence imaging of human tissues and tumors using t-CyCIF and conventional optical microscopes. Elife [Internet]. 2018. Jul 11;7. Available from: 10.7554/eLife.31657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkert M, Rueden CT, Allan C, Burel JM, Moore W, Patterson A, et al. Metadata matters: access to image data in the real world. J Cell Biol [Internet]. 2010. May 31;189(5):777–82. Available from: 10.1083/jcb.201004104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomax J. Using ontologies to unlock the full potential of your scientific data - Part 1 [Internet]. SciBite. 2019. [cited 2023 Oct 8]. Available from: https://scibite.com/news/using-ontologies-to-unlock-the-your-scientific-data-1/ [Google Scholar]
- Ludin A, Korir PK, Salavert-Torres J, Kleywegt GJ, Patwardhan A. EMPIAR: a public archive for raw electron microscopy image data. Nat Methods [Internet]. 2016. May;13(5):387–8. Available from: 10.1038/nmeth.3806 [DOI] [PubMed] [Google Scholar]
- Marconato L, Palla G, Yamauchi KA, Virshup I, Heidari E, Treis T, et al. SpatialData: an open and universal data framework for spatial omics [Internet]. bioRxiv. 2023. Available from: 10.1101/2023.05.05.539647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marqués G, Pengo T, Sanders MA. Imaging methods are vastly underreported in biomedical research. Elife [Internet]. 2020. Aug 11;9. Available from: 10.7554/eLife.55133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx V. Imaging standards to ease reproducibility and the everyday. Nat Methods [Internet]. 2022aJul;19(7):784–8. Available from: 10.1038/s41592-022-01544-w [DOI] [PubMed] [Google Scholar]
- Marx V. The making of microscope camera standards. Nat Methods [Internet]. 2022b. Jul;19(7):788–91. Available from: 10.1038/s41592-022-01545-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCafferty S, Poger D, Yvette W, Seal C, Burgess R, Kenna E. Best Practices: PIDs for Instruments [Internet].Zenodo. Zenodo; 2023. Available from: https://zenodo.org/records/7759201 [Google Scholar]
- Meyn SM, Ramirez-Aguilar KA, Gregory CW, Mische S, Ott AW, Sol-Church K, et al. Addressing the Environmental Impact of Science Through a More Rigorous, Reproducible, and Sustainable Conduct of Research. J Biomol Tech [Internet]. 2022. Dec 31;33(4). Available from: 10.7171/3fc1f5fe.d085ce95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Australia Microscopy. Microscopy Australia. 2016. [cited 2023 Oct 24]. Available from: https://micro.org.au [Google Scholar]
- Mitra-Behura S, Fiolka RP, Daetwyler S. Singularity Containers Improve Reproducibility and Ease of Use in Computational Image Analysis Workflows. Front Bioinform [Internet]. 2021;1:757291. Available from: 10.3389/fbinf.2021.757291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Nørrelykke SF. Reproducible image handling and analysis. EMBO J [Internet]. 2021. Feb1;40(3):e105889. Available from: 10.15252/embj.2020105889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt JR, Bambah-Mukku D, Eichhorn SW, Vaughn E, Shekhar K, Perez JD, et al. Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science [Internet]. 2018. Nov 16;362(6416). Available from: 10.1126/science.aau5324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt JR, Lundberg E, Heyn H. The emerging landscape of spatial profiling technologies. Nat Rev Genet[Internet]. 2022. Dec;23(12):741–59. Available from: 10.1038/s41576-022-00515-3 [DOI] [PubMed] [Google Scholar]
- Montero Llopis P, Senft RA, Ross-Elliott TJ, Stephansky R, Keeley DP, Koshar P, et al. Best practices and tools for reporting reproducible fluorescence microscopy methods. Nat Methods [Internet].2021;18(12):1463–76. Available from: https://idp.nature.com/authorize/casa?redirect_uri=https://www.nature.com/articles/s41592-021-01156-w&casa_token=PAcH9_Q1bXIAAAAA:Q_-qCzfAZ2uUL0fpWS9u9ctY-jiNHseLcwvQztqPIqmj0-ybbccjPe2POB0gv8JinC4PiAILnahjjjsE9Q [DOI] [PubMed] [Google Scholar]
- Moore J. The Case forNext-Generation Metadata [Internet]. 2022 Open Microscopy Environment Community Meeting; figshare; 2022a. Available from: https://figshare.com/articles/presentation/OME2022_Day3_Moore_20221115_pdf/21556998/1 [Google Scholar]
- Moore J. The Case for Next-Generation Metadata - Presentation at the OME2022 - Day 3 [Internet]. 2022 OME Community Meeting ; 2022b. Available from: https://figshare.com/articles/presentation/OME2022_Day3_Moore_20221115_pdf/21556998 [Google Scholar]
- Moore J, Allan C, Besson S, Burel JM, Diel E, Gault D, et al. OME-NGFF: a next-generation file format for expanding bioimaging data-access strategies. Nat Methods [Internet]. 2021. Dec;18(12):1496–8. Available from: 10.1038/s41592-021-01326-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J, Basurto-Lozada D, Besson S, Bogovic J, Bragantini J, Brown EM, et al. OME-Zarr: a cloud-optimized bioimaging file format with international community support. Histochem Cell Biol [Internet]. 2023. Jul 10; Available from: 10.1007/s00418-023-02209-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mund A, Coscia F, Kriston A, Hollandi R, Kovács F, Brunner AD, et al. Deep Visual Proteomics defines single-cell identity and heterogeneity. Nat Biotechnol [Internet]. 2022. Aug;40(8):1231–40. Available from: 10.1038/s41587-022-01302-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustra M, Delac K, Grgic M. Overview of the DICOM standard. In: 2008 50th International Symposium ELMAR [Internet]. IEEE; 2008. p. 39–44. Available from: https://ieeexplore.ieee.org/abstract/document/4747434/ [Google Scholar]
- Nagaraj A, Shears E, de Vaan M. Improving data access democratizes and diversifies science. Proc Natl Acad Sci U S A [Internet]. 2020. Sep 22;117(38):23490–8. Available from: 10.1073/pnas.2001682117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakane T, Kotecha A, Sente A, McMullan G, Masiulis S, Brown PMGE, et al. Single-particle cryo-EM at atomic resolution. Nature [Internet]. 2020. Nov;587(7832):152–6. Available from: 10.1038/s41586-020-2829-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nature Editorial Staff. Better research through metrology. Nat Methods [Internet]. 2018. Jun;15(6):395.Available from: 10.1038/s41592-018-0035-x [DOI] [PubMed] [Google Scholar]
- Nelson G. Monitoring the point spread function for quality control of confocal microscopes v1 [Internet].Protocols.io; 2022. Jul. Available from: https://www.protocols.io/view/monitoring-the-point-spread-function-for-quality-ccsuswew [Google Scholar]
- Nelson G, Boehm U, Bagley S, Bajcsy P, Bischof J, Brown CM, et al. QUAREP-LiMi: A community-driven initiative to establish guidelines for quality assessment and reproducibility for instruments and images in light microscopy. J Microsc ( 10.1111/jmi13041) [Internet]. 2021. Jul 2;284(1):56–73. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- NFDI4Plants Consortium. Swate: Excel Add-In for annotation of experimental data and computational workflows [Internet]. Github; 2022. [cited 2023 Oct 8]. Available from: https://github.com/nfdi4plants/Swate [Google Scholar]
- NIH Strategic Plan for Data Science. [cited 2023 Oct 27]. Available from:https://datascience.nih.gov/nih-strategic-plan-data-science [Google Scholar]
- NOT-OD-21–013: Final NIH Policy for Data Management and Sharing. [cited 2023 Oct 24]. Available from: https://grants.nih.gov/grants/guide/notice-files/NOT-OD-21-013.html
- Ong E, Sarntivijai S, Jupp S, Parkinson H, He Y. Comparison, alignment, and synchronization of cell line information between CLO and EFO. BMC Bioinformatics [Internet]. 2017. Dec 21;18(Suppl 17):557. Available from: 10.1186/s12859-017-1979-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines J. Image integrity and standards. Open Biol [Internet]. 2020. Jun;10(6):200165. Available from: 10.1098/rsob.200165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porciani PF, Cherici G, Novelli C. [The action on bacterial plaque of the mastication of a sorbitol chewing gum]. Minerva Stomatol [Internet]. 1992. Oct;41(10):445–9. Available from: https://www.ncbi.nlm.nih.gov/pubmed/1293493 [PubMed] [Google Scholar]
- Preparing your data management plan. NSF - National Science Foundation. [cited 2023 Oct 24]. Available from: https://new.nsf.gov/funding/data-management-plan [Google Scholar]
- PyTorch Consortium. Reproducibility — PyTorch 2.1 documentation [Internet]. PyTorch. 2023. [cited 2023 Oct10]. Available from: https://pytorch.org/docs/stable/notes/randomness.html [Google Scholar]
- Quardokus EM, Saunders DC, McDonough E, Hickey JW, Werlein C, Surrette C, et al. Organ Mapping Antibody Panels: a community resource for standardized multiplexed tissue imaging. Nat Methods[Internet]. 2023. Aug;20(8):1174–8. Available from: 10.1038/s41592-023-01846-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radtke AJ, Kandov E, Lowekamp B, Speranza E, Chu CJ, Gola A, et al. IBEX: A versatile multiplex optical imaging approach for deep phenotyping and spatial analysis of cells in complex tissues. Proc Natl Acad Sci U S A [Internet]. 2020. Dec 29;117(52):33455–65. Available from: 10.1073/pnas.2018488117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R, Bugbee K, Murphy K. From open data to open science. Earth Space Sci [Internet]. 2021. May;8(5). Available from: 10.1029/2020EA001562 [DOI] [Google Scholar]
- RCSB Consortium. Protein Data Bank (PDB) [Internet]. Research Collaboratory for Structural Bioinformatics PDB. 2019. Available from: https://www.rcsb.org [Google Scholar]
- Registration overview — SimpleITK documentation. [cited 2023 Oct 27]. Available from: 10.5281/zenodo.7693278 [DOI] [Google Scholar]
- Rigano A, Ehmsen S, Öztürk SU, Ryan J, Balashov A, Hammer M, et al. Micro-Meta App: an interactive tool for collecting microscopy metadata based on community specifications. Nat Methods [Internet]. 2021. Dec;18(12):1489–95. Available from: 10.1038/s41592-021-01315-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivest F, Eroglu D, Pelz B, Kowal J, Kehren A, Procopio MG, et al. Fully Automated Sequential Immunofluorescence (seqIF) for Hyperplex Spatial Proteomics [Internet]. bioRxiv. 2023. [cited 2023 Oct 9]. p. 2023.07.07.548135. Available from: 10.1101/2023.07.07.548135v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouillard JM, Zuker M, Gulari E. OligoArray 2.0: Design of oligonucleotide probes for DNA microarrays using a thermodynamic approach. Nucleic Acids Res [Internet]. 2003;31(12):3057–62. Available from: 10.1093/nar/gkg426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenblatt-Rosen O, Regev A, Oberdoerffer P, Nawy T, Hupalowska A, Rood JE, et al. The Human Tumor Atlas Network: Charting Tumor Transitions across Space and Time at Single-Cell Resolution. Cell [Internet]. 2020. Apr 16;181(2):236–49. Available from: 10.1016/j.cell.2020.03.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- RTmfm. RTmfm – Reseau Technologique Microscopie de Fluorescence Multidimensionnelle de la Mission pour les Initiatives Transverses et Interdisciplinaires (MITI) du CNRS [Internet]. RTmfm - CNRS. 2022. [cited 2023 Oct 9]. Available from: https://rtmfm.cnrs.fr/ [Google Scholar]
- Rustici G, Kapushesky M, Kolesnikov N, Parkinson H, Sarkans U, Brazma A. Data storage and analysis in ArrayExpress and Expression Profiler. Curr Protoc Bioinformatics [Internet]. 2008. Sep;Chapter 7:Unit 7.13. Available from: 10.1002/0471250953.bi0713s23 [DOI] [PubMed] [Google Scholar]
- Ryan J, Pengo T, Rigano A, Montero Llopis P, Itano MS, Cameron LC, et al. MethodsJ2: A Software Tool to Capture and Generate Comprehensive Microscopy Methods Text and Improve Reproducibility. Nature Methods ( 10.1038/s41592-021-01290-5) [Internet]. 2021. Oct 15;18:1414–5. Available from: 10.1038/s41592-021-01290-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka SK, Wang Y, Kishi JY, Zhu A, Zeng Y, Xie W, et al. Immuno-SABER enables highly multiplexed and amplified protein imaging in tissues. Nat Biotechnol [Internet]. 2019. Sep;37(9):1080–90. Available from: 10.1038/s41587-019-0207-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansone SA, Rocca-Serra P, Field D, Maguire E, Taylor CF, Hofmann O, et al. Toward interoperable bioscience data. Nat Genet [Internet]. 2012. Jan 27;44(2):121–6. Available from: http://www.nature.com/ng/journal/v44/n2/full/ng.1054.html [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkans U, Chiu W, Collinson L, Darrow MC, Ellenberg J, Grunwald D, et al. REMBI: Recommended Metadata for Biological Images-enabling reuse of microscopy data in biology. Nat Methods [Internet]. 2021. Dec;18(12):1418–22. Available from: 10.1038/s41592-021-01166-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schapiro D, Sokolov A, Yapp C, Chen YA, Muhlich JL, Hess J, et al. MCMICRO: a scalable, modular image-processing pipeline for multiplexed tissue imaging. Nat Methods [Internet]. 2021. Nov 25 [cited 2022 Mar 18];19(3):311–5. Available from: https://www.nature.com/articles/s41592-021-01308-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schapiro D, Yapp C, Sokolov A, Reynolds SM, Chen YA, Sudar D, et al. MITI minimum information guidelines for highly multiplexed tissue images. Nat Methods [Internet]. 2022. Mar;19(3):262–7. Available from: 10.1038/s41592-022-01415-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmied C, Nelson MS, Avilov S, Bakker GJ, Bertocchi C, Bischof J, et al. Community-developed checklists for publishing images and image analyses. Nat Methods [Internet]. 2023. Sep 14; Available from: 10.1038/s41592-023-01987-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen MR, Fields JL, Northan B, Lacoste J, Ang LH, Fiering S, et al. Replication Study: Biomechanical remodeling of the microenvironment by stromal caveolin-1 favors tumor invasion and metastasis. Elife [Internet]. 2019. Dec 17;8. Available from: 10.7554/eLife.45120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shillum C, Petro JA, Demeranville T, Wijnbergen I, Hershberger S, Simpson W. From Vision to Value: ORCID’s 2022–2025 Strategic Plan [Internet]. ORCID; 2021. Available from: https://orcid.figshare.com/articles/online_resource/From_Vision_to_Value_ORCID_s_2022_2025_Strategic_Plan/16687207/1 [Google Scholar]
- Sickle Cell Disease Ontology Working Group. The Sickle Cell Disease Ontology: enabling universal sickle cell-based knowledge representation. Database [Internet]. 2019. Jan 1;2019. Available from: 10.1093/database/baz118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solbrig H, Moxon S, Unni D, Vaidya G, Hegde H, Duncan B, et al. LinkML [Internet]. Zenodo; 2023. Available from: 10.5281/ZENODO.8084083 [DOI] [Google Scholar]
- Steeleworthy M. Research Data Management and the Canadian Academic Library: An Organizational Consideration of Data Management and Data Stewardship. NISTIR 8297 [Internet]. 2014. [cited 2023 Oct 10]; Available from: https://scholars.wlu.ca/lib_pub/29/ [Google Scholar]
- Stirling DR, Swain-Bowden MJ, Lucas AM, Carpenter AE, Cimini BA, Goodman A. CellProfiler 4:improvements in speed, utility and usability. BMC Bioinformatics [Internet]. 2021. Sep 10;22(1):433. Available from: 10.1186/s12859-021-04344-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker M, Darroch L, Krahl R, Habermann T, Devaraju A, Schwardmann U, et al. Persistent identification of instruments. Data Sci J [Internet]. 2020. May 5;19. Available from: 10.5334/dsj-2020-018/ [DOI] [Google Scholar]
- Strambio-De-Castillia C, Bajcsy P, Boehm U, Chambers J, Corbett AD, Faklaris O, et al. BioImaging North America (BINA): Quality Control and Data Management WG (QC-DM-WG) [Internet]. Bioimaging North America Site. 2019. [cited 2020 Jan 16]. Available from: https://www.bioimagingnorthamerica.org/qc-dm-wg/ [Google Scholar]
- Swedlow JR, Goldberg IG, Brauner E, Sorger PK. Informatics and Quantitative Analysis in Biological Imaging.Science [Internet]. 2003. Apr 4;300(5616):100–2. Available from: 10.1126/science.1082602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swedlow JR, Kankaanpää P, Sarkans U, Goscinski W, Galloway G, Sullivan RP, et al. A Global View of Standards for Open Image Data Formats and Repositories. Nature Methods ( 10.1038/s41592-021-01113-7) [Internet]. 2021. May 4;18:1440–6. Available from: 10.1038/s41592-021-01113-7 [DOI] [PubMed] [Google Scholar]
- Swedlow JR, Lewis SE, Goldberg IG. Modelling data across labs, genomes, space and time. Nat Cell Biol [Internet]. 2006. Nov 1;8(11):1190–4. Available from: 10.1038/ncb1496 [DOI] [PubMed] [Google Scholar]
- Takei Y, Yun J, Zheng S, Ollikainen N, Pierson N, White J, et al. Integrated spatial genomics reveals global architecture of single nuclei. Nature [Internet]. 2021. Feb;590(7845):344–50. Available from: 10.1038/s41586-020-03126-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegunov D, Xue L, Dienemann C, Cramer P, Mahamid J. Multi-particle cryo-EM refinement with M visualizes ribosome-antibiotic complex at 3.5 Å in cells. Nat Methods [Internet]. 2021. Feb;18(2):186–93. Available from: 10.1038/s41592-020-01054-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- TensorFlow Development Team. TensorFlow _ Configures TensorFlow ops to run deterministically (ConfigExperimentalEnable_op_determinism) [Internet]. 2023. [cited 2023 Oct 10]. Available from:https://www.tensorflow.org/versions/r2.8/api_docs/python/tf/config/experimental/enable_op_determinism
- Tideman LEM, Migas LG, Djambazova KV, Patterson NH, Caprioli RM, Spraggins JM, et al. Automated biomarker candidate discovery in imaging mass spectrometry data through spatially localized Shapley additive explanations. Anal Chim Acta [Internet]. 2021. Sep 8;1177:338522. Available from: 10.1016/j.aca.2021.338522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tromans-Coia C, Jamali N, Abbasi HS, Giuliano KA, Hagimoto M, Jan K, et al. Assessing the performance of the Cell Painting assay across different imaging systems. Cytometry A [Internet]. 2023. Oct 3; Available from: 10.1002/cyto.a.24786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- UNESCO. UNESCO Recommendation on Open Science [Internet]. Unesco. 2022. [cited 2023 Oct 9]. Available from: https://www.unesco.org/en/open-science/about?hub=686 [Google Scholar]
- UNESCO, Canadian Commission for UNESCO. An introduction to the UNESCO Recommendation on Open Science. In UNESCO; 2022. [cited 2023 Oct 9]. p. 11. Available from: https://unesdoc.unesco.org/ark:/48223/pf0000383771 [Google Scholar]
- Vandereyken K, Sifrim A, Thienpont B, Voet T. Methods and applications for single-cell and spatial multi-omics. Nat Rev Genet [Internet]. 2023. Aug;24(8):494–515. Available from: 10.1038/s41576-023-00580-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Various Authors. Map of HighEnd CryoEM Worldwide [Internet]. Google Maps. 2023. [cited 2023 Oct 30].Available from: https://www.google.com/mymaps/viewer?mid=1eQ1r8BiDYfaK7D1S9EeFJEgkLggMyoaT&hl=en_US [Google Scholar]
- Viana MP, Chen J, Knijnenburg TA, Vasan R, Yan C, Arakaki JE, et al. Integrated intracellular organization and its variations in human iPS cells. Nature [Internet]. 2023. Jan;613(7943):345–54. Available from: 10.1038/s41586-022-05563-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierdag WMAM., Saka SK. A perspective on FAIR quality control in multiplexed imaging data processing. 2024 Front. Bioinform. Sec. Computational BioImaging, Volume 4 - 2024, doi: 10.3389/fbinf.2024.1336257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkmann N. The joys and perils of flexible fitting. Adv Exp Med Biol [Internet]. 2014;805:137–55. Available from: 10.1007/978-3-319-02970-2_6 [DOI] [PubMed] [Google Scholar]
- Wang S, Su JH, Beliveau BJ, Bintu B, Moffitt JR, Wu CT, et al. Spatial organization of chromatin domains and compartments in single chromosomes. Science [Internet]. 2016;353(6299):598–602. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27445307%5Cnhttp://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC4991974%5Cn 10.1126/science.aaf8084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Eddison M, Fleishman G, Weigert M, Xu S, Wang T, et al. EASI-FISH for thick tissue defines lateral hypothalamus spatio-molecular organization. Cell [Internet]. 2021. Dec;184(26):6361–77.e24. Available from: 10.1016/j.cell.2021.11.024 [DOI] [PubMed] [Google Scholar]
- Wang Y, Woehrstein JB, Donoghue N, Dai M, Avendaño MS, Schackmann RCJ, et al. Rapid Sequential in Situ Multiplexing with DNA Exchange Imaging in Neuronal Cells and Tissues. Nano Lett [Internet]. 2017. Oct 11;17(10):6131–9. Available from: 10.1021/acs.nanolett.7b02716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Chen Z, Shi L, Long R, Anzalone AV, Zhang L, et al. Super-multiplex vibrational imaging. Nature [Internet]. 2017. Apr 27;544(7651):465–70. Available from: 10.1038/nature22051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson MD, Dumontier M, Aalbersberg IJJ, Appleton G, Axton M, Baak A, et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data [Internet]. 2016. Mar 15;3:160018. Available from: 10.1038/sdata.2016.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windhager J, Bodenmiller B, Eling N. An end-to-end workflow for multiplexed image processing and analysis [Internet]. bioRxiv. 2021. Available from: 10.1101/2021.11.12.468357 [DOI] [PubMed] [Google Scholar]
- Wollmann T, Erfle H, Eils R, Rohr K, Gunkel M. Workflows for microscopy image analysis and cellular phenotyping. J Biotechnol [Internet]. 2017. Nov 10;261:70–5. Available from: 10.1016/j.jbiotec.2017.07.019 [DOI] [PubMed] [Google Scholar]
- Wollmann T, Hiltemann S, Kostrykin L. Introduction to image analysis using Galaxy. 2023. Aug 14 [cited 2023 Oct 10]; Available from:https://training.galaxyproject.org/training-material/topics/imaging/tutorials/imaging-introduction/tutorial.html [Google Scholar]
- Wolstencroft K, Owen S, Goble C, Nguyen Q, Krebs O, Muller W. RightField: Semantic enrichment of Systems Biology data using spreadsheets. In: 2012 IEEE 8th International Conference on E-Science [Internet]. IEEE; 2012. Available from: http://ieeexplore.ieee.org/document/6404412/ [Google Scholar]
- Xue L, Lenz S, Zimmermann-Kogadeeva M, Tegunov D, Cramer P, Bork P, et al. Visualizing translation dynamics at atomic detail inside a bacterial cell. Nature [Internet]. 2022. Oct;610(7930):205–11. Available from: 10.1038/s41586-022-05255-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagnik G, Liu Z, Rothschild KJ, Lim MJ. Highly Multiplexed Immunohistochemical MALDI-MS Imaging of Biomarkers in Tissues. J Am Soc Mass Spectrom [Internet]. 2021. Apr 7;32(4):977–88. Available from: 10.1021/jasms.0c00473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaniv Z, Anidi I, Arakkal L, Arroyo-Mejias A, Beuschel RT, Börner K, et al. Iterative Bleaching Extends Multiplexity (IBEX) Knowledge-Base [Internet]. 2023. Available from: https://zenodo.org/record/7693279 [Google Scholar]
- Yip KM, Fischer N, Paknia E, Chari A, Stark H. Atomic-resolution protein structure determination by cryo-EM. Nature [Internet]. 2020. Nov;587(7832):157–61. Available from: 10.1038/s41586-020-2833-4 [DOI] [PubMed] [Google Scholar]
- Zhang T, Liu G, Zhao H, Braz GT, Jiang J. Chorus2: design of genome-scale oligonucleotide-based probes for fluorescence in situ hybridization. Plant Biotechnol J [Internet]. 2021. Oct;19(10):1967–78. Available from: 10.1111/pbi.13610 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.