Abstract
Corn cob is a major waste mass-produced in corn agriculture. Corn cob hydrolysate containing xylose, arabinose, and glucose is the hydrolysis product of corn cob. Herein, a recombinant Escherichia coli strain BT-10 was constructed to transform corn cob hydrolysate into 1,2,4-butanetriol, a platform substance with diversified applications. To eliminate catabolite repression and enhance NADPH supply for alcohol dehydrogenase YqhD catalyzed 1,2,4-butanetriol generation, ptsG encoding glucose transporter EIICBGlc and pgi encoding phosphoglucose isomerase were deleted. With four heterologous enzymes including xylose dehydrogenase, xylonolactonase, xylonate dehydratase, α-ketoacid decarboxylase and endogenous YqhD, E. coli BT-10 can produce 36.63 g/L 1,2,4-butanetriol with a productivity of 1.14 g/[L·h] using xylose as substrate. When corn cob hydrolysate was used as the substrate, 43.4 g/L 1,2,4-butanetriol was generated with a productivity of 1.09 g/[L·h] and a yield of 0.9 mol/mol. With its desirable characteristics, E. coli BT-10 is a promising strain for commercial 1,2,4-butanetriol production.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12934-024-02317-0.
Keywords: Corn cob hydrolysate; 1,2,4-butanetriol; Xylose; Metabolic engineering; Escherichia coli
Introduction
Corn is one of the most important sources of starch in the world. Corn cob is inevitably and massively generated as a by-product of the corn agriculture. Nowadays, corn cob is either burnt as fuel or treated as a waste causing environmental pollution [1,2]. Corn cob can be hydrolyzed into corn cob hydrolysate (CCH) with xylose, arabinose and glucose [1,2]. Numerous efforts have been made to obtain efficient routes for resource utilization of CCH [3–5]. Many value-added chemicals including xylitol, [6] xylonate, [7] and ethanol [8] can be produced from CCH by metabolic engineered microorganisms.
1,2,4-Butanetriol (1,2,4-BT) is a platform chemical with versatile applications [9]. The most important application of 1,2,4-BT is to produce 1,2,4-butanetriol trinitrate (BTTN). BTTN has the advantages of low impact sensitivity, high energy level, high thermal stability, no brittleness at low temperature and high reaction safety, and thus is an excellent substitute for nitroglycerin [10]. In addition, 1,2,4-BT can also be used as the precursor for production of plastic monomers or medicines [11,12]. Currently, 1,2,4-BT is commercially produced through malate reduction with NaBH4 under high temperature and high pressure [13]. Harsh reaction conditions, serious pollutions, and difficult downstream purification process restricted the application of this chemical method [13]. Therefore, microbial 1,2,4-BT synthesis from renewable sources has attracted considerable attentions [14–16].
Researchers have designed different unnatural pathways for 1,2,4-BT biosynthesis from xylose, glucose, arabinose or malate [14–16]. Niu et al. established the first fermentation process for 1,2,4-BT synthesis using two-step fermentation [17]. Xylose or arabinose, the two major pentoses in CCH, are transformed into 1,2,4-BT through a five-enzyme process [17]. The five successive enzymatic reactions can also be introduced into a recombinant strain to realize 1,2,4-BT production [18–22]. Jing et al. blocked the branching pathway of xylose metabolism, fine-tuned the expression of xylose isomerase in Escherichia coli, screened an efficient 2-keto-3-deoxyxylonate (2-KDX) decarboxylase KivD, and produced 10.03 g/L 1,2,4-BT from xylose in Luria–Bertani medium [19]. Recently, the group of Sutiono reported the enzymatic synthesis of 1,2,4-BT using xylose as the substrate. 1,2,4-BT production with titers beyond 100 g/L can be acquired under the most suitable conditions [23]. CCH is an ideal substrate for fermentative production of 1,2,4-BT. However, glucose in CCH may induce carbon catabolite repression and inhibit the transport and biotransformation of xylose and arabinose [24]. In addition, glucose catabolism through Embden-Meyerhof-Parnas (EMP) pathway mainly generates NADH but reduction of 3,4-dihydroxybutanal to produce 1,2,4-BT catalyzed by alcohol dehydrogenase YqhD requires NADPH as the cofactor [25]. Herein, E. coli W3110 (DE3) was systematically metabolic engineered for generation of 1,2,4-BT from CCH. Briefly, the key genes for production of 1,2,4-BT were overexpressed while the competing pathways related to xylose catabolism were blocked. The ptsG encoding the glucose-specific transporter EIICBGlc was knocked out to eliminate carbon catabolite repression and enhance xylose utilization. The pgi encoding glucose-6-phosphate isomerase PGI in EMP pathway was deleted to strengthen pentose phosphate pathway and NADPH supply (Fig. 1). Finally, the production of 1,2,4-BT with high concentration, yield and productivity was achieved by the constructed strain E. coli BT-10 with CCH as substrate.
Fig. 1.
Engineering strategies for 1,2,4-BT production from CCH by recombinant E. coli. Red crosses meant that these genes were deleted. Blue typefaces meant that these genes (xylB, xylC and xylD from Caulobacter crescentus and kdcA from Lactococcus lactis) were overexpressed. xylB, xylose dehydrogenase coding gene. xylC, xylonolactonase coding gene. xylD, xylonate dehydratase coding gene. kdcA, α-ketoacid decarboxylase coding gene. xylA, xylose isomerase coding gene. yjhG/yagF, xylonate dehydratase coding gene. yjhH/yagE, 2-keto-3-deoxy-xylonate aldolase coding gene. yqhD, NADPH-dependent alcohol dehydrogenase coding gene. ptsG, glucose-specific transporter EIICBGlc coding gene. glK, glucose kinase coding gene. pgi, glucose-6-phosphate isomerase coding gene. zwf, glucose-6-phosphate dehydrogenase coding gene. pfk, 6-phosphofructokinase coding gene. gnd, 6-phosphogluconate dehydrogenase coding gene. EMP, Embden-Meyerhof-Parnas pathway. HMP, Hexose Monophosphate pathway. xynR, regulator of xylonate catabolism coding gene. XylE, xylose transporter. AraFGH, arabinose ABC transporter. XylFGH, xylose ABC transporter. GalP, galactose H+ symporter. PTS, phosphotransferase system
Methods
Chemicals
Xylose (99%) and lactose were purchased from Shandong Xitang Biotech Co., Ltd. (Jinan, China). 1,2,4-BT was bought from J&K Scientific Ltd (Beijing, China). Xylonate, 2-keto-3-deoxyxylonate (2-KDX), and 3,4-dihydroxybutyrate (3,4-DHBA) were purchased from Sigma-Aldrich (Louis, Missouri, USA). Restriction enzymes were purchased from Thermo Fisher (USA). Polymerase chain reaction (PCR) primers were provided by Beijing Tsingke Biotech Co., Ltd (Qingdao, China). T4 DNA ligase and FastPfu DNA polymerase were purchased from Thermo Fisher (USA) and TransGen Biotech (China), respectively. Whey powder containing 77.0% lactose, 11.2% protein, 1.1% fat, 1.9% moisture, and 8.2% ash was purchased from Kuoquan Biotech (Jinan, China). Detoxified CCH containing 118.5 g/L xylose, 11.8 g/L arabinose, 11.5 g/L glucose, 1.4 g/L formate, 0.34 g/L acetate, 0.83 g/L ethanol and 13.5 mg/L furfural was a kindly gift from Shandong Futase Co., Ltd (Dingtao, China). Other chemicals were analytical grade and commercially available.
Bacterial strains, plasmids and growth conditions
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli strains were generally cultivated in Luria–Bertani (LB) medium (10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl) at 180 rpm and 37 °C. The pTKRED and pCP20 were used for the gene knock-out or knock-in of E. coli W3110 (DE3). Kanamycin, chloramphenicol and spectinomycin were added at a concentration of 50, 40, and 50 μg/mL when necessary.
Table 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Relevant characteristicsa | Origin |
|---|---|---|
| Strain | ||
| E. coli DH5α | F– φ80lacZ∆M15 ∆(lacZYA-argF) U169 deoR recA1 endA1 hsdR17(rK–, mK+) phoA supE44 λ– thi-1 gyrA96 relA1 | Novagen |
| E. coli 0K | E. coli W3110 (DE3) | [27] |
| E. coli BT-1 | E. coli 0K containing plasmid pETPtac-xylBC | This study |
| E. coli 1K | E. coli W3110 (DE3)ΔxylA | [27] |
| E. coli BT-2 | E. coli 1K containing plasmid pETPtac-xylBC | This study |
| E. coli 3K | E. coli 1KΔyjhHΔyagE | [27] |
| E. coli BT-3 | E. coli 3K containing plasmid pETPtac-xylBC | This study |
| E. coli 4K | E. coli 3KΔxynR | [27] |
| E. coli BT-4 | E. coli 4K containing plasmid pETPtac-xylBC | This study |
| E. coli 4KI | E. coli W3110 (DE3)ΔxylAΔyjhHΔyagEΔxynR with PT7-xylD-kdcA expression cassette knocking in the position of xynR in chromosome | [27] |
| E. coli BT-5 | E. coli 4KI containing plasmid pETPtac-xylBC | This study |
| E. coli 5KI | E. coli 4KIΔptsG | This study |
| E. coli BT-6 | E. coli 5KI containing plasmid pETPtac-xylBC | This study |
| E. coli BT-7 | E. coli 5KI with PT7-xylBC knocking in the position of ptsG in chromosome | This study |
| E. coli BT-8 | E. coli 5KI with PT7-xylBC knocking in the position of mgsA in chromosome | This study |
| E. coli BT-9 | E. coli 5KI with xylBC knocking in the position of xylA in chromosome | This study |
| E. coli BT-10 | E. coli BT-9Δpgi | This study |
| Plasmid | ||
| pETPtac-xylBC | Expression vector, pETPtac with gene xylBC | [7] |
| pTKRED | Plasmid expressing Red recombinase genes; Sper | Addgene |
| pCP20 | Plasmid expressing Flp recombinase to remove kan cassette; Cmr | CGSCb |
| pKD4 | Template for kanamycin resistance cassette; Kanr | CGSCb |
aCmr chloramphenicol resistant, Kmr kanamycin resistant, Sper spectinomycin resistant
bCGSC Coli Genetic Stock Center, Yale university plasmid gene preservation center
DNA manipulation in E. coli W3110 (DE3)
The primers used are listed in Table S1. Vector isolation, restriction enzyme digestion, agarose gel electrophoresis, and other DNA manipulations were carried out using standard protocols as in our previous study [7]. Plasmid was transformed into chemically competent cells by heat shock, and transformants were isolated by plating on antibiotic LB-agar plates. The genes xylB, xylC, xylD and kdcA were synthesized by Tongyong Biosystem Co., Ltd. (Chuzhou, Anhui, China) in our previous study [7,27]. The Red recombination technology was applied for knockout of genes pgi and ptsG, and knock-in of genes xylBC from Caulobacter crescentus in E. coli W3110 (DE3) [26]. The fragment ΔptsG used for knockout of ptsG was obtained by directly amplify the fragment with primers ΔptsG-F1/ΔptsG-R1 from E. coli K12ΔptsG::kan from Coli Genetic Stock Center. The fragment ΔxylA::xylBC used for knock-in of genes xylBC at the position of gene xylA was obtained as follows. Primers ΔxylA::xylBC-F1/ΔxylA::xylBC-R1, ΔxylA::xylBC-F2/ΔxylA::xylBC-R2, ΔxylA::xylBC-F3/ΔxylA::xylBC-R3, and ΔxylA::xylBC-F4/ΔxylA::xylBC-R4 (Additional file 1: Table S1) were used to clone up homologous arm of xylA, the genes of xylBC, kanamycin resistance gene, and down homologous arm of xylA. These four fragments were recombined to form fragment ΔxylA::xylBC. The fragments Δpgi for knockout of pgi, ΔptsG::PT7-xylBC and ΔmgsA::PT7-xylBC for knock-in of genes xylBC at the position of genes ptsG and mgsA were obtained through similar process. The fragments for knockout or knock-in of different genes were transformed into E. coli cells containing pTKRED plasmid by electrotransformation. Positive transformants were selected by relevant antibiotics and confirmed by PCR and subsequent DNA sequencing. Then, the plasmid pCP20 was transformed to eliminate the kanamycin resistance gene kan from chromosome. The two temperature-sensitive plasmids, pTKRED and pCP20, were removed by culture at 42 °C overnight.
Batch fermentations and fed-batch fermentation
Batch fermentation was carried out in 300 mL shake flasks containing 50 mL of LB medium with 10 g/L xylose and 2 g/L lactose at 180 rpm and 37 °C for 24 h. Xylose at the concentration of 10 g/L was added in the medium for fed-batch fermentation in shake flask when necessary. Fed-batch fermentation was also conducted in 1.0-L bioreactor (Infors AG, Bottmingen, Switzerland) with an operating volume of 0.8 L or 7.5-L (B. Braun Biotech International GmbH, Germany) bioreactor with an operating volume of 5 L, respectively. The seed was inoculated directly into a 35 mL test tube containing 5 mL of the LB medium and then was cultivated at 180 rpm in a rotary shaker at 37 °C overnight. The seed culture was prepared in a 500 mL shake-flask containing 100 mL LB medium, and then seed culture was inoculated (10%, v/v) into the fermentation medium. The LB fermentation medium contained 10 g/L lactose, 10 g/L glucose and 30 g/L xylose. Alternatively, the detoxified CCH (25%, v/v) and whey powder (12.99 g/L) were added into the broth to make the xylose concentration at about 30 g/L and lactose concentration at about 10 g/L. Fermentation was performed at 30 °C with an aeration rate of 1.5 vvm and an agitation of 400 rpm. Samples were withdrawn periodically to determine the cell density, concentrations of glucose, xylose, arabinose, lactose, 1,2,4-BT and by-products. When xylose concentration was lower than 10 g/L, xylose or CCH was added to make xylose concentration to 30 g/L. The pH was maintained at 7.0 by automatic addition of 10 M NaOH.
Analytical methods
Cell density was determined by monitoring the absorbance at 600 nm using a visible spectrophotometer (V-5100H, METASH, China). The glucose concentration was enzymatically measured using a bioanalyzer (SBA-40D, Shandong Academy of Sciences, China) after appropriate dilution. The concentrations of xylose, arabinose, xylonate, 2-KDX, 3,4-DHBA, 3,4-DHB, 1,2,4-BT and lactose were analyzed using HPLC system (LC-20AT, Shimadzu, Japan) equipped with Aminex HPX-87H column (300 × 7.8 mm, Bio-Rad, USA) linked with an Aminex fast acid analysis column (100 × 7.8 mm, BioRad, USA). The samples withdrawn periodically were centrifuged at 13,000 g for 10 min and the supernatants were filtered using a 0.22 μm Millipore filter (Millipore Corp, Bedford, MA, USA) before HPLC analysis. The 0.1% formic acid was used as the mobile phase at a flow rate of 0.4 mL/min, and the column temperature was 30 °C [27].
Results and discussion
Construction of 1,2,4-butanetriol biosynthesis pathway in E. coli
Biosynthesis of 1,2,4-BT from xylose and arabinose involves five identical enzymic steps including dehydrogenation, hydrolysis, dehydration, decarboxylation and reduction (Fig. 1). In this study, the synthesis pathway for production of 1,2,4-BT from xylose, the most abundant carbohydrate in corn cob, was firstly constructed in E. coli W3110 (DE3) (referred to herein as 0K). E. coli 1K, 3K, 4K and 4KI were four derivative strains of E. coli 0K constructed previously [27]. The xylA encoding xylose isomerase was deleted in E. coli 1K while the yjhH and yagE encoding 2-KDX aldolases were further deleted in E. coli 3K. To enhance the utilization of xylonate, the xynR encoding the regulator of xylonate catabolism was deleted in strain 3K to obtain E. coli 4K. A gene cassette for expressing of xylonate dehydratase XylD (YP_002516235.1) of C. crescentus and α-ketoacid decarboxylase KdcA (WP_095586306.1) of Lactococcus lactis was knocked in E. coli 4K at position of xynR, resulting in E. coli 4KI [27]. Xylose dehydrogenase XylB (YP_002516237.1) and xylonolactonase XylC (YP_002516236.1) from C. crescentus can efficiently catalyze xylose into xylonate. The plasmid pETPtac-xylBC with xylB and xylC genes from C. crescentus was previously constructed and introduced into E. coli for efficient production of xylonate from xylose [7]. In this study, the plasmid pETPtac-xylBC was transformed into E. coli 0K, 1K, 3K, 4K and 4KI, resulting in strains E. coli BT-1, BT-2, BT-3, BT-4 and BT-5, respectively. Expression of xylB and xylC in pETPtac-xylBC is under control of Ptac and lactose inducible. These recombinant E. coli strains were cultivated in LB medium containing 10 g/L xylose and 2 g/L lactose at 37 ℃ and 180 rpm for 24 h. As shown in Fig. 2, E. coli BT-1, BT-2, BT-3, BT-4 and BT-5 with xylB and xylC exhibited obviously xylose utilization and xylonate production. Endogenous alcohol dehydrogenase YqhD in E. coli could reduce 3,4-DHB to 1,2,4-BT [28]. However, only the strain E. coli BT-5, in which the genes xylD and kdcA responsible for 3,4-DHB production from xylonate were knocked in, has the ability for 3,4-DHB production from xylonate. Thus, no accumulation of 1,2,4-BT was observed during culture of E. coli BT-1, BT-2, BT-3, and BT-4. 1,2,4-BT at a concentration of 2.06 g/L and a yield of 0.29 mol/mol xylose was accumulated within 24 h by E. coli BT-5 (Fig. 2).
Fig. 2.
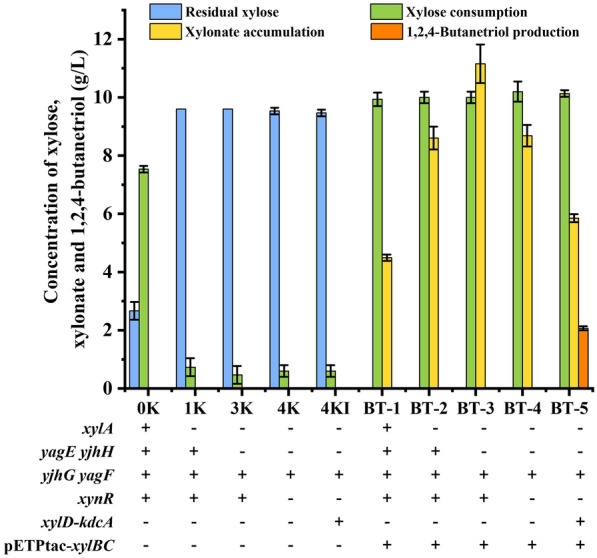
Performance of different recombinant E. coli strains in 1,2,4-BT production from xylose. The experiment was carried out in the 300 mL shake flask containing 50 mL LB broth with 10 g/L xylose and 2 g/L lactose at 37 °C and 180 rpm for 24 h. Values are the average ± SD (n = 3 independent experiments)
Knockout of ptsG to increase 1,2,4-butanetriol production
Besides xylose and arabinose, glucose which can support E. coli growth is also present in CCH. Thus, E. coli BT-5 was cultured in LB with 5 g/L glucose and 10 g/L xylose where glucose was utilized to support E. coli BT-5 growth and xylose was used for 1,2,4-BT synthesis. When xylose concentration was lower than 5 g/L, 10 g/L xylose was added in the broth. As shown in Fig. 3A, 2.62 g/L 1,2,4-BT was obtained from 18.67 g/L xylose by E. coli BT-5 with a yield of 0.20 mol/mol xylose.
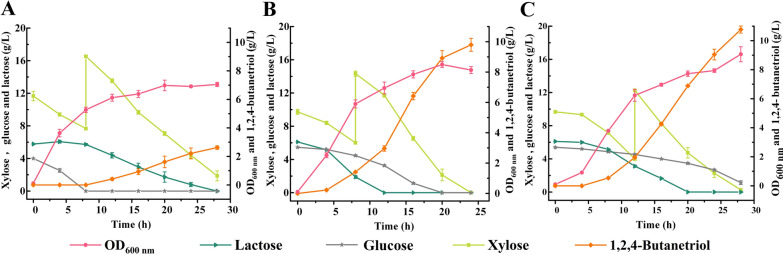
Fig. 3.
Effect of gene ptsG deletion on production of 1,2,4-BT from xylose. A Time-course of fed-batch fermentation in the 300 mL shake flask by E. coli BT-5 at 180 rpm and 37 °C. B Time-course of fed-batch fermentation in the 300 mL shake flask by E. coli BT-6 at 180 rpm and 37 °C. C Time-course of fed-batch fermentation in 300 mL shake flask by E. coli BT-6 at 180 rpm and 30 °C. Xylose at the concentration of 10 g/L was added in the medium when necessary. Values are the average ± SD (n = 3 independent experiments)
E. coli preferentially utilizes glucose in medium containing glucose and other utilizable sugars like xylose and lactose [24]. Knocked out the ptsG encoding the glucose-specific transporter EIICBGlc can eliminate carbon catabolite repression of glucose. Thus, E. coli 5KI was constructed by deleting ptsG in E. coli 4KI and then the plasmid pETPtac-xylBC was transformed, resulting in strain E. coli BT-6. Glucose can also be transported by galactose symporter after deletion of ptsG gene but the glucose utilization rate of the constructed strain E. coli BT-6 decreased due to the knocked out of ptsG (Fig. 3B). The major byproduct of E. coli BT-5 was xylonate (12.30 g/L) (Additional file 1: Figure S1), which can be transformed by lactose induced XylD and KdcA into 3,4-DHB and then reduced to 1,2,4-BT. Expression of xylD and kdcA is under control of PT7 in E. coli BT5 and BT-6 and lactose inducible. Besides xylose utilization, the catabolism of lactose in E. coli is also repressed by glucose due to carbon catabolite repression [7]. Delayed response to induction often occurs during fermentation using lactose as inducer and glucose as carbon source. Although the consumption of xylose was not improved obviously, the inactivation of ptsG could enhance the utilization of lactose by E. coli BT-6 (Fig. 3B) and thus may also increase the expression of xylD and kdcA. As shown in Fig. 3B, 9.80 g/L 1,2,4-BT was obtained by E. coli BT-6 with a yield of 0.77 mol/mol xylose and accumulation of xylonate decreased to 4.14 g/L (Additional file 1: Figure S1). The enhanced production of 1,2,4-BT by E. coli BT-6 may due to the higher expression of xylD and kdcA by higher lactose utilization. The α-ketoacid decarboxylase KdcA used in this study was from L. lactis, whose optimum growth temperature is 30 ℃. The fermentation temperature for 1,2,4-BT production was tentatively adjusted to 30 ℃ and a slightly higher 1,2,4-BT concentration of 10.76 g/L was obtained with a yield of 0.86 mol/mol xylose from 17.67 g/L xylose (Fig. 3C). Thus, the 1,2,4-BT fermentation was conducted at 30 °C in subsequent experiments.
Integration of xylonate synthesis genes into E. coli genome to increase 1,2,4-butanetriol production
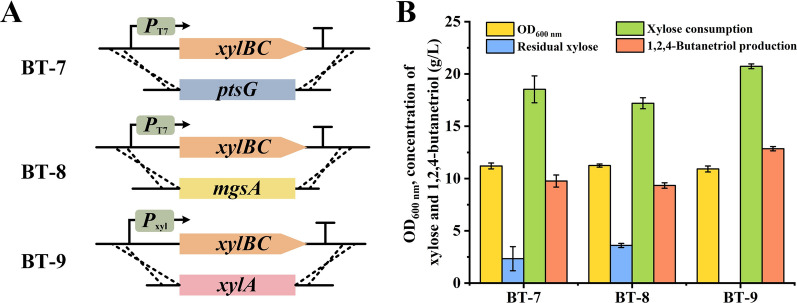
Expression of exogenous gene based on plasmid may increase the metabolic burden of recombinant E. coli [29]. Expression of xylB and xylC based on plasmid pETPtac-xylBC may increase the metabolic burden of recombinant E. coli. As shown in Fig. 3B, the relief of carbon catabolite repression through deletion of ptsG enhanced lactose utilization and increased the production of 1,2,4-BT. It was reported that deletion of mgsA, the methylglyoxal synthase coding gene, can also effectively weaken the carbon catabolite repression [30]. Thus, xylB and xylC under the control of PT7 were knocked into the position of ptsG and mgsA in E. coli 5KI genome for simultaneously relieving catabolite repression and expression of key enzymes for 1,2,4-BT production, resulting in E. coli BT-7 and E. coli BT-8, respectively. The endogenous xylose isomerase coding gene xylA is under the control of Pxyl and can be induced by xylose in E. coli [31]. The xylB and xylC were also directly knocked into the position of xylA and under the control of Pxyl in E. coli 5KI, resulting in strain E. coli BT-9 (Fig. 4A). As shown in Fig. 4B, the strain E. coli BT-9 exhibited the best performance in 1,2,4-BT production. 1,2,4-BT at a concentration of 12.85 g/L was obtained from 20.7 g/L xylose with a yield of 0.88 mol/mol xylose in 24 h (Additional file 1: Figure S2).
Fig. 4.
Selection of integration site of xylBC to increase 1,2,4-BT production. A Scheme of the integration mode of xylBC in E. coli BT-7, E. coli BT-8, and E. coli BT-9. B Performance of E. coli BT-7, E. coli BT-8, and E. coli BT-9 in transforming xylose into 1,2,4-BT. The fed-batch fermentation was carried out in 300 mL shake flask at 180 rpm and 30 °C. When xylose concentration was lower than 5 g/L, 10 g/L xylose was added. Values are the average ± SD (n = 3 independent experiments)
Fed-batch fermentation of E. coli BT-9 to produce 1,2,4-butanetriol in 1-L bioreactor
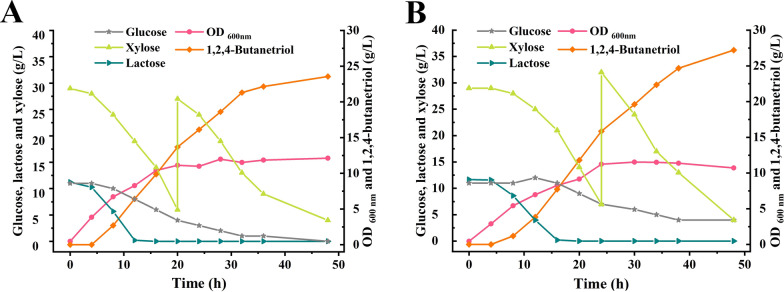
Then, fed-batch fermentation was carried out to produce 1,2,4-BT with E. coli BT-9. E. coli BT-9 was cultured in a 1-L bioreactor with 30 g/L xylose, 10 g/L lactose and 10 g/L glucose. Solid xylose was added under non-sterile condition when xylose concentration was lower than 10 g/L. After 48 h fed-batch fermentation, 1,2,4-BT at a concentration of 23.55 g/L was obtained from 46 g/L xylose with a yield of 0.72 mol/mol xylose. (Fig. 5A).
Fig. 5.
Effect of pgi deletion on 1,2,4-BT production from xylose. A Time-course of fed-batch fermentation by E. coli BT-9 in 1-L bioreactor. B Time-course of fed-batch fermentation by E. coli BT-10 in 1-L bioreactor. The fed-batch fermentation was conducted in 1-L bioreactor containing 0.8 L LB broth with 30 g/L xylose, 10 g/L glucose and 10 g/L lactose at 400 rpm, 1.5 vvm and 30 °C. Xylose concentration was adjusted to 30 g/L when lower than 10 g/L. The experiments were conducted in triplicate. Two representative time-courses of E. coli BT-9 A and E. coli BT-10 (B) are reported herein
Knockout of pgi to strengthen NADPH supply and 1,2,4-butanetriol production
XylB catalyzes xylose into xylonolactone and generate NADH. However, YqhD required for 1,2,4-BT production is NADPH-dependent. Glucose can enter into EMP pathway to produce NADH or enter into pentose phosphate pathway to produce NADPH. Thus, the pgi encoding glucose phosphate isomerase was deleted in E. coli BT-9 to block the EMP pathway and enhance glucose metabolism via the HMP pathway to provide more NADPH required for 1,2,4-BT synthesis. The obtained E. coli strain BT-10 was used for 1,2,4-BT production in 1-L bioreactor. As shown in Fig. 5B, the 1,2,4-BT production of E. coli BT-10 increased to 27.2 g/L with a yield of 0.77 mol/mol xylose. Compared with E. coli BT-9, the accumulation of 3,4-DHB decreased 14.58% in E. coli strain BT-10 (Additional file 1: Figure S3). Then, the fed-batch fermentation of 1,2,4-BT by E. coli BT-10 was conducted in a 7.5-L bioreactor. Both the growth of E. coli BT-10 and the xylose consumption rate increased in the 7.5-L bioreactor, which may due to the improved dissolved oxygen during the enlargement of fermentation volume. As shown in Fig. 6A, 36.63 g/L of 1,2,4-BT was produced from 79 g/L xylose with a productivity of 1.14 g/[L·h] and a yield of 0.66 mol/mol xylose.
Fig. 6.
1,2,4-BT production through fed-batch fermentation with E. coli BT-10 using xylose or CCH as the substrate. A Time-course of fed-batch fermentation by E. coli BT-10 in 7.5-L bioreactor with xylose as the substrate. The experiment was conducted in 7.5-L bioreactor with 5 L LB broth containing 30 g/L xylose, 10 g/L glucose and 10 g/L lactose at 400 rpm, 1.5 vvm and 30 °C. Xylose concentration was adjusted to 30 g/L when lower than 10 g/L. The experiment was conducted in triplicate. A representative time-course is reported herein. B Time-course of fed-batch fermentation by E. coli BT-10 in 7.5-L bioreactor with CCH as the substrate. The experiment was conducted in 7.5-L bioreactor containing 5 L LB broth with CCH (the final concentration of xylose was adjusted to 30 g/L) and whey powder (the final concentration of lactose was adjusted to 10 g/L) at 400 rpm, 1.5 vvm and 30 °C. Xylose concentration was adjusted to 30 g/L when lower than 10 g/L. The experiment was conducted in triplicate. A representative time-course is reported herein
Fed-batch fermentation of 1,2,4-BT with detoxified CCH as the substrate
Finally, CCH was used as the substrate for fed-batch fermentation of 1,2,4-BT by E. coli BT-10 in a 7.5-L bioreactor. To further reduce the cost for 1,2,4-BT production, lactose was replaced by whey powder (lactose concentration was adjusted to 10 g/L) as the inducer of the genes for 1,2,4-BT production. The protein and fat in whey powder can also be utilized by E. coli BT-10 for growth. The biomass of E. coli BT-10 increased in the fermentation system with whey powder (Fig. 6A and B), which may improve the transform of xylose and arabinose in CCH for 1,2,4-BT production. As shown in Fig. 6B, E. coli BT-10 consumed 62 g/L xylose, 6.16 g/L arabinose and 6.4 g/L glucose in CCH within 40 h. 1,2,4-BT at a concentration of 43.4 g/L was obtained with a productivity of 1.09 g/[L·h]. The yield of 1,2,4-BT from xylose and arabinose was 90% of the theoretical value.
Many biotechnological routes have been developed for the fermentative production of 1,2,4-BT. The synthesis of 1,2,4-BT from xylose or arabinose requires relatively few steps and results in low carbon loss, and thus has been intensively investigated in recent years [16–22,28,32–36]. A series of metabolic strategies such as screening enzymes with high activities, blocking the branch pathways, and enhancing the expression of the key enzymes, have been applied to improve 1,2,4-BT production from xylose or arabinose. CCH containing xylose, arabinose and glucose is an ideal substrate for 1,2,4-BT production. In this work, the encoding genes of XylB, XylC, XylD and KdcA were integrated into the genome of E. coli W3110 (DE3). Besides xylose, xylonolactone and xylonate, XylB, XylC and XylD from C. crescentus are also active on arabinose, arabinolactone and arabinonate, respectively [37]. KdcA from L. lactis catalyzes the decarboxylation of both 2-keto-3-deoxy-xylonate and 2-keto-3-deoxy-arabinonate [38]. Although the catabolic genes for arabinose utilization were not deleted in E. coli BT-10, these four heterologous enzymes and endogenous alcohol dehydrogenase YqhD may efficiently redirect the metabolic flux of arabinose from central metabolism to 1,2,4-BT production (Fig. 1). To eliminate catabolite repression and enhance the supply of NADPH for YqhD catalyzed 1,2,4-BT production with glucose, the ptsG and pgi genes were also deleted in E. coli W3110 (DE3). The final recombinant strain E. coli BT-10 can produce 43.4 g/L 1,2,4-BT from CCH with a productivity of 1.09 g/[L·h]. Compared with other strains constructed for 1,2,4-BT production (Table 2), E. coli BT-10 has significant advantages of high 1,2,4-BT concentration, productivity, yield and efficient utilization of cheap substrate CCH.
Table 2.
Comparison of 1,2,4-butanetriol production by different microorganisms
| Strain | Fermentation method | Substrate | Concentration (g/L) | Yield (mol/mol) | Productivity (g/[L·h]) | Reference |
|---|---|---|---|---|---|---|
| E. coli BL21(DE3)/pWN6.222A | Batch fermentation in 1-L bioreactor | l-Arabinonic acid | 2.4 | 0.35 | NFa | [17] |
| E. coli DH5α/pWN6.186A | Batch fermentation in 1-L bioreactor | d-Xylonic acid | 1.6 | 0.25 | NFa | [17] |
| E. coli EWBT304 | Batch fermentation in 300 mL shake flasks | Xylose | 0.88 | 0.13 | 0.02 | [18] |
| E. coli as3KXW004 | Batch fermentation in 50 mL LB medium | Xylose | 10.03 | 0.47 | 0.21 | [19] |
| Saccharomyces cerevisiae BDδK6035 | Fed-batch fermentation in 1.0-L bioreactors | Glucose and xylose | 6.60 | 0.57 | 0.05 | [20] |
| S. cerevisiae BDδD-2tkdcA-ΔBOL2-tTYW1 | Batch fermentation in 200 mL-baffled erlenmeyer flask | Glucose and xylose | 1.70 | 0.25 | 0.02 | [21] |
| E. coli R1 | Fed-batch fermentation in 50 mL fermentation medium | Glucose and xylose | 14.4 | NFa | NFa | [28] |
| E. coli BL21-14 | Batch fermentation in 500-mL shake flasks | Xylose | 5.1 | 0.18 | 0.07 | [32] |
| E. coli MJ134k-1 | Batch fermentation in 50 mL medium of 250 mL shake flasks | Glucose and xylose | 3.96 | 0.24 | 0.11 | [33] |
| E. coli BL21ΔxylAB/pE-mdlCxylBC&pA-adhPyjhG | Fed-batch fermentation in 5 L-scale bioreactor | Glycerol and xylose | 3.92 | 0.28 | 0.07 | [34] |
| E. coli (pEtac-mdlC-tac-xylB) | Batch fermentation in 20 mL LB medium | Xylose | 0.90 | 0.04 | 0.02 | [35] |
| E. coli W031 | Batch fermentation in 5-L bioreactor | Glucose and xylose | 3.9 | 0.3 | NFa | [36] |
| E. coli BT-10 | Fed-batch fermentation in 7.5-L bioreactor | Glucose and xylose | 36.63 | 0.66 | 1.14 | This study |
| E. coli BT-10 | Fed-batch fermentation in 7.5-L bioreactor | Corn cob hydrolysate | 43.40 | 0.90 | 1.09 | This study |
aNF not found
Conclusion
In summary, a systematically metabolic engineered strain E. coli BT-10 was constructed to produce 1,2,4-BT. 1,2,4-BT at a concentration of 36.63 g/L and a productivity 1.14 g/[L·h] was produced by E. coli BT-10 with xylose as the substrate. E. coli BT-10 can also use glucose in CCH for growth and transform xylose and arabinose into 1,2,4-BT. 1,2,4-BT of 43.4 g/L was produced from CCH with a productivity of 1.09 g/[L·h]. The process presented here is both a good example for efficient resource utilization of CCH and a promising alternative for industrial 1,2,4-BT production.
Supplementary Information
Additional file1: Table S1. The primers used in this study. Figure S1. Effect of gene ptsG deletion on byproducts generation during 1,2,4-BT production from xylose. Figure S2. Selection of integration site of xylBC to increase 1,2,4-BT production. Figure S3. Effect of pgi deletion on byproducts generation during 1,2,4-BT production from xylose.
Acknowledgements
We thank Xiangmei Ren from Core Facilities for Life and Environmental Sciences (State Key Laboratory of Microbial Technology, Shandong University) for assistance in HPLC analysis and Chengjia Zhang and Nannan Dong from Core Facilities for Life and Environmental Sciences (State Key Laboratory of Microbial Technology, Shandong University) for assistance in microbial fermentation.
Abbreviations
- 1,2,4-BT
1,2,4-Butanetriol
- CCH
Corn cob hydrolysate
- BTTN
1,2,4-Butanetriol trinitrate
- 2-KDX
2-Keto-3-deoxyxylonate
- 3,4-DHB
3,4-Dihydroxybutanal
- 3,4-DHBA
3,4-Dihydroxybutyrate
- EMP
Embden-meyerhof-parnas pathways
- HMP
Hexose monophosphate pathway
Author contributions
PL: Writing original draft, Investigation, Data curation. MW: Formal analysis, Software, Validation. HD: Software, Validation. QD: Software, Funding acquisition. YZ: Resources. XT: Software, Validation. PX: Software, Supervision. CG: Software, Funding acquisition. TJ: Software, Funding acquisition. CL: Software, Funding acquisition. CM: Conceptualization, Supervision, Writing – review, Funding acquisition. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (31970056 and 31970055), the National Key R&D Program of China (2022YFC2106000), Natural Science Foundation of Shandong Provincial (ZR2022QC092), State Key Laboratory of Microbial Technology Open Projects Fund (M2023-02), and State Key Laboratory of Microbial Technology (SKLMTFCP-2023–03).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Chuanjuan Lü, Email: chuanjuanlv@sdu.edu.cn.
Cuiqing Ma, Email: macq@sdu.edu.cn.
References
- 1.Wakudkar H, Jain S. A holistic overview on corn cob biochar: a mini-review. Waste Manag Res. 2022;40(8):1143–1155. doi: 10.1177/0734242X211069741. [DOI] [PubMed] [Google Scholar]
- 2.Islam F, Imran A, Afzaal M, Saeed F, Asghar A, Shahid S, Shams A, Zahra SnM, Biswas S, Aslam MA. Nutritional, functional, and ethno-medical properties of sweet corn cob: a concurrent review. Int J Food Sci Technol. 2023;58(5):2181–2188. doi: 10.1111/ijfs.16338. [DOI] [Google Scholar]
- 3.Kim M, Singhvi MS, Kim BS. Eco-friendly and rapid one-step fermentable sugar production from raw lignocellulosic biomass using enzyme mimicking nanomaterials: A novel cost-effective approach to biofuel production. Chem Eng J. 2023;465:142879. doi: 10.1016/j.cej.2023.142879. [DOI] [Google Scholar]
- 4.Zhou H, Mao Y, Zheng Y, Liu T, Yang Y, Si C, Wang L, Dai L. Complete conversion of xylose-extracted corncob residues to bioplastic in a green and low carbon footprint way. Chem Eng J. 2023;471:144572. doi: 10.1016/j.cej.2023.144572. [DOI] [Google Scholar]
- 5.Li B, Wang P, Feng Y, Xu X, Zhang Y, Zou X. Bio-refinery of xylose processing wastes for green polymalic acid production and l-malic acid recovery by engineered Aureobasidium pullulans in a non-waste-disposal system. Chem Eng J. 2023;454:140533. doi: 10.1016/j.cej.2022.140533. [DOI] [Google Scholar]
- 6.Baptista SL, Cunha JT, Romani A, Domingues L. Xylitol production from lignocellulosic whole slurry corn cob by engineered industrial Saccharomyces cerevisiae PE-2. Bioresour Technol. 2018;267:481–491. doi: 10.1016/j.biortech.2018.07.068. [DOI] [PubMed] [Google Scholar]
- 7.Zhang YP, Guo ST, Wang YX, Liang XP, Xu P, Gao C, Ma CQ. Production of d-xylonate from corn cob hydrolysate by a metabolically engineered Escherichia coli strain. ACS Sustain Chem Eng. 2019;7(2):2160. doi: 10.1021/acssuschemeng.8b04839. [DOI] [Google Scholar]
- 8.Cai D, Dong Z, Wang Y, Chen C, Li P, Qin P, Wang Z, Tan T. Biorefinery of corn cob for microbial lipid and bio-ethanol production: an environmental friendly process. Bioresour Technol. 2016;211:677–684. doi: 10.1016/j.biortech.2016.03.159. [DOI] [PubMed] [Google Scholar]
- 9.Li Y, Yao P, Zhang S, Feng J, Su H, Liu X, Sheng X, Wu Q, Zhu D, Ma Y. Creating a new benzaldehyde lyase for atom-economic synthesis of chiral 1,2,4-butanetriol and 2-aminobutane-1,4-diol from formaldehyde. Chem Catal. 2023;3(1):100467. doi: 10.1016/j.checat.2022.11.006. [DOI] [Google Scholar]
- 10.Zhu W, Yan Q, Pang A, Chi X, Du X, Xiao H. A DFT study of the unimolecular decomposition of 1,2,4-butanetriol trinitrate. J Mol Model. 2014;20(2):2081. doi: 10.1007/s00894-014-2081-3. [DOI] [PubMed] [Google Scholar]
- 11.Tan R, Liu D. Synthesis of cationic lipids from 1,2,4-butanetriol. Tetrahedron Lett. 1999;40(2):209–212. doi: 10.1016/S0040-4039(98)02381-8. [DOI] [Google Scholar]
- 12.Liu M, Liu C. Comparative simulation study of chemical synthesis of energetic (R)-1,2,4-butanetriol trinitrate plasticizer. Int J Quantum Chem. 2017;117(16):e25402. doi: 10.1002/qua.25402. [DOI] [Google Scholar]
- 13.Hanessian S, Ugolini A, Dubé D, Glamyan A. Facile access to (S)-1,2,4-butanetriol and its derivatives. Can J Chem. 2011;62(11):2146–2147. doi: 10.1139/v84-367. [DOI] [Google Scholar]
- 14.Shen X, Xu H, Wang T, Zhang R, Sun X, Yuan Q, Wang J. Rational protein engineering of a ketoacids decarboxylase for efficient production of 1,2,4-butanetriol from arabinose. Biotechnol Biofuels Bioprod. 2023;16(1):172. doi: 10.1186/s13068-023-02414-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X, Cai Z, Li Y, Zhang Y. Design and construction of a non-natural malate to 1,2,4-BT pathway creates possibility to produce 1,2,4-butanetriol from glucose. Sci Rep. 2014;4:5541. doi: 10.1038/srep05541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao Q, Wang X, Hu S, Xu N, Jiang M, Ma C, Yang J, Xu S, Chen K, Ouyang P. High-yield production of d-1,2,4-butanetriol from lignocellulose-derived xylose by using a synthetic enzyme cascade in a cell-free system. J Biotechnol. 2019;292:76–83. doi: 10.1016/j.jbiotec.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 17.Niu W, Molefe MN, Frost JW. Microbial synthesis of the energetic material precursor 1,2,4-BT. J Am Chem Soc. 2003;125(43):12998–12999. doi: 10.1021/ja036391+. [DOI] [PubMed] [Google Scholar]
- 18.Valdehuesa KNG, Liu H, Ramos KRM, Park SJ, Nisola GM, Lee W, Chung W. Direct bioconversion of d-xylose to 1,2,4-butanetriol in an engineered Escherichia coli. Process Biochem. 2014;49(1):25–32. doi: 10.1016/j.procbio.2013.10.002. [DOI] [Google Scholar]
- 19.Jing P, Cao X, Lu X, Zong H, Zhuge B. Modification of an engineered Escherichia coli by a combined strategy of deleting branch pathway, fine-tuning xylose isomerase expression, and substituting decarboxylase to improve 1,2,4-butanetriol production. J Biosci Bioeng. 2018;126(5):547–552. doi: 10.1016/j.jbiosc.2018.05.019. [DOI] [PubMed] [Google Scholar]
- 20.Yukawa T, Bamba T, Guirimand G, Matsuda M, Hasunuma T, Kondo A. Optimization of 1,2,4-butanetriol production from xylose in Saccharomyces cerevisiae by metabolic engineering of NADH/NADPH balance. Biotechnol Bioeng. 2021;118(1):175–185. doi: 10.1002/bit.27560. [DOI] [PubMed] [Google Scholar]
- 21.Bamba T, Yukawa T, Guirimand G, Inokuma K, Sasaki K, Hasunuma T, Kondo A. Production of 1,2,4-butanetriol from xylose by Saccharomyces cerevisiae through Fe metabolic engineering. Metab Eng. 2019;56:17–27. doi: 10.1016/j.ymben.2019.08.012. [DOI] [PubMed] [Google Scholar]
- 22.Hu S, Gao Q, Wang X, Yang J, Xu N, Chen K, Xu S, Ouyang P. Efficient production of d-1,2,4-butanetriol from d-xylose by engineered Escherichia coli whole-cell biocatalysts. Front Chem Sci Eng. 2018;12(4):772–779. doi: 10.1007/s11705-018-1731-x. [DOI] [Google Scholar]
- 23.Sutiono S, Zachos I, Paschalidism L, Pick A, Burger J, Sieber V. Biocatalytic Production of 1,2,4-butanetriol beyond a titer of 100 g/L: Boosting by intermediates. ACS Sustain Chem Eng. 2023;11(17):6592–6599. doi: 10.1021/acssuschemeng.2c07418. [DOI] [Google Scholar]
- 24.Yuan X, Tu S, Lin L, Yang J, Shen H, Wu M. Combination of the CRP mutation and ptsG deletion in Escherichia coli to efficiently synthesize xylitol from corncob hydrolysates. Appl Microbiol Biotechnol. 2020;104(5):2039–2050. doi: 10.1007/s00253-019-10324-0. [DOI] [PubMed] [Google Scholar]
- 25.Jarboe R. YqhD: a broad-substrate range aldehyde reductase with various applications in production of biorenewable fuels and chemicals. Appl Microbiol Biotechnol. 2011;89:249–257. doi: 10.1007/s00253-010-2912-9. [DOI] [PubMed] [Google Scholar]
- 26.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang YP, Liu YD, Zhu JN, Xiao D, Xu P, Ma CQ, Gao C, Lu CJ. Coculture of Gluconobacter oxydans and Escherichia coli for 3,4-dihydroxybutyric acid production from xylose. ACS Sustain Chem Eng. 2021;9(32):10809–10817. doi: 10.1021/acssuschemeng.1c02511. [DOI] [Google Scholar]
- 28.She D, Wang S, Zong H, Lu X, Zhuge B. Genetic modification of Escherichia coli to improve 1,2,4-butanetriol production from cellulose hydrolysate. Syst Microbiol Biomanufactur. 2023;9:1–9. [Google Scholar]
- 29.Ow DS, Nissom PM, Philp R, Oh SK, Yap MG. Global transcriptional analysis of metabolic burden due to plasmid maintenance in Escherichia coli DH5α during batch fermentation. Enzyme Microb Tech. 2006;39:391–398. doi: 10.1016/j.enzmictec.2005.11.048. [DOI] [Google Scholar]
- 30.Yomano LP, York SW, Shanmugam KT, Ingram LO. Deletion of methylglyoxal synthase gene (mgsA) increased sugar co-metabolism in ethanol-producing Escherichia coli. Biotechnol Lett. 2009;31:1389–1398. doi: 10.1007/s10529-009-0011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song SG, Park C. Organization and regulation of the d-xylose operons in Escherichia coli K-12: XylR acts as a transcriptional activator. J Bacteriol. 1997;179(22):7025–7032. doi: 10.1128/jb.179.22.7025-7032.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X, Xu N, Hu S, Yang J, Gao Q, Xu S, Chen K, Ouyang P. d-1,2,4-butanetriol production from renewable biomass with optimization of synthetic pathway in engineered Escherichia coli. Bioresour Technol. 2018;250:406–412. doi: 10.1016/j.biortech.2017.11.062. [DOI] [PubMed] [Google Scholar]
- 33.Ma PF, Meng J, Zhou J, Gao HJ. Biosynthesis of d-1,2,4-butanetriol from d-xylose by recombinant Escherichia coli. CIESC J. 2015;66:2620–2627. [Google Scholar]
- 34.Cao Y, Niu W, Guo J, Xian M, Liu H. Biotechnological production of 1,2,4-butanetriol: an efficient process to synthesize energetic material precursor from renewable biomass. Sci Rep. 2016;5(1):18149. doi: 10.1038/srep18149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun WL, Lu XY, Zong H, Fang HY, Zhu G, Song J. Biosynthesis of d-1,2,4-butanetriol by an engineered Escherichia coli. Microbiol China. 2014;41(10):1948–1954. [Google Scholar]
- 36.Zu JS, Xu SW, Lu XY, Zong H, Zhuge B. Construction of a 1,2,4-butanetriol-integrated strain and fermentation of its co-substrate. Chin J Appl Environ Biol. 2019;25(4):966–971. [Google Scholar]
- 37.Stephens C, Christen B, Fuchs T, Sundaram V, Watanabe K, Jenal U. Genetic analysis of a novel pathway for d-xylose metabolism in Caulobacter crescentus. J Bacteriol. 2007;189(5):2181–2185. doi: 10.1128/JB.01438-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang J, Chen Q, Wang X, Chen K, Ouyang P. The biosynthesis of d-1,2,4-butanetriol from d-arabinose with an engineered Escherichia coli. Front Bioeng Biotechnol. 2022;10:2620–2627. doi: 10.3389/fbioe.2022.844517. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file1: Table S1. The primers used in this study. Figure S1. Effect of gene ptsG deletion on byproducts generation during 1,2,4-BT production from xylose. Figure S2. Selection of integration site of xylBC to increase 1,2,4-BT production. Figure S3. Effect of pgi deletion on byproducts generation during 1,2,4-BT production from xylose.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.