Abstract
Background
Epidemiological nowcasting traditionally relies on count surveillance data. The availability and quality of such count data may vary over time, limiting representation of true infections. Wastewater data correlates with traditional surveillance data and may provide additional value for nowcasting disease trends.
Methods
We obtained SARS-CoV-2 case, death, wastewater, and serosurvey data for Jefferson County, Kentucky (USA), between August 2020 and March 2021, and parameterized an existing nowcasting model using combinations of these data. We assessed the predictive performance and variability at the sewershed level and compared the effects of adding or replacing wastewater data to case and death reports.
Findings
Adding wastewater data minimally improved the predictive performance of nowcasts compared to a model fitted to case and death data (Weighted Interval Score (WIS) 0.208 versus 0.223), and reduced the predictive performance compared to a model fitted to deaths data (WIS 0.517 versus 0.500). Adding wastewater data to deaths data improved the nowcasts agreement to estimates from models using cases and deaths data. These findings were consistent across individual sewersheds as well as for models fit to the aggregated total data of 5 sewersheds. Retrospective reconstructions of epidemiological dynamics created using different combinations of data were in general agreement (coverage > 75%).
Interpretation
These findings show wastewater data may be valuable for infectious disease nowcasting when clinical surveillance data are absent, such as early in a pandemic or in low-resource settings where systematic collection of epidemiologic data is difficult.
Funding
CDC, Louisville-Jefferson County Metro Government, and other funders.
Keywords: epidemiological nowcasting, infectious disease, predictive performance, public health, SARS-CoV-2, wastewater surveillance
Introduction
Epidemiological nowcasting is an important tool for understanding infectious disease trends, and could be used to produce real-time estimates of the transmission rate and reproduction number (Rt) of an infectious pathogen.1 This information is critical for informing risk assessment and the subsequent public health response.
To-date, nowcasting methods rely on surveillance count data (e.g., case reports, hospitalizations, vaccination records) that have limitations for describing disease trends.2 First, asymptomatic and mild infections may not be diagnosed, and so may be missing from clinical surveillance data. Second, variation in healthcare access or sampling methods can mean that available data are not representative of the whole population.3, 4 A third limitation is the lag between the time transmission occurs and when the consequences of transmission become apparent in measured quantities. For instance, a coronavirus disease 2019 (COVID-19) diagnosis typically occurs after symptoms have developed, around five days after infection with severe acute respiratory syndrome coronavirus 2 (SARS-COV-2).5 There may be additional lags between the development of symptoms and the time of diagnosis, and between diagnosis and reporting. Finally, surveillance data are often aggregated over large geographical areas or time periods, which may mask heterogeneities in transmission patterns and prevent prompt geographic targeting of mitigation measures. For example, in the initial stages of the COVID-19 pandemic, states and counties generally reported daily surveillance data by city, county and state. Over time reporting frequencies decreased to weekly or biweekly, reducing the temporal resolution of the data and increasing the lag between infection and reporting.
Wastewater-based epidemiology (WBE) has been used for infectious disease surveillance of for example polio6 and was increasingly relied upon during the COVID-19 pandemic.6, 7 Samples from wastewater provide passively collected information from a population group linked geographically to the sewer network, and if collected at a regular frequency, wastewater data can have a strong temporal and geographical correlation with changes in disease incidence.8, 9 The promise of improved temporal signal in the wastewater data is a major potential benefit given the importance of accurate information on current transmission dynamics in informing prompt action or policy responses. In the United States (USA), several large, publicly accessible wastewater databases have been developed during the SARS-CoV-2 pandemic.10–12
Wastewater data has been used in nowcasts of large metropolitan areas, but there is no definitive agreement on a case count to wastewater concentration quantification.8, 13–15 Nevertheless, WBE for surveillance has its own unique challenges, including variation in sampling and quantification methods, and challenges mapping wastewater concentration to estimates of local disease burden.16–18 As a result, the value of wastewater data for nowcasting disease transmission in subpopulations in addition to, or instead of, traditional surveillance data remains unclear. To assess the added utility of using WBE to count-based nowcasting, we examine SARS-CoV-2 surveillance data from Louisville, KY, which were available for five distinct sewersheds and could be combined for an aggregate countywide view. We used these data and a published nowcasting model to evaluate the utility of wastewater data for improving the accuracy of nowcasts.
Methods
Data
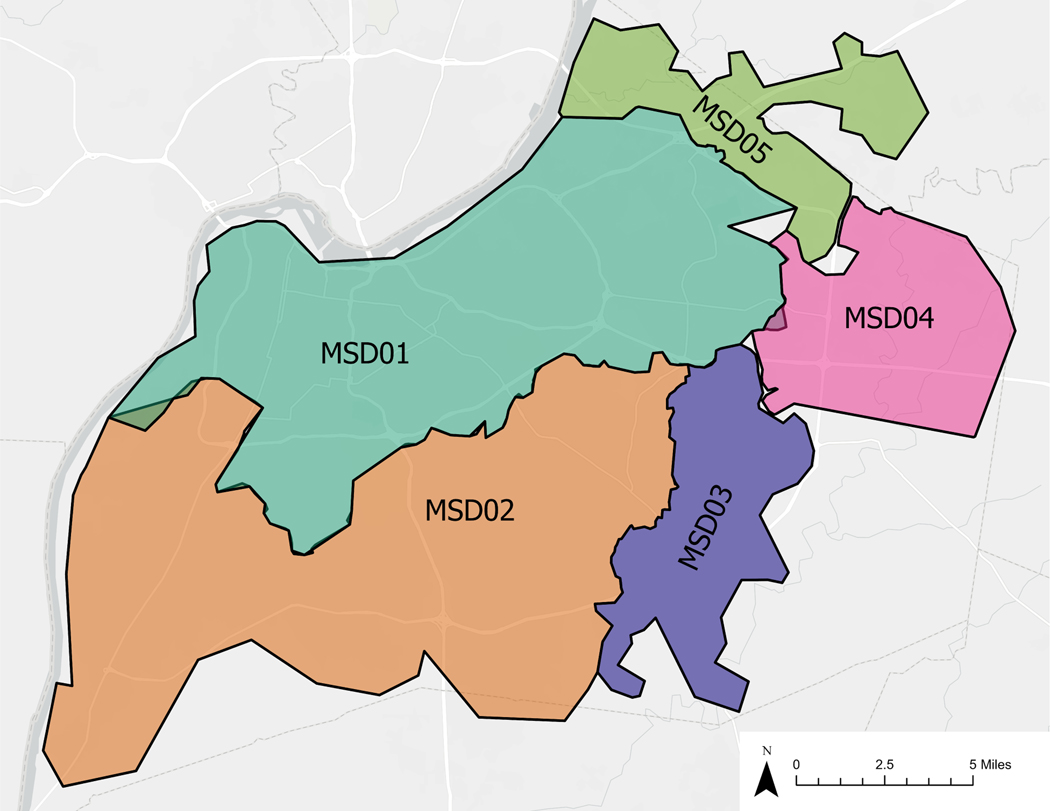
We used temporally and spatially paired data from Louisville/Jefferson County, Kentucky, covering the period between August 2020 and March 2021, prior to public vaccine access, across clinical case and death reports, wastewater concentration data, and four stratified simple random serosurvey waves.8, 19 Data were geocoded to five wastewater treatment plant catchment areas or “sewersheds”, named MSD0[1–5], that cover 97% of the county population (Figure 1 and Table S1).20 Data were additionally aggregated for a total countywide level comparison. Finally, all data sources were aggregated to a weekly level, to match the lowest sampling frequency of the wastewater data.
Figure 1.
Five studied wastewater treatment plant zones (sewersheds), Jefferson County, KY (USA).
Case reports.
Louisville Metro Department of Public Health and Wellness (LMPHW) provided daily positive case reports, that were then geocoded to sewersheds based on reported address or zip code. Due to irregularities in the reporting frequency a weekly aggregate of daily positive case reports was used for the analysis. The first reliable reports were available for the week starting on July 6, 2020. The last available weekly report included was for the week starting on March 29, 2021 (ending April 4, 2021).
Death reports.
LMPHW also provided death reports, including date and reported address or zip code. Data were filtered for August 1, 2020 to March 31, 2021. Total deaths for this period were 1090. Nearly 90% (978/1090) of the death records had address information able to be geocoded to the studied sewersheds. For the 112 reports that could not be assigned, due to either absence of address and/or zip code information or a recorded address outside a treatment plant area but still reported within the county, we probabilistically assigned these deaths to one of the five sewersheds, weighted by the relative population sizes. We assumed the risk of death from SARS-CoV-2 was the same across the sewersheds (Table S3). Deaths were aggregated by week and geocoded to sewersheds, matching the frequency of the observed case data.
Wastewater data.
Wastewater samples were collected two to four times a week, between August 2020 through December 2020, and once a week from January 2021 through March 2021. The five studied sewersheds include both combined and separated network pipes. Samples were typically from a 24-hour time-weighted composite sampler in an ice bath, though in the event of a composite sampler equipment malfunction, a grab sample was collected. All wastewater samples were analyzed at the University of Louisville, using previously published methods20–22 for SARS-CoV-2 (N1) and the fecal indicator pepper mild mottle virus (PMMoV).20 In brief, samples were concentrated with polyethylene glycol precipitation, and quantified in triplicate by reverse transcription polymerase chain reaction (RT-PCR) and reported as copies/milliliter (mL). The threshold value of SARS-CoV-2 (N1) assays was 7.5 copies per ml.
Weekly average COVID-19 concentrations were computed from quantifiable data. We used raw data (copies of SARS-CoV-2 (N1)/mL) for our main analyses. To account for possible wastewater system dilution we used the normalized ratio of copies of SARS-CoV-2 (N1)/mL divided by the copies of PMMoV/mL in a supplementary analysis. Previous studies investigated calibration of the statistical noise arising from changes in PMMoV and flow rate in these five sewersheds.22, 23
Wastewater data for the last two weeks of December 2020 were not available due to a laboratory holiday closure.
Serosurvey data.
We used aggregated data from a stratified simple random sampling serosurvey that was executed in the study area over four discrete time periods, for which 18,000 to 36,000 invitations were mailed in each wave, with an overall response rate of around 3%.19 Data were aggregated by wave and sewershed area at the four dates within the current study period. These data contain the number of serosurvey samples taken in a wave for each sewershed, the number of seropositive positive samples, and a weighted estimated percentage (with 95% CI) of the seropositivity in the population. We used these data to validate the cumulative infection estimates from the nowcasting models.
Ethics.
For the seroprevalence and data on COVID-19 deaths and infected individuals provided by the LMPHW under a Data Transfer Agreement, the University of Louisville Institutional Review Board approved this as Human Subjects Research (IRB number: 20.0393). For the wastewater data, the University of Louisville Institutional Review Board classified this as non-human subjects research (reference #: 717950).
Model
Nowcasting model.
We used a published Bayesian mathematical back-calculation model that estimates SARS-CoV-2 infections and effective reproduction number (Rt) from observed case and death data.24 The model is anchored on the observed deaths, and back-calculates the transmission rates and infections from the observed cases and the assumed progression probabilities and delays. This model includes a time varying probability of diagnosis if infected to account for changes in testing numbers and testing behavior over time. We modified this model to a weekly timeframe, in line with the changes made in Klaassen et al.25 (Supplementary Methods). With these adjustments, the basic version of this nowcasting model estimates weekly infections and transmission using reported case and reported death counts.
Four variations of nowcasting model
To compare nowcasts produced with or without wastewater data, we made additional adjustment to the nowcasting model to fit to four combinations of input data: (1) a ‘Cases-Deaths Model’, using cases and deaths data, consistent with the published version of the nowcasting model; (2), an ‘Additive Model’, using wastewater data in addition to cases and deaths data; (3) a ‘Substitutive Model’, using wastewater and deaths data; and (4) a ‘Deaths-Only Model’, using only deaths data, representing a worst-case scenario where no wastewater or case data are available. For the models without case data (Substitutive and Deaths-Only Models), we simply omitted the likelihood evaluation of the case data. For the models that include wastewater (Additive and Substitutive Models), we added an additional likelihood to the model. We used a sequential approach, where we first determined the transformation of wastewater data with the strongest correlation with the modeled infection estimates from the Cases-Deaths Model across the five sewersheds. We then correlated the raw measurements of the SARS-CoV-2 (N1) (copies per mL) with the infection estimates, and correlated the first-order differenced wastewater data (rate of change in the wastewater levels) with the estimates of Rt. The best fitting model informed our implementation of the model including wastewater. We also assessed the same correlations using the SARS-CoV-2 (N1)/PMMoV ratio and the first order difference of this ratio. The strongest correlation existed for the raw SARS-CoV-2 (N1) concentration to the infection estimates, resulting in our decision to model the wastewater data using a Student’s T distribution with 10 degrees of freedom and a linear relationship to the modeled infection estimates.
Analytic approach.
We fitted the four models to the timeseries data for each of the five sewersheds as well as their combined aggregate (Total), rendering six geographies. We ran the four models for each of the six geographies for each cumulative month of data after an initial first two months of data. There were 8.5 months of data available, and we created monthly snapshots of the data from month 2 to month 8, as well as the complete 8.5 months. We compared the Cases-Deaths Model to the Additive Model to assess the effects of adding wastewater data to existing case and death data. We compared the relative performance of the Substitutive Model and the Deaths-Only Model to the Cases-Deaths Model to assess the effects of including wastewater data when case data are absent. Finally, we compared the deviation of each of the five sewersheds from the Total to assess geographical variation.
Outcomes
For each of the model runs, we extracted the estimated infections and Rt timeseries. We compared predictive accuracy of these estimates between models, using four measures used by the COVID-19 Forecast Hub to evaluate the accuracy of forecasts26, 27: (1) the Absolute Difference between the point estimates (median of posterior distribution) of two models where a smaller Absolute Difference indicates a better agreement of two models; (2) the Sharpness, which is the weighted average of the widths of Credible intervals across K=11 coverages (10%, 20%, 30%, …, 90%, as well as 95% and 98%), with weights of where a smaller Sharpness indicates a more precise prediction; (3) the Coverage, defined here as the coverage of the 95% credible interval from one nowcasting model of the posterior distribution of the nowcasts from the comparison model; and (4) the Weighted Interval Score (WIS), which is the weighted penalized average of the Absolute Difference of the K Credible intervals from the median estimate of the target model (Supplementary Methods). We adapted these measures to allow for the comparison between two sets of predictions, or estimated quantities, rather than a prediction against a ground truth (Supplementary Methods). To assess the predictive value of the wastewater compared to or in addition to the case data, we computed these measures across two dimensions of our analyses. First, we computed the predictive within models across the snapshots of data, that is, assessing the predictive value of the nowcasts on the last date of each of the snapshot nowcasts to the estimates on that date from the complete data. Second, we calculated the predictive performance between models, that is, the difference between the two models, to quantify the agreement between models. We used the Cases-Deaths Model as a reference model, as this is representing the current standard in nowcasting infections, and this allows us to compare the precision of a model that adds wastewater data or replaces cases data with wastewater data. Finally, we assessed the historic reconstruction of each model by comparing the historic estimates qualitatively, by visual inspection, and quantitatively, by computing the overlap of the 95% credible intervals of the two posterior distributions. For models that included wastewater data (Substitutive and Additive models), we also assessed the fit of the model to the observed SARS-CoV-2 (N1) wastewater concentration.
Sensitivity analysis.
We tested the sensitivity of the results to outliers in the wastewater data, by refitting nowcasting models after removing outliers. To identify outliers, we fitted a smoothing spline to the daily wastewater timeseries data, and classified observations as outliers if their deviation from the spline was greater than three times the interquartile range of the deviations. Between 2% and 19% of the data points were marked as outliers (Table S2). We removed outliers, re-calculated weekly averages, and re-ran analyses on the adjusted data (Figure S1).
We also tested the sensitivity of our results to our implementation of probabilistically assigning deaths with an unknown sewershed using the relative frequency of reported SARS-CoV-2 deaths instead of the relative population size (Table S3). While the relative probabilities are slightly different, the estimated infection estimates are not strongly subject to either implementation (Figure S5).
Validation.
To validate the nowcast estimates, we compared the cumulative infection estimates against the serosurvey data and against the estimates produced by the published nowcasting model using daily case and deaths data compiled by Johns Hopkins University, since the beginning of reporting up until December 2021.28, 29
We present results estimated for the Total sewershed, without outliers removed, and focus on the infections outcome. By default, we present nowcast estimates for the complete timeseries, and refer to other snapshots where relevant. The results for individual sewersheds and each snapshot are available for both the infections and the Rt outcomes in the supplementary materials and referenced where relevant.
Software.
Data were analyzed using R (version 4.1.0) and the rstan package (version 2.21.5).30, 31 Figures were rendered using ggplot (version 3.3.6).32 Model code and documentation is available on GitHub (https://github.com/fayetteklaassen/ww-nowcasting). Figure 1 was made with ArcGIS Pro 2.5.2.
Results
Correlation of wastewater with epidemiological outcomes
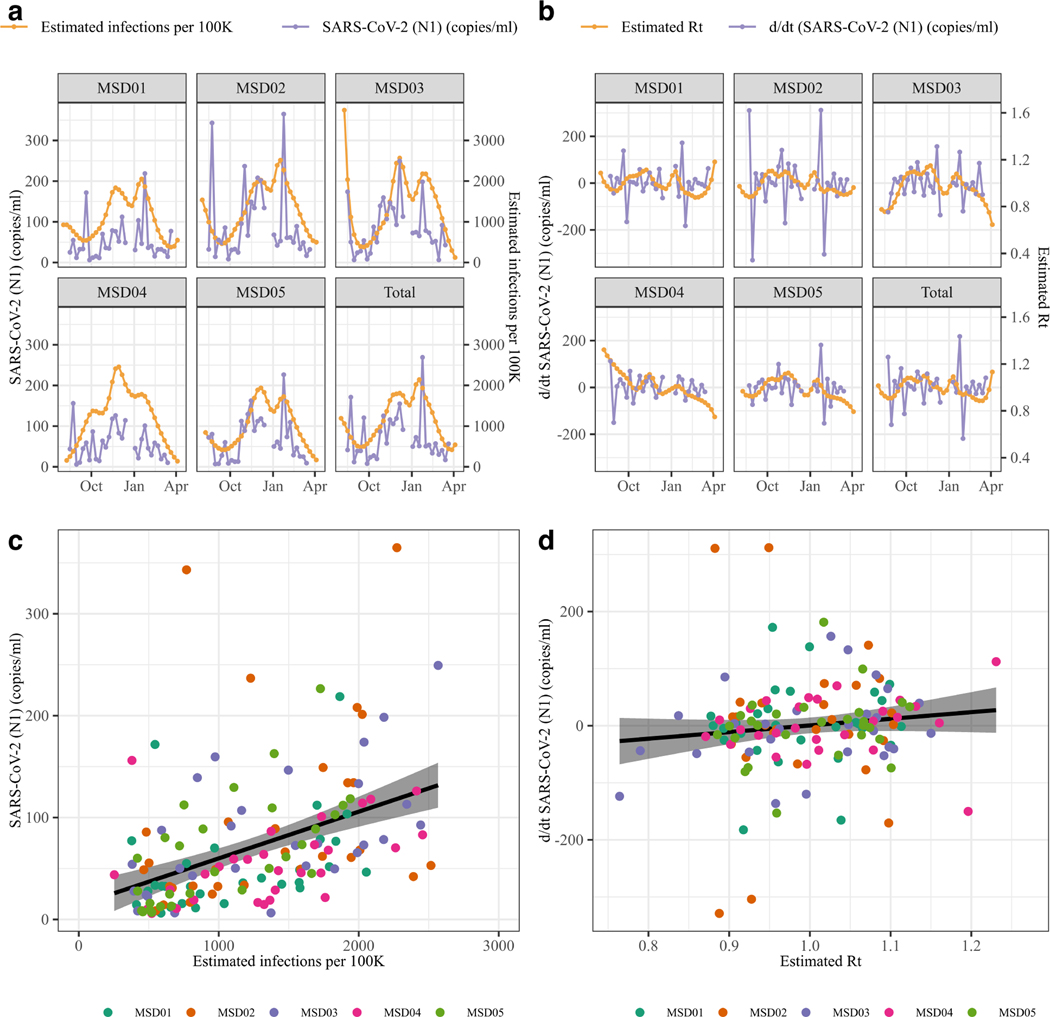
The timeseries of wastewater data SARS-CoV-2 (N1) (copies/ml) correlated positively with reported cases (r = 0.393, [955%CI, 0.261 – 0.511], Figure S2 a-d) and with the estimated infections from the Cases-Deaths Model (r = 0.362, [95% CI, 0.285 – 0.434], outliers removed r = 0.486, [0.415 – 0.551], Figure 2 a-b). The first order difference of the SARS-CoV-2 (N1) timeseries had no correlation with Rt estimates (r = 0.032 [−0.057 – 0.121], outliers SARS-CoV-2 (N1) removed r = 0.092, [−0.003 – 0.185], Figure 1 c-d). Based on these results, we modeled the wastewater data assuming a linear relationship to the infection estimates (Supplementary Methods).
Figure 2. Relationship between wastewater SARS-CoV-2 (N1) concentration and modeled estimates of infections and Rt.
(ab) Relationship between estimated infections per 100,000 inhabitants and SARS-CoV-2 (N1) (copies/mL); (cd) Relationship between estimated Rt versus SARS-CoV-2 (N1) (copies/mL, first order difference). (ac) Overlapping timeseries of wastewater (purple dots and lines) and estimated epidemic outcomes (orange points and lines); (bd) Scatterplot and fitted linear model, for the full dataset. Datapoints are colored by sewershed.
Predictive performance of each model: within model comparison
In comparison to the Deaths-Only model, the addition of case data (Cases-Deaths model) resulted in smaller Sharpness, Absolute Deviation and WIS for within model predictive performance (Figure S3, Figure S4, Table 1). Similarly, a model fit to cases, deaths and wastewater data had improved performance metrics relative to a model using only deaths and wastewater data (Additive versus Substitutive Models). The addition of wastewater data resulted in improved performance metrics when case data were present (Additive Model versus Cases-Deaths Model), but not when case data were absent (Substitutive Model versus Deaths-Only Model). The relative reductions in Absolute Deviation, Sharpness and WIS were larger for the addition of case data than for the addition of wastewater data. The coverage across the snapshots of the complete data was similar for each of the four models.
Table 1:
Predictive and historic performance of log(infections) nowcasts, within and between models
| Comparison | Statistic | Cases-Deaths | Additive | Substitutive | Deaths-Only |
|---|---|---|---|---|---|
| Within model predictive performance | Absolute Deviation Sharpness Coverage WIS |
0.325 0.0168 99.3% 0.223 |
0.291 0.0165 99.3% 0.208 |
0.757 0.0435 99.3% 0.517 |
0.667 0.0477 99.3% 0.500 |
| Between model predictive performance | Absolute Deviation Sharpness Coverage WIS |
- 0.0168 - 0.149 |
0.246 0.0165 93.9% 0.188 |
1.18 0.0435 98.7% 0.691 |
1.35 0.0477 96.9% 0.789 |
| Between model historic reconstruction |
Absolute Deviation Sharpness Coverage WIS |
- 0.0069 - 0.059 |
0.280 0.0064 76.8% 0.156 |
0.321 0.0112 91.2% 0.185 |
0.397 0.0125 92.5% 0.231 |
For within-model comparisons, the last estimates from each model using the snapshots are compared against the timeseries estimates from the same model using the complete data. For the between model comparison of predictive performance, each the last estimates from each snapshot from each model are compared against the last estimates from the same snapshot of the Cases-Deaths Model. For the between model historic reconstruction, the timeseries estimates from each model using the complete data are compared against the estimates from the Cases-Deaths Model using the complete data.
Addition of wastewater: between model comparison
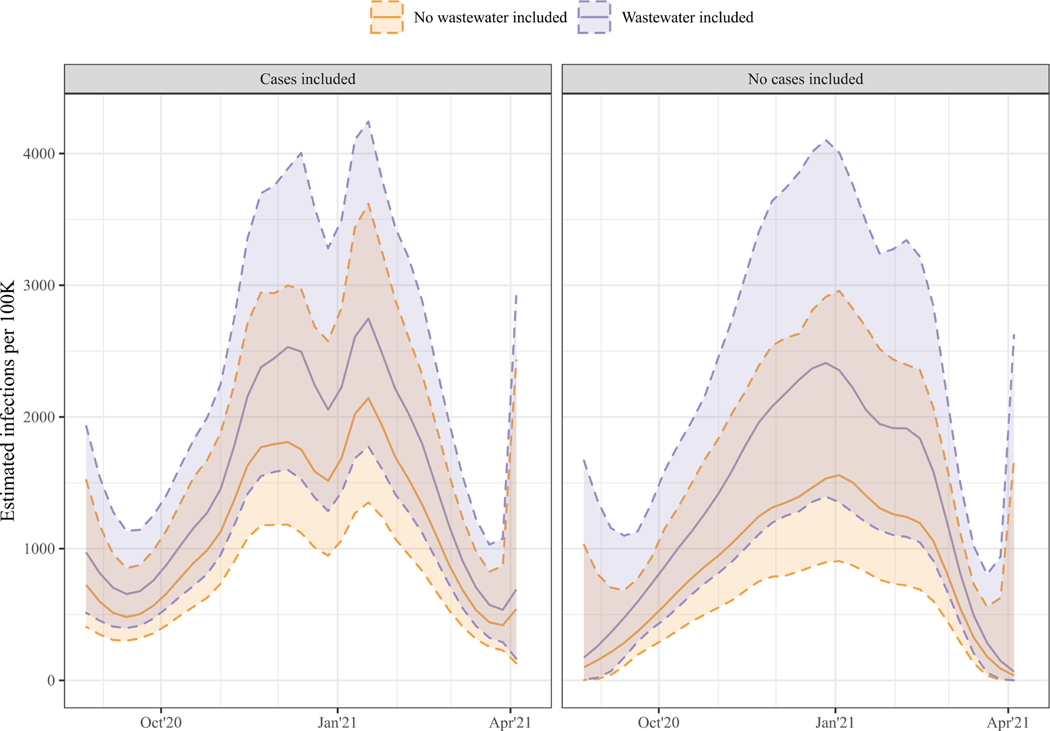
Using wastewater data in the nowcasting model in addition to the cases and deaths data or to the deaths data did not result in any qualitative differences in the timeseries of infection estimates (Figure 3). While the 95% credible intervals of the Additive Model’s historic reconstruction of estimated infections covered only 76.8% of the Cases-Deaths Model’s posterior distribution, across the snapshots, the 95% credible intervals of the last date’s nowcasts covered on average 93.9% of the Cases-Deaths Model’s posterior distribution. The Additive Model estimated a higher overall incidence of infections, but this difference was not statistically significant. Similarly, in the Substitutive Model the estimated incidence of infections was higher than in the Deaths-Only Model.
Figure 3. Timeseries of estimated infections per 100K for the total sewershed.
The left panel shows results from models including case data (Cases-Deaths Model in orange and Additive Model in purple), and the right panel shows results from models without case data (Deaths-Only Model in orange and Substitutive Model in purple). The solid lines mark the median of the posterior distribution, and the shaded area, marked by the dashed lines, the 95% Credible Interval.
We found no strong differences in the within-model predictive performance of the Additive Model and the Cases-Deaths Model. The Additive Model had slightly lower scores on the Absolute Deviance, Sharpness and WIS, indicating stronger internal consistency of the estimates as data accrued. Out of the three alternatives to the Cases-Deaths Model, the Additive Model has the lowest WIS scores, indicating the best tradeoff in precision and certainty relative to the Cases-Deaths Model.
Substitution of wastewater: between model comparison
The Deaths-Only Model outperformed the Substitutive Model in the within-model assessment. While the average Coverage of the historic last estimates to the complete estimates was similar, Absolute Difference, Sharpness and WIS were less for the Deaths-Only Model (Table 1), indicating a more consistent prediction when only deaths data were used compared to wastewater and deaths data. However, relative to the Cases-Deaths Model, the Substitutive Model had slightly better predictive and historic reconstruction power. The Absolute Difference of the median estimated log(infections) from the Substitutive Model to the Cases-Deaths Model across historic runs was 0.321, while the Deaths-Only Model has an Absolute Deviation of 0.397 (Table 1). The WIS was lower for the Substitutive Model than the Deaths-Only Model both when comparing to the Cases-Deaths Model across the snapshots and in the historic reconstruction.
Geographic granularity and wastewater data: between sewershed comparison
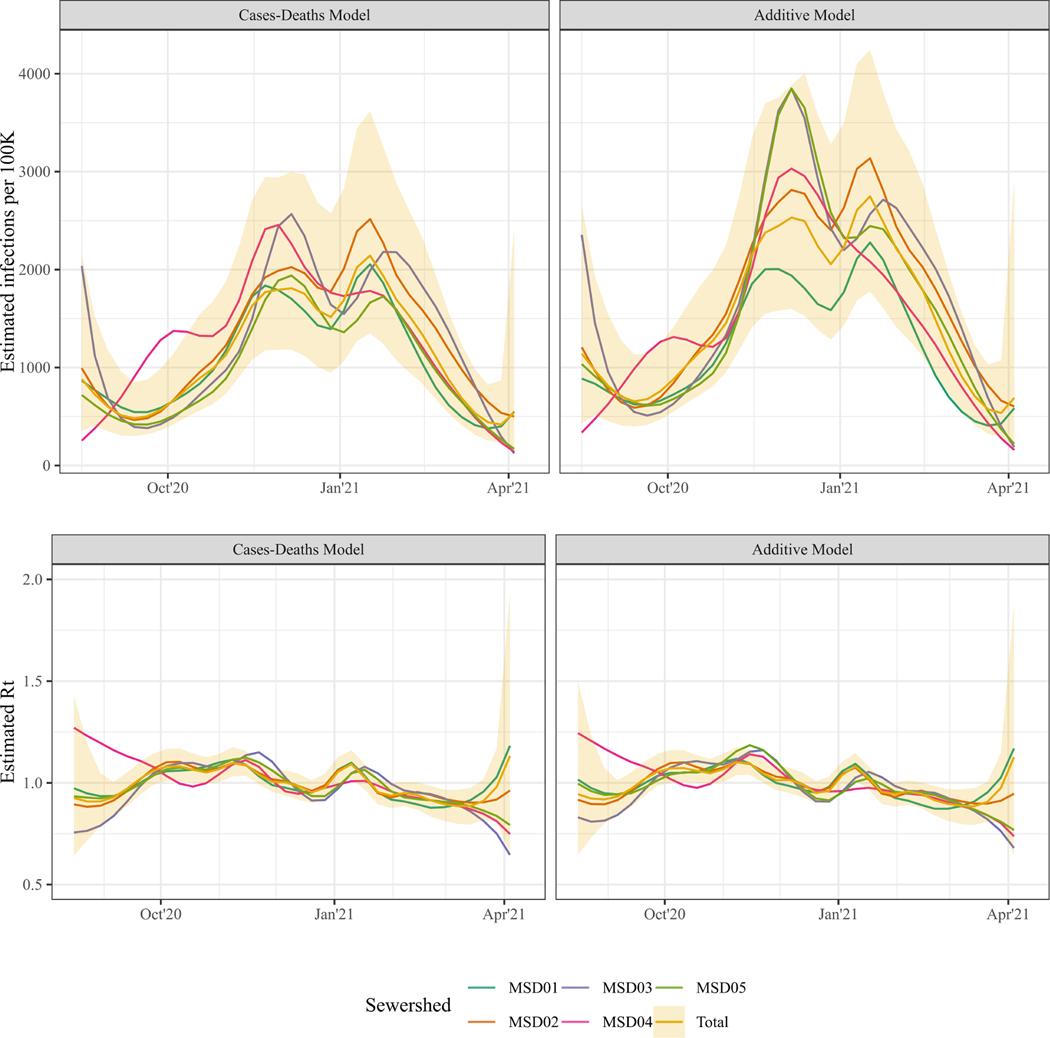
The 95% credible interval of the timeseries estimates for the complete county covered MSD02 best (Coverage of 89.8% for the Cases-Deaths Model, and 89.6% for the Additive model), and MSD04 worst (Coverage of 69.7% and 72.1% respectively; Table 2, Figure 4). While the historic reconstruction of the timeseries estimates of infections and Rt was on average not statistically different, smaller population level sewersheds had larger Absolute Deviations and lower Coverage from the Total estimates. Notably, for MSD01, the largest sewershed in terms of population and area, the Coverage of the Total estimates was lower and the WIS was higher for the Additive Model compared to the Cases-Deaths Model, indicating that the wastewater data increased the variability in estimates between the sewersheds.
Table 2.
Historic overlap of log(infections) estimates for each sewershed to the estimates for the total sewershed.
| Model | Statistic | MSD01 | MSD02 | MSD03 | MSD04 | MSD05 |
|---|---|---|---|---|---|---|
| Cases-Deaths Model | Absolute Deviation | 0.106 | 0.157 | 0.178 | 0.330 | 0.206 |
| Sharpness | 0.0092 | 0.0093 | 0.0113 | 0.0121 | 0.0142 | |
| Coverage | 89.5% | 89.8% | 82.2% | 69.7% | 70.4% | |
| WIS | 0.086 | 0.098 | 0.127 | 0.206 | 0.131 | |
| Additive Model | Absolute Deviation | 0.270 | 0.136 | 0.185 | 0.289 | 0.213 |
| Sharpness | 0.0088 | 0.0085 | 0.0097 | 0.0108 | 0.0110 | |
| Coverage | 77.1% | 89.6% | 80.9% | 72.1% | 75.5% | |
| WIS | 0.156 | 0.089 | 0.127 | 0.185 | 0.131 |
All statistics are calculated comparing the historic estimates from the complete data for each sewershed to the historic estimates from the complete data for the total area. Coverage is defined as the average percentage of the posterior distribution from each sewershed covered by the 95% Credible Interval from the Total dataset.
Figure 4. Infection and Rt estimates for each sewershed.
Median estimates of the infection timeseries (top panes) and Rt timeseries (bottom panels) of the Cases-Deaths Model (left panels) and the Additive Model (right panel) for each sewershed. For the total sewershed (yellow), the 95% Credible Interval is plotted as a shaded interval.
Validation
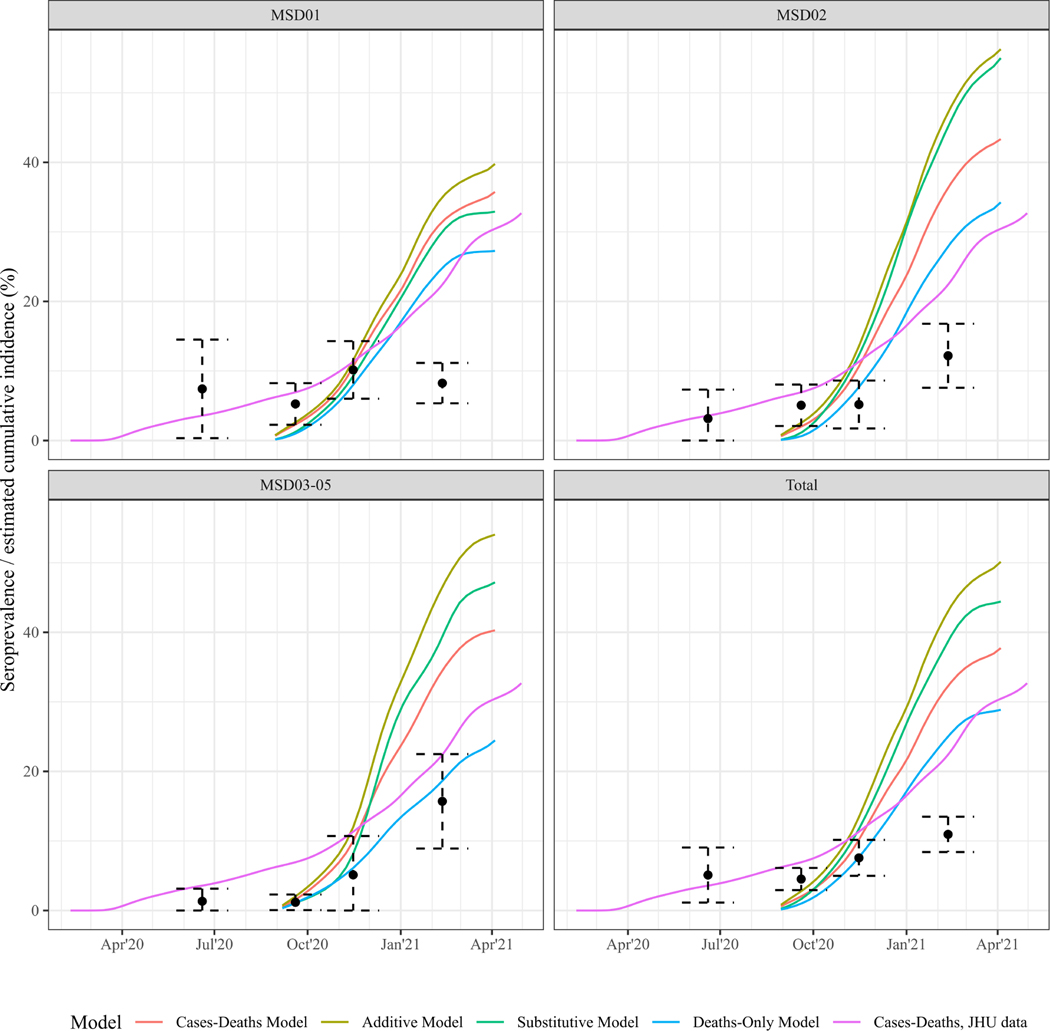
Modelled cumulative infection estimates from the available data were higher than the cumulative infection estimates for the entire county rendered using the JHU data from March 2020 until December 2021. Comparing the modeled cumulative infection estimate to the serosurvey data from Keith et al shows that for Wave 2 and 3, the serosurvey data and error bars overlap with the cumulative infection estimates, and for Wave 4, all cumulative infection estimates exceed the seroprevalence (Figure 5). For MSD02–05 the serosurvey data and the cumulative infection estimates follow a similar trend, while for MSD01 the serosurvey data flattens out over the last two observations, while the modelled estimates continue to increase.
Figure 5. Serosurvey and cumulative incidence estimates for Jefferson County, KY (USA).
Colored lines show the estimated cumulative incidence (% of the population infected) for MSD01, MSD02 and for MSD03–05 jointly, for each of the four models As a reference, in pink, estimates of the nowcasting model are included, that were rendered using the Johns-Hopkins daily case and death data28, 29, starting the first reported case on March 2020 up until December 2021, to render estimates based on a more complete set of surveillance data. Black dots and 95% error bars show the seroprevalence estimates and uncertainty bounds at four time points from data serosurvey conducted by Keith et al.19
Discussion
This study assessed the value of wastewater data for infectious disease nowcasting. We used SARS-CoV-2 case, death and wastewater data from Jefferson County, Kentucky and an adapted version of a published nowcasting model to evaluate the predictive and historic performance of wastewater data if they were used in addition or as a replacement for case data.
The results of our study showed a positive association between wastewater concentration and infection estimates from the nowcasting model fit to cases and death data. There were no significant differences in the within-model predictive performance, other than the expected improvements in Sharpness and WIS as more data were included. The addition of case data resulted in a greater relative improvement in the performance than the addition of wastewater data. The model containing all three data sources had the highest predictive performance. Nonetheless, the additive benefit of wastewater data was limited, as these data did not improve the predictive performance substantially.
This work demonstrates that wastewater data may be a viable substitute for case data, as estimates using death and wastewater data approximated the estimates from a cases and deaths model closer than when only deaths were used. As the availability of traditional clinical surveillance data deteriorates during an ongoing pandemic, the possibility of using wastewater data in nowcasts and monitoring transmission is of high public health interest and underlines the potential utility of these data for future pandemic preparedness. This approach will have additional relevance in low-resource settings where systematic collection of epidemiologic data is likely to be severely limited regardless of the disease being studied. Such low-resource areas are likely not only low- and middle-income countries, but rural areas within both the United States and Europe. Further research is needed to examine the relationship between wastewater and infection estimates across multiple locations and longer timeseries to confirm our findings and to evaluate their applicability in different social and geographic contexts.
We found that the models that include wastewater concentration consistently estimated higher infections than the models without wastewater data included, which may be an artefact of a different estimated probability of progressing to symptomatic and severe disease. Additionally, we considered the question of the potential use of wastewater in smaller populations (sewersheds), where count surveillance data (e.g., case, death, hospitalizations) may not be as readily available as in larger county or state areas, yet wastewater data can still be obtained. The epidemiological trends estimated for the smaller sewersheds in our study area deviated more from the aggregate estimates than the larger sewersheds. This highlights the importance of high frequency local (sub-countywide) surveillance data for guiding future public health responses, as transmission may differ by area, and local granular data might help monitor disease trends more closely.
This study is a first attempt to assess the value of adding wastewater data in a nowcasting model. Several limitations should be considered. First, the study had a limited scope in time and geography, and we only considered a single nowcasting model. When we compared estimates of cumulative infection generated by this study to other estimates, we found sewershed-specific estimates as well as the aggregated countywide estimates exceeded the estimates from the nowcasting model using a longer case and death timeseries, indicating the contribution of historical transmission to cumulative infection estimates. At the end of the period under consideration, the estimated cumulative infections were around 2–3 times higher than the estimates from the seroprevalence survey. Seroprevalence data only included the population older than 18 years whereas wastewater captures a wider portion of the population. Furthermore, the low response rate of 3% could have impacted the comparability of these data to estimates of cumulative infections. The quality of these data is discussed in more detail in Keith et al.19 We assumed a positive antibody test could be from one or more infections, whereas the nowcasting model did not account for multiple infections, and the count surveillance could have double counted individuals. It is unclear whether the differences in the estimates of modeled cumulative infections and the seroprevalence estimates reflects overestimation on the part of the model, incomparability of the data to the estimates, or losses in seropositivity among previously infected individuals.
Another potential set of limitations is linked to our decisions regarding outliers and not normalizing the wastewater data by flow or a fecal indicator. Past WBE research demonstrates a wide range of quantification and normalization methods, and normalization may not improve the signal in the wastewater data.33, 34 The correlations of the normalized wastewater data SARS-CoV-2 (N1) (copies/ml) divided by PMMoV (copies/ml) with infection estimates (r = 0.393, [95% CI, 0.261 – 0.511]) and of the normalized wastewater data SARS-CoV-2 (N1) (copies/ml) divided by flow (millions of gallons per day, MGD) with the infection estimates (r = 0.244, [95% CI, 0.100 – 0.379]) were similar to the correlation between the raw wastewater data SARS-CoV-2 (N1) (copies/ml) and the infection estimates (Figure S5). Across the sewersheds, up to 19% of the wastewater observations could be marked as outliers using a spline timeseries. Wastewater data may detect local events, such as festivals or conferences, which may be an indication of transmission at a short timescale, but not of infections or sustained transmission in the population. This could have been a factor in the current study, as the Kentucky Derby took place in Louisville, Kentucky, on September 5, 2020, which coincides with some of the extreme datapoints in the wastewater data (Figure S1). Nonetheless, the sensitivity analyses that excluded wastewater data outliers did not result in different conclusions in this study.
One potential reason for the limited additive value of the wastewater timeseries is that these data were much less smooth compared to the case data (Figure S2a). In other words, while adding information, the wastewater data also introduced further uncertainty, and consequently add less information in the nowcasting than the case data. The option of considering what smoothing function would be appropriate to use wastewater data in infectious disease surveillance is beyond the scope of this article. For future use of wastewater data in infectious disease surveillance, it is important to investigate the sampling frequency and data quality needed.
The results from this study of a strong correlation between the wastewater data and estimated infections correspond to other studies where wastewater data were used to predict case or hospitalization reports.8, 13, 15 The strength of this relationship appears to be stronger when daily moving averages of the wastewater data are available. 25 Despite potential bias introduced by using the wastewater data to inform the parameterization of those data in the model, the additive and substitutive power in this study were not very strong, when compared with a study in nowcasting SARS-CoV-2 infections in the Boston metropolitan area, covering a much larger population.14 This further supports the need for future research into the conditions required for using wastewater data in infectious disease nowcasting. Finally, our models included a simple linear relationship between wastewater data and modeled infections. There is a range of additional assumptions and more complex modeling choices that can be made to further support the use of wastewater in nowcasts.35 This might complicate models and might make them less versatile across various infectious diseases, as these assumptions are disease specific, or require much additional data, like temperature and flow rates. The presence and level of viral load in wastewater data is a function of many other variables (such as temperature, sample type, flow rates, distance from households), and the association between the viral load and the transmission in the population is a function of the amount and length of viral shedding at various disease stages.36
In conclusion, in this case study, we found that the use of wastewater data improved the performance of COVID-19 nowcasts for Jefferson County, Kentucky. However, these improvements were modest, particularly when case data were available. Future research on the value of wastewater data for nowcasting when case data are absent or unreliable would be beneficial. For public health officials this is critical information in balancing the focus of surveillance efforts – while wastewater may offer early detection, it’s incremental value for nowcasting may be limited if high-quality case reports are available, providing a reminder of the value of investments in traditional surveillance data. It is also possible that wastewater data will provide stronger evidence in other settings where case data is absent, and research on approaches to strengthen the value of these data for surveillance proposes is important for ongoing pandemic preparedness.
Supplementary Material
Highlights.
Wastewater infectious disease measurements correlated with count surveillance data
The predictive power of adding wastewater data to case data was minimal
Using wastewater data in addition to deaths data improved the nowcast performance
Wastewater may be valuable when traditional surveillance is limitedly available
Acknowledgements
This work was supported by contracts from the Centers for Disease Control and Prevention (75D30121C10273) and the Louisville-Jefferson County Metro Government as a component of the Coronavirus Aid, Relief, and Economic Security Act, as well as grants from the James Graham Brown Foundation, Owsley Brown II Family Foundation, and the Welch Family. Serological work was also supported in part by the Jewish Heritage Fund and the Center for Predictive Medicine for Biodefense and Emerging Infectious Diseases. This project has also been funded (in part) by contract 200-2016-91779 with the Centers for Disease Control and Prevention, the Centers for Disease Control and Prevention though the Council of State and Territorial Epidemiologists (NU38OT000297-03) and NIH National Cancer Institute (1U01CA261277). We thank the Louisville/Jefferson County Metropolitan Sewer District for their valuable collaboration with the wastewater sample collection and Louisville Metro Department of Public Health and Wellness for geocoding the death reports.
Fayette Klaassen reports financial support was provided by Centers for Disease Control and Prevention. Fayette Klaassen reports financial support was provided by Council of State and Territorial Epidemiologists. Nicolas Menzies reports financial support was provided by Centers for Disease Control and Prevention. Nicolas Menzies reports financial support was provided by Council of State and Territorial Epidemiologists. Ted Cohen reports financial support was provided by Council of State and Territorial Epidemiologists. Rochelle H Holm reports financial support was provided by Centers for Disease Control and Prevention. Ted Smith reports financial support was provided by Centers for Disease Control and Prevention. Aruni Bhatnagar reports financial support was provided by Centers for Disease Control and Prevention. Rochelle H Holm reports financial support was provided by Louisville-Jefferson County Metro Government. Ted Smith reports financial support was provided by Louisville-Jefferson County Metro Government. Aruni Bhatnagar reports financial support was provided by Louisville-Jefferson County Metro Government. Rochelle Holm reports financial support was provided by James Graham Brown Foundation. Ted Smith reports financial support was provided by James Graham Brown Foundation. Aruni Bhatnagar reports financial support was provided by James Graham Brown Foundation. Rochelle H Holm reports financial support was provided by Owsley Brown II Family Foundation. Ted Smith reports financial support was provided by Owsley Brown II Family Foundation. Aruni Bhatnagar reports financial support was provided by Owsley Brown II Family Foundation. Rochelle H Holm reports financial support was provided by Welch Family. Ted Smith reports financial support was provided by Welch Family. Aruni Bhatnagar reports financial support was provided by Welch Family. Rochelle H Holm reports financial support was provided by Jewish Heritage Fund. Ted Smith reports financial support was provided by Jewish Heritage Fund. Aruni Bhatnagar reports financial support was provided by Jewish Heritage Fund. Rochelle H Holm reports financial support was provided by University of Louisville Center for Predictive Medicine for Biodefense and Emerging Infectious Diseases. Ted Smith reports financial support was provided by University of Louisville Center for Predictive Medicine for Biodefense and Emerging Infectious Diseases. Aruni Bhatnagar reports financial support was provided by University of Louisville Center for Predictive Medicine for Biodefense and Emerging Infectious Diseases.
Footnotes
Data sharing
The seroprevalence, wastewater concentration, and case by sewershed area as used in the study, and the code and instructions to recreate the results will be made available immediately after publication on GitHub. Death data are not publicly available; contact Louisville Metro Department of Public Health and Wellness if interested in establishing data sharing arrangements.
Disclaimer: The findings, conclusions, and views expressed are those of the author(s) and do not necessarily represent the official position of the Centers for Disease Control and Prevention (CDC), Council of State and Territorial Epidemiologists (CSTE), or National Institutes of Health (NIH). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Author Credit Statement of:
RHH, TS, TC, NAM and FK conceptualized the project and developed the analysis plan.
RHH, TS and AB curated and verified the data sources.
FK executed the analyses, visualized the results and drafted the original manuscript and figures.
All authors reviewed and edited the original manuscript and figures and their revision.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Fayette Klaassen, Department of Global Health and Population, Harvard TH Chan School of Public Health, Boston MA, USA.
Rochelle H. Holm, Christina Lee Brown Envirome Institute, School of Medicine, University of Louisville, Louisville, KY, USA
Ted Smith, Christina Lee Brown Envirome Institute, School of Medicine, University of Louisville, Louisville, KY, USA.
Ted Cohen, Department of Epidemiology of Microbial Diseases and Public Health Modeling Unit, Yale School of Public Health, New Haven, Connecticut, USA.
Aruni Bhatnagar, Christina Lee Brown Envirome Institute, School of Medicine, University of Louisville, Louisville, KY, USA.
Nicolas A. Menzies, Center for Health Decision Science, Harvard TH Chan School of Public Health, Boston MA, USA Department of Global Health and Population, Harvard TH Chan School of Public Health, Boston MA, USA.
References
- 1.Wu JT, Leung K, Lam TTY, Ni MY, Wong CKH, Peiris JSM, et al. Nowcasting epidemics of novel pathogens: lessons from COVID-19. Nat Med. 2021;27(3):388–95. doi: 10.1038/s41591-021-01278-w. [DOI] [PubMed] [Google Scholar]
- 2.Rossman H, Segal E. Nowcasting the spread of SARS-CoV-2. Nature Microbiology. 2022;7(1):16–7. doi: 10.1038/s41564-021-01035-2. [DOI] [PubMed] [Google Scholar]
- 3.Pitzer VE, Chitwood M, Havumaki J, Menzies NA, Perniciaro S, Warren JL, et al. The Impact of Changes in Diagnostic Testing Practices on Estimates of COVID-19 Transmission in the United States. Am J Epidemiol. 2021;190(9):1908–17. doi: 10.1093/aje/kwab089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bilal U, Tabb LP, Barber S, Diez Roux AV. Spatial Inequities in COVID-19 Testing, Positivity, Confirmed Cases, and Mortality in 3 U.S. Cities. Ann Intern Med. 2021;174(7):936–44. doi: 10.7326/M20-3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lauer SA, Grantz KH, Bi Q, Jones FK, Zheng Q, Meredith HR, et al. The Incubation Period of Coronavirus Disease 2019 (COVID-19) From Publicly Reported Confirmed Cases: Estimation and Application. Ann Intern Med. 2020;172(9):577–82. doi: 10.7326/m20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kilaru P, Hill D, Anderson K, Collins MB, Green H, Kmush BL, et al. Wastewater Surveillance for Infectious Disease: A Systematic Review. Am J Epidemiol. 2022;192(2):305–22. doi: 10.1093/aje/kwac175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shah S, Gwee SXW, Ng JQX, Lau N, Koh J, Pang J. Wastewater surveillance to infer COVID-19 transmission: A systematic review. Sci Total Environ. 2022;804:150060. doi: 10.1016/j.scitotenv.2021.150060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith T, Holm RH, Keith RJ, Amraotkar AR, Alvarado CR, Banecki K, et al. Quantifying the relationship between sub-population wastewater samples and community-wide SARS-CoV-2 seroprevalence. Sci Total Environ. 2022;853:158567. doi: 10.1016/j.scitotenv.2022.158567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu F, Xiao A, Zhang J, Moniz K, Endo N, Armas F, et al. SARS-CoV-2 RNA concentrations in wastewater foreshadow dynamics and clinical presentation of new COVID-19 cases. Sci Total Environ. 2022;805:150121. doi: 10.1016/j.scitotenv.2021.150121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pulicharla R, Kaur G, Brar SK. A year into the COVID-19 pandemic: Rethinking of wastewater monitoring as a preemptive approach. Journal of Environmental Chemical Engineering. 2021;9(5):106063. doi: 10.1016/j.jece.2021.106063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Centers for Disease Control and Prevention. COVID Data Tracker: Wastewater Surveillance [webpage]. Atlanta, GA: US Department of Health and Human Services, CDC; 2023. [Accessed: April 10, 2023]. Available from: https://covid.cdc.gov/covid-data-tracker. [Google Scholar]
- 12.Biobot Analytics. Data on Covid-19 and Mpox Wastewater Monitoring [webpage]. Cambridge, MA: [Accessed: May 12, 2023]. Available from: https://biobot.io/. [Google Scholar]
- 13.Schenk H, Heidinger P, Insam H, Kreuzinger N, Markt R, Nägele F, et al. Prediction of hospitalisations based on wastewater-based SARS-CoV-2 epidemiology. Sci Total Environ. 2023;873:162149. doi: 10.1016/j.scitotenv.2023.162149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phan T, Brozak S, Pell B, Gitter A, Xiao A, Mena KD, et al. A simple SEIR-V model to estimate COVID-19 prevalence and predict SARS-CoV-2 transmission using wastewater-based surveillance data. Sci Total Environ. 2023;857:159326. doi: 10.1016/j.scitotenv.2022.159326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeng HA, Singh R, Diawara N, Curtis K, Gonzalez R, Welch N, et al. Application of wastewater-based surveillance and copula time-series model for COVID-19 forecasts. Sci Total Environ. 2023;885:163655. doi: 10.1016/j.scitotenv.2023.163655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farkas K, Hillary LS, Malham SK, McDonald JE, Jones DL. Wastewater and public health: the potential of wastewater surveillance for monitoring COVID-19. Current Opinion in Environmental Science & Health. 2020;17:14–20. doi: 10.1016/j.coesh.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahmed W, Simpson SL, Bertsch PM, Bibby K, Bivins A, Blackall LL, et al. Minimizing errors in RT-PCR detection and quantification of SARS-CoV-2 RNA for wastewater surveillance. Sci Total Environ. 2022;805:149877. doi: 10.1016/j.scitotenv.2021.149877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rondeau Nicole C, Rose Oliver J, Alt Ellen R, Ariyan Lina A, Elikan Annabelle B, Everard Jenna L, et al. Building-Level Detection Threshold of SARS-CoV-2 in Wastewater. Microbiology Spectrum. 2023;11(2):e0292922. doi: 10.1128/spectrum.02929-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keith RJ, Holm RH, Amraotkar AR, Bezold MM, Brick JM, Bushau-Sprinkle AM, et al. Stratified Simple Random Sampling Versus Volunteer Community-Wide Sampling for Estimates of COVID-19 Prevalence. Am J Public Health. 2023:e1–e10. doi: 10.2105/AJPH.2023.307303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holm RH, Mukherjee A, Rai JP, Yeager RA, Talley D, Rai SN, et al. SARS-CoV-2 RNA abundance in wastewater as a function of distinct urban sewershed size. Environmental Science: Water Research & Technology. 2022;8(4):807–19. doi: 10.1039/D1EW00672J. [DOI] [Google Scholar]
- 21.Rouchka EC, Chariker JH, Saurabh K, Waigel S, Zacharias W, Zhang M, et al. The Rapid Assessment of Aggregated Wastewater Samples for Genomic Surveillance of SARS-CoV-2 on a City-Wide Scale. Pathogens. 2021;10(10). doi: 10.3390/pathogens10101271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holm RH, Nagarkar M, Yeager RA, Talley D, Chaney AC, Rai JP, et al. Surveillance of RNase P, PMMoV, and CrAssphage in wastewater as indicators of human fecal concentration across urban sewer neighborhoods, Kentucky. FEMS Microbes. 2022;3:xtac003. doi: 10.1093/femsmc/xtac003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanneganti D, Reinersman LE, Holm RH, Smith T. Estimating sewage flow rate in Jefferson County, Kentucky, using machine learning for wastewater-based epidemiology applications. Water Supply. 2022;22(12):8434–9. doi: 10.2166/ws.2022.395. [DOI] [Google Scholar]
- 24.Chitwood MH, Russi M, Gunasekera K, Havumaki J, Klaassen F, Pitzer VE, et al. Reconstructing the course of the COVID-19 epidemic over 2020 for US states and counties: Results of a Bayesian evidence synthesis model. PLoS Comput Biol. 2022;18(8):e1010465. doi: 10.1371/journal.pcbi.1010465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klaassen F, Chitwood MH, Cohen T, Pitzer VE, Russi M, Swartwood NA, et al. Changes in Population Immunity Against Infection and Severe Disease From Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Omicron Variants in the United States Between December 2021 and November 2022. Clin Infect Dis. 2023:ciad210. doi: 10.1093/cid/ciad210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bracher J, Ray EL, Gneiting T, Reich NG. Evaluating epidemic forecasts in an interval format. PLoS Comput Biol. 2021;17(2):e1008618. doi: 10.1371/journal.pcbi.1008618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cramer EY, Huang Y, Wang Y, Ray EL, Cornell M, Bracher J, et al. The United States COVID-19 Forecast Hub dataset. Scientific Data. 2022;9(1):462. doi: 10.1038/s41597-022-01517-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dong E, Du H, Gardner L. An interactive web-based dashboard to track COVID-19 in real time. The Lancet Infectious Diseases. 2020;20(5):533–4. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.covidestim: COVID-19 nowcasting [webpage]. 2022. [Accessed: March 13, 2023]. Available from: https://legacy.covidestim.org/.
- 30.R Core Team. R: A language and environment for statistical computing. 2021. R Foundation for Statistical Computing:Vienna, Austria. [Google Scholar]
- 31.Stan Development Team. RStan: the R interface to Stan. 2020. R package version 2.21.2 [Google Scholar]
- 32.Wickham H. ggplot2: Elegant Graphics for Data Analysis. 2016. Springer-Verlag, New York. [Google Scholar]
- 33.Maal-Bared R, Qiu Y, Li Q, Gao T, Hrudey SE, Bhavanam S, et al. Does normalization of SARS-CoV-2 concentrations by Pepper Mild Mottle Virus improve correlations and lead time between wastewater surveillance and clinical data in Alberta (Canada): comparing twelve SARS-CoV-2 normalization approaches. Sci Total Environ. 2023;856(Pt 1):158964. doi: 10.1016/j.scitotenv.2022.158964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Greenwald HD, Kennedy LC, Hinkle A, Whitney ON, Fan VB, Crits-Christoph A, et al. Tools for interpretation of wastewater SARS-CoV-2 temporal and spatial trends demonstrated with data collected in the San Francisco Bay Area. Water Research X. 2021;12:100111. doi: 10.1016/j.wroa.2021.100111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang G, Wu J, Weidhaas J, Li X, Chen Y, Mueller J, et al. Artificial neural network-based estimation of COVID-19 case numbers and effective reproduction rate using wastewater-based epidemiology. Water Res. 2022;218:118451. doi: 10.1016/j.watres.2022.118451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arts Peter J, Kelly JD, Midgley Claire M, Anglin K, Lu S, Abedi Glen R, et al. Longitudinal and quantitative fecal shedding dynamics of SARS-CoV-2, pepper mild mottle virus, and crAssphage. mSphere. 2023;0(0):e00132–23. doi: 10.1128/msphere.00132-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.